Abstract
Success rates for genomic analyses of highly heterogeneous disorders can be greatly improved if a large cohort of patient data is assembled to enhance collective capabilities for accurate sequence variant annotation, analysis, and interpretation. Indeed, molecular diagnostics requires the establishment of robust data resources to enable data sharing that informs accurate understanding of genes, variants, and phenotypes. The “Mitochondrial Disease Sequence Data Resource (MSeqDR) Consortium” is a grass-roots effort facilitated by the United Mitochondrial Disease Foundation to identify and prioritize specific genomic data analysis needs of the global mitochondrial disease clinical and research community. A central Web portal (https://mseqdr.org) facilitates the coherent compilation, organization, annotation, and analysis of sequence data from both nuclear and mitochondrial genomes of individuals and families with suspected mitochondrial disease. This Web portal provides users with a flexible and expandable suite of resources to enable variant-, gene-, and exome-level sequence analysis in a secure, Web-based, and user-friendly fashion. Users can also elect to share data with other MSeqDR Consortium members, or even the general public, either by custom annotation tracks or through use of a convenient distributed annotation system (DAS) mechanism. A range of data visualization and analysis tools are provided to facilitate user interrogation and understanding of genomic, and ultimately phenotypic, data of relevance to mitochondrial biology and disease. Currently available tools for nuclear and mitochondrial gene analyses include an MSeqDR GBrowse instance that hosts optimized mitochondrial disease and mitochondrial DNA (mtDNA) specific annotation tracks, as well as an MSeqDR locus-specific database (LSDB) that curates variant data on more than 1,300 genes that have been implicated in mitochondrial disease and/or encode mitochondria-localized proteins. MSeqDR is integrated with a diverse array of mtDNA data analysis tools that are both freestanding and incorporated into an online exome-level dataset curation and analysis resource (GEM.app) that is being optimized to support needs of the MSeqDR community. In addition, MSeqDR supports mitochondrial disease phenotyping and ontology tools, and provides variant pathogenicity assessment features that enable community review, feedback, and integration with the public ClinVar variant annotation resource. A centralized Web-based informed consent process is being developed, with implementation of a Global Unique Identifier (GUID) system to integrate data deposited on a given individual from different sources. Community-based data deposition into MSeqDR has already begun. Future efforts will enhance capabilities to incorporate phenotypic data that enhance genomic data analyses. MSeqDR will fill the existing void in bioinformatics tools and centralized knowledge that are necessary to enable efficient nuclear and mtDNA genomic data interpretation by a range of shareholders across both clinical diagnostic and research settings. Ultimately, MSeqDR is focused on empowering the global mitochondrial disease community to better define and explore mitochondrial disease.
INTRODUCTION
Mitochondrial disease is highly heterogeneous in cause and features, where traditional single-gene testing strategies have had limited diagnostic success [1]. Newer genomics technologies enable comprehensive and efficient testing for all known genetic causes in both genomes, which currently include greater than 200 nuclear genes and all 37 genes in the mitochondrial DNA (mtDNA) genome [2]. Indeed, it is now recognized that more than half of patients with suspected mitochondrial disease can be diagnosed in a single test that includes whole exome and mtDNA sequencing [3, 4]. This relatively recent ability to increasingly establish specific genetic etiologies for the complex group of individuals with suspected mitochondrial disease has greatly enhanced our collective understanding of the molecular pathways that drive biochemical dysfunction and multi-organ system disease [1]. However, many novel deleterious variants and disease-related genes have yet to be discovered, as indicated by the fact that thirty to fifty percent of individuals with suspected mitochondrial disorders remain undiagnosed despite use of the latest technological advances. There are also many variants that remain of uncertain clinical significance that have been identified but not yet shared with the larger community, as would likely facilitate more definitive ultimate classification. Furthermore, potential modifying factors that mediate disease severity within mitochondrial disease subtypes remain largely unknown or poorly understood, where collecting, storing, and sharing information from entire datasets rather than only deleterious variants is likely to enable recognition of even small effects of combined variants. Thus, much of the difficulty in better understanding these complex diseases is now largely attributable to the relatively rare frequency of each individual gene disorder and lack of mechanisms to share information on rare and private mutations rather than a lack of robust genomic analysis methodologies. It is also imperative to be able to identify cases that may exist throughout the world to better understand the etiologies and natural history of individual disease subtypes. Such capability would also facilitate the identification of patients throughout the global mitochondrial disease patient community who may be suitable for emerging clinical trials.
