Abstract
Awareness on antioxidants and its significance in human healthcare has increased many folds in recent time. Increased demand requisite on welcoming newer and alternative resources for natural antioxidants. Seaweed associated pigmented bacteria screened for its antioxidant potentials reveals 55.5% of the organisms were able to synthesize antioxidant compounds. DPPH assay showed 20% of the organisms to reach a antioxidant zone of 1 cm and 8.3% of the strains more than 3 cm. Pseudomonas koreensis (JX915782) a Sargassum associated yellowish brown pigmented bacteria have better activity than known commercial antioxidant butylated hydroxytoluene (BHT) against DPPH scavenging. Serratia rubidaea (JX915783), an associate of Ulva sp. and Pseudomonas argentinensis (JX915781) an epiphyte of Chaetomorpha media , were also contributed significantly towards ABTS (7.2% ± 0.03 to 15.2 ± 0.09%; 1.8% ± 0.01 to 15.7 ± 0.22%) and FRAP (1.81 ± 0.01 to 9.35 ± 0.98; 7.97 ± 0.12 to 18.70 ± 1.84 μg/mL of AsA Eq.) respectively. 16S rRNA gene sequence analysis revealed bacteria that have higher antioxidant activity belongs to a bacterial class Gammaproteobacteria. Statistical analysis of phenolic contents in relation with other parameters like DPPH, ABTS, reducing power and FRAP are well correlated (p < 0.05). Results obtained from the current study inferred that the seaweed associated pigmented bacteria have enormous potential on antioxidant compounds and need to be extracted in a larger way for clinical applications.
Keywords: antioxidant, epiphytic, pigmented bacteria, seaweeds
Introduction
Oxidants are commonly known as free radicals that are chemically reactive and unstable species which need to be controlled to avoid a chain of unwanted reactions in the living system. The most important free radicals in the body are the reactive oxygen species (ROS). They try to react with the surrounding macromolecules like lipids, proteins, deoxyribonucleic acids (DNA) and certain carbohydrates in order to stabilise ( Cheeseman and Slater, 1993 ; Dekkers et al. , 1996 ), which leads to oxidative stress and finally results in cell death ( Halliwell and Gutteridge, 1999 ). However, in order to cope up with oxidative stress the human body has an elaborate antioxidant defence system ( Alscher et al. , 1997 ). Antioxidants are the bio-molecules manufactured within the body as well as extracted from the food that we consume where a variety of vitamins, minerals and enzymes helps to protect the body by scavenging free radicals ( Dekkers et al. , 1996 ).
Coastal inhabitants like seaweeds and associated bacteria are continuously exposed to various environmental stresses like desiccation, freezing, ultraviolet radiation, and heavy metal fluctuations which result in gradual production and accumulation of ROS ( Dring, 2005 ). Overproduction of ROS hampers not only the process of photosynthesis and growth in plants ( Dring, 2005 ) but also affects the cellular structures and physiology in microbes ( Fridovich, 1986 ). In order to withstand such stress conditions, these organisms activate antioxidant defence mechanisms to neutralize ROS.
Seaweeds are well evaluated in the context of radical scavenging properties and are reported to possess natural antioxidants such as carotenoids, pigments, polyphenols, enzymes and diverse functional polysaccharides ( Vinayak et al. , 2011 ). Common seaweeds like, Sargassum , ( Garcia-Casal et al. , 2009 ) and Chaetomorpha ( Kelman et al. , 2012 ) species are found to possess strong radical scavenging activities. Not only seaweeds, some of the marine pigmented bacteria and their derivatives are also known to produce high antioxidant efficiency compounds ( Dufosse, 2006 ; Konzen et al. , 2006 ; Shindo et al. , 2007 ; Correa-Llanten et al. , 2012 ).
Seaweeds and the pigmented bacteria are independently studied and reported to exhibit various antioxidant responses towards excess of ROS. But pigmented bacteria and their symbiotic interactions to produce antioxidant compounds remain unexplored for quite a long time. Seaweeds can host many heterotrophic bacteria which play an important role in maintaining the health of the host organism ( Bolinches et al. , 1998 ) by producing unique bioactive secondary metabolites. Hence, epibiotic bacteria become an incredible source of new natural products ( Holmstrom and Kjelleberg, 1999 ) for the biomedical and biopharmaceutical industry. Thus, seaweed-associated bacteria may be useful in the development and production of bioactive compounds and biocontrol agent. The initial-step in the development of any bioactive compounds involves isolation of appropriate bacteria from natural sources and optimization of its antagonistic activity. In this background, the present study is initiated to explore the antioxidant potential of marine culturable pigmented bacteria associated with seaweeds from the central west coast of India (CWCI) as a suitable source for antioxidant compounds.
Materials and Methods
Chemicals
Sodium carbonate (Na 2 CO 3 ), butylated hydroxy toluene (BHT), 2,2 diphenyl-1-picraylhydrozyl (DPPH), dimethyl sulfoxide (DMSO), ascorbic acid, trichloro acetic acid (TCA), 2,2 azinobis (3-ethylbenzothiozoline-6-sulfonic acid) diammonium salt (ABTS), quercetin, potassium ferric cyanide, ferric chloride (FeCl 3 ), thiobarbituric acid (TBA), ethylenediaminetetraacetic acid (EDTA) and gallic acid were purchased from Sigma-Aldrich chemical Pvt. Ltd., Bangalore. For molecular analysis, the gel extraction and purification kit was purchased from QIAGEN India Pvt. Ltd., New Delhi. All other chemicals and reagents used for experimental work were of extra pure quality.
Sample collection and isolation of marine bacteria
The seaweed samples ( Table 1 ) were picked with sterile forceps from the intertidal region along the central west coast of India during 2010–2011 ( Figure 1 ). All the samples were stored in sterile plastic bottles and brought to the laboratory in ice box. Isolation of bacteria was carried out by spread plate method ( Lakshmanaperumalsamy and Purushothaman, 1982 ) using Zobell marine agar 2216 media (Hi-Media, M384, Mumbai). Seaweed samples were rinsed with sterile seawater to remove debris and unattached bacteria. Approximately 10 g (wet weight) of seaweed sample was suspended in 100 mL of sterile seawater in conical flask. This was kept in a rotary shaker at 150 rpm for 30 min. Then the water samples were serially diluted and plated in duplicates. Plates were incubated at 28 ± 2 °C for 48–72 h. Distinct pigmented colonies were selected and purified. These cultures were stored in glycerol vials at −80 °C until further analysis.
Table 1.
