Abstract
G protein-coupled receptor kinases (GRKs) play an important role in the desensitization of G protein-mediated signaling of G protein-coupled receptors (GPCRs). The level of interest in mapping their phosphorylation sites has increased because recent studies suggest that the differential pattern of receptor phosphorylation has distinct biological consequences. In vitro phosphorylation experiments using well-controlled systems are useful for deciphering the complexity of these physiological reactions and understanding the targeted event. Here, we report on the phosphorylation of the class A GPCR neurotensin receptor 1 (NTSR1) by GRKs under defined experimental conditions afforded by nanodisc technology. Phosphorylation of NTSR1 by GRK2 was agonist-dependent, whereas phosphorylation by GRK5 occurred in an activation-independent manner. In addition, the negatively charged lipids in the immediate vicinity of NTSR1 directly affect phosphorylation by GRKs. Identification of phosphorylation sites in agonist-activated NTSR1 revealed that GRK2 and GRK5 target different residues located on the intracellular receptor elements. GRK2 phosphorylates only the C-terminal Ser residues, whereas GRK5 phosphorylates Ser and Thr residues located in intracellular loop 3 and the C-terminus. Interestingly, phosphorylation assays using a series of NTSR1 mutants show that GRK2 does not require acidic residues upstream of the phospho-acceptors for site-specific phosphorylation, in contrast to the β2-adrenergic and μ-opioid receptors. Differential phosphorylation of GPCRs by GRKs is thought to encode a particular signaling outcome, and our in vitro study revealed NTSR1 differential phosphorylation by GRK2 and GRK5.
G protein-coupled receptors (GPCRs) make up the largest superfamily of eukaryotic integral membrane proteins.1 Despite their diversity, a common mechanism involving a class of GPCR kinases (GRKs) regulates their desensitization.2,3 Termination of GPCR signaling is caused by receptor phosphorylation at intracellular Ser or Thr residues, followed by arrestin binding that blocks the interaction between activated GPCRs and G proteins. Recent studies have suggested that individual GRKs contribute differently to the phosphorylation of a given GPCR, and that a distinct pattern of phosphorylation established by each GRK may trigger distinct signaling events.4−7 Determining the pattern of GPCR phosphorylation by each GRK is therefore important, as these signaling mechanisms are potential pharmacological targets. Although mapping phosphorylation sites on various GPCRs has been reported using in vivo and in vitro methods,8−11 inherent complexities have made it difficult to map the phospho-acceptor residues used by individual GRKs.12−14 Overexpressed receptors in the in vivo assays can differ from endogenous receptors, as GRK regulation of GPCRs is likely to vary between cell and tissue types and GPCRs can be phosphorylated by other Ser/Thr kinases. In vitro GRK phosphorylation assays usually need to be executed on a GPCR embedded in a lipid bilayer, as lipid–GRK interactions are typically required for GRK function.2,15 However, the GPCR oligomeric state and topology of insertion cannot be controlled with most methods.16 A detergent-free environment is also typically required, as GRK activity can be inhibited by detergents.17
Neurotensin (NTS) is a 13-residue neuropeptide18 with diverse biological activities, ranging from cancer growth19 to Parkinson’s disease.20 Most effects of NTS are mediated by NTS receptor 1 (NTSR1), a class A GPCR.21 Although the mechanism of desensitization of NTSR1 by binding of arrestin to phosphorylated NTSR1 in HEK293 and COS7 cells was demonstrated,22−24 these studies could not identify the specific residues phosphorylated or the GRKs responsible for that phosphorylation. Therefore, we designed this study to identify GRK-specific phosphorylation of NTSR1 in vitro under well-controlled conditions offered by nanodisc technology. One advantage of using GPCRs incorporated into nanodiscs with respect to GRK function is that the nanodisc lipid bilayer offers a platform by which GRK–lipid interactions required for GRK activity can be preserved25−28 while maintaining a nativelike lipid environment for the receptor.29,30 Using a combination of studies of receptor mutants and liquid chromatography followed by tandem mass spectrometry (LC/MS/MC),7,31 this method allows for identification of the phosphorylation pattern provided by each GRK and offers insight into the complexity of these physiological reactions. Here we report the unique phosphorylation of NTSR1 by GRK2 and GRK5, which belong to the GRK2 and GRK4 subfamilies, respectively. We found that (i) GRK2 showed agonist-dependent phosphorylation whereas GRK5 showed activation-independent phosphorylation, (ii) the negatively charged lipids in the vicinity of the receptor are required for phosphorylation, and (iii) GRK2 and GRK5 produced different phosphorylation patterns. In addition, we demonstrated that the activity of GRK2 did not require acidic residues upstream of the phospho-acceptors, in contrast to the β2-adrenergic (β2AR)8 and μ-opioid (μOP)31 receptors.
Materials and Methods
Materials
[3H]NTS {[3,11-tyrosyl-3,5-3H(N)]-pyroGlu-Leu-Tyr-Glu-Asn-Lys-Pro-Arg-Arg-Pro-Tyr-Ile-Leu} and [γ-32P]ATP were purchased from PerkinElmer. Unlabeled NTS was synthesized by the Center for Biologics Evaluation and Research (Food and Drug Administration, Silver Spring, MD). SR48692 {2-[(1-(7-chloro-4-quinolinyl)-5-(2,6-dimethoxyphenyl)pyrazol-3-yl)carbonylamino]tricyclo(3.3.1.1.3.7)decan-2-carboxylic acid} was purchased from Sigma-Aldrich (St. Louis, MO). 1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) and 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-1′-rac-glycerol (POPG) were purchased from Avanti Polar Lipids (Alabaster, AL). Tobacco etch virus (TEV) protease mutant His-TEV(S219V)-Arg was prepared as previously reported.32 The expression and purification of MSP1E3D1 were performed as described in the Supporting Information.
Preparation of NTSR1 Embedded in Nanodiscs
The NTSR1 wild type (WT) and mutants were expressed in Escherichia coli as fusion proteins (NTSR1f) consisting of the E. coli maltose-binding protein (MBP), followed by a TEV protease recognition site, the rat NTSR1 (the N-terminally truncated rat NTSR1 starting at Thr43), a second TEV protease recognition site at the receptor C-terminus, followed by E. coli thioredoxin (TrxA), and a decahistidine (H10) tag, and purified as described previously.33 Nanodisc reconstitutions were conducted as previously reported with modification.30,33 A brief description is provided in the Supporting Information.
Preparation of GRKs
GRK2S670A-H6 was expressed as a human GRK2 S670A mutant containing a C-terminal hexahistidine tag. Bovine GRK5-H6 was expressed in a truncated form ending at residue 561, followed by a hexahistidine tag. Both GRKs were expressed in baculovirus-infected High-5 cells and purified using a common procedure consisting of Ni-NTA affinity, followed by Source15S cation exchange, and tandem S200 size exclusion chromatography as previously described.34−37 The purity of each GRK variant was judged by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE).
Sedimentation Velocity Analytical Ultracentrifugation
Sedimentation velocity analytical ultracentrifugation (SV-AUC) was performed to characterize the nanodisc preparations and establish the receptor and scaffold protein to lipid stoichiometry as described in the Supporting Information.
Radiolabeled Ligand Binding Assay
NTSR1f nanodiscs were treated with TEV protease prior to ligand binding experiments, to cleave off the MBP and TrxA-H10 fusion proteins and generate NTSR1 with near authentic N- and C-termini (see reconstitution and purification of NTSR1f nanodiscs in the Supporting Information). Single-point ligand binding experiments and saturation binding experiments were conducted as described previously.30 The Kd values were unchanged except for that of WT in 100% POPC nanodiscs. Therefore, the amount of specifically bound [3H]NTS was corrected for fractional occupancy using a Kd of 1.0 nM for NTSR1 WT in 100% POPC nanodisc and a Kd of 0.5 ± 0.2 nM for all other cases.
