Abstract
The bacterial pathogen most commonly associated with endemic forms of childhood diarrhoea is Escherichia coli . Studies of epidemiological characteristics of HEp-2 cell-adherent E. coli in diarrhoeal disease are required, particularly in developing countries. The aim of this study was evaluate the presence and significance of adherent Escherichia coli from diarrhoeal disease in children. The prevalence of LA, AA, and DA adherence patterns were determined in HEp-2 cells, the presence of virulence genes and the presence of the O serogroups in samples obtained from 470 children with acute diarrhoea and 407 controls in Porto Velho, Rondônia, Brazil. E. coli isolates were identified by PCR specific for groups of adherent E. coli . Out of 1,156 isolates obtained, 128 (11.0%) were positive for eae genes corresponding to EPEC, however only 38 (29.6%) of these amplified bfpA gene . EAEC were isolated from 164 (14.1%) samples; of those 41(25%), 32 (19%) and 16 (9.7%) amplified eagg , aggA or aafA genes, respectively and aggA was significantly associated with diarrhoea ( P = 0.00006). DAEC identified by their adhesion pattern and there were few isolates. In conclusion, EAEC was the main cause of diarrhoea in children, especially when the aggA gene was present, followed by EPEC and with a negligible presence of DAEC.
Keywords: Enteroadherent Escherichia coli, diarrhoea, children
Introduction
Diarrhoea continues to be one of the most common causes of morbidity and mortality among infants and children in developing countries ( Nakhjavani et al. , 2013 ). The bacterial pathogen most commonly associated with endemic forms of childhood diarrhoea is Escherichia coli , which can be identified in about 50% of cases as presented by Chandra et al. (2012) . At least six categories of diarrhoeagenic E. coli strains are recognized on the basis of distinct epidemiological and clinical features, specific virulence determinants, and an association with certain serotypes: enteropathogenic E. coli (EPEC), enterotoxigenic E. coli (ETEC), enteroinvasive E. coli (EIEC), enterohemorrhagic E. coli (EHEC), enteroaggregative E. coli (EAEC), and diffusely adherent E. coli (DAEC) ( Torres et al. , 2005 ). EPEC, EAEC, and DAEC isolates are characterized by their distinct patterns of adherence to cultured epithelial cells in vitro . EPEC strains are responsible for a large number of cases of infantile diarrhoea in several developing countries ( Abba et al. , 2009 ; Chandra et al. , 2012 ; Contreras et al. , 2012 ; Nakhjavani et al. , 2013 ). These strains belong to specific serotypes within different E. coli serogroups (O groups) and produce a characteristic adherence pattern in tissue culture cells called localized adherence (LA) ( Humphries and Armstrong, 2010 ).
In the LA pattern, bacteria bind to localized areas of the cell surface, forming compact microcolonies (bacterial clusters) that can be visualized after 3 h of contact between the bacteria and the cells ( Humphries and Armstrong, 2010 ). This phenomenon is associated with the presence of the EPEC plasmid adherence factor (EAF) ( Bardiau et al. , 2010 ). The central mechanism of EPEC pathogenesis is a lesion called attaching and effacing (A/E) characterized by microvilli destruction, intimate adherence of bacteria to the intestinal epithelium, pedestal formation, and aggregation of polarized actin and other elements of the cytoskeleton at sites of bacterial attachment ( Humphries and Armstrong, 2010 ; Torres et al. , 2005 ). Atypical EPEC strains that do not carry the EAF plasmid can exhibit different adherence patterns including localized adherence like (LAL) pattern, and have been isolated from acute infantile diarrhoea in Brazil ( Arenas-Hernández et al. , 2012 ; Bardiau et al. , 2010 ; Ochoa and Contreras, 2011 ).
EAEC strains are defined by their characteristic aggregative adherence (AA) to HEp-2 cells in bacterial culture ( Weintraub, 2007 ). EAEC produce at least three fimbrial adhesins encoded on a 60- to 65-MDa virulence plasmid required for expression of the AA pattern: aggA (AAF/I), aafA (AAF/II), and agg-3 (AAF/III) ( Huang et al. , 2006 ).
