Abstract
Background and Aims Sorghum is an essential grain crop whose evolutionary placement within the Andropogoneae has been the subject of scrutiny for decades. Early studies using cytogenetic and morphological data point to a poly- or paraphyletic origin of the genus; however, acceptance of poly- or paraphyly has been met with resistance. This study aimed to address the species relationships within Sorghum, in addition to the placement of Sorghum within the tribe, using a phylogenetic approach and employing broad taxon sampling.
Methods From 16 diverse Sorghum species, eight low-copy nuclear loci were sequenced that are known to play a role in morphological diversity and have been previously used to study evolutionary relationships in grasses. Further, the data for four of these loci were combined with those from 57 members of the Andropogoneae in order to determine the placement of Sorghum within the tribe. Both maximum likelihood and Bayesian analyses were performed on multilocus concatenated data matrices.
Key Results The Sorghum-specific topology provides strong support for two major lineages, in alignment with earlier studies employing chloroplast and internal transcribed spacer (ITS) markers. Clade I is composed of the Eu-, Chaeto- and Heterosorghum, while clade II contains the Stipo- and Parasorghum. When combined with data from the Andropogoneae, Clade II resolves as sister to a clade containing Miscanthus and Saccharum with high posterior probability and bootstrap support, and to the exclusion of Clade I.
Conclusions The results provide compelling evidence for a two-lineage polyphyletic ancestry of Sorghum within the larger Andropogoneae, i.e. the derivation of the two major Sorghum clades from a unique common ancestor. Rejection of monophyly in previous molecular studies is probably due to limited taxon sampling outside of the genus. The clade consisting of Para- and Stiposorghum resolves as sister to Miscanthus and Saccharum with strong node support.
Keywords: Sorghum, phylogeny, Andropogoneae, Poaceae, low-copy nuclear loci, polyphyly
INTRODUCTION
Sorghum L. Moench consists of approx. 25 species of C4 sub-tropical grasses that are widely distributed throughout the Americas, Africa, Asia and Australia (Garber, 1950). Given that Sorghum is an agronomically important crop used for food, fibre and fuel, it is not surprising that a large part of the taxonomic literature focuses on the domestication of wild African Sorghum and its subsequent migration to the Americas during the early 20th century (Snowden, 1935, 1936; de Wet and Huckabay, 1967; de Wet and Harlan, 1971; Dillon et al., 2007). The cytology and morphology of Sorghum species endemic to Australia received little attention until much later (Lazarides et al., 1991; Spangler, 2003). Current classification divides Sorghum into five sub-sections: Eusorghum, Chaetosorghum, Heterosorghum, Parasorghum and Stiposorghum (Garber, 1950; Dahlberg, 2000). Ambiguous relationships between these sub-sections, however, have been disputed for over half a century (Garber, 1950; Celarier, 1959). Early work by Garber (1950) distinguished Eu-, Chaeto- and Heterosorghum from Stipo- and Parasorghum based on the presence/absence of bearded culm nodes and the prominence of awns. The sub-sections were further distinguished by panicle branching, callus morphology, the presence of pedicellate or sessile spikelets and several other morphological, geographic and cytological characteristics. More recent herbarium-based specimen analyses by Spangler (2003) provided support for the use of bearded culm nodes, spikelet morphology and lemma awn characteristics for classification, yet many traits were variable within sub-sections. Such difficulties in taxonomic distinction are probably caused by environmentally malleable morphology that contributes to ambiguities not only in Sorghum classification, but also throughout the larger Andropogoneae (Mathews et al., 2002).
Before the widespread use of molecular markers, several studies examined the relationships within Sorghum via cytological methods. Sorghum species contain a variable range (from ten to 40) of diploid chromosomes (Doggett, 1988; Lazarides et al., 1991). An early observation showed that species of Sorghum display different mean chromosome sizes, with a sub-set of species displaying larger mean chromosome sizes (Magoon and Shambulingappa, 1961). This observation has since been verified genus wide (Price et al., 2005). Furthermore, there was no correlation between geographical distribution and mean chromosome size. The authors found that Sorghum species clustered into two distinct lineages, one consisting of the chromosomes of larger size of n = 5 and their polyploid derivatives (sub-sections Parasorghum and Stiposorghum) and the other of the smaller sized chromosomes of n = 10 and their polyploid derivatives (sub-sections Eusorghum, Chaetosorghum and Heterosorghum). As with molecular markers, these cytological studies suggest a polyphyletic origin of Sorghum (i.e. a different ancestor for each mean chromosome size). To date, this work may best explain the two distinct lineages of Sorghum; however, the study did not include a large sampling of sister taxa to test this hypothesis.