Type of seaweeds used for isolation of epiphytic pigmented bacteria.
| Sr. no. | Type of algae used | Month, year and site of sample collection * | Number of pigmented bacteria isolated |
|---|---|---|---|
| Chlorophyta (green algae) | |||
| 1. | Caulerpa racemosa | Nov-10-Cabo De Rama | 1 |
| 2. | Chaetomorpha media and Chaetomorpha antennina | Mar-10-Malvan, Mar-10-Vagator, May-10-Cabo De Rama, July-10-Malvan, July-10-Kunkeshwar, Sep-10-Malvan | 8 |
| 3. | Ulva sp. and Ulva fasciata | Mar-10-Malvan, Sep-10-Vagator, July-10-Malvan, July-10-Kunkeshwar | 6 |
| Phaeophyta (brown algae) | |||
| 4. | Padina boergesenii and Padina tetrastromatica | Sep-10-Kunkeshwar, Jan-11-Malvan | 3 |
| 5. | Sargassum sp., Sargassum vulgare and Sargassum ilicifolium | May-10-Cabo De Rama, Sep-10-Cabo De Rama, Sep-10-Kunkeshwar, Nov-10-Cabo De Rama, Jan-11-Vagator, May-10-Vagator | 7 |
| 6. | Stechospermum marginatum | Jan-11-Kunkeshwar | 1 |
| Rhodophyta (red algae) | |||
| 7. | Ceramium sp. | Mar-10-Kunkeshwar | 3 |
| 8. | Gelidium micropterum | Mar-10-Malvan, May-10-Cabo De Rama, July-10-Malvan, Nov-10-Malvan | 4 |
| 9. | Gracilaria corticata | Mar-10-Malvan, July-10-Kunkeshwar, Sep-10-Malvan, Sep-10-Kunkeshwar, Sep-10-Cabo De Rama, Jan-11-Vagator | 3 |
Example: Jan-11-Vagator indicates the respective sample was collected during January-2011 from sampling site Vagator of CWCI.
Figure 1.
Sampling locations along the central west coast (CWC) of India.
Primary screening of antioxidant producing strains
The simple primary DPPH-antioxidant screening method applied by Takao et al. (1994) was followed for initial screening. Pure culture suspension broth (2 μL) was drop inoculated on nutrient agar plates and incubated at 28 °C for 24 h. A sterilized filter paper (Whatman no. 1) was then placed on the agar plate, so that colonies and their extracellular metabolites get replicated on the paper. Incubation was further continued at 28 °C for another 24 h. Then the filter paper was taken out, dried and sprayed with a DPPH solution (80 μg mL −1 in methanol). Strains showing a white on purple spot were considered as antioxidant producing strains. Colony size and zones of decolorization were noted for each analysed culture ( Table 2 ).
Table 2.
Primary antioxidant screening results of DPPH decolorization assay.
| Sr. no. | Culture code | Pigmentation | Antioxidant zone size (cm) * | Source | ||
|---|---|---|---|---|---|---|
|
|
||||||
| a | b | b-a | ||||
| 1. | PIGB 1 | Orange | 0.4 | 3.1 | 2.7 | Gelidium micropterum |
| 2. | PIGB 10 | Yellow | 0.3 | 0.9 | 0.6 | Gracilaria corticata |
| 3. | PIGB 12 | Yellow | 0.3 | 0.3 | 0 | Chaetomorpha media |
| 4. | PIGB 13 | Yellow | 0.3 | 0.3 | 0 | Chaetomorpha media |
| 5. | PIGB 16 | Yellow | 0.2 | 0.2 | 0 | Ceramium sp. |
| 6. | PIGB 18 | Red | 0.6 | 2.5 | 1.9 | Ceramium sp. |
| 7. | PIGB 20 | Orange | 0.3 | 0.3 | 0 | Ceramium sp. |
| 8. | PIGB 28 | Pale yellow | 0.3 | 0.9 | 0.6 | Chaetomorpha antennina |
| 9. | PIGB 29 | Pale yellow | 0.2 | 1.1 | 0.9 | Sargassum sp. |
| 10. | PIGB 30 | Pale yellow | 0.4 | 0.13 | 0.9 | Gelidium micropterum |
| 11. | PIGB 33 | Pale yellow | 0.4 | 0.9 | 0.5 | Sargassum ilicifolium |
| 12. | PIGB 44 | Pale yellow | 1.0 | 2.6 | 1.6 | Ulva fasciata |
| 13. | PIGB 45 | Yellow | 0.3 | 0.8 | 0.5 | Ulva fasciata |
| 14. | PIGB 46 | Pale yellow | 0.5 | 4.0 | 3.5 | Chaetomorpha media |
| 15. | PIGB 47 | Red/orange | 0.5 | 2.8 | 2.3 | Chaetomorpha media |
| 16. | PIGB 48 | Yellow | 0.3 | 0.3 | 0 | Chaetomorpha media |
| 17. | PIGB 54 | Pale yellow | 0.2 | 0.2 | 0 | Gracilaria corticata |
| 18. | PIGB 55 | Pale orange | 0.3 | 0.3 | 0 | Chaetomorpha media |
| 19. | PIGB 57 | Pale yellow | 0.3 | 0.3 | 0 | Ulva fasciata |
| 20. | PIGB 62 | Orange | 0.2 | 0.2 | 0 | Chaetomorpha antennina |
| 21. | PIGB 76 | Creamy | 0.4 | 1.5 | 1.1 | Sargassum vulgare |
| 22. | PIGB 77 | Yellowish brown | 0.7 | 5.0 | 4.3 | Sargassum vulgare |
| 23. | PIGB 78 | Peach | 0.3 | 0.3 | 0 | Gracilaria corticata |
| 24. | PIGB 79 | Orange | 0.7 | 3.5 | 2.8 | Padina boergesenii |
| 25. | PIGB 80 | Orange/Red | 0.3 | 1.7 | 1.4 | Padina boergesenii |
| 26. | PIGB 86 | Pale yellow | 0.4 | 0.4 | 0 | Ulva sp. |
| 27. | PIGB 87 | Creamy | 0.5 | 1.3 | 0.8 | Ulva sp. |
| 28. | PIGB 88 | Pink | 0.5 | 5.4 | 4.9 | Ulva sp. |
| 29. | PIGB 110 | Yellow | 0.4 | 0.4 | 0 | Gelidium micropterum |
| 30. | PIGB 112 | Creamy | 0.2 | 0.2 | 0 | Gelidium micropterum |
| 31. | PIGB 115 | Creamy | 0.3 | 0.3 | 0 | Sargassum vulgare |
| 32. | PIGB 116 | Yellow | 0.2 | 0.2 | 0 | Sargassum vulgare |
| 33. | PIGB 117 | Creamy | 0.4 | 2.3 | 1.9 | Sargassum vulgare |
| 34. | PIGB 119 | Brown | 0.2 | 2.8 | 2.6 | Caulerpa racemosa |
| 35. | PIGB 126 | Pale orange | 0.3 | 0.3 | 0 | Stechospermum marginatum |
| 36. | PIGB 140 | Pale orange | 0.5 | 1.0 | 0.5 | Padina tetrastromatica |
a: Colony size, b: Zone of DPPH decolorization, b-a: Final of zone of antioxidant.
