Abstract
Genome-wide expression and genotyping technologies have uncovered the genetic bases of complex diseases at unprecedented rates. However, despite its heavy burden and high prevalence, the molecular characterization of major depressive disorder (MDD) has lagged behind. Transcriptome studies report multiple brain disturbances but are limited by small sample sizes. Genome-wide association studies (GWAS) report weak results but suggest an overlapping genetic risk with other neuropsychiatric disorders. We performed a systematic molecular characterization of altered brain function in MDD using meta-analysis of differential expression of 8 gene array studies across 3 corticolimbic brain regions in 101 subjects. The identified ‘metaA-MDD’ genes suggest altered neurotrophic support, brain plasticity and neuronal signaling in MDD. Notably, metaA-MDD genes display a low connectivity and hubness in coexpression networks as well as a uniform genomic distribution, which is consistent with diffuse polygenic mechanisms. We have integrated these findings with results from over 1,800 published GWAS and show that genetic variations nearby metaA-MDD genes predict a greater risk for neuropsychiatric disorders, and notably for age-related phenotypes, but not for other medical illnesses (including those frequently co-occurring with depression) or body characteristics. Collectively, the intersection of unbiased investigations of gene function (transcriptome) and structure (GWAS) provides novel leads to investigate molecular mechanisms of MDD and suggests common biological pathways between depression, other neuropsychiatric diseases and brain aging.
Key Words: Depression, Transcriptome, Meta-analysis, Genetic association, Medical illnesses
Introduction
Major depressive disorder (MDD), a common complex polygenic disease with moderate heritability (∼31-42%) [1], is the leading cause of years lost due to disability worldwide in women and men [2], reflecting in many individuals a lifelong trajectory of recurrent episodes, increasing severity and progressive treatment resistance [3]. Gene expression profiles (transcriptomes) in MDD have reported changes in several biological pathways across brain areas, affecting multiple cell types (principal neurons, interneurons, oligodendrocytes or astrocytes), signaling systems [glutamate, γ-aminobutyric acid (GABA), brain-derived neurotrophic factor (BDNF) and fibroblast growth factor] and biological functions (plasticity and inflammation) [4,5,6,7,8,9,10,11,12]. However, the moderate sample sizes of postmortem cohorts have limited the interpretability of these studies. Searches for genetic variants associated with MDD using genome-wide association studies (GWAS) and candidate gene studies have not yielded robust leads for MDD [1], although few significant loci were identified when combined with other major psychiatric disorders [13], suggesting a partially shared genetic structure. Similarly, biological and epidemiological studies report common pathologies and risks with other psychiatric disorders and cardiovascular, metabolic and inflammatory diseases [14,15,16,17,18] as well as accelerated patterns of lifelong changes in peripheral biological markers and brain gene expression naturally occurring with aging [19], suggesting accelerated aging processes in MDD [19,20,21]. However, overall, it is not clear whether these comorbidity patterns reflect shared genetic risks (i.e. a common etiology) or whether the presence of one disorder influences the development of another.
Here, we predicted that combining two large-scale, unbiased and independent investigations of gene function (transcriptome) and structure (GWAS) would bypass the limitations of single approaches and identify robust and informative gene changes in MDD. We further predicted that integrating the analysis of MDD transcriptomes with that of GWAS for multiple traits and disorders would contribute to delineating biological links between MDD and other sets of diseases and phenotypes.
Subjects and Methods
Human Postmortem Samples
Brain samples were obtained after consent from next of kin during autopsies conducted at the Allegheny County Medical Examiner's Office (Pittsburgh, Pa., USA) using procedures approved by the Institutional Review Board and Committee for Oversight of Research Involving the Dead at the University of Pittsburgh. Consensus DSM-IV diagnoses were made by an independent committee of experienced clinical research scientists using information from clinical records, toxicology results and a standardized psychological autopsy [22]. This latter incorporates a structured interview, conducted by a licensed clinical psychologist with family members of the index subject, to assess the diagnosis, psychopathology and medical, social and family histories as well as the history of substance abuse.
A total of 51 MDD and 50 control subjects were included in the 8 studies (online suppl. table S1; for all online suppl. material, see www.karger.com/doi/10.1159/000369974). Samples from the dorsolateral prefrontal cortex (dlPFC), subgenual anterior cingulate cortex (sgACC) or rostral amygdala enriched in lateral, basolateral and basomedial nuclei had been previously collected and processed on Affymetrix HG-U133 Plus 2 or Illumina HT12 gene arrays (online suppl. table S1). Four studies were performed in the sgACC, 2 studies in the amygdala and 2 in the dlPFC. Half of the studies had been performed in female subjects in each brain region. Details on subjects, areas investigated and other parameters are available in online supplementary table S2. See also previous reports on the cohorts and datasets [5,10,23,24]. Group means for age, postmortem interval and brain pH were not statistically different for all studies.
Gene Array Data Preprocessing, Gene Matching and Data Filtering
The microarrays were scanned and summarized as by the manufacturers' default settings. Data from Affymetrix arrays were processed by a robust multiarray (RMA) method, and data from Illumina arrays were processed with the manufacturer's BeadArray software for probe analysis. Batch effects were evaluated and normalized. Oligonucleotide probes (or probe sets) were matched to gene symbols using the hgu133plus2.db and illuminaHumanv4.db Bioconductor packages. When multiple probes matched with an identical gene symbol, the probe that generated the best disease association (by paired t test) was selected. The potential bias caused by this selection was addressed by permutation analysis in the following analytical steps.
A total of 16,689 unique genes were matched across the 8 studies. Two sequential steps of gene filtering were then performed. First, the sum of ranks across the 8 studies of each gene was calculated, and genes with the lowest 20% rank sum were considered nonexpressed and filtered out. Second, genes displaying very small variation in expression (lowest 20% rank sum of standard deviations) were filtered out. Together, this left 10,680 unique genes for further analysis (online suppl. table S3).
Meta-Analysis of Transcriptome Datasets
A summary is provided here, and details and formulas are available in the supplementary material. In short, we adopted a hypothesis-free data mining approach, applied linear models to account for potential confounding covariates and employed a meta-analysis pipeline to combine 8 studies using methods we have recently described [25].