Early studies employing molecular markers suggested that Sorghum contains two major clades; one exclusive to Stiposorghum and Parasorghum, and the other composed of the remaining sub-sections (Duvall and Doebley, 1990; Sun et al., 1994). Both of these studies included the sister taxon Cleistachne sorghoides, and both suggested that C. sorghoides is more closely related to one of the Sorghum clades than the two Sorghum clades are to each other, indicating that the genus may be para- or polyphyletic. A subsequent study, using the same gene sequences from a larger taxonomic sampling, affirmed the ambiguous relationships among Sorghum sub-sections (Dillon et al., 2001). This study supported the previous evidence that Sorghum is divided into two major lineages, with the Eu-, Chaeto- and Heterosorghum sections making up one lineage and the Para- and Stiposorghum sections making up the second. Additionally, Saccharum officinarum and C. sorghoides failed to resolve outside the Sorghum genus, again indicating para- or polyphyletic relationships.
Suggestions of paraphyly are not confined to Sorghum, but found throughout the Andropogoneae. While molecular phylogenetic data support the monophyly of the tribe, the relationships between genera have been difficult to resolve (Spangler et al., 1999; Mathews et al., 2002). Spangler et al. suggested that rapid radiation and the presence of continuous morphological characters are proving problematic in delineating relationships between genera. The authors used chloroplast ndhF sequences from 13 Sorghum species, and observed that Sorghum is split into three lineages most easily explained by geographical distribution. Again, these results indicated para- or polyphyly of Sorghum.
Despite the consistency of data suggesting Sorghum is para- or polyphyletic, several studies have argued the contrary. One found that S. officinarum resolved outside of Sorghum; however, as with the earlier studies based on internal transcribed spacer (ITS) sequence, C. sorghoides again resolved within the Para- and Stiposorghum lineage (Dillon et al., 2004). The authors argued for monophyly of the genus, but they supported a reduction of the sub-sections from five to three. The second study resolved Sorghum as one distinct genus composed of two major lineages; however, the authors determined that C. sorghoides resolved within their Eu-, Chaeoto- and Heterosorghum clade (Dillon et al., 2007). Cleistachne sorghoides has previously been reported as 2n = 36 chromosomes (Celarier, 1959), which is incongruent with 2n = 20 chromosomes of Eusorghum, and thus its inclusion within Sorghum may not be the most parsimonious explanation. Both studies included a limited number of Andropogoneae taxa outside of Sorghum and may, therefore, lack the power to evaluate monophyly of the genus. Further, all of these studies employed the ITS gene, which has been suggested to be problematic for resolving genus-level relationships due to rapid duplication and gene conversion events (Alvarez and Wendel, 2003). Recently, several low-copy nuclear gene sequences have proven to be phylogenetically informative in the Panicoideae (Estep et al., 2012). These sequences were used to reconstruct evolutionary relationships within the Andropogoneae in an effort to resolve genera-level relationships within the tribe (Estep et al., 2014). Here, we use the same low-copy nuclear genes from several Sorghum species, and combine these data with those of Estep et al. to test explicitly the generic limits of Sorghum.
MATERIALS AND METHODS
Plant material
We included 16 Sorghum species representative of all five sub-sections: Eusorghum, Heterosorghum, Chaetosorghum, Stiposorghum and Parasorghum (Table 1). Sorghum propinquum seed for accession PI653737 was obtained from the USDA Agricultural Research Service Plant Genetic Resources Conservation Unit (Griffin, GA, USA), and seed for the unnamed S. propinquum accessions was provided courtesy of Dr Bill Rooney (Texas A&M University, College Station, TX, USA). All Australian accessions were obtained from the Australian Tropical Grains Germplasm Centre (Biloela, Central Queensland, Australia). Verification of ploidy in S. halepense was determined via flow cytometry, performed in triplicate, in the Flow Cytometry Core Lab at the Benaroya Research Institute at Virginia Mason (Seattle, WA, USA). The genome sizes of all other accessions are reported in Price et al. (2005). Plants were grown in the WVU Life Sciences greenhouse under normal greenhouse conditions. Leaves were flash frozen in liquid nitrogen and stored at –80 °C.
Table 1.
