Abstract
Recently, Mendelian disorders of the DNA methylation machinery have been described which demonstrate the complex roles of epigenetics in neurodevelopment and disease. For example, defects of DNMT1, the maintenance methyltransferase, lead to adult-onset progressive neurologic disorders, whereas defects of the de novo methyltransferases DNMT3A and DNMT3B lead to nonprogressive neurodevelopmental conditions. Furthermore, patients with DNMT3A deficiency demonstrate overgrowth, a feature common to disorders of histone machinery and imprinting disorders, highlighting the interconnectedness of the many epigenetic layers. Disorders of the DNA methylation machinery include both the aforementioned “writers” and also the “readers” of the methyl mark, such as MeCP2, the cause of Rett syndrome. Any dosage disruption, either haploinsufficiency or overexpression of DNA methylation machinery leads to wide-spread gene expression changes in trans, disrupting expression of a subset of target genes that contribute to individual disease phenotypes. In contrast, classical imprinting disorders such as Angelman syndrome have been thought generally to cause epigenetic dysregulation in cis. However, the recent description of multilocus methylation disorders challenges this generalization. Here, in addition to summarizing recent developments in identifying the pathogenesis of these diseases, we highlight clinical considerations and some unexpected therapeutic opportunities, such as to poisomerase inhibitors for classical imprinting disorders.
Keywords: intellectual disability, imprinting, Rett syndrome, Angelman syndrome
Mendelian Disorders of the Epigenetic Machinery
The study of epigenetic modifications of genes with crucial roles in the nervous system is an expanding field within neurogenetics.1 Epigenetics refers to a heritable change in gene expression through modifications of DNA or proteins without a change in the DNA sequence itself.2 There are several molecular mechanisms by which gene expression can be influenced by epigenetic alterations, for instance, DNA cytosine methylation and histone modifications.3
DNA methylation occurs when a methyl group is chemically attached to the 5th position of a cytosine nucleotide.4 This generally occurs on cytosines that are followed by guanines, so called CpG dinucleotides.5 Traditionally, methylation of a cytosine residue is thought to be associated with gene silencing,6 although we now know that in certain contexts it can be associated with gene activation as well.7 Recent research has indicated that in mammalian neurons, DNA methylation also occurs on cytosine residues followed by nonguanine nucleotides (non-CpG methylation), which can also be associated with transcriptional regulation.8 Like DNA itself, histones can be modified and these modifications also affect expression of specific genes. Histone modifications and the diseases caused by abnormalities of the histone machinery have been reviewed elsewhere.9–11
In the last several years, several conditions have been discovered that are caused by inherited defects in the methylation and histone machinery, and these uniformly appear to lead to neurologic dysfunction.10 Although many epigenetic disorders, such as the classical imprinting disorders, generally occur due to epimutations in cis,12 the newly emerging Mendelian disorders of epigenetic machinery are likely to have widespread effects in trans.10 In some cases, the target genes dysregulated by the missing or dysfunctional epigenetic machinery have been identified, and for some, genetic variation surrounding the target gene itself can also modify the phenotype, highlighting the complexity of these interactions.13 In this review, we will focus our discussion on disorders of the DNA methylation mark and machinery (►Table 1).
Table 1.
Human Mendelian disorders of the DNA methylation machinery
| Protein | Function | Mutation | Disease name or phenotype |
|---|---|---|---|
| DNMT1 | Maintenance DNA methyltransferase | Heterozygous missense mutations in exon 20 or 21 | HSAN1E |
| Heterozygous missense mutations in exon 21 | ADCA:DN | ||
| DNMT3A | De novo DNA methyltransferase | Heterozygous missense, frameshift, in-frame deletion | Syndrome of overgrowth and intellectual disability |
| DNMT3B | De novo DNA methyltransferase | Homozygous or compound heterozygous missense | ICF1 syndrome |
| MBD5 | Methyl-CPG binding protein (MBD) | Gene deletion or duplication on one allele, autosomal dominant nonsense | MBD5 deficiency |
| MeCP2 | Methyl-CPG binding protein (MBD) | X-linked dominant deletion or missense or nonsense or frameshift | Rett syndrome |
| Gene duplication | MeCP2 duplication syndrome | ||
| ZBTB24 | Methyl-CPG binding protein (Zinc finger) | Homozygous or compound heterozygous missense, nonsense or frameshift→ stop codon | ICF2 |
| ZFP57 | Methyl-CPG binding protein (Zinc finger) | Homozygous or compound heterozygous missense, nonsense or frameshift | Transient neonatal diabetes mellitus |
Note: MBD6 has been associated with autism spectrum disorder, but not known to be definitively causal.