DAEC strains are characterized by their dispersing adherence (DA) pattern on cultured epithelial cells ( Servin, 2005a ). Two adhesins capable of mediating the diffuse-adherence phenotype have been characterized for DAEC strains. F1845 is a fimbrial adhesin that mediates the adherence of the DAEC strain C1845 to epithelial cells ( Servin, 2005a , b ). F1845 fimbriae are encoded by five genes, designated daaABCDE . The significance of F1845 in virulence remains unclear because DAEC strains of faecal origin rarely express F1845 adhesin ( Campos et al. , 1999 ; Torres et al. , 2005 ). The adhesin involved in diffuse adherence (AIDA-I) is a plasmid-encoded protein of the clinical DAEC strain 2787. The AIDA-I precursor protein is encoded by the aidA gene and its mature form mediates diffuse adherence to HeLa cells ( Benz and Schmidt, 1992a , b ).
Studies evaluating the epidemiological characteristics of HEp-2 cell-adherent E. coli in diarrhoeal disease are required, particularly in developing countries. For this reason, and in order to determine the significance of the EAEC, EPEC, and DAEC strains as possible pathogenic microorganisms causing infantile diarrhoea, we determined the prevalence of the LA, AA, and DA patterns, the presence of virulence genes and the presence of O serogroups in samples obtained from children with acute diarrhoea and controls in Porto Velho, Rondônia, Brazil.
Material and Methods
Search for pathogens in the samples
Faecal samples from 470 diarrheic children (ages 0 to 72 months) and 407 children without diarrhoea (controls) were collected. The samples were collected at the Infantile Hospital Cosme and Damião in Porto Velho, Rondonia, Brazil, from March 2000 to March 2002.
Faecal samples were collected after natural excretion or through stimulation with a glycerin suppository. Samples were divided into two fractions: one fraction was used for parasitological examination for helminths eggs and protozoa cysts. The second aliquot was processed by routine microbiological and biochemical studies to identify E. coli . Five lactose-fermenting colonies and up to three lactose-negative colonies from each child were selected from McConkey plates to be tested by conventional and PCR procedures. Reference strains used as positive controls in the PCR tests included the E. coli strains O44H18, EDL933, 6085, O157:H7, H19 and C600PEB1 provided by Pasteur Institute (Paris, France). The non-pathogenic E. coli strain HB101 was used as a negative control and to monitor for PCR contamination.
PCR analysis
All strains were evaluated by PCR for identification of virulence genes from a distinct category of E. coli : eaeA and bfpA for EPEC; astA (encoding the toxin EAST1), eagg , aafA and aggA for EAEC; and daaE for DAEC ( Pass et al. , 2000 ), using primer sequences presented in Table 1 . PCR amplifications were performed as follows: 5.0 μL of bacterial extract was added to a reaction mixture (Life Technologies, Carlsbad, CA, USA) with a final volume of 50 μL containing: 0.1 mM each of dATP, dCTP, dGTP and dTTP PCR buffer (10 mM Tris HCL, pH 8.3; 50 mM KCl; 2 mM MgCl 2 ); 10 pMol of each PCR primer, and 1U of Taq DNA polymerase. Amplification was performed in a BioRad thermal cycler. The conditions used for amplification of the target genes are presented in Table 2 . After the amplification process the tubes were cooled rapidly at 4 °C. The amplified DNA products were resolved by agarose gel electrophoresis (2%, Life Technologies, Carlsbad, CA, USA) and visualized by UV transillumination after ethidium bromide staining.
Table 1.
Target genes and primers used to identify adherent Escherichia coli obtained from children diarrhoea in the Brazilian Amazon.
| Target | Gene | Primer | EP | Reference |
|---|---|---|---|---|
| EPEC | eaeA |
fp : 5′-TGAGCGGCTGGCATGAGTCATAC-3′
bp : 5′-TCGATCCCCATCGTCACCAGAGG-3′ |
241 | Pass et al. (2000) |
| EPEC | bfp |
fp : 5′-AATGGTGCTTGCGCTTGCTGC-3′
bp : 5′-GCCGCTTTATCCAACCTGGTA-3′ |
324 | Gunzburg et al. (1995) |
| EAEC | eagg |
fp : 5′-AGACTCTGGCGAAAGACTGTATC-3′
bp : 5′-ATGGCTGTCTGTAATAGATGAGAAC-3′ |
194 | Pass et al. (2000) |
| EAEC | aggA |
fb : 5′-GCTAACGCTGCGTTAGAAAGACC-3′
bp : 5′-GGAGTATCATTCTATATTCGCC-3′ |
352 | Piva et al. (2003) |
| EAEC | aafA |
fp : 5′ - GACAACCGCAACGCTGCGCTG-3′
p : 5′-GATAGCCGGTGTAATTGAGCC-3′ |
307 | Piva et al. (2003) |
Note - EP: expected product; fp, forward primer; bp, backward primer.