MSeqDR Consortium
The Mitochondrial Disease Sequence Data Resource (MSeqDR) Consortium was initiated at the United Mitochondrial Disease Foundation (UMDF) Annual Symposium in June 2012 to create an international source of genomic information in individuals with suspected mitochondrial diseases. Recognizing the vast quantities and mixed types of genomic data being generated in clinical and research labs world-wide, including both large-scale sequencing panels of several to hundreds of nuclear genes, as well as mtDNA, whole exome, and whole genome datasets, MSeqDR participants agreed that even following completion of initial data analysis, each dataset itself remains highly valuable. Firstly, pooled exome datasets inform the frequency of around 22,000 coding variants in each person [5]. If it were possible to link variant data to phenotypic information, pooling these data across patients from all laboratory testing settings and ethnic origins would prove highly valuable to enable more efficient interpretation of allele pathogenicity in the disease population. Such capability might even allow diagnostic identification of additional patients sharing specific alleles and/or additional mutations in a given known or novel disease-related gene that may have been overlooked at the time of initial analysis. Second, collective exome analysis in specific subgroups or among multiple individuals with single gene disorders may reveal variants that modify phenotypes or predict response to specific therapies. Thirdly, making genomic data available in a more organized and collaborative fashion would allow each dataset to „stay alive” and be used for future medical and/or research purposes by each patient's own team or the broader mitochondrial disease community to accelerate and facilitate understanding of mitochondrial disease etiology and mechanism. Finally, making genomic data available enables ready identification of individuals with specific genetic etiologies, modifier variants, and/or pharmacogenomically important variants, for example, which might enable more precise clinical trials to be designed to maximize efficacy and minimize toxicity in the disease population. To fully realize these important opportunities, MSeqDR Consortium participants recognized that development of a common exome-level, and eventually genome-level, sequence data repository and array of readily accessible bioinformatics analysis tools would be essential in order to maximize data utility across the global mitochondrial disease community of clinicians and researchers alike.
The MSeqDR Consortium initially brought together more than 100 expert clinicians, researchers, and bioinformaticians to determine what already existed and what was needed to develop such a community resource for collective genomic data deposition, curation, annotation, and analysis. Facilitated by Web-based conferences organized by the UMDF staff, three general introductory meetings were organized in July and August 2012 to learn about existing genomics resources including NCBI resources, the Human Variome Project and the Locus Specific Database initiative. Subsequently, working groups were established that met by Web-based meetings on three to four occasions throughout the Autumn of 2012. Divided into “Technology and Bioinformatics”, “Phenotyping, Databasing, Institutional Review Board Concerns, Security, and Data Access”, and “Mitochondrial DNA specific concerns”, each working group was co-chaired by multi-disciplinary clinical and research teams to determine the major challenges and community needs within each domain. General MSeqDR Consortium Web meetings held on a monthly basis were used to inform the broader group about the major discussion points and consensus opinions from each working group.
MSeqDR Prototype Development
Based on the outcome of the working group discussions, it was determined that development of a prototype was needed to spearhead the goals of the MSeqDR Consortium. Funded by the UMDF and the North American Mitochondrial Disease Consortium (NAMDC), a bioinformatician, Lishuang Shen, PhD, began to build the MSeqDR prototype in July 2013 under the co-direction of Marni J. Falk, MD, Xiaowu Gai, PhD, and Stephan Zuchner, MD. Monthly prototype development Web meetings were held with up to 75 participants from 7 countries to resolve specific challenges regarding best ways to meet overall consortium goals. Major topics of discussion included bioinformatics development, integration with public and other available genomic resources, and establishment of an MSeqDR portal website and suite of bioinformatics analysis tools. Technical issues addressed included optimization of exome data file handling and file server issues, assessment of the community value versus cost of cloud computing, optimization of a common variant annotation pipeline for data deposition into common visualization and analysis tool(s), and coordination of a comparative analysis project of variant calling and annotation pipelines. Practical usage discussions focused on extraction and linkage of phenotypic data from patient registries including NAMDC to generate a defined ontology for mitochondrial disease that might be readily linked at variable levels of detail to genomic data, optimization of user-friendly Web interfaces that enable users to efficiently analyze nuclear and mtDNA genomic sequence data, and establishment of an effective and streamlined informed consent process to enable individual-level genomic and/or phenotypic data to be deposited into MSeqDR.
MSeqDR Functionality
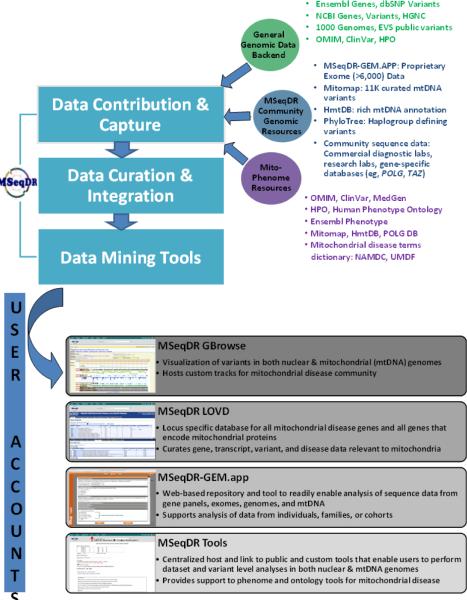
Hosted at a secure Web portal (https://mseqdr.org), MSeqDR currently has a landing page from which can be accessed all MSeqDR informational content and tools (Figure 1). The MSeqDR Web portal facilitates community participation in data deposition, curation, and sharing, while allowing each user to establish data sharing groups and control access to which datasets they wish to share. Registration is required to access all data deposited in MSeqDR, unless otherwise specified by the data owner. Registration is free and open to all academic users. Registration is also open to commercial entities including clinical diagnostic laboratories and pharmaceutical companies, although their use is on a fee-based structure designed to support the ongoing activities of the MSeqDR Consortium.
Figure 1. MSeqDR overview flowchart.
Curated data captured or contributed from a variety of public and mitochondrial disease community genomic and phenotype data resources are bioinformatically integrated to enable end-users to harness in a centralized fashion a variety of online data mining tools that are organized into four major functional domains. All MSeqDR functions can be accessed from a common home page at https://mseqdr.org.