Phosphorylation Assay
Because of aggregation and loss of material during concentration and dialysis, NTSR1f nanodiscs were used without further treatment in the elution buffer containing 50 mM Tris (pH 7.4), 200 mM NaCl, and 200 mM imidazole. Phosphorylation assays were conducted as described previously38 with slight modifications. Briefly, NTSR1 nanodiscs with various POPC:POPG ratios were phosphorylated in 20 mM Tris (pH 7.4), 2 mM MgCl2, 0.4 mM dithiothreitol, 40 mM NaCl, and 40 mM imidazole (final concentration) for 10 min at 30 °C. Analytical reactions were conducted using 95 nM receptor and 310 nM GRK2 or 350 nM GRK5 in a volume of 20 μL with [γ-32P]ATP (final specific activity of 500–1000 cpm/pmol) in the absence of ligand and in the presence of NTS (60 nM) or SR48692 (60 nM) and stopped by the addition of 7 μL of NuPage 4×LDS sample buffer (Invitrogen), and the mixtures were resolved via SDS–PAGE (NuPAGE 4–12% Bis-Tris gel, 1× MES running buffer, Invitrogen). Gels were stained with SimplyBlue SafeStain (Invitrogen), dried, and exposed to X-ray film (Kodak Bio XAR Film, Kodak, Rochester, NY) for 14–17 h. NTSR1 bands were then excised, and the radioactivity was quantified in a liquid scintillation counter. Means ± the standard deviation (SD, error bars) from two experiments performed in duplicate are shown. The data were statistically analyzed using one-way analysis of variance (ANOVA) with a Dunnett test or two-way ANOVA with a Holm-Šídák test for multiple comparisons in GraphPad Prism (version 6.05, GraphPad Software).
Identification of Phosphorylated Amino Acids by Mass Spectrometry
NTSR1 in nanodiscs prepared using 75% POPC and 25% POPG were phosphorylated by GRKs in a volume of 200–250 μL, as described in the previous section, except for the addition of [γ-32P]ATP. Two 10 μL aliquots were treated with [γ-32P]ATP and incubated in parallel to confirm the phosphorylation stoichiometry. After separation of the sample by SDS–PAGE and staining of the gel, the bands of interest were excised. The following steps were performed by the Taplin Biological Mass Spectrometry Facility (Harvard Medical School, Boston, MA). The excised bands were subjected to in-gel digestion with trypsin or chymotrypsin. Phosphopeptides were separated by in-house packed C18 reverse phase HPLC columns (25–30 cm in length with an internal diameter of 100 μm, Accucore beads with a 2.6 μm diameter). Peptide sequencing analysis was performed by the LTQ-Orbitrap-Pro mass spectrometer (Thermo Fisher Scientific, Inc.). The LC/MS/MS data were searched against the sequence of NTSR1. The TMSFcore based on Ascore algorithm was used on the internal server of the Taplin Biological Mass Spectrometry Facility to determine whether the phosphorylation could be confidently assigned to a specific residue. The Ascore algorithm is a probability-based score that measures the probability of correct phosphorylation site localization based on the presence and the intensity of site-determining ions in the phosphopeptide MS/MS spectra.39 Phosphopeptides with an Ascore greater than or equal to 19 always produced >99% certainty, and those with an Ascore of 15–19 produced a >90% success rate. These were used for the identification of phosphorylation sites.
Results
NTSR1f Is Monomeric in Nanodiscs
We utilized a zwitterionic POPC and monovalent negatively charged POPG for NTSR1 nanodisc reconstitutions. Both POPC and POPG have a phase transition temperature below 0 °C,40,41 and a cylindrical molecular shape.42 Although POPG is a minor constituent in eukaryotic membranes,42 it was chosen instead of the more abundant 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-l-serine because the latter has a higher transition temperature of 14 °C.43 All reconstitution experiments were conducted at 4 °C or on ice, to preserve NTSR1 activity and remain above the phase transition temperatures of POPC and POPG. Nanodisc reconstitution was performed using NTSR1f, POPC along with 0–50% of POPG, and MSP1E3D1. The NTSR1f nanodiscs were characterized by SV-AUC to establish their biophysical properties, receptor stoichiometry, and protein:lipid stoichiometry.30,33 SV-AUC experiments showed the presence of a major species at 6.6–7.0 S, representing 35–50% of the loading absorbance (Table 1 and Figure 1). We noted that the sedimentation coefficient increases slightly with an increasing POPG content, reflecting in part the smaller partial specific volume of POPG.33 The best-fit frictional ratios f/f0 of ∼1.3 were consistent with the expected globular shape of the receptor–nanodisc assembly. An analysis of the absorbance and interference signal intensities for the major species leads to a lipid stoichiometry of 120–150 molecules per receptor, confirming the presence of one NTSR1f and two MSP1E3D1 molecules per receptor–nanodisc assembly. Simple volume and shape calculations yielded an expected sedimentation coefficient of ∼6.8 S for a receptor in nanodiscs having a 150:2:1 lipid:MSP1E3D1:NTSR1f stoichiometry. The same analysis was performed for empty nanodiscs (Table S1 and Figure S1 of the Supporting Information). It revealed 190–220 phospholipid molecules per nanodisc, confirming the presence of two MSP1E3D1 molecules per empty nanodisc. On the basis of the experimentally determined lipid stoichiometries, monomeric NTSR1f in MSP1E3D1 nanodiscs displaces ∼65–70 phospholipid molecules, in close agreement with observations made for MSP1D1 nanodiscs containing one receptor.30,33
Table 1. Characterization of NTSR1f–Nanodisc Complexes by Sedimentation Velocity.
| nanodisc lipid compositiona | s20,w (S) | Mexp (kDa) | % load, c (μM)b | f/f0c | lipidd (εJ/ε280) | Rhe (nm) |
|---|---|---|---|---|---|---|
| POPC (2) | 6.65 ± 0.03 | 330 ± 40 | 35 (1.6) | 1.32 | 152 | 5.8 |
| 25% POPG (3) | 7.0 ± 0.2 | 325 ± 25 | 47 (2.0–2.8) | 1.26 | 120 | 5.7 |
Experimental sedimentation coefficients and molar masses represent average values for the major species observed in the c(s) distribution. Averages are obtained from independent experiments (numbers parentheses) and are based on both the absorbance and interference data.
Percent of the loading absorbance that represents the major species of interest. The corresponding concentration of this species is indicated in parentheses.
Best-fit frictional ratios from the continuous c(s) distribution in SEDFIT.
Lipid stoichiometries per single nanodisc based on the presence of two MSP1E3D1 molecules, and in the case of the receptor nanodisc, one molecule of NTSR1f. Data are based on signal contributions of the major species to the absorbance (protein alone) and interference (protein and lipid) data.
Hydrodynamic radii based on the sedimentation coefficient and calculated values for the molar mass and partial specific volume.
Figure 1.
