Abstract
Dietary polyphenols protect against metabolic syndrome, despite limited absorption and digestion, raising questions about their mechanism of action. We hypothesized that one mechanism may involve the gut microbiota. To test this hypothesis, C57BL/6J mice were fed a high-fat diet (HFD) containing 1% Concord grape polyphenols (GP). Relative to vehicle controls, GP attenuated several effects of HFD feeding, including weight gain, adiposity, serum inflammatory markers (tumor necrosis factor [TNF]α, interleukin [IL]-6, and lipopolysaccharide), and glucose intolerance. GP lowered intestinal expression of inflammatory markers (TNFα, IL-6, inducible nitric oxide synthase) and a gene for glucose absorption (Glut2). GP increased intestinal expression of genes involved in barrier function (occludin) and limiting triglyceride storage (fasting-induced adipocyte factor). GP also increased intestinal gene expression of proglucagon, a precursor of proteins that promote insulin production and gut barrier integrity. 16S rRNA gene sequencing and quantitative PCR of cecal and fecal samples demonstrated that GP dramatically increased the growth of Akkermansia muciniphila and decreased the proportion of Firmicutes to Bacteroidetes, consistent with prior reports that similar changes in microbial community structure can protect from diet-induced obesity and metabolic disease. These data suggest that GP act in the intestine to modify gut microbial community structure, resulting in lower intestinal and systemic inflammation and improved metabolic outcomes. The gut microbiota may thus provide the missing link in the mechanism of action of poorly absorbed dietary polyphenols.
Introduction
Metabolic syndrome (MetS), characterized by concurrence of at least three of five risk factors (i.e., obesity, hypertension, dyslipidemia, insulin resistance, and hyperglycemia), is a global epidemic that increases the risk of developing type 2 diabetes (T2D) and cardiovascular disease (1). Evidence strongly suggests that chronic low-grade inflammation promoted by complex interactions between an individual’s diet and their gut microbiota is an important factor underlying chronic disorders such as MetS (2). In addition to adipose tissue, the intestine has emerged as an important source of inflammatory mediators that disrupt insulin signaling leading to whole-body insulin resistance and hyperglycemia (3). Mice fed a high-fat diet (HFD) showed increased levels of inflammatory cytokines (tumor necrosis factor [TNF]α and interleukin [IL]-6) in ileum, colon, and surrounding mesenteric fat but not in other fat depots, even before the development of obesity (4,5). Gnotobiotic or germ-free mice are generally protected from HFD-induced obesity, insulin resistance, and intestinal inflammation; however, when colonized with the microbiota of obese mice, germ-free mice rapidly developed these features of MetS (6–9), indicating a critical role for the gut microbiota in the development of metabolic disease.
Obesity-related MetS is also associated with chronically higher levels of proinflammatory and gut microbiota–derived lipopolysaccharide (LPS) in circulation, an event defined as metabolic endotoxemia (10). An HFD decreases expression of intestinal tight junction proteins, leading to greater intestinal epithelium permeability and increased leakage of LPS into circulation (10,11). Transport of LPS by gut enterocyte-derived chlyomicrons also contributes to the increased levels of systemic LPS (12). HFD-induced metabolic endotoxemia provided a key concept linking diet-induced changes in the gut microbiota and intestinal barrier function with the chronic low-grade inflammation that ultimately leads to insulin receptor dysfunction, insulin resistance, and glucose intolerance (13). No single or combination drug therapy has been effective in curtailing the prevalence of MetS, signifying the need for new approaches.
Numerous epidemiological, clinical, and preclinical studies indicate that dietary polyphenols can protect against MetS (14,15). Grapes and grape products are a major source of dietary polyphenols that have been shown to attenuate many symptoms of obesity-related MetS, including chronic low-grade inflammation (16). Anthocyanins (ACNs) comprise the most abundant class of polyphenols in Concord grape berries and juice (17), while monomeric flavan-3-ols and their oligomers, B-type proanthocyanidins (PACs), are the major classes contained in grape seeds (18). We have previously demonstrated that Concord grape (Vitis labrusca) polyphenols can be stably sorbed to a protein-rich food matrix and that this complex induces antihyperglycemic effects in HFD-fed mice (19,20).
ACNs and PACs confer protection against symptoms of MetS despite their limited absorption in circulation (21–23). In rodent studies, 88–94% of the administered radiolabeled ACN or PAC compounds were recovered in the gastrointestinal tract and feces (21,24). While polyphenols are known to be biotransformed by gut microbiota into simpler phenolic compounds that may be absorbed (25), the levels and bioactivities of circulating metabolites may not be sufficient to explain the pharmacological effects of polyphenols. More than 75% of PACs in grapes are polymers having more than four degrees of polymerization; however, the ability of microbes to catabolize PACs declines with increased molecular size (26). For example, the yield of phenolic acids in rat gut was 10% and 7% for catechin monomer and PAC dimers but just 0.7% and 0.5% for PAC trimers and polymers (27). These data indicate that polyphenol absorption is not a requirement for bioactivity. One possibility is that grape polyphenols (GP) act by remodeling the gut microbiota, leading to reduced inflammation and improved metabolic function. The current study provides compelling evidence in support of this hypothesis.
Research Design and Methods
Preparation of GP Stabilized on a Protein Matrix
We previously demonstrated that, due to a natural affinity of polyphenols and proteins, dietary polyphenols can be stabilized for at least 24 weeks (and up to 1 year [unpublished data]) at 37°C when sorbed to a protein-rich food matrix, such as soy protein isolate (SPI), without compromising subsequent release from the matrix or efficacy (19,28,29). Studies performed in models of the human intestine have also demonstrated that protein-polyphenol particles provide an efficient vehicle for delivering intact polyphenols, e.g., ACN, to the lower part of the intestine (30). We therefore used this protein sorption process to maintain the ex situ and in situ stability of extracted Concord GP incorporated into the rodent diets. While GP alone are somewhat crystalline and sticky when dried, the GP-SPI complex is a free-flowing powder that provides a convenient delivery vehicle for uniform incorporation of GP into rodent diets. The biochemical composition of the grape pomace extract used in this study and production of GP sorbed and stabilized to the SPI matrix (i.e., GP-SPI) have previously been described in detail (19). Concentration of total polyphenols in the grape pomace extract was quantified using a modified Folin-Ciocalteu method, and a calculated amount of SPI was mixed with the extract. The mixture was tray dried at 50°C under vacuum until moisture was <5% to produce the GP-SPI complex containing 10% GP.
Preparation of Diets
Nutritional composition of GP-SPI and SPI was determined by Medallion Labs (Minneapolis, MN) in accordance with the Association of Analytical Communities methods for ash, moisture, proteins, dietary fiber, and total fat; total carbohydrates were determined by difference and calories by calculation (Supplementary Table 1). The HFD (D12492; Research Diets, NJ) derived 61% of kilocalories from fat, and the low-fat diet (LFD) (D12450B; Research Diets) derived 10% of kilocalories from fat. Nutritional analysis data were used by Research Diets to formulate an HFD containing 10% SPI (defined as SPI diet) or an HFD containing 10% GP-SPI (defined as GP-SPI diet). The GP-SPI powder contained less protein (57.6%) than the SPI alone (89.3%) and small amounts of glucose (1.8%) and fructose (2.6%) as well as other pomace-derived carbohydrates (26.7%) (Supplementary Table 1). Glucose and fructose were added to the HFD formulation supplemented with 10% SPI (i.e., SPI diet), while more SPI and less cellulose were added to the HFD formulation supplemented with 10% GP-SPI (i.e., GP-SPI diet) so that HFD, GP-SPI diet, and SPI diet were all comparable in the percentage of kilocalories contributed by protein, fat, and carbohydrates as well as total energy (kilocalories per gram) (Supplementary Table 2). GP-SPI contained 10% GP; therefore, the GP-SPI diet containing 10% GP-SPI contained 1% GP.
