Abstract
Islet autoantibodies detected at disease onset in patients with type 1 diabetes are signs of an autoimmune destruction of the insulin-producing β-cells. To further investigate the genetic determinants of autoantibody positivity, we performed dense immune-focused genotyping on the Immunochip array and tested for association with seven disease-specific autoantibodies in a large cross-sectional cohort of 6,160 type 1 diabetes–affected siblings. The genetic association with positivity for GAD autoantibodies (GADAs), IA2 antigen (IA-2A), zinc transporter 8, thyroid peroxidase, gastric parietal cells (PCAs), tissue transglutaminase, and 21-hydroxylase was tested using a linear mixed-model regression approach to simultaneously control for population structure and family relatedness. Four loci were associated with autoantibody positivity at genome-wide significance. Positivity for GADA was associated with 3q28/LPP, for IA-2A with 1q23/FCRL3 and 11q13/RELA, and for PCAs with 2q24/IFIH1. The 3q28 locus showed association after only 3 years duration and might therefore be a marker of persistent GADA positivity. The 1q23, 11q13, and 2q24 loci were associated with autoantibodies close to diabetes onset and constitute candidates for early screening. Major susceptibility loci for islet autoantibodies are separate from type 1 diabetes risk, which may have consequences for intervention strategies to reduce autoimmunity.
Introduction
Diabetes-associated insulin autoantibodies, IA2 antigen (IA-2A), 65-kDa GAD (GADAs), and zinc transporter 8 (ZnT8A), are detected in a majority of patients at the time of diagnosis of type 1 diabetes (1), and are a distinctive signature of the autoimmune process that precedes the disease (2–4). Their role in the pathogenesis, however, is unclear. Autoantibodies present at onset are associated with age, sex, and specific HLA genotypes (5–7). Apart from the HLA genes, only a limited number of loci have been robustly associated with autoantibodies in type 1 diabetes. Two recent genome-wide association scans (GWAS) identified the FCRL3 gene locus on chromosome 1q23 as being associated with IA-2A and ZnT8A in European type 1 diabetes patients (8,9).
Recent GWAS studies have highlighted shared susceptibility loci between type 1 diabetes and autoimmune diseases such as celiac disease (CD), autoimmune thyroid disease (AITD), and rheumatoid arthritis (RA) (10–12). This is likely due to an overlap in the causal genes and pathways that control general autoimmunity. To identify genetic factors controlling autoimmunity, we took advantage of Immunochip genotyping data available for 6,160 type 1 diabetes–affected siblings from multiplex families collected by the Type 1 Diabetes Genetics Consortium (T1DGC) (13). The Immunochip is a custom genotyping array designed to capture the overlap of susceptibility loci identified across 12 immune-mediated diseases. Hence, in addition to investigating the genetic association of the diabetes-specific islet autoantibodies IA-2A, GADA, and ZnT8A, we also investigated the genetic susceptibility of autoantibodies in four additional autoimmune diseases known to cosegregate in type 1 diabetes families directed against the following: thyroid peroxidase (TPOA) associated with AITD, gastric parietal cell antibodies (PCAs) associated with autoimmune gastritis, tissue transglutaminase (TGA) associated with CD, and 21-hydroxylase (OH21A) associated with autoimmune hypoadrenalism.
Research Design and Methods
Type 1 Diabetes Cohort
An overview of the study can be seen in Supplementary Fig. 1. A total of 7,077 type 1 diabetes–affected siblings from 4,134 multiplex families were available from the T1DGC, and data were collected by four regional networks. Eighty-four percent of individuals were of Caucasian ethnicity; 10.4% of black or African American ethnicity; 5.4% of Asian ethnicity; and 0.3% of Native American, Native Alaskan, Hawaiian, or Pacific Islander ethnicity. The study was approved by review boards for all contributing institutions, and informed consent was obtained from all families. Inclusion criteria have been described previously (13). The median age at diagnosis was 9 years (SD 7.52), and the median disease duration at blood sampling was 7 years (SD 10.06), with 25% of the samples taken within 3 years.
Autoantibody Measurements
Autoantibodies were measured in serum that had been stored at −80°C. GADA, IA-2A, TPOA, TGA, and OH21A were measured by radiobinding assays using labeled recombinant proteins according to standard protocols. GADAs and IA-2A were measured in two different laboratories (Denver, CO, and Bristol, U.K.), while TPOA, TGA, OH21A, PCAs, and ZnT8A were all measured in the Denver laboratory. The threshold values were obtained from the Diabetes Autoantibody Harmonization Program of the National Institute of Diabetes and Digestive and Kidney Diseases (14), following conversion of local assay values (15) to digestive and kidney units (DK units) per milliliter after reassay of a large number of samples using the harmonized method in both laboratories. The thresholds for GADAs were 20 DK units/mL in Denver and 33 DK units/mL in Bristol. The sensitivity for GADAs in samples from 100 patients with recent-onset type 1 diabetes was 83% in Denver and 81% in Bristol, at a specificity of 97% in 974 control samples. For IA-2A, the threshold was 5 DK units/mL (Denver) and 2 DK units/mL (Bristol). The sensitivity in 50 recent-onset type 1 diabetes patients was 64% at specificities of 99.4% (Denver) and 99.2% (Bristol) in 500 control samples. TPOA was measured with a commercial kit from Kronus (Star, ID). TPOA levels were divided into the following three groups; negative (<1.0 World Health Organization [WHO] unit/mL), indeterminate (1–5 WHO units/mL), and positive (≥5 WHO units/mL). For TGA, the upper limits of normal (0.050) were established as the 100th percentile from receiver operating characteristic curves in 184 healthy control subjects, and European Medicines Agency standards for positivity and negativity among the patients with diabetes (n = 859). For OH21A, the upper limits of normal (0.150) were established as the 100th percentile of 241 healthy control subjects.
PCA assays were performed with a construct of the major intracellular domain of the human ATPase 4A subunit. 35S Met–labeled translation products were generated from pCDNA3.1 directional TOPO vector and incorporated into an overnight immunoprecipitation/filtration assay.
The sensitivity for the assay determined a series of 230 sera from nondiabetic healthy subjects alongside 100 recent-onset diabetic sera was 14.6% at 100% specificity, and 23.4% at 96%. In the DASP 2010 samples, the assay achieved a sensitivity of 20% at 100% specificity, and a sensitivity of 36% at 96% specificity. The latter corresponds to a cutoff index of 0.02 as applied in current analyses.
ZnT8 immunoprecipitation assays were performed with dimeric constructs of the COOH-terminal cytosolic domain of the human ZnT8 cDNA (amino acids 275–369) incorporating the two polymorphic variants at amino acid 325 separated by a flexible linker, namely CR/CW heterodimer (JH6.2), and the homodimers CRCR (JH6.4) and CWCW (JH6.5). 35S Met–labeled translation products were generated from pCMVTNT vector and incorporated into an overnight immunoprecipitation/filtration assay. The sensitivity for each assay at 99% specificity was determined for a series of 100 sera from recent-onset type 1 diabetes patients and 230 sera from nondiabetic subjects, and was 65% for CWCR, 60% for CRCR, and 50% for CWCW. The assays have been validated in 2009 and 2010 DASP workshops (16). All 7,077 case patients were measured for GADA and IA-2A, while 4 case patients had missing measurements for TPOA, TGA, and OH21A; and 5 case patients had missing measurements for PCA. A subset of 1,504 case patients had ZnT8A measurements available. Their disease duration was <2 years and a median age at disease onset of 11 years.