Table 2.
Amplification conditions for PCR reactions to identify adherent Escherichia coli obtained from children diarrhoea in the Brazilian Amazon.
| Target gene | First cycle | Intermediaries conditions | Final cycle |
|---|---|---|---|
| eaeA | 94 °C, 5 min | 25 cycles [94 °C, 2 min; 68 °C 1 min; 72 °C, 2 min] | 72 °C, 5 min |
| eagg | 94 °C, 5 min | 25 cycles [94 °C, 2 min; 68 °C 1 min; 72 °C, 2 min] | 72 °C, 5 min |
| bfpA | 94 °C, 5 min | 30 cycles [94 °C, 1 min; 56 °C 2 min; 72 °C, 1 min] | 72 °C, 5 min |
| daaE | 94 °C, 5 min | 30 cycles [94 °C, 1 min; 56 °C 2 min; 72 °C, 1 min] | 72 °C, 5 min |
| aggA | 94 °C, 5 min | 35 cycles [94 °C, 1 min; 60 °C 40 s; 72 °C, 30 s] | 72 °C, 5 min |
| aafA | 94 °C, 5 min | 35 cycles [94 °C, 1 min; 60 °C 20 s; 72 °C, 15 s] | 72 °C, 5 min |
| astA | 94 °C, 5 min | 30 cycles [94 °C, 1 min; 58 °C 1 min; 72 °C, 30 s] | 72 °C, 5 min |
HEp-2 adherence test
All E. coli isolates were subjected to HEp-2 adherence tests ( Cravioto et al. , 1991 ). HEp-2 cells were grown overnight to 50% confluence in Dulbecco’s Modified Eagle’s medium (Gibco BRL, Gaithersburg-MD, USA) containing penicillin, streptomycin, and 2% foetal bovine serum on eight-well chamber slides. Bacteria were grown for 16 h in Luria broth without shaking. The HEp-2 cells were washed five times with Phosphate Buffered Saline and then the medium was replaced with Dulbecco’s Modified Eagle’s medium containing 1% mannose. A volume of 10 μL of bacterial suspension was added per well and the slides were incubated at 37 °C in 5% CO2 for 3 h. The monolayers were washed five times with PBS, then fixed with 70% methanol and Giemsa. The strains were stained with solutions provided in the Panoptic Quick staining kit for one minute. Each strain was tested in duplicate, and appropriate controls were included in the test. Strains that adhered to the monolayers were recorded as adhering in localized, diffuse, or aggregative patterns.
Serotype methods
Enteroadherent E. coli isolates grown on nutrient agar were used for the identification of somatic (O) antigens by standard agglutination methods using specific polyvalent and monovalent antisera (Biomerieux, Craponne, Auvergne, France).
Statistical analysis
The prevalence of diarrhoeagenic E. coli in patient and control samples was compared by a two-tailed X2 test with Yates correction and Fisher’s exact test. When analysing the association of two or three pathogens in the same diarrhoeal patient, the same test was used to evaluate the probability of association by random chance as a function of their respective individual frequencies in the population.
Results
A total of 1.156 E. coli strains were isolated from faecal specimens from 470 diarrheic children (patients) and 407 children without diarrhoea (controls). The PCR screening results for virulence factors of enteroadherent diarrhoeagenic E. coli are presented in Table 3 .
Table 3.
Occurrence of enteropathogenic E. coli among children with and without diarrhoea from Porto Velho, RO, Brazil.
| Virulence genes associated with Enteroadherent E. coli | ||||||
|---|---|---|---|---|---|---|
|
|
||||||
| Pathogen (n ∘ of colonies) | eaeA | bfp | eagg | aggA | aafA | daaE |
| Typical EPEC (38) | 38 (100%) | 38 (100%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) |
| Atypical EPEC (90) | 90 (100%) | 0 (0%) | 10 (11.1%) | 0 (0%) | 0 (0%) | 0 (0%) |
| EAEC (164) | 0 (0%) | 0 (0%) | 41 (25%) | 32 (19.5%) | 16 (9.7%) | 0 (0%) |
| DAEC (68) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 6 (8.8%) |
Screening for the eae sequence, which is specific for both EPEC and EHEC, there were 128 positive isolates comprising 11.7% of the total. Only 38 eae -positive patient isolates amplified with bfpA primers, indicating that they belonged to typical EPEC. Thus 90 isolates were considered as atypical EPEC, once all isolates were negative for verotoxin genes in preview work ( Orlandi et al. , 2006 ). EAEC were isolated from 164 samples; of those 41(25%), 32 (19%) and 16 (9.7%) amplified for eagg , aggA and aafA, respectively. Of the isolated EAEC, 75 (45.7%) exhibited the AA adhesion pattern in HEp-2 cells, not amplifying for any specific markers for EAEC tested in that State. DAEC were mainly characterized by the pattern of HEp2 cell adhesion. Among the 68 DAEC isolates, only 6 (8.8%) presented the daaE gene that encodes biogenesis of the F1845 adhesin. The enteroadherent E. coli analysed had higher frequencies in children with acute gastroenteritis ( Table 4 ).