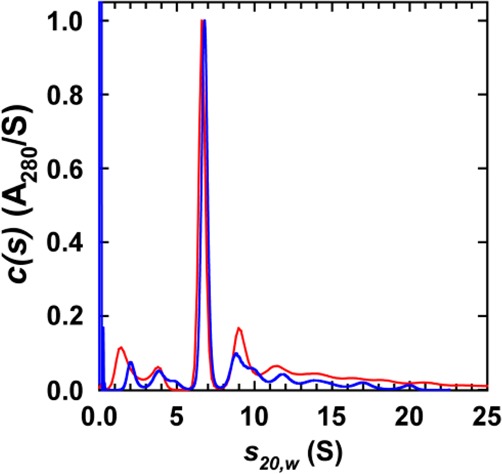
Sedimentation velocity-analytical ultracentrifugation of NTSR1f nanodiscs. Absorbance sedimentation velocity c(s) distributions obtained for NTSR1f nanodiscs reconstituted with POPC (red) and 75% POPC with 25% POPG (blue). Similar profiles were obtained using interference data. In all cases, data were collected at 10 °C and 40000 rpm in a Beckman Coulter An50 Ti rotor on a Beckman Coulter ProteomeLab XL-I analytical ultracentrifuge, as described previously.33
GRK2 Shows Agonist-Dependent Phosphorylation, but GRK5 Also Phosphorylates Inactive NTSR1
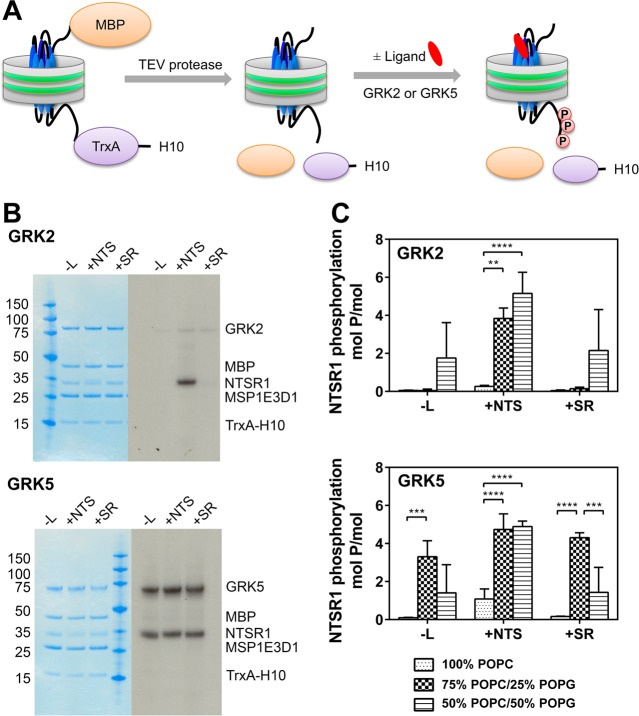
We first tested the effects of lipid and ligand on monomeric NTSR1 phosphorylation by GRK2 and GRK5. NTSR1f nanodiscs were treated with TEV protease to generate NTSR1 with near authentic N- and C-termini30 (Figure 2A). This preparation was assayed for phosphorylation in the absence of ligand, in the presence of NTS, and with added antagonist SR4869244 (Figure 2B,C). In the case of GRK2, only agonist-activated NTSR1 was phosphorylated to yield ∼4–5 mol of phosphate/mol of receptor in the presence of either 25 or 50% POPG. In contrast, GRK5 phosphorylated NTSR1 with the same yield as GRK2 in the presence of POPG, but in an activation-independent manner. GRK5 has a highly basic region in the C-terminal domain, allowing it to bind phospholipid surfaces.45 However, soluble GRK2 is thought to interact with Gβγ2,46 to facilitate binding to the membrane. To test whether membrane association affected phosphorylation by GRK2, NTSR1 phosphorylation by GRK2 was tested in the presence of Gβ1γ1, an isoform that was shown to interact with NTSR1 in combination with Gαq,30 and that can be purified in the absence of detergent, unlike Gβ1γ2.47 We observed no clear effect of Gβ1γ1 on GRK2 activity toward agonist-activated NTSR1 embedded in 75% POPC/25% POPG nanodiscs when Gβ1γ1 was titrated with agonist-activated NTSR1/GRK2 mixtures, even at high ratios of Gβ1γ1 to GRK2 (Figure S2 of the Supporting Information). This result also shows that activation-independent phosphorylation by GRK5 may not depend on its constitutive membrane association and may in fact reflect the inherent functional properties of GRK5 as observed for the dopamine D1 receptor,48 β2AR, and M2 muscarinic receptor.49
Figure 2.
NTSR1 phosphorylation by GRK2 and GRK5. (A) NTSR1f nanodiscs were treated with TEV protease to remove the N-terminal maltose-binding protein (MBP) and C-terminal E. coli thioredoxin-decahistidine (TrxA-H10) tail. NTSR1 nanodiscs were then phosphorylated by GRKs in the presence or absence of ligand. (B) Phosphorylation of NTSR1 by GRK2 and GRK5. Monomeric NTSR1 embedded in nanodiscs prepared using MSP1E3D1 and a 75% POPC/25% POPG mixture (95 nM) was phosphorylated by GRK2 (310 nM) and GRK5 (350 nM) in the absence of ligand (-L) or in the presence of agonist (neurotensin; NTS) or antagonist (SR48692; SR), as described in Materials and Methods, and then resolved by SDS–PAGE (left). The gel was stained with SimplyBlue SafeStain, dried, and then exposed to X-ray film for 14 h (right). (C) NTSR1 nanodiscs prepared with various lipid mixtures were phosphorylated with GRK2 and GRK5 in the absence of ligand or in the presence of agonist NTS or antagonist SR48692. The stoichiometry of phosphorylation was determined, as described in Materials and Methods. Means ± SD (error bars) from two experiments performed in duplicate are shown. **p < 0.01, ***p < 0.001, and ****p < 0.0001 compared with WT (two-way ANOVA followed by a Holm-Šídák test).
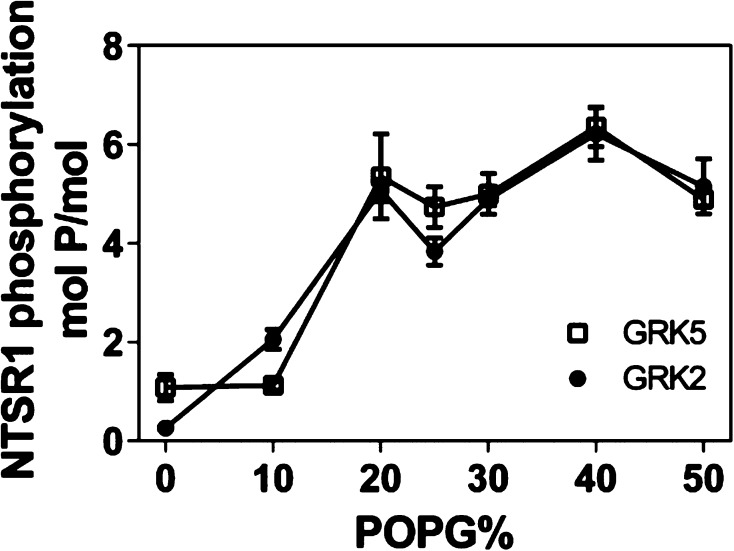
To investigate the effect of negatively charged lipids, we conducted phosphorylation studies of agonist-activated NTSR1 using nanodiscs containing varying amounts of POPG (Figure 3). The level of phosphorylation increased with an increasing level of POPG, up to 20%, and subsequently remained constant up to 50% POPG. The results indicate that negatively charged lipids in the vicinity of the receptor are required for phosphorylation by GRK2 and GRK5. On the basis of these observations, along with the negatively charged phospholipid headgroup composition of the plasma membrane (∼24%, mol % of total phosphate),50 NTSR1 was reconstituted into 75% POPC/25% POPG nanodiscs for subsequent studies.
Figure 3.
Negatively charged lipids promote phosphorylation of NTSR1 by GRK2 and GRK5. NTS-bound NTSR1 in nanodiscs prepared using POPC with the indicated fractions of POPG was phosphorylated by GRK2 (●) or GRK5 (□), and the stoichiometry of phosphorylation was determined as described in Materials and Methods. Means ± SD from two experiments performed in duplicate are shown.
GRK2 and GRK5 Target Different Residues on Activated NTSR1
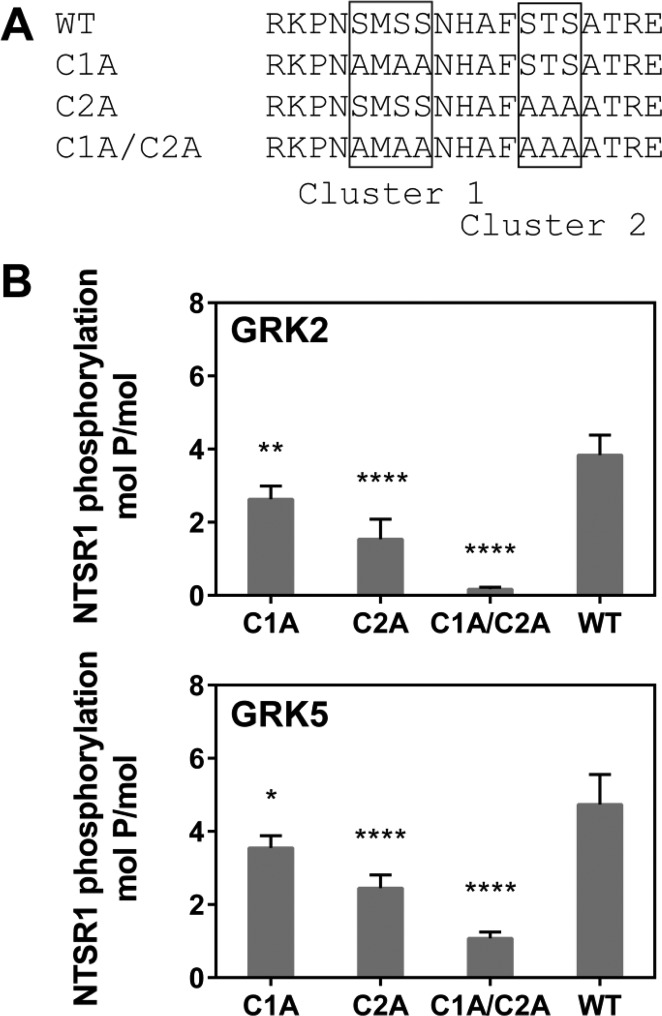
GRKs preferentially phosphorylate Ser/Thr residues located in the C-terminal tail and/or intracellular loops of GPCRs.7,8,31 In the case of NTSR1, two Ser/Thr clusters within the C-terminal tail [cluster 1 (C1), S407, S409, and S410; cluster 2 (C2), S415, T416, and S417] have been proposed as GRK phosphorylation sites.22 We constructed three cluster mutants, C1A (S407A, S409A, and S410A), C2A (S415A, T416A, and S417A), and C1A/C2A (S407A, S409A, S410A, S415A, T416A, and S417A) (Figure 4A), and reconstituted these mutants into 75% POPC/25% POPG nanodiscs to test agonist-activated phosphorylation of NTSR1 by GRK2 or GRK5 (Figure 4B). In the case of GRK2, the C1A and C2A mutants showed a diminished level of phosphorylation with 2.1 and 1.2 mol of phosphate/mol of receptor, respectively. The C1A/C2A mutant was not significantly phosphorylated. Similar observations were made using GRK5, with the C1A, C2A, and C1A/C2A mutants leading to 3.2, 2.0, and 1.1 mol of phosphate/mol of receptor, respectively. These results suggest that clusters C1 and C2 are phosphate acceptors for both GRK2 and GRK5 in vitro, as previously reported in vivo,22 and that GRK5 phosphorylates at least one site outside of clusters C1 and C2.