Genotyping and Quality Control
Of the 7,077 affected siblings available, 7,039 were genotyped for 183,546 single nucleotide polymorphisms (SNPs) on the Immunochip. Genotypes were called using GenomeStudio (Illumina). Initial sample quality control metrics included sample call rate, and concordance check of reported gender versus genotyped sex (using ∼2,000 SNPs on X- and Y-chromosomes to infer the sex). Relationship inference was performed using the relationship inference method implemented in KING (17) and was compared with reported pedigree data. After cryptic relatedness was identified, pedigree errors were resolved by removing problematic individuals (within families) and/or by reconstructing the pedigree (both within and across families) incorporating the newly identified first-degree and second-degree relationships. Ten case patients were removed for having a missing rate >5%. Eighteen case patients were removed for gender errors. At the SNP level, monomorphic markers (∼15,000) were first removed, a total of 1,387 SNPs were rejected due to a genotyping call rate of <95%, and 2,939 SNPs failed the Mendelian inconsistency (MI) errors (with a standard MI error rate of >0.5% or an adjusted MI error rate of >5% for rarer variants). A total of 165,118 variants passed quality control analysis. Additional quality control was performed for the 6,160 case patients with autoantibody data, removing SNPs with a call rate of <95% and a minor allele frequency (MAF) of <1% in this subset. All individuals had a call rate of >95%. The SNPs were not filtered for deviations from the Hardy-Weinberg equilibrium (HWE). Selection due to disease association can cause deviation from the HWE in affected offspring, as can population stratification, both of which were present in the data. Instead of excluding SNPs based on HWE, and risking missing the true association signals in regions associated with type 1 diabetes, all SNPs were included but were checked carefully for deviations among the loci that were significantly associated with autoantibody positivity. After filtering, 136,078 SNPs were retained in the analysis, and the mean genotyping success rate in individuals was 99.9%. The quality filtering was performed in PLINK version 1.07 (18).
Statistical Analyses
As the samples were collected from a multiethnic cohort, the population structure was investigated by principal component (PC) analysis and multidimensional scaling in PLINK (18) (Supplementary Fig. 2). The projection of the first two PCs corresponded to the self-reported primary ethnicity with one additional subgroup situated between the Caucasian and Asian populations. We then corrected for the confounding effects of family relatedness and population structure by applying a linear mixed model developed for GWAS (factored spectrally transformed linear mixed model; FaST-LMM) (19) that makes use of the estimated pairwise genetic similarity between all individuals. The association was tested, with autoantibody positivity as the outcome variable and SNP genotypes with additive allelic effects as predictors. Age at onset, diabetes duration, and sex were included as covariates in the regression model. P values below 5 × 10−08 were considered to have genome-wide significance. After the FaST-LMM analysis, there was no evidence of inflation of the test statistics (GADA λLMM = 0.95; IA-2A λLMM = 0.92; ZnT8A λLMM = 0.98; PCA λLMM = 0.94; TPOA λLMM = 0.96; TGA λLMM = 0.95; and OH21A λLMM = 0.98). Quantile-quantile plots of the −log10 P value distribution from the FaST-LMM analysis (PLMM) distribution showed deviations from the expected at the tail of the (Supplementary Fig. 3). To obtain odds ratios (ORs) and 95% CIs, logistic regression was performed and adjusted for family relatedness using generalized estimating equations (GEEs) (20) in the R package geepack (21). Family identification (ID) was used to identify clusters, an exchangeable working correlation matrix, and robust variance was used to test for association using the Wald test. The GEE models were adjusted for significant covariates and ethnicity by correcting for the first four PCs from the multidimensional scaling analysis. To test for independent association signals logistic regression with GEE conditional on the lead SNPs in each associated region was performed. Calculations of linkage disequilibrium (LD) was performed in PLINK using only 5,067 nondiabetic unrelated founders in data from the full family cohort available from the T1DGC. MAFs were compared with data from the 1000 Genomes project CEU population (Utah residents with Northern and Western European ancestry from the CEPH collection) (release 2011). After closer inspection, the following two association signals were discarded as being spurious findings: the SNP rs8072874 on chromosome 17 with a P value of 3.18 × 10−10 for TGA positivity, and seq-VH-3453 (rs78773831) on chromosome 4 with a P value of 6.19 × 10−9 for OH21A positivity. No other SNPs within the two loci showed signs of association with the traits (uncorrected P value >0.01). Both SNPs passed the HWE, but had low MAFs of 0.027 and 0.031, and only one, versus two, antibody-positive individuals were homozygous for the minor allele of rs8072874 and seq-VH-3453, respectively. Data from 2,572 T1DGC families with IA-2A measurements have previously been included in the study by Plagnol et al. (8), and data from 855 individuals with ZnT8A measurements were included in the study by Howson et al. (9).
Results
Autoantibody Positivity
The autoantibody prevalence and associated covariates in 7,077 affected siblings are summarized in Table 1. Briefly, positivity for GADAs was detected in 45.4%, for IA-2A in 47.1%, for TPOA in 25.2%, for PCA in 19.9%, for TGA in 7.1%, and for OH21A in 1.5% of the case patients, respectively. ZnT8A measurements obtained within 2 years after diagnosis were available for a subset of 1,504 cases, of which 57.3% were positive. A higher age at onset correlated positively with GADA, TPOA, and PCA positivity, but negatively with TGA positivity. The duration of diabetes correlated negatively with GADA, IA-2A, ZnT8A, and TGA, but positively with TPOA, PCA, and OH21A. Female sex was positively correlated with GADA, TPOA, PCA, and TGA (Table 1). Pairwise correlations between autoantibodies are shown in Supplementary Table 1.
Table 1.
Frequency of positivity for the seven autoantibodies and their association with age at onset, disease duration, and female sex
| % Positive | Age at onset |
Disease duration |
Female sex |
||||
|---|---|---|---|---|---|---|---|
| P value | OR (95% CI)* | P value | OR (95% CI)* | P value | OR (95% CI) | ||
| GADA (n = 7,077) | 45.4 | <2.0 × 10−16 | 1.92 (1.78–2.08) | <2.0 × 10−16 | 0.68 (0.64–0.72) | 3.0 × 10−15 | 1.53 (1.37–1.70) |
| IA-2A (n = 7,077) | 47.1 | NS | NS | <2.0 × 10−16 | 0.53 (0.49–0.57) | NS | NS |
| ZnT8A (n = 1,504) | 57.3 | NS | NS | 9.80 × 10−6 | 0.72 (0.62–0.83) | NS | NS |
| TPOA (n = 7,074) | 25.2 | 2.75 × 10−7 | 1.22 (1.13–1.32) | 1.83 × 10−11 | 1.21 (1.15–1.28) | <2.0 × 10−16 | 2.21 (1.97–2.49) |
| PCA (n = 7,073) | 19.9 | 1.62 × 10−8 | 1.27 (1.17–1.38) | <2.0 × 10−16 | 1.38 (1.30–1.46) | <2.0 × 10−16 | 1.75 (1.54–1.98) |
| TGA (n = 7,074) | 7.1 | 2.67 × 10−11 | 0.55 (0.47–0.66) | 0.00016 | 0.80 (0.71–0.90) | 0.0056 | 1.31 (1.08–1.58) |
| OH21A (n = 7,074) | 1.5 | NS | NS | 0.0088 | 1.22 (1.05–1.41) | NS | NS |
P value, OR, and 95% CI were calculated from logistic regression models with GEEs (20) to correct for family relatedness, and were adjusted for each covariate and population structure using the first four PCs calculated from the genetic data.
*ORs and CIs are reported for a 10-year difference for age at onset and disease duration, except for ZnT8A.
Genetic Associations of Autoantibody Positivity
The association of 136,078 high-quality SNPs with autoantibody positivity in 6,160 affected siblings was tested with the FaST-LMM statistic for GWAS data (19), which simultaneously controls for the genetic confounding due to population structure and relatedness. The −log10 PLMM values across all chromosomes for each autoantibody are shown in Supplementary Fig. 3. In total, four non-MHC loci were associated with autoantibody positivity at a stringent genome-wide significance level (P < 5 × 10−8), and one locus almost reached the Bonferroni correction limit (P < 3.7 × 10−07) (Table 2 and Fig. 1).