Defective Writing of the DNA Methylation Mark
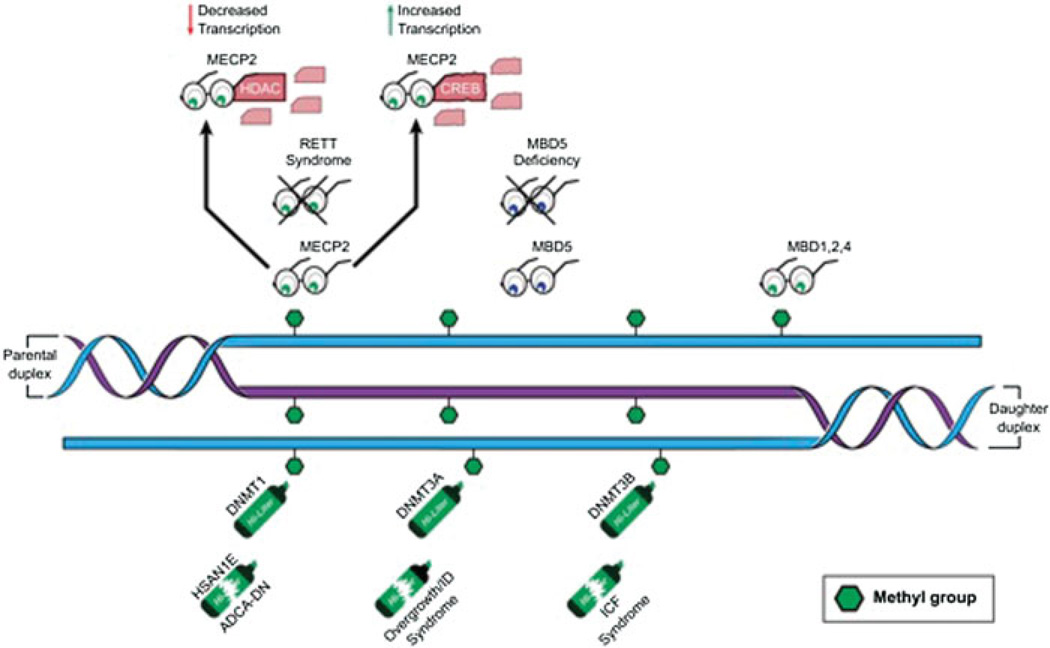
In mammals, methyl groups are added to CpG dinucleotides by several enzymes, known as DNA methyltransferases (the “writers” of the DNA methylation code).3 DNA methyltransferase 1 (DNMT1) is the only known DNA methyltransferase that does not have a major role in de novo DNA methylation, but whose primary role is thought to be to maintain methylation patterns through replication by copying the methylation pattern from the parent strand to the daughter strand (►Fig. 1).14,15 Mutations in the chromatin binding domains of DNMT1 have been shown to cause two separate progressive autosomal dominant adult-onset neurologic disorders (►Fig. 1).16,17 Hereditary sensory and autonomic neuropathy type 1with dementia and hearing loss (HSAN1E) is a disorder in which individuals have normal development, followed by sensory neuropathy and hearing loss in their teens to thirties, and eventually dementia in their thirties or forties.16 HSAN1E is caused by mutations in exon 20 of DNMT1,which codes for a portion of the targeting sequence (TS) domain and allows DNMT1 to attach to heterochromatin.16 In vitro studies of human cells with this exon 20 DNMT1 mutation demonstrate abnormal DNMT1 binding to heterochromatin, premature degradation of DNMT1 transcripts and global hypomethylation with specific areas of hypermethylation.16 When mutations are found in exon 21 of DNMT1, which also encodes a portion of the TS domain, a different syndrome called autosomal dominant cerebellar ataxia, deafness and narcolepsy (ADCA-DN) occurs. ADCA-DN is characterized by adult-onset of narcolepsy—usually with cataplexy—followed by onset of sensorineural deafness, cerebellar ataxia, and eventually dementia.17 The phenotypes of ADCA-DN and HSAN1E are progressive and both have late occurring neurologic manifestations. This perhaps relates to a gradual loss of DNA methylation marks over time, potentially explaining the late-onset and the progressive nature of the symptoms. It has previously been proposed that such age-related degradation of the DNA methylation mark may be a general mechanism accounting for the late onset of many common complex disease phenotypes.18
Fig. 1.