Figure 4.
Phosphorylation of NTSR1 mutants by GRKs. (A) The C-terminus of NTSR1 contains two Ser/Thr clusters, C1 (S407, S409, and S410) and C2 (S415, T416, and S417). Ser and Thr residues in these clusters were all mutated to Ala to make C1A and C2A. (B) NTS-bound NTSR1 WT and cluster mutants, C1A, C2A, and C1A/C2A, embedded in 75% POPC/25% POPG nanodiscs, were phosphorylated by GRK2 and GRK5, and the stoichiometry of phosphorylation was determined as described in Materials and Methods. Means ± SD from two experiments performed in duplicate are shown. *p < 0.05, **p < 0.01, and ****p < 0.001 compared with WT (one-way ANOVA followed by Dunnett’s test).
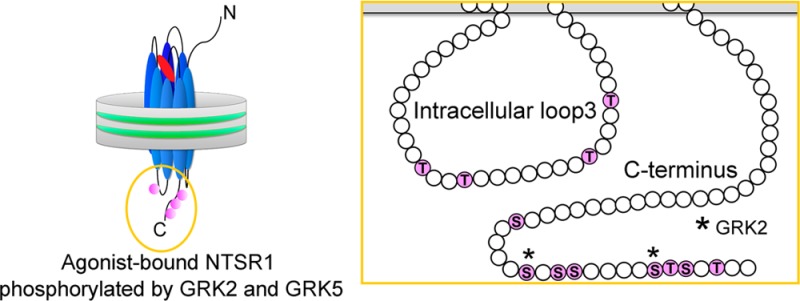
Because we cannot rule out the possibility that these cluster mutations may influence protein function and/or structure resulting in phosphorylation artifacts,51 we directly identified phosphorylated residues in NTSR1 WT by LC/MS/MS (Figure 5 and Figure S3 and Table S2 of the Supporting Information). In the case of GRK2, we identified residues S407 in C1 and S415 in C2 as the principal phosphorylation targets. In contrast, treatment with GRK5 resulted in 12 phosphorylation sites: four residues within IL3 (T279, T282, T290, and T294) and eight residues on the C-terminal tail, including C1 and C2 (S402, S407, S409, S410, S415, T416, S417, and T419). This demonstrates that GRK2 and GRK5 catalyze the formation of distinct phosphorylation patterns on NTSR1.
Figure 5.
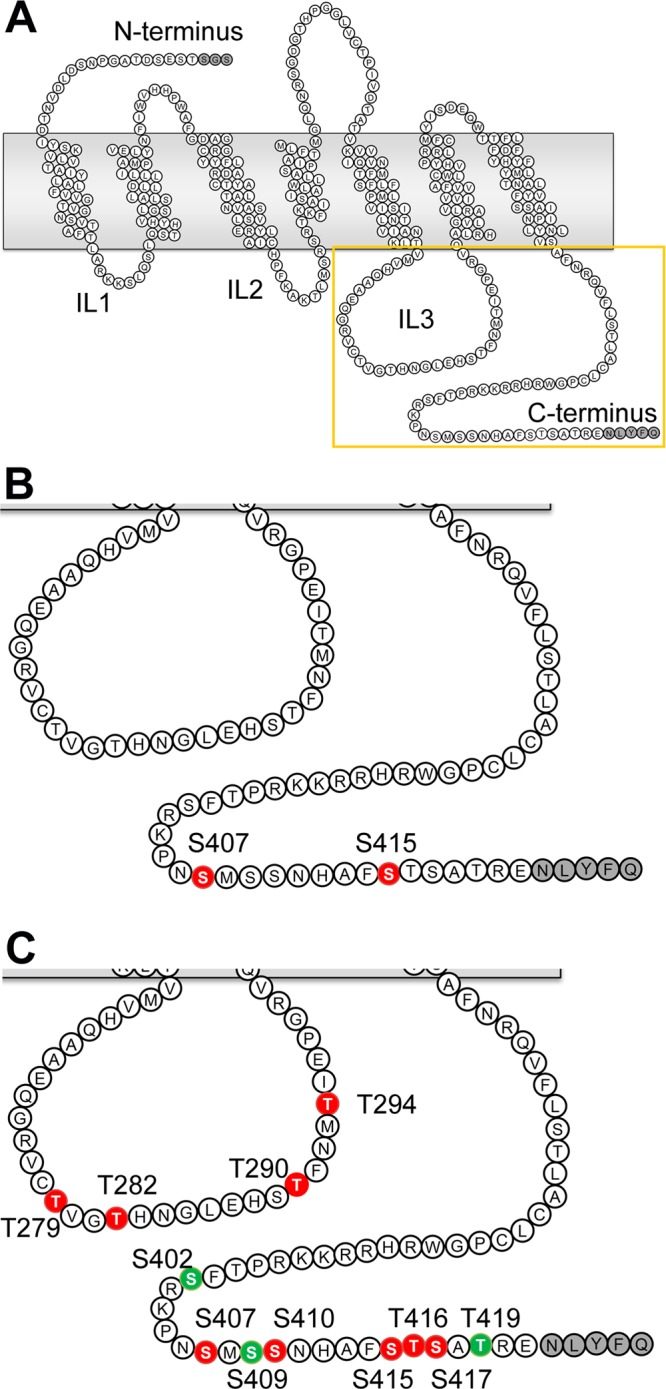
Mapping of phosphorylation sites on NTSR1 WT phosphorylated by GRKs. NTS-bound NTSR1 in 75% POPC/25% POPG nanodiscs was phosphorylated by GRKs and then analyzed by LC/MS/MS. Spectra were analyzed using the Ascore algorithm. (A) The square indicates the region of the NTSR1 depicted in panels B and C. Mapping of positions phosphorylated by (B) GRK2 and (C) GRK5. Residues that have >99 and 90% success rates with Ascores of ≥19 and 15–19 are colored red and green, respectively. The amino acid residues at the N- and C-termini derived from the TEV cleavage sites and a spacer are colored gray.