Mass culture, extraction and dry weight determination
An aliquot of 100 μL of selected pure cultures (selection based on highest zone of DPPH decolorization) were inoculated in sterile nutrient broth (300 mL). It was incubated on shaker incubator at 28 °C with 120 rpm for 48 h. It was then centrifuged (10 °C, 10 min at 6153 × g) and supernatant was collected in another sterile collection tube. The supernatant was filtered (0.45 μm) to remove all the bacterial cells from broth and extracted with ethyl acetate (30% v/v). The extracts were evaporated and concentrated with rotary evaporator (Equitron, Roteva (63) R-V) until complete dryness and finally retained in minimum HPLC grade methanol. 0.5–1.0 mL of sample was taken in a pre-weighed aluminium foil and allowed to dry for weight determination ( Ekanayake et al. , 2004 ). The known concentrations of sample viz. 1.0, 5.0, 10 and 15 mg/mL were prepared in methanol, and used for quantitative antioxidant analysis.
Antioxidant analysis
DPPH radical scavenging activity
Radical scavenging activity of different bacterial extracts against DPPH radicals was assessed according to the method of Larrauri et al. (1998) . Briefly, 0.8 mL (0.05 mM) DPPH-methanol solution was mixed with different concentration of samples and final volume was made up to 1.0 mL. Reaction mixture was incubated for 30 min at 25 °C in dark and absorbance was measured at 517 nm. Control was maintained with DPPH and methanol instead of antioxidant solution while blanks contained only methanol instead of DPPH solution throughout the experiment. BHT was used as positive control. The inhibition of DPPH radicals by the samples was calculated according to the following equation,
Ferric reducing antioxidant power (FRAP) assay
FRAP analysis was done with the modified method of Benzie and Strain’s (1996) . The working FRAP reagent was prepared by mixing 300 mM acetate buffer (pH 3.6), 10 mM TPTZ solution (10 mM TPTZ in 40 mM HCl) and 20 mM FeCl 3 6H 2 O in a 10:1:1 ratio just before use and heated to 37°C. A total of 150 μL of working FRAP reagent was added to each well of a 96 well microtiter plate. A blank reading was then taken at 600 nm using an ELISA plate reader (Biorad 680 XR, Biosciences). A total of 20 μL of sample was then added to each well. After addition of sample to the FRAP reagent, a second reading at 595 nm was performed after 8 min. Ascorbic acid was used as standard and the antioxidant content of the extract was expressed as ascorbic acid equivalents (AsA Eq.) μg/mL of crude sample extract.
ABTS assay
The ABTS radical cation decolourization assay ( Re et al. , 1999 ) was used to evaluate antioxidant power of sample extracts. ABTS was dissolved in water to a 7 mM concentration. ABTS radical cations were produced by reacting ABTS stock solution with 2.45 mM potassium per sulphate (final concentration) and allowing the mixture to stand in the dark for 12–16 h before use. The stock solution was diluted with ethanol (1:89 v/v) to an absorbance of 0.70 (± 0.02) at 734 nm and equilibrated at 30°C exactly 6 min after initial mixing. One millilitre of diluted ABTS solution was mixed with 10 μL of sample extracts of different strength (1.0 to 15 mg/mL). The decrease of absorbance was measured at 734 nm and the antioxidant capacity was expressed in percent inhibition (%I). Quercetin was used as a standard antioxidant and IC 50 value was calculated from regression analysis.
Determination of reducing power
The reducing abilities of sample extracts were determined according to the method of Ferreira (2007) . Briefly, 0.25 mL aliquots of various concentrations of sample were mixed with 2.5 mL of 200 mM sodium phosphate buffer (pH 6.6) and 2.5 mL of 1% potassium ferricyanide. The mixture was then incubated at 50 °C for 20 min. Later, 2.5 mL of 10% TCA (w/v) was added to the reaction mixture and centrifuged at 4062 × g for 10 min. Finally, 5 mL aliquot of the upper layer was mixed with 5 mL of distilled water and 1.0 mL of 0.1% ferric chloride. The absorbance was measured at 700 nm.
Determination of total phenolic contents
Total Phenolic compounds in extracellular extracts were quantified by using Folin-ciocalteu’s colorimetric method described by Shan et al. (2005). Different concentrations of extracted samples (0.2 mL) were initially oxidized with 1 mL of 0.5 M of Folin-ciocalteu’s reagent for 4 min. The reaction was finally neutralized with 1 mL of saturated sodium carbonate (75 g/L) after 30 min of incubation and absorbance of the resulting blue color was measured at 760 nm. Quantification was done from standard of gallic acid curve and results were expressed as mg of gallic acid equivalents (GAE) per mL of sample extract. All tests were performed in triplicate.
Identification of antioxidant potent strains by 16S rRNA gene
Genomic DNA was extracted from all the potent isolates by using DNeasy Blood and Tissue kit (Qiagen) according to manufacturer’s instructions. The extracted DNA was subjected to polymerase chain reaction (PCR) with universal primers 27F and 1492R ( Lane, 1991 ). DNA sequencing was performed with an automatic sequencer (Applied Biosystems 3130xl Genetic Analyzer) with the bacterial primers 27F, 518F and 1492R. The 16S rRNA gene sequences obtained were submitted to the GenBank and the accession numbers were assigned from JX915781 to JX915783.
Statistical analysis
Each sample was run in triplicate for statistical analysis. Results are expressed as mean ± standard deviation (SD). Changes in biochemical parameters were tested using one-way ANOVA and post hoc tests (Newman-Keuls) to discriminate between means of values. Differences were considered statistically significant when p < 0.05.
Results
Pigmented bacteria from the seaweeds
A total of 36 pigmented bacterial strains isolated from 14 different seaweeds are listed in Table 1 . Highest numbers of pigmented bacteria were associated with Chaetomorpha and Sargassum species. While, the seaweeds like Caulerpa racemosa and Stechospermum marginatum indicated the poor association with the pigmented bacteria. These two species could associate only one pigmented bacterial colony throughout the year. Moreover, high numbers of pigmented bacteria were retrieved from the algal surface which belongs to Chlorophyta (green algae) while, Phaeophyta (brown algae) and Rhodophyta (red algae) were seen to be loaded with near about equal numbers of bacteria. Seaweeds ( Chaetomorpha , Sargassum and Ulva spp.) possessing highest number of pigmented bacteria i.e. 58.34% of the total population were observed to be dominating during pre-monsoon and monsoon seasons. Another interesting finding is that though the Gracilaria corticata was dominating most of the sampling stations throughout the year it was able to give only limited number of pigment producers.