Single-Study Analysis of MDD-Related Differential Gene Expression
The individual study analysis to detect candidate genes involved two major components: a random intercept model (RIM) and variable selection. Real data analysis and simulation showed improved statistical power and accuracy when applying the two techniques [25]. To account for the paired design and for the existence of several MDD-related potential covariates (e.g. alcohol dependence, antidepressant drug use and death by suicide as well as numerical covariates for age, brain pH, postmortem interval, etc.), we applied an RIM with parameter selection using the smallest Bayesian information criterion (BIC) [26]. To correct the potential bias of the variable selection procedure, we performed a permutation analysis that randomly shuffled the disease labels within each pair to generate a null distribution for p value assessment (B = 500). Subsequently, the permutation-corrected and unbiased p values were adjusted by the Benjamini-Hochberg procedure [27] for multiple comparisons within each study to control the false discovery rate (FDR). Online supplementary table S4 shows the number of differentially expressed genes detected under the raw p value threshold at 0.001 and FDR = 0.05, 0.1, 0.15 and 0.2 using the RIM-BIC model in individual analyses compared to a naïve paired t test. For instance, at the p < 0.001 threshold, the RIM-BIC model detected many more genes than paired t tests. However, when controlled by FDR at a 20% threshold, very few genes were detected, hence motivating the following meta-analysis across the 8 studies.
Meta-Analysis of MDD-Related Differential Gene Expression in the Eight Gene Array Studies
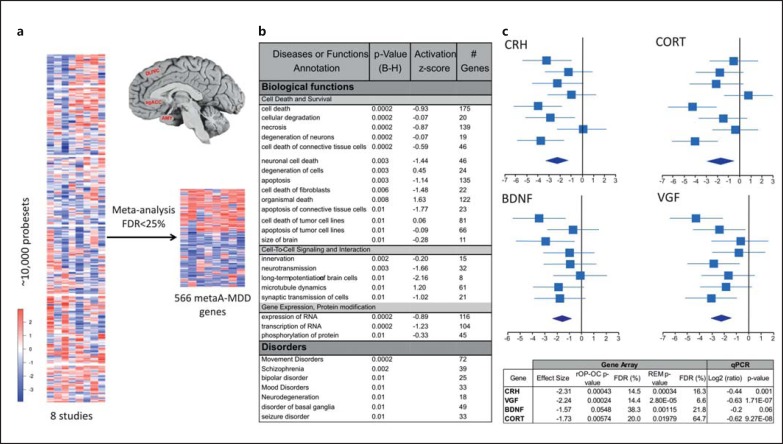
Many microarray meta-analytical methods are available, and the best choice of method for their application depends on the data structure and biological goal [28,29]. In this study, we combined two approaches based on their complement biological assumptions: the rth ordered p value with one-sided correction (rOP-OC) method [28] detected genes that were differentially expressed in at least 5 out of the 8 studies and with a consistent direction of changes (up- or downregulated), and the random effects model (REM) detected genes by combining effect sizes across all studies. Moreover, a permutation analysis was conducted, since some studies were performed in different brain regions of the same subjects. The results are shown in online supplementary table S4, including the number of differentially expressed genes detected from single-study analyses and meta-analyses. Finally, we applied a loose FDR threshold at 25% for downstream exploratory analysis and took the union of differentially expressed genes identified with rOP-OC and REM (online suppl. fig. S2), forming a 566-candidate gene set denoted ‘metaA-MDD’ genes. Forest plots for all the analyzed genes are available at an interactive website (https://research.psychiatry.upmc.com/SibilleMDD8/).
Real-Time Quantitative PCR
PCR products were amplified in quadruplets on a Mastercycler real-time PCR machine (Eppendorf, Hamburg, Germany) using universal PCR conditions. cDNA samples were obtained from independent cryostat-cut samples adjacent to sections used for the microarray analyses. Results were calculated as the geometric mean of relative intensities compared to internal controls (actin, glyceraldehyde-3-phosphate dehydrogenase and cyclophilin G). Absences of MDD-related changes in the internal controls were tested in individual cohorts. Diagnosis-dependent gene expression differences were assessed by ANCOVA using SPSS (SPSS Inc., Chicago, Ill., USA). To determine covariates to include in the gene-specific models, nominal factors were each tested as the main factor by ANOVA, scale covariates were tested by Pearson's correlation, and repeated measures were corrected by the modified Holm-Bonferroni test. ANCOVA models including significant cofactors were then applied. Analyses of quantitative PCR (qPCR) results across cohorts were performed using Stouffer's z-score method [30] in the cohorts that contributed to the identification of differential expression using the rOP-OC method [31], i.e. in at least 5 out of the 8 studies where genes of interest displayed expression changes [corticotrophin-releasing hormone (CRH): studies 1, 2 and 4-8; BDNF: studies 2, 3 and 5-8; VGF: studies 1, 3, 4 and 6-8; the study numbers are from online suppl. table S1]. In addition, the qPCR results obtained in the original single-cohort studies were combined, and a Pearson correlation factor with array results was calculated.
Functional Analysis and Gene Set Enrichments
An analysis was performed for the 566 metaA-MDD genes to investigate biological pathways potentially affected by the disease using Ingenuity Pathway Analysis (IPA; www.ingenuity.com) as well as using manual annotations of gene sets enriched in various cellular compartments. To create a list of excitatory and inhibitory synaptic markers, glutamate and GABA receptor signaling pathway-related molecules were downloaded from IPA. Genes exclusively expressed on the pre- or postsynaptic compartment were retained based on the location of gene expression shown in the knowledge base (i.e. presynaptic nerve terminal, postsynaptic neuron and glia). The significance of changes across gene groups was obtained by comparing the combined p values using Fisher's method with results obtained in gene sets of the same size randomly sampled from the background (13,350 genes) using 10,000 resamplings.
Meta-Analysis of Gene Coregulation Network Properties
To further investigate the 566 MDD disease candidate genes and to quantify their network properties compared to random subnetworks in the genome, we constructed unweighted gene coexpression networks based on the Pearson correlation in disease samples only, in control samples only as well as in combined disease and control samples. In gene coexpression networks, nodes represent genes, and nodes are connected if the genes are significantly coexpressed (e.g. measured by correlation) above a certain threshold. Based on the literature, one study [32] found that most inferred biological networks have a connectivity of ∼1% out of all connections; thus, we applied the same threshold, enabling comparable evaluations across networks.
We first overlaid the 566 genes on the full networks of all genes covered in our study after preprocessing (10,680 genes) and investigated whether the metaA-MDD genes displayed increased degrees (i.e. more connections) compared to other genes. We calculated the mean degree of the 566 genes, i.e. the average number of coexpressed links of the 566 genes among all 10,680 investigated genes. For comparison purposes, we randomly sampled 566 genes from the same 10,680 genes and calculated the mean degree of that gene set. This procedure was repeated 1,000 times to generate a null distribution, with the null hypothesis stating that the mean connectivity of the metaA-MDD genes is the same as that of randomly selected gene sets of the same size. p values were assessed using one-sided tests, with the null hypothesis being that the disease network has a larger network property (e.g. mean degree) than random networks for each of the 8 transcriptome studies. The 8 results were further combined using Fisher's meta-analysis method (online suppl. table S6). We also calculated three other network measures, namely mean clustering coefficient, mean betweenness centrality and assortativity, for the network constructed from the 566 genes (online suppl. tables S7S8S9). The clustering coefficient measures the local community structure. Betweenness centrality measures the extended influence of a network node and quantifies how many shortest network paths intersect a given node. Assortativity quantifies the tendency that an edge will connect two nodes of similar degree (online suppl. table S9).