Even though nanodiscs expose the extra- and intracellular surfaces of the receptor for both the interacting partner protein and ligand, GRK2 and GRK5 phosphorylated Ser/Thr residues located only at the intracellular region of the receptor. It is therefore presumed that specific sequences on NTSR1 provide recognition targets for the GRKs. Earlier studies have shown that GRK2 actively phosphorylates Ser/Thr near acidic residues,8,31,52 and that GRK5 prefers a nonacidic peptide as its substrate.53 The GRK2 substrates β2AR and μOP both have pairs of acidic residues (Asp and Glu) near their phospho-acceptor sites8,31 (Figure 6A). Interestingly, the relative position of the charged amino acid residues near the C-terminal Ser/Thr residues of NTSR1 is distinct, in that there are positively charged clusters (residues 392–398, 403, and 404) rather than pairs of acidic residues. To elucidate whether GRKs recognize particular residues near the phospho-acceptor site, we tested the activity of GRKs using NTSR1 variants in which two adjacent basic amino acid residues were mutated to neutral Ala or the negatively charged Glu. These NTSR1 mutants were reconstituted into 75% POPC/25% POPG nanodiscs and then activated by agonist binding for phosphorylation assays. As shown in Figure 6B, all mutants behaved much like WT with respect to GRK2 activity, indicating that GRK2 is not influenced by these residues. That is, site-specific phosphorylation of NTSR1 by GRK2 does not require acidic residues near phospho-acceptor sites, in contrast to β2AR and μOP. In the case of GRK5, all mutants except for R403E/K404E also showed the same behavior as WT. For unknown reasons, the R403E/K404E mutant exhibited nonphysiological phosphorylation of the extracellular part of the receptor by LS/MS/MS analysis (data not shown). Although we did not identify a motif for site-specific GRK phosphorylation for NTSR1, these results indicate that the GRKs distinguish the specific phospho-acceptor sites of the receptor in vitro.
Figure 6.

Effect of the charged amino acid residues in the vicinity of phospho-acceptors in the C-terminus of NTSR1. (A) Comparison of the C-terminal region of class A GPCRs. Sequences are shown after the highly conserved NPXXY motif at the cytoplasmic end of the seventh transmembrane domain. β2AR and μOP have acidic residues (blue boxes) near the phosphorylated Ser and Thr residues (red). In the case of NTSR1, there are many basic amino acid residues (pink boxes) and no acidic residues in the C-terminus. (B) Comparison of the phosphorylation efficiency of NTSR1 WT and double mutants by GRK2. Agonist-activated NTSR1 double mutants (AA or EE) in 75% POPC/25% POPG nanodiscs were phosphorylated by GRK2.
Discussion
GPCR desensitization is initiated by phosphorylation of its C-terminal tail and/or intracellular loops by GRKs. Recent studies have suggested that individual GRKs contribute differently to this process and that the distinct pattern of phosphorylation by each GRK triggers distinct signaling events.4−7 It is therefore important to elucidate the pattern of GPCR phosphorylation on a given receptor by each GRK. There are a number of examples of GPCRs whose phosphorylation sites have been mapped.7−9,11,54 GRK knockdowns in HEK293 cells have been used to study the action of specific GRKs in vivo.4,7,55,56 However, as GPCRs can be phosphorylated by other Ser/Thr kinases, it is hard to unambiguously identify specific GRK sites using such systems. In vitro GRK phosphorylation assays usually need to be executed on a GPCR embedded in a lipid bilayer or membrane and in a detergent-free environment, as lipid–GRK interactions are required for GRK function,2,15 and both GRK and GPCR activity can be inhibited by detergents.17 Although phospholipid vesicles and bicelles offer a lipid environment for GPCR reconstitution, these methods cannot control the GPCR oligomeric state and topology of insertion.16 Nanodisc technology, on the other hand, allows for the preparation of essentially homogeneous and monodisperse receptor–nanodisc samples, in which the oligomeric state of the receptor is controlled. It is also possible to control the lipid composition and operate in a detergent-free environment.33,38,57 Purified NTSR1 was incorporated into nanodiscs using MSP1E3D1 and zwitterionic POPC with various ratios of negatively charged POPG. In these nanodiscs, NTSR1 was monomeric and surrounded by ∼150 lipid molecules (Table 1), likely providing a cushion of two or three lipid layers around the receptor. Thus, this preparation provides an opportunity to observe the interaction of monomeric NTSR1 with GRKs and to study the effect of lipid charge on NTSR1 phosphorylation with a defined lipid composition.
Previous studies reported the trafficking of an arrestin complex with the phosphorylated NTSR1 in HEK293 and COS7 cells.22−24 Even though these studies suggest that the Ser/Thr clusters on the C-terminal tail of NTSR1 were phosphorylated by GRKs, the use of a whole cell extract does not allow for the identification of the GRK(s) responsible. Here we observed that (i) GRK2 and GRK5 phosphorylate monomeric NTSR1 in an activation-dependent and -independent manner in vitro, respectively, (ii) the negatively charged lipid POPG in the immediate vicinity of NTSR1 has a direct effect on phosphorylation, and (iii) GRK2 and GRK5 have different phosphorylation patterns on NTSR1.
GRK2 phosphorylated NTSR1 in an agonist-dependent manner (Figure 2), consistent with the hypothesis that GRKs serve as regulators for agonist-activated GPCRs in the desensitization pathway.2,3 In the cell, Gβγ recruits cytosolic GRK2 to the cell membrane and adjusts its orientation to promote interaction with an activated receptor and its phosphorylation.58,59 Under the conditions used in the in vitro experiments, GRK2 does not require Gβγ for membrane association. Nanodisc-embedded NTSR1 is surrounded by a cushion of two or three lipid layers and likely does not offer GRK2 and Gβγ a sufficient area to interact with lipids and form a GRK2–Gβγ complex;58 nonetheless, GRK2 exhibits site-specific phosphorylation. Gβγ may be required only in cells, where mass action and/or competition with other targets does not favor association of GRK2 with the active receptor unless specifically targeted by Gβγ to membranes containing agonist-activated receptors. In contrast, GRK5 phosphorylates NTSR1 in an activation-independent manner (Figure 2). There have been a number of reports that GRKs of the GRK4 subfamily phosphorylate inactive GPCRs, which is thus perhaps an inherent characteristic of the GRK4 subfamily. The α isoform of GRK4 phosphorylates receptors in the absence of agonist activation, resulting in an increase in the level of receptor internalization, and a decrease in the total number of receptors.48 GRK5 phosphorylates β2AR and M2 muscarinic receptor in the absence of agonist.49,60 In the case of β2AR, this agonist-independent receptor phosphorylation by GRK5 promotes arrestin recruitment. Li et al. have shown that inactive receptor phosphorylation by GRK5 is not caused by the constitutive activity of the receptor or the membrane association of the kinase.49 Thus, the agonist-independent phosphorylation of NTSR1 by GRK5 is not an exception. The differential phosphorylation observed for NTSR1 indicates that NTSR1 targeting by a specific GRK may affect arrestin binding,61 leading to the activation of distinct signaling pathways, which may be agonist-dependent and/or tissue- and disease-specific.
GRKs are phospholipid-dependent enzymes, and in the case of GRK2, negatively charged lipids promote the phosphorylation of GPCRs.26,46,62 In the case of GRK5, acidic phospholipids such as phosphatidylinositol 4,5-bisphosphate are responsible for its constitutive membrane association62,63 and promote its autophosphorylation,27 which enhances GRK5 activity.64 In this study, we used POPG as a negatively charged lipid. Both GRKs required POPG for receptor phosphorylation (Figures 2 and 3). Interestingly, efficient NTSR1 phosphorylation is observed for both GRK2 and GRK5 at a POPG content of >20%, a value similar to the proportion of negatively charged lipids in native plasma membranes.50 These results are thus consistent with the notion that negatively charged lipids in the immediate vicinity of NTSR1 have a direct effect on the phosphorylation by GRKs, and that it is indeed important to mimic the native lipid composition when probing GPCR phosphorylation by GRK in vitro.
We indirectly identified residues targeted for phosphorylation by GRK2 and GRK5 using NTSR1 mutants, identifying six residues within the two Ser/Thr clusters at the C-terminus. These results are consistent with previously reported in vivo assays22 (Figure 4). However, it remains controversial whether mutational studies using C1A, C2A, and C1A/C2A mutants reflect the phosphorylation of WT receptor by GRKs. For example, there are disagreements between the phosphorylation sites identified and the role that such residues play in the regulation of μOP.65−67 To identify the phosphorylated sites on NTSR1 directly, agonist-activated NTSR1 WT embedded in 75% POPC/25% POPG nanodiscs was exposed to GRK2 and GRK5 and analyzed by LC/MS/MS (Figure 5 and Figure S3 and Table S2 of the Supporting Information). We identified two sites within the C-terminal C1 and C2 clusters that are phosphorylated by GRK2. In the case of GRK5, 12 phosphorylation sites were identified: four located within IL3 and the others in the C-terminal Ser/Thr clusters. Because of the easy loss of phosphoric acid in these analyses, it is difficult to identify all phosphorylated species.68 However, the results strongly suggest that GRK2 and GRK5 phosphorylate NTSR1 specifically and that we can observe differential phosphorylation by different GRKs in vitro.