Mice and Intervention Protocol
Protocols were approved by the Rutgers University Institutional Care and Use Committee and followed federal and state laws. Five-week-old male C57BL/6J mice (10–20 g) were purchased from The Jackson Laboratory (Bar Harbor, ME) and fed a standard chow diet ad libitum (cat. no. 5015; Purina) during their 1-week acclimatization period. Animals were housed, five per cage, with free access to water in a room with a temperature of 24 ± 1°C and a 12:12-h light:dark cycle (7:00 a.m.–7:00 p.m.). At 6 weeks of age, oral glucose tolerance tests (OGTTs) were performed on 45 mice. The area under the curve (AUC) corresponding to the OGTT data from each mouse was calculated, and a mean AUC for each cage of five mice was determined. The nine cages were separated into three groups based on the average AUCs calculated for each cage so that each group of 15 mice would be similar at baseline with respect to oral glucose tolerance. This method of assignment was used as a way to normalize oral glucose tolerance at baseline and also keep mice in their original cage placements, as switching the animals around can sometimes lead to aggressive behavior in the new group. Mice were fed GP-SPI diet, SPI diet, or HFD (n = 15 mice/diet group) for a total of 13 weeks. The HFD group was used mainly as a control to monitor body weight gain and food intake between groups. Various end points were measured during the intervention period as described below. A second group of 5-week-old male C57BL/6J mice (10–20 g) (n = 10) was purchased at a later time to have an LFD cohort with which to compare body weights, food intake, and microbiome samples. These LFD-fed mice were similarly housed (five per cage) in the same experimental room and space. Mice were initially fed a regular chow diet ad libitum for 1 week and then switched to the LFD for 12 weeks with OGTT performed at the same intervals.
Body Weights, Food Intake, and Body Composition
Body weights of mice were recorded weekly. Food intake per mouse per day was calculated as follows: [total food intake per cage]/[mice per cage]/[days of food consumption]. Body composition (fat mass, lean mass, and total water) was evaluated by quantitative nuclear MRI (EchoMRI 3-in-1 Analyzer; EchoMRI, Houston, TX).
OGTT
Mice were fasted in the morning for 6 h, and body weights were measured. A glucometer (AlphaTRAK 32004-02; Abbott Animal Health) was used to measure fasting blood glucose levels prior to glucose administration (T = 0). Mice were then gavaged with 2 g/kg glucose (500 mg/mL); blood glucose was tested every 30 min up to 120 min. OGTTs performed prior to the introduction of intervention diets (week 0) were used for assigning animals into GP-SPI diet, SPI diet, and HFD groups, and OGTTs were then repeated during intervention at indicated weeks.
Blood Serum Analysis
Mice were sacrificed 13 weeks postintervention by CO2 asphyxiation. GP-SPI, SPI, and HFD groups of mice were sacrificed over a 3-day period at the same time of day. The LFD group was later added as a control and sacrificed at the same time of day in a single day. Trunk blood was collected into microfuge tubes and allowed to clot. Samples were centrifuged at 5,000 rpm for 10 min, and serum was collected and frozen at −80°C until biochemical analysis, which was performed by the Clinical Chemistry Laboratory at Pennington Biomedical Research Center (Baton Rouge, LA). Serum samples obtained from mice in the SPI and GP-SPI diet groups (n = 10 per group) were measured for triglyceride, total cholesterol, and total antioxidant activity (ferric reducing antioxidant power method). Triglycerides, total cholesterol, and total antioxidant activity were quantified by Beckman Coulter DXC 600 Pro (Beckman Coulter, Inc., Brea, CA) using standard spectrophotometric assays. Serum insulin (Crystal Chem, Inc.) was quantified by ELISA. Adiponectin was measured using Milliplex map single-plex adiponectin kit (Millipore). The concentrations of inflammatory cytokines IL-6, IL-1β, and TNFα were quantified using the Milliplex map mouse cytokine/chemokine kit (Millipore). Serum LPS levels were analyzed using a mouse LPS ELISA kit (Cusabio, Wuhan, China).
Tissue Collections
On the day of sacrifice, each mouse was placed in an empty cage without bedding for 10–15 min to allow collection of fresh stool samples that were snap frozen in liquid nitrogen. The liver, jejunum, ileum, cecum, and colon were accurately dissected from each mouse and snap frozen within 5–7 min postmortem. Liver and cecum tissues were weighed. Intestinal tissues were carefully stripped of mesentery or fat. The lumina of the jejunum, ileum, and colon segments were thoroughly flushed with cold PBS (pH 7.4) using a ball tip dosing needle attached to a syringe to remove feces; then, PBS and contents were pushed out with blunt forceps. Cleaned tissues were subsequently placed in individual cryogenic tubes, snap frozen in liquid nitrogen, and stored at −80°C until analysis.
Liver Lipid Analysis
Total lipids from liver samples (n = 10 each from GP-SPI and SPI groups) were isolated as previously described (31). Briefly, 300 mg liver tissue was homogenized with 20 volumes (6 mL) of chloroform:methanol (2:1). Homogenate was filtered through Whatman #1 paper into preweighed 15-mL glass tubes. Filtrate was washed once with 2 mL chloroform:methanol (2:1) and 0.4 mL NaCl 0.9%. Samples were centrifuged for 1 min, and the upper layer of the biphasic system was removed. The entire lower phase was evaporated to dryness in a speed vacuum. Tubes containing dried lipid were weighed, and lipid weight was calculated by subtracting empty tube weight.
Real-Time PCR
Individual tissue samples (50–100 mg) were homogenized with 2.5-mm stainless steel beads and Qiazol reagent using the 2010 GenoGrinder (Spex Sample Prep, Metuchen, NJ), and RNA was isolated according to the manufacturer’s instructions (RNeasy Plus Universal kits; QIAGEN). RNA was quantified by Nanodrop (Thermo Fisher Scientific, Inc.). Oligo dT primers were used to reverse transcribe 5 µg mRNA to cDNA (ABI High-Capacity cDNA Reverse Transcription kit). RT-PCR was performed on an ABI 7300 machine. The following primers were validated for PCR efficiency and a unique melt curve with SYBR Green chemistry: forward, TNFα AGACCCTCACACTCAGATCA; reverse, TNFα TCTTTGAGATCCATGCCGTTG; forward, fasting-induced adipocyte factor (Fiaf) CAATGCCAAATTGCTCCAATT; reverse, Fiaf TGGCCGTGGGCTCAGT; forward, hydroxymethylbilane synthase (HMBS) CCGGGTGGGCCAGATT; and reverse, HMBS GCTCCCTGACCCACAGCATA. Inventoried TaqMan primer sets (Life Technologies) were used for analyses of IL-6 (Mm00446190_m1), inducible nitric oxide synthase (iNOS) (Mm00440502_m1), occludin (Mm00500912_m1), ZO-1 (Mm00493699_m1), Glut2 (Mm00446229_m1), and GCG/proglucagon (Mm01269055_m1), using HMBS (Mm01143545_m1) or GAPDH (Mm99999915_g1) as control. RT-PCR conditions were 2 min at 50°C and 10 min at 95°C followed by 40 cycles of two-step PCR denaturation at 95°C for 15 s and annealing extension at 60°C for 1 min. Duplicate assay samples contained 50–200 ng cDNA and 6 μmol/L primers in 1× Power SYBR Green PCR Master Mix (ABI) or 1× TaqMan gene expression assay (probe and primers) in 1× TaqMan Gene Expression Master Mix (ABI) in a final volume of 20 or 25 μL. Means of duplicates were taken, and relative amount of target mRNA was normalized to HMBS or GAPDH levels as an endogenous control gene, data were analyzed according to the 2−ΔΔCT method, and fold difference was calculated between SPI and GP-SPI diet groups.
