Abstract
An Aspergillus niger UFV-1 phytase was characterized and made available for industrial application. The enzyme was purified via ultrafiltration followed by acid precipitation, ion exchange and gel filtration chromatography. This protein exhibited a molecular mass of 161 kDa in gel filtration and 81 kDa in sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), indicating that it may be a dimer. It presented an optimum temperature of 60 °C and optimum pH of 2.0. The K M for sodium phytate hydrolysis was 30.9 mM, while the k cat and k cat / K M were 1.46 ×10 5 s −1 and 4.7 × 10 6 s −1 .M −1 , respectively. The purified phytase exhibited broad specificity on a range of phosphorylated compounds, presenting activity on sodium phytate, p-NPP, 2- naphthylphosphate, 1- naphthylphosphate, ATP, phenyl-phosphate, glucose-6-phosphate, calcium phytate and other substrates. Enzymatic activity was slightly inhibited by Mg 2+ , Cd 2+ , K + and Ca 2+ , and it was drastically inhibited by F − . The enzyme displayed high thermostability, retaining more than 90% activity at 60 °C during 120 h and displayed a t 1/2 of 94.5 h and 6.2 h at 70 °C and 80 °C, respectively. The enzyme demonstrated strong resistance toward pepsin and trypsin, and it retained more than 90% residual activity for both enzymes after 1 h treatment. Additionally, the enzyme efficiently hydrolyzed phytate in livestock feed, liberating 15.3 μmol phosphate/mL after 2.5 h of treatment.
Keywords: phosphatase, phytic acid, dephosphorylation
Introduction
Phytic acid ( myo -inositol hexakisphosphate) accounts for 60–90% of the total phosphorus content in plants, where it serves as a major storage form of phosphorus and divalent cations that can be hydrolyzed by several enzymes during seed germination ( Vats et al. , 2009 ). Phytic acid acts as an antinutritional factor in animals because of its ability to complex with ions such as calcium, magnesium, iron and zinc as well as proteins, thus decreasing its bioavailability ( Bohn et al. , 2008 ; Fugthong et al. , 2010 ). Additionally, phytic phosphorus cannot be absorbed by monogastric animals including humans because of phytate degrading enzyme insufficiency or absence. Thus, unutilized phytate phosphorus from plant feed is excreted, which becomes an environmental pollutant in areas of intensive livestock units ( Yao et al. , 2011 ). One of the alternatives for solving this problem may be supplementation of animal feed with phytase ( Maguire et al. , 2005 ).
Phytases (myo-inositol hexakisphosphate phosphohydrolases (EC 3.1.3.8, 3.1.3.26 and 3.1.3.72)) are phosphatase enzymes that catalyze the hydrolysis of phosphate moieties from phytate, resulting in stepwise formation of myo -inositol pentakis-, tetrakis-, tris-, bis- and monophosphate isomers as well as liberation of inorganic phosphate and potentially chelated minerals ( Bohn et al. , 2008 ; Azeke et al. , 2011 ). Therefore, the benefits of phytase are at least three-fold: increase bio-availability of phosphorus and others minerals in livestock feed, preserve non-renewable phosphorus sources by reducing the need for supplementing diets with phosphorus and reduce environmental pollution ( Yao et al. , 2011 ).
This paper describes the purification and characterization of a novel phytase produced from a Aspergillus niger UFV-1 isolate and its application in a commercial livestock feed.
Materials and Methods
Materials
Substrates including sodium phytate, calcium phytate, p-nitrophenyl phosphate (pNPP), adenosine triphosphate (ATP), adenosine diphosphate (ADP), adenosine monophosphate (AMP), glucose-6-phosphate, glucose-1-phosphate, 1-naphtyl phosphate, 2-naphtyl phosphate, phenyl phosphate and glycerol 2-phosphate as well as the chemical reagents Tween-20, calcium chloride, monopotassium phosphate, sodium carbonate, sodium dodecyl sulfate (SDS), β-mercaptoethanol, dithiothreitol (DTT), urea, guanidine chloride, ethylenediaminetetraacetic acid (EDTA) and potato dextrose agar (PDA) were purchased from Sigma Chemical Co. (St. Louis, MO, USA). Peptone was obtained from Himedia Laboratories Co. (Mumbai, Maharashtra, India), and sodium acetate, sucrose, hydrochloric acid and trichloroacetic acid was acquired from Vetec Fine Chemical (Duque de Caxias, RJ, Brazil). Glycine, 2-Morpholinoethanesulfonic acid (MES) and tris(hydroxymethyl)aminomethane (TRIS) were purchased from Merck (Darmstadt, Germany). Standard molecular weight markers were obtained from Fermentas (USA), and protein standards, trypsin and pepsin were also acquired from Sigma. Potato and commercial pig feed was obtained from the local market. All other reagents used in this study were of analytical grade.
Strain isolation, culture conditions and enzyme production
Fungus was isolated from sugarcane bagasse samples that had been collected at a sugar-alcohol plant located at Urucânia, Minas Gerais, Brazil. Subsequently, the isolates were identified by morphology and physiology ( Klich, 2002 ; Samson et al. , 2010 ) as Aspergillus niger . The identification was performed at the Phytopathology Department of the Federal University of Viçosa. The fungus was maintained in the culture collection of the Applied Enzymology Laboratory, Federal University of Viçosa (UFV), Brazil. In this study, it is designated as Aspergillus niger UFV-1.
Induction of phytase was achieved in liquid medium according to Gunashree and Venkateswaran (2008) with some modifications. Its content per liter potato broth was: sucrose 10 g, peptone 5 g, Tween-20 2.5 mL and CaCl 2 0.10 g. Potato broth was obtained by boiling 200 g fresh potato in 1 L distilled water. The final medium pH was adjusted to 5.5 by addition of a 0.1 M HCl solution. Aliquots of 50 mL medium were dispensed in 250 mL Erlenmeyer flasks and autoclaved at 121 °C for 20 min. Seven day-old spores grown on solid potato dextrose agar media were harvested by washing the plate surface with a distillated water solution containing 0.2% (v/v) Tween-20. The medium was inoculated with a spore suspension (10 6 spores mL −1 ). The flasks were incubated for 8 days at 28 °C on an orbital shaker at 180 RPM. The culture broth was then harvested and submitted to initial filtration through a nylon filter, and the filtrate was centrifuged (10,000 × g , 10 min at 4 °C). The supernatant was stored at 4 °C until purification.
Phytase assay
Phytase activity was determined by measuring the inorganic phosphate released from sodium phytate. For analysis, 600 μL 1.5 mM sodium phytate in 100 mM sodium acetate buffer, pH 2.5 and 150 μL enzymatic extract were used. The reaction was conducted in a water bath at 50 °C, and it was stopped after 30 min by adding 250 μL 10% (v/v) trichloroacetic acid solution. This was followed by the addition of 1000 μL colorimetric reagent to the tubes ( Harland and Harland, 1980 ). Absorbance was measured at 700 nm, and the measured values were correlated with a standard curve that had been constructed using monopotassium phosphate. All of the enzyme activity analyses were performed in triplicate; one unit of phytase activity was defined as the amount of enzyme that released 1 μmol phosphate per minute under the assay conditions.