The DNA methylation machinery consists of DNA methyltransferases (highlighters), readers of the methylation mark (glasses), and erasers (not shown). Broken highlighters indicate diseases known to be caused by dysfunction of these proteins. Crossed out glasses indicate diseases caused by dysfunction of these proteins. DNMT1—DNA methyltransferase 1, is the maintenance methyltransferase that copies the signal from the parental strand; DNMT3A—DNA methyltransferase 3A and DNMT3B—DNA methyltransferase 3B are the de novo methyltransferases and are not limited to hemimethylated sites; MECP2—methyl-CPG binding protein reads the DNA methylation mark and can either lead to gene activation or repression depending on partners; MBD1, 2, 4—methyl-binding proteins 1, 2, and 4 also read methyl-CpGs; MBD5—methyl-binding protein 5 does not read a CpG methylation, but associates with heterochromatin; HSAN1E—hereditary sensory and autonomic neuropathy type 1 with dementia and hearing loss syndrome; ADCA-DN—autosomal dominant cerebellar ataxia, deafness, and narcolepsy syndrome; ICF—immunodeficiency, centromeric instability, and facial anomalies syndrome; HDAC—histone deacetylase protein; CREB—cAMP-binding response element-binding protein.
Two other DNA methyltransferase proteins are known in mammals—DNMT3A and DNMT3B (►Fig. 1). These enzymes are thought to be primarily responsible for de novo methylation of DNA.19 They also have a role in maintenance methylation as they show ability to methylate both unmethylated and hemi-methylated CpGs.4,14,15 DNMT3A is also thought to be responsible for the aforementioned non-CpG DNA methylation.8 Recently, mutations in highly conserved domains of DNMT3A have been shown to cause overgrowth associated with intellectual disability and facial dysmorphisms.20 In contrast, biallelic mutations in DNMT3B cause ICF syndrome: immunodeficiency, centromeric instability, and facial anomalies, which are characterized by severe immunodeficiency with reduction in multiple immunoglobulin subtypes, a genomic instability of the pericentromeric heterochromatin (particularly chromosomes 1,9, and 16), and specific facial anomalies.21 ICF syndrome is inherited in an autosomal recessive pattern, which is notable because most of the Mendelian disorders of methylation machinery are dominantly inherited (►Table 1). Molecular studies in mice and in vitro studies in human cells indicate that mutations that cause ICF syndrome alter highly conserved regions in the methyltransferase domains of the protein but DNMT3B still retains partial activity.22 Complete loss of function of DNMT3B would likely be incompatible with life, as is seen in mice with homozygous loss of function mutations in Dnmt3b.23,24 Patients with ICF syndrome show significantly decreased global methylation (< 50%), reminiscent of DNMT1 defects.16,25 However, unlike phenotypes associated with DNMT1 mutations, this disorder is fully penetrant in early life and nonprogressive.21 The DNA methylation abnormalities present in ICF have demonstrable functional consequences, with expression of over 700 genes altered in samples from patients with ICF syndrome.26 The overgrowth seen in DNMT3A deficiency is a feature shared with some of the Mendelian disorders of histone machinery and classical imprinting disorders, highlighting the interconnectedness of the different epigenetic layers10 and ICF provides an excellent example of how defects of the DNA methylation machinery can have many farreaching trans effects on gene expression.