Statistical Analysis of Mouse Phenotypes
Mouse phenotypes were analyzed with STATISTICA, version 9.1 (StatSoft). One-way ANOVA was used to determine significance among three or more groups followed by the indicated post hoc test. Paired t tests were performed within groups (before versus after treatment), and unpaired t tests were used for independent groups.
16S rRNA Gene Sequencing and Analysis
16S rRNA gene sequencing was performed on 80 paired cecal and fecal samples from 40 C57BL/6J mice consuming HFD, SPI diet, GP-SPI diet, or LFD for 13 weeks (n = 10 mice per diet group). For HFD, SPI diet, and GP-SPI diet groups, three to four paired cecal and fecal samples were randomly chosen from each of three treatment cages. The LFD group contained a total of 10 mice (in two cages), and we used all paired fecal and cecal samples from the LFD group for 16S sequencing. DNA was extracted using the PowerSoil bacterial DNA extraction kit (MoBio, Carlsbad, CA) and PCR-amplified using barcoded universal bacterial primers targeting variable region 4 of the 16S rRNA gene: 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) (Integrated DNA Technologies, Coralville, IA). The following thermocycler protocol was used: denature at 94°C for 3 min and 35 cycles of 94°C for 45 s, 50°C for 30 s, and 72°C for 90 s, with a final extension at 72°C for 10 min (32). Triplicate reactions for each sample were pooled and amplification was confirmed by 1.5% gel electrophoresis. Amplicons were cleaned with the Ampure XP kit (Agencourt, Danvers, MA) and quantified using the Quant-iT Picogreen dsDNA Assay kit (Invitrogen, Carlsbad, CA). Barcoded amplicons from all 80 samples were pooled and sequenced on one lane of an Illumina HiSeq, resulting in >2 × 107 150-bp single-end reads. We obtained 253,135 ± 9,112 sequences per sample (range 138,728–460,521). To avoid bias owing to differences in sampling depth, we conducted analyses at a subsampled depth of 100,000 sequences. Sequences were analyzed on the Harvard Odyssey computational cluster using the Quantitative Insights Into Microbial Ecology (QIIME) software package (33). Operational taxonomic units were picked at 97% similarity against the Greengenes database (34) (constructed by the “nested_gg_workflow.py” script), which we trimmed to span only the 16S rRNA region flanked by our sequencing primers (positions 521–773). Bray-Curtis principal coordinate analysis was performed using the QIIME script “beta_diversity_through_plots.py.” Statistical analyses of microbial community structure (permutational multivariate ANOVA [PERMANOVA] and analysis of similarities [ANOSIM]) were performed using the QIIME script “compare_categories.py.” Microbial biomarker discovery was performed using the LEfSe algorithm (35), with an LDA score of three or above set as the threshold for significance.
Quantitative PCR Analysis of Microbial DNA
Quantitative PCR (qPCR) was performed on the 40 fecal samples employed in 16S rRNA gene sequencing. To quantify Akkermansia muciniphila (A. muciniphila) abundance, we used validated primers specific for A. muciniphila (AM1: 5′-CAGCACGTGAAGGTGGGGAC-3′; AM2: 5′-CCTTGCGGTTGGCTTCAGAT-3′) (36). To quantify total microbial DNA, we used universal bacterial primers 515F and 806R—the same used for 16S sequencing. For each reaction, template DNA was diluted to 0.5 ng/μL in 0.1% Tween 20, and then 2 µL (1 ng) was combined with 12.50 µL SYBR Green qPCR Mix (Applied Biosystems, Carlsbad, CA), 6 µL nuclease-free H2O, and 2.25 µL of each primer, for a total reaction volume of 25 µL. Standard curves were created using serial twofold dilutions of pure culture A. muciniphila genomic DNA. For qPCR with AM1/AM2, the standard curve employed genomic DNA in the following amounts per reaction (in picograms): 100, 50, 25, 12.5, 6.25, 3.13, 1.56, and 0.78, plus a nontemplate control (R2 = 0.9304). For qPCR with 515F/806R, the standard curve used genomic DNA in the following amounts per reaction (in nanograms): 10, 5, 2.5, 1.25, 0.63, 0.31, 0.16, and 0.08, plus a nontemplate control (R2 = 0.9949). The following RT-PCR protocol was run on a Stratagene MX3000P qPCR system (Agilent Technologies, Santa Clara, CA): 95°C for 15 min, followed by 40 cycles of 95°C for 15 s, 76°C (AM1/AM2) or 50°C (515F/806R) for 40 s, and 72°C for 30 s. A melting curve was performed after amplification to distinguish between the targeted and nontargeted PCR products. All reactions were performed in duplicate, with the mean value used for statistical analyses. Bacterial abundances were analyzed as genome equivalents, where A. muciniphila was assigned a multiplier of 3.42 × 105 genome equivalents per nanogram of DNA based on its genome size (2.66 Mbp; ATCC BAA-835), and the gut microbial community as a whole was assigned a multiplier of 2.03 × 105 based on a mean genome size of 4.50 Mbp. For qPCR analyses, sample-specific relative abundance of A. muciniphila was determined as genome equivalents amplified by AM1/AM2 divided by genome equivalents amplified by 515F/806R. Absolute abundance per gram of feces was determined by adjusting the concentrations of A. muciniphila and microbial DNA determined by qPCR for the dilutions performed during DNA extraction (1:50), normalization (dilution to 5 ng/μL), and qPCR set-up (1:10), and dividing this starting concentration by the total grams of feces utilized for the original DNA extraction. Relationships between qPCR and 16S rRNA results were determined by linear regression on log10-transformed data. DNA abundances across diet groups were compared by nonparametric Kruskal-Wallis ANOVA with Dunn correction for multiple comparisons. All analyses were conducted in Prism 6 (GraphPad Software, La Jolla, CA).