Sorghum species included in this analysis with their respective accession ID, chromosome number and section
| Species | Accession | Chromosome number | Section |
|---|---|---|---|
| Sorghum angustum S. T. Blake | ausTRC 302605 | 10 | Stiposorghum |
| S. brachypodum Lazarides | ausTRC 302481 | 10 | Stiposorghum |
| S. ecarinatum Lazarides | ausTRC 302661 | 10 | Stiposorghum |
| S. exstans Lazarides | ausTRC 302557 | 10 | Stiposorghum |
| S. halepense (L.) Pers. | PI 663976 | 40 | Eusorghum |
| S. halepense (L.) Pers. | PI 271241 | 40 | Eusorghum |
| S. halepense (L.) Pers. | PI 302268 | 40 | Eusorghum |
| S. halepense (L.) Pers. | PI 663975 | 40 | Eusorghum |
| S. interjectum Lazarides | ausTRC 302445 | 30 | Stiposorghum |
| S. intrans F. Muell. Ex Benth. | ausTRC 302389 | 10 | Stiposorghum |
| S. laxiflorum Bailey | ausTRC 302510 | 40 | Heterosorghum |
| S. leiocladum (Hack.) C E. Hubb | ausTRC 300170 | 10 | Parasorghum |
| S. macrospermum Garber | ausTRC 302367 | 40 | Chaetosorghum |
| S. matarankense Garber & Snyder | ausTRC 302637 | 10 | Parasorghum |
| S. plumosum (R. Br.) P. Beauv. | ausTRC 302635 | 40 | Stiposorghum |
| S. propinquum (Kunth) Hitch. | unnamed | 20 | Eusorghum |
| S. propinquum (Kunth) Hitch. | PI 653737 | 20 | Eusorghum |
| S. purpureosericeum (A. Rich) | ausTRC 318068 | 10 | Parasorghum |
| S. stipoideum (Ewart & Jean White) | ausTRC 302614 | 10 | Stiposorghum |
Molecular techniques
Frozen leaf tissue was ground with a mortar and pestle, and DNA was extracted using either the Promega Wizard Genomic DNA Purification Kit (Madison, WI, USA) or the QIAGEN DNeasy Plant Mini Kit (Germantown, MD, USA) following the manufacturer’s protocol. Eight low-copy nuclear gene sequences were employed in this study and are as follows: Erect Panicle 2 (EP2_ex7 and EP2_ex8), Liguleless 1 (LIG1), Vanishing Tassel 2 (VT2), Ramosa 1 (RA1), Ramosa 2 (RA2), Dwarf 8 (D8) and Aberrant Panicle Organization 1 (APO1). The PCR primer sequences used are as described in table 1 of Estep et al. (2012), with the exception of those for RA1 and RA2. Both RA1 and RA2 were unreliably amplified, probably due to sequence divergence in the species more distantly related to S. bicolor. Therefore, new primer sequences for RA1 (F, AGCTCAGCTTTGGTGTATAT; R, TAAGCTGAAGATCCAGACG) and RA2 (F, CACCAGCAACAACTCGGCC; R, GAGGCGCTGATGGCATTCAC) were designed by aligning conserved coding sequences for the same gene regions from other grasses.
Polymerase chain reactions contained 50 ng of template DNA, dimethylsulphoxide (DMSO), 0·5 m betaine, 1× Mg-included Taq buffer, 0·8 mm dNTPs, 0·4 pm each forward and reverse primer, and 0·05 U µL–1 of Taq polymerase (Denville Scientific, South Plainfield, NJ, USA). Reaction conditions were as follows: initial denaturation at 94 °C for 5 min, 32 cycles of denaturation at 94 °C for 1 min, annealing at 5 °C less than the melting temperature for 1 min, and elongation of 72 °C for 1 min, followed by a final extension of 72 °C for 5 min. A touchdown protocol was used for reactions that were difficult to amplify. The touchdown protocol included three rounds of five cycles per round beginning with an annealing temperature of the primer-specific melting temperature in the first round and reducing the annealing temperature by 2 °C in each subsequent round. These initial 15 cycles were followed by 25 cycles with an annealing temperature 10 °C less than the primer melting temperature.
Amplification products were resolved on 1·5 % agarose gels. Bands were excised and purified using the Invitrogen DNA Pure Link Quick Gel Extraction kit (Carlsbad, CA, USA), following the manufacturer’s protocol. Purified PCR products were cloned using the Invitrogen TOPO TA Cloning Kit and transformed into One Shot Top 10 electrocompetent Escherichia coli cells. Cells were plated on LB agar containing kanamycin, and positive transformants were selected via blue/white screening. Plasmid DNA from eight clones for diploids and 16 clones for polyploids was extracted for sequencing. Each sequencing reaction contained approx. 300–500 ng of template, 1/16× BigDye [BigDye Terminator v.3.1. Cycle Sequencer Kit (Austin, TX, USA), 1× buffer, 0·4 pm M13 forward or 0·4 pm M13 reverse primer and 0·5 m betaine. Sequencing reaction conditions were as follows: 45 cycles of 1 min at 96 °C, 30 s at 50 °C and 4 min at 60 °C. Sequencing reactions were purified by ethanol precipitation and sequenced at the West Virginia University Genomics Core Facility using the ABI 3130XL.
Data analysis
Sequence data were vector screened and trimmed of both vector and primer sequences, and aligned using MUSCLE (Edgar, 2004). Alignments were manually inspected and adjusted in Bioedit v7.3.5 (Hall, 1999) to ensure that the amino acid alignment was maintained and that no aberrant alignment errors were included. Redundant (identical) alleles were removed for each gene from each taxon. All trimmed and inspected reads used in this study are available via GenBank (accession nos KR493932–KR494220). Data statistics, as reported in Table 2, were generated in PAUP* v.4.0b10 (Swofford, 2003). Parsimony analyses on single gene alignments were performed in PAUP using a heuristic search. All nucleotide characters were included, unweighted gaps were treated as missing data, and trees were rooted with outgroup sequences from Zea mays. Likelihood analyses on single gene alignments were performed in RAxML v.8 (Stamatakis, 2006), as described below. The resulting parsimony and likelihood trees were used to determine genome-specific paralogues in polyploid and heterozygous diploid taxa for informing concatenation of alleles into a single multigene matrix for further analysis, as described in Estep et al. (2014). Accessions that were homozygous for all genes (e.g. S. propinquum, S. bicolor and S. leiocladum) were represented by a single allele. Further, the Sorghum sequences for EP2_ex7, EP2_ex8, D8 and APO were combined with that of Andropogoneae from Estep et al. (2014) (courtesy of Dr Elizabeth Kellogg, Danforth Plant Science Center) for analyses aimed at delineating the placement of Sorghum within the tribe.