Chromosome Localization of MetaA-MDD Genes
We overlaid the 566 metaA-MDD genes onto the chromosomes and assessed their chromosomal enrichment compared to the respective background gene density using a 1-Mb overlapping moving window on each of the 24 chromosomes. The number of genes among the metaA-MDD genes was counted in each 1-Mb interval on each chromosome as well as the number of background genes (10,680 genes). The significance of enrichment of the metaA-MDD genes in each chromosome region was assessed by Fisher's exact test.
Intersection of MetaA-MDD Genes with Disease-Associated Single Nucleotide Polymorphisms from GWAS
We compared the 566 metaA-MDD genes with genes nearby single nucleotide polymorphisms (SNPs) as identified in the GWAS Catalog database (http://www.genome.gov/gwastudies/). The database (as of February 4, 2014) contained 15,579 entries of disease- or trait-associated SNPs with p values <1/10−5 in 1,796 GWAS. We manually regrouped the disorders and traits into 11 categories: (1) neurological disorders and other brain phenotypes; (2) neuropsychiatric disorders; (3) MDD-related disorders; (4) medical illnesses related to MDD; (5) cardiovascular disease and related traits; (6) aging; (7) metabolic syndrome and related traits; (8) immune system, inflammation (excluding AIDS); (9) other medical illnesses not related to MDD; (10) cancer, and (11) miscellaneous body measures. Due to the nesting and overlap in some of the categories, we further tested the following gene groups: ‘(1) - (2)’ [i.e. studies in category (1) but not in category (2)] and ‘(6) - (2)’ (see legend for table 1). The enrichment p value by Fisher's exact test for each of the 1,796 studies was calculated and combined by Fisher's meta-analysis method for each of the 13 categories as the test statistics. The corrected p value was calculated from randomly selecting the same number of studies and calculating combined p values 10,000 times to generate a null distribution. Analyses were performed using 20-, 50-, 100- and 200-kb upstream or downstream windows to assign genes to SNPs.
Table 1.
Intersection of transcriptome meta-analysis and GWAS investigations for MDD, comorbid disorders and other disorders and traits
| Categories | Studies, n | 20 kb |
50 kb |
100 kb |
200 kb |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p value | genes, n | metaA-MDD genes, n | p value | genes, n | metaA-MDD genes, n | p value | genes, n | metaA-MDD genes, n | p value | genes, n | metaA-MDD genes, n | ||
| Neurological disorders and other brain phenotypes | 221 | 0.065 | 1,453 | 53 | 0.032 | 1,823 | 60 | 0.023 | 2,168 | 70 | 0.032 | 2,483 | 75 |
| Neuropsychiatric disorders | 97 | 0.021 | 720 | 31 | 0.007 | 913 | 35 | 0.006 | 1,106 | 42 | 0.010 | 1,267 | 42 |
| MDD-related disorders | 22 | 0.199 | 273 | 9 | 0.214 | 338 | 10 | 0.304 | 415 | 11 | 0.248 | 489 | 13 |
| Aging | 30 | 0.030 | 215 | 14 | 0.007 | 271 | 17 | 0.007 | 321 | 18 | 0.008 | 360 | 19 |
| Medical illnesses related to MDD | 315 | 0.214 | 2,608 | 89 | 0.163 | 3,198 | 98 | 0.298 | 3,676 | 103 | 0.285 | 4,060 | 115 |
| Cardiovascular diseases and related traits | 113 | 0.545 | 762 | 21 | 0.516 | 961 | 26 | 0.597 | 1,125 | 30 | 0.681 | 1,264 | 33 |
| Metabolic syndrome and related traits | 98 | 0.272 | 1,281 | 43 | 0.143 | 1,566 | 49 | 0.265 | 1,802 | 52 | 0.131 | 2,034 | 61 |
| Immune system, inflammation | 94 | 0.601 | 847 | 33 | 0.733 | 1,028 | 34 | 0.795 | 1,207 | 38 | 0.825 | 1,336 | 40 |
| Other medical illnesses not related to MDD | 444 | 0.825 | 2,264 | 78 | 0.870 | 2,685 | 85 | 0.745 | 3,076 | 97 | 0.747 | 3,419 | 98 |
| Cancer | 110 | 0.96 | 549 | 15 | 0.975 | 660 | 16 | 0.878 | 771 | 23 | 0.829 | 879 | 26 |
| Miscellaneous body measures | 230 | 0.567 | 1,309 | 45 | 0.708 | 1,569 | 46 | 0.723 | 1,812 | 52 | 0.782 | 2,027 | 51 |
| (Neurological disorders) – (neuropsychiatric disorders) | 118 | 0.550 | 792 | 24 | 0.596 | 995 | 28 | 0.508 | 1,185 | 33 | 0.473 | 1,373 | 38 |
| (Aging) – (neuropsychiatric disorders) | 27 | 0.016 | 213 | 14 | 0.009 | 269 | 17 | 0.010 | 319 | 18 | 0.009 | 358 | 19 |
Genes identified nearby significant genetic variants in each GWAS (at p < 1 × 10–5) of the ~1,800 studies in the GWAS Catalog were sorted into categories based on clinical evidence for comorbidity (or not) with MDD, and the significance of overlap with metaA-MDD genes was calculated for each category. The table includes the number of studies in the respective category as well as the p value of the significance for the overrepresentation of the metaA-MDD genes compared to the total number of genes in that category. In the bottom two analyses, GWAS related to both categories were removed. Results from the ‘aging’ category are not due to overlap in gene content with neuropsychiatry-related genes, whereas the ‘neurological disorders and other brain phenotypes’ category results are driven by neuropsychiatry-related genes. The results are presented for four genomic windows represented by the distance of genes from GWAS SNPs (numbers at the top of the table). Values in italics indicate significant results. Gene categories in boldface are significant.