Enzyme purification
Crude extracellular phytase was concentrated using an Amicon ultrafiltration cell model 8400 (Bedford, MA, USA) with a 30 kDa molecular cutoff PM 30 Amicon membrane at 4 °C. The resulting retentate was acidified with 0.1 M HCl until it reached pH 2.0, it was maintained under slow agitation for 30 min at 4 °C and centrifuged (10,000 × g , 10 min at 4 °C). A 10 mL aliquot of the supernatant was loaded onto a 20 mL DEAE-Sepharose CL-6B column that had been pre-equilibrated with 50 mM sodium acetate buffer, pH 5.0 containing 0.5% (w/v) sucrose. Unbound protein was removed by washing with four bed volumes of equilibration buffer. Bound protein was then eluted using a linear salt gradient (0–0.5 M NaCl in 50 mM sodium acetate buffer pH 5.0 containing 0.5% (w/v) sucrose) at 1 mL/min; 4 mL fractions were collected. Pooled activity (3 mL) was loaded onto a Sephacryl S-300 HR (Amersham Biosciences, Uppsala, Sweden) column (2.6 × 60 cm) that had been pre-equilibrated with a 25 mM sodium acetate buffer, pH 5.0, containing 0.5% (w/v) sucrose at 0.8 mL/min using a AKTA Purifier fast protein liquid chromatography device (Amersham Biosciences), from which 2.5 mL fractions were collected. Ion exchange chromatography was performed at 4 °C, and gel filtration was performed at room temperature. Protein concentration was determined by the Bradford method using bovine serum albumin as the standard at 0–20 μg/mL.
Electrophoretic analysis and molecular mass determination
One dimensional SDS gel electrophoresis was performed using a 12.5% (w/v) acrylamide gel in a vertical electrophoresis system, and the proteins bands were visualized by silver staining.
For zymogram analysis, one dimensional SDS gel electrophoresis was conducted using a 12.5% (w/v) acrylamide gel, where SDS and β-mercaptoethanol were omitted from the gel and buffer, respectively, and the sample was not heated prior to analysis. After electrophoresis, the gel was equilibrated with 0.2 M sodium acetate buffer, pH 2.5 for 30 min and then incubated in the same buffer containing 0.04% (w/v) pNPP at 50 °C for 30 min. After incubation, the gel was rinsed with distilled water, and then a 0.5 M Na 2 CO 3 solution was added for visualization of yellow zones, indicating substrate hydrolysis.
Native enzyme molecular mass was determined by gel filtration chromatography using a Sephacryl S-200 (Amersham Biosciences) column (100 × 2.5 cm) and 25 mM sodium acetate buffer, pH 5.0, containing 0.5% (w/v) sucrose as a running buffer. Void volume was determined using blue dextran (2 mg/mL), and the protein standards utilized were albumin (Bovine serum, MW 66,000), alcohol dehydrogenase (yeast, MW 150,000), β-amylase (Sweet potato, MW 200,000), carbonic anhydrase (bovine erythrocytes, MW 29,000) and cytochrome C (Horse heart, MW 12,400).
Effect of temperature and pH on enzyme activity
The temperature profile of the purified enzyme was determined within the range of 20 to 80 °C using the standard phytase assay. The pH profile was determined by measuring phytase activity using the following buffers: 0.2 M glycine-HCl (pH 1.0–3.5), 0.2 M sodium acetate (pH 4.0–5.5), 0.2 M MES (pH 6.0–6.5) and 0.2 M Tris-HCl (pH 7.0–8.0).
Enzyme thermal stability and proteolysis resistance
Purified phytase was incubated at 60, 70 and 80 °C for periods of up to 144, 120 and 12 hours, respectively, cooled to 4 °C and assayed in accordance with the phytase assay above.
Phytase resistance to pepsin and trypsin was assessed in accordance with Zhang et al. (2010) with modifications. The purified enzyme (0.2 μg/mL) was incubated in 0.1 M glycine-HCl buffer (pH 2.5) containing pepsin (0.2 μg/mL, pepsin/enzyme ratio of 1:1 (v/v)) or 0.1 M Tris-HCl buffer (pH 8.0) containing trypsin (0.2 μg/mL, trypsin/enzyme ratio of 1:1 (v/v)) for 10, 20, 40 and 60 min at 37 °C. As a control, the purified enzyme was incubated at the same conditions as above but without pepsin or trypsin addition. Residual phytase activity was assessed in accordance with the phytase assay above. The positive control of pepsin and trypsin was performed with azocasein as the substrate, in accordance with Morgavi et al. (2001) , with modifications. For analysis, 50 μL 1% azocasein solution and 50 μL 0.2 μg/mL pepsin in 100 mM glycine-HCl buffer, pH 2.5 or 0.2 μg/mL trypsin in 100 mM Tris-HCl buffer, pH 8.0 were used. The reaction was performed at 37 °C and was stopped after 60 min by adding 100 μL 10% (v/v) trichloroacetic acid solution, and the mixture was centrifuged at 12,000 × g for 10 min. An aliquot of the supernatant was mixed with an equal volume of 500 mM NaOH solution, and the concentration of digested azocasein was determined spectrophotometrically at 440 nm.
Effect of metal ions and others reagents on phytase activity from Aspergillus niger UFV-1
The effects of Mg 2+ , Cd 2+ , Mn 2+ , Cu 2+ , Zn 2+ , Ca 2+ , Hg 2+ , Na + and F − ions at 5, 10 and 20 mM, EDTA, β-mercaptoethanol and DTT at 1 and 5 mM, SDS at 0.1 and 0.5% (v/v) and chaotropic agents (urea and guanidine chloride) at 0.5, 1.0 and 2.0 M on enzyme activity were investigated by incorporating these compounds into the reaction mixtures. Residual phytase activity was then obtained in accordance with the phytase assay above.
Determination of substrate specificity and kinetic parameters
Purified phytase substrate specificity was evaluated by replacing phytic acid in the reaction mixture with other phosphate compounds. The substrates used were sodium phytate, calcium phytate, pNPP, ATP, ADP, AMP, glucose-6-phosphate, glucose-1-phosphate, 1-naphtyl phosphate, 2-naphtyl phosphate, phenyl phosphate and glycerol 2-phosphate at final concentrations of 1.2 mM. Phytase activity was obtained in accordance with the phytase assay above, except for pNPP, in which the reaction mixture was composed of 400 μL 3.0 mM pNPP in 0.2 M glycine-HCl (pH 2.5), 400 μL distilled water and 150 μL purified phytase, which was incubated at 50 °C for 15 min. The reaction was stopped by the addition of 1 mL 0.5 mM Na 2 CO 3 , and the p-nitrophenyl (pNP) concentration was measured at 410 nm.