Table 2.
Phylogenetic summary statistics and parsimony or likelihood results for the eight low-copy nuclear loci used in the analysis
| Locus | Aligned length (bp) | Variable characters | Parsimony informative characters | No. of best trees | Score of best tree |
|---|---|---|---|---|---|
| APO1 | 766 | 90 | 70 | 1498 | 134 |
| D8 | 1018 | 150 | 96 | 190 | 207 |
| EP2_ex7 | 991 | 184 | 82 | 14 | 225 |
| EP2_ex8 | 809 | 130 | 59 | 30 | 166 |
| LG1 | 833 | 163 | 99 | N/A | N/A |
| RA1 | 402 | 108 | 60 | 1152 | 133 |
| RA2 | 755 | 86 | 46 | 12 | 105 |
| VT2 | 999 | 329 | 206 | N/A | N/A |
The number of best trees and score of best tree were determined using a heuristic search in PAUP*. Due to computational intensity, single gene trees for LG1 and VT2 were generated in RAxML.
A maximum likelihood analysis on both the Sorghum and Andropogoneae concatenated alignments was performed in RAxML v.8 using the GTR + gamma substitution model over two threads, and employing the autoMRE function for bootstrap replication. For the Sorghum alignment, Z. mays was designated as the outgroup, and for the Andropogoneae alignment, Paspalum and Plagiantha were designated as outgroups. Bayesian analysis of both alignments was performed in MrBayes v3.2.3 using rates = invgamma and nst = 6 (Ronquist and Huelsenbeck, 2003). Two separate runs of 50 million generations were performed, sampling each run every 1000 generations. Trees were visualized in FigTree v1.4.2.
RESULTS
Amplification of the eight loci from the accessions listed in Table 1 resulted in a total of 6573 characters, of which 724 (11 %) are parsimony informative. These genes, physically located on nine of the ten maize chromosomes in the B73 reference, are known to play a role in shaping morphological and inflorescence diversity and have been previously identified as useful loci for the determination of evolutionary relationships among grasses (Estep et al., 2012). The percentage of parsimony informative characters ranged from 6·1 to 20·6 %; a particularly high number of informative characters was obtained for VT2, which contains two diverse introns. Details for each of the genes are listed individually in Table 2.
Individual gene trees were constructed in PAUP for the determination of genome-specific paralogues. Seven of the eight individual gene trees supported two distinct clades, one containing Eu/Chaeto/Heterosorghum (henceforth called Clade I), and a second containing Stipo/Parasorghum (Clade II). The gene tree for RA1 placed half of the S. macrospermum and S. laxiflorum alleles in Clade I with strong bootstrap support (BS = 93) and the other half in Clade II with weak bootstrap support (BS = 42). Interestingly, two of the S. macrospermum alleles (a polyploid) always resolved with two of the S. laxiflorum alleles (also polyploid), suggesting orthologous relationships. General relationships within Eusorghum were consistent across all trees, resolving one of the S. propinquum accessions (PI 653737) sister to S. bicolor, and placing the second S. propinquum (unnamed) basal to all other Eusorghum taxa. Placement of various S. halepense alleles differed slightly among trees, but they were always sister to the S. bicolor + S. propinquum PI 653737 clade. Relationships within Clade II were more difficult to distinguish and less consistent across trees, probably due to limited phylogenetic signal leading to very short branch lengths; however, a few relationships were well supported. In all trees, one allele from S. matarankense clustered with one allele of S. interjectum; S. intrans and S. exstans were sister to one another. and the S. plumosum alleles were dispersed throughout the entirety of Clade II.
