Summary
In 1964, Kundig, Ghosh and Roseman reported the discovery of the phosphoenolpyruvate:sugar phosphotransferase system (PTS), which they subsequently proposed might catalyze sugar transport as well as sugar phosphorylation. What we have learned in the past 50 years, since its discovery, is that in addition to these primary functions, the PTS serves as a complex protein kinase system that regulates a wide variety of transport, metabolic and mutagenic processes as well as the expression of numerous genes. Recent operon- and genome-sequencing projects have revealed novel PTS protein-encoding genes, many of which have yet to be functionally defined. The current picture of the PTS is that of a complex system with ramifications in all aspects of cellular physiology. Moreover, its mosaic evolutionary history is unusual and intriguing. The PTS can be considered to serve many prokaryotes in capacities of communication and coordination as do the nervous systems of animals.
Introduction
Fifty years ago, a novel sugar-phosphorylating system was discovered in Escherichia coli [Kundig et al., 1964]. The unique features of this phosphotransferase system (PTS) included the use of phosphoenolpyruvate (PEP) as the phosphoryl donor for sugar phosphorylation and the presence of three essential catalytic entities, termed Enzyme I, Enzyme II and HPr (heat-stable, histidinephosphorylatable protein). The discovery of this system provided an explanation for pleiotropic carbohydrate-negative mutants of E. coli described as early as 1949 [Doudoroff et al., 1949].
As noted above, the three initially recognized activities of the E. coli PTS were presumed to correspond to three proteins. Now, several dozen PTS proteins are known in E. coli [Tchieu et al., 2001] with even more in some Firmicutes [Barabote and Saier, 2005; Comas et al., 2008]. More recently, genes encoding proteins of the PTS have been identified and characterized in archaea [Cai et al., 2014; Pickl et al., 2012], but no eukaryote has yet been shown to possess a protein constituent of the system. It therefore appears to be a prokaryote-specific system, a potential target of antimicrobial agents.
Known PTS functions
In 1964, a single function for the PTS, namely sugar phosphorylation, was known. Fifty years later, we find that this system plays roles in many surprising aspects of bacterial physiology (Table 1). Established primary functions of the system include sugar transport and phosphorylation as well as sugar reception for chemotactic responses, whereas secondary functions include various ramifications of metabolic and transcriptional regulation [Lengeler and Jahreis, 2009; Vastermark and Saier, 2014]. Targets of PTS-mediated regulation include (i) carbohydrate catabolic enzymes, sugar permeases, and the cyclic AMP biosynthetic enzyme, adenylate cyclase, all regulated allosterically by the IIAG1c PTS protein in enteric bacteria; (ii) a variety of transcription factors, non-PTS transport systems and even the PTS itself, regulated by HPr(ser-P)-dependent allostery in many bacteria including Gram-positive firmicutes; (iii) transcriptional activators and antiterminators regulated by direct PTS-mediated phosphorylation in both Gram-negative and Gram-positive bacteria; (iv) other transcriptional regulators controlled indirectly by PTS-mediated phosphorylation by virtue of direct interactions with PTS proteins; (v) metabolism of nitrogen- and phosphorous-containing compounds; (vi) intracellular ionic (K+) homeostasis; (vii) carbon (glycogen, β-hydroxybutyrate) storage; (viii) biofilm formation; (ix) virulence and (x) directed transposon insertion for the purpose of operon activation (directed mutation) [Barabote and Saier, 2005; Krausse et al., 2009; Le Bouguenec and Schouler, 2011; Lopes et al., 2011; Pickering et al., 2014; Zhang and Saier, 2009a, b]. Moreover, the detection of novel, functionally uncharacterized PTS proteins in bacteria as diverse as E. coli, Acholeplasma laidlawii, Listeria monocytogenes and several antibiotic-producing species of Streptomyces, suggests the involvement of PTS proteins in cellular processes distinct from those listed in Table 1 [Barabote and Saier, 2005; Deutscher et al., 2014; Rigali et al., 2006].