Primary screening of extracellular antioxidants
DPPH screening method indicates that 20 different pigmented bacteria were able to produce extracellular antioxidants. Cultures showing decolonization zone above 3 cm were considered as high potent strains and below this zone were excluded for further investigation. Bacterial associates of Ulva , Sargassum and Chaetomorpha spp. exhibited high DPPH activity in terms of the hollow zones of 4.9, 4.3 and 3.5 cm respectively. Isolates obtained from Caulerpa racemosa , Gelidium micropterum and Padina boergesenii expressed second level of activity and exhibited the zones of 2.6–2.8 cm ( Table 2 ).
Antioxidant potential of pigmented strains
Of the total pigmented isolates tested ~55.5% bacteria showed antioxidant properties. The bacteria coloured yellow, yellowish brown and pink were able to produce prominent antioxidant zones. The second level of activities were obtained with near about 20% of strains having orange, yellowish-brown, pink and brown pigmentation whereas creamy and pale yellow coloured colonies (44.5%) exhibited nil or negligible amount of activity. This clearly indicates variation in pigment coloration contributes significantly towards radical scavenging activities. Table 2 shows that, among the total antioxidant producers 27% were associated with green algae (Chlorophyta) and 8.3% of the pigmented bacteria (PIGB 46, PIGB 77 and PIGB 88) exhibiting highest activities were selected for quantitative analysis.
DPPH radical scavenging assay on selected bacteria
Free radical scavenging activities of selected bacterial species tested against commercial DPPH reagent showed the linear relation over concentration. Increased concentration indicates the raise over DPPH scavenging effect. P. koreensis exhibited 47.3 ± 0.77% of scavenging activities for 1 mg of crude extract and ascended up to 95.6 ± 5.61% when the concentrations were increased to 15 mg. These activities were much higher than that of known synthetic BHT used in the study ( Table 3 ). Serratia rubidaea and P. argentinensis also exhibited 57.5 ± 3.03 and 28.7 ± 0.92% of DPPH scavenging while increasing the sample concentrations ( Table 3 ).
Table 3.
Antioxidant activities of extracellular bacterial extracts by DPPH and ABTS radical scavenging assay. Values are mean ± SD (n = 3).
| Sample conc. (mg/mL) | DPPH scavenging assay (%) | EC 50 (mg/mL) | ABTS scavenging assay (%) | EC 50 (mg/mL) |
|---|---|---|---|---|
| Pseudomonas argentinensis (PIGB 46) | ||||
| 1 | 10.9 ± 0.28 | 31.13 ± 4.15 | 1.8 ± 0.01 | 46.14 ± 3.49 |
| 5 | 13.7 ± 0.39 | 4.7 ± 0.01 | ||
| 10 | 22.4 ± 0.33 | 8.4 ± 0.03 | ||
| 15 | 28.7 ± 0.92 | 15.7 ± 0.22 | ||
| Pseudomonas koreensis (PIGB 77) | ||||
| 1 | 47.3 ± 0.77 | 1.17 ± 0.06 | 19.2 ± 0.05 | 9.41 ± 0.09 |
| 5 | 76.1 ± 2.61 | 38.9 ± 1.98 | ||
| 10 | 93.4 ± 6.54 | 53.4 ± 2.75 | ||
| 15 | 95.6 ± 5.61 | 69.5 ± 1.00 | ||
| Serratia rubidaea (PIGB 46) | ||||
| 1 | 18.0 ± 0.17 | 10.31 ± 0.48 | 7.2 ± 0.03 | 88.34 ± 2.15 |
| 5 | 35.5 ± 0.32 | 9.3 ± 0.03 | ||
| 10 | 53.0 ± 0.56 | 11.3 ± 0.03 | ||
| 15 | 57.5 ± 3.03 | 15.2 ± 0.09 | ||
| Standards | ||||
| Sample conc. (μg/mL) | DPPH i Scavenging assay (%) | EC 50 (μg/mL) | * ABTS ii scavenging assay (%) | Sample conc. (μM/mL) |
| 40 | 56.6 ± 1.63 | 24.33 ± 1.82 | 78.0 ± 1.14 | 5.74 ± 1.26 |
| 80 | 80.3 ± 0.61 | 97.6 ± 1.25 | ||
| 120 | 82.8 ± 0.72 | 99.6 ± 1.18 | ||
| 160 | 84.3 ± 1.34 | 99.5 ± 1.06 | ||
Standards used for various assays:
DPPH scavenging assay - BHT,
ABTS scavenging assay -Quercetin.
Sample concentrations used for ABTS standard were 1.0, 5.0, 10.0 and 15.0 μM/mL respectively.
ABTS assay
P. argentinensis and S. rubidaea exhibited poor ABTS scavenging activity 1.8 ± 0.01 to 15.7 ± 0.22% and 7.2 ± 0.03 to 15.2 ± 0.09% respectively at 1 to 15 mg of sample concentration. Highest activities by P. koreensis (19.2 ± 0.05 to 69.5 ± 1.00%) with lowest EC 50 value of 9.41 ± 0.09 mg/mL indicate its potential role towards extracellular antioxidant ( Table 3 ). Other strains, though having different EC 50 values ( P. argentinensis , EC 50 = 46.14 ± 3.49 and S. rubidaea , EC 50 = 88.34 ± 2.15 mg/mL) were able to exhibit the same ABTS scavenging effects ( Table 3 ). However, higher concentrations of the extracts were more effective in quenching free radicals in the system.
FRAP assay
Results of FRAP analysis indicated significant levels of AsA Eq. in P. koreensis which was followed by P . argentinensis and S. rubidaea . Increasing orders of FRAP values were observed in case of P. koreensis from 5.90 ± 0.13 for 1 mg and 47.01 ± 0.27 μg AsA Eq./mL for 15 mg of samples ( Table 4 ). Contrastingly, S. rubidaea was found to possess low levels of AsA Eq. though it exhibited better DPPH and ABTS scavenging than that of P . argentinensis. Moreover, correlation results of FRAP were significant with total phenolic contents.
Table 4.