Results
Meta-Analysis of Altered Gene Expression in MDD
We performed a meta-analysis of 8 transcriptome studies across 3 brain regions that are engaged in emotion regulation and implicated in MDD (dlPFC, sgACC and amygdala) in a combined cohort of 51 MDD and 50 control subjects to identify genes with consistent changes in expression in MDD (online suppl. tables S1S2S3). For within-study comparison, we used a permutation-based RIM with best-fit variable selection to assess the MDD effect and to regress for clinical, technical and demographic parameters at the individual gene level [25], hence ensuring that the results were not confounded by any of these parameters. To increase the detection power of capturing MDD effects across studies, we applied two meta-analytical methods based on complementary hypotheses for disease effects, namely consistent effect size (REM) and expression changes (rOP-OC; see Subjects and Methods and Wang et al. [25]). The FDR was set at 25% to maintain a discovery component in the study and to retain sufficient genes for combining with results from GWAS in the second phase of the study. This combined approach resulted in the identification of 566 metaA-MDD genes (56.18% genes downregulated) (fig. 1a, online suppl. fig. S1, S2, online suppl. tables S4, S5). We also applied a metaregression approach for sex/gender effect across studies. These studies were considered exploratory, since metaregression usually requires larger number of studies. In all, 39 metaA-MDD genes also displayed a significant sex effect by metaregression (<20% FDR; ‘metaR’ in online suppl. table S5), and 50 additional genes were identified by metaregression only. Forest plots and metaA and metaR results are available online for all tested genes (https://research.psychiatry.upmc.com/SibilleMDD8/).
Fig. 1.
a Heat map of the meta-analysis of gene expression across 8 datasets in 3 brain regions frequently affected in MDD. The color scale is in effect size. AMY = Amygdala. b Diseases and biological pathways associated with metaA-MDD genes extracted from the IPA. p values are Bonferroni-Holms (B-H) corrected. Activation scores indicate the putative direction of the biological effect. c Forest plots and qPCR validations of representative metaA-MDD genes. In the forest plots, the mean effect size (and 95% confidence interval) for each study are plotted and summarized in the bottom diamond. The x-axis is in variance units. Cortistatin (CORT) codes for a GABA neuron marker previously associated with MDD [5]. The gene array p values are from two alternate meta-analysis strategies (i.e. rOP-OC and REM; see Subjects and Methods). qPCR values are given in average log values of MDD versus control signal ratios. Significant qPCR values were obtained for CRH, acronymic VGF and CORT, and a trend significance level was found for BDNF. Other qPCR validation of array results performed in the original studies [5,10,25,47] yielded a combined array-qPCR Pearson correlation factor of 0.94 (n = 17 genes, p < 0.001; online suppl. fig. S3).
To identify biological pathways represented by the metaA-MDD genes and potentially affected in the disease, we performed an unbiased analysis of overrepresented gene annotations using IPA. The top disorders associated with metaA-MDD genes included mood disorders, other major mental illnesses and brain disorders (fig. 1b). Biological functions overrepresented in metaA-MDD genes centered on two main themes, specifically cell death and survival as well as cell-to-cell signaling, with predicted activation z-scores showing mostly reduced activity (fig. 1b). These results include a decreased expression of genes coding for signaling neuropeptides (CORT, VEGFA and TGFB2) as well as CRH, BDNF and acronymic VGF, three genes with ample prior evidence for involvement in stress-related disorders and MDD (fig. 1c) [33,34,35]. Notably, these findings from large-scale unbiased approaches are consistent with current hypotheses for depression, i.e. the stress and low neurotrophin hypotheses of MDD [36], and further suggest resistance to cell death and reduced signaling, potentially leading to loss of homeostasis in brain regions associated with mood regulation.
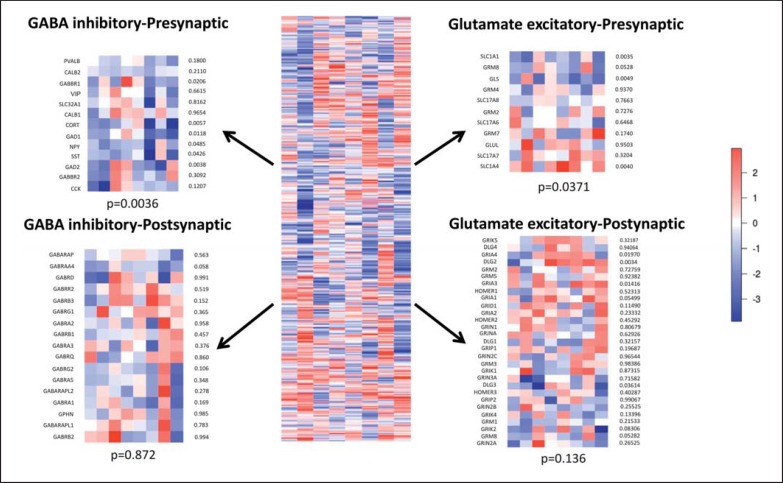
Since these results were obtained from combined gray matter samples, we sought to investigate in silico the putative cellular sites of reduced signaling suggested by the pathway analysis. For this purpose, we returned to the main dataset and examined MDD effects on groups of genes known to be expressed in the inhibitory and excitatory pre- and postsynaptic compartments. We observed a significant pattern of downregulated expression of presynaptic GABA neuron markers, followed by weaker and heterogeneous changes in presynaptic glutamatergic excitatory components. No group-wise changes were observed for genes whose products comprise postsynaptic components (fig. 2).
Fig. 2.
Heat maps of the differential expression of genes expressed in the pre- and postsynaptic excitatory glutamatergic and related inhibitory GABAergic genes. The statistical significance of observing the combined changes is indicated for each compartment. A significant pattern of downregulated expression of presynaptic GABA neuron markers was observed, followed by weaker and heterogeneous changes in presynaptic glutamatergic excitatory components. No group-wise changes were observed for genes whose products comprise postsynaptic components. The color scale is in effect size.