Kinetic properties were investigated using pNPP as a substrate at 0–95 mM. The kinetic enzyme parameters were investigated using a Lineweaver-Burk plot.
Evaluation of phytate hydrolysis in livestock feed
To evaluate the effect of phytase on phytic acid hydrolysis, a commercial pig feed was used that was composed of corn grain, corn gluten meal, soybean meal, wheat bran, bovine meat and bone meal, dicalcium phosphate, calcium carbonate, sodium chloride, mineral premix and vitamin premix. Autoclaved (121 °C/20 min) feed samples (1 g) were suspended in 5 mL 0.2 M glycine-HCl buffer (pH 2.5) in a 50 mL conical flask and supplemented with crude phytase (4 U/mL). An autoclaved feed suspension without phytase was used as a control. The flasks were then incubated at 50 °C and 200 rpm in a rotary shaker. Samples were withdrawn at intervals up to 8 h (test and control flasks) and immediately cooled to 4 °C and centrifuged (10000 × g , 10 min, at 4 °C), and the supernatant was used for quantification of colorimetrically released phosphate at 700 nm in accordance with the phytase assay above.
Results and Discussion
Enzyme purification
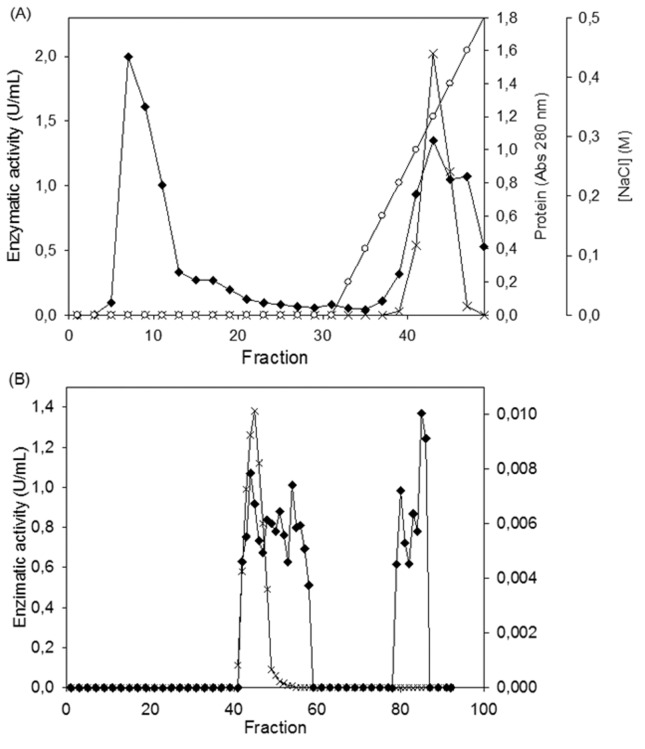
Enzyme purification was achieved by a combination of ultrafiltration followed by acid precipitation and ion exchange and gel filtration chromatography. After the ultrafiltration and precipitation steps, the fractions adsorbed on DEAE-Sepharose demonstrated phytase activity ( Figure 1A ). Subsequently, only 1 peak with phytase activity was obtained when this fraction was submitted to gel filtration chromatography ( Figure 1B ). A purification factor of 64.34 was recorded with 11% yield ( Table 1 ).
Figure 1.
Elution profiles of phytase from Aspergillus niger UFV-1. (A) Ion exchange chromatography of fractions adsorbed on DEAE-Sepharose. The column was initially eluted with sodium acetate buffer (50 mM, pH 5.0) containing 0.5% (w/v) sucrose and subsequently with a linear gradient of 0–0.5 M NaCl in the same buffer. The flow rate was 1.0 mL/min. (B) Gel filtration of fractions displayed phytase activity from DEAE-Sepharose on a Sephacryl S-300 HR column (2.6 × 60 cm), which was eluted with sodium acetate buffer (25 mM, pH 5,0) containing 0.5% (w/v) sucrose. The flow rate was 0.8 mL/min. Protein (-♦-), NaCl gradient (-○-), Phytase activity (-x-).
Table 1.
Summary of the steps for phytase purification from Aspergillus niger UFV-1.
| Purification step | Total protein (mg) | Total activity (U) | Specific activity (U/mg) | Purification (fold) | Yield (%) |
|---|---|---|---|---|---|
| Crude extract | 47.70 | 138.6 | 2.9 | 1 | 100 |
| Ultra-filtration | 41.30 | 129.6 | 3.14 | 1.08 | 93.5 |
| Acid treatment | 15.36 | 103.7 | 6.75 | 2.33 | 74.8 |
| DEAE-Sepharose CL-6B | 0.988 | 24.50 | 24.80 | 8.55 | 17.7 |
| Sephacryl S-300 HR | 0.082 | 15.30 | 186.6 | 64.34 | 11.0 |
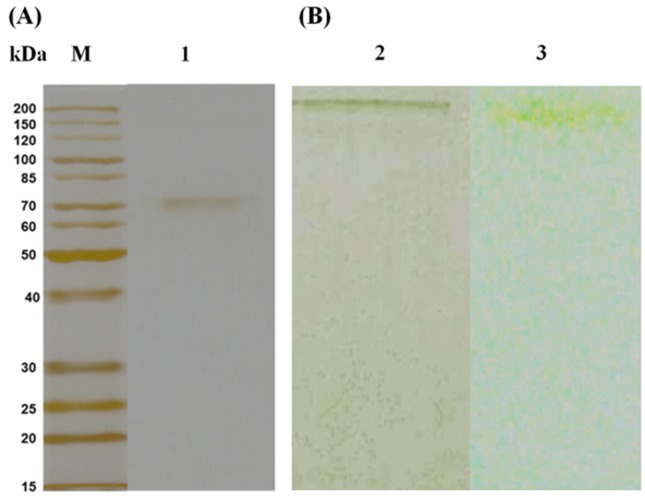
Purification to homogeneity was confirmed by SDS PAGE analysis, and a single band was observed on the non-denaturing electrophoresis gels. Identification of this band as phytase on the non-denaturing gels was confirmed by zymogram analysis, where the band stained for protein and enzymatic activity displayed identical retention factor ( R f ) values ( Figure 2 ). Previous studies on fungal phytase purifications present similar purification factors and yields. The A. niger 11T53A9 phytase was purified 51 times with a 20.3% yield ( Greiner et al. , 2009 ), and Thermomyces lanuginosus TL-7 phytase purification demonstrated a purification factor of 40.75-fold and 3.44% yield ( Gulati et al. , 2007 ).
Figure 2.