FRAP and total phenolic contents in extracellular ethyl acetate extracts of selected bacterial strains.
| Sample | Sample conc. (mg/mL) | FRAP assay (μg/mL of AsA Eq.) | Total phenolic content assay (mg/mL of GAE) |
|---|---|---|---|
| Pseudomonas argentinensis (PIGB 46) | 1 | 7.97 ± 0.12 | 0.09 ± 0.01 |
| 5 | 10.80 ± 0.13 | 0.17 ± 0.01 | |
| 10 | 14.11 ± 2.3 | 0.24 ± 0.02 | |
| 15 | 18.70 ± 1.84 | 0.31 ± 0.00 | |
| Pseudomonas koreensis (PIGB 77) | 1 | 5.90 ± 0.13 | 0.21 ± 0.01 |
| 5 | 23.26 ± 4.96 | 0.34 ± 0.02 | |
| 10 | 35.17 ± 0.36 | 0.65 ± 0.02 | |
| 15 | 47.01 ± 0.27 | 0.83 ± 0.05 | |
| Serratia rubidaea (PIGB 88) | 1 | 1.81 ± 0.01 | 0.08 ± 0.01 |
| 5 | 4.76 ± 0.05 | 0.21 ± 0.01 | |
| 10 | 7.46 ± 0.12 | 0.32 ± 0.01 | |
| 15 | 9.35 ± 0.98 | 0.48 ± 0.07 |
AsA Eq.: Ascorbic acid equivalents, GEA: Gallic acid equivalents.
Reducing power assay
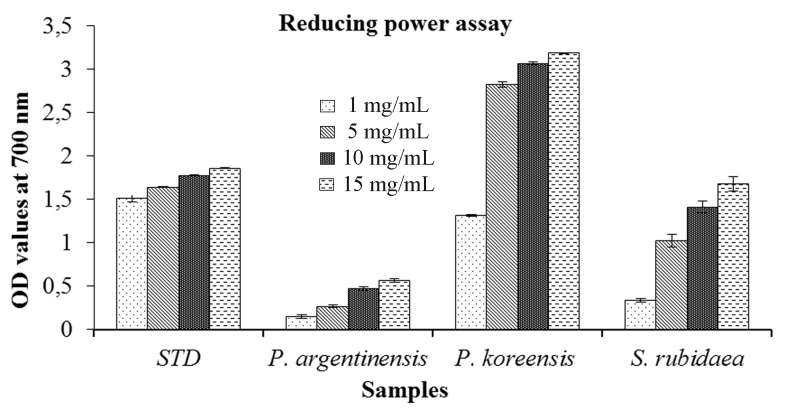
The reducing capacities of ethyl acetate extracts were determined based on the change in absorbance i.e. in terms of increasing OD values and results were represented in Figure 2 . P. koreensis showed highest absorbance. The activity tested with P. koreensis was much higher than that of standard compound i.e. , ascorbic acid. However, these reducing abilities were also shown linear with the increase in concentrations.
Figure 2.
Reducing potentials of selected bacteria.*Concentrations of the sample: 1, 5, 10, 15 mg/mL, STD: standard compound used i.e. ascorbic acid.
Total phenolic content assay
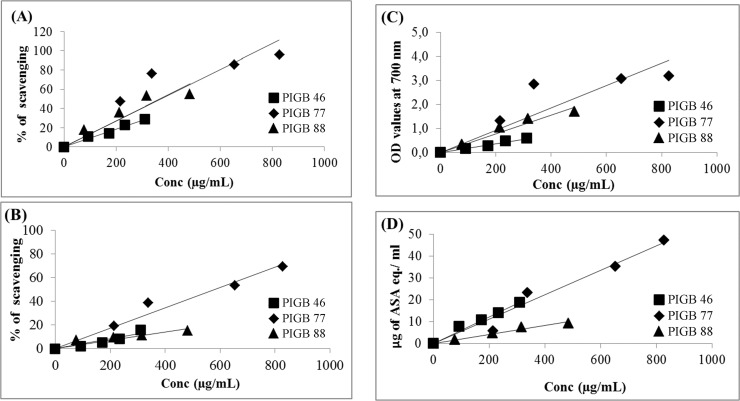
The TPCs in all the tested samples showed less than 1 mg/mL of GAE. Analysis results clearly showed that P . koreensis extracts had the highest phenolic content (0.83 ± 0.05 mg GAE/mL). While, other species like P. argentinensis (0.31 ± 0.00 mg GAE/mL) and S. rubidaea (0.48 ± 0.07 mg GAE/mL) were observed to possess low TPCs ( Table 4 ). Results of correlation analysis between each antioxidant parameter and total phenolic content are shown ( Figure 3 ). TPC exhibited significant r 2 values over all the other antioxidant tests performed in this investigation (p < 0.005).
Figure 3.
Correlation analysis of antioxidant parameteres. PIGB 46; PIGB 77; PIGB 88. A. TPC Vs DPPH (PIGB 46; r 2 : 0.976, PIGB 77; r 2 : 0.741, PIGB 88; r 2 : 0.855) p = 0.004, B. TPC Vs ABTS (PIGB 46; r 2 : 0.872, PIGB 77; r 2 : 0.964, PIGB 88; r 2 : 0.790) p = 0.009, C. TPC Vs Reducing power (PIGB 46; r 2 : 0.979, PIGB 77; r 2 : 0.725, PIGB 88; r 2 : 0.943) p = 0.01, D. TPC Vs FRAP (PIGB 46; r 2 : 0.971, PIGB 77; r 2 : 0.962, PIGB 88; r 2 : 0.975) p = 0.01.
High potent strains and their source
Considering the source of the potent strains, the highest antioxidant producing strain PIGB77 was found to be associated with Sargassum species (Phaeophyta) whereas the other two strains belonged to Chaetomorpha and Ulva spp. (Chlorophyta) ( Table 5 ). This shows a bacterium living in the vicinity of seaweeds belonging to Phaeophyta and Chlorophyta supports more bioactive flora than Rhodophyta.
Table 5.
16S rRNA identity for high potent antioxidant strains.
| Isolate | Isolation source | Closest match (accession number) | Phylogenetic group | Similarity (%) | Pigmentation |
|---|---|---|---|---|---|
| PIGB 46 | Chaetomorpha media | Pseudomonas argentinensis (JX915781) | γ-proteobacteria | 97 | Yellow |
| PIGB 77 | Sargassum vulgare | Pseudomonas Koreensis (JX915782) | γ-proteobacteria | 100 | Yellow/brown |
| PIGB 88 | Ulva sp. | Serratia rubidaea (JX915783) | γ-proteobacteria | 99 | Pink |
High potent strains and their molecular identity (16S rRNA)
The sequence data obtained was visualised by CromasPro 1.41. Closest matches assessed using BLASTn and accession numbers obtained from GenBank were given in Table 5 . Out of 3 selected strains two belonged to Pseudomonas genus while the other was from Serretia . Sequence match for species identity revealed selected strains as, P. argentinensis (97%), P. koreensis (100%), and S. rubidaea (99%). Moreover, 16S rRNA results highlights Gammaproteobacteria group as a common origin for antioxidant potent bacterial flora ( Table 5 ).