Early studies suggested that GRK2 targets peptides that have pairs of acidic residues preceding their phospho-acceptor sites like β2AR8 and μOP31 (Figure 6A) and that GRK5 prefers a nonacidic peptide as its substrate.53 In the case of NTSR1, there are positively charged clusters instead of acidic ones. However, the mutational study did not show a charge effect for NTSR1 phosphorylation by GRK2 and GRK5. In the case of GRK2, it did not require acidic residues in the proximity of phospho-acceptors, in contrast to β2AR and μOP (Figure 6B). Thus, it is difficult to define a consensus motif for site-specific GRK phosphorylation because the intracellular regions of GPCRs vary markedly and each GRK has a distinct pattern for a given GPCR.
It was hypothesized that differential phosphorylation of GPCRs by different GRKs acts as a “barcode”, regulating their interaction with arrestins, thus encoding a particular signaling outcome.4−7,11,54 Nobles et al. reported that agonist-activated β2AR phosphorylation by GRK2 and GRK6 resulted in a distinct phosphorylation pattern, and that this distinct pattern induced different β-arrestin functions by inducing altered conformations of the β2AR-bound β-arrestin.7 Even though the extension of this hypothesis to the NTSR1 will require elucidation of a GRK-dependent NTSR1−β-arrestin interaction, differential phosphorylation patterns of the two GRKs in vitro in this study using nanodiscs are consistent with the idea that the action of different GRKs at the same receptor can lead to significantly different physiological outcomes.
Acknowledgments
We thank Dr. Ross Tomaino at the Taplin Biological Mass Spectrometry Facility of Harvard Medical School for helpful discussions and for conducting the mass spectrometric analysis.
Glossary
Abbreviations
- μOP
μ-opioid receptor
- C1
cluster 1
- C1A
C1 residues mutated to alanine
- C2
cluster 2
- C2A
C2 residues mutated to alanine
- GPCR
G protein-coupled receptor
- GRK
G protein-coupled receptor kinase
- H10
decahistidine
- IL3
intracellular loop 3
- LC/MS/MS
liquid chromatography and tandem mass spectrometry
- MBP
maltose-binding protein
- MSP
membrane scaffold protein
- NTS
neurotensin
- NTSR1
neurotensin receptor 1
- NTSR1f
neurotensin receptor 1 fusion protein
- POPC
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- POPG
1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-1′-rac-glycerol
- β2AR
β2-adrenergic receptor
- SV-AUC
sedimentation velocity analytical ultracentrifugation
- TEV
tobacco etch virus
- TrxA
E. coli thioredoxin
- WT
wild-type.
Supporting Information Available
Additional methods, tables, and figures. The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biochem.5b00285.
This research was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases (R. Ghirlando), and National Institute of Neurological Disorders and Stroke (S.I., J.F.W., and R. Grisshammer), and by National Institutes of Health Grants HL071818 (J.J.G.T.) and GM077561 (V.V.G.).
The authors declare no competing financial interest.
Supplementary Material
References
- Fredriksson R.; Lagerstrom M. C.; Lundin L. G.; Schioth H. B. (2003) The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol. Pharmacol. 63, 1256–1272 10.1124/mol.63.6.1256. [DOI] [PubMed] [Google Scholar]
- Pitcher J. A.; Freedman N. J.; Lefkowitz R. J. (1998) G protein-coupled receptor kinases. Annu. Rev. Biochem. 67, 653–692 10.1146/annurev.biochem.67.1.653. [DOI] [PubMed] [Google Scholar]
- Moore C. A.; Milano S. K.; Benovic J. L. (2007) Regulation of receptor trafficking by GRKs and arrestins. Annu. Rev. Physiol. 69, 451–482 10.1146/annurev.physiol.69.022405.154712. [DOI] [PubMed] [Google Scholar]
- Kim J.; Ahn S.; Ren X. R.; Whalen E. J.; Reiter E.; Wei H.; Lefkowitz R. J. (2005) Functional antagonism of different G protein-coupled receptor kinases for beta-arrestin-mediated angiotensin II receptor signaling. Proc. Natl. Acad. Sci. U. S. A. 102, 1442–1447 10.1073/pnas.0409532102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X. R.; Reiter E.; Ahn S.; Kim J.; Chen W.; Lefkowitz R. J. (2005) Different G protein-coupled receptor kinases govern G protein and beta-arrestin-mediated signaling of V2 vasopressin receptor. Proc. Natl. Acad. Sci. U. S. A. 102, 1448–1453 10.1073/pnas.0409534102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zidar D. A.; Violin J. D.; Whalen E. J.; Lefkowitz R. J. (2009) Selective engagement of G protein coupled receptor kinases (GRKs) encodes distinct functions of biased ligands. Proc. Natl. Acad. Sci. U. S. A. 106, 9649–9654 10.1073/pnas.0904361106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobles K. N.; Xiao K.; Ahn S.; Shukla A. K.; Lam C. M.; Rajagopal S.; Strachan R. T.; Huang T. Y.; Bressler E. A.; Hara M. R.; Shenoy S. K.; Gygi S. P.; Lefkowitz R. J. (2011) Distinct phosphorylation sites on the beta(2)-adrenergic receptor establish a barcode that encodes differential functions of beta-arrestin. Sci. Signaling 4, ra51. 10.1126/scisignal.2001707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredericks Z. L.; Pitcher J. A.; Lefkowitz R. J. (1996) Identification of the G protein-coupled receptor kinase phosphorylation sites in the human beta2-adrenergic receptor. J. Biol. Chem. 271, 13796–13803 10.1074/jbc.271.23.13796. [DOI] [PubMed] [Google Scholar]
- Trester-Zedlitz M.; Burlingame A.; Kobilka B.; von Zastrow M. (2005) Mass spectrometric analysis of agonist effects on posttranslational modifications of the beta-2 adrenoceptor in mammalian cells. Biochemistry 44, 6133–6143 10.1021/bi0475469. [DOI] [PubMed] [Google Scholar]
- Bouvier M.; Hausdorff W. P.; De Blasi A.; O’Dowd B. F.; Kobilka B. K.; Caron M. G.; Lefkowitz R. J. (1988) Removal of phosphorylation sites from the beta 2-adrenergic receptor delays onset of agonist-promoted desensitization. Nature 333, 370–373 10.1038/333370a0. [DOI] [PubMed] [Google Scholar]
- Butcher A. J.; Prihandoko R.; Kong K. C.; McWilliams P.; Edwards J. M.; Bottrill A.; Mistry S.; Tobin A. B. (2011) Differential G-protein-coupled receptor phosphorylation provides evidence for a signaling bar code. J. Biol. Chem. 286, 11506–11518 10.1074/jbc.M110.154526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godovac-Zimmermann J.; Soskic V.; Poznanovic S.; Brianza F. (1999) Functional proteomics of signal transduction by membrane receptors. Electrophoresis 20, 952–961. [DOI] [PubMed] [Google Scholar]
- Tran T. M.; Friedman J.; Qunaibi E.; Baameur F.; Moore R. H.; Clark R. B. (2004) Characterization of agonist stimulation of cAMP-dependent protein kinase and G protein-coupled receptor kinase phosphorylation of the beta2-adrenergic receptor using phosphoserine-specific antibodies. Mol. Pharmacol. 65, 196–206 10.1124/mol.65.1.196. [DOI] [PubMed] [Google Scholar]
- Clark R. B.; Rich T. C. (2003) Probing the roles of protein kinases in g-protein-coupled receptor desensitization. Mol. Pharmacol. 64, 1015–1017 10.1124/mol.64.5.1015. [DOI] [PubMed] [Google Scholar]
- Homan K. T.; Glukhova A.; Tesmer J. J. (2013) Regulation of G protein-coupled receptor kinases by phospholipids. Curr. Med. Chem. 20, 39–46. [PubMed] [Google Scholar]
- Serebryany E.; Zhu G. A.; Yan E. C. (2012) Artificial membrane-like environments for in vitro studies of purified G-protein coupled receptors. Biochim. Biophys. Acta, Biomembr. 1818, 225–233 10.1016/j.bbamem.2011.07.047. [DOI] [PubMed] [Google Scholar]