Table 4.
Adhesion pattern of E. coli isolated from children with and without diarrhea in Porto Velho, RO, Brazil.
| No. of isolates (%) in children | |||
|---|---|---|---|
|
|
|||
| Adhesion pattern | Total (n = 877) | With diarrhoea (n = 470) | Without diarrhoea (n = 407) |
| Typical EPEC (AL) | 38 (4.3) | 26 (5.5) | 12 (2.9) |
| Atypical EPEC (ALL) | 90 (10.2) | 55 (11.7) | 35 (8.5) |
| EAEC (AA) | 164 (18.7) | 91 (19.3) | 73 (17.9) |
| DAEC (AD) | 68 (7.7) | 35 (7.4) | 33 (8.1) |
| Total | 360 (41.0) | 207 (44.0) | 153 (37.5) |
The results show us that all samples belonging to the typical EPEC showed the LA pattern of adhesion after 3 h of incubation; they were distributed in 26 (68.4%) patients and 12 (31.5%) of the control samples. Of the 90 isolated atypical EPEC, we found that 55 (61.1%) of the cases presented the LAL adhesion pattern after 6 h of incubation ( Figure 1 ), while in the control samples 35 (38.9%) showed this adhesion profile. EAEC showed the AA adhesion pattern in 91 (55.5%) of the patients and 73 (44.5%) of the control samples ( Figure 1 ). Diffuse adhesion was characterized in 35 (51.4%) of the patients and 33 (48.6%) of the control samples ( Figure 1 ). When in association with EAEC, we observed that atypical EPEC showed an atypical phenotype (LAL) at a high frequency ( Figure 1 ). We also found that the presence of the aggA (AAF/I) gene was significantly associated with diarrhoea, because 31 isolates having the aggA gene (6.5%) were related to patients, while only (0.2%) of controls had this gene (p = 0.00006). The presence of the astA gene in samples of EAEC showed no correlation with diarrhoea, since it occurred in both patient and control isolates at almost the same frequency ( Table 5 ).
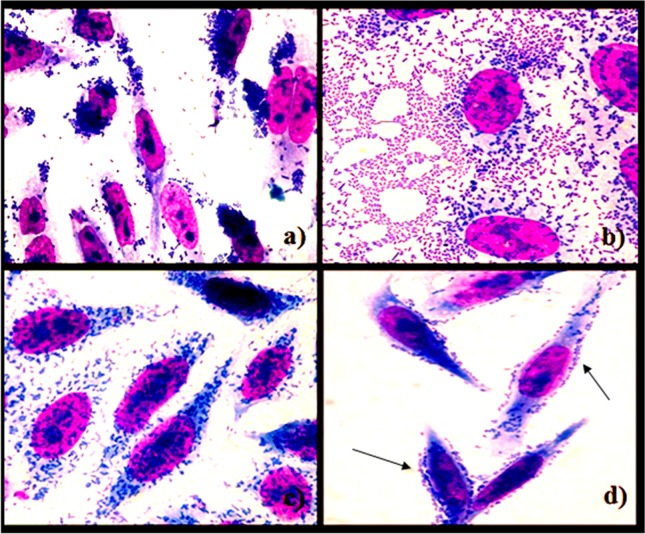
Figure 1.
Micrographs of adherence patterns on HEp2 cells of Escherichia coli obtained from faeces of children with diarrhoea and controls from Porto Velho-RO, Brazil: a) LA pattern after 3 h of incubation with typical EPEC; b) AA pattern after 3 h of incubation with EAEC; c) DA pattern after 3 h of incubation with DAEC; d) LAL pattern after 6 h of incubation with atypical EPEC (arrows).
Table 5.