Discussion
Bacteria are well known to be associated with seaweed surfaces. Earlier studies had revealed distinct diversities among the seaweed epiphytic bacterial flora ( Oberbeckmann, 2007 ; Menezes et al. , 2010 ). These bacteria have recently gained attention of the scientific community due to their source for novel compounds of industrial importance. Pigmented bacteria are of similar concern observed to have distinct physiological and biological roles. We retrieved prominent numbers of such bacteria from the surfaces of selected seaweeds ( Table 1 ). Supporting growth for pigmented bacteria had been reported by Shiba et al. (1979) , who found thalli of Enteromorpha linza and Sargassum horneri were dominant with sixteen strains of aerobic pink or orange bacteria. Moreover, novel species of pigmented bacteria such as Arenibacter and Flavobacter are also found to be associated with seaweeds ( Nedashkovskaya et al. , 2004 , 2006 ). This clearly shows that pigmented bacteria also form a major part of the epiphytic bacterial flora. The reason behind their attached lifestyle may be the utilization of various substances secreted by living or dead algae and sometimes vice versa; these associations play an important role in host (algae) growth ( Soria-Mercado et al. , 2012 ).
Takao et al. (1994) , first gave a simple and efficient method for screening antioxidants of bacterial origin using DPPH decolorization assay. Our investigation followed a similar method for primary screening and selection of antioxidant potent pigmented strains. Of the total strains tested, 55.5% showed positive results which may be due to the scavenging properties of extracellular compounds including pigments. Few of the recent studies on bacterial pigments of marine realm e.g. violacein, astaxanthin, canthaxanthin, zeaxanthin, rubrolone and carotenoid derivatives evidence their efficient radical scavenging activities ( Dufosse, 2006 ; Shindo et al. , 2007 ). These studies clearly indicate that variation in pigment coloration contributes towards their radical scavenging activities.
The DPPH activity is a measure of the reactivity where decrease in absorbance is estimated as the reaction between DPPH and antioxidant progresses. All the bacterial extracellular extracts under the study exhibited increasing DPPH scavenging activities with increase in sample concentration ( Table 2 ). These scavenging effects may be due to the donation of hydrogen atoms to quench free radicals by extracellularly released compounds like carotenoid pigments by selected bacterial strains. The same kind of quenching effects of hydrogen donation by an antioxidant sample were also observed by Kahkonen et al. (1999) . Carotenoids of bacterial origin are well reported to possess DPPH scavenging and or antioxidant properties ( Kuo et al. , 2011 , Shindo et al. , 2007 ). Miki et al. (1994) , showed the presence of carotenoid derived compounds in Pseudomonas sp. isolated from marine environment. In addition, Ye et al. (2012) had recently reported strong DPPH scavenging by exopolysaccharides isolated from Pseudomonas PF-6. So the overall scavenging effects in the present study may be due to the combined quenching effect of carotenoids and polysaccharide like reductants. Moreover, positive correlation with reducing power and phenolic contents also indicates their responsible roles ( Figure 3 ).
Scavenging of proton radicals is an important attribute of natural antioxidants. ABTS is a synthetic protonated radical which exhibits decrease in absorbance with the scavenging of the proton radicals (Mathew and Abraham, 2004). Ethyl acetate extracts of P. koreensis were able to scavenge these radicals efficiently, indicating higher responses towards their activity and comparable with standard compound (Quercetin) used ( Table 3 ). Various factors like stereo-selectivity of the radicals, the presence of functional groups in the bioactive compound, solubility of the extract in different testing systems and polarity of the solvent are reported to react and quench free radicals ( Yu et al. , 2002 ). Wang et al. (1998) , had reported that some compounds having ABTS scavenging property may not show similar DPPH scavenging effects. In our study Serratia rubidaea extracts also showed increased DPPH and decreased ABTS scavenging ( Table 3 ). Whereas, P. koreensis showed strong scavenging effects towards both DPPH and ABTS radicals. These results indicate that the radical scavenging activities may be in relation to properties of a compound excreted by the bacteria.
Increased FRAP value represents higher content of antioxidants in the sample extract responsible for reduction of ferric ion to the ferrous ion ( Smet et al. , 2006 ). We observed similar kind of expression in FRAP with P. koreensis and P. argentinensis ( Table 4 ). Although S. rubidaea exhibited good radical scavenging activities, the FRAP values were found to be lower in this case. Synergistic effects of the substances like carotenoids, vitamins and minerals are responsible in antioxidant activity variations ( Ratnam et al. , 2006 ) and this type of material might have contributed radical scavenging activity of the above species.
All the sample extracts showed increase in absorbance with increasing sample concentration ( Figure 2 ). The reducing potential of the sample involves various reactions such as prevention of chain initiation, binding of transition metal ion catalysts, decomposition of peroxides, prevention of continued hydrogen abstraction and radical scavenging ( Diplock, 1997 ). Higher reducing powers with Sargassum extract were reported at 10 mg/mL ( Cho et al. , 2007 ) and until now there are no reports in associated bacteria which can yield similar results. Further P. koreensis , associated with Sargassum in our study could show the highest reducing abilities within a concentration of 5 mg/mL ( Figure 3 ). This result suggests that, reducing capabilities of samples could be enhanced by the associated bacteria and it can be used as an alternative source for antioxidative compounds.
Generally, plants are well known source of phenolic and flavonoid compounds and till now limited amount of information is available on their presence in bacterial species. The current investigation on seaweed associated bacteria shows a positive indication of producing significant levels of TPCs ( Table 4 ). Polyphenolics and their derivatives appear to function as good electron and hydrogen atom donors and therefore, cause termination of radical chain reaction by converting free radicals to more stable products ( Shahidi and Wanasundara, 1992 ). Earlier studies on TPCs were strongly correlated with antioxidant capacities ( Kalt et al. , 1999 ). Our studies also showed positive correlations of TPC (p < 0.05) towards the other assessed antioxidant activities. As the prominent levels of phenolic contents were observed in the P. koreensis extracts, it may be assumed that this species may possess one or more bioactivities of similar kind.
In the current study we have come across one of the bacterial species Pseudomonas koreensis (JX915782) a Sargassum associated yellowish brown pigmented bacterium with high antioxidant activity while comparing with a known commercial antioxidant butylated hydroxytoluene (BHT) against DPPH scavenging. BHT is primarily used as an antioxidant additive in food, cosmetics, pharmaceuticals, rubber electrical transformer oil, and embalming fluid ( Malhotra, 2010 ). BHT also finds uses in hydraulic fluids, turbine and gear oils, jet fuels ( Michael and Irene, 2004 ). Keeping the various application of BHT listed above by many researchers we could suggest here our material on the above isolate Pseudomonas koreensis (JX915782) will definitely form a better natural resource in terms of biotechnological prospects and may be looked for its future applications.