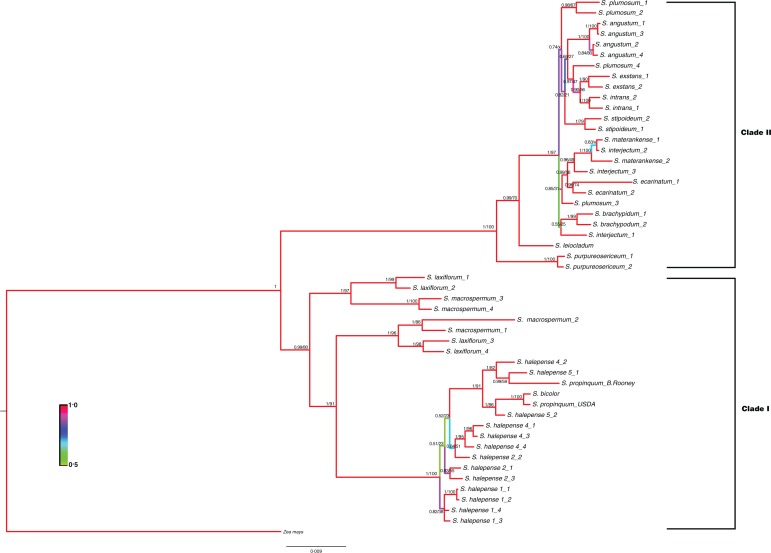
Upon determination of genome-specific paralogues from the individual gene trees, all loci were concatenated into a single data matrix for both maximum likelihood and Bayesian analyses (Fig. 1). Both analyses revealed very strong support for Clade 1 [posterior probability (PP) = 0·99, BS = 80] and Clade II (PP = 1, BS = 100). Sorghum macrospermum and S. laxiflorum are divided into two basal groups in Clade I, each containing half of the alleles from each species. One of the alleles from an S. halepense accession (PI 663975) resolved with S. propinquum (PP = 0·99, BS = 59), while the other is sister to S. bicolor (PP = 1, BS = 86), as would be expected given the widespread assumption that these are the genome donors to the polyploid S. halepense (Paterson et al., 1995). In Clade II, there is strong support for the basal placement of S. purpureosericeum (PP = 1, BS = 100), a member of the subgenus Parasorghum. The only other Parasorghum species used in this study, S. matarankense, resolved firmly within the Stiposorghum and sister to one of the S. interjectum alleles (PP = 1, BS = 100). Although support on internal branches throughout Clade II is weak, some relationships were still apparent, such as the sister relationship between S. intrans and S. exstans, and the resolution of S. ecarinatum as sister to the S. matarankense + S. interjectum clade.
Fig. 1.
Sorghum phylogenetic tree derived from Bayesian analysis of eight low-copy nuclear loci. For each branch, Bayesian posterior probabilities (PP) and maximum likelihood bootstrap (BS) scores are given in the form of PP/BS. Branch colours indicate higher (red) to lower (green) posterior probability. An asterisk, *, indicates bootstrap scores <50. An × indicates that the maximum likelihood analysis did not support the same topology.
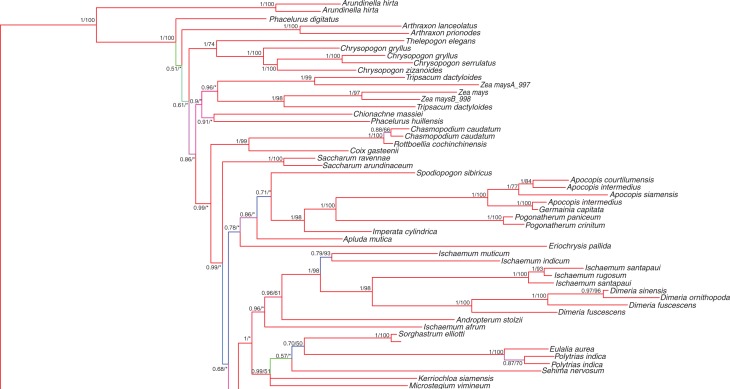
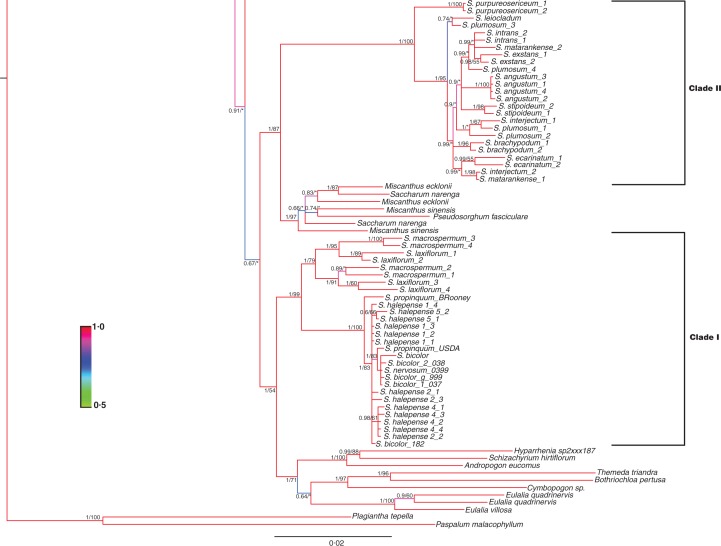
The Sorghum data for APO1, the two exons from EP2, and D8 were then combined with the same sequence data for numerous members of the Andropogoneae (Estep et al., 2014). The topology for the basal Andropogoneae was highly congruent with that of Estep et al. (see fig. S2 in Estep et al., 2014) with a few minor exceptions (Fig. 2), probably due to the exclusion of sequence data for Retarded palea1 (REP1), which was part of the Estep et al. (2014) analysis; however, the inclusion of the additional Stipo- and Parasorghum species improved internal branch support in this part of the Andropogoneae tree. A sister relationship between Sorghum Clade II and the Miscanthus + Saccharum clade resolves with strong support (PP = 1, BS = 93). Sorghum Clades I and II are polyphyletic, with posterior probability support for their separation at 0·67, although bootstrap support at this internal node was <50 %. Nevertheless, there is strong evidence that the Saccharum +Miscanthus clade is firmly nested within Sorghum, and evidence for a sister relationship between Clade I and several members of the core Andropogoneae.