MSeqDR hosts a rich backend of curated genome, exome, and phenotype data. In addition, MSeqDR is organized to support several complementary bioinformatics needs of relevance to the mitochondrial disease community (Figure 1): [1] MSeqDR GBrowse enables integrated visualization and analysis of heterogeneous variation and other genomic data in a classic genome browser interface; [2] MSeqDR-LSDB links the data subsets for mitochondrial diseases, phenotypes, genes, and pathogenic variants, and enables data sharing to external LOVD instances (http://www.LOVD.nl/GeneSymbol) and genome browsers; [3] MSeqDR-GEM.app supports gene panel, exome, genome, and phenotype data archiving as well as phenotype-based mining of patient-, family-, and cohort-level genomic and phenotypic data; and [4] centralized access is provided to a suite of Web-based bioinformatics tools that have been tailored to support genomic and/or phenotypic investigations relevant to mitochondrial function and disease, in addition to generalized genome and phenotype data browsing. A “search box” accessible from the home page enables MSeqDR users to quickly survey all datasets and links to specific domains to pursue on any gene or variant of interest. Details of these major features are provided below.
MSeqDR GBrowse
MSeqDR GBrowse (https://mseqdr.org/gbrowse_bridge.php) is a human mitochondrial-disease focused instance of a genome browser to readily enable bioinformatics analyses of genomic data via a user-friendly graphical interface. Users can select specific „tracks” of data to readily show, hide, re-order or zoom from chromosome to single basepair level resolution, which conveniently allows interpretation of genetic variants in specific genomic contexts and for any specific genomic region of interest. Key standard reference gene and variant annotation tracks are available to enable users to visualize all variant-level data in public exome and genome datasets, including those in the National Heart Lung and Blood Institute (NHLBI) exome variant server (“EVS Exome Variants” track) and 1000 Genomes Project (“1000 Genomes” and “1000 Genomes Whole Exome Sequencing Mitochondrial Variants” tracks [6, 7].
MSeqDR further aims to host custom tracks designed by and for the mitochondrial disease community. mtDNA specific open source tracks that are currently available in MSeqDR GBrowse include HmtDB [8], Mitomap [9], PhyloTree [10], and the MitoBreak database [11], with individual functionality of these resources explained below. Individual tracks are also available to display all variants in a given exome (“MSeqDR Exome Variants” track), pathogenicity predictions of all possible non-synonymous mitochondrial variants (“Mitochondrial Pathogenicity Predictions” track) and annotations of all known mtDNA variations reported to date and derived from Mitomap (“Mitomap Variants” and “Mitomap Variation Disease” tracks) and HmtDB (“HmtDB rCRS Variants” and “HmtDB RSRS Variants” tracks). In addition, custom tracks are available to improve analysis of relevant nuclear genome regions, including tracks to indicate whether a given region harbors a NumtS (Nuclear mitochondrial sequence) generated by translocation of mitochondrial genome fragments into the nuclear genome (“Human NumtS” track)[12, 13], whether a gene product is known to localize to mitochondria or cause a mitochondrial disease, and detail which commercial diagnostic laboratory tests analyze a given gene (“Mitochondria Localized/Disease Candidate Genes” track). Individual tracks are also available on specific highly studied and important mitochondrial disease-related genes or loci, such as the Human DNA POLG variants that are curated at NIEHS (http://tools.niehs.nih.gov/polg) [14] and the TAZ Mutation Database that is curated by Dr. Iris Gonzalez at the Barth Syndrome Foundation (http://www.barthsyndrome.org/science--medicine/human-tafazzin-taz-gene-mutation--variation-database), with direct links provided to the originating website to enable users to readily obtain additional levels of information.
Similarly, a given laboratory or researcher can show the aggregate of all sequence variants within a given gene, gene panel, or exome obtained within their group, where MSeqDR users can then contact that group for additional information about a specific variant or individual of interest to them within that cohort. A Distributed Annotation System (DAS) protocol is supported to facilitate sharing of aggregate data even further, such that the original data files may remain remotely with the laboratory of origin but enable data sharing at various investigator and institutional levels of comfort. Indeed, sharing of all custom tracks remains at the discretion of the depositing laboratory or researcher, such that they can choose to share the data with only their group, registered MSeqDR users, or the public and change that decision at any time. For example, the publication criteria for some journals requiring data deposition of gene set or exome level data into a public repository may be satisfied by depositing that data as a track in MSeqDR. Finally, MSeqDR will continue to work with public resources such as NCBI dbGaP and ClinVar to coordinate large-scale genomic data transfer and two-way communication of variant annotations relevant to mitochondrial biology and disease.
MSeqDR LOVD
The MSeqDR Locus Specific Database is an instance of the Leiden Open Variant Database, which is developed by Leiden University Medical Center (http://www.lovd.nl/3.0/home) and has been employed for pathogenic variant databases of many human diseases [15]. The MSeqDR instance is unique in supporting mitochondrial genome data entries and has been optimized to support curation of more than 1,300 nuclear and mtDNA genes relevant to mitochondrial biology and disease. Indeed, the MSeqDR LOVD database provides continually updated information for these genes at the level of each gene, transcript, variant, phenotype, and disease. Data is accessible in table format, with embedded links to connect users to all available information on each variant. An MSeqDR user blog-like feedback function enables users to comment on the putative pathogenicity of specific variants, and clinically relevant variant data from ClinVar is visible when available. A broader goal of the MSeqDR Consortium over time is to review and refine the predicted pathogenicity of all variants in mitochondrial disease-related genes through a community-based effort, collect variant annotations in a format that conforms to ClinVar specifications, and serve as a mitochondrial domain resource by regular communication of these data with the administrators of the NCBI ClinVar database.