16S rRNA gene sequencing results of the high potent strains, PIGB46, PIGB77 and PIGB88 indicates their common origin from the phylogenetic group Gammaproteobacteria. Among which two of the antioxidant producing bacteria PIGB46, PIGB77 belonged to the genera Pseudomonas and the other was Serratia . The variation in radical scavenging activities expressed by two different species of Pseudomonas indicates responsive role of genetic variability and its significance on synthesizing antioxidant compounds. All the three high potent strains worked out in the present study are expected to have similar or related genes in their nucleotide sequences for their synthesis; however analysis of such specific genes for quantifying these compounds remains the objective of future study.
Acknowledgments
Authors like to thank the Director of NIO, Dr. P. Vethamony, Dr. N. Ramaiah and Mr. Ram Murti Meena for their support in this study. The authors declare that there are no conflicts of interest. This is NIO contribution number 5592.
References
- Alscher RG, Donahue JL, Cramer CL. Reactive oxygen species and antioxidants: relationships in green cells. Physiol Plant. 1997;100:224–233. [Google Scholar]
- Benzie IFF, Strain JJ. The ferric reducing ability of plasma (FRAP) as a measure of ‘antioxidant power’: the FRAP assay. Anal Biochem. 1996;239:70–76. doi: 10.1006/abio.1996.0292. [DOI] [PubMed] [Google Scholar]
- Bolinches J, Lemos ML, Barja JL. Population dynamics of heterotrophic bacterial communities associated with Fucus vesiculosus and Ulva rigida in an estuary . Microb Ecol. 1988;15:345–357. doi: 10.1007/BF02012647. [DOI] [PubMed] [Google Scholar]
- Cheeseman KH, Slater TF. An introduction to free radical biochemistry. Bri Med Bull. 1993;49:481–493. doi: 10.1093/oxfordjournals.bmb.a072625. [DOI] [PubMed] [Google Scholar]
- Cho SH, Kang SE, Cho JY, Kim AR, Park SM, Hong YK, Ahn DH. The antioxidant properties of brown seaweed ( Sargassum siliquastrum ) extracts . J Med Food. 2007;10:479–485. doi: 10.1089/jmf.2006.099. [DOI] [PubMed] [Google Scholar]
- Correa-Llanten DN, Amenabar MJ, Blamey JM. Antioxidant capacity of novel pigments from an Antarctic bacterium. J Microbiol. 2012;50:374–379. doi: 10.1007/s12275-012-2029-1. [DOI] [PubMed] [Google Scholar]
- Dekkers JC, Van Doornen LJPH, Kemper CG. The role of antioxidant vitamins and enzymes in the prevention of exercise-induced muscle damage. Sports Med. 1996;21:213–238. doi: 10.2165/00007256-199621030-00005. [DOI] [PubMed] [Google Scholar]
- Diplock AT. Will the ‘good fairies’ please proves to us that vitamin E lessens human degenerative of disease? Free Rad Res. 1997;27:511–532. doi: 10.3109/10715769709065791. [DOI] [PubMed] [Google Scholar]
- Dring MJ. Stress resistance and disease resistance in seaweeds: the role of reactive oxygen metabolism. Adv Bot Res. 2005;43:175–207. [Google Scholar]
- Dufosse L. Microbial production of food grade pigments. Food Technol Biotechnol. 2006;44:313–321. [Google Scholar]
- Ekanayake P, Lee YD, Lee J. Antioxidant activity of flesh and skin of Eptatretus burgeri (Hag Fish) and Enedrias nebulosus (White Spotted Eel) . Food Sci Tech Int. 2004;10:171–177. [Google Scholar]
- Ferreira ICFR, Baptista P, Vilas-Boas M, Barros L. Free-radical scavenging capacity and reducing power of wild edible mushrooms from northeast Portugal: individual cap and stipe activity. Food Chem. 2007;100:1511–1516. [Google Scholar]
- Fridovich I. Biological effects of the superoxide radical. Arch Biochem Biophys. 1986;247:1–11. doi: 10.1016/0003-9861(86)90526-6. [DOI] [PubMed] [Google Scholar]
- Garcia-Casal MN, Ramirez J, Leets I, Pereira AC, Quiroga MF. Antioxidant capacity, polyphenol content and iron bioavailability from algae ( Ulva sp., Sargassum sp. and Porphyra sp.) in human subjects . Br J Nutr. 2009;101:79–85. doi: 10.1017/S0007114508994757. [DOI] [PubMed] [Google Scholar]
- Halliwell B, Gutteridge JM. Free Radicals in Biology and Medicine. 3rd ed. Oxford University Press; Oxford; 1999. [Google Scholar]
- Holmstrom C, Kjelleberg S. Marine Pseudoalteromonas species are associated with higher organisms and produce biologically active extracellular agents . FEMS Microbiol Ecol. 1999;30:285–293. doi: 10.1111/j.1574-6941.1999.tb00656.x. [DOI] [PubMed] [Google Scholar]
- Kahkonen MP, Hopia AI, Vuorela HJ, Rauha JP, Pihlaja K, Kujala TS, Heinonen M. Antioxidant activity of plant extracts containing phenolic compounds. J Agric Food Chem. 1999;47:3954–3962. doi: 10.1021/jf990146l. [DOI] [PubMed] [Google Scholar]
- Kalt W, Forney CF, Martin A, Prior RL. Antioxidant capacity, vita-min C, phenolics, and anthocyanins after fresh storage of small fruits. J Agr Food Chem. 1999;47:4638–4644. doi: 10.1021/jf990266t. [DOI] [PubMed] [Google Scholar]
- Kelman D, Posner EK, McDermid KJ, Tabandera NK, Wright PR, Wright AD. Antioxidant activity of Hawaiian marine algae. Mar Drugs. 2012;10:403–416. doi: 10.3390/md10020403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konzen M, De Marco D, Cordova CA, Vieira TO, Antonio RV, Creczynski-Pasa TB. Antioxidant properties of violacein: possible relation on its biological function. Bioorg Med Chem. 2006;14:8307–8313. doi: 10.1016/j.bmc.2006.09.013. [DOI] [PubMed] [Google Scholar]
- Kuo YH, Liang TW, Liu KC, Hsu YW, Hsu HC, Wang SL. Isolation and identification of a novel antioxidant with antitumor activity from Serratia ureilytica using squid pen as fermentation substrate . Marine Biotechnol. 2011;13:451–461. doi: 10.1007/s10126-010-9316-9. [DOI] [PubMed] [Google Scholar]
- Lakshmanaperumalsamy P, Purushothaman A. Heterotrophic bacteria associated with seaweed. Proc Indian Acad Sci. 1982;91:487–493. [Google Scholar]