Table 2.
Loci demonstrating significant associations with autoantibody positivity
| SNP | Chr. | Locus | Nearest gene | MAF1000G | MAFT1D | PLMM | SNPweight | OR (95% CI)* | HWE† | |
|---|---|---|---|---|---|---|---|---|---|---|
| GADA | rs9815073 C>A | 3 | 3q28 | LPP | 0.385 | 0.334 | 1.30 × 10−8 | −0.038 | 0.78 (0.72–0.85) | 0.018 |
| rs59822814 A>T | 3 | 3q28 | LPP | 0.420 | 0.410 | 1.33 × 10−8 | 0.038 | 1.27 (1.17–1.37) | 0.016 | |
| rs1559810 C>A | 3 | 3q28 | LPP | 0.420 | 0.408 | 2.81 × 10−8 | 0.037 | 1.26 (1.17–1.36) | 0.025 | |
| rs6444284 T>C | 3 | 3q28 | LPP | 0.420 | 0.413 | 4.48 × 10−8 | 0.036 | 1.26 (1.16–1.36) | 0.031 | |
| IA-2A | rs3761959 G>A | 1 | 1q23 | FCRL3 | 0.477 | 0.435 | 1.53 × 10−19 | −0.056 | 0.70 (0.65–0.76) | 0.018 |
| rs7522061 T>C | 1 | 1q23 | FCRL3 | 0.477 | 0.457 | 7.96 × 10−18 | −0.054 | 0.72 (0.67–0.78) | 0.0065 | |
| rs2050568 C>T | 1 | 1q23 | FCRL1 | 0.483 | 0.458 | 6.97 × 10−14 | −0.048 | 0.74 (0.69–0.80) | 0.00016 | |
| rs2777965 T>G | 1 | 1q23 | FCRL4 | 0.385 | 0.353 | 3.66 × 10−10 | −0.039 | 0.77 (0.71–0.84) | 0.11 | |
| rs7538531 T>C | 1 | 1q23 | FCRL1 | 0.483 | 0.404 | 3.18 × 10−9 | −0.038 | 0.79 (0.73–0.86) | 0.015 | |
| rs6696137 T>C | 1 | 1q23 | FCRL1 | 0.483 | 0.397 | 4.77 × 10−9 | −0.037 | 0.79 (0.74–0.86) | 0.070 | |
| rs12749630 T>C | 1 | 1q23 | FCRL1 | 0.483 | 0.397 | 4.91 × 10−9 | −0.037 | 0.79 (0.74–0.86) | 0.066 | |
| rs6689427 A>G | 1 | 1q23 | FCRL1 | 0.483 | 0.397 | 5.00 × 10−9 | −0.037 | 0.79 (0.74–0.86) | 0.066 | |
| rs11264825 A>G | 1 | 1q23 | FCRL1 | 0.483 | 0.397 | 5.44 × 10−9 | −0.037 | 0.79 (0.74–0.86) | 0.062 | |
| rs2210911 A>G | 1 | 1q23 | FCRL3 | 0.397 | 0.491 | 4.91 × 10−8 | −0.034 | 0.80 (0.74–0.87) | 0.16 | |
| rs568617 C>T | 11 | 11q13 | FIBP | 0.230 | 0.204 | 3.25 × 10−8 | 0.034 | 1.35 (1.22–1.48) | 0.013 | |
| PCA | rs1990760 T>C | 2 | 2q24 | IFIH1 | 0.362 | 0.379 | 2.05 × 10−10 | −0.036 | 0.73 (0.66–0.81) | 6.18 × 10−5 |
| rs2111485 G>A | 2 | 2q24 | IFIH1 | 0.385 | 0.380 | 2.31 × 10−9 | −0.033 | 0.74 (0.67–0.82) | 2.78 × 10−5 | |
| TPOA | rs6679677 C>A | 1 | 1p13 | PTPN22 | 0.103 | 0.160 | 4.77 × 10−7 | 0.034 | 1.33 (1.18–1.48) | 2.30 × 10−5 |
N = 6,160. Chr., chromosome; HWE, P value from analysis of the HWE; MAF1000G, MAF from the 1000 Genomes project; MAFT1D, MAF estimated from the data; SNPweight, SNP effects from the FaST-LMM analysis adjusted for age at onset, duration of diabetes, and sex. As association with disease can cause deviation from the HWE for SNPs in risk loci in a case patient–only cohort, instead of removing SNPs based on HWE, all SNPs with significant associations were checked carefully for deviations. The 2q24 and 1p13 loci showed deviations from HWE and are both robustly associated with the risk of type 1 diabetes.
*Logistic regression analysis was performed to obtain ORs and 95% CIs of the association detected in the LMM analysis. The models were adjusted for age at onset, duration of diabetes, sex, ancestry using the first four PCs calculated from the genetic data, and family relatedness using GEE.
†A P value for HWE was calculated for genotype frequencies in case patients.
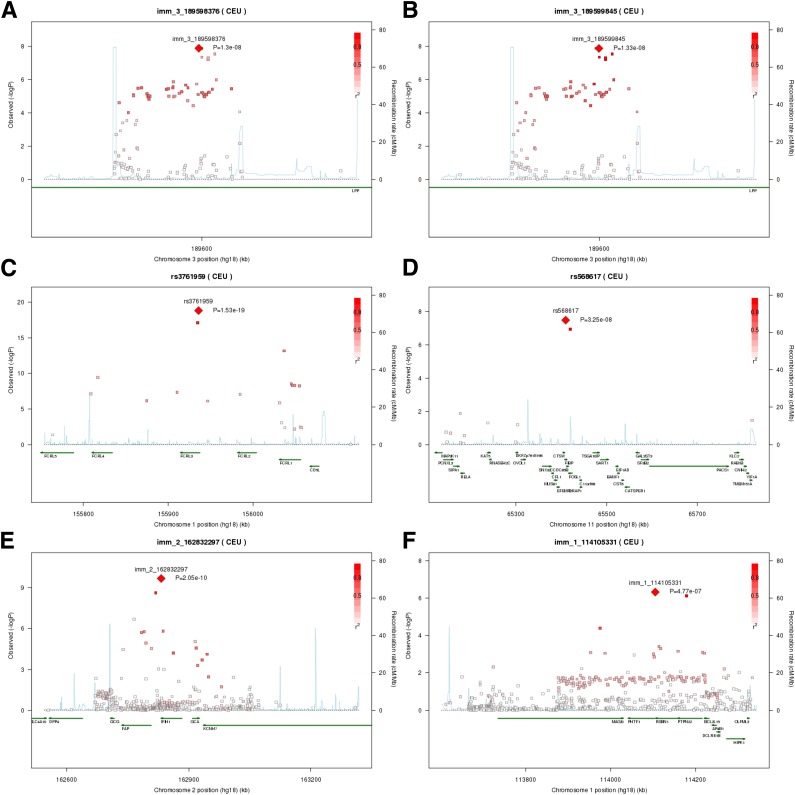
Figure 1.
Chromosomal regions of significant associations with autoantibody positivity. A: Association signals in the LPP gene on 3q28 for GADA positivity, with LD calculated for the lead SNP rs9815073 (Immunochip ID: imm_3_189598376). B: The same data as in A, but with LD calculated for the SNP rs59822814 (Immunochip ID: imm_3_189588845). C: Association signals in the FCRL3 region on 1q23 for IA-2A positivity. D: Association signals in the RELA region on 11q13 for IA-2A positivity. E: Association signal in the IFIH1 region on 2q24 for PCA positivity. F: Association signals in the PTPN22 region on 1p13 for TPOA positivity. In each plot, the −log10(PLMM) values of the SNPs are plotted against their chromosomal position based on National Center for Biotechnology Information Build 36 coordinates. The most associated SNP in each region is shown as a red diamond and is labeled with its Immunochip ID. The color intensity of the symbols represents the extent of LD with the lead SNP. The r2 values were estimated from nondiabetic unrelated control subjects. Genetic recombination rates, shown in light blue, were estimated from HapMap CEU samples. Plots were produced in SNAP (http://www.broadinstitute.org/mpg/snap/).