Fig. 2.
Combined Bayesian tree including four low-copy nuclear loci showing the placement of Sorghum within Andropogoneae. Branch support is represented by the Bayesian posterior probability (PP)/maximum likelihood bootstrap (BS) score. Branch colours indicate higher (red) to lower (green) posterior probability. An asterisk, *, indicates bootstrap scores <50. An × indicates that the maximum likelihood analysis did not support the same topology. Sorghum Clades I and II are delineated on the right.
DISCUSSION
Maximum likelihood and Bayesian analyses of the combined eight-locus Sorghum-specific data set support interspecific relationships as previously described using various morphological and molecular markers (Garber, 1950; Duvall and Doebley, 1990; Spangler, 2003; Dillon et al., 2004, 2007; Price et al., 2005; Ng’uni et al., 2010; Liu et al., 2014). Specifically, Sorghum is divided into two major clades, one containing Eu/Chaeto/Heterosorghum (Clade I) and the other composed of the Stipo/Parasorghum (Clade II). Sub-section Eusorghum, which includes the diploids S. bicolor and S. propinquum and the polyploid S. halepense, is strongly supported. Sorghum halepense, better known as ‘Johnsongrass’, is thought to have originated via hybridization between S. bicolor and S. propinquum, subsequently followed by polyploidization (Paterson et al., 1995). Cytogenetic evidence also supports S. propinquum as one of the S. halepense progenitors (Magoon and Shambulingappa, 1961). We included four S. halepense accessions in this study, only one of which clusters with S. propinquum with strong support. It should be noted, however, that we originally obtained ten geographically distinct S. halepense accessions from the USDA-GRIN seed repository, but during our grow-out we noticed a wide range in both seed and plant morphologies. Therefore, we measured the genome size of each of these accessions and found that only the four accessions used in this study contained the nuclear DNA content expected for S. halepense (1C approx. 1600 Mb). This suggests that the phenotypic characteristics associated with S. halepense may also easily arise in diploid genotypes, and therefore particular care should be taken in species identification before use in phylogenetic analyses. Further, given that only one of the four S. halepense accessions showed a strongly supported relationship with S. propinquum, it is also possible that polyploid S. halepense has arisen more than once via disparate pathways, and that this convergence may confound inferences regarding its origin.
Sorghum macrospermum and S. laxiflorum, the single species belonging to Chaeto- and Heterosorghum, respectively, are closely related polyploids of n = 20 belonging to Clade I. In the work presented here, we show clear genome-specific association of orthologous alleles. Specifically, two of the alleles from S. macrospermum cluster with two of the alleles from S. laxiflorum with higher support than with the remaining S. macrospermum alleles. This implies that these species may have originated from a single polyploidization event or from separate polyploidization events involving the same or similar parental species, and that it is appropriate to merge them into a single subgenus. Although several studies have suggested the merger of these two subgenera (Sun et al., 1994; Ng’uni et al., 2010), our results provide compelling evidence to support such a reclassification. Further, given the clear interspecific pairing of orthologues, our data do not support the proposal to retain these species as distinct sections within a single sub-section, as suggested by Liu et al. (2014). In addition, our results provide strong evidence for the sister relationship of these species with Eusorghum (PP = 1, BS = 99), and therefore do not support the proposal by Spangler to classify these species as a distinct genus (Vacoparis).
Clade II, composed of Para- and Stiposorghum, is also strongly supported. Due to low seed viability, only two Parasorghum species were included in this study. Sorghum purpureosericeum resolved as the basal lineage of Clade II in all of our analyses. This result was expected because (1) Parasorghum is considered ancestral to Stiposorghum, and (2) S. purpureosericeum is the only Clade II taxon included in this study that is endemic to an area outside of Australia. The second Parasorghum species, S. matarankense, resolved within the Stiposorghum with strong support in all of our analyses. Indeed, the two heterozygous S. matarankense alleles consistently clustered with S. interjectum in all individual locus trees and with S. interjectum and the S. intrans + S. exstans clade in both of our combined loci analyses, suggesting either that S. matarankense belongs to Stiposorghum or that Parasorghum is paraphyletic. We note that S. matarankense was originally circumscribed within Stiposorghum (Garber, 1950), was included within Stiposorghum by Spangler (2003) and was only more recently placed within Parasorghum.