Results
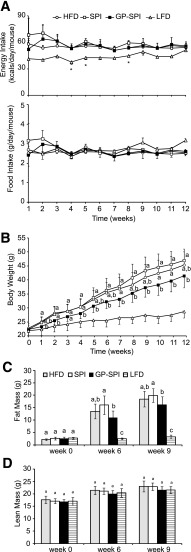
Effect of GP on Food Intake, Body Weight, and Body Composition
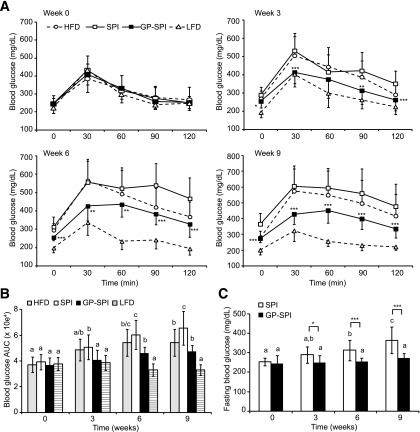
Based on the results of baseline OGTTs (Fig. 2A) (week 0) performed at 6 weeks of age, mice were assigned to three groups (n = 15 per group) to receive intervention diets consisting of HFD, HFD supplemented with 10% SPI (i.e., SPI diet), or HFD supplemented with 10% GP-SPI (i.e., GP-SPI diet). These three high fat–based diets were equivalent in terms of the percentage of kilocalories contributed by protein, carbohydrate, fiber, and fat; therefore, calorie consumption (56 ± 2.1 kcal/day/mouse) was similar between the high fat–based diet groups (Fig. 1A, top). Mice fed a diet matched for the same ingredients, but with 10% of kilocalories from fat (n = 10), were added as an additional control (i.e., LFD) and consumed an average of 44 ± 4 kcal/day/mouse (Fig. 1A, top). Diet formulation details are presented in Supplementary Table 2. Diet consumption was stable and was not significantly different for any of the groups when considering average weight of daily diet consumed per mouse (Fig. 1A, bottom), indicating the diets had equivalent palatability. Mice in the GP-SPI group ingested 26.2 ± 1.7 mg polyphenols (gallic acid equivalents) per day. Compared with the SPI diet group, mice ingesting the GP-SPI diet had significantly lower body weight at weeks 1 and 2 and then from 4 to 12 weeks (Fig. 1B), as well as significantly less adiposity (Fig. 1C). Adiposity in the HFD group was higher than in the GP-SPI group and lower than in the SPI group but not significantly different from either (Fig. 1C). Lean mass (Fig. 1D) and total body water (data not shown) were similar between all four groups. Mice consuming the LFD or GP-SPI diet had similar liver weights, which were significantly lower than those of mice fed the SPI diet or HFD (Table 1). In line with these findings, the lipid content of livers collected from GP-SPI diet–fed mice (112 ± 54 µg/mg of tissue) was significantly lower (P = 0.016) than that of SPI diet–fed mice (170 ± 58 µg/mg of tissue). We also observed that the ceca of mice fed the GP-SPI diet were enlarged and nearly double the mass of ceca among mice fed the SPI diet (Table 1), due to increased water retention (Table 2) (n = 5 mice per group). There was no significant difference in dry cecal weights between groups, although the GP-SPI group trended toward the highest mass (Table 2).
Figure 2.
Mice fed GP-SPI diet show improved fasting glucose and oral glucose tolerance. A: Blood glucose concentrations (mg/dL) expressed as mean ± SD (n = 15 for HFD, SPI diet, and GP-SPI diet; n = 10 for LFD) were measured at the indicated time points (0–120 min) after administration of 2 g/kg glucose to mice after they had consumed HFD, SPI diet, GP-SPI diet, or LFD for 0, 3, 6, or 9 weeks. LFD and HFD groups are shown for reference, and main analyses were performed on SPI and GP-SPI groups to assess the effect of GP supplementation. At each time point, a two-tailed t test was performed to evaluate the significance of differences between SPI and GP-SPI groups: *P < 0.05; **P < 0.01; ***P < 0.001. B: AUC representation of HFD, SPI, GP-SPI, and LFD group data in A; each bar represents the mean ± SD (n = 15 for HFD, SPI, and GP-SPI; n = 10 for LFD) for each group at indicated weeks. Between-group analyses were performed with one-way ANOVA followed by unequal honestly significantly different post hoc test. Significant differences between diet groups at the indicated week are signified by letters, where different letters indicate difference (P < 0.05) between groups, while the same letter indicates no difference. C: Blood glucose measurements (mg/dL) taken after a 4-h fast at indicated weeks. Each bar represents mean ± SD (n = 15) of SPI (white bars) and GP-SPI (black bars) diet groups. Within-group analyses comparing data at 0, 3, 6, and 9 weeks were performed with one-way ANOVA followed by Tukey post hoc test. Significant differences within group are signified by letters, where different letters indicate difference (P < 0.05) within group, while the same letter indicates no difference. Between-group differences at each week were determined by t test (two tailed): *P < 0.05; ***P < 0.001.
Figure 1.
GP-SPI diet reduces weight gain and adiposity of mice but not food intake or lean mass. A: Calorie consumption (mean ± SD) of HFD-based groups was not significantly different during intervention period, but LFD group showed lower calorie intake that was significant at weeks indicated by the asterisks (one-way ANOVA followed by unequal honestly significantly different [HSD] post hoc test, P < 0.05) (top). Food intake (mean ± SD) of mice on indicated diets during the intervention period was not significantly different between groups (one-way ANOVA followed by unequal HSD post hoc test) (bottom). B: Body weights (g) of mice (mean ± SD) consuming the indicated diets for the 12-week intervention period. One-way ANOVA followed by Tukey HSD post hoc test was performed on data at each time point for HFD, SPI diet, and GP-SPI diet groups. LFD group is shown as reference. C: Echo MRI data showing percentage of whole-body fat mass (mean ± SD) for each group. D: Echo MRI data showing percentage of whole-body lean mass (mean ± SD) for each group. C and D: One-way ANOVA followed by unequal HSD post hoc test was performed on data from the four diet groups at each time point. Significant difference between groups for each week is signified by letter a, b, or c; different letters indicate significant difference (P < 0.05) between groups, while the same letter indicates no difference.
Table 1.
Liver and cecum weights
| HFD (n = 15) | SPI (n = 15) | GP-SPI (n = 15) | LFD (n = 10) | |
|---|---|---|---|---|
| Liver (g) | 1.77 ± 0.463b | 2.05 ± 0.505b | 1.36 ± 0.441a | 1.22 ± 0.225a |
| Cecum (g) | 0.22 ± 0.057a | 0.21 ± 0.034a | 0.37 ± 0.061b | 0.23 ± 0.041a |
Data are expressed as mean and SD. One-way ANOVA followed by unequal N HSD test. Different letter superscripts indicate significant difference between diet groups (P < 0.05). HSD, honestly significantly different.
Table 2.
Cecal fresh weight, dry weight, and water content
| HFD (n = 5) | SPI (n = 5) | GP-SPI (n = 5) | |
|---|---|---|---|
| Fresh weight (g) | 0.20 ± 0.04b | 0.21 ± 0.04b | 0.35 ± 0.05a |
| Dry weight (g) | 0.06 ± 0.02a | 0.07 ± 0.01a | 0.09 ± 0.02a |
| Water weight (g) | 0.14 ± 0.02b | 0.14 ± 0.03b | 0.26 ± 0.04a |
Data are expressed as mean and SD. One-way ANOVA followed by Tukey HSD test. Different letter superscripts indicate significant difference between diet groups (P < 0.05). HSD, honestly significantly different.
GP Improve Glucose Tolerance
OGTTs were repeated at 3, 6, and 9 weeks. With the exception of week 3 at 60 min, the GP-SPI diet showed significantly better oral glucose tolerance at individual time points (Fig. 2A) and with respect to area under the blood glucose response curve (AUC) (Fig. 2B) in comparison with mice consuming the ingredient-matched SPI diet. The LFD group is presented as a reference for a normal OGTT profile (Fig. 2A and B). HFD group AUC values were not significantly different from those of the SPI and GP-SPI diet groups at weeks 3 and 6 (Fig. 2B). At week 9, the AUC of the SPI group was significantly higher than that of both HFD and GP-SPI (Fig. 2B). Fasting glucose levels in the SPI group also increased over the 9-week period (Fig. 2C); however, fasting glucose in the GP-SPI diet group remained unchanged and significantly lower than that in the SPI group over the same time period (Fig. 2C).
GP Attenuate Metabolic Endotoxemia and Systemic Inflammation
Compared with the SPI group, mice fed the GP-SPI diet had significantly lower blood serum levels of TNFα and undetectable levels of IL-6, indicating that GP attenuate systemic inflammation (Table 3). Compared with the SPI group, mice fed the GP-SPI diet also had significantly lower serum levels of bacterial LPS (Table 3), a potent inducer of systemic inflammation, which could explain the lower levels of host-derived inflammatory mediators. Total antioxidant status was significantly lower in the GP-SPI group compared with the SPI group, although the magnitude of this difference (i.e., 4.5%) was relatively small. Levels of serum cholesterol, triglycerides, and IL-1β in the GP-SPI group trended lower but were not significantly different from those in the SPI group.