A new risk locus for GADA positivity was identified on chromosome 3q28 in a region of strong LD in the first intron of the gene LPP, which was originally identified as a susceptibility locus for CD (22,23). The lead SNP rs9815073 conferred protection from GADA (PLMM = 1.30 × 10−8, SNPeffect = −0.038), while a second association signal tagged by rs59822814 was associated with susceptibility to GADA development (PLMM = 1.33 × 10−8, SNPeffect = 0.038; Fig. 1A and B). The independent signals were confirmed by conditional logistic regression analysis (Supplementary Table 2). The majority of associations across the region was explained by rs59822814, while conditioning on rs9815073 revealed residual association at SNPs tagged by rs59822814. The 3q28/LPP locus also demonstrated an association with TGA, which is associated with CD, although only for rs9815073 (PLMM = 4.21 × 10−6, SNPeffect = −0.017; Supplementary Table 3 and Supplementary Fig. 5), suggesting heterogeneity in allelic effects between GADA and TGA.
IA-2A positivity was strongly associated with the previously reported 1q23/FCRL3 locus (rs3761959: PLMM = 1.53 × 10−19, SNPeffect = −0.056). The lead SNP was located in intron 3 of FCRL3 in a region of high LD that spans across several genes (Fig. 1C). The SNP rs3761959 is in high LD (r2 = 0.92) with a nonsynonymous SNP (nsSNP) rs7522061 (Asn28Asp), in exon 4 of FCRL3 (PLMM = 7.96 × 10−18, SNPeffect = −0.054), which was recently associated with protection from ZnT8A positivity in type 1 diabetes (9). In the conditional analysis, rs3761959 fully explained the association across the locus for IA-2A, while conditioning on rs7522061 revealed residual association at rs3761959 (Supplementary Table 2). The reported negative association with ZnT8A was replicated in our subset of 1,221 case patients with genotyping and ZnT8A measurements available (rs3761959: PLMM = 4.10 × 10−6, SNPeffect = −0.064; Supplementary Table 3 and Supplementary Fig. 5). The FCRL3 locus was also positively associated with PCA (rs3761959: PLMM = 1.16 × 10−3, SNPeffect = 0.17; Supplementary Table 3).
A second signal of genome-wide significance for IA-2A positivity was identified at the 11q13/RELA locus, which has previously been associated with Crohn disease (24). The lead SNP was located in intron 4 of the gene FIBP (rs568617: PLMM = 3.25 × 10−8, SNPeffect = 0.034). Additional association in this region was found for only one SNP, rs62261, in high LD with rs568617 (r2 = 0.89; Fig. 1D). The region was otherwise characterized by low LD and low coverage (Supplementary Table 2).
Association with PCA positivity was demonstrated with the 2q24/IFIH1 locus (rs1990760: PLMM = 2.05 × 10−10, SNPeffect = −0.036; Fig. 1E). The lead SNP is an nsSNP (Ala946Thr) in IFIH1, which is a known susceptibility locus for several autoimmune diseases. The same SNP also demonstrated suggestive evidence of association with TPOA positivity (P = 5.27 × 10−6, SNPeffect = −0.028; Supplementary Table 3 and Supplementary Fig. 5). After conditioning on rs1990760, suggestive independent associations with PCA positivity were demonstrated for three SNPs in low LD with the lead SNP (Supplementary Table 2). The most associated of these SNPs (rs76714048: P = 3.39 × 10−4, r2 = 0.08) was located in the gene FAP downstream of IFIH1.
No association reached genome-wide significance for TPOA positivity, but a signal in the 1p13/PTPN22 locus almost reached the Bonferroni correction limit (P < 3.7 × 10−7) (rs6679677: PLMM = 4.77 × 10−7, SNPeffect = 0.034; Fig. 1F). This locus is an established susceptibility locus for type 1 diabetes and other autoimmune diseases, and also demonstrated suggestive evidence of association with PCA (PLMM = 6.55 × 10−6, SNPeffect = 0.028), IA-2A (PLMM = 6.85 × 10−4, SNPeffect = −0.026; Supplementary Table 3 and Supplementary Fig. 5), and OH21A (PLMM = 9.42 × 10−3, SNPeffect = 0.005; Supplementary Table 3). The rs6679677 SNP explained the association signal for TPOA in the conditional analysis (Supplementary Table 2) and is in almost perfect LD with the nsSNP rs2476601 (Arg620Trp) in PTPN22.
Effects on Long-term Persistent Autoantibody Positivity
To investigate whether the associated SNPs were predictive of autoantibody positivity at different stages of diabetes duration, the association was tested within quartiles according to duration. The LPP locus (tagged by rs59822814) was not associated with GADA positivity in the quartile closest to diabetes onset (≤3 years) (Table 3). In contrast to GADA positivity, which decreased with longer duration, the association between LPP and GADA increased in strength with duration, and had the largest effect observed in the fourth quartile. The effect of FCRL3/rs3761959 for IA-2A positivity revealed strong effects regardless of diabetes duration, while RELA/rs568617 demonstrated the strongest association with IA-2A in the first two quartiles (Table 3). IFIH1/rs1990760 demonstrated the largest effect on PCA positivity close to onset and in the fourth quartile, while PTPN22/rs6679677 was most strongly associated with TPOA positivity closest to diabetes onset (Table 3).
Table 3.