The topology within Stiposorghum resolved with high posterior probability scores but low bootstrap values, making relationships more difficult to delineate. Nevertheless, some relationships were apparent even in the single gene analyses, and were not only strikingly congruent with the classification of Australian endemics by Lazarides et al., and Spangler based on morphology and geographic distribution, but also with the taxonomic treatment of the genus by Garber (Garber, 1950; Lazarides et al., 1991, Spangler, 2003). For example, Lazarides discusses polyploid S. interjectum and considers it similar to S. plumosum, while Spangler considered S. interjectum synonymous with S. plumosum. Our results show alleles from both species clustering with high support. Lazarides also lists an S. plumosum × S. intrans hybrid from the Northern Territory (see Table 2), and we also see clustering of one of the S. plumosum alleles with the S. exstans + S. intrans clade. Sorghum plumosum, S. extans and S. intrans are narrowly distributed in the northern part of Australia’s Northern Territory, providing further support for these species relationships (see figs 7 and 8 of Spangler, 2003). Finally, Lazarides suggested that S. intrans, S. exstans and S. angustum are the most geographically restricted and morphologically specialized, and that S. intrans was probably derived from S. stipoideum. Our Bayesian analysis places S. intrans + S. exstans + S. angustum in a clade sister to S. stipoideum.
The monophyly of Sorghum and its placement within the Andropogoneae has been a contentious topic for several decades, in part due to difficulties in phylogenetic reconstruction in the face of rapid radiation of the tribe leading to continuity in morphological variation (Mathews et al., 2002; Estep et al., 2014). Early molecular and cytogenetic evidence provided weak support for a para- or polyphyletic origin of Sorghum, and suggested that the genus should possibly be reclassified to accommodate distinctions among the subgenera (Duvall and Doebley, 1990; Sun et al., 1994; Spangler et al., 1999; Dillon et al., 2001). Duvall and Doebley noted that the Australian species are highly diverged from the other Sorghum species in comparison with divergence rates among other angiosperms, implying that the Australian species may warrant distinct generic status. This idea was echoed by Spangler et al. almost 10 years later when analyses of NDHF sequences led them to conclude that the Australian group is ‘distinct enough to be proposed as a separate taxon’. Shortly thereafter, however, several studies employing a larger number of molecular markers disputed these early findings, and indicated that Sorghum is indeed monophyletic, although these studies included a very limited number of non-Sorghum species for comparison (Dillon et al., 2004, 2007; Price et al., 2005; Ng’uni et al., 2010; Liu et al., 2014).
Our results are congruent with those of Duvall and Doebley, as well as with Spangler et al., indicating that Sorghum is polyphyletic and supporting reclassification of Clade II (Para/Stiposorghum) as a distinct genus, Sarga (Duvall and Doebley, 1990; Spangler et al., 1999; Spangler, 2003). In both our Bayesian and maximum likelihood analyses, there exists strong support for a distinction between these clades. The Sarga clade is sister to Saccharum + Miscanthus with a posterior probability score of 1 and bootstrap score of 87 % (Fig. 2). Branch support for the split between this group and the clade containing Eu/Chaeto/Heterosorghum is less convincing, with a posterior probability score of 0·67 but bootstrap score of <50 %. Nevertheless, the sister relationship of Sarga with Miscanthus + Saccharum is evident, and demonstrates a clear polyphyletic relationship within Sorghum.
Concluding remarks
The work presented here represents the most comprehensive study of Sorghum placement within Andropogoneae to date. Our results are congruent with those of early morphological, cytogenetic and molecular studies arguing that Sorghum is polyphyletic. Given the clear polyphyletic placement of Sorghum, the distinct base chromosome number for Clade II (n = 5) and the strong node support for the sister relationship between Clade II and Miscanthus + Saccharum, our results support the proposal by Spangler to adopt ‘Sarga’ as a distinct genus composed of Para- and Stiposorghum (Spangler, 2003). Within Sorghum, our data suggest paraphyly of Parasorghum, but, given the historical variation in placement of S. matarankense within the Stiposorghum, we can neither support nor refute the monophyly of this sub-section. Our results do, however, suggest that reclassification of Hetero- and Chaetosorghum as a single sub-section is warranted, but do not support its circumscription as a distinct genus (e.g. Vacoparis).
ACKNOWLEDGEMENTS
The authors wish to thank Elizabeth Kellogg, Matt Estep and Mike McKain for providing the Andropogoneae sequence alignment, Bill Rooney for providing Sorghum propinquum seed, and Jonah Joffee for his involvement in generating some sequence data for his REU research project. We acknowledge the use of the Super Computing System Spruce Knob at West Virginia University, which is funded in part by the National Science Foundation EPSCoR Research Infrastructure Improvement Cooperative Agreement 1003907, the state of West Virginia (WVEPSCoR via the Higher Education Policy Commission) and West Virginia University. This work was partially funded by NSF-REU DBI 0849917, Plant Responses to the Environment: From Genes to Ecosystems, to the Department of Biology at West Virginia University.