MSeqDR-GEM.app
Datasets of gene panels, mtDNA, exomes, and even genomes that are deposited by FTP server to MSeqDR will be uploaded to an MSeqDR-optimized secure instance of Genome Management Application (GEM.app, https://genomics.med.miami.edu) [16] that already hosts more than 4,500 exomes and genomes from individuals and families with neurologic diseases. The general utility of this data resource is to function both as a data archive and enable registered users to readily explore genomic variant data from patients and families having specific clinical phenotypes. Users are provided an updated log of all datasets they have deposited or that have been shared with them, with detailed information provided about data capture and analysis methods as well as data quality. Users control access to each of their datasets, and can readily share or revoke specific analyses, datasets, or groups of data at their discretion. All data is de-identified and assigned a „global unique identifier” (GUID, http://ndar.nih.gov/ndarpublicweb/tools.html#GUID), which is associated with the data at the time of submission by each user. GEM.app enables users to interrogate their own submitted datasets to query genes or variants that meet a pre-defined or custom-set filter within individuals, families, or cohorts in a secure and user-friendly Web interface. Pedigrees can be stored and displayed if submitted by the user. All query output provides extensive variant annotations including genomic and protein coordinates and links to public resources, allele frequencies in both the general public and disease communities, multiple pathogenicity prediction and conservation scores, and data quality parameters within the datasets queried. All annotation tracks in MSeqDR for both nuclear and mtDNA genes are also viewable at the level of individual variants in a specific genomic region being analyzed within GEM.app. Thus, clinicians, laboratorians, and researchers who have distinct hypotheses can readily visualize all potential rare variants that fit different disease models within their patient or family to fully assess all levels of information necessary to prioritize a candidate variant as potentially supportive of disease association. Users have the option to make their data “discoverable” at the individual variant level, thereby enabling other users to contact them to discuss a specific variant of interest that may facilitate collaborations that will eventually lead to the elucidation of shared etiologies or cases in different cohorts.
For purposes of MSeqDR-deposited sequence data, all reads from a sequenced individual's gene panel, exome, or genome are reannotated using a custom variant annotation pipeline that is rooted in Ensembl (http://www.ensembl.org/index.html) to define gene coordinates and then deposited both for gene and variant analysis in GEM.app as well as for variant visualization throughout a specific exome in MSeqDR GBrowse. To prevent potential variant overrepresentation from a given individual's data being submitted in the form of more than one test type or sequence data depositor, all data deposited into MSeqDR for a given individual will be linked to the same GUID throughout the data resource. GEM.app specific features also enable users to search their genomic datasets for variants within subcategories such as specific gene ontology groupings, pharmacogenomic variant sets, or known mitochondrial disease-related genes. Future efforts will build improved phenotypic data annotation of individuals with suspected mitochondrial disease on whom genomic data is available in MSeqDR-GEM.app.
Other MSeqDR genome analysis tools
The MSeqDR Consortium participants recognize that users may need to access different bioinformatics tools at different times and for different purposes. Thus, MSeqDR provides access to a wide range of Web-based bioinformatics tools and databases (Table 1), which their developers have agreed to share to enable genomic analysis of both the nuclear genome and mtDNA. For example, MSeqDR users can access the human basepair codon resource (HBCR), which is a Web-based interface to a custom pipeline developed by Dr. Xiaowu Gai for real-time Ensembl-based annotation of exome datasets. Access is also provided to multiple mtDNA analysis tools that enable users to perform sequence annotation and analysis, haplogroup determination, and heteroplasmy quantitation in their own datasets. These include Mitomap that shares a manually-curated database of all mtDNA variants [9]; Mitomaster to perform mtDNA sequence analysis [9]; MToolBox [17], an automated pipeline that enables genome assembly of mtDNA from next generation sequencing data with variant calling, annotation and prioritization, heteroplasmy detection, as well as haplogroup prediction by using custom developed algorithms and information extracted from publicly available mtDNA datasets hosted in the HmtDB database [8, 17]; MT.AT that enables mtDNA sequence files to be readily annotated with prioritization of disease variants using custom algorithms developed by Alphons Stassen; PhyloTree (http://www.phylotree.org) that reports the complete classification of mtDNA haplogroups [10]; Phy-mer that enables mtDNA haplogroup analysis in an alignment-free and reference-independent fashion using a custom algorithm developed by Drs. Daniel Navarro-Gomez, Xiaowu Gai and Mannis van Oven; as well as MitoBreak that provides a database of all reported mtDNA breakpoints [11]. MSeqDR also hosts the Transcriptome of Mitochondrial Dysfunction (ToMD), which provides users a means to assess gene expression data stored in a commonly reannotated repository of genome-wide transcriptome data from a variety of cell, animal, and human tissue studies in mitochondrial disease [18]. Further information about the capabilities and use of each program are provided both on the MSeqDR website. The MSeqDR Consortium is hopeful that additional tools will be made available as they emerge to the greater mitochondrial disease community through central access at the MSeqDR portal. In addition, custom tools were developed by Dr. Lishuang Shen and hosted within MSeqDR that map all data elements currently used by the North American Mitochondrial Disease Consortium (NAMDC) to assess phenotypic symptoms and signs in patients with suspected mitochondrial disease to corresponding Human Phenotype Ontology (HPO) terms [19]. When ultimately linked to genomic data from the same individual using a GUID system, phenotypic data described through HPO will improve the ability to determine whether specific genetic variants are the cause for specific cases or subtypes of mitochondrial disease. Future efforts will build improved phenotypic data annotation of individuals with suspected mitochondrial disease on whom genomic data is available to enable data discovery at the global level [20] in the MSeqDR environment.