Table 1.
Primary (metabolic) and secondary (regulatory) functions currently recognized for the PTS.
| Primary Metabolic Functions |
| Catalysis of: |
| Sugar transport |
| Sugar phosphorylation |
| Sugar chemoreception |
| Secondary Regulatory Functions |
| Control of the activities of: |
| PTS permeases |
| Non-PTS permeases |
| Catabolic enzymes |
| Adenylate cyclase |
| Transcription factors (many) |
| Control of physiological processes including: |
| Ionic homeostasis (potassium transport) |
| Glycogen accumulation |
| Polyhydroxybutyrate storage |
| Nitrogen utilization |
| Phosphorous metabolism |
| Biofilm formation |
| Virulence |
| Transposon-mediated directed mutation |
Historical background
In his first insightful review on the PTS, Saul Roseman, who died in 2011 at the age of 90, noted a potential analogy between the PTS-catalyzed coupled transport and phosphorylation of sugars and Peter Mitchell's postulated mechanism of group translocation, a process in which penetration of the membrane by a solute occurs concomitantly with a chemical reaction that modifies the transported substrate [Mitchell, 1954; Roseman, 1969]. Evidence for the involvement of the PTS in sugar transport had been published in 1966 [Kundig et al., 1966]. Although the experimental design utilized in the reported studies proved to be both conceptually and technically faulty, the conclusion that the PTS is a transport system proved to be correct [Postma et al., 1993]. Thus, in this instance, intuition provided a more accurate view of reality than did empiricism.
The first PTS permease to be successfully purified [Jacobson et al., 1983; Jacobson et al., 1979], reconstituted in an artificial membrane [Leonard and Saier, 1983], and sequenced [Lee and Saier, 1983] was the mannitol Enzyme II of E. coli. Since then, several other PTS permeases have been purified and characterized, and recently, the high-resolution 3-d structure of an integral membrane PTS Enzyme IIC was determined [Cao et al., 2011; McCoy et al., 2015]. The high-resolution structures of several IIA and IIB proteins (see below) have also been solved, alone or complexed to other PTS or non-PTS proteins. These basic scientific breakthroughs allow us to understand the PTS-mediated phosphoryl transfer and transport mechanisms in structural detail [McCoy et al., 2015].
PTS structure and function
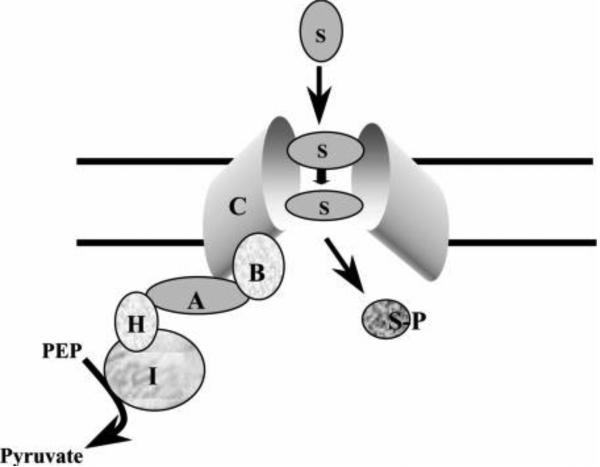
PTS permeases consist of two peripheral cytoplasmic membrane proteins or domains (IIA and IIB; sugar-specific energy coupling proteins) as well as one (or sometimes two) integral membrane proteins or domains (IIC, the actual sugar permease, with or without IID, depending on the system) (See Figure 1 and Table 2). These protein domains can be detached or fused together so that the Enzyme II complex can consist of one, two, three or four polypeptide chains encoded by the same number of genes. The IIA, IIB and IIC domains are frequently covalently linked to each other via gene-encoded flexible peptide linkers in a variable but non-random fashion. Thus, for example, a PTS permease containing a IIC domain may have the corresponding IIB domain either preceding or following it, but the IIA domain, when present in the same polypeptide chain, is usually linked at the C-terminal end of the permease.