LITERATURE CITED
- Alvarez I, Wendel JF. 2003. Ribosomal ITS sequences and plant phylogenetic inference. Molecular Phylogenetics and Evolution 29: 417–434. [DOI] [PubMed] [Google Scholar]
- Celarier RP. 1959. Cytotaxonomy of the Andropogoneae. III. Subtribe Sorgeae, genus Sorghum. Cytologia 23: 395–418. [Google Scholar]
- Dahlberg J. 2000. Classification and characterization of Sorghum. In: Smith CW, Frederiksen RA, eds. Crop Science. Sorghum: origin, history, technology, and production. Chichester, UK: John Wiley & Sons, 99–130. [Google Scholar]
- De Wet JMJ, Harlan JR. 1971. The origin and domestication of Sorghum bicolor. Economic Botany 25: 128–135. [Google Scholar]
- De Wet JMJ, Huckabay JP. 1967. The origin of Sorghum bicolor. II. Distribution and domestication. Evolution 21: 787–802. [DOI] [PubMed] [Google Scholar]
- Dillon SL, Lawrence PK, Henry RJ. 2001. The use of ribosomal ITS to determine phylogenetic relationships within Sorghum. Plant Systematics and Evolution 230: 97–110. [Google Scholar]
- Dillon SL, Lawrence PK, Henry RJ, Ross L, Price HJ, Johnston JS. 2004. Sorghum laxiflorum and S. macrospermum, the Australian native species most closely related to the cultivated S. bicolor based on ITS1 and ndhF sequence analysis of 25 Sorghum species. Plant Systematics and Evolution 249: 233–246. [Google Scholar]
- Dillon SL, Shapter FM, Henry RJ, Cordeiro G, Izquierdo L, Lee LS. 2007. Domestication to crop improvement: genetic resources for Sorghum and Saccharum (Andropogoneae). Annals of Botany 100: 975–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doggett H. 1988. Sorghum, 2nd edn New York: John Wiley & Sons. [Google Scholar]
- Duvall MR, Doebley JF. 1990. Restriction site variation in the chloroplast genome of Sorghum (Poaceae). Systematic Botany 15: 472–480. [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estep MC, Diaz DV, Zhong J, Kellogg EA. 2012. Eleven diverse nuclear-encoded phylogenetic markers for the subfamily Panicoideae (Poaceae). American Journal of Botany 99: e443–e446. [DOI] [PubMed] [Google Scholar]
- Estep MC, McKain MR, Diaz DV, et al. 2014. Allopolyploidy, diversification, and the Miocene grassland expansion. Proceedings of the National Academy of Sciences, USA 111: 15149–15154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber ED. 1950. Cytotaxonomic studies in the genus Sorghum. University of California Publications in Botany 23: 283–361. [Google Scholar]
- Hall TA. 1999. Bioedit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98. [Google Scholar]
- Lazarides M, Hacker JB, Andrew MH. 1991. Taxonomy, cytology, and ecology of indigenous Australian Sorghums (Sorghum Moench: Andropogoneae: Poaceae). Australian Systematic Botany 4: 591–635. [Google Scholar]
- Liu Q, Liu H, Wen J, Peterson PM. 2014. Infrageneric phylogeny and temporal divergences in Sorghum (Andropogonodae, Poaceae) based on low-copy nuclear and plastid sequences. PLoS One 9: e104933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magoon ML, Shambulingappa KG. 1961. Karyomorphology of Sorghum propinquum and its bearing on the origin of 40-chromosome Sorghum. Chromosoma 12: 460–465. [DOI] [PubMed] [Google Scholar]
- Mathews S, Spangler RE, Mason-Garner RJ, Kellogg EA. 2002. Phylogeny of Andropogoneae inferred from Phytochrome B, GBSSI, and ndhF. International Journal of Plant Sciences 163: 441–450. [Google Scholar]
- Ng’uni D, Geleta M, Fatih M, Bryngelsson T. 2010. Phylogenetic analysis of the genus Sorghum based on combined sequence data from cpDNA regions and ITS generate well-supported trees with two major lineages. Annals of Botany 105: 471–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH, Schertz KF, Lin Y-R, Liu S-C, Chang Y-L. 1995. The weediness of wild plants: molecular analysis of genes influencing dispersal and persistence of johnsongrass, Sorghum halepense (L.) Pers. Proceedings of the National Academy of Sciences, USA 92: 6127–6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price HJ, Dillon SL, Hodnett G, Rooney WL, Ross L, Johnston S. 2005. Genome evolution in the genus Sorghum (Poaceae). Annals of Botany 95: 219–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- Snowden JD. 1935. A classification of the cultivated Sorghum. Kew: Royal Botanic Gardens. [Google Scholar]
- Snowden JD. 1936. The cultivated races of Sorghum. London: Allard and Son. [Google Scholar]
- Spangler RE. 2003. Taxonomy of Sarga, Sorghum, and Vacoparis (Poaceae: Andropogoneae). Australian Systematic Botany 16: 279–299. [Google Scholar]
- Spangler RE, Zaitchik B, Russo E, Kellogg EA. 1999. Andropogoneae evolution and generic limits in Sorghum (Poaceae) using ndhF sequences. Systematic Botany 24: 267–281. [Google Scholar]
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. [DOI] [PubMed] [Google Scholar]
- Sun Y, Skinner DZ, Liang GH, Hulbert SH. 1994. Phylogenetic analysis of Sorghum and related taxa using internal transcribed spacers of nuclear ribosomal DNA. Theoretical and Applied Genetics 89: 26–32. [DOI] [PubMed] [Google Scholar]
- Swofford DL. 2003. Phylogenetic analysis using parsimony (*and other methods). Version 4. Sunderland, MA: Sinauer Associates. [Google Scholar]