Phenotypic and genotypic frequencies found in enteroaggregative E. coli analysed among children with (470) and without (407) diarrhoea from Porto Velho, RO, Brazil.
| Virulence Markers | Target | With diarrhoea | Without diarrhoea | Value p |
|---|---|---|---|---|
| AA | Adherent Pattern | 91 (19.4%) | 73 (17.8%) | 0.5 |
| aggA | Fimbrial subunity AAF/I | 31 (6.5%) | 1 (0.2%) | 0.00006 |
| aafA | Fimbrial subunity AAF/II | 8 (1.7) | 8 (1.9%) | 0.7 |
| astA | East Toxin | 9 (1.9%) | 8 (1.9%) | 0.9 |
| Total | 174 (37.0) | 121 (29.7%) | 0.02 |
A total of 205/425 (48.2%) isolates were submitted to serotyping of somatic antigen (O), and the results are presented in Table 6 . We observed that the classic serogroups of EPEC and EAEC were the most frequently isolated, in agreement with the results obtained with the virulence factors and especially with the phenotypes in cellular assays. The serogroups O125, O111, O44, O55 and O127 appeared as possibly associated with diarrhoea, because we found that most of these serogroups were isolated from children with acute gastroenteritis, in agreement with the distribution of categories of diarrhoeagenic E. coli isolated ( Table 6 ). EAEC isolates were mostly in the O44, O55, O111, O125 and O127 serogroups ( Table 6 ).
Table 6.
Serotyping of somatic antigen (O) of enteroaggregative E. coli isolated from children with and without diarrhoea from Porto Velho, RO, Brazil.
| Pathogen | Serotype | With diarrhoea (n = 252) % | Without diarrhoea (n = 173) % | Total (n = 425) % |
|---|---|---|---|---|
| Typical EPEC | O111 | 3 (1.2) | - | 3 (0.7) |
| O119 | 2 (0.8) | - | 2 (0.5) | |
| O127 | 2 (0.8) | 3 (1.6) | 5 (1.2) | |
| O44 | 2 (0.8) | 1 (0.6) | 3 (0.7) | |
| O55 | 4 (1.5) | 1 (0.6) | 5 (1.2) | |
| O126 | 1 (0.4) | - | 1 (0.2) | |
| O86 | - | 1 (0.6) | 1 (0.2) | |
| Subtotal | 14 (5.5) | 6 (3.5) | 20 (4.7) | |
| Atypical EPEC | O1 | 4 (1.5) | 1 (0.6) | 5 (1.2) |
| O111 | 2 (0.8) | - | 2 (0.5) | |
| O125 | 5 (1.9) | 2 (1.1) | 7 (1.6) | |
| O166 | 6 (2.4) | - | 6 (1.4) | |
| O44 | 7 (2.8) | 5 (2.9) | 12 (2.8) | |
| O6 | 4 (1.5) | - | 4 (0.9) | |
| O86 | 2 (0.8) | 1 (0.6) | 3 (0.7) | |
| Subtotal | 30 (11.9) | 9 (5.2) | 39 (9.1) | |
| EAEC | O1 | 7 (2.8) | 10 (5.8) | 17 (4.0) |
| O111 | 7 (2.8) | 6 (3.4) | 13 (3.0) | |
| O125 | 6 (2.4) | 4 (2.3) | 10 (2.2) | |
| O127 | 10 (3.9) | 3 (1.7) | 13 (3.0) | |
| O166 | 6 (2.3) | 4 (2.3) | 10 (2.2) | |
| O44 | 12 (4.8) | 9 (5.2) | 21 (4.8) | |
| O55 | 6 (2.3) | 3 (1.7) | 12 (2.8) | |
| O8 | 4 (1.5) | 1 (0.6) | 5 (1.2) | |
| O169 | 5 (1.9) | 1 (0.6) | 6 (1.4) | |
| O26 | 2 (0.8) | 5 (2.9) | 7 (1.6) | |
| Subtotal | 65 (25.8) | 46 (26.6) | 111 (26.2) | |
| DAEC | O111 | 2 (0.8) | 1 (0.6) | 3 (0.7) |
| O1 | 3 (1.2) | 1 (0.6) | 4 (0.9) | |
| O125 | 4 (1.5) | 2 (1.1) | 6 (1.4) | |
| O127 | 5 (1.9) | 6 (3.4) | 11 (2.5) | |
| O44 | 5 (1.9) | 4 (2.3) | 9 (2.1) | |
| O6 | 2 (0.8) | - | 2 (0.5) | |
| Subtotal | 21 (8.3) | 14 (8.1) | 35 (8.2) | |
| Total | 130 (51.6) | 75 (43 4) | 205 (48.2) |
Discussion
The pathogenic profile of enteroadherent E. coli has been studied throughout the world. We studied the frequencies of EPEC, EAEC and DAEC in faecal samples from children with and without diarrhoea in Porto Velho, Rondonia Brazil. Screening for the genetic eaeA marker showed that 128 E. coli isolates possessed this characteristic factor of both typical and atypical EPEC. This number of EPEC represented 11.4% of the total isolates, showing a presence of EPEC approximately twice that found preview study ( Nakhjavani et al. , 2013 ) in a screen for this E. coli group among 412 isolates obtained from 612 stool samples of children with diarrhoea in Tehran, Iran. The 90 samples that only had the eaeA gene belonged to the group of atypical EPEC, representing about 70% of all EPEC, corroborating the proportion of atypical EPEC presented by preview study ( Nakhjavani et al. , 2013 ).