Figure 1. Schematic depiction of the protein constituents of a typical PTS permease.
A PTS permease is a sugar transporting Enzyme II complex of the bacterial PEP-dependent phosphotransferase system. The sugar substrate (S) is transported from the extracellular medium through the membrane in a pathway determined by the integral membrane permease-like Enzyme IIC (C) constituent, a homodimer in the membrane as shown. The sequentially acting energy-coupling proteins transfer a phosphoryl group from the initial phosphoryl donor, phosphoenolpyruvate (PEP), to the ultimate phosphoryl acceptor, sugar, yielding a sugar-phosphate (S-P). These enzymes are: Enzyme I (I), HPr (H), Enzyme IIA (A) and Enzyme IIB (B). I is the first general energy-coupling protein; H is the second general energy-coupling protein; A is the family-specific phosphoryl donor; B is the permease-specific phosphoryl donor; and C is the permease/receptor that mediates transport and catalyzes concomitant phosphorylation of the sugar substrate. A given bacterial cell may possess multiple PTS Enzyme II complexes, each specific for a different sugar or set of sugars. Some bacteria also possess multiple sets of PTS energy-coupling proteins (Enzymes I, HPr, IIA and IIB) that may play regulatory roles dependently or independently of sugar transport.
Table 2.
Four evolutionarily distinct families of PTS Enzyme IIC proteins and their characteristics
| Enzyme family1 | Characteristics1 |
|---|---|
| A) The Glc-Fru-Lac Superfamily | |
| 1 | Fru has been proposed to be the original PTS, but other IIC proteins diverged in function, sequence and possibly structure. |
| 2 | A basic dimeric 10 TMS topology, with two functionally different 5TMS halves per promoter is established for one member, but topological variations for other family members are possible. |
| 3 | IIAs and IIBs have mosaic origins; e.g. IIAGlc is not homologous to IIAMtl, IIANtr or IIALac; IIBGlc is not homologous to IIBChb. |
| 4 | IIAs, IIBs and IICs did not coevolve. |
| B) The Asc-Gat Superfamily | |
| 1 | IICAsc homologues are often fused to IIA and IIB homologues, but IICGat homologues never are. The IIC proteins have an 11 or 12 TMS topology, probably with a basic internal 6 TMS repeat unit, with an N-terminal TMS deleted for the 11 TMS proteins. |
| 2 | IICAsc homologues are always encoded by genes in operons with IIA and IIB genes, but IICGat homologues can be encoded in operons lacking IIA and IIB genes. |
| 3 | Some IICGat homologues are found in organisms that lack all other PTS proteins, suggesting roles as secondary carriers. |
| 4 | Asc and Gat IIA and IIB constituents are distantly related to IIA and IIB constituents of the Glc-Fru-Lac Superfamily |
| C) The Man Family | |
| 1 | All constituents (IIA, IIB, IIC and IID) differ structurally from all other PTS permease proteins. |
| 2 | All members, but only members of this family, have IID constituents. |
| 3 | The IIB constituents are phosphorylated on His rather than Cys residues as is true for other IIB proteins. |
| 4 | Members often exhibit broad specificity for aldo- and keto- sugars. |
| D) The Dha Family | |
| 1 | DhaK and DhaL are homologous to the N- and C-terminal domains of ATP-dependent dihydroxyacetone (DHA) kinases. |
| 2 | DhaM consists of three domains: IIAMan-HPr-I Δ2. |
| 3 | The three domains of DhaM are phosphorylated by PEP, EI and HPr, but DhaK and L are not phosphorylated. |
| 4 | DhaK binds DHA covalently to a His residue and transfers the phosphoryl group from IIA of DhaM via DhaL-ADP to DHA. |
Abbreviations: Glc, glucose; Fru, fructose; Lac, lactose; Asc, L-ascorbate; Gat, galactitol; Man, mannose; Chb, diacelylchitobiose; Dha or DHA, dihydroxyacetone. See text and Figure 1 for protein designations.