Defective Reading of the DNA Methylation Mark
The effects of DNA cytosine methylation on gene transcription are performed in multiple ways. GC-rich motifs can act as binding sites for transcription factors, and CpG methylation can prevent binding of these factors, which can lead to repression of transcription.27 Additionally, gene expression can be modulated through the action of proteins that specifically bind to methylated DNA.28 These “readers” of the DNA methylation signal are known as methyl-CpG-binding proteins.29,30 These proteins are classified by the type of domains they contain that bind methyl-CpG. For example, the zinc finger protein family preferentially binds to methylated CpGs contained in a specific target sequence,31 and these proteins are thought to repress gene expression through their subsequent interaction with histone deacetylases.32,33 One zinc finger protein, ZBTB24, has been found to be a cause of ICF syndrome—ICF type 2 (►Table 2),34,35 which shares most of the phenotypic characteristics of ICF syndrome resulting from DNMT3B mutations.36 ZBTB24 does not appear to directly bind methylated DNA, but is thought to modify transcription of genes through participation in epigenetic modifier complexes, thus producing a similar phenotype to ICF type 1.34–36
Table 2.
Human imprinting disorders
| Disorder | Chromosome region involved |
Possible gene(s) involved |
Mutation or epimutation category | Phenotype | Associated with MLMD |
|---|---|---|---|---|---|
| Beckwith-Wiedemann syndrome | 11p15 | IGF2, H19, KCNQ1, CDKN1C | Overexpression of paternal alleles at 2 ICRs | Overgrowth, macroglossia, abdominal wall defects, hemi-hypertrophy, variable cognitive issues | Yes |
| Silver-Russell syndrome | 11p15 | IGF2, H19 | Overexpression of maternal alleles at 2 ICRs | Growth restriction, triangular facies, limb length asymmetry, variable cognitive issues | Yes |
| 7p12.2 | GRB10? | ||||
| 7q32.2 | PEG1? | ||||
| Transient neonatal diabetes mellitus | 6q24 | ZFP57, ZAC1, HYMA1, PLAGL1 | Loss of maternal allelic expression | Transient diabetes, macroglossia, omphalocele | Always |
| Angelman syndrome | 15q11–13 | UBE3A | Loss of maternal UBE3A expression | ID, seizures, language delay, microcephaly, ataxia | No |
| Prader-Willi syndrome | 15q11–13 | SNRPN | Loss of paternal gene allelic expression | Prenatal growth restriction, hypotonia, ID, hypogonadism, hyperphagia, obesity | Yes |
| Maternal UPD 14 | 14q32 | DLK1, RTL1 | Loss of paternal gene allelic expression | Growth restriction, hypotonia, scoliosis, precocious puberty, learning disabilities | No |
| Paternal UPD 14 | 14q32 | DLK1, RTL1, DIO3 | Loss of maternal gene allelic expression | Growth restriction, skeletal anomalies, abdominal wall defects, learning disabilities | No |
| PHP1A | 20q13 | GNAS | Loss of maternal GNAS allele | AHO, resistance to multiple hormones | No |
| PHP1B | 20q13 | STX16 | Loss of maternal allele → no GNAS expression | Resistance to hormones | |
| P-PHP | 20q13 | GNAS | Loss of paternal GNAS allele | AHO |
Abbreviations: AHO, Albright hereditary osteodystrophy41; ID, intellectual disability; MLMD, multilocus methylation defects; PHP1A, pseudohypoparathyroidism type 1A; PHP1B, pseudohypoparathyroidism type 1B; P-PHP, pseudo-pseudohypoparathyroidism; UPD, uniparental disomy.