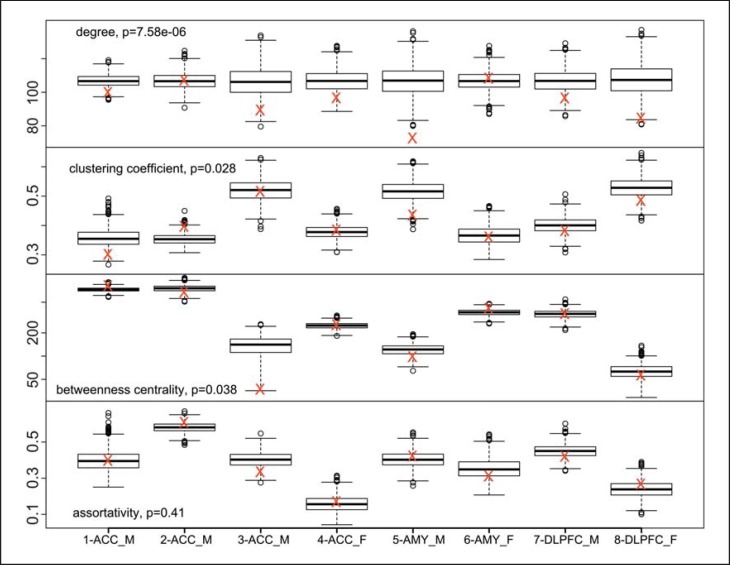
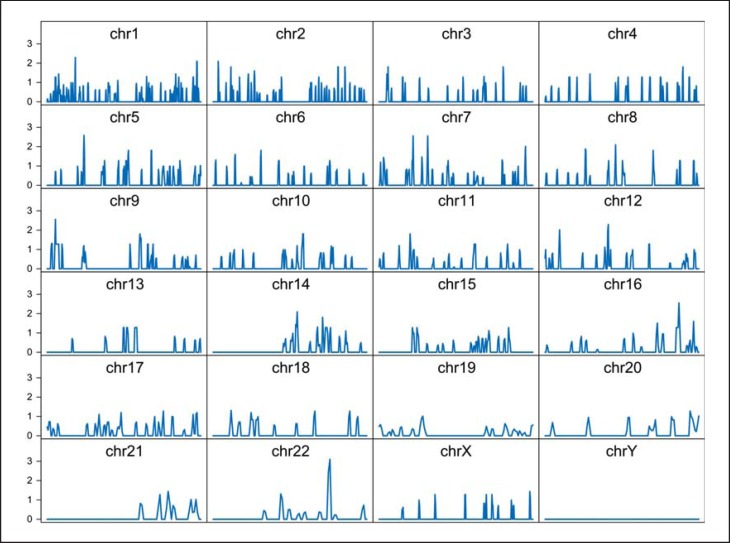
Based on the observations in organ and system diseases, where disease-related genes have often been associated with highly connected hub genes in the coexpression networks [37], we investigated the integration of the metaA-MDD genes within the broader gene coexpression landscape formed by the brain transcriptome [38]. We constructed unweighted gene coexpression networks for each of the 8 studies and performed meta-analyses of network properties. The results show that the metaA-MDD genes displayed a significantly lower mean connectivity (p < 0.0001; fig. 3) and a modestly lower clustering coefficient (measure of local community structure; p = 0.025), but unchanged betweenness centrality (measure of influence of a gene on the network; p = 0.99) and mean assortativity (likelihood of links connecting genes with similar connectivity; p > 0.9; fig. 2; online suppl. tables S6S7S8S9). The lack of changes in the latter two measures shows that the overall structure of the gene network has remained intact in MDD, whereas the results of a lower connectivity and clustering coefficient indicate that metaA-MDD genes are generally located more peripherally on gene networks and not enriched in more central, highly connected hub genes. These results are consistent with an earlier report using expression datasets across psychiatric disorders [39]. Relatedly, we tested for the presence of chromosomal enrichment of the metaA-MDD genes. When accounting for the local gene density, the metaA-MDD genes displayed an even distribution across the genome (fig. 4), ruling out the presence of local chromosomal enrichment or genomic hot spots associated with depression.
Fig. 3.
Gene network properties of metaA-MDD genes. The mean network properties of metaA-MDD genes (red crosses) are presented for the individual studies and compared to the box plot results of 1,000 results from randomly selected gene sets of the same size. The included p values represent the results of Fisher's meta-analysis of the 8 results. The y-axes are in units for the respective network properties. AMY = Amygdala.
Fig. 4.
Chromosome enrichment (1-Mb interval regions) of the 566 metaA-MDD genes. The significance of enrichment of the metaA-MDD genes in each chromosome region was assessed by Fisher's exact test. The y-axis represents -log10 of p values. chr = Chromosome.
These combined results (metaA-MDD gene identification, biological pathways, network analyses and chromosomal localizations) are consistent with a disease model implicating multiple genes with weak effects rather than few key genes with dominant effects, and together they underscore the complex molecular structure for MDD.
From Expression Meta-Analysis in MDD to GWAS in MDD and Other Disorders
In the second phase of the study, we sought to independently assess the validity of metaA-MDD genes as candidate genes implicated in polygenic mechanisms not only for MDD but also for other related brain disorders and comorbid medical illnesses and phenotypes. For this, we predicted that the metaA-MDD gene set would significantly and selectively overlap with gene findings obtained via an unrelated and unbiased approach, namely from independent GWAS for neuropsychiatric disorders and other disorders and for phenotypes showing a continuum of risk with MDD. We manually sorted results from ∼1,800 studies that are included in the GWAS Catalog into several categories based on clinical evidence for comorbidity (or not) with MDD and as described in table 1 and online supplementary table S10. We then identified genes located within 100 kb of significant genetic variants in each GWAS (at p < 1 × 10−5) and sought overrepresentation of metaA-MDD genes within those gene sets compared to random gene sets of similar sizes; 167 of the 566 metaA-MDD genes were identified by at least 1 GWAS. These 167 metaA-MDD genes were most significantly overrepresented in the category of ‘neuropsychiatric disorders’, followed by ‘neurological disorders and other brain phenotypes’ and ‘aging’ (table 1; online suppl. table S11). Since these categories are not fully independent, we removed overlapping neuropsychiatry-related studies included in the ‘neurological disorders’ and ‘aging’ categories and retested for significant overlaps with metaA-MDD genes. After this subtraction, the results in the ‘aging’ category remained at the same significance level (table 1, bottom 2 rows), demonstrating that the age-related findings are related to metaA-MDD genes independently of other neuropsychiatry-related genes. On the other hand, the ‘neurological disorders and other brain phenotypes’ category results were not significant anymore, indicating that those results were driven by genes associated with neuropsychiatric disorders. There was no significant overlap between metaA-MDD genes in the category of diseases and phenotypes for which there is low or no evidence for a shared biological risk. Note that GWAS for MDD per se have reported too few significant findings for formal statistical enrichment analysis. Interestingly, we observed no overlap with gene sets associated with cardiovascular, metabolic and inflammatory diseases. This indicates that the clinical comorbidity between MDD and these disorders may be mediated by disease-/MDD-induced biological changes rather than through a shared genetic risk. Similar results were obtained with 20- to 200-kb sliding genomic windows around GWAS-identified SNPs and with several versions of the GWAS Catalog (table 1). Details on metaA-MDD and GWAS-identified genes as well as overlap are shown in online supplementary tables S11 and S12.
Discussion
To begin addressing the major challenge of identifying and characterizing the scope and boundaries of the molecular pathology of MDD, a heterogeneous disorder with expected complex polygenic mechanisms, we first performed a meta-analysis of large-scale transcriptome surveys in corticolimbic brain areas using human postmortem samples. We then used the results from this meta-analysis as an enriched set of candidate genes to perform a top-down-directed analysis for overlap with findings extracted from independent large-scale genetic studies in MDD and multiple other disorders and traits. If successful, we predicted that the integration of these two independent and unbiased investigations of gene function (transcriptome) and structure (GWAS) would (1) identify and confirm some prior findings obtained in MDD using single approaches, (2) bypass some of the limitations of individual approaches (e.g. low power, multiple repeated testing and false positives/negatives) and lead to the identification of robust and informative gene changes in MDD and (3) contribute to detangling the genetic structure of the depression syndrome from its patterns of clinical comorbidities.