The screen for the EAEC adherence factor showed that among 164 samples with the AA pattern, only 41 isolates were presented positive for at least one EAEC virulence marker; however, those 123 that were negative for these markers, were positive for the AA adhesion pattern in HEp-2 cells, demonstrating the specificity of the test cells. Within the EAEC samples, 32 possessed the aggA gene and 16 possessed the aafA gene.
We did not find an association between DAEC and diarrhoea. In contrast, studies conducted in the state of Espirito Santo and Northeast Brazil showed the diarrhoeagenic E. coli most prevalent in children with acute diarrhoea was DAEC ( Scaletsky et al. , 2002b ; Spano et al. , 2008 ).
Regarding the DA adhesion pattern, the results showed low specificity for the daaE gene. Studies have compared different methods to characterize enteroadherent E. coli . With regard to tests carried out for detection of DAEC, the use of a probe for detection of the daaC gene sequence showed low sensitivity (64.3%), confirming frequencies found by other investigators ( Scaletsky et al. , 2002a ).
This study identified a large number of isolates that did not amplify with specific markers; however, the adhesion phenotypic test was able to identify the different adherence profiles, although more tests must be performed to identify possible adhesins associated with this pattern of adherence. The association between atypical EPEC and EAEC indicates a possible phenotype only observed when the two groups are associated with diarrhoea. In this study, the presence of the aggA (AAF/I) gene was associated with diarrhoea. We found 31 isolates positive for the aggA gene (6.5%) linked to patient cases, while only one patient (0.2%) of controls had this gene (p = 0.00006). A recent study has shown similar results, finding a higher frequency of the aggA (AAF/I) gene isolated from cases of diarrhoea ( Piva et al. , 2003 ). We observed that the presence of the fimbriae AAF/I in our EAEC isolates is statistically associated with diarrhoea. Studies performed in Spain showed that the presence of fimbriae AAF/I was not observed in EAEC samples; however, AAF/II was detected in 8.7% of the EAEC isolated ( Vila et al. , 2000 ). Studies in southwest Nigeria showed the presence of AAF/I in 63% of isolated EAEC; 35% had AAF/II ( Okeke et al. , 2000 ). Similar results were found in studies in India ( Kahali et al. , 2004 ). In contrast other study ( Elias et al. , 1999 ) detected AAF/I and AAF/II in 19% and 8%, respectively, of EAEC samples in São Paulo. Studies performed in Gabon showed that the presence of the AAF I and AAF II fimbriae was more likely in EAEC than non-EAEC isolates ( Presterl et al. , 2003 ). From these data we conclude that the prevalence of AAF seems to vary in relation to geographic region ( Flores and Okhuysen, 2009 ).
The distribution of the serogroups found revealed a diversity of strains, which did not allow us to infer the actual prevalence of a specific serogroup in the population studied. In this study, we observed a higher frequency of the O111, O125, O44, O55 and O127 serogroups ( Table 6 ); we emphasize the importance of serogroups since they are described as associated with pathogenic strains of E. coli .
In conclusion, EAEC are the main cause of diarrhoea in children from Porto Velho, RO, Brazil and the presence of aggA is highly correlated with this disease. EPEC is the second most common cause and when the same child is infected by both EAEC and atypical EPEC, a higher number of isolates of the latter present the LAL adherence pattern in HEp2 cells. In contrast to other regions in Brazil, DAEC was a significant contributor to diarrhoea cases in this city in the Brazilian western Amazon. In the studied city a more specific prevalent serogroup in paediatric diarrhoea was not identified. Therefore, the need to understand the mechanisms involved in the pathogenicity of these strains should be emphasized, especially when we consider the associations identified, recalling that bacterial pathogenicity is a property contributed to by multiple factors that are expressed in their natural environment, including horizontal gene transmission. Going forward, other experimental models should be used for complementary phenotypic and genomic studies.