SDS-PAGE (A) and zymogram (B) analysis of Aspergillus niger UFV-1 phytase. Lane M: molecular mass marker; Lane 1: Purified phytase; Lane 2: Non-denaturing gel electrophoresis; Lane 3: Phytase activity on a non-denaturing gel.
Molecular mass
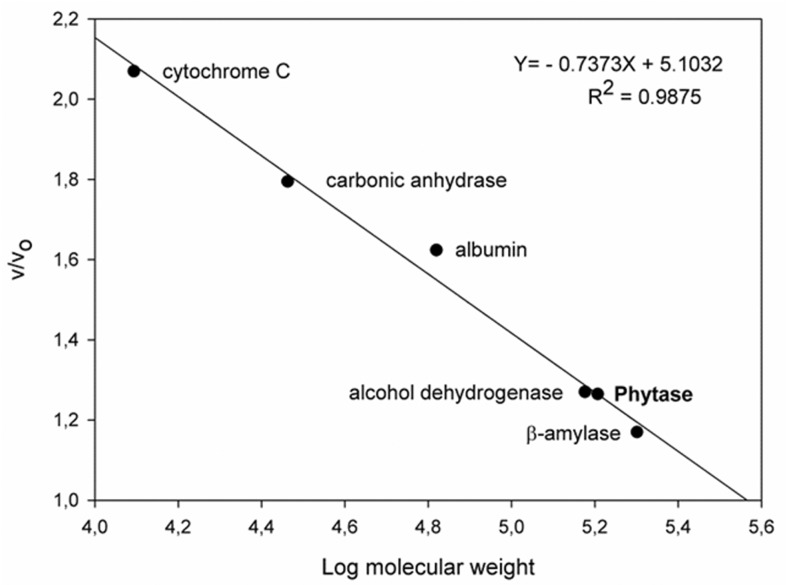
Molecular mass was determined by gel filtration chromatography using a standard curve ( Figure 3 ) and SDS-PAGE. The native phytase molecular mass was determined to be 161 kDa by gel filtration, while SDS-PAGE analysis demonstrated a single band of 81 kDa ( Figure 2A ). A single band of phytase activity was observed on a non-denaturating gel ( Figure 2B ), which suggests that the enzyme could be a dimer, and the molecular mass is in the same range as that observed in previous studies on fungal phytase. The molecular mass of phytase from A. ficuum as determined by gel filtration analysis was 130 kDa, while SDS-PAGE resulted in approximately 68 kDa ( Ullah and Cummins, 1987 ). In another study, the molecular mass of phytase from A. niger van Teighem was demonstrated to be a pentamer of 353 kDa with 66 kDa monomers ( Vats and Banerjee, 2005 ). The molecular mass of A. niger NCIM 563 phytases (I, II, III and IV) as determined by SDS-PAGE and gel filtration were 66, 264; 150, 148; 87, 85 and 120, 120 kDa, respectively, indicating that phytase I consists of four subunits and others are monomers ( Soni et al. , 2010 ; Bhavsar et al. , 2013 ). Bhavsar et al. (2011) identified a phytase from A. niger NCIM 563 at 87 kDa. Spier et al. (2011) studied a 108 kDa phytase from A. niger FS3, while another A. niger 11T53A9 phytase was identified at 85 kDa ( Greiner et al. , 2009 ).
Figure 3.
Native molecular mass determination of purified phytase by Sephacryl S-200 gel filtration chromatography.
Effects of temperature and pH on enzyme activity
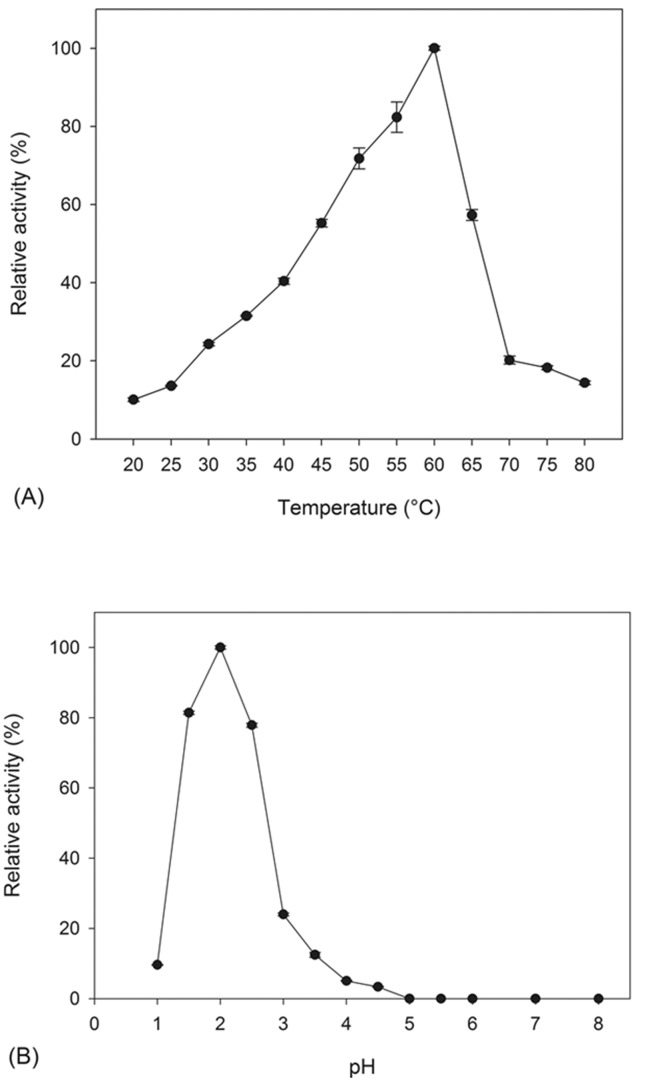
Purified phytase from A. niger UFV-1 demonstrated optimum activity at 60 °C ( Figure 4 A ), similar to other fungal phytases ( Fugthong et al. , 2010 ; Bhavsar et al. , 2013 ), and its optimum pH was 2.0 with a decrease in enzyme activity as the pH approached neutral ( Figure 4 B ).
Figure 4.
Effects of temperature (A) and pH (B) on purified phytase activity from Aspergillus niger UFV-1. Phytase activity was expressed as the mean ± SD (n = 3).