Table 3.
Serum biochemistry
| SPI | GP-SPI | |
|---|---|---|
| Cholesterol (mg/dL) | 140.2 ± 6.6 | 134.0 ± 19.9 |
| Triglycerides (mg/dL) | 68.4 ± 18.0 | 49.6 ± 26.8 |
| TAS (mmol/L) | 1.63 ± 0.04 | 1.56 ± 0.06** |
| IL-1β (pg/mL) | 21.3 ± 24.5 | 11.3 ± 17.2 |
| IL-6 (pg/mL) | 1,080.6 ± 592.9 | nd*** |
| TNFα (pg/mL) | 19.8 ± 9.1 | 11.1 ± 2.7* |
| LPS (ng/mL) | 2.9 ± 1.1 | 1.6 ± 0.7** |
Data are expressed as mean and SD. nd, not detected (levels were <73.2 pg/mL detection limit of assay); TAS, total antioxidant status. Two-tailed t test,
*P < 0.05,
**P < 0.01, ***P < 0.001.
GP Counteract HFD-Induced Effects in the Gastrointestinal Tract
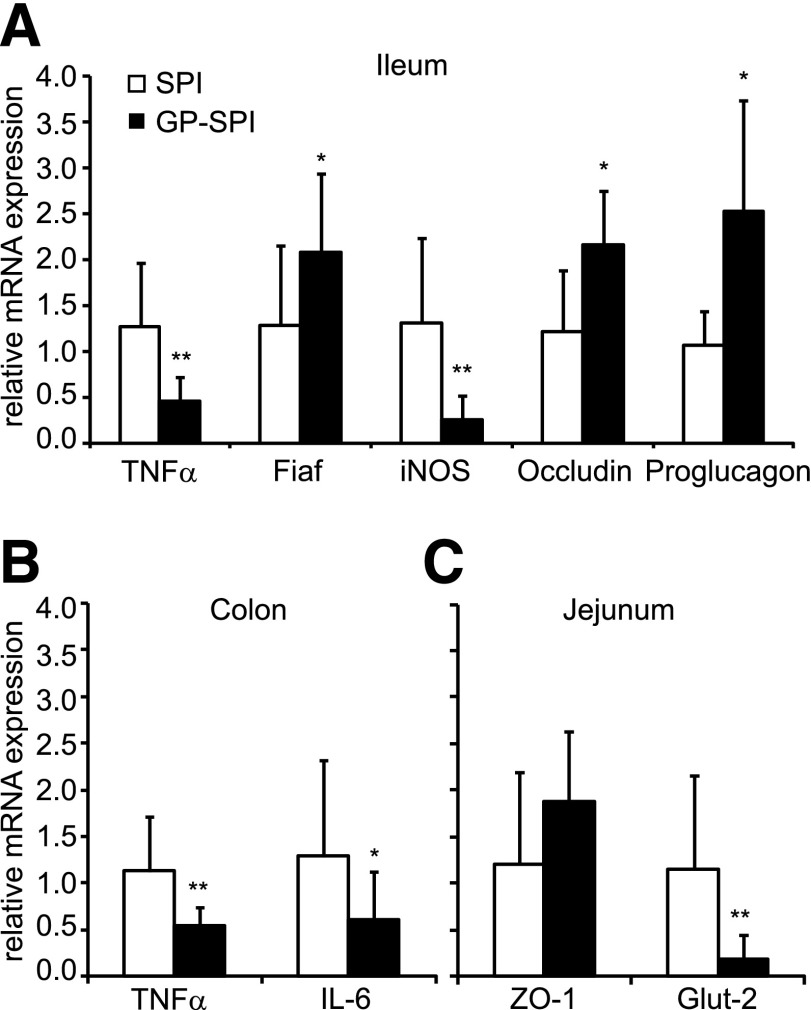
Interaction between the gut microbiota and HFD was reported to promote expression of proinflammatory cytokines in intestinal tissues before development of obesity and insulin resistance (4,5). Compared with the SPI diet group, mice fed the GP-SPI diet had significantly lower expression of the inflammatory mediators TNFα and iNOS in ileum tissue (Fig. 3A) as well as lower expression of TNFα and IL-6 in colon tissue (Fig. 3B).
Figure 3.
Intestinal tissues of mice fed GP-SPI diet show gene expression changes consistent with attenuated inflammation, increased gut barrier integrity, and improved metabolic function. A: Ileum tissues of mice fed GP-SPI diet have significantly decreased gene expression of TNFα and iNOS and increased Fiaf, occludin, and proglucagon (GCG) expression compared with control. B: Colon tissues of GP-SPI diet–fed mice have significantly higher expression of TNFα and IL-6. C: Jejunum tissues of GP-SPI diet–fed mice have significantly lower levels of Glut2. Data are means ± SD (n = 10 samples per group) for each RT-PCR experiment and were attained using the average of technical duplicates for each sample. Between-group differences were determined by t test (two tailed): *P < 0.05; **P < 0.01.
Fiaf is secreted by the ileum and functions as a circulating inhibitor of LPL, restraining its ability to import and store fatty acids in peripheral tissues (37). Elevated Fiaf levels have previously been shown to protect against diet-induced obesity (6,38). Compared with SPI diet–fed mice, Fiaf expression was significantly increased by twofold in ileum tissue of the GP-SPI mice, indicating that GP may suppress fatty acid storage (Fig. 3A).
HFD has been reported to reduce expression of tight junction proteins that control the permeability of the intestinal epithelium (13). Compared with the SPI diet–fed group, mice fed the GP-SPI diet showed a significant increase in occludin gene expression consistent with GP playing a role in maintaining intestinal barrier integrity (Fig. 3A). Compared with the SPI diet group, jejunal expression of the plaque protein ZO-1 trended higher but was not significantly increased in the GP-SPI group (Fig. 3C).
Proglucagon (GCG), expressed in enteroendocrine L cells of the ileal epithelium, is the precursor of glucagon-like peptide 1 (GLP-1) and GLP-2 proteins (39). GLP-1 promotes insulin production and secretion from pancreatic β-cells, while GLP-2 promotes mucosal and gut barrier integrity (39). The GP-SPI diet group showed significantly higher proglucagon expression in ileum tissue compared with the SPI diet group (Fig. 3A), suggesting higher GLP-1 and GLP-2 protein levels, which could contribute to the improved glucose tolerance and decreased presence of LPS in serum, respectively.
Finally, compared with the SPI diet group, jejunum tissue of the GP-SPI diet group showed significantly lower gene expression of Glut2 (Fig. 3C), the main GLUT of the small intestine (40), suggesting an additional mechanism for the observed improvement in glucose tolerance and lower body weight in these animals.
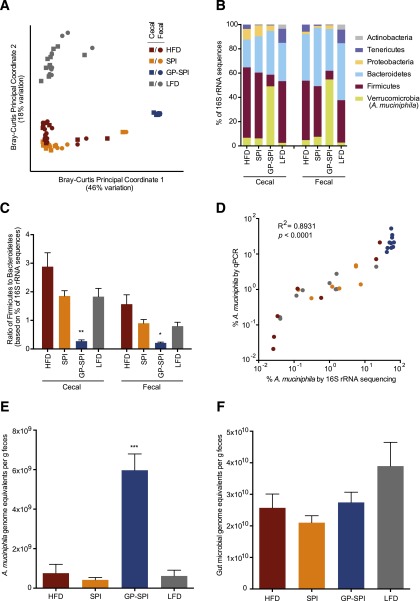
GP Alter the Gut Microbiota
16S rRNA gene sequencing of paired cecal and fecal samples (n = 10 per group) revealed that overall, microbial communities were strongly shaped by diet (PERMANOVA; F = 47.0543; P < 0.001). Remarkably, principal coordinate analysis showed that microbial community structure was more sensitive to polyphenol supplementation than to dietary fat (Fig. 4A), a conclusion also supported by quantitative variance partitioning (PERMANOVA; polyphenol F = 62.2547; fat F = 21.0509; both P < 0.001).