I Δ, a truncated (partially deleted) Enzyme I.
The linking of IIA domains to general or sugar-inducible energy-coupling protein domains can occur either N- or C-terminal to the latter proteins. In a particular PTS permease, a domain may be either deleted, as is true of the IIA domains of the sucrose and trehalose permeases of E. coli, duplicated, as is true of the IIB domains of the fructose permeases of E. coli and Rhodobacter capsulatus [Wu et al., 1990], or spliced in half, as seems to be true of the IIC domain of the glucitol permease of E. coli [Yamada and Saier, 1987]. If a IIA domain is deleted, then another, such as IIAGlc, must substitute for it since IIA, IIB and IIC are all required for PTS-mediated sugar phosphorylation.
The mosaic origins of PTS permeases (Enzyme II complexes)
PTS Enzyme IIC proteins/domains fall into four families with members of each family being homologous, deriving from a common ancestral protein (Table 2; see subclass 4.A in the Transporter Classification Database, TCDB; www.tcdb.org) [Saier et al., 2006; 2009; 2014]. (1) The first of these four families is the Glucose-Fructose-Lactose (Glc-Fru-Lac) family, the largest of the four families. One member of this family, the diacetylchitobiose IIC protein, is now known to have a basic 10 transmembrane α-helical segment (TMS) topology consisting of two 5 TMS domains serving different functions [Cao et al., 2011; McCoy et al., 2015]. The split glucitol IIC system is probably a distant member of this superfamily. (2) The Ascorbate-Galactitol (Asc-Gat) family has members with 11 or 12 TMSs with a primordial internal duplication of 6 TMSs. The evidence summarized in Table 2 suggests that these IIC proteins may derive from secondary carriers, and some may still be capable of catalyzing transport in the absence of sugar phosphorylation, although this possibility has not been adequately tested [Hvorup et al., 2003; Saier et al., 2005]. (3) The Mannose (Man) family, with IIC domains probably of 6 TMSs, as well as a second integral membrane constituent unique to this family (IID), appears to have evolved four protein constituents (IIA, IIB, IIC and IID), all of which arose independently of all other PTS proteins [Erni, 2006; Garcia-Alles et al., 2002]. (4) The Dihydroxyacetone (Dha) family apparently derived relatively recently from a cytoplasmic primordial ATP-dependent dihydroxyacetone kinase [Bächler et al., 2005; Erni et al., 2006; Saier et al., 2005] (Table 2).
As emphasized in Table 2, the IIA and IIB PTS proteins did not coevolve with the IIC constituents, and it is possible that different IIC members of the Glc-Fru-Lac family actually have different toplogies [McCoy et al., 2015]. These observations reveal that the PTS is truly a mosaic system, derived from many ancestral sources that probably evolved independently of each other and later came together to form functional Enzyme II complexes [Saier and Reizer, 1994; Saier et al., 2005]. Thus, the PTS may have evolved late, after the appearance of established universal metabolic pathways such as glycolysis, gluconeogenesis and the Krebs cycle.
Overview of this PTS50 Written Symposium
The PTS scientific community celebrated the 50th anniversary of the discovery of the PTS in the summer of 2014 in Göttingen, Germany. The meeting was organized by Jörg Stülke, Josef Deutscher and myself. Over 100 people, most of them actively researching the PTS, attended the meeting, and many of them presented talks or posters describing their recent research efforts. All in attendance agreed it was a most informative and enjoyable event with old friends meeting new members of the PTS community while exchanging scientific and personal information.