Table 1.
Web-based bioinformatics tools and databases related to mitochondrial biology, variation, and disease that are accessible from MSeqDR.
| Tool | Link/Reference | Description |
|---|---|---|
| GEM.APP | https://genomics.med.miami.edu/gem-app/ [16] | Explore variation data from patients and families with defined diseases. Search and filter by phenotypes and inheritance patterns |
| HmtDB | http://www.hmtdb.uniba.it/hmdb/ [8] | A human mitochondrial genomic resource based on variability studies to support population genetics and biomedical research |
| HBCR | https://mseqdr.org/HBCR.php | Human basepair codon resource: annotation of exome variant datasets, in command line or Web-based |
| MITOMAP & MITO Master | http://mitomap.org/ [9] | A compendium of over 10,000 mainly manually curated polymorphisms in human mtDNA in MITOMAP, and mtDNA variation analysis tools in MITO Master |
| MToolBox | https://mseqdr.Org/mtoolbox.php [17] | Mitochondrial genome assembly, haplogroup assignment, variant prioritization, and heteroplasmy analysis from sequencing data |
| MT.AT | https://mseqdr.org/mt_at.php | Annotates mtDNA sequences with prioritization of disease variants |
| MitoBreak | http://mitobreak.portugene.com/ [11] | A database of all reported mtDNA rearrangement breakpoints |
| NAMDC | http://www.rarediseasesnetwork.org/namdc/ | Maintains a contact registry and tissue biorepository for patients with mitochondrial disorders, and developing a diagnostic and phenotyping dictionary for human mitochondrial disease |
| PhyloTree | http://www.phylotree.org/ [10] | Provides the complete classification of mtDNA haplogroups and a comprehensive phylogenetic tree of global human mitochondrial DNA variations |
| Phy-mer | https://github.com/danielnavarrogomez/phy-mer | Tool for mtDNA haplogroup analysis in an alignment-free and reference-independent fashion |
| ToMD | https://mseqdr.org/data/tomd/ [18] | An online platform in which to access data and interrogate gene expression results from transcriptome studies of mitochondrial dysfunction and disease |
Supporting individual patient preferences for genomic data analysis and sharing
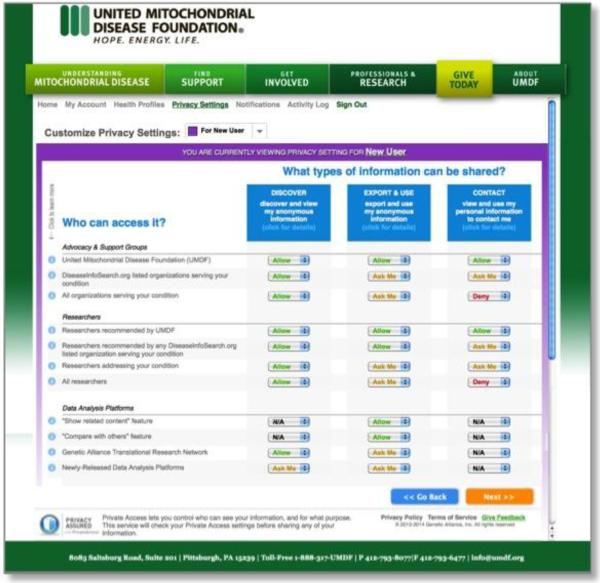
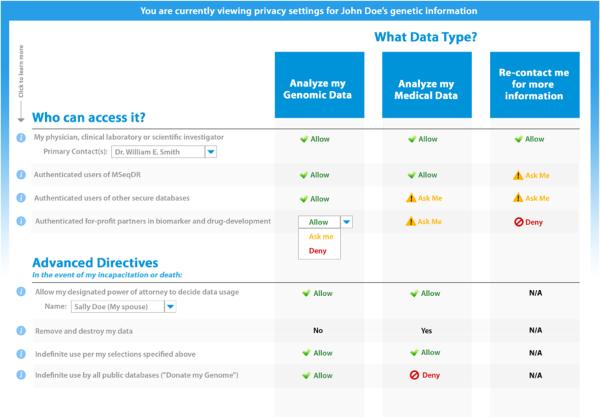
MSeqDR is integrally connected to the mitochondrial disease patient community, as it is a global effort to compile genomic, and ultimately phenotype, information on individual patients under the stewardship of the United Mitochondrial Disease Foundation (www.umdf.org). MSeqDR aims to support individual patient autonomy in selecting their own privacy settings for analyses and sharing of their genomic and/or phenotypic data with clinical caregivers, enrolled research studies, the larger MSeqDR community, or even the broader research community (http://gds.nih.gov). A similar platform has recently been implemented by the UMDF for enabling patient control of their medical data, as enacted using custom privacy settings that are selected on a Web-based platform designed by Private Access [21] and shown in Figure 2a. To address the inherent challenges in communicating the unique nature of genomic data to the patient community and readily implement their choices of how their data should be used over time, MSeqDR partnered with the Genetic Alliance (http://www.geneticalliance.org) to develop custom privacy settings specific for individual-level genomic data that will be deposited in a deidentified fashion within MSeqDR (Figure 2b). Future work will aim to obtain centralized Institutional Review Board approval for this patient consent platform. The MSeqDR Consortium will also focus on translating the Web-based patient consent process into different languages and incorporating flexibility in the specific privacy setting options offered to respect different cultural norms, as will be necessary to enable MSeqDR to become a truly global repository and resource for patient-determined genomic data utilization throughout the international mitochondrial disease community.