Certain methyl-CpG binding proteins contain a specific methyl-CpG binding domain and are known as MBDs. The proteins in this family are MBD1–6 and methyl-CpG-binding protein 2 (MeCP2). Most of these bind to methyl-CpG (►Fig. 1), with the exception of MBD3, MBD5, and MBD6.28,32,37 MBD1, MBD2, MBD4, and MeCP2 are thought to, among other functions, attract several other proteins to form chromatin remodeling corepressor complexes, which can repress gene expression at the corresponding loci.6 MBD5 and MBD6 are expressed in human brain and associate with heterochromatin, but do not bind to methylated DNA (►Fig. 1).38 MBD5 is located at 2q23.1, and partial or complete deletions of this gene are associated with specific dysmorphic features, intellectual disability with language and speech particularly affected, seizures, and autistic features. These patients also demonstrate sleep disturbances, short stature, and brachycephaly.38 In vitro studies show that when MBD5 is deleted, there is a change in expression of RAI1 (implicated in Smith-Magenis pathophysiology), NR1D2 (a gene important for circadian rhythms), and MBD1.39 Some of the changes in expression of these putative target genes may therefore explain some of the subphenotypes of this disease. Patients with nonsense mutations or intragenic rearrangements of MBD5 have a very similar phenotype to those with a deletion.40 Interestingly, patients who have duplications in the region containing MBD5 have very similar neurodevelopmental phenotypes to patients with deletions, although on average less severe.41 The similarities between phenotypes in MBD5 deletion and duplication syndromes highlight the idea that concentrations of readers of DNA methylation, like most other components of the epigenetic machinery, are tightly controlled. Thus the associated neurologic phenotypes demonstrate a dose dependence, such that a disruption in either direction can cause disease. In fact, this dosage sensitivity appears to be a general feature of the Mendelian disorders of the epigenetic machinery.10
Methyl-CpG-binding protein 2 (MeCP2) is the methyl-binding protein that has been studied most extensively.28,42 Although MeCP2 does have activity as a transcriptional repressor, extensive research has shown that it can also act as an activator depending on the other proteins to which MeCP2 binds while remaining bound to methylated CpGs (►Fig. 1).7,43
Deficiency of MeCP2 causes Rett syndrome–a neurodevelopmental disorder characterized by acquired microcephaly, progressive intellectual disability, loss of motor skills, and epilepsy.44 Rett syndrome occurs in 1 out of every 10,000 to 20,000 live births.45 Children with Rett syndrome have normal growth, head size and development until approximately 5 to 6 months of age when they demonstrate developmental stagnation and then regression, acquired microcephaly, and seizures.46,47 The majority (90%) of classical Rett syndrome cases are caused by loss of function mutations in MeCP2, located at Xq28.44 Those with classical Rett syndrome are generally girls who are heterozygous for the loss of function mutation.44 When boys with a MeCP2 mutation or deletion survive until birth, they exhibit a severe infantile encephalopathy with seizures.48,49 Rare cases of males with more classical Rett syndrome have been reported, and these cases either show significant somatic mosaicismor a 47XXY karyotype.50 Recently, Xq28 duplication (including MeCP2) has been found to be associated with a phenotype similar to classical Rett.51 Additionally, Rett syndrome severity correlates with the type of MeCP2 mutation,52 indicating that presence of a partially functional protein can ameliorate the disease phenotype.
MeCP2 is expressed in many tissues, but shows the greatest preference for mature neurons.42,46,53 When Mecp2 is knocked out either systemically or exclusively in neurons, the mouse Rett phenotype is present.54,55 Neurons in humans with Rett syndrome, neurons in Mecp2 knockout mice, and neurons derived from human stem cells with absent function of MeCP2 show reduced size of the cellular soma with decreased dendritic spine density and dendritic arborization.56,57 These abnormal cells have genome-wide downregulation of gene expression coupled with downregulation of protein synthesis,58 including some crucial for neuronal maturation and synaptic plasticity.59,60 Girls with Rett syndrome as well as Mecp2 knockout mice show significantly impaired synaptic function.61,62 Autopsy specimens of girls with Rett syndrome show increased NMDA receptor density at early ages, but decreased density thereafter in specific areas of the brain,63,64 which could explain some of the temporal developmental variation seen in Rett syndrome.46,47 Interestingly, mouse neurons show similar synaptic structural abnormalities when Mecp2 is duplicated, which is consistent with the phenotypic similarities in humans with the two conditions.65 This re-emphasizes the importance of tight regulation and dose dependence of MeCP2 activity (and DNA methylation “writing,” “erasing,” and “reading” in general) in neurologic functioning.