The first session of the meeting was opened by Joseph Lengeler of Germany who presented a historical view of the PTS in a talk which formed the basis for the first full-length article in this JMMB written symposium. The second article in the symposium, by Joyet et al., describes the recent research conducted within the research group of Josef Deutscher in France. This article concerns the involvement of the PTS in transcriptional regulation in Gram-positive firmicutes. The next three articles in this symposium deal with different aspects of the PTS in firmicutes, the first by Van der Heiden et al. (Belgium), dealing with the newly characterized PTS-mediated D-tagatose catabolic pathway in Bacillus licheniformis, the second, by Kuipers et al. (the Netherlands), concerning PTS-mediated fucose (6-deoxygalactose) metabolism in the pathogen, Streptococcus pneumoniae, and the third, by W.J. Mitchell (United Kingdom), dealing with genomic analyses of the PTS in solventogenic Clostridia, fairly close relatives of the Bacilli.
The sixth symposium article, by Gordon et al., from the research group of Orna Amster-Chloder in Isreal, deals with the bioinformatic identification and functional characterization of PTS-regulated RNA antiterminator-like sites in bacterial genomes, a paper that suggests the occurrence of novel functions for these sites. In the seventh article, Jacqueline Plumbridge (France) contrasts the regulation of amino sugar utilization by the proteobacterium, E. coli, and the firmicute, Bacillus subtilis. Then two papers, one by Luettman et al. from Boris Görke's laboratory (Austria), and the second by Wolf et al. from Andreas Kremling's lab (Germany), deal with the involvement of the nitrogen regulatory PTS (EINtr, NPr and IIANtr) in the control of potassium ion uptake in two distantly related γ-proteobacteria, Pseudomonas putida and E. coli, respectively. These papers describe a novel function of the PTS in maintaining ionic homeostasis.
In the tenth article, Carmona et al. (Guillermo Gosset's lab in Mexico) discuss the use of pts mutants of E. coli to facilitate the industrial production of various compounds including aromatic metabolites and biofuels. As also noted in Wilf Mitchell's article, the biotechnological use of bacteria for useful metabolite production is of growing interest and utility. Sarah Sutrina of the West Indies, in symposium article #11, summarizes the work executed in her laboratory defining the characteristics of E. coli biofilms and how these “differentiated” multicellular structures are organized and regulated during their biogenesis by the PTS and cyclic AMP.
The last article, by my research associate, Zhongge Zhang, and myself (U.S.A.), deals with the novel and surprising observation that the PTS regulates the process of directed mutation of the glycerol catabolic operon, glpFK, in E. coli. The gene products of this operon are essential for normal growth of these bacteria with this compound as the sole carbon source. The PTS controls cyclic AMP synthesis and inducer uptake, as established in previous publications from several laboratories. In this case, glycerol uptake via the glycerol facilitator, GlpF, is followed by its glycerol kinase (GlpK)-dependent phosphorylation to glycerol 3-phosphate, the true inducer of the glycerol (glp) utilization regulon. These two small molecules, cyclic AMP and glycerol 3-P, interact with their cognate transcription factors, the cyclic AMP receptor protein (Crp), an activator of transcription, and the glycerol repressor (GlpR), a negative effector of transcription, respectively. Binding of either of these two proteins to their DNA binding sites in the glpFK operon inhibits, and thereby regulates, insertion of a small transposon, IS5, into a single upstream site of the glpFK promoter, thereby activating this promoter and allowing the bacteria to overcome the inhibitory effects of non-metabolizable PTS sugars on glycerol utilization [Saier and Zhang, 2014; Zhang and Saier, 2009b, a]. This novel discovery represents the most recently identified regulatory function of the PTS.
Concluding Remarks
The studies reported in this comprehensive PTS50 written symposium illustrate the fact that our understanding of the PTS is still expanding with respect to its structure, mechanism, evolution and physiological functions. Since the last 50 years of PTS research have been so revealing, we wonder what the next 50 years will yield. Structural diversity in the proteins that comprise the system, novel mechanisms of catalysis, and undreamt of functions are likely to come to light. But only a few of us may live to see that day. Nevertheless, our collective efforts will hopefully live on if we humans don't destroy the only habitat we have, the planet, Earth.