Figure 2. Individual patient privacy settings.
Privacy settings are used in a Web-based environment to determine patient preferences for sharing [A] Medical data entered into the Mitochondrial Disease Patient Registry, which went live in September 2014 at http://umdf.org, and [B] Deidentified genomic data deposited and analyzed within MSeqDR, which is now under active development.
Data security and access
The MSeqDR Consortium considers data security and privacy protections as critical elements in the design and utilization of MSeqDR. A data access and use oversight committee with membership comprised of mitochondrial disease physicians, researchers, bioinformaticians, diagnostic lab directors, advocacy group, and patient representation will collectively determine data utilization and download access rights. Respecting the unique perspectives of different countries, membership will be sought for at least one member of each participating nation in MSeqDR. The MSeqDR portal itself is currently hosted from a secure server at Massachusetts Eye and Ear Infirmary (MEEI) at Harvard University, whereas GEM.app is hosted in a cloud computing environment. Future focus will consider moving all MSeqDR data and computation to a cloud environment to balance cost and security concerns as they are likely to change over time.
Getting started
MSeqDR user needs will vary depending on multiple factors: [a] type of analysis, [b] genomic region of interest, [c] level of bioinformatics needs, and [d] security concerns and comfort levels. As to the type of analysis, variant data can be analyzed at individual, family, and/or cohort levels. The region of interest can be a gene, other specific region, or across the mitochondrial or nuclear genomes. The level of bioinformatics needs varies greatly among users, where some might have variants of interest previously detected using other tools that they desire to interpret in the context of MSeqDR GBrowse or LOVD annotations, while others have raw sequence data that they would like to analyze from scratch. Lastly, whereas sharing aggregate variant data of all output from a given laboratory's gene, panel, or exome test will benefit delineation of allele frequencies in the broader community and is generally associated with less security concern, sharing individual-level variant data without patient informed consent would not be permissible. MSeqDR tries to address these varied needs by providing a variety of different data input and sharing mechanisms.
A Web-based general tutorial demonstration of how to get started and navigate through the main features of MSeqDR is available at https://mseqdr.org/tutorial.php. All MSeqDR users should register for a login ID and password at https://mseqdr.org. Logging in enables users to deposit data, share settings and searches, participate in a user-based blog to comment on variant data, and access all data available throughout the resource. For users who are interested in depositing and analyzing individual-, family-, or cohort-level variant data across the genome, they can upload their data to MSeqDR in either the standard VCF format if variant calls have been made, or in the raw-data FASTQ format. The MSeqDR Consortium supports use of VCF format for mtDNA data regardless of originating sequencing methodology, with a generic template of this suggested format provided as Table 2. Users should contact the MSeqDR administrator, Dr. Lishuang Shen at Lishuang_Shen@meei.harvard.edu, to create an FTP account for data transfer on the MSeqDR secure FTP server. The FTP server is currently hosted using ExpeDat on a server at the Massachusetts Eye and Ear Infirmary (MEEI), but may be migrated to cloud-based computing depending on future community needs. Deposited data will be made available for investigator-level nuclear or mtDNA genomic analysis through GEM.app (which uses a standard BWA-GATK variant calling and annotation pipeline), and also as individual custom annotation tracks on MSeqDR GBrowse (which annotates variation files using the custom HBCR pipeline). If a user is interested in sharing aggregate variant data of a cohort across the genome or for a specific gene or region, such as the entire mtDNA or POLG gene, they should also contact the MSeqDR administrator or organizers to identify a data sharing method that is most convenient for the user; appropriate credits will be given in MSeqDR to all data providers, as well as contact information to facilitate potential collaborations. If a user is interested in simply visualizing and interpreting variant data for a specific region or across the genome, they can directly upload their data as custom track(s) through the MSeqDR GBrowse website using any of several commonly used formats including BED, GFF, GFF3, WIG, and BAM. Such custom tracks are only accessible to the specific user, or those whom the user specifically invites to share, and can be removed anytime per user discretion.
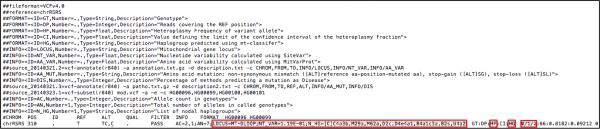
Table 2. Generic VCF template for mtDNA.