Enzyme thermal stability and proteolysis resistance
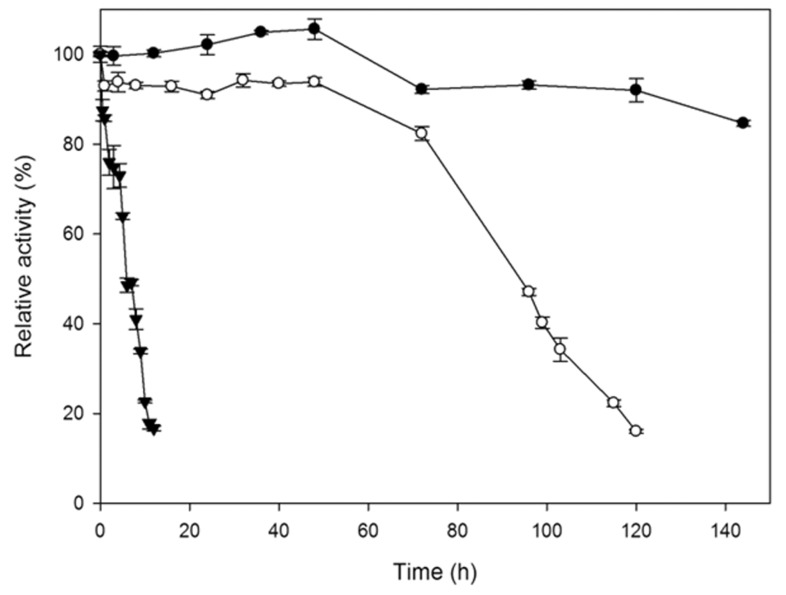
Purified phytase of A. niger UFV-1 retained more than 90% activity at 60 °C during 120 h. At 70 °C, approximately 80% activity was retained after 72 h with a t 1/2 of 94.5 h. The enzyme maintained more than 70% activity at 80 °C after 4 h with t 1/2 of 6.2 h ( Figure 5 ). Thermostability and resistance to proteolysis in the animal digestive tracts are important and useful criteria for industrial phytase application ( Yao et al. , 2011 ). The purified phytase from A. ficuum NTG-23 demonstrated complete activity loss when maintained at 80 °C for 10 min ( Zhang et al. , 2010 ), while the purified phytase from A. niger FS3 maintained 25.1% residual activity after 15 min at 80 °C ( Spier et al. , 2011 ).
Figure 5.
Thermal stability of the purified Aspergillus niger UFV-1 phytase at 60 °C (●), 70 °C (○) and 80 °C (▼). The activity of an unheated phytase sample was defined as 100%. Phytase activity was expressed as the mean ± SD (n = 3).
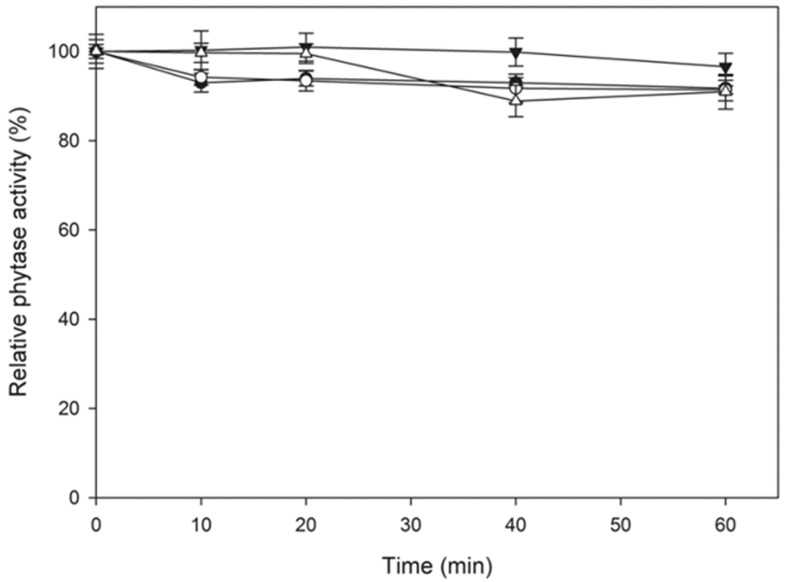
Purified phytase from A. niger UFV-1 demonstrated high stability in the presence of proteolytic enzymes at a 1:1 ratio (pepsin or trypsin/phytase, w/w), while retaining more than 90% residual activity for both proteolytic enzymes ( Figure 6 ). The positive control of pepsin and trypsin demonstrated that the enzymes exhibited proteolytic activity under the experimental conditions (data not shown). The recombinant phytase from Eupenicillium parvum BCC 17694 was expressed in Pichia pastoris and presented low trypsin resistance at a protease/phytase ratio of 1:500 or higher ( Fugthong et al. , 2010 ). The A. fumigatus WY-2 phytase was also evaluated for proteolysis resistance, and it was very sensitive to trypsin at a protease/phytase ratio of 1:200, thus maintaining approximately 30% of its initial activity after 2 h of treatment ( Wang et al. , 2007 ).
Figure 6.
Proteolysis resistance of the purified Aspergillus niger UFV-1 phytase at 37 °C, phytase control, pH 2.5 (●); phytase + pepsin, pH 2.5 (○); phytase control, pH 8.0 (▼) and phytase + trypsin, pH 8.0 (Δ). Phytase activity was expressed as the mean ± SD (n = 3).
Effect of metal ions and other reagents on phytase activity from Aspergillus niger UFV-1
The effects of metal ions and other reagents are shown in Table 2 . At 5 mM, Mg 2+ , Cd 2+ and Ca 2+ ions slightly inhibited phytase activity, while Mn 2+ , Cu 2+ , Zn 2+ , Hg 2+ and Na + had a moderate inhibitory effect. However, the F − ion highly inhibited enzyme activity. Fluoride is a strong competitive inhibitor of several acid bacterial, fungal and plant phytases ( Konietzny and Greiner, 2002 ). Other than the F − ion, the other ions evaluated inhibited enzyme activity at 10 and 20 mM. Metal ions modulate phytase activity, while it is suggested that the inhibitory effect of various metal ions results from the formation of metal ion-phytate complexes that display low solubility. Moreover, most phytases that have been characterized to date are greatly inhibited by Cu 2+ and Zn 2+ ( Konietzny and Greiner, 2002 ). The Sporotrichum thermophile phytase demonstrates 30.0, 12.4, 16.6, 58.2 and 10.5% inhibition when evaluated in the presence of 5 mM Ca 2+ , Mn 2+ , Cu 2+ , Zn 2+ and Na + ions, respectively ( Singh and Satyanarayana, 2009 ). In the presence of 1 mM Zn 2+ , Mg 2+ , Ca 2+ , Mn 2+ , K + and Na + ions, Cladosporium sp. FP-1 phytase was inhibited by 46.0, 45.0, 44.0, 54.0, 39.0 and 34.0%, respectively ( Quan et al. , 2004 ). The phytase from A. niger NCIM 563 was moderately stimulated in the presence of 5 mM Ca 2+ , Fe 2+, Fe 3+ , Ba 2+ and Pb 2+ and was inhibited in the presence of 1 mM Hg 2+ , Ni 2+ , Zn 2+ , Cu 2+ , Ag 2+ , Fe 2+ , Fe 3+ , Pb 2+ and Ca 2+ ( Bhavsar et al. , 2011 ; Bhavsar et al. , 2013 ).
Table 2.