Diseases Caused by Abnormalities of Genomic Imprinting
In addition to the Mendelian disorders of epigenetic machinery discussed above that affect gene expression in trans, there are conditions in which specific epimutations appear limited to a particular locus, causing dysfunction in cis.18 Genomic imprinting occurs when two alleles at a specific locus are not functionally equivalent, but rather differentially expressed depending on the parent of origin.66 Maternal and paternal alleles of imprinted genes have different DNA methylation patterns, leading to parent-specific gene expression at the locus. Imprinted genes tend to be clustered together and their transcription is regulated by control centers known as imprinting control regions (ICRs). There are at least 70 imprinted genes currently identified in humans, and disrupted expression is associated with a wide range of phenotypes in the nervous system (►Table 2).67
One of the most studied ICRs is located at 15q11–q13. This ICR regulates both maternally and paternally expressed genes, and is typically methylated on the maternal allele only.68 Prader-Willi syndrome (PWS) is characterized by severe neonatal hypotonia with poor growth in infancy followed by significant hyperphagia and rapid and excessive weight gain in childhood and adulthood. These individuals have mild intellectual disability or learning disabilities, obsessive– compulsive features, and manipulative behaviors. PWS is caused by loss of the paternal allele at 15q11–q13 either through a paternal deletion or through maternal uniparental disomy.66 In rare cases, an imprinting defect in which both alleles carry the methylation pattern normally seen only on the maternal allele is the cause of PWS. This imprinting defect occurs because of incomplete erasure of the paternal grandmother imprinting pattern on the paternally inherited allele.69 Similar to what is seen in Rett syndrome, individuals with PWS demonstrate temporal variation in the phenotype, first exhibiting failure to thrive and later polyphagia and obesity, indicating that epigenetic disruption, such as that seen in PWS and Rett syndrome, interact with normal developmental programming in some fundamental yet currently unexplained manner.
The other syndrome caused by abnormal imprinting at 15q11.13 is Angelman syndrome (AS): a neurodevelopmental disorder characterized by severe global developmental delay leading to intellectual disability, near or complete absence of language, gait ataxia, microcephaly, and seizures.70 Angelman syndrome occurs in approximately 1 out of every 15,000 births and equally affects males and females.70 Children typically appear normal at birth, but from an early age have persistent and severe developmental delay with head growth slowing over time.71 Seizures typically develop between 12 and 36 months of age, and electroencephalogram (EEG) patterns are often specific to AS. In contrast to the seizures that improve with age, the motor dysfunction in AS is progressive over time with hypotonia progressing to hypertonia, ataxia, and tremulousness. The cognitive and language impairments, however, are not progressive and children with AS do not typically experience regressions.71,72
Investigations of the PWS/AS ICR revealed that AS is caused by disruption of expression of the maternal copy of the ubiquitin-protein ligase E3A (UBE3A) gene.73 UBE3A is an enzyme that targets proteins for degradation, and also has a role as a transcriptional activator.74 UBE3A is important for proper neuronal function, and neurons in both human and mouse brains preferentially express the maternally derived UBE3A.75 Mouse models with knockout of maternal Ube3a show truncated dendritic processes, defects in long-term potentiation, and postsynaptic signaling.76–78 These findings emphasize the importance of UBE3A in synaptic function, and are reminiscent of what is seen in MeCP2, likely accounting for known phenotypic overlap.
In humans, maternal UBE3A can be disrupted in several ways to cause AS. Maternal allelic deletions of 15q11.2–q13 are the most common cause of AS.70 In addition to deletions of the region containing UBE3A, mutations within the maternal copy of the UBE3A gene can also cause AS, as can paternal uniparental disomy of chromosome 15. Imprinting defects in which the maternal allele carries the paternal methylation pattern are the least frequent cause of AS.79 Interestingly, MeCP2 has been shown to be involved in maintaining the methylation of the PWS/AS ICR, and this may explain some of the phenotypic overlap and similar developmental time courses of Rett syndrome and AS.80
Patients with large deletions containing UBE3A have the classic and most severe phenotype of AS, and mouse models with maternal deletions of Ube3a also have the most severe phenotype with decreased Ube3a expression in all examined neurons.76,79 Angelman syndrome patients with UPD have fewer seizures and motor abnormalities as well as better language and growth, whereas patients with imprinting defects or mutations of UBE3A are also relatively severe, but have better language than deletion patients.81–83 Mice with paternal duplication of Ube3a also show a less severe phenotype.76
Several studies suggest that in cohorts of patients with AS and other imprinting disorders —particularly Beckwith-Wiedemann syndrome (►Table 2)—there is an increased frequency of use of assisted reproductive technology (ART), such as in vitro fertilization (IVF) or intracytoplasmic sperm injection (ICSI).84 At this point, it is unclear whether the increased risk is related to the ART itself or to epigenetic defects in the parents leading to fertility problems that became unmasked by the ART intervention.84 However, given this association, clinicians should inquire about ART whenever an imprinting disorder is suspected.