Acknowledgments
The authors thank the Brazilian National Council for Scientific and Technological Development (CNPq).
References
- Abba K, Sinfield R, Hart CA, et al. Pathogens associated with persistent diarrhoea in children in low and middle income countries: Systematic review. BMC Infect Dis. 2009;9:88. doi: 10.1186/1471-2334-9-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenas-Hernández MMP, Martínez-Laguna Y, Torres AG. Clinical implications of enteroadherent escherichia coli. Curr Gastroen Rep. 2012;14:386–394. doi: 10.1007/s11894-012-0277-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardiau M, Szalo M, Mainil JG. Initial adherence of EPEC, EHEC and VTEC to host cells. Vet Res. 2010;41:57. doi: 10.1051/vetres/2010029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benz I, Schmidt MA. AIDA-I, the adhesin involved in diffuse adherence of the diarrhoeagenic Escherichia coli strain 2787 (O126:H27), is synthesized via a precursor molecule. Mol Microbiol. 1992a;6:1539–1546. doi: 10.1111/j.1365-2958.1992.tb00875.x. [DOI] [PubMed] [Google Scholar]
- Benz I, Schmidt MA. Isolation and serologic characterization of AIDA-I, the adhesin mediating the diffuse adherence phenotype of the diarrhea-associated Escherichia coli strain 2787 (O126:H27) Infect Immun. 1992b;60:13–18. doi: 10.1128/iai.60.1.13-18.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos LC, Vieira MA, Trabulsi LR, et al. Diffusely adhering Escherichia coli (DAEC) strains of fecal origin rarely express F1845 adhesin. Microbiol Immunol. 1999;43:167–170. doi: 10.1111/j.1348-0421.1999.tb02388.x. [DOI] [PubMed] [Google Scholar]
- Chandra BK, Singh G, Taneja N, et al. Diarrhoeagenic Escherichia coli as a predominant cause of paediatric nosocomial diarrhoea in India. J Med Microbiol. 2012;61:830–836. doi: 10.1099/jmm.0.041848-0. [DOI] [PubMed] [Google Scholar]
- Contreras CA, Ochoa TJ, Ruiz J, et al. Genetic diversity of locus of enterocyte effacement genes of enteropathogenic Escherichia coli isolated from Peruvian children. J Med Microbiol. 2012;61:1114–1120. doi: 10.1099/jmm.0.045443-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cravioto A, Tello A, Navarro A, et al. Association of Escherichia coli HEp-2 adherence patterns with type and duration of diarrhoea. Lancet. 1991;337:262–264. doi: 10.1016/0140-6736(91)90868-p. [DOI] [PubMed] [Google Scholar]
- Elias WP, Suzart S, Trabulsi LR, et al. Distribution of aggA and aafA gene sequences among Escherichia coli isolates with genotypic or phenotypic characteristics, or both, of enteroaggregative E. coli. J Med Microbiol. 1999;48:597–599. doi: 10.1099/00222615-48-6-597. [DOI] [PubMed] [Google Scholar]
- Flores J, Okhuysen PC. Enteroaggregative Escherichia coli infection. Curr Opin Gastroen. 2009;25:8–11. doi: 10.1097/MOG.0b013e32831dac5e. [DOI] [PubMed] [Google Scholar]
- Gunzburg ST, Tornieporth NG, Riley LW. Identification of enteropathogenic Escherichia coli by PCR-bases detection of the bundle forming pilus gene . J Clin Microbiol. 1995;33:1375–1377. doi: 10.1128/jcm.33.5.1375-1377.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DB, Mohanty A, DuPont HL, et al. A review of an emerging enteric pathogen: Enteroaggregative Escherichia coli . J Med Microbiol. 2006;55:1303–1311. doi: 10.1099/jmm.0.46674-0. [DOI] [PubMed] [Google Scholar]
- Humphries RM, Armstrong GD. Sticky situation: Localized adherence of enteropathogenic Escherichia coli to the small intestine epithelium. Future Microbiol. 2010;5:1645–1661. doi: 10.2217/fmb.10.124. [DOI] [PubMed] [Google Scholar]