Figure 4.
GP supplementation has a dramatic impact on the gut microbiota. A: Bray-Curtis principal coordinate analysis of the 16S rRNA gene in cecal samples and fecal samples collected from mice after 13 weeks of consuming HFD, SPI diet, GP-SPI diet, or LFD. Note the strong separation of GP-SPI samples along PC1, which explains 46% of variation in the sample pool. Diets with a high-fat base (HFD, SPI diet, GP-SPI diet) separate from LFD along PC2, which explains an additional 18% of variation. B: Relative abundance of bacterial phyla. In both cecal and fecal samples, the GP-SPI diet is associated with a significantly higher abundance of Verrucomicrobia, specifically, A. muciniphila (Kruskal-Wallis ANOVA with Dunn correction for multiple comparisons; all GP-SPI pairwise P < 0.05). C: Ratio of the percentage of 16S rRNA gene sequences assigned to Firmicutes versus Bacteroidetes. In both cecal and fecal samples, the GP-SPI diet is associated with a significantly lower ratio (mean ± SEM) (Kruskal-Wallis ANOVA with Dunn correction for multiple comparisons; all GP-SPI pairwise P < 0.05). D: Strong relationship between the relative abundance of A. muciniphila in fecal samples analyzed by 16S rRNA gene sequencing (x-axis) and by qPCR (y-axis), where qPCR relative abundance was quantified by amplifying fecal DNA with primers specific for A. muciniphila (AM1/AM2) and universal bacterial V4 primers (515F/806R). R2 and P value reflect linear regression on log10-transformed data. E: Absolute abundance of A. muciniphila per gram of feces was higher in the GP-SPI group, with significant differences observed between GP-SPI and all other diets (mean ± SEM) (Kruskal-Wallis ANOVA with Dunn correction for multiple comparisons; all GP-SPI pairwise P < 0.05). F: qPCR revealed a trend toward lower microbial DNA on HFD-based diets (HFD, SPI diet, and GP-SPI diet) versus LFD, but total microbial DNA per gram of feces did not differ significantly among groups (mean ± SEM) (Kruskal-Wallis ANOVA; P = 0.2634). *P < 0.05; **P < 0.01; ***P < 0.001.
Cecal and fecal samples from the same host clustered together, although they could be distinguished statistically by permutation methods (ANOSIM; R2 = 0.0401; P = 0.042). To avoid pseudoreplication, we conducted additional analyses on the cecal and fecal subsets separately. Analysis of bacterial relative abundance confirmed prior reports that gut microbial communities of mice are dominated by bacteria from the Firmicutes and Bacteroidetes phyla (Fig. 4B) and that consumption of a nonsupplemented HFD increases the proportion of Firmicutes versus Bacteroidetes (7,41) (Fig. 4C). The addition of the SPI vehicle to the HFD had limited effects on community composition (Fig. 4A–C), although the SPI diet and HFD could still be distinguished by permutation methods (ANOSIM; cecal: R2 = 0.2144, P = 0.020; fecal: R2 = 0.2720, P = 0.010).
By contrast, the effects of adding GP were profound. Supplementation with GP dramatically increased the relative abundance of A. muciniphila within the Verrucomicrobia phylum (cecal: 6.2 ± 4.6% on SPI vs. 49.1 ± 2.0% on GP-SPI; fecal: 7.5 ± 4.7% on SPI vs. 54.8 ± 2.5% on GP-SPI [Fig. 4B]; cecal or fecal samples, considered separately: Kruskal-Wallis ANOVA with Dunn correction for multiple comparisons, P < 0.0001; all GP-SPI pairwise comparisons, P < 0.05). For both cecal and fecal samples, linear discriminant analysis of microbial biomarkers distinguishing the SPI and GP-SPI diets using LEfSe (35) ranked the increase in A. muciniphila as the strongest biomarker of GP supplementation when analyzed at all taxonomic levels from phylum to species (Supplementary Tables 3 and 4). Although other microbial changes also emerged as strong biomarkers of GP supplementation, including an increase in the genus Alistipes and decreases in several taxa within the order Clostridiales (Supplementary Tables 3 and 4), the increase in A. muciniphila was the only strong biomarker consistently detectable across taxonomic levels and sample type. Notably, supplementation with GP also reduced the ratio of Firmicutes to Bacteroidetes (Fig. 4C) (cecal or fecal samples, considered separately: Kruskal-Wallis ANOVA with Dunn correction for multiple comparisons, P < 0.0001; all GP-SPI pairwise comparisons, P < 0.05), a microbial profile consistent with the relatively lean phenotypes observed in these subjects.
Given the dramatic bloom of A. muciniphila within the GP-SPI diet group, we conducted qPCR on fecal DNA to validate and extend the 16S rRNA gene sequencing results. We analyzed qPCR-based bacterial abundance in terms of genome equivalents (see research design and methods). Relative abundance of A. muciniphila by qPCR was determined as genome equivalents amplified by primers specific to A. muciniphila versus those amplified by universal 16S rRNA gene primers. Absolute abundance per gram of feces was determined by adjusting genome equivalents for dilutions made during analysis and then dividing by the grams of feces in the original DNA extraction. We observed a strong and significant correlation between qPCR and 16S measurements of the fraction of microbial DNA attributable to A. muciniphila (linear regression on log10-transformed data; R2 = 0.8931; P < 0.0001) (Fig. 4D). Quantitative PCR confirmed that DNA attributable to A. muciniphila was uniquely elevated in the GP-SPI group (Fig. 4E), with significant differences observed between GP-SPI and all other diets (Kruskal-Wallis ANOVA with Dunn correction for multiple comparisons, P = 0.0002). Although there was a trend toward lower microbial DNA on high fat–based diets (HFD, SPI diet, and GP-SPI diet) versus LFD (Fig. 4F), total microbial DNA per gram of fecal material did not differ among groups (Kruskal-Wallis ANOVA, P = 0.2634). Together, these data suggest that the GP-SPI diet was associated with a bloom of A. muciniphila in absolute numbers as well as a contraction in the absolute abundance of other bacterial taxa.
Discussion
Dietary supplementation of an HFD with GP led to dramatic changes in gut microbial community structure, including a reduction in the ratio of Firmicutes to Bacteroidetes and a bloom of A. muciniphila. These changes may confer some degree of protection from the negative consequences of an HFD. HFD and high-fat, high-sugar diets have repeatedly been shown to increase the proportion of Firmicutes to Bacteroidetes (36), a microbial community structure that is sufficient to induce increased host body fat upon transplantation into germ-free mouse recipients (7,42). Furthermore, administration of A. muciniphila, but not heat-killed cells, to HFD-fed mice can reduce host adiposity, inflammatory markers, and insulin resistance (43). Similarly, consumption of oligofructose by ob/ob mice deficient for the leptin gene leads to a marked increase in A. muciniphila abundance and similar metabolic improvements (44). A. muciniphila increases in abundance after gastric bypass surgery in humans (45) and mice (42); the postsurgery gut microbiota is sufficient to cause weight loss and decreased adiposity upon transplantation into germ-free recipients (42).