Effect of metal ions and other reagents on phytase activity from Aspergillus niger UFV-1
| Metal ions and others reagents | Relative activity a (%) | ||
|---|---|---|---|
|
|
|||
| Control | 100 | ||
|
|
|||
| 5 mM | 10 mM | 20 mM | |
| MgSO 4 | 95.3 ± 1.2 | 58.1 ± 0.9 | 28.9 ± 1.0 |
| (CH 3 COO) 2 Mg | 100.0 ± 0.6 | 26.6 ± 0.3 | 24.2 ± 1.5 |
| C 4 H 6 CdO 4 | 95.5 ± 0.9 | 59.6 ± 2.5 | 27.3 ± 0.7 |
| MnSO 4 | 74.9 ± 1.5 | 38.3 ± 1.6 | 21.4 ± 1.5 |
| CuSO 4 | 81.2 ± 0.2 | 52.8 ± 1.8 | 18.9 ± 0.5 |
| ZnSO 4 | 87.7 ± 2.7 | 52.2 ± 0.4 | 30.0 ± 1.1 |
| ZnCl 2 | 81.0 ± 2.6 | 60.9 ± 1.1 | 38.0 ± 1.1 |
| CaCl 2 | 96.3 ± 2.5 | 85.7 ± 0.5 | 64.6 ± 2.7 |
| HgCl 2 | 75.6 ± 2.4 | 59.3 ± 1.7 | 49.8 ± 2.6 |
| NaCl | 76.1 ± 1.1 | 68.0 ± 1.8 | 51.4 ± 1.1 |
| KCl | 101.8 ± 1.4 | 98.0 ± 1.3 | 87.5 ± 1.5 |
| Na 2 SO 4 | 76.8 ± 1.2 | 63.2 ± 0.9 | 38.4 ± 1.6 |
| NaF | 0 | 0 | 0 |
| 1 mM | 5 mM | 20 mM | |
| EDTA | 218.7 ± 0.7 | 227.2 ± 2.1 | 234.4 ± 2.5 |
| β-mercaptoethanol | 94.9 ± 1.8 | 64.3 ± 1.6 | |
| DTT | 97.5 ± 1.8 | 81.9 ± 2.0 | |
| 0.5 M | 1 M | 2 M | |
| Urea | 100.3 ± 2.7 | 75.8 ± 1.6 | 63.2 ± 1.8 |
| Guanidine-HCl | 42.0 ± 1.8 | 0 | 0 |
| 0.1% (v/v) | 0.5% (v/v) | ||
| SDS | 0 | 0 | |
Metal ions and other reagent concentrations were calculated as 5 mM, 10 mM and 20 mM of Mg 2+ , Cd 2+ (C 4 H 6 CdO 4 = cadmium acetate), Mn 2+ , Cu 2+ , Zn 2+ , Ca 2+ , Hg 2+ , Na + , K + and F − ; 1 mM, 5 mM and 20 mM EDTA, β-mercaptoethanol and DTT; 0.5 M, 1 M and 2 M Urea, Guanidine-HCl and SDS.
Phytase activity was expressed as the mean ± SD (n = 3).
The purified phytase of A. niger UFV-1 was moderately inhibited by 5 mM of the reducing agents β-mercaptoethanol (35.7% decrease) and DTT (18.1% decrease), which is different from other phytases that were not inhibited by these agents ( Quan et al. , 2004 ; Singh and Satyanarayana, 2009 ). The phytases (I and II) from A. niger NCIM 563 were not significantly affected by DTT and β-mercaptoethanol at 1 mM and 0.1% concentrations, respectively ( Soni et al. , 2010 ). However, the phytase from E. parvum (BCC17694) demonstrated a similar inhibitory effect when evaluated in the presence of β-mercaptoethanol and DTT at 5 mM ( Fugthong et al. , 2010 ). This outcome suggested that the sulfhydryl group may be involved in the catalytic activity of the enzyme. Most phytases have a number of cysteine residues, which may be implicated in disulfide bonds, as described in A. ficuum ( Kostrewa et al. , 1997 ) and Escherichia coli ( Lim et al. , 2000 ). The chelating agent EDTA did not inhibit enzyme activity, suggesting that this enzyme does not require metal ions for its activity. EDTA enhanced phytase activity at 1–20 mM by approximately 130%. This behavior is similar to that of most phytases except the alkaline phytases of Bacillus spp., for example, which are calcium dependent ( Oh et al. , 2004 ). The enzyme activity of A. ficuum NTG-23 phytase was stimulated by EDTA concentrations of 1.25–10 mM, increasing enzyme activity by approximately 10% ( Zhang et al. , 2010 ). A similar result was demonstrated for the phytase from A. niger van Teighem; enzymatic activity was enhanced by approximately 50% with 0.1–2.0 mM EDTA ( Vats and Banerjee, 2005 ). However, this result differs from other studies where the presence of EDTA at 1–5 mM inhibited phytase enzyme activity ( Quan et al. , 2004 ; Gulati et al. , 2007 ; Fugthong et al. , 2010 ). Chaotropic agents such as urea and guanidinium chloride inhibited phytase activity, particularly guanidinium chloride at 1–2 M, which severely inhibited phytase activity. It is suggested that non-covalent forces such as H-bonds and van der Waals interactions play a role in maintaining the active conformation of the enzyme ( Vats and Banerjee, 2005 ; Gulati et al. , 2007 ). The anionic detergent SDS, even at low concentrations, fully inhibited enzymatic activity. The phytase from A. niger van Teighem was also inhibited by SDS as a concentration of 0.1% (v/v), resulting in a 92% loss in enzymatic activity ( Vats and Banerjee, 2005 ). Anionic detergents can bind to proteins and induce structural changes that inhibit enzymatic activity ( Singh and Satyanarayana, 2009 ).
Substrate specificity and kinetic parameters
The purified phytase exhibited broad substrate specificity on a range of phosphorylated compounds ( Table 3 ), presenting activity on pNPP, 2-naphthyl phosphate, 1-naphthyl phosphate, and ATP of more than 3-fold greater enzymatic activity than sodium phytate. In vitro experiments with livestock feed suggest that phytate degrading enzymes with broad substrate specificity are better suited for animal nutrition purposes than phytate-degrading enzymes with narrow substrate specificity ( Wyss et al. , 1999 ; Escobin-Mopera et al. , 2012 ). Fungal phytases demonstrated different substrate specificity. The phytase of A. ficuum NTG-23 displayed a broad substrate specificity, with enzyme activity on ATP, fructose-6-phosphate, and pNPP more than 2-fold higher than sodium phytate ( Zhang et al. , 2010 ). A study of the catalytic properties of fungal phytase demonstrated that A. niger acid phosphatase and A. fumigatus phytase displayed rather broad substrate specificity, while phytases from A. niger , A. terreus CBS, A. terreus 9A1 and E. coli were more specific to phytic acid ( Wyss et al. , 1999 ). The phytase from Rhizopus oligosporus had a rather broad substrate specificity but was more specific to p-NPP than sodium phytate ( Azeke et al. , 2011 ). Similar results were shown for the R. oligosporus ATCC 22959 phytase, which displayed rather broad substrate specificity; however, it was more specific to phytate than other phosphate compounds ( Casey and Walsh, 2004 ).