Acknowledgements
I thank the many participants of the PTS50 congress for their insightful oral and written contributions as well as discussions. Jörg Stülke and Josef Deutscher were instrumental in insuring the smooth flow of events at the meeting, making it most memorable for the PTS research community. Research in the Saier laboratory was supported by NIH grants GM077402, GM109859 and GM094610.
References
- Bächler C, Flukiger-Bruhwiler K, Schneider P, Bahler P, Erni B. From ATP as substrate to ADP as coenzyme: functional evolution of the nucleotide binding subunit of dihydroxyacetone kinases. J Biol Chem. 2005;280:18321–18325. doi: 10.1074/jbc.M500279200. [DOI] [PubMed] [Google Scholar]
- Barabote RD, Saier MH., Jr. Comparative genomic analyses of the bacterial phosphotransferase system. Microbiol Mol Biol Rev. 2005;69:608–634. doi: 10.1128/MMBR.69.4.608-634.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L, Cai S, Zhao D, Wu J, Wang L, Liu X, Li M, Hou J, Zhou J, Liu J, Han J, Xiang H. Analysis of the transcriptional regulator GlpR, promoter elements, and posttranscriptional processing involved in fructose-induced activation of the phosphoenolpyruvate-dependent sugar phosphotransferase system in Haloferax mediterranei. Appl Environ Microbiol. 2014;80:1430–1440. doi: 10.1128/AEM.03372-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Jin X, Huang H, Derebe MG, Levin EJ, Kabaleeswaran V, Pan Y, Punta M, Love J, Weng J, Quick M, Ye S, Kloss B, Bruni R, Martinez-Hackert E, Hendrickson WA, Rost B, Javitch JA, Rajashankar KR, Jiang Y, Zhou M. Crystal structure of a potassium ion transporter. TrkH. Nature. 2011;471:336–340. doi: 10.1038/nature09731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comas I, Gonzalez-Candelas F, Zuniga M. Unraveling the evolutionary history of the phosphoryl-transfer chain of the phosphoenolpyruvate:Phosphotransferase system through phylogenetic analyses and genome context. BMC Evol Biol. 2008;8:147. doi: 10.1186/1471-2148-8-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher J, Ake FM, Derkaoui M, Zebre AC, Cao TN, Bouraoui H, Kentache T, Mokhtari A, Milohanic E, Joyet P. The bacterial phosphoenolpyruvate:Carbohydrate phosphotransferase system: Regulation by protein phosphorylation and phosphorylation-dependent protein-protein interactions. Microbiol Mol Biol Rev. 2014;78:231–256. doi: 10.1128/MMBR.00001-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudoroff M, Hassid WZ, et al. Direct utilization of maltose by Escherichia coli. J Biol Chem. 1949;179:921–934. [PubMed] [Google Scholar]
- Erni B. The mannose transporter complex: An open door for the macromolecular invasion of bacteria. J Bacteriol. 2006;188:7036–7038. doi: 10.1128/JB.01074-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erni B, Siebold C, Christen S, Srinivas A, Oberholzer A, Baumann U. Small substrate, big surprise: Fold, function and phylogeny of dihydroxyacetone kinases. Cell Mol Life Sci. 2006;63:890–900. doi: 10.1007/s00018-005-5518-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Alles LF, Zahn A, Erni B. Sugar recognition by the glucose and mannose permeases of Escherichia coli. Steady-state kinetics and inhibition studies. Biochemistry. 2002;41:10077–10086. doi: 10.1021/bi025928d. [DOI] [PubMed] [Google Scholar]
- Hvorup R, Chang AB, Saier MH., Jr. Bioinformatic analyses of the bacterial L-ascorbate phosphotransferase system permease family. J Mol Microbiol Biotechnol. 2003;6:191–205. doi: 10.1159/000077250. [DOI] [PubMed] [Google Scholar]