- Loudon R. P.; Benovic J. L. (1997) Altered activity of palmitoylation-deficient and isoprenylated forms of the G protein-coupled receptor kinase GRK6. J. Biol. Chem. 272, 27422–27427 10.1074/jbc.272.43.27422. [DOI] [PubMed] [Google Scholar]
- Carraway R.; Leeman S. E. (1973) The isolation of a new hypotensive peptide, neurotensin, from bovine hypothalami. J. Biol. Chem. 248, 6854–6861. [PubMed] [Google Scholar]
- Carraway R. E.; Plona A. M. (2006) Involvement of neurotensin in cancer growth: evidence, mechanisms and development of diagnostic tools. Peptides 27, 2445–2460 10.1016/j.peptides.2006.04.030. [DOI] [PubMed] [Google Scholar]
- Schimpff R. M.; Avard C.; Fenelon G.; Lhiaubet A. M.; Tenneze L.; Vidailhet M.; Rostene W. (2001) Increased plasma neurotensin concentrations in patients with Parkinson’s disease. J. Neurol., Neurosurg. Psychiatry 70, 784–786 10.1136/jnnp.70.6.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K.; Masu M.; Nakanishi S. (1990) Structure and functional expression of the cloned rat neurotensin receptor. Neuron 4, 847–854 10.1016/0896-6273(90)90137-5. [DOI] [PubMed] [Google Scholar]
- Oakley R. H.; Laporte S. A.; Holt J. A.; Barak L. S.; Caron M. G. (2001) Molecular determinants underlying the formation of stable intracellular G protein-coupled receptor-beta-arrestin complexes after receptor endocytosis. J. Biol. Chem. 276, 19452–19460 10.1074/jbc.M101450200. [DOI] [PubMed] [Google Scholar]
- Oakley R. H.; Laporte S. A.; Holt J. A.; Caron M. G.; Barak L. S. (2000) Differential affinities of visual arrestin, beta arrestin1, and beta arrestin2 for G protein-coupled receptors delineate two major classes of receptors. J. Biol. Chem. 275, 17201–17210 10.1074/jbc.M910348199. [DOI] [PubMed] [Google Scholar]
- Law I. K.; Murphy J. E.; Bakirtzi K.; Bunnett N. W.; Pothoulakis C. (2012) Neurotensin-induced proinflammatory signaling in human colonocytes is regulated by beta-arrestins and endothelin-converting enzyme-1-dependent endocytosis and resensitization of neurotensin receptor 1. J. Biol. Chem. 287, 15066–15075 10.1074/jbc.M111.327262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman C. V.; Barak L. S.; Chen C.; Liu-Chen L. Y.; Onorato J. J.; Kennedy S. P.; Caron M. G.; Benovic J. L. (2000) Mutational analysis of Gbetagamma and phospholipid interaction with G protein-coupled receptor kinase 2. J. Biol. Chem. 275, 10443–10452 10.1074/jbc.275.14.10443. [DOI] [PubMed] [Google Scholar]
- Onorato J. J.; Gillis M. E.; Liu Y.; Benovic J. L.; Ruoho A. E. (1995) The beta-adrenergic receptor kinase (GRK2) is regulated by phospholipids. J. Biol. Chem. 270, 21346–21353 10.1074/jbc.270.36.21346. [DOI] [PubMed] [Google Scholar]
- DebBurman S. K.; Ptasienski J.; Boetticher E.; Lomasney J. W.; Benovic J. L.; Hosey M. M. (1995) Lipid-mediated regulation of G protein-coupled receptor kinases 2 and 3. J. Biol. Chem. 270, 5742–5747 10.1074/jbc.270.11.5742. [DOI] [PubMed] [Google Scholar]
- Premont R. T.; Koch W. J.; Inglese J.; Lefkowitz R. J. (1994) Identification, purification, and characterization of GRK5, a member of the family of G protein-coupled receptor kinases. J. Biol. Chem. 269, 6832–6841. [PubMed] [Google Scholar]
- Whorton M. R.; Jastrzebska B.; Park P. S.; Fotiadis D.; Engel A.; Palczewski K.; Sunahara R. K. (2008) Efficient coupling of transducin to monomeric rhodopsin in a phospholipid bilayer. J. Biol. Chem. 283, 4387–4394 10.1074/jbc.M703346200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki S.; Ghirlando R.; White J. F.; Gvozdenovic-Jeremic J.; Northup J. K.; Grisshammer R. (2012) Modulation of the interaction between neurotensin receptor NTS1 and Gq protein by lipid. J. Mol. Biol. 417, 95–111 10.1016/j.jmb.2012.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. J.; Oldfield S.; Butcher A. J.; Tobin A. B.; Saxena K.; Gurevich V. V.; Benovic J. L.; Henderson G.; Kelly E. (2013) Identification of phosphorylation sites in the COOH-terminal tail of the mu-opioid receptor. J. Neurochem. 124, 189–199 10.1111/jnc.12071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapust R. B.; Tozser J.; Fox J. D.; Anderson D. E.; Cherry S.; Copeland T. D.; Waugh D. S. (2001) Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng., Des. Sel. 14, 993–1000 10.1093/protein/14.12.993. [DOI] [PubMed] [Google Scholar]
- Inagaki S.; Ghirlando R.; Grisshammer R. (2013) Biophysical characterization of membrane proteins in nanodiscs. Methods 59, 287–300 10.1016/j.ymeth.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P.; Wang B.; Maeda T.; Palczewski K.; Tesmer J. J. (2008) Structures of rhodopsin kinase in different ligand states reveal key elements involved in G protein-coupled receptor kinase activation. J. Biol. Chem. 283, 14053–14062 10.1074/jbc.M708974200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C. C.; Yoshino-Koh K.; Tesmer J. J. (2009) A surface of the kinase domain critical for the allosteric activation of G protein-coupled receptor kinases. J. Biol. Chem. 284, 17206–17215 10.1074/jbc.M809544200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodowski D. T.; Tesmer V. M.; Benovic J. L.; Tesmer J. J. (2006) The structure of G protein-coupled receptor kinase (GRK)-6 defines a second lineage of GRKs. J. Biol. Chem. 281, 16785–16793 10.1074/jbc.M601327200. [DOI] [PubMed] [Google Scholar]
- Yang P.; Glukhova A.; Tesmer J. J.; Chen Z. (2013) Membrane orientation and binding determinants of G protein-coupled receptor kinase 5 as assessed by combined vibrational spectroscopic studies. PLoS One 8, e82072. 10.1371/journal.pone.0082072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayburt T. H.; Vishnivetskiy S. A.; McLean M. A.; Morizumi T.; Huang C. C.; Tesmer J. J.; Ernst O. P.; Sligar S. G.; Gurevich V. V. (2011) Monomeric rhodopsin is sufficient for normal rhodopsin kinase (GRK1) phosphorylation and arrestin-1 binding. J. Biol. Chem. 286, 1420–1428 10.1074/jbc.M110.151043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beausoleil S. A.; Villen J.; Gerber S. A.; Rush J.; Gygi S. P. (2006) A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat. Biotechnol. 24, 1285–1292 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- Boggs J. M.; Tummler B. (1993) Interdigitated gel phase bilayers formed by unsaturated synthetic and bacterial glycerolipids in the presence of polymyxin B and glycerol. Biochim. Biophys. Acta, Biomembr. 1145, 42–50 10.1016/0005-2736(93)90379-E. [DOI] [PubMed] [Google Scholar]
- Davis P. J.; Fleming B. D.; Coolbear K. P.; Keough K. M. (1981) Gel to liquid-crystalline transition temperatures of water dispersions of two pairs of positional isomers of unsaturated mixed-acid phosphatidylcholines. Biochemistry 20, 3633–3636 10.1021/bi00515a051. [DOI] [PubMed] [Google Scholar]
- Dowhan W., Bogdanov M., and Mileykovskaya E. (2008) Functional roles of lipids in membranes. In Biochemistry of Lipids, Lipoproteins and Membranes (Vance E. D., and Vance J. E., Eds.) 5th ed., pp 1–37, Elsevier Science, New York. [Google Scholar]
- Mattai J.; Hauser H.; Demel R. A.; Shipley G. G. (1989) Interactions of metal ions with phosphatidylserine bilayer membranes: effect of hydrocarbon chain unsaturation. Biochemistry 28, 2322–2330 10.1021/bi00431a051. [DOI] [PubMed] [Google Scholar]