It remains to be determined whether the interaction between dietary polyphenols and the gut microbiota is direct or indirect (i.e., mediated through altered host physiology). Multiple findings suggest that direct interactions are critical. We did not observe significant differences in mucin (Muc2) gene expression in jejunum or colon tissues from SPI and GP-SPI groups (data not shown), suggesting that A. muciniphila is not simply responding to increased host mucin production, its preferred carbon and nitrogen source (46). Consistent with this, a recent in vitro study revealed that polyphenols from a grape juice/red wine mixture can directly increase the abundance of A. muciniphila (47).
Two potential mechanisms for the observed impact of polyphenols on the gut microbiota involve their reported antimicrobial and antioxidant (free radical scavenging) effects (48,49). We had originally considered that the enhanced cecal size in the GP-SPI diet group was indicative of the former mechanism, due to the fact that multiple antibiotics are known to increase cecum size, as in germ-free mice (50–53). Surprisingly, we found that microbial abundance was similar across feeding groups whether measured by qPCR (Fig. 4F) or as total DNA isolated per gram of gut contents extracted (data not shown). These findings suggest that GP are not exerting a strong antimicrobial effect; however, it remains possible that they do selectively suppress the growth of some members of the gut microbiota, for example, microbes associated with maintaining normal cecum size (51,52). A. muciniphila is an obligate anaerobe, lacking protection against free oxygen radicals (46); therefore, it is tempting to speculate that strong oxygen radical scavenging capacity of poorly absorbed GP can provide a survival advantage to A. muciniphila and possibly other beneficial obligate anaerobes. If confirmed, this explanation may clarify the mechanism by which various classes of dietary antioxidants benefit human health.
Our results are strikingly consistent with a recent study in which cranberry polyphenol supplementation decreased body and liver weight gain in mice; improved insulin and glucose tolerance; lowered hepatic, intestinal, and plasma triglycerides; attenuated intestinal and hepatic inflammation; reduced serum LPS; and increased relative abundance of A. muciniphila (54). GP are typically a mixture of ACNs, type-B PACs with up to 13 degrees of polymerization, hydroxycinnamic acids, monomeric flavan-3-ols, and flavonols (19), while cranberry polyphenols are made up of type-A PACs in addition to the mentioned classes of compounds (55). The similarity of the host response to grape and cranberry polyphenols suggests that diverse types of polyphenols may have similar effects on gut microbiome. Additional studies are needed to clarify which compounds are responsible for the various metabolic outcomes.
The observed change in gut microbial community structure and the associated local and systemic effects reported in this study are consistent with protection against HFD-induced metabolic dysfunctions. We propose that this altered gut microbiota is, in part, responsible for the altered intestinal gene expression, epithelial integrity, and inflammatory markers, which then leads to decreased fat deposition and glucose absorption, along with increased insulin secretion. Elucidating the mechanisms linking dietary bioactive compounds and our gut microbiota will likely be an essential tool in our arsenal against the increasing global burden of metabolic disease.
Article Information
Acknowledgments. The authors thank Joshua Bergman (Rutgers) and Amy Tsou (Harvard) for technical assistance. Blood serum chemistry was performed by the Clinical Chemistry Laboratory at Pennington Biomedical Research Center. Sequencing reads are in the MG-RAST (56) under the accession number 13326.
Funding. This work was partly supported by P50-AT-002776-01, a pilot award to D.E.R. from the Botanical Research Center (027693-001-003), and R01-AT-008618-01 from the National Center for Complementary and Alternative Medicine and the Office of Dietary Supplements. R.N.C. was supported by the National Institutes of Health (F32-DK-101154). P.J.T. was supported by the Harvard Bauer Fellows program and University of California, San Francisco, Departmental Funds.
Duality of Interest. This work was supported in part by SBIR Phase I grant 2011-33610-30489 and SBIR Phase II grant 2012-33610-20106 awarded to Nutrasorb, LLC, from the U.S. Department of Agriculture. D.E.R. and I.R. have equity interests in and K.M. is employed by Nutrasorb, LLC. No other potential conflicts of interest relevant to this article were reported.
Author Contributions. D.E.R. designed experiments and performed statistical analysis. D.E.R., R.N.C., and P.J.T. wrote the manuscript. D.E.R., P.K., K.M., and P.R.-S. performed animal studies. R.N.C. performed 16S amplicon sequencing, qPCR, and microbiota data analysis. I.R. provided oversight for the work. All authors read and approved the final manuscript. D.E.R. and P.J.T. are the guarantors of this work and, as such, had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db14-1916/-/DC1.
References
- 1.Wilson PWF, D’Agostino RB, Parise H, Sullivan L, Meigs JB. Metabolic syndrome as a precursor of cardiovascular disease and type 2 diabetes mellitus. Circulation 2005;112:3066–3072 [DOI] [PubMed] [Google Scholar]
- 2.Cani PD, Osto M, Geurts L, Everard A. Involvement of gut microbiota in the development of low-grade inflammation and type 2 diabetes associated with obesity. Gut Microbes 2012;3:279–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ding S, Lund PK. Role of intestinal inflammation as an early event in obesity and insulin resistance. Curr Opin Clin Nutr Metab Care 2011;14:328–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lam YY, Ha CW, Campbell CR, et al. Increased gut permeability and microbiota change associate with mesenteric fat inflammation and metabolic dysfunction in diet-induced obese mice. PLoS ONE 2012;7:e34233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ding S, Chi MM, Scull BP, et al. High-fat diet: bacteria interactions promote intestinal inflammation which precedes and correlates with obesity and insulin resistance in mouse. PLoS ONE 2010;5:e12191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bäckhed F, Manchester JK, Semenkovich CF, Gordon JI. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc Natl Acad Sci U S A 2007;104:979–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turnbaugh PJ, Bäckhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 2008;3:213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006;444:1027–1031 [DOI] [PubMed] [Google Scholar]
- 9.Vijay-Kumar M, Aitken JD, Carvalho FA, et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science 2010;328:228–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cani PD, Amar J, Iglesias MA, et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 2007;56:1761–1772 [DOI] [PubMed] [Google Scholar]
- 11.Cani PD, Possemiers S, Van de Wiele T, et al. Changes in gut microbiota control inflammation in obese mice through a mechanism involving GLP-2-driven improvement of gut permeability. Gut 2009;58:1091–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghoshal S, Witta J, Zhong J, de Villiers W, Eckhardt E. Chylomicrons promote intestinal absorption of lipopolysaccharides. J Lipid Res 2009;50:90–97 [DOI] [PubMed] [Google Scholar]
- 13.Geurts L, Neyrinck AM, Delzenne NM, Knauf C, Cani PD. Gut microbiota controls adipose tissue expansion, gut barrier and glucose metabolism: novel insights into molecular targets and interventions using prebiotics. Benef Microbes 2014;5:3–17 [DOI] [PubMed] [Google Scholar]
- 14.Visioli F, Davalos A. Polyphenols and cardiovascular disease: a critical summary of the evidence. Mini Rev Med Chem 2011;11:1186–1190 [DOI] [PubMed] [Google Scholar]