- Kahali S, Sarkar B, Rajendran K, et al. Virulence characteristics and molecular epidemiology of enteroaggregative Escherichia coli isolates from hospitalized diarrheal patients in Kolkata, India . J Clin Microbiol. 2004;42:4111–4120. doi: 10.1128/JCM.42.9.4111-4120.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakhjavani FA, Emaneini M, Hosseini H, et al. Molecular analysis of typical and atypical enteropathogenic Escherichia coli (EPEC) isolated from children with diarrhoea. J Med Microbiol. 2013;62:191–195. doi: 10.1099/jmm.0.046516-0. [DOI] [PubMed] [Google Scholar]
- Ochoa TJ, Contreras CA. Enteropathogenic Escherichia coli infection in children. Curr Opin Infect Dis. 2011;24:478–483. doi: 10.1097/QCO.0b013e32834a8b8b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okeke IN, Lamikanra A, Czeczulin J, et al. Heterogeneous virulence of enteroaggregative Escherichia coli strains isolated from children in Southwest Nigeria. J Infect Dis. 2000;181:252–260. doi: 10.1086/315204. [DOI] [PubMed] [Google Scholar]
- Orlandi PP, Magalhães GF, Matos NB, et al. Etiology of diarrheal infections in children of Porto Velho (Rondonia, Western Amazon region, Brazil) Braz J Med Biol Res. 2006;39:507–517. doi: 10.1590/s0100-879x2006000400011. [DOI] [PubMed] [Google Scholar]
- Pass MA, Odedra R, Batt RM. Multiplex PCRs for identification of Escherichia coli virulence genes. J Clin Microbiol. 2000;38:2001–2004. doi: 10.1128/jcm.38.5.2001-2004.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piva IC, Pereira AL, Ferraz LR, et al. Virulence markers of enteroaggregative Escherichia coli isolated from children and adults with diarrhea in Brasília, Brazil. J Clin Microbiol. 2003;41:1827–1832. doi: 10.1128/JCM.41.5.1827-1832.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presterl E, Zwick RH, Reichmann S, et al. Frequency and virulence properties of diarrheagenic Escherichia coli in children with diarrhea in Gabon. Am J Trop Med Hyg. 2003;69:406–410. [PubMed] [Google Scholar]
- Scaletsky ICA, Fabbricotti SH, Aranda KR, et al. Comparison of DNA hybridization and PCR assays for detection of putative pathogenic enteroadherent Escherichia coli. J Clin Microbiol. 2002a;40:1254–1258. doi: 10.1128/JCM.40.4.1254-1258.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaletsky ICA, Fabbricotti SH, Carvalho RLB, et al. Diffusely adherent Escherichia coli as a cause of acute diarrhea in young children in northeast Brazil: A case-control study. J Clin Microbiol. 2002b;40:645–648. doi: 10.1128/JCM.40.2.645-648.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servin AL. Pathogenesis of Afa/Dr Diffusely Adhering Escherichia coli. Clin Microbiol Rev. 2005a;18:264–292. doi: 10.1128/CMR.18.2.264-292.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servin AL. Pathogenesis of Afa/Dr diffusely adhering Escherichia coli. Clin Microbiol Rev. 2005b;18:264–292. doi: 10.1128/CMR.18.2.264-292.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spano LC, Sadovsky ADI, Segui PN, et al. Age-specific prevalence of diffusely adherent Escherichia coli in Brazilian children with acute diarrhoea. J Med Microbiol. 2008;57:359–363. doi: 10.1099/jmm.0.47660-0. [DOI] [PubMed] [Google Scholar]
- Torres AG, Zhou X, Kaper JB. Adherence of diarrheagenic Escherichia coli strains to epithelial cells. Infect Immun. 2005;73:18–29. doi: 10.1128/IAI.73.1.18-29.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vila J, Vargas M, Henderson IR, et al. Enteroaggregative Escherichia coli virulence factors in traveler’s diarrhea strains . J Infect Dis. 2000;182:1780–1783. doi: 10.1086/317617. [DOI] [PubMed] [Google Scholar]
- Weintraub A. Enteroaggregative Escherichia coli: Epidemiology, virulence and detection. J Med Microbiol. 2007;56:4–8. doi: 10.1099/jmm.0.46930-0. [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Echeverria P. Detection of the enteroaggregative Escherichia coli heat-stable enterotoxin 1 gene sequences in enterotoxigenic E. coli strains pathogenic for humans . Infect Immun. 1996;61:1441–1445. doi: 10.1128/iai.64.4.1441-1445.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]