The results from the transcriptome meta-analysis identified a set of differentially expressed genes in MDD that is enriched for changes in components of synaptic machinery, specifically suggesting reduced neurotrophic support and presynaptic GABA function (fig. 1, 2; online suppl. table S5). These results are consistent with prior findings in MDD and from individual datasets included here [5,40], and together they provide unbiased evidence in support of several current hypotheses about biological mechanisms of MDD [41], specifically the stress, GABA and low neurotrophin hypotheses of MDD [35,36,42]. Note that a low CRH expression in MDD subjects was unexpected, considering evidence from human and animal studies suggesting that elevated CRH function and activity of the neuroendocrine stress axis may be engaged in MDD [35]. A potential explanation is that low CRH levels may result from a feedback loop on central CRH due to peripheral neuroendocrine overactivation.
The transcriptome meta-analysis results do not provide evidence in support of several other biological hypotheses about mechanisms of MDD (for instance related to mitochondrial dysfunction, increased inflammation and activated immune functions) [41]. There may be several explanations for these negative results. First, a lack of positive findings may reflect the reactive nature of induced depressive states through some of these mechanisms (e.g. inflammation). These phenotypes may occur in subgroups of MDD cases, and these sporadic cases would not be captured by meta-analytical approaches. Second, the current approach tested for similar and consistent changes across brain regions, and alternative brain region-specific hypotheses were not evaluated here. For instance, no evidence for changes in monoamine-related genes was found (with the exception of reduced HTR2C expression; online suppl. table S5), but we did not investigate the presynaptic midbrain compartment of those systems, which is more likely dysregulated in MDD [43]. Moreover, associations of changes with several parameters, including death by suicide and drug treatments, were not investigated here, since our meta-analytical methods regressed for the putative effects of these factors. We did, however, perform exploratory analyses on metaregression for sex, although these studies were underpowered due to the limited number of studies and since the results were corrected for simultaneous testing on 10,680 genes (see Subjects and Methods). Nonetheless, the data are available on a per gene basis investigation (https://research.psychiatry.upmc.com/SibilleMDD8/), and uncorrected exploratory and a priori analyses of the dataset may yield further information. For instance, we previously showed a reduced expression of somatostatin, specifically in women with MDD, in the same cohort [40,44,45], which here yielded metaA and metaregression p values of 0.03 and 0.006 (FDR = 0.25), respectively. Together, we interpret the current sex-/gender-related analyses as suggesting variability in effect sizes rather than distinct pathologies between men and women with MDD.
An inspection of forest plots for single genes (fig. 1; online resources) shows variability in MDD effects across regions and cohorts, which is consistent with the known heterogeneity of clinical cohorts and putative disease mechanisms. Thus, the current findings provide converging results at the level of selected biological pathways as described above, but results at the level of single genes should be interpreted in the light of the caveats mentioned here. Similarly, gene coexpression network and chromosomal localization studies can identify key genes with central roles or critical genomic DNA loci implicated in illnesses; however, consistent with the expected heterogeneity of the illnesses and the current lack of identified dominant causal genes or mutations, the results from these hub-seeking studies were mostly negative. Instead, the results from the coexpression network studies suggest a small and distributed contribution of multiple genes and gene products rather than central and causal disturbances. These findings extend results from an earlier report using expression datasets across psychiatric disorders (including some of the datasets from this study) [39]. Therefore, we refrained from overinterpreting single gene findings here. Instead, we provide forest plots and meta-analysis results for all tested genes, so that researchers can directly investigate genes with a priori hypotheses.
To address this limitation of the transcriptome-only analysis, we next performed a more global analysis and specifically tested whether metaA-MDD genes overlapped with genes identified by genetic studies in depression and other disorders and traits. This top-down-directed analysis revealed a significant overlap between metaA-MDD genes and genes identified by nearby genetic variants in GWAS for neuropsychiatric disorders as a whole and for aging, but not for other medical illnesses for which MDD is a significant risk factor or for other brain phenotypes (table 1). Although our approach was based on genomic proximity or guilt by association (i.e. genes nearby GWAS-identified SNPs), these results were robust, since almost identical results were obtained with genomic windows ranging from 20 to 200 kb from GWAS-identified SNPs (table 1). On the other hand, the negative results for overlap with GWAS for MDD and related phenotypes can be explained by too few significant findings obtained in these studies (i.e. underpowered analysis), as illustrated by the low number of genes in that category (table 1).
Collectively, these findings contribute to delineating biological pathways for risk, or a broad vulnerability to psychopathology across psychiatric disorders, consistent with meta-analytical GWAS across major mental illnesses [13]. This may occur most likely through concerted changes in genes collectively mediating critical biological pathways, including presynaptic GABA inhibitory and neurotrophic functions as well as cell survival (fig. 1b). This may not necessarily translate into a common pathology across those disorders. Rather, pathways corresponding to those genetic risks may represent biological substrates for moderation by multiple external and/or environmental factors in MDD, and for interactions with other disease-specific pathologies occurring in other psychiatric disorders, together giving rise to the observed diversity in pathophysiological and clinical presentations.
However, our approach also substantially departed from a GWAS across psychiatric disorders [13], since it sought convergence between two unbiased and independent investigations of gene function (transcriptome) and structure (GWAS) and since it extended the analysis to multiple groups of disorders and phenotypes. The overlap between metaA-MDD genes and genes identified by nearby genetic variants in GWAS for age-related phenotypes is intriguing (table 1). We are separately investigating the potential biological sources of the increased risk for neurodegenerative and neuropsychiatric disorders that is associated with aging [21]. These parallel studies are mostly prompted by observations that age-dependent gene expression changes in the human brain occur along a continuous lifelong trajectory and affect many of the same genes and in the same directions as in depression and other brain disorders [21,46]. These observations have collectively suggested a pattern of accelerated, or at least anticipated, brain aging in psychiatric disorders. Hence, the prediction would have been that the significant overlap between metaA-MDD genes and aging would have disappeared after removing genes in the ‘aging’ category that were also identified through GWAS for psychiatric disorders. This was clearly not the result observed, as the age-related findings remained at a very similar statistical significance after that exclusion (table 1), suggesting that the pathways may be independent. These results support the presence of a more profound biological link between depression and aging, which will need to be further investigated in follow-up studies, specifically in terms of putative overlap and links between implicated biological pathways.