- Gully D.; Canton M.; Boigegrain R.; Jeanjean F.; Molimard J. C.; Poncelet M.; Gueudet C.; Heaulme M.; Leyris R.; Brouard A. (1993) Biochemical and pharmacological profile of a potent and selective nonpeptide antagonist of the neurotensin receptor. Proc. Natl. Acad. Sci. U. S. A. 90, 65–69 10.1073/pnas.90.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pronin A. N.; Carman C. V.; Benovic J. L. (1998) Structure-function analysis of G protein-coupled receptor kinase-5. Role of the carboxyl terminus in kinase regulation. J. Biol. Chem. 273, 31510–31518 10.1074/jbc.273.47.31510. [DOI] [PubMed] [Google Scholar]
- DebBurman S. K.; Ptasienski J.; Benovic J. L.; Hosey M. M. (1996) G protein-coupled receptor kinase GRK2 is a phospholipid-dependent enzyme that can be conditionally activated by G protein betagamma subunits. J. Biol. Chem. 271, 22552–22562 10.1074/jbc.271.37.22552. [DOI] [PubMed] [Google Scholar]
- Lodowski D. T.; Barnhill J. F.; Pitcher J. A.; Capel W. D.; Lefkowitz R. J.; Tesmer J. J. (2003) Purification, crystallization and preliminary X-ray diffraction studies of a complex between G protein-coupled receptor kinase 2 and Gbeta1gamma2. Acta Crystallogr., Sect. D: Biol. Crystallogr. 59, 936–939 10.1107/S0907444903002622. [DOI] [PubMed] [Google Scholar]
- Rankin M. L.; Marinec P. S.; Cabrera D. M.; Wang Z.; Jose P. A.; Sibley D. R. (2006) The D1 dopamine receptor is constitutively phosphorylated by G protein-coupled receptor kinase 4. Mol. Pharmacol. 69, 759–769. [DOI] [PubMed] [Google Scholar]
- Li L.; Homan K. T.; Vishnivetskiy S. A.; Manglik A.; Tesmer J. J.; Gurevich V. V.; Gurevich E. V. (2015) G Protein-coupled Receptor Kinases of the GRK4 Protein Subfamily Phosphorylate Inactive G Protein-coupled Receptors (GPCRs). J. Biol. Chem. 290, 10775–10790 10.1074/jbc.M115.644773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boesze-Battaglia K.; Schimmel R. (1997) Cell membrane lipid composition and distribution: implications for cell function and lessons learned from photoreceptors and platelets. J. Exp. Biol. 200, 2927–2936. [DOI] [PubMed] [Google Scholar]
- Kim C. H.; Braud S.; Isaac J. T.; Roche K. W. (2005) Protein kinase C phosphorylation of the metabotropic glutamate receptor mGluR5 on Serine 839 regulates Ca2+ oscillations. J. Biol. Chem. 280, 25409–25415 10.1074/jbc.M502644200. [DOI] [PubMed] [Google Scholar]
- Onorato J. J.; Palczewski K.; Regan J. W.; Caron M. G.; Lefkowitz R. J.; Benovic J. L. (1991) Role of acidic amino acids in peptide substrates of the beta-adrenergic receptor kinase and rhodopsin kinase. Biochemistry 30, 5118–5125 10.1021/bi00235a002. [DOI] [PubMed] [Google Scholar]
- Kunapuli P.; Onorato J. J.; Hosey M. M.; Benovic J. L. (1994) Expression, purification, and characterization of the G protein-coupled receptor kinase GRK5. J. Biol. Chem. 269, 1099–1105. [PubMed] [Google Scholar]
- Hinkle P. M.; Gehret A. U.; Jones B. W. (2012) Desensitization, trafficking, and resensitization of the pituitary thyrotropin-releasing hormone receptor. Front. Neurosci. 6, 180. 10.3389/fnins.2012.00180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwood B. K.; Lopez J.; Wager-Miller J.; Mackie K.; Straiker A. (2011) Expression of G protein-coupled receptors and related proteins in HEK293, AtT20, BV2, and N18 cell lines as revealed by microarray analysis. BMC Genomics 12, 14. 10.1186/1471-2164-12-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J.; Busillo J. M.; Benovic J. L. (2008) M3 muscarinic acetylcholine receptor-mediated signaling is regulated by distinct mechanisms. Mol. Pharmacol. 74, 338–347 10.1124/mol.107.044750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie T. K.; Grinkova Y. V.; Bayburt T. H.; Denisov I. G.; Zolnerciks J. K.; Atkins W. M.; Sligar S. G. (2009) Reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol. 464, 211–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodowski D. T.; Pitcher J. A.; Capel W. D.; Lefkowitz R. J.; Tesmer J. J. (2003) Keeping G proteins at bay: a complex between G protein-coupled receptor kinase 2 and Gbetagamma. Science 300, 1256–1262 10.1126/science.1082348. [DOI] [PubMed] [Google Scholar]
- Pitcher J. A.; Inglese J.; Higgins J. B.; Arriza J. L.; Casey P. J.; Kim C.; Benovic J. L.; Kwatra M. M.; Caron M. G.; Lefkowitz R. J. (1992) Role of beta gamma subunits of G proteins in targeting the beta-adrenergic receptor kinase to membrane-bound receptors. Science 257, 1264–1267 10.1126/science.1325672. [DOI] [PubMed] [Google Scholar]
- Baameur F.; Morgan D. H.; Yao H.; Tran T. M.; Hammitt R. A.; Sabui S.; McMurray J. S.; Lichtarge O.; Clark R. B. (2010) Role for the regulator of G-protein signaling homology domain of G protein-coupled receptor kinases 5 and 6 in beta 2-adrenergic receptor and rhodopsin phosphorylation. Mol. pharmacol. 77, 405–415 10.1124/mol.109.058115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich V. V.; Gurevich E. V. (2006) The structural basis of arrestin-mediated regulation of G-protein-coupled receptors. Pharmacol. Ther. 110, 465–502 10.1016/j.pharmthera.2005.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitcher J. A.; Fredericks Z. L.; Stone W. C.; Premont R. T.; Stoffel R. H.; Koch W. J.; Lefkowitz R. J. (1996) Phosphatidylinositol 4,5-bisphosphate (PIP2)-enhanced G protein-coupled receptor kinase (GRK) activity. Location, structure, and regulation of the PIP2 binding site distinguishes the GRK subfamilies. J. Biol. Chem. 271, 24907–24913 10.1074/jbc.271.40.24907. [DOI] [PubMed] [Google Scholar]
- Thiyagarajan M. M.; Stracquatanio R. P.; Pronin A. N.; Evanko D. S.; Benovic J. L.; Wedegaertner P. B. (2004) A predicted amphipathic helix mediates plasma membrane localization of GRK5. J. Biol. Chem. 279, 17989–17995 10.1074/jbc.M310738200. [DOI] [PubMed] [Google Scholar]
- Kunapuli P.; Gurevich V. V.; Benovic J. L. (1994) Phospholipid-stimulated autophosphorylation activates the G protein-coupled receptor kinase GRK5. J. Biol. Chem. 269, 10209–10212. [PubMed] [Google Scholar]
- Chu J.; Zheng H.; Loh H. H.; Law P. Y. (2008) Morphine-induced mu-opioid receptor rapid desensitization is independent of receptor phosphorylation and beta-arrestins. Cell. Signalling 20, 1616–1624 10.1016/j.cellsig.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Kouhen R.; Burd A. L.; Erickson-Herbrandson L. J.; Chang C. Y.; Law P. Y.; Loh H. H. (2001) Phosphorylation of Ser363, Thr370, and Ser375 residues within the carboxyl tail differentially regulates mu-opioid receptor internalization. J. Biol. Chem. 276, 12774–12780 10.1074/jbc.M009571200. [DOI] [PubMed] [Google Scholar]
- Pak Y.; O'Dowd B. F.; George S. R. (1997) Agonist-induced desensitization of the mu opioid receptor is determined by threonine 394 preceded by acidic amino acids in the COOH-terminal tail. J. Biol. Chem. 272, 24961–24965 10.1074/jbc.272.40.24961. [DOI] [PubMed] [Google Scholar]
- Steen H.; Jebanathirajah J. A.; Rush J.; Morrice N.; Kirschner M. W. (2005) Phosphorylation analysis by mass spectrometry: myths, facts, and the consequences for qualitative and quantitative measurements. Mol. Cell. Proteomics 5, 172–181 10.1074/mcp.M500135-MCP200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.