- Lane DJ. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic Acid Techniques in Bacterial Systematic. John Wiley and Sons Incorporation; New York: 1991. pp. 115–175. [Google Scholar]
- Larrauri JA, Sanchez-Moreno C, Saura-Calixo F. Effect of temperature on the free radical scavenging capacity of extracts from red and white grape pomace peels. J Agri Food Chem. 1998;46:2694–2697. [Google Scholar]
- Malhotra GK. Chemical Process Simplification: Improving Productivity and Sustainability. Wiley-Blackwell (John Wiley & Sons Ltd); Chicester: 2010. [Google Scholar]
- Mathew S, Abraham TE. In-vitro antioxidant activity and scavenging effect of Cinnamomum verum leaf extract assayed by different methodologies . Food Chem Toxicol. 2006;44:198–206. doi: 10.1016/j.fct.2005.06.013. [DOI] [PubMed] [Google Scholar]
- Menezes CB, Bonugli-Santos RC, Miqueletto PB, Passarini MR, Silva CH, Justo MR, Leal RR, Fantinatti-Garboggini F, Oliveira VM, Berlinck RG, Sette LD. Microbial diversity associated with algae, ascidians and sponges from the north coast of Sao Paulo state. Brazil Microbiol Res. 2010;165:466–482. doi: 10.1016/j.micres.2009.09.005. [DOI] [PubMed] [Google Scholar]
- Michael A, Irene A. Handbook of Preservatives. Synapse Information Resources Inc.; Endicott, NY: 2004. [Google Scholar]
- Miki W, Otaki N, Yokoyama A, Izumida H, Shimidzu N. Okadaxanthin, a novel C50-carotenoid from a bacterium, Pseudomonas sp. KK10206C associated with marine sponge, Halichondria okadai . Experientia. 1994;50:684–686. [Google Scholar]
- Nedashkovskaya OI, Kim SB, Han SK, Lysenko A, Mikhailov VV, Bae KS, Swings J. Arenibacter palladensis sp. nov., a novel marine bacterium isolated from the green alga Ulva fenestrata , and emended description of the genus Arenibacter . Int J Syst Evol Microbiol. 2006;56:155–160. doi: 10.1099/ijs.0.63893-0. [DOI] [PubMed] [Google Scholar]
- Nedashkovskaya OI, Kim SB, Han SK, Rhee MS, Lysenko AM, Falsen E, Frolova GM, Mikhailov VV, Bae KS. Ulvibacter litoralis gen. nov., sp. nov., a novel member of the family Flavobacteriaceae isolated from the green alga Ulva fenestrata . Int J Syst Evol Microbiol. 2004;54:119–123. doi: 10.1099/ijs.0.02757-0. [DOI] [PubMed] [Google Scholar]
- Oberbeckmann S, Gerdts G, Wichels A, Laatsch H, Schumann G. Diversity of epiphytic marine bacteria associated with fronds of the brown algae Laminaria . Annual Conference of the Association for General and Applied Microbiology; Osnabruck, Germany. 2007. [Google Scholar]
- Ratnam DV, Ankola DD, Bhardwaj V, Sahana DK, Kumar MN. Role of antioxidants in prophylaxis and therapy: A pharmaceutical prospective. J Control Release. 2006;113:189–207. doi: 10.1016/j.jconrel.2006.04.015. [DOI] [PubMed] [Google Scholar]
- Re R, Pellegrini N, Proteggente A, Pannala A, Yang M, Rice-Evans C. Antioxidant activity applying an improved ABTS radical cation decolorization assay. Free Radic Biol Med. 1999;26:1231–1237. doi: 10.1016/s0891-5849(98)00315-3. [DOI] [PubMed] [Google Scholar]
- Shahidi F, Wanasundara PKJPD. Phenolic antioxidants. Crit Rev Food Sci Nutr. 1992;32:67–103. doi: 10.1080/10408399209527581. [DOI] [PubMed] [Google Scholar]
- Shan B, Cai YZ, Brooks JD, Corke H. The in vitro antibacterial activity of dietary spice and medicinal herb extracts. Int J Food Microbiol. 2007;117:112–119. doi: 10.1016/j.ijfoodmicro.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Shiba T, Simidu U, Taga N. Distribution of aerobic bacteria which contain bacteriochlorophyll a . Appl Environ Microbiol. 1979;38:43–45. doi: 10.1128/aem.38.1.43-45.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo K, Kikuta K, Suzuki A, Katsuta A, Kasai H, Yasumoto-Hirose M, Matsuo Y, Misawa N, Takaichi S. Rare carotenoids, (3R)-saproxanthin and (3R,2’S)-myxol, isolated from novel marine bacteria (Flavobacteriaceae) and their antioxidative activities. Appl Microbiol Biotechnol. 2007;74:1350–1357. doi: 10.1007/s00253-006-0774-y. [DOI] [PubMed] [Google Scholar]
- Smet K, Raes K, De Smet S. Novel approaches in measuring the antioxidative potential of animal feeds: the FRAP and DPPH methods. J Sci Food Agric. 2006;86:2412–2416. [Google Scholar]
- Soria-Mercado IE, Villarreal-Gomez LJ, Rivas GG, Sanchez NEA. Bioactive compounds from bacteria associated to marine algae. In: Sammour RH, editor. Biotechnology - Molecular Studies and Novel Applications for Improved Quality of Human Life. Croatia: 2012. pp. 25–44. [Google Scholar]
- Takao T, Kitatani F, Watanabe N, Yagi A, Sakata K. A simple screening method for antioxidants and isolation of several antioxidant produced by marine bacteria from fish and shellfish. Biosci Biotech Biochem. 1994;58:1780–1783. [Google Scholar]
- Vinayak RC, Sabu AS, Chatterji A. Bio-prospecting of a few brown seaweeds for their cytotoxic and antioxidant activities. Evid Based Complement Alternat Med. 2011;7:1–9. doi: 10.1093/ecam/neq024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Li J, Rangarajan M, Shao Y, La Voie EJ, Huang TC, Ho CT. Antioxidative phenolic compounds from sage ( Salvia officinalis ) . J Agric Food Chem. 1998;46:4869–4873. [Google Scholar]
- Ye S, Liu F, Wang J, Wang H, Zhang M. Antioxidant activities of an exopolysaccharide isolated and purified from marine Pseudomonas PF-6 . Carbohydrate Polymers. 2012;87:764–770. doi: 10.1016/j.carbpol.2011.08.057. [DOI] [PubMed] [Google Scholar]
- Yu L, Haley S, Perret J, Harris M, Wilson J, Qian M. Free radical scavenging properties of wheat extracts. J Agric Food Chem. 2002;50:1619–1624. doi: 10.1021/jf010964p. [DOI] [PubMed] [Google Scholar]