Association of autoantibody positivity within quartiles of diabetes duration
| Variable | 0–3 years (n = 1,664) |
3–7 years (n = 1,353) |
7–14 years (n = 1,476) |
Over 14 years (n = 1,667) |
||||
|---|---|---|---|---|---|---|---|---|
| P | OR (95% CI)* | P | OR (95% CI)* | P | OR (95% CI)* | P | OR (95% CI)* | |
| GADA | ||||||||
| % positive | 58.5 | 45.5 | 38.3 | 34.1 | ||||
| Female sex* | 8.26 × 10−5 | 1.50 (1.22–1.83) | NS | NS | 7.53 × 10−8 | 1.82 (1.46–2.66) | 9.54 × 10−6 | 1.60 (1.30–1.97) |
| Onset*† | 2.62 × 10−8 | 1.60 (1.36–1.89) | <2 × 10−16 | 2.39 (1.98–2.89) | <2 × 10−16 | 1.97 (1.71–2.26) | <2 × 10−16 | 1.82 (1.59–2.09) |
| Duration* | 1.82 × 10−5 | 0.81 (0.74–0.89) | NS | NS | 0.038 | 0.94 (0.89–0.997) | 0.0074 | 0.98 (0.97–0.995) |
| rs59822814‡ | 0.05 | NS | 1.05 × 10−4 | 1.37 (1.17–1.61) | 0.0050 | 1.28 (1.09–1.50) | 9.71 × 10−5 | 1.36 (1.17–1.59) |
| IA-2A | ||||||||
| % positive | 62.9 | 50.8 | 42.1 | 28.2 | ||||
| Female sex* | NS | NS | NS | NS | NS | NS | NS | NS |
| Onset*† | 5.65 × 10−5 | 0.74 (0.64–0.86) | NS | NS | NS | NS | 0.0020 | 1.23 (1.08–1.41) |
| Duration* | 0.016 | 0.89 (0.81–0.98) | 0.0040 | 0.87 (0.79–0.96) | 1.98 × 10−4 | 0.90 (0.86–0.95) | 6.04 × 10−7 | 0.97 (0.95–0.98) |
| rs3761959‡ | 5.68 × 10−9 | 0.64 (0.56–0.75) | 0.0011 | 0.78 (0.67–0.92) | 1.29 × 10−4 | 0.74 (0.64–0.85) | 2.79 × 10−7 | 0.66 (0.56–0.78) |
| rs568617‡ | 0.0012 | 1.39 (1.14–1.70) | 9.54 × 10−4 | 1.44 (1.18–1.75) | 0.016 | 1.23 (1.02–1.49) | 0.0068 | 1.29 (1.07–1.55) |
| PCA | ||||||||
| % positive | 13.8 | 15.7 | 20.1 | 29.0 | ||||
| Female sex* | 0.0030 | 1.54 (1.16–2.05) | 1.77 × 10−7 | 2.27 (1.67–3.09) | 8.06 × 10−4 | 1.56 (1.20–2.03) | 1.77 × 10−8 | 1.84 (1.49–2.27) |
| Onset*† | 6.32 × 10−5 | 1.43 (1.20–1.71) | 3.59 × 10−6 | 1.60 (1.33–1.91) | 0.0023 | 1.26 (1.09–1.47) | NS | NS |
| Duration* | 0.017 | 1.18 (1.03–1.34) | NS | NS | NS | NS | NS | NS |
| rs1990760‡ | 5.35 × 10−4 | 1.50 (1.21–1.86) | 0.0012 | 1.54 (1.20–1.97) | 0.033 | 1.20 (0.99–1.46) | 1.20 × 10−4 | 1.36 (1.15–1.61) |
| TPOA | ||||||||
| % positive | 19.1 | 25.5 | 28.3 | 32.3 | ||||
| Female sex* | 5.10 × 10−9 | 2.11 (1.64–2.71) | 3.76 × 10−9 | 2.14 (1.66–2.78) | 1.64 × 10−11 | 2.21 (1.75–2.78) | 6.33 × 10−15 | 2.38 (1.91–2.96) |
| Onset*† | 1.82 × 10−6 | 1.47 (1.25–1.71) | 0.012 | 1.22 (1.05–1.43) | 0.0088 | 1.20 (1.05–1.38) | NS | NS |
| Duration* | 0.036 | 1.13 (1.01–1.26) | NS | NS | NS | NS | NS | NS |
| rs6679677‡ | 1.89 × 10−4 | 1.54 (1.23–1.92) | 0.056 | 1.29 (1.02–1.63) | 0.019 | 1.31 (1.05–1.63) | 0.023 | 1.25 (1.03–1.52) |
*ORs and 95% CIs were calculated from logistic regression models with age at disease onset, duration of diabetes, and sex as predictors; and were adjusted for population structure using the first four PCs calculated from the genetic data, and for family relatedness using GEE.
†ORs and CIs for age at disease onset are reported for a 10-year difference.
‡P value from the FaST-LMM analysis adjusted for age at disease onset, duration of diabetes, and sex, while ORs and 95% CIs were calculated from logistic regression models.
Association Within the MHC Region
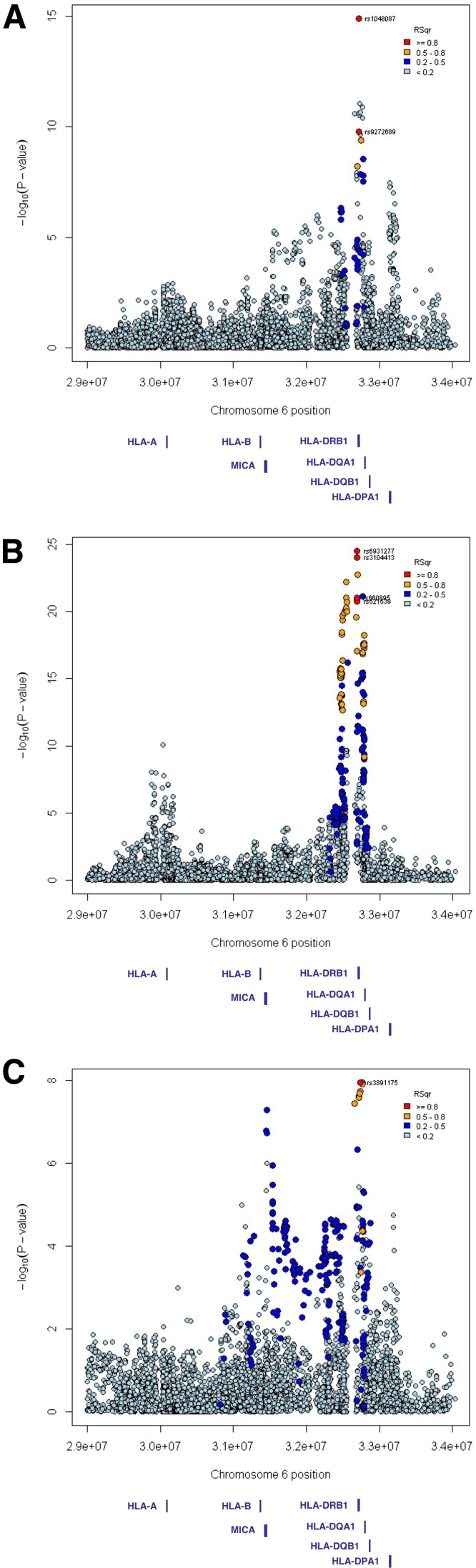
The peak of association in the MHC region was located within the HLA class II region for all autoantibodies, but considerable heterogeneity was observed (Table 4, Fig. 2, and Supplementary Fig. 6). Genome-wide significant associations were demonstrated for IA-2A, GADA, and TGA. Three independent signals were observed for GADA positivity. The most strongly associated SNP was rs1048087 located in exon 2 of HLA-DQA1 (PLMM = 1.21 × 10−15, SNPeffect = −0.059; Fig. 2A). The association was independent of the type 1 diabetes high-risk alleles HLA-DRB1*03 and HLA-DRB1*04 (tagged by rs2187668 and rs7454108, respectively; Supplementary Table 4). A second signal was tagged by rs9273327 (PLMM = 8.79 × 10−12, SNPeffect = 0.060), which is located between HLA-DRB1 and HLA-DQA1, but the association was dependent on HLA-DRB1*03 (Supplementary Table 4). A third independent signal was tagged by rs2301226 in the last intron of HLA-DPA1 (PLMM = 3.35 × 10−8, SNPeffect = 0.038). Two major association peaks were observed for IA-2A (Fig. 2B). The most strongly associated SNP was rs6931277 (PLMM = 2.97 × 10−25, SNPeffect = 0.096). It was located between HLA-DRB1 and HLA-DQA1, and tagged most of the association signal in the HLA class II region. The SNP rs6931277 was in moderate LD with the HLA-DRB1*04 allele (Table 4), but nevertheless conferred risk independent of HLA-DRB1*04 and HLA-DRB1*03 (Supplementary Table 4). The second peak was located in the HLA class I region and was tagged by rs3893538 (PLMM = 7.69 × 10−11, SNPeffect = −0.054). The rs3893538 variant is in almost perfect LD with the HLA-A*24 allele (25) and showed an effect independent of the HLA class II variants (Supplementary Table 4). The most associated SNP for TGA was rs3891175 (PLMM = 1.11 × 10−8, SNPeffect = 0.025) located upstream of HLA-DQB1 (Fig. 2C). The association was dependent on HLA-DRB1*03 (Supplementary Table 4). A second independent signal was observed in the class I region between HLA-B and MICA (rs2596565, PLMM = 1.14 × 10−8, SNPeffect = 0.030; Supplementary Table 4). The HLA-DRB1*03 allele was associated with positivity for GADA and TGA, but conferred protection from IA-2A, while HLA-DRB1*04 was associated with positivity for IA-2A, ZnT8A, TPOA, PCA, and OH21A (Table 4).
Table 4.