In summary, the results from the transcriptome meta-analysis and from the analyses of gene function, the coexpression network and the chromosomal localization of metaA-MDD genes together support a molecular pathology for MDD that is distributed across multiple genes, with some functional convergence for genes coding for stress and neurotrophic factors and suggesting reduced neurotrophic support, neuronal signaling and GABA function. These findings are consistent not only with selected prior knowledge about putative mechanisms of MDD but also with past difficulties in identifying singular molecular changes. Nonetheless, the current study provides numerous new gene leads for further analysis. The integration of transcriptome meta-analysis with genetic studies led to critical new perspectives. Specifically, the convergence of these two unbiased and independent investigations of gene function and structure supports the presence of pathophysiologically relevant and genetically linked biological modules that are implicated across psychiatric disorders and that are continuous with aging. Our results also show that this genetic and biological overlap does not extend to medical illnesses that are frequently co-occurring with MDD. Accordingly, this report significantly contributes to the debate on the singularity versus continuum of pathological substrates underlying common psychiatric and brain disorders.
Disclosure Statement
D.A.L. currently receives investigator-initiated research support from Bristol-Myers Squibb and Pfizer and in 2012-2014 served as a consultant in the areas of target identification and validation and new compound development to Autifony, Bristol-Myers Squibb, Concert Pharmaceuticals and Sunovion. The other authors have no conflicts of interest to disclose.
Supplementary Material
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Acknowledgements
This work was supported by the National Institute of Mental Health (NIMH) grants No. MH084060 (E.S.), MH085111 (E.S.), MH094862 (G.C.T.) and MH084053 (D.A.L.).
References
- 1.Flint J, Kendler KS. The genetics of major depression. Neuron. 2014;81:484–503. doi: 10.1016/j.neuron.2014.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO . The global burden of disease - 2004 update. Geneva: WHO; 2008. [Google Scholar]
- 3.Moylan S, Maes M, Wray NR, Berk M. The neuroprogressive nature of major depressive disorder: pathways to disease evolution and resistance, and therapeutic implications. Mol Psychiatry. 2013;18:595–606. doi: 10.1038/mp.2012.33. [DOI] [PubMed] [Google Scholar]
- 4.Sequeira A, Mamdani F, Ernst C, Vawter MP, Bunney WE, Lebel V, Rehal S, Klempan T, Gratton A, Benkelfat C, Rouleau GA, Mechawar N, Turecki G. Global brain gene expression analysis links glutamatergic and GABAergic alterations to suicide and major depression. PLoS One. 2009;4:e6585. doi: 10.1371/journal.pone.0006585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guilloux JP, Douillard-Guilloux G, Kota R, Wang X, Gardier AM, Martinowich K, Tseng GC, Lewis DA, Sibille E. Molecular evidence for BDNF- and GABA-related dysfunctions in the amygdala of female subjects with major depression. Mol Psychiatry. 2012;17:1130–1142. doi: 10.1038/mp.2011.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evans SJ, Choudary PV, Neal CR, Li JZ, Vawter MP, Tomita H, Lopez JF, Thompson RC, Meng F, Stead JD, Walsh DM, Myers RM, Bunney WE, Watson SJ, Jones EG, Akil H. Dysregulation of the fibroblast growth factor system in major depression. Proc Natl Acad Sci USA. 2004;101:15506–15511. doi: 10.1073/pnas.0406788101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miguel-Hidalgo JJ, Jiang W, Konick L, Overholser JC, Jurjus GJ, Stockmeier CA, Steffens DC, Krishnan KR, Rajkowska G. Morphometric analysis of vascular pathology in the orbitofrontal cortex of older subjects with major depression. Int J Geriatr Psychiatry. 2013;28:959–970. doi: 10.1002/gps.3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karolewicz B, Maciag D, O'Dwyer G, Stockmeier CA, Feyissa AM, Rajkowska G. Reduced level of glutamic acid decarboxylase-67 kDa in the prefrontal cortex in major depression. Int J Neuropsychopharmacol. 2010;13:411–420. doi: 10.1017/S1461145709990587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aston C, Jiang L, Sokolov BP. Transcriptional profiling reveals evidence for signaling and oligodendroglial abnormalities in the temporal cortex from patients with major depressive disorder. Mol Psychiatry. 2005;10:309–322. doi: 10.1038/sj.mp.4001565. [DOI] [PubMed] [Google Scholar]
- 10.Sibille E, Wang Y, Joeyen-Waldorf J, Gaiteri C, Surget A, Oh S, Belzung C, Tseng GC, Lewis DA. A molecular signature of depression in the amygdala. Am J Psychiatry. 2009;166:1011–1024. doi: 10.1176/appi.ajp.2009.08121760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choudary PV, Molnar M, Evans SJ, Tomita H, Li JZ, Vawter MP, Myers RM, Bunney WE Jr, Akil H, Watson SJ, Jones EG. Altered cortical glutamatergic and GABAergic signal transmission with glial involvement in depression. Proc Natl Acad Sci USA. 2005;102:15653–15658. doi: 10.1073/pnas.0507901102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kang HJ, Adams DH, Simen A, Simen BB, Rajkowska G, Stockmeier CA, Overholser JC, Meltzer HY, Jurjus GJ, Konick LC, Newton SS, Duman RS. Gene expression profiling in postmortem prefrontal cortex of major depressive disorder. J Neurosci. 2007;27:13329–13340. doi: 10.1523/JNEUROSCI.4083-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cross-Disorder Group of the Psychiatric Genomics Consortium Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381:1371–1379. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ciechanowski PS, Katon WJ, Russo JE. Depression and diabetes: impact of depressive symptoms on adherence, function, and costs. Arch Intern Med. 2000;160:3278–3285. doi: 10.1001/archinte.160.21.3278. [DOI] [PubMed] [Google Scholar]
- 15.Steptoe A, Whitehead DL. Depression, stress, and coronary heart disease: the need for more complex models. Heart. 2005;91:419–420. doi: 10.1136/hrt.2004.045310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Evans DL, Charney DS, Lewis L, et al. Mood disorders in the medically ill: scientific review and recommendations. Biol Psychiatry. 2005;58:175–189. doi: 10.1016/j.biopsych.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 17.McIntyre RS, Soczynska JK, Konarski JZ, Woldeyohannes HO, Law CW, Miranda A, Fulgosi D, Kennedy SH. Should depressive syndromes be reclassified as ‘metabolic syndrome type II’? Ann Clin Psychiatry. 2007;19:257–264. doi: 10.1080/10401230701653377. [DOI] [PubMed] [Google Scholar]