- Jacobson GR, Lee CA, Leonard JE, Saier MH., Jr. Mannitol-specific enzyme II of the bacterial phosphotransferase system. I. Properties of the purified permease. J Biol Chem. 1983;258:10748–10756. [PubMed] [Google Scholar]
- Jacobson GR, Lee CA, Saier MH., Jr. Purification of the mannitol-specific enzyme II of the Escherichia coli phosphoenolpyruvate:sugar phosphotransferase system. J Biol Chem. 1979;254:249–252. [PubMed] [Google Scholar]
- Krausse D, Hunold K, Kusian B, Lenz O, Stulke J, Bowien B, Deutscher J. Essential role of the hprk gene in ralstonia eutropha h16. J Mol Microbiol Biotechnol. 2009;17:146–152. doi: 10.1159/000233505. [DOI] [PubMed] [Google Scholar]
- Kundig W, Ghosh S, Roseman S. Phosphate bound to histidine in a protein as an intermediate in a novel phospho-transferase system. Proc Natl Acad Sci U S A. 1964;52:1067–1074. doi: 10.1073/pnas.52.4.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundig W, Kundig FD, Anderson B, Roseman S. Restoration of active transport of glycosides in Escherichia coli by a component of a phosphotransferase system. J Biol Chem. 1966;241:3243–3246. [PubMed] [Google Scholar]
- Le Bouguenec C, Schouler C. Sugar metabolism, an additional virulence factor in enterobacteria. Int J Med Microbiol. 2011;301:1–6. doi: 10.1016/j.ijmm.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CA, Saier MH., Jr. Mannitol-specific enzyme ii of the bacterial phosphotransferase system. Iii. The nucleotide sequence of the permease gene. J Biol Chem. 1983;258:10761–10767. [PubMed] [Google Scholar]
- Lengeler JW, Jahreis K. Bacterial PEP-dependent carbohydrate: phosphotransferase systems couple sensing and global control mechanisms. Contrib Microbiol. 2009;16:65–87. doi: 10.1159/000219373. [DOI] [PubMed] [Google Scholar]
- Leonard JE, Saier MH., Jr. Mannitol-specific enzyme ii of the bacterial phosphotransferase system. Ii. Reconstitution of vectorial transphosphorylation in phospholipid vesicles. J Biol Chem. 1983;258:10757–10760. [PubMed] [Google Scholar]
- Lopes MS, Gosset G, Rocha RC, Gomez JG, Ferreira da Silva L. PHB biosynthesis in catabolite repression mutant of Burkholderia sacchari. Curr Microbiol. 2011;63:319–326. doi: 10.1007/s00284-011-9981-6. [DOI] [PubMed] [Google Scholar]
- McCoy JG, Levin EJ, Zhou M. Structural insight into the PTS sugar transporter EIIC. Biochim Biophys Acta. 2015;1850:577–585. doi: 10.1016/j.bbagen.2014.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P. Transport of phosphate across the osmotic barrier of Micrococcus pyogenes; specificity and kinetics. J Gen Microbiol. 1954;11:73–82. doi: 10.1099/00221287-11-1-73. [DOI] [PubMed] [Google Scholar]
- Pickering BS, Lopilato JE, Smith DR, Watnick PI. The transcription factor Mlc promotes Vibrio choleraebiofilm formation through repression of phosphotransferase system components. J Bacteriol. 2014;196:2423–2430. doi: 10.1128/JB.01639-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickl A, Johnsen U, Schonheit P. Fructose degradation in the haloarchaeon Haloferax volcanii involves a bacterial type phosphoenolpyruvate-dependent phosphotransferase system, fructose-1-phosphate kinase, and class II fructose-1,6-bisphosphate aldolase. J Bacteriol. 