The most associated SNPs within the MHC region on 6p21 for each autoantibody
| SNP | MAFT1D | PLMM | SNPweight | OR(95% CI)* | Nearest gene | HWE† | LD with lead SNP | |
|---|---|---|---|---|---|---|---|---|
| GADA | rs1048087 T>C | 0.1765 | 1.21 × 10−15 | −0.059 | 0.61 (0.54–0.68) | HLA-DQA1 | 0.0484 | 1.0 |
| rs9273327 A>C | 0.3244 | 8.79 × 10−12 | 0.060 | 1.53 (1.37–1.70) | HLA-DRB1 | 1.80 × 10−25 | 0.10 | |
| rs2301226 C>T | 0.1806 | 3.35 × 10−8 | 0.038 | 1.36 (1.23–1.50) | HLA-DPA1 | 0.131 | 0.000 | |
| rs2187668 G>A | 0.3257 | 2.92 × 10−11 | 0.059 | 1.52 (1.37–1.70) | DR3 | 9.78 × 10−28 | 0.11 | |
| rs7454108 T>C | 0.3735 | NS | NS | NS | DR4 | 2.44 × 10−104 | 0.11 | |
| IA-2A | rs6931277 A>T | 0.419 | 2.97 × 10−25 | 0.096 | 2.38 (2.13–2.66) | HLA-DQA1 | 6.09 × 10−124 | 1.0 |
| rs3893538 A>G | 0.2368 | 7.69 × 10−11 | −0.054 | 0.66 (0.60–0.73) | HLA-A | 0.1038 | 0.003 | |
| rs2187668 G>A | 0.3257 | 2.93 × 10−7 | −0.051 | 0.65 (0.58–0.73) | DR3 | 9.78 × 10−28 | 0.19 | |
| rs7454108 T>C | 0.3735 | 6.22 × 10−19 | 0.082 | 2.16 (1.93–2.41) | DR4 | 2.44 × 10−104 | 0.75 | |
| ZnT8A | rs522308 G>A | 0.4516 | 5.39 × 10−5 | 0.068 | 1.57 (1.28–1.94) | HLA-DQA1 | 1.50 × 10−97 | 1.0 |
| rs2187668 G>A | 0.3257 | NS | NS | NS | DR3 | 9.78 × 10−28 | 0.21 | |
| rs7454108 T>C | 0.3735 | 9.63 × 10−4 | 0.055 | 1.53 (1.24–1.90) | DR4 | 2.44 × 10−104 | 0.55 | |
| PCA | rs7745040 C>T | 0.4981 | 5.21 × 10−6 | −0.031 | 0.70 (0.62–0.79) | HLA-DQB1 | 5.92 × 10−93 | 1.0 |
| rs2187668 G>A | 0.3257 | NS | NS | NS | DR3 | 9.78 × 10−28 | 0.27 | |
| rs7454108 T>C | 0.3735 | 2.85 × 10−3 | 0.029 | 1.38 (1.22–1.57) | DR4 | 2.44 × 10−104 | 0.39 | |
| TPOA | rs7745040 C>T | 0.4981 | 5.55 × 10−8 | −0.041 | 0.73 (0.66–0.82) | HLA-DQB1 | 5.92 × 10−93 | 1.0 |
| rs2187668 G>A | 0.3257 | NS | NS | NS | DR3 | 9.78 × 10−28 | 0.27 | |
| rs7454108 T>C | 0.3735 | 1.52 × 10−3 | 0.035 | 1.27 (1.13–1.42) | DR4 | 2.44 × 10−104 | 0.39 | |
| TGA | rs3891175 G>A | 0.377 | 1.11 × 10−8 | 0.025 | 1.85 (1.55–2.21) | HLA-DQB1 | 7.38 × 10−39 | 1.0 |
| rs2596565 C>T | 0.1836 | 1.14 × 10−8 | 0.030 | 2.46 (1.81–3.36) | MICA | 1.40 × 10−15 | 0.25 | |
| rs2187668 G>A | 0.3257 | 2.39 × 10−8 | 0.026 | 1.88 (1.57–2.25) | DR3 | 9.78 × 10−28 | 0.66 | |
| rs7454108 T>C | 0.3735 | NS | NS | NS | DR4 | 2.44 × 10−104 | 0.20 | |
| OH21A | rs9275599 C>T | 0.3309 | 2.79 × 10−6 | 0.009 | 2.64 (1.83–3.82) | HLA-DQA2 | 4.49 × 10−61 | 1.0 |
| rs2187668 G>A | 0.3257 | NS | NS | NS | DR3 | 9.78 × 10−28 | 0.13 | |
| rs7454108 T>C | 0.3735 | 2.24 × 10−5 | 0.008 | 2.68 (1.82–3.93) | DR4 | 2.44 × 10−104 | 0.74 |
HWE, P value from analysis of HWE; MAFT1D, MAF estimated from the data; SNPweight, SNP effect from the FaST-LMM analysis adjusted for age at onset, duration of diabetes, and sex. As association with disease can cause deviation from HWE for SNPs in risk loci in a case patient–only cohort, instead of removing SNPs based on HWE, all SNPs with significant associations were checked carefully for deviations. The 6p21 locus showed deviations from HWE and is robustly associated with the risk of type 1 diabetes and other autoimmune diseases.
*ORs and 95% CIs were calculated from logistic regression models adjusted for age at onset, duration of diabetes, sex, population structure using the first four PCs calculated from the genetic data, and family relatedness using GEE.
†A P value for HWE was calculated for genotype frequencies in case patients.
Figure 2.
MHC regional association plots. Association signals in the MHC region for GADA (A), IA-2A (B), and TGA (C). In each plot, the −log10(PLMM) values of the SNPs are plotted against their chromosomal position based on National Center for Biotechnology Information Build 36 coordinates. The most associated SNP in each region is shown in red and is labeled with its Immunochip ID. The color intensity of the symbols represents the extent of LD with the lead SNP. The r2 values were estimated from nondiabetic unrelated control subjects.
Discussion
We have identified three novel non-HLA susceptibility loci for autoantibody positivity, and confirmed the strong association with 1q23/FCRL3 for IA-2A and ZnT8A. In line with emerging evidence, we found a limited overlap between the known susceptibility loci for type 1 diabetes and loci controlling islet autoimmunity. We cannot, however, rule out that the present subphenotyping approach may have uncovered previously unrecognized genetic contributors to type 1 diabetes susceptibility with individually weak pathogenic effects not detected by previous GWAS. The stringent genome-wide significance threshold applied here has most likely excluded candidate loci with smaller individual effects, which could be investigated more carefully given the higher prior probability to find associations with autoimmunity in loci covered on the Immunochip. The association between 2q24/IFIH1 and 1p13/PTPN22 with PCA and TPOA, on the other hand, extends the observed commonality of risk loci for immune-mediated diseases and highlights shared etiological factors, and may constitute early markers for the identification of subsets of individuals with type 1 diabetes who are at increased risk for autoimmune comorbidities. 3q28/LPP is not associated with type 1 diabetes risk (11), but pinpoints a novel genetic link between GADA autoimmunity and the risk of CD (22,23), vitiligo (26), and AITD (27), which should prove valuable for understanding the biology and mechanistic triggers of GADA. Little is known about the function of LPP in autoimmunity, but it is highly expressed in the small intestine (22), where its role in CD presumably is exerted. It has been suggested that gluten consumption, gut inflammation, and altered gut permeability could be involved in the pathogenesis of type 1 diabetes (28,29). Apart from the small intestine, LPP is also expressed in pancreatic islets (30), so the role of LPP in autoimmunity against the β-cell might be independent of its function in the gut.