- 18.Dantzer R, O'Connor JC, Freund GG, Johnson RW, Kelley KW. From inflammation to sickness and depression: when the immune system subjugates the brain. Nat Rev Neurosci. 2008;9:46–56. doi: 10.1038/nrn2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Douillard-Guilloux G, Guilloux JP, Lewis DA, Sibille E. Anticipated brain molecular aging in major depression. Am J Geriatr Psychiatry. 2013;21:450–460. doi: 10.1016/j.jagp.2013.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wolkowitz OM, Reus VI, Mellon SH. Of sound mind and body: depression, disease, and accelerated aging. Dialogues Clin Neurosci. 2011;13:25–39. doi: 10.31887/DCNS.2011.13.1/owolkowitz. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sibille E. Molecular aging of the brain, neuroplasticity, and vulnerability to depression and other brain-related disorders. Dialogues Clin Neurosci. 2013;15:53–65. doi: 10.31887/DCNS.2013.15.1/esibille. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glantz LA, Austin MC, Lewis DA. Normal cellular levels of synaptophysin mRNA expression in the prefrontal cortex of subjects with schizophrenia. Biol Psychiatry. 2000;48:389–397. doi: 10.1016/s0006-3223(00)00923-9. [DOI] [PubMed] [Google Scholar]
- 23.Chang LC, Jamain S, Lin CW, Rujescu D, Tseng GC, Sibille E. A Conserved BDNF, glutamate- and GABA-enriched gene module related to human depression identified by coexpression meta-analysis and DNA variant genome-wide association studies. PLoS One. 2014;9:e90980. doi: 10.1371/journal.pone.0090980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sibille E, Arango V, Galfalvy HC, Pavlidis P, Erraji-Benchekroun L, Ellis SP, John Mann J. Gene expression profiling of depression and suicide in human prefrontal cortex. Neuropsychopharmacology. 2004;29:351–361. doi: 10.1038/sj.npp.1300335. [DOI] [PubMed] [Google Scholar]
- 25.Wang X, Lin Y, Song C, Sibille E, Tseng GC. Detecting disease-associated genes with confounding variable adjustment and the impact on genomic meta-analysis: with application to major depressive disorder. BMC Bioinformatics. 2012;13:52. doi: 10.1186/1471-2105-13-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwarz GE. Estimating the dimension of a model. Ann Stat. 1978;6:461–464. [Google Scholar]
- 27.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Methodol. 1995;57:289–300. [Google Scholar]
- 28.Chang LC, Lin HM, Sibille E, Tseng GC. Meta-analysis methods for combining multiple expression profiles: comparisons, statistical characterization and an application guideline. BMC Bioinformatics. 2013;14:368. doi: 10.1186/1471-2105-14-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tseng GC, Ghosh D, Feingold E. Comprehensive literature review and statistical considerations for microarray meta-analysis. Nucleic Acids Res. 2012;40:3785–3799. doi: 10.1093/nar/gkr1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Whitlock MC. Combining probability from independent tests: the weighted Z-method is superior to Fisher's approach. J Evol Biol. 2005;18:1368–1373. doi: 10.1111/j.1420-9101.2005.00917.x. [DOI] [PubMed] [Google Scholar]
- 31.Song C, Tseng GC. Hypothesis setting and order statistic for robust genomic meta-analysis. Ann Appl Stat. 2014;8:777–800. doi: 10.1214/13-aoas683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee HK, Hsu AK, Sajdak J, Qin J, Pavlidis P. Coexpression analysis of human genes across many microarray data sets. Genome Res. 2003;14:1085–1094. doi: 10.1101/gr.1910904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sen S, Duman R, Sanacora G. Serum brain-derived neurotrophic factor, depression, and antidepressant medications: meta-analyses and implications. Biol Psychiatry. 2008;64:527–532. doi: 10.1016/j.biopsych.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hunsberger JG, Newton SS, Bennett AH, Duman CH, Russell DS, Salton SR, Duman RS. Antidepressant actions of the exercise-regulated gene VGF. Nat Med. 2007;13:1476–1482. doi: 10.1038/nm1669. [DOI] [PubMed] [Google Scholar]
- 35.De Kloet ER, Joels M, Holsboer F. Stress and the brain: from adaptation to disease. Nat Rev Neurosci. 2005;6:463–475. doi: 10.1038/nrn1683. [DOI] [PubMed] [Google Scholar]
- 36.Duman RS, Monteggia LM. A neurotrophic model for stress-related mood disorders. Biol Psychiatry. 2006;59:1116–1127. doi: 10.1016/j.biopsych.2006.02.013. [DOI] [PubMed] [Google Scholar]
- 37.Albert R, Jeong H, Barabasi AL. Error and attack tolerance of complex networks. Nature. 2000;406:378–382. doi: 10.1038/35019019. [DOI] [PubMed] [Google Scholar]
- 38.Gaiteri C, Ding Y, French B, Tseng GC, Sibille E. Beyond modules and hubs: the potential of gene coexpression networks for investigating molecular mechanisms of complex brain disorders. Genes Brain Behav. 2014;13:13–24. doi: 10.1111/gbb.12106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gaiteri C, Sibille E. Differentially expressed genes in major depression reside on the periphery of resilient gene coexpression networks. Front Neurosci. 2011;5:95. doi: 10.3389/fnins.2011.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tripp A, Oh H, Guilloux JP, Martinowich K, Lewis DA, Sibille E. Brain-derived neurotrophic factor signaling and subgenual anterior cingulate cortex dysfunction in major depressive disorder. Am J Psychiatry. 2012;169:1194–1202. doi: 10.1176/appi.ajp.2012.12020248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Belmaker RH, Agam G. Major depressive disorder. N Engl J Med. 2008;358:55–68. doi: 10.1056/NEJMra073096. [DOI] [PubMed] [Google Scholar]
- 42.Luscher B, Shen Q, Sahir N. The GABAergic deficit hypothesis of major depressive disorder. Mol Psychiatry. 2011;16:383–406. doi: 10.1038/mp.2010.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arango V, Underwood MD, Mann JJ. Serotonin brain circuits involved in major depression and suicide. Prog Brain Res. 2002;136:443–453. doi: 10.1016/s0079-6123(02)36037-0. [DOI] [PubMed] [Google Scholar]
- 44.Tripp A, Kota RS, Lewis DA, Sibille E. Reduced somatostatin in subgenual anterior cingulate cortex in major depression. Neurobiol Dis. 2011;42:116–124. doi: 10.1016/j.nbd.2011.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guilloux JP, Douillard-Guilloux G, Kota R, Wang X, Gardier AM, Martinowich K, Tseng GC, Lewis DA, Sibille E. Molecular evidence for BDNF- and GABA-related dysfunctions in the amygdala of female subjects with major depression. Mol Psychiatry. 2012;17:1130–1142. doi: 10.1038/mp.2011.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Glorioso C, Oh S, Douillard GG, Sibille E. Brain molecular aging, promotion of neurological disease and modulation by Sirtuin5 longevity gene polymorphism. Neurobiol Dis. 2011;41:279–290. doi: 10.1016/j.nbd.2010.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sibille E, Morris HM, Kota RS, Lewis DA. GABA-related transcripts in the dorsolateral prefrontal cortex in mood disorders. Int J Neuropsychopharmacol. 2011;14:721–734. doi: 10.1017/S1461145710001616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data