Table 3.
Substrate specificity of the purified phytase from Aspergillus niger UFV-1.
| Substrate | Relative activity a (%) |
|---|---|
| Sodium phytate | 100.0 ± 1.3 |
| p -NPP | 384.1 ± 2.3 |
| 2-naphthyl phosphate | 377.2 ± 3.4 |
| 1-naphthyl phosphate | 359.1 ± 3.3 |
| ATP | 343.6 ± 3.6 |
| Phenyl phosphate | 272.8 ± 2.5 |
| Glucose 6-phosphate | 189.2 ± 2.4 |
| ADP | 101.7 ± 1.7 |
| Calcium phytate | 97.0 ± 1.2 |
| Glycerol 2-phosphate | 67.3 ± 0.3 |
| Glucose 1-phosphate | 58.2 ± 2.8 |
| AMP | 11.0 ± 0.3 |
Phytase activity was expressed as the mean ± SD (n = 3).
Upon incubation of the purified phytase with various pNPP concentrations (up to 95.0 mM), its kinetic properties were determined from a Lineweaver-Burk plot. The K M and V max values were 30.9 mM and 7.48 μmol/min, respectively. The estimated k cat of the enzyme was 1.46 × 10 5 s −1 , and the catalytic efficient ( k cat / K M ) was 4.7 × 10 6 s −1 .M −1 . Generally, enzymatic hydrolysis presents classical Michaelis-Menten kinetics, whereas relatively low K M values have been reported for phytate-degrading enzymes from A. niger (10–40 μM), A. terreus (11–23 μM), A. fumigatus (< 10 μM), Schwanniomyces castelli (38 μM), Klebsiella aerogenes (62 μM), cattail pollen (17 μM), maize root (24–43 μM), tomato root (38 μM), oat (30 μM), wheat bran (PHY1: 48 μM, PHY2: 77 μM), barley (P1: 72 μM), soybean (48–61 μM), and lupine (L11: 80 μM) ( Konietzny and Greiner, 2002 ; Bohn et al. , 2008 ).
Evaluation of phytate hydrolysis in livestock feed
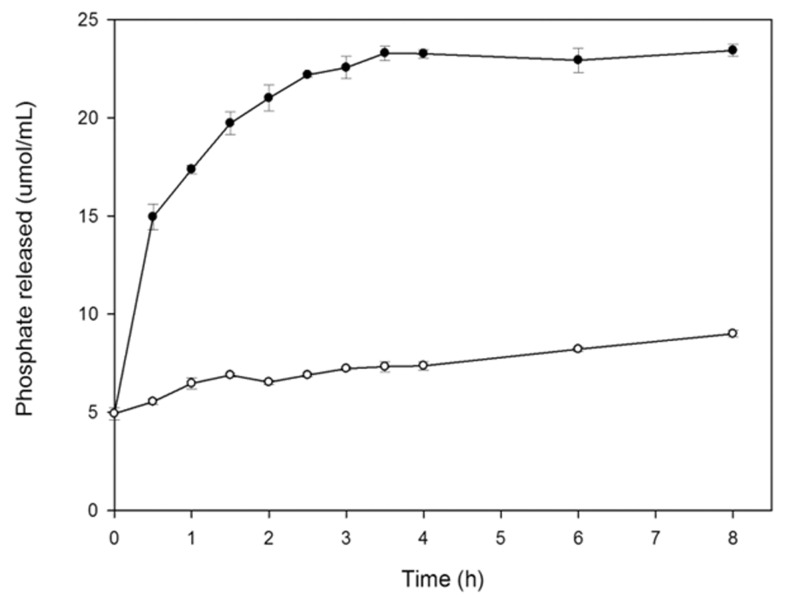
Crude phytase extract from A. niger UFV-1 was able to effectively hydrolyze phytate and other phosphate compounds in the animal feed tested. After 2.5 h, 15.3 μmol phosphate/mL had already been liberated, and this rate was maintained up to 8 h ( Figure 7 ).
Figure 7.
In vitro phosphate liberation from commercial livestock feed at 50 °C, enzymatic hydrolysis (●) and control (○). Phytase activity was expressed as the mean ± SD (n = 3).
Because of the amount of phosphorus released in a short time, this result is very significant. Dephosphorylation of phytic acid in livestock feed using phytase from A. niger van Teighem at 8.4 U/mL demonstrated a maximum liberation of 0.048 μmol Pi/mL at 55 °C after 48 h treatment ( Vats et al. , 2009 ).
Conclusions
Because of the excellent features displayed, this phytase appears to be a promising enzyme for use in animal feed. Its high thermostability suggests that the enzyme is suitable for industrial use because high temperatures are usually encountered in industrial animal food processing. Its high ability to hydrolyze phytate in acidic conditions as well as its high resistance to proteolytic enzymes also suggest that it may effectively release phytic phosphorus in the animal digestive tract.
Acknowledgments
This study was supported by grants from the Fundação de Amparo à Pesquisa do Estado de Minas Gerais - FAPEMIG and Conselho Nacional de Desenvolvimento Científico e Tecnológico - CNPq, Brazil.