We hypothesized that distinct mechanisms control phenotypic differences between autoimmunity at onset and persistent autoimmunity. Interestingly, the LPP locus was associated with GADA only after a duration of 3 years, strongly suggesting that it is not associated with the development of autoantibodies per se, but instead with persistent GADA positivity. Even after >14 years of diabetes duration, 34% of the patients were positive for GADA, and carriers of the LPP risk variant had a 36% increased probability for GADA positivity. This frequency of GADA positivity is comparable to other cohorts of patients with long-standing diabetes (31,32). Although type 1 diabetes is thought to result from an irreversible destruction of β-cells, some long-standing patients have detectable functioning β-cells (33–35), indicating either surviving β-cells or continued renewal due to ongoing autoimmune destruction. Our data suggest that LPP together with GADA positivity could be markers of the latter.
FCRL3 encodes a member of the Ig receptor superfamily with a presumed role in the regulation of the immune response, but its exact function in autoimmunity is unknown. Our lead SNP is in perfect LD with rs7528684, which has been associated with the risk of RA (36,37), systemic lupus erythematosus (36), and AITD (38), and protection from autoimmune hypoadrenalism (39) and multiple sclerosis (40), but not type 1 diabetes (8,9). rs7528684 alters a binding site for the transcription factor nuclear factor-κB, and associates with increased FCRL3 mRNA expression and autoantibody production in RA patients (36). Intriguingly, this allele would be associated with protection from IA-2A and ZnT8A, which confirms previous findings (8,9). Our results also suggest an association with PCA and autoimmune gastritis, which requires further replication. The association between 11q13/RELA and IA-2A positivity has not been reported previously, but this locus is associated with a risk of Crohn disease (24). Our lead SNP is in high LD (r2 > 0.9) with the Crohn disease risk variant rs2231884 and, interestingly, is positively correlated with IA-2A, in contrast with the opposite direction of the effects for all other IA-2A–associated loci.
IFIH1 and PTPN22 represent robust susceptibility loci for several autoimmune diseases. For both loci, the type 1 diabetes risk allele correlated with autoantibody positivity. IFIH1 has been associated with AITD, but convincing evidence from large GWAS is lacking (41,42), and no association with autoimmune gastritis has so far been reported. IFIH1 encodes an RNA helicase that functions as an early sensor for double-stranded RNA that detects viral infections and triggers an immune response (43). It is highly expressed in activated immune cells, but also in β-cells, where it contributes to local release of proinflammatory cytokines and apoptosis (44). The PTPN22 locus did not reach genome-wide significance, but, given that it constitutes a known risk locus for several autoimmune diseases including type 1 diabetes and AITD (45), the effect on TPOA and PCA positivity is likely to be true. PTPN22 encodes a lymphoid-specific intracellular phosphatase involved in T-cell activation (46). The strong effect on autoantibody positivity suggests that IFIH1 and PTPN22 have specific functions in autoimmunity, which should be investigated further. We did not replicate the ABO locus that has been associated with PCA (8), even though the sample size of our cohort should make it possible to detect such an effect.
The MHC region is a major determinant of autoantibody positivity, but the exact alleles and their effects varied among the autoantibodies. For GADA positivity, some of the association was explained by LD with HLA-DRB1*03, but two additional independent signals were observed in the HLA class II region, making it unlikely that the type 1 diabetes risk variant alone explains the genetic susceptibility in this region. We did not find evidence for any major class I association, which has previously been reported (6). For IA-2A, the major association in the HLA class II region was independent of HLA-DRB1 high-risk alleles. We replicated the previously reported class II independent association between HLA-A*24 and protection from IA-2A (6,7), although HLA-A*24 is associated with a risk of type 1 diabetes. Association with TGA in the class II region was explained by LD with HLA-DRB1*03, which is associated with a risk of CD (47), but there was also evidence for an independent signal in the class I region. Even though the MHC region did not reach genome-wide significance for all autoantibodies, suggestive evidence of association was found. It is interesting to note that for PCA the strength of association with the MHC region was weaker than that for the IFIH1 locus.
This study supports the hypothesis that islet autoantibodies in type 1 diabetes are not pathogenic but, rather, are a consequence of the autoimmune destruction of the β-cells. Rather than affecting general B-cell function, it appears that each autoantibody profile is controlled by a distinct set of genetic markers that alters the B-cell specificity. Our data also suggest that markers for persistent autoimmunity and/or disease progression are most likely independent of disease susceptibility. It remains to be clarified whether LPP also correlates with residual β-cell mass or function; however, such a genetic marker should be useful for stratifying individuals for intervention trials aimed at lowering autoimmune stress to preserve β-cell function and improve endogenous insulin production.
Article Information
Funding. This research used resources provided by the Type 1 Diabetes Genetics Consortium, a collaborative clinical study sponsored by the National Institute of Diabetes and Digestive and Kidney Diseases, the National Institute of Allergy and Infectious Diseases, the National Human Genome Research Institute, the National Institute of Child Health and Human Development, and the JDRF and was supported by National Institutes of Health (NIH) grant U01-DK-062418. Specific parts of the projects were funded by the NIH (grant 1-DP3-DK-085678-01).
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. C.A.B. researched the data and wrote the manuscript. S.O., W.-M.C., J.W., L.Y., P.B., A.J.K.W., and P.J.B. researched the data, reviewed and edited the manuscript, and contributed to the discussion. J.C.H., G.S.E., P.C., S.S.R., and F.P. reviewed and edited the manuscript, and contributed to the discussion. F.P. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db14-1730/-/DC1.
A full list of Type 1 Diabetes Genetics Consortium members appears in the Supplementary Appendix online.
Deceased.
References
- 1.Atkinson MA, Eisenbarth GS. Type 1 diabetes: new perspectives on disease pathogenesis and treatment. Lancet 2001;358:221–229 [DOI] [PubMed] [Google Scholar]
- 2.Achenbach P, Warncke K, Reiter J, et al. Stratification of type 1 diabetes risk on the basis of islet autoantibody characteristics. Diabetes 2004;53:384–392 [DOI] [PubMed] [Google Scholar]
- 3.Kulmala P, Savola K, Petersen JS, et al.; The Childhood Diabetes in Finland Study Group . Prediction of insulin-dependent diabetes mellitus in siblings of children with diabetes. A population-based study. J Clin Invest 1998;101:327–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Verge CF, Gianani R, Kawasaki E, et al. Prediction of type I diabetes in first-degree relatives using a combination of insulin, GAD, and ICA512bdc/IA-2 autoantibodies. Diabetes 1996;45:926–933 [DOI] [PubMed] [Google Scholar]
- 5.Graham J, Hagopian WA, Kockum I, et al.; Diabetes Incidence in Sweden Study Group; Swedish Childhood Diabetes Study Group . Genetic effects on age-dependent onset and islet cell autoantibody markers in type 1 diabetes. Diabetes 2002;51:1346–1355 [DOI] [PubMed] [Google Scholar]