Variant Call Format (VCF v.4.0) file template for mtDNA, as supported by MSeqDR Consortium consensus. Red boxes highlight recommendations of where to place certain features specific to mtDNA. Alleles may be placed in the GT field of the VCF file to display complex variants. The VCF file header should also specify the mtDNA reference genome used (i.e., either rCRS or RSRS). This format is recommended to upload mtDNA sequences generated by any sequencing methodology or analysis tool. To best inform future analyses that may use data generated by different platforms and methodologies, the MSeqDR Consortium recommends, but does not require, submission of calls at all nucleotide positions in the mtDNA rather than only variant calls.
|
CONCLUSIONS
The MSeqDR Consortium is an international mitochondrial disease community collaborative effort to create a unified genomic data resource that facilitates accurate diagnosis and enables improved understanding of individual mitochondrial diseases. MSeqDR at https://mseqdr.org offers a centralized entry portal for clinicians, diagnostic labs, and researchers, enabling genomic data sharing and analyses in suspected mitochondrial disease. MSeqDR provides a flexible, updated suite of Web-based and open access software tools accessible from a user's office or clinic personal computer to securely interrogate all genetic variant data or entire exome datasets in real-time. This approach exploits collective information of variant allele frequencies in the general public as well as a large cohort of individuals with suspected mitochondrial disease. The MSeqDR Consortium believes that providing common data resources and tools to the mitochondrial biology and disease community will accelerate pace and accuracy of diagnosing both known and novel genes underlying mitochondrial diseases. MSeqDR also enables genomic data deposition for community archiving, sharing of aggregate level data to individual investigator or institutional comfort levels, and supports individual patient-determined autonomy in selecting their own privacy settings for genomic data analysis and sharing within their clinical caregivers, enrolled research studies, or larger MSeqDR community. Future work will focus on enabling linkage of relevant phenotype and laboratory data to genomic data at various analysis levels, from individuals to specific phenotypes or larger mitochondrial disease cohorts.
HIGHLIGHTS.
MSeqDR is a genomic data repository and analysis resource for mitochondrial disease
MSeqDR GBrowse enhances interrogation of both genomes with custom annotation tracks
MSeqDR LSDB curates variation in >1,300 genes relevant to mitochondrial biology
MSeqDR GEM.app enables Web-based exome or genome dataset curation and analysis
MSeqDR hosts bioinformatics & phenotyping tools for mitochondrial disease analyses
ACKNOWLEDGMENTS
We are grateful to the outstanding leadership and staff of the United Mitochondrial Disease Foundation, including Chuck Mohan, Dan Wright, Cliff Gorski, and Janet Owens for their tireless efforts to organize and provide ongoing financial support for the MSeqDR Consortium activities. This work was also supported in part by the National Institutes of Health (U54-NS078059 - North American Mitochondrial Disease Consortium pilot award #NAMDC7407 to MJF and XG; and U41-HG006834) and the Netherlands Genomic Initiative (NGI)/Netherlands Organization for Scientific Research (NWO) within the framework of the Forensic Genomics Consortium Netherlands (FGCN) to MvO. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
MSeqDR Consortium Participants: Sherri Bale, Jirair Bedoyan, Doron Behar, Penelope Bonnen, Lisa Brooks, Claudia Calabrese, Sarah Calvo, Patrick Chinnery, John Christodoulou, Deanna Church, Rosanna Clima, Bruce Cohen, Richard G. Cotton, IFM de Coo, Olga Derbenevoa, Johan T. den Dunnen, David Dimmock, Gregory Enns, Giuseppe Gasparre, Amy Goldstein, Katrina Gwinn, Sihoun Hahn, Richard Haas, Hakon Hakonarson, Michio Hirano, Douglas Kerr, Dong Li, Maria Lvova, Finley Macrae, Donna Maglott, Elizabeth McCormick, Grant Mitchell, Vamsi Mootha, Iris Gonzalez, Yasushi Okazaki, Aurora Pajul, Melissa Parisi, Juan Carlos Perin, Eric Pierce, Vincent Procaccio, Shamima Rahman, Honey Reddi, Heidi Rehm, Erin Riggs, Richard Rodenburg, Yaffa Rubinstein, Russell Saneto, Mariangela Santorsola, Curt Scharfe, Claire Sheldon, Eric Shoubridge, Domenico Simone, Bert Smeets, Jan Smeitink, Christine Stanley, Anu Suomalainen-Waartiovaara, Mark Tarnopolsky, Isabelle Thiffault, David Thorburn, Johan Van Hove, Lynne Wolfe, Lee-Jun Wong
REFERENCES
- 1.McCormick E, Place E, Falk MJ. Molecular genetic testing for mitochondrial disease: from one generation to the next Neurotherapeutics : the journal of the American Society for Experimental. NeuroTherapeutics. 2013;10:251–261. doi: 10.1007/s13311-012-0174-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koopman WJ, Willems PH, Smeitink JA. Monogenic mitochondrial disorders. The New England journal of medicine. 2012;366:1132–1141. doi: 10.1056/NEJMra1012478. [DOI] [PubMed] [Google Scholar]