2012;194:3088–3097. doi: 10.1128/JB.00200-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postma PW, Lengeler JW, Jacobson GR. Phosphoenolpyruvate:Carbohydrate phosphotransferase systems of bacteria. Microbiol Rev. 1993;57:543–594. doi: 10.1128/mr.57.3.543-594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigali S, Nothaft H, Noens EE, Schlicht M, Colson S, Muller M, Joris B, Koerten HK, Hopwood DA, Titgemeyer F, van Wezel GP. The sugar phosphotransferase system of Streptomyces coelicolor is regulated by the GntR-family regulator DasR and links N-acetylglucosamine metabolism to the control of development. Mol Microbiol. 2006;61:1237–1251. doi: 10.1111/j.1365-2958.2006.05319.x. [DOI] [PubMed] [Google Scholar]
- Roseman S. The transport of carbohydrates by a bacterial phosphotransferase system. J Gen Physiol. 1969;54:138–184. doi: 10.1085/jgp.54.1.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier MH, Jr., Tran CV, Barabote RD. Tcdb: The transporter classification database for membrane transport protein analyses and information. Nucleic Acids Res. 2006;34:D181–186. doi: 10.1093/nar/gkj001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier MH, Jr., Yen MR, Noto K, Tamang DG, Elkan C. The transporter classification database: Recent advances. Nucleic Acids Res. 2009;37:D274–278. doi: 10.1093/nar/gkn862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier MH, Jr., Reddy VS, Tamang DG, Vastermark A. The transporter classification database. Nucleic Acids Res. 2014;42:D251–258. doi: 10.1093/nar/gkt1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier MH, Hvorup RN, Barabote RD. Evolution of the bacterial phosphotransferase system: From carriers and enzymes to group translocators. Biochem Soc Trans. 2005;33:220–224. doi: 10.1042/BST0330220. [DOI] [PubMed] [Google Scholar]
- Saier MH, Jr., Reizer J. The bacterial phosphotransferase system: New frontiers 30 years later. Mol Microbiol. 1994;13:755–764. doi: 10.1111/j.1365-2958.1994.tb00468.x. [DOI] [PubMed] [Google Scholar]
- Saier MH, Jr., Zhang Z. Transposon-mediated directed mutation controlled by DNA binding proteins in Escherichia coli. Front Microbiol. 2014;5:390. doi: 10.3389/fmicb.2014.00390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchieu JH, Norris V, Edwards JS, Saier MH., Jr. The complete phosphotransferase system in Escherichia coli. J Mol Microbiol Biotechnol. 2001;3:329–346. [PubMed] [Google Scholar]
- Vastermark A, Saier MH., Jr. The involvement of transport proteins in transcriptional and metabolic regulation. Curr Opin Microbiol. 2014;18:8–15. doi: 10.1016/j.mib.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu LF, Tomich JM, Saier MH., Jr. Structure and evolution of a multidomain multiphosphoryl transfer protein. Nucleotide sequence of the frub(hi) gene in Rhodobacter capsulatus and comparisons with homologous genes from other organisms. J Mol Biol. 1990;213:687–703. doi: 10.1016/S0022-2836(05)80256-6. [DOI] [PubMed] [Google Scholar]
- Yamada M, Saier MH., Jr. Glucitol-specific enzymes of the phosphotransferase system in Escherichia coli. Nucleotide sequence of the gut operon. J Biol Chem. 1987;262:5455–5463. [PubMed] [Google Scholar]
- Zhang Z, Saier MH., Jr. A novel mechanism of transposon-mediated gene activation. PLoS Genet. 2009a;5:e1000689. doi: 10.1371/journal.pgen.1000689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Saier MH., Jr. A mechanism of transposon-mediated directed mutation. Mol Microbiol. 2009b;74:29–43. doi: 10.1111/j.1365-2958.2009.06831.x. [DOI] [PMC free article] [PubMed] [Google Scholar]