- 6.Howson JM, Stevens H, Smyth DJ, et al. Evidence that HLA class I and II associations with type 1 diabetes, autoantibodies to GAD and autoantibodies to IA-2, are distinct. Diabetes 2011;60:2635–2644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qu HQ, Polychronakos C. The effect of the MHC locus on autoantibodies in type 1 diabetes. J Med Genet 2009;46:469–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Plagnol V, Howson JM, Smyth DJ, et al.; Type 1 Diabetes Genetics Consortium . Genome-wide association analysis of autoantibody positivity in type 1 diabetes cases. PLoS Genet 2011;7:e1002216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Howson JM, Krause S, Stevens H, et al. Genetic association of zinc transporter 8 (ZnT8) autoantibodies in type 1 diabetes cases. Diabetologia 2012;55:1978–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pociot F, Akolkar B, Concannon P, et al. Genetics of type 1 diabetes: what’s next? Diabetes 2010;59:1561–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smyth DJ, Plagnol V, Walker NM, et al. Shared and distinct genetic variants in type 1 diabetes and celiac disease. N Engl J Med 2008;359:2767–2777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cotsapas C, Voight BF, Rossin E, et al.; FOCiS Network of Consortia . Pervasive sharing of genetic effects in autoimmune disease. PLoS Genet 2011;7:e1002254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rich SS, Concannon P, Erlich H, et al. The Type 1 Diabetes Genetics Consortium. Ann N Y Acad Sci 2006;1079:1–8 [DOI] [PubMed] [Google Scholar]
- 14.Bonifacio E, Yu L, Williams AK, et al. Harmonization of glutamic acid decarboxylase and islet antigen-2 autoantibody assays for national institute of diabetes and digestive and kidney diseases consortia. J Clin Endocrinol Metab 2010;95:3360–3367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bingley PJ, Williams AJ, Colman PG, et al.; T1DGC . Measurement of islet cell antibodies in the Type 1 Diabetes Genetics Consortium: efforts to harmonize procedures among the laboratories. Clin Trials 2010;7(Suppl.):S56–S64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lampasona V, Schlosser M, Mueller PW, et al. Diabetes antibody standardization program: first proficiency evaluation of assays for autoantibodies to zinc transporter 8. Clin Chem 2011;57:1693–1702 [DOI] [PubMed] [Google Scholar]
- 17.Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen WM. Robust relationship inference in genome-wide association studies. Bioinformatics 2010;26:2867–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81:559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lippert C, Listgarten J, Liu Y, Kadie CM, Davidson RI, Heckerman D. FaST linear mixed models for genome-wide association studies. Nat Methods 2011;8:833–835 [DOI] [PubMed] [Google Scholar]
- 20.Liang KY, Zeger SL. Longitudinal data-analysis using generalized linear-models. Biometrika 1986;73:13–22 [Google Scholar]
- 21.Halekoh U, Hojsgaard S, Yan J. The R package geepack for generalized estimating equations. J Stat Softw 2006;15:1–11 [Google Scholar]
- 22.Hunt KA, Zhernakova A, Turner G, et al. Newly identified genetic risk variants for celiac disease related to the immune response. Nat Genet 2008;40:395–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dubois PC, Trynka G, Franke L, et al. Multiple common variants for celiac disease influencing immune gene expression. Nat Genet 2010;42:295–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jostins L, Ripke S, Weersma RK, et al.; International IBD Genetics Consortium (IIBDGC) . Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 2012;491:119–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Bakker PI, McVean G, Sabeti PC, et al. A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat Genet 2006;38:1166–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin Y, Birlea SA, Fain PR, et al. Variant of TYR and autoimmunity susceptibility loci in generalized vitiligo. N Engl J Med 2010;362:1686–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cooper JD, Simmonds MJ, Walker NM, et al.; Wellcome Trust Case Control Consortium . Seven newly identified loci for autoimmune thyroid disease. Hum Mol Genet 2012;21:5202–5208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vaarala O, Atkinson MA, Neu J. The “perfect storm” for type 1 diabetes: the complex interplay between intestinal microbiota, gut permeability, and mucosal immunity. Diabetes 2008;57:2555–2562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frisk G, Hansson T, Dahlbom I, Tuvemo T. A unifying hypothesis on the development of type 1 diabetes and celiac disease: gluten consumption may be a shared causative factor. Med Hypotheses 2008;70:1207–1209 [DOI] [PubMed] [Google Scholar]
- 30.Kutlu B, Burdick D, Baxter D, et al. Detailed transcriptome atlas of the pancreatic beta cell. BMC Med Genomics 2009;2:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keenan HA, Sun JK, Levine J, et al. Residual insulin production and pancreatic β-cell turnover after 50 years of diabetes: Joslin Medalist Study. Diabetes 2010;59:2846–2853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Richardson CC, Dromey JA, McLaughlin KA, et al. High frequency of autoantibodies in patients with long duration type 1 diabetes. Diabetologia. 20 August 2013. [Epub ahead of print]. DOI: 10.1007/s00125-013-3017-7 [DOI] [PubMed] [Google Scholar]
- 33.Liu EH, Digon BJ 3rd, Hirshberg B, et al. Pancreatic beta cell function persists in many patients with chronic type 1 diabetes, but is not dramatically improved by prolonged immunosuppression and euglycaemia from a beta cell allograft. Diabetologia 2009;52:1369–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gianani R, Campbell-Thompson M, Sarkar SA, et al. Dimorphic histopathology of long-standing childhood-onset diabetes. Diabetologia 2010;53:690–698 [DOI] [PubMed] [Google Scholar]
- 35.Meier JJ, Bhushan A, Butler AE, Rizza RA, Butler PC. Sustained beta cell apoptosis in patients with long-standing type 1 diabetes: indirect evidence for islet regeneration? Diabetologia 2005;48:2221–2228 [DOI] [PubMed] [Google Scholar]
- 36.Kochi Y, Yamada R, Suzuki A, et al. A functional variant in FCRL3, encoding Fc receptor-like 3, is associated with rheumatoid arthritis and several autoimmunities. Nat Genet 2005;37:478–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maehlen MT, Nordang GB, Syversen SW, et al. FCRL3 -169C/C genotype is associated with anti-citrullinated protein antibody-positive rheumatoid arthritis and with radiographic progression. J Rheumatol 2011;38:2329–2335 [DOI] [PubMed] [Google Scholar]
- 38.Chu X, Pan CM, Zhao SX, et al.; China Consortium for Genetics of Autoimmune Thyroid Disease . A genome-wide association study identifies two new risk loci for Graves’ disease. Nat Genet 2011;43:897–901 [DOI] [PubMed] [Google Scholar]
- 39.Owen CJ, Kelly H, Eden JA, Merriman ME, Pearce SH, Merriman TR. Analysis of the Fc receptor-like-3 (FCRL3) locus in Caucasians with autoimmune disorders suggests a complex pattern of disease association. J Clin Endocrinol Metab 2007;92:1106–1111 [DOI] [PubMed] [Google Scholar]
- 40.Matesanz F, Fernández O, Milne RL, et al. The high producer variant of the Fc-receptor like-3 (FCRL3) gene is involved in protection against multiple sclerosis. J Neuroimmunol 2008;195:146–150 [DOI] [PubMed] [Google Scholar]
- 41.Sutherland A, Davies J, Owen CJ, et al. Genomic polymorphism at the interferon-induced helicase (IFIH1) locus contributes to Graves’ disease susceptibility. J Clin Endocrinol Metab 2007;92:3338–3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang N, Shen N, Vyse TJ, et al. Selective IgA deficiency in autoimmune diseases. Mol Med 2011;17:1383–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meylan E, Tschopp J, Karin M. Intracellular pattern recognition receptors in the host response. Nature 2006;442:39–44 [DOI] [PubMed] [Google Scholar]
- 44.Colli ML, Moore F, Gurzov EN, Ortis F, Eizirik DL. MDA5 and PTPN2, two candidate genes for type 1 diabetes, modify pancreatic beta-cell responses to the viral by-product double-stranded RNA. Hum Mol Genet 2010;19:135–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smyth D, Cooper JD, Collins JE, et al. Replication of an association between the lymphoid tyrosine phosphatase locus (LYP/PTPN22) with type 1 diabetes, and evidence for its role as a general autoimmunity locus. Diabetes 2004;53:3020–3023 [DOI] [PubMed] [Google Scholar]
- 46.Bottini N, Musumeci L, Alonso A, et al. A functional variant of lymphoid tyrosine phosphatase is associated with type I diabetes. Nat Genet 2004;36:337–338 [DOI] [PubMed] [Google Scholar]
- 47.van Heel DA, Franke L, Hunt KA, et al. A genome-wide association study for celiac disease identifies risk variants in the region harboring IL2 and IL21. Nat Genet 2007;39:827–829 [DOI] [PMC free article] [PubMed] [Google Scholar]