References
- Azeke MA, Greiner R, Jany K-D. Purification and characterization of two intracellular phytases from the tempeh fungus Rhizopus oligosporus . J Food Biochem. 2011;35:213–227. [Google Scholar]
- Bhavsar K, Buddhiwant P, Soni SK, et al. Phytase isozymes from Aspergillus niger NCIM 563 under solid state fermentation: Biochemical characterization and their correlation with submerged phytases . Process Biochem. 2013;48:1618–1625. [Google Scholar]
- Bhavsar K, Kumar VR, Khire JM. High level phytase production by Aspergillus niger NCIM 563 in solid state culture: response surface optimization, up-scaling, and its partial characterization . J Ind Microbiol Biotechnol. 2011;38:1407–1417. doi: 10.1007/s10295-010-0926-z. [DOI] [PubMed] [Google Scholar]
- Bohn L, Meyer A, Rasmussen S. Phytate: impact on environment and human nutrition. A challenge for molecular breeding. J Zhejiang Univ Sci B. 2008;9:165–191. doi: 10.1631/jzus.B0710640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey A, Walsh G. Identification and characterization of a phytase of potential commercial interest. J Biotechnol. 2004;110:313–322. doi: 10.1016/j.jbiotec.2004.03.001. [DOI] [PubMed] [Google Scholar]
- Escobin-Mopera L, Ohtani M, Sekiguchi S, et al. Purification and characterization of phytase from Klebsiella pneumoniae 9-3B . J Biosci Bioeng. 2012;113:562–567. doi: 10.1016/j.jbiosc.2011.12.010. [DOI] [PubMed] [Google Scholar]
- Fugthong A, Boonyapakron K, Sornlek W, et al. Biochemical characterization and in vitro digestibility assay of Eupenicillium parvum (BCC17694) phytase expressed in Pichia pastoris . Protein Expr Purif. 2010;70:60–67. doi: 10.1016/j.pep.2009.10.001. [DOI] [PubMed] [Google Scholar]
- Greiner R, Silva LG, Couri S. Purification and characterization of an extracellular phytase from Aspergillus niger 11T53A9 . Braz J Microbiol. 2009;40:795–807. doi: 10.1590/S1517-838220090004000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulati HK, Chadha BS, Saini HS. Production, purification and characterization of thermostable phytase from thermophilic fungus Thermomyces lanuginosus TL-7 . Acta Microbiol Immunol Hung. 2007;54:121–138. doi: 10.1556/AMicr.54.2007.2.3. [DOI] [PubMed] [Google Scholar]
- Gunashree B, Venkateswaran G. Effect of different cultural conditions for phytase production by Aspergillus niger CFR 335 in submerged and solid-state fermentations . J Ind Microbiol Biotechnol. 2008;35:1587–1596. doi: 10.1007/s10295-008-0402-1. [DOI] [PubMed] [Google Scholar]
- Harland BF, Harland J. Fermentative reduction of phytate in rye, white, and whole wheat breads. Cereal Chem. 1980;57:226–229. [Google Scholar]
- Klich MA. Identification of common Aspergillus species . Centraalbureau voor Schimmelcultures; Utrecht, The Netherlands: 2002. [Google Scholar]
- Konietzny U, Greiner R. Molecular and catalytic properties of phytate-degrading enzymes (phytases) Int J Food Sci Technol. 2002;37:791–812. [Google Scholar]
- Kostrewa D, Gruninger-Leitch F, D’Arcy A, et al. Crystal structure of phytase from Aspergillus ficuum at 2.5 Ä resolution . Nat Struct Mol Biol. 1997;4:185–190. doi: 10.1038/nsb0397-185. [DOI] [PubMed] [Google Scholar]
- Lim D, Golovan S, Fosberg CW, et al. Crystal structures of Escherichia coli phytase and its complex with phytate . Nat Struct Mol Biol. 2000;7:108–113. doi: 10.1038/72371. [DOI] [PubMed] [Google Scholar]
- Maguire RO, Dou Z, Sims JT, et al. Dietary strategies for reduced phosphorus excretion and improved water quality. J Environ Qual. 2005;34:2093–2103. doi: 10.2134/jeq2004.0410. [DOI] [PubMed] [Google Scholar]
- Morgavi DP, Beauchemin KA, Nsereko VL, et al. Resistance of feed enzymes to proteolytic inactivation by rumen microorganisms and gastrointestinal proteases. J Anim Sci. 2001;79:1621–1630. doi: 10.2527/2001.7961621x. [DOI] [PubMed] [Google Scholar]
- Oh B, Choi W, Park S, et al. Biochemical properties and structure specificities of alkaline and histidine acid phytases. Appl Microbiol Biotechnol. 2004;63:362–372. doi: 10.1007/s00253-003-1345-0. [DOI] [PubMed] [Google Scholar]
- Quan C, Tian W, Fan S, et al. Purification and properties of a low-molecular-weight phytase from Cladosporium sp. FP-1 . J Biosci Bioen. 2004;97:260–266. doi: 10.1016/S1389-1723(04)70201-7. [DOI] [PubMed] [Google Scholar]
- Samson RA, Houbraken J, Thrane U, et al. Food and indoor fungi. Centraalbureau voor Schimmelcultures; Utrecht, The Netherlands: 2010. [Google Scholar]
- Singh B, Satyanarayana T. Characterization of a HAP-phytase from a thermophilic mould Sporotrichum thermophile . Bioresour Technol. 2009;100:2046–2051. doi: 10.1016/j.biortech.2008.10.025. [DOI] [PubMed] [Google Scholar]
- Soni SK, Magdum A, Khire JM. Purification and characterization of two distinct acidic phytases with broad pH stability from Aspergillus niger NCIM 563 . World J Microbiol Biotechnol. 2010;26:2009–2018. doi: 10.1007/s11274-010-0385-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spier MR, Fendrich RC, Almeida PC, et al. Phytase produced on citric byproducts: purification and characterization. World J Microbiol Biotechnol. 2011;27:267–274. [Google Scholar]
- Ullah AH, Cummins BJ. Purification, N-terminal amino acid sequence and characterization of pH 2.5 optimum acid phosphatase (E.C. 3.1.3.2) from Aspergillus ficuum . Prep Biochem. 1987;17:397–422. doi: 10.1080/00327488708062504. [DOI] [PubMed] [Google Scholar]
- Vats P, Banerjee UC. Biochemical characterization of extracellular phytase (myo-inositol hexakisphosphate phosphohydrolase) from hyper-producing strain of Aspergillus niger van Teighem . J Ind Microbiol Biotechnol. 2005;32:141–147. doi: 10.1007/s10295-005-0214-5. [DOI] [PubMed] [Google Scholar]
- Vats P, Bhushan B, Banerjee UC. Studies on the dephosphorylation of phytic acid in livestock feed using phytase from Aspergillus niger van Teighem . Bioresour Technol. 2009;100:287–291. doi: 10.1016/j.biortech.2008.06.021. [DOI] [PubMed] [Google Scholar]
- Wang Y, Gao X, Su Q, et al. Cloning, expression, and enzyme characterization of an acid heat-stable phytase from Aspergillus fumigatus WY-2 . Curr Microbiol. 2007;55:65–70. doi: 10.1007/s00284-006-0613-5. [DOI] [PubMed] [Google Scholar]
- Wyss M, Brugger R, Kronenberger A, et al. Biochemical Characterization of fungal phytases (myo-inositol hexakisphosphate phosphohydrolases): Catalytic properties. Appl Environ Microbiol. 1999;65:367–373. doi: 10.1128/aem.65.2.367-373.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao M-Z, Zhang Y-H, Lu W-L, et al. Phytases: crystal structures, protein engineering and potential biotechnological applications. J Appl Microbiol. 2011;112:1–14. doi: 10.1111/j.1365-2672.2011.05181.x. [DOI] [PubMed] [Google Scholar]
- Zhang GQ, Dong XF, Wang ZH, et al. Purification, characterization, and cloning of a novel phytase with low pH optimum and strong proteolysis resistance from Aspergillus ficuum NTG-23 . Bioresour Technol. 2010;101:4125–4131. doi: 10.1016/j.biortech.2010.01.001. [DOI] [PubMed] [Google Scholar]