- 3.Taylor RW, Pyle A, Griffin H, Blakely EL, Duff J, He L, Smertenko T, Alston CL, Neeve VC, Best A, Yarham JW, Kirschner J, Schara U, Talim B, Topaloglu H, Baric I, Holinski-Feder E, Abicht A, Czermin B, Kleinle S, Morris AA, Vassallo G, Gorman GS, Ramesh V, Turnbull DM, Santibanez-Koref M, McFarland R, Horvath R, Chinnery PF. Use of whole-exome sequencing to determine the genetic basis of multiple mitochondrial respiratory chain complex deficiencies. JAMA : the journal of the American Medical Association. 2014;312:68–77. doi: 10.1001/jama.2014.7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lieber DS, Calvo SE, Shanahan K, Slate NG, Liu S, Hershman SG, Gold NB, Chapman BA, Thorburn DR, Berry GT, Schmahmann JD, Borowsky ML, Mueller DM, Sims KB, Mootha VK. Targeted exome sequencing of suspected mitochondrial disorders. Neurology. 2013;80:1762–1770. doi: 10.1212/WNL.0b013e3182918c40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, Lee C, Shaffer T, Wong M, Bhattacharjee A, Eichler EE, Bamshad M, Nickerson DA, Shendure J. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Genomes Project C, Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diroma MA, Calabrese C, Simone D, Santorsola M, Calabrese FM, Gasparre G, Attimonelli M. Extraction and annotation of human mitochondrial genomes from 1000 Genomes Whole Exome Sequencing data. BMC genomics. 2014;15(Suppl 3):S2. doi: 10.1186/1471-2164-15-S3-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rubino F, Piredda R, Calabrese FM, Simone D, Lang M, Calabrese C, Petruzzella V, Tommaseo-Ponzetta M, Gasparre G, Attimonelli M. HmtDB, a genomic resource for mitochondrion-based human variability studies. Nucleic acids research. 2012;40:D1150–1159. doi: 10.1093/nar/gkr1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lott MT, Leipzig JN, Derbeneva O, Xie HM, Chalkia D, Sarmady M, Procaccio V, Wallace DC. mtDNA variation and analysis using MITOMAP and MITOMASTER. Current Protocols in Bioinformatics. 2013;44:1.23.21–21.23.26. doi: 10.1002/0471250953.bi0123s44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Oven M, Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Human mutation. 2009;30:E386–394. doi: 10.1002/humu.20921. [DOI] [PubMed] [Google Scholar]
- 11.Damas J, Carneiro J, Amorim A, Pereira F. MitoBreak: the mitochondrial DNA breakpoints database. Nucleic acids research. 2014;42:D1261–1268. doi: 10.1093/nar/gkt982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lascaro D, Castellana S, Gasparre G, Romeo G, Saccone C, Attimonelli M. The RHNumtS compilation: features and bioinformatics approaches to locate and quantify. Human NumtS BMC genomics. 2008;9:267. doi: 10.1186/1471-2164-9-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simone D, Calabrese FM, Lang M, Gasparre G, Attimonelli M. The reference human nuclear mitochondrial sequences compilation validated and implemented on the UCSC genome browser. BMC genomics. 2011;12:517. doi: 10.1186/1471-2164-12-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Longley MJ, Graziewicz MA, Bienstock RJ, Copeland WC. Consequences of mutations in human DNA polymerase gamma. Gene. 2005;354:125–131. doi: 10.1016/j.gene.2005.03.029. [DOI] [PubMed] [Google Scholar]
- 15.Fokkema IF, Taschner PE, Schaafsma GC, Celli J, Laros JF, den Dunnen JT. LOVD v.2.0: the next generation in gene variant databases. Human mutation. 2011;32:557–563. doi: 10.1002/humu.21438. [DOI] [PubMed] [Google Scholar]
- 16.Gonzalez MA, Lebrigio RF, Van Booven D, Ulloa RH, Powell E, Speziani F, Tekin M, Schule R, Zuchner S. GEnomes Management Application (GEM.app): a new software tool for large-scale collaborative genome analysis. Human mutation. 2013;34:842–846. doi: 10.1002/humu.22305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Calabrese C, Simone D, Diroma MA, Santorsola M, Gutta C, Gasparre G, Picardi E, Pesole G, Attimonelli M. MToolBox: a highly automated pipeline for heteroplasmy annotation and prioritization analysis of human mitochondrial variants in high-throughput sequencing. Bioinformatics. 2014 doi: 10.1093/bioinformatics/btu483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Z, Falk MJ. Integrated transcriptome analysis across mitochondrial disease etiologies and tissues improves understanding of common cellular adaptations to respiratory chain dysfunction. The international journal of biochemistry & cell biology. 2014;50:106–111. doi: 10.1016/j.biocel.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kohler S, Doelken SC, Mungall CJ, Bauer S, Firth HV, Bailleul-Forestier I, Black GC, Brown DL, Brudno M, Campbell J, FitzPatrick DR, Eppig JT, Jackson AP, Freson K, Girdea M, Helbig I, Hurst JA, Jahn J, Jackson LG, Kelly AM, Ledbetter DH, Mansour S, Martin CL, Moss C, Mumford A, Ouwehand WH, Park SM, Riggs ER, Scott RH, Sisodiya S, Van Vooren S, Wapner RJ, Wilkie AO, Wright CF, Vulto-van Silfhout AT, de Leeuw N, de Vries BB, Washingthon NL, Smith CL, Westerfield M, Schofield P, Ruef BJ, Gkoutos GV, Haendel M, Smedley D, Lewis SE, Robinson PN. The Human Phenotype Ontology project: linking molecular biology and disease through phenotype data. Nucleic acids research. 2014;42:D966–974. doi: 10.1093/nar/gkt1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masino AJ, Dechene ET, Dulik MC, Wilkens A, Spinner NB, Krantz ID, Pennington JW, Robinson PN, White PS. Clinical phenotype-based gene prioritization: an initial study using semantic similarity and the human phenotype ontology. BMC bioinformatics. 2014;15:248. doi: 10.1186/1471-2105-15-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shelton RH. Electronic consent channels: preserving patient privacy without handcuffing researchers. Science translational medicine. 2011;3:69cm64. doi: 10.1126/scitranslmed.3002037. [DOI] [PubMed] [Google Scholar]