- 15.Visioli F, De La Lastra CA, Andres-Lacueva C, et al. Polyphenols and human health: a prospectus. Crit Rev Food Sci Nutr 2011;51:524–546 [DOI] [PubMed] [Google Scholar]
- 16.Chuang CC, McIntosh MK. Potential mechanisms by which polyphenol-rich grapes prevent obesity-mediated inflammation and metabolic diseases. Annu Rev Nutr 2011;31:155–176 [DOI] [PubMed]
- 17.Xu Y, Simon JE, Welch C, et al. Survey of polyphenol constituents in grapes and grape-derived products. J Agric Food Chem 2011;59:10586–10593 [DOI] [PubMed] [Google Scholar]
- 18.Liang Z, Yang Y, Cheng L, Zhong GY. Characterization of polyphenolic metabolites in the seeds of Vitis germplasm. J Agric Food Chem 2012;60:1291–1299 [DOI] [PubMed] [Google Scholar]
- 19.Roopchand DE, Kuhn P, Krueger CG, Moskal K, Lila MA, Raskin I. Concord grape pomace polyphenols complexed to soy protein isolate are stable and hypoglycemic in diabetic mice. J Agric Food Chem 2013;61:11428–11433 [DOI] [PubMed] [Google Scholar]
- 20.Roopchand DE, Kuhn P, Poulev A, et al. Biochemical analysis and in vivo hypoglycemic activity of a grape polyphenol-soybean flour complex. J Agric Food Chem 2012;60:8860–8865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Felgines C, Krisa S, Mauray A, et al. Radiolabelled cyanidin 3-O-glucoside is poorly absorbed in the mouse. Br J Nutr 2010;103:1738–1745 [DOI] [PubMed] [Google Scholar]
- 22.Czank C, Cassidy A, Zhang Q, et al. Human metabolism and elimination of the anthocyanin, cyanidin-3-glucoside: a (13)C-tracer study. Am J Clin Nutr 2013;97:995–1003 [DOI] [PubMed] [Google Scholar]
- 23.Choy YY, Jaggers GK, Oteiza PI, Waterhouse AL. Bioavailability of intact proanthocyanidins in the rat colon after ingestion of grape seed extract. J Agric Food Chem 2013;61:121–127 [DOI] [PubMed] [Google Scholar]
- 24.Abia R, Fry SC. Degradation and metabolism of 14C-labelled proanthocyanidins from carob (Ceratonia siliqua) pods in the gastrointestinal tract of the rat. J Sci Food Agric 2001;81:1156–1165 [Google Scholar]
- 25.Selma MV, Espín JC, Tomás-Barberán FA. Interaction between phenolics and gut microbiota: role in human health. J Agric Food Chem 2009;57:6485–6501 [DOI] [PubMed] [Google Scholar]
- 26.Ou KQ, Gu LW. Absorption and metabolism of proanthocyanidins. J Funct Foods 2014;7:43–53 [Google Scholar]
- 27.Gonthier MP, Donovan JL, Texier O, Felgines C, Remesy C, Scalbert A. Metabolism of dietary procyanidins in rats. Free Radic Biol Med 2003;35:837–844 [DOI] [PubMed] [Google Scholar]
- 28.Roopchand DE, Grace MH, Kuhn P, et al. Efficient sorption of polyphenols to soybean flour enables natural fortification of foods. Food Chem 2012;131:1193–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roopchand DE, Kuhn P, Rojo LE, Lila MA, Raskin I. Blueberry polyphenol-enriched soybean flour reduces hyperglycemia, body weight gain and serum cholesterol in mice. Pharmacol Res 2013;68:59–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ribnicky DM, Roopchand DE, Oren A, et al. Effects of a high fat meal matrix and protein complexation on the bioaccessibility of blueberry anthocyanins using the TNO gastrointestinal model (TIM-1). Food Chem 2014;142:349–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cequier-Sánchez E, Rodríguez C, Ravelo AG, Zárate R. Dichloromethane as a solvent for lipid extraction and assessment of lipid classes and fatty acids from samples of different natures. J Agric Food Chem 2008;56:4297–4303 [DOI] [PubMed] [Google Scholar]
- 32.Caporaso JG, Lauber CL, Walters WA, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 2012;6:1621–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010;7:335–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DeSantis TZ, Hugenholtz P, Larsen N, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 2006;72:5069–5072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Segata N, Izard J, Waldron L, et al. Metagenomic biomarker discovery and explanation. Genome Biol 2011;12:R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carmody RN, Gerber GK, Luevano JM, et al. Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe 2015;17:72–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoshida K, Shimizugawa T, Ono M, Furukawa H. Angiopoietin-like protein 4 is a potent hyperlipidemia-inducing factor in mice and inhibitor of lipoprotein lipase. J Lipid Res 2002;43:1770–1772 [DOI] [PubMed] [Google Scholar]
- 38.Bäckhed F, Ding H, Wang T, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci U S A 2004;101:15718–15723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sinclair EM, Drucker DJ. Proglucagon-derived peptides: mechanisms of action and therapeutic potential. Physiology (Bethesda) 2005;20:357–365 [DOI] [PubMed] [Google Scholar]
- 40.Kellett GL, Brot-Laroche E, Mace OJ, Leturque A. Sugar absorption in the intestine: the role of GLUT2. Annu Rev Nutr 2008;28:35–54 [DOI] [PubMed] [Google Scholar]
- 41.Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med 2009;1:6ra14 [DOI] [PMC free article] [PubMed]
- 42.Liou AP, Paziuk M, Luevano JM Jr, Machineni S, Turnbaugh PJ, Kaplan LM. Conserved shifts in the gut microbiota due to gastric bypass reduce host weight and adiposity. Sci Transl Med 2013;5:178ra141 [DOI] [PMC free article] [PubMed]
- 43.Everard A, Belzer C, Geurts L, et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci U S A 2013;110:9066–9071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Everard A, Lazarevic V, Derrien M, et al. Responses of gut microbiota and glucose and lipid metabolism to prebiotics in genetic obese and diet-induced leptin-resistant mice. Diabetes 2011;60:2775–2786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang H, DiBaise JK, Zuccolo A, et al. Human gut microbiota in obesity and after gastric bypass. Proc Natl Acad Sci U S A 2009;106:2365–2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Derrien M, Vaughan EE, Plugge CM, de Vos WM. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol 2004;54:1469–1476 [DOI] [PubMed] [Google Scholar]
- 47.Kemperman RA, Gross G, Mondot S, et al. Impact of polyphenols from black tea and red wine/grape juice on a gut model microbiome. Food Res Int 2013;53:659–669 [Google Scholar]
- 48.El Gharras H. Polyphenols: food sources, properties and applications – a review. Int J Food Sci Technol 2009;44:2512–2518 [Google Scholar]
- 49.Daglia M. Polyphenols as antimicrobial agents. Curr Opin Biotechnol 2012;23:174–181 [DOI] [PubMed] [Google Scholar]
- 50.Reikvam DH, Erofeev A, Sandvik A, et al. Depletion of murine intestinal microbiota: effects on gut mucosa and epithelial gene expression. PLoS ONE 2011;6:e17996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Loesche WJ. Effect of bacterial contamination on cecal size and cecal contents of gnotobiotic rodents. J Bacteriol 1969;99:520–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Savage DC, Dubos R. Alterations in the mouse cecum and its flora produced by antibacterial drugs. J Exp Med 1968;128:97–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Savage DC, McAllister JS. Cecal enlargement and microbial flora in suckling mice given antibacterial drugs. Infect Immun 1971;3:342–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anhê FF, Roy D, Pilon G, et al. A polyphenol-rich cranberry extract protects from diet-induced obesity, insulin resistance and intestinal inflammation in association with increased Akkermansia spp. population in the gut microbiota of mice. Gut 2015;64:872–883 [DOI] [PubMed] [Google Scholar]
- 55.Côté J, Caillet S, Doyon G, Sylvain JF, Lacroix M. Bioactive compounds in cranberries and their biological properties. Crit Rev Food Sci Nutr 2010;50:666–679 [DOI] [PubMed] [Google Scholar]
- 56.Meyer F, Paarmann D, D’Souza M, et al. The metagenomics RAST server - a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 2008;9:386. [DOI] [PMC free article] [PubMed] [Google Scholar]