Multilocus Methylation Disorders
Recently, in addition to the classical imprinting disorders, patients have been described with hypomethylation at multiple imprinted loci. This group of conditions has been called multilocus methylation defects (MLMD).85 These patients typically have similar features to one of the classical imprinting disorders, but often with extra features. These patients frequently have developmental delay as a cardinal feature.34,85–87 Classical imprinting disorders are caused by epigenetic disruption of a single gene or locus in cis. However, MLMDs can affect both maternally and paternally imprinted loci on different chromosomes that implicate necessary trans acting factors.85 ZFP57 is a zinc finger methyl-CpG binding protein thought to be important in establishment and maintenance of DNA methylation, and homozygous mutations in this gene have been identified as a cause of transient neonatal diabetes mellitus associated with multiple hypomethylated loci (►Tables 1 and 2).34 It is likely that other trans acting genes that control methylation and imprinting are yet to be found.
Diagnostic Workup for Imprinting Disorders and MDMM
When a disorder of methylation or imprinting is suspected, there are several clinically available molecular methods which clinicians can use to assist in making the diagnosis. When suspicion of a single gene disorder of the DNA methylation machinery such as Rett syndrome is high, specific sequencing of that gene with or without deletion and duplication testing of that gene can be performed clinically. Deletions or duplications of larger chromosomal regions can be assessed through high-resolution chromosomal microarrays, and this can also reveal some cases of uniparental disomy (isodisomy). When an imprinting disorder is suspected, methylation testing of the relevant area is warranted. If methylation tests are abnormal, a diagnosis of an imprinting disorder is made. If multiple methylation tests of a locus are abnormal, segregation of DNA polymorphisms can be analyzed to investigate for uniparental disomy (heterodisomy). Imprinting control regions can also be sequenced. For MLMD, multiple methylation assays can be sent, although in the future it may be possible to explore this on a more global scale.70,88
Outlook for Therapeutic Development
Several recent observations yield new hope for the group of disorders with defects of the DNA methylation mark and its machinery. A recent study demonstrated that when Mecp2 was systemically delivered to 10- to 12-month-old mice carrying a defective Mecp2 copy, the mice demonstrated an improvement in neurologic function.89 This provides hope for approaches aimed at increasing the expression of MeCP2 in reversing the phenotype of Rett syndrome even postnatally. Another surprising study suggested that in mice with a maternal Ube3a deletion, systemic administration of the topoisomerase inhibitor topotecan led to detectable levels of paternally derived but completely functional Ube3a in several brain areas, including neurons of the hippocampus, neocortex, striatum, and cerebellum.90 Uncovering the disease pathogenesis of disorders with abnormalities of the DNA methylation mark and its machinery may thereby provide therapeutic options for this expanding group of genetically based neurodevelopmental disorders.
Acknowledgments
This work was supported by a grant to H.T.B. by the NIH Director’s Early Independence Award (DP5OD017877).We would like to acknowledge Catherine Kiefe for help with artwork, and Jill Fahrner and Carolyn Applegate for carefully reading the manuscript.
References
- 1.Urdinguio RG, Sanchez-Mut JV, Esteller M. Epigenetic mechanisms in neurological diseases: genes, syndromes, and therapies. Lancet Neurol. 2009;8(11):1056–1072. doi: 10.1016/S1474-4422(09)70262-5. [DOI] [PubMed] [Google Scholar]
- 2.Bird A. Perceptions of epigenetics. Nature. 2007;447(7143):396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- 3.Weichenhan D, Plass C. The evolving epigenome. Hum Mol Genet. 2013;22(Suppl 1):R1–R6. doi: 10.1093/hmg/ddt348. [DOI] [PubMed] [Google Scholar]
- 4.Conerly M, Grady WM. Insights into the role of DNA methylation in disease through the use of mouse models. Dis Model Mech. 2010;3(5–6):290–297. doi: 10.1242/dmm.004812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eckhardt F, Lewin J, Cortese R, et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet. 2006;38(12):1378–1385. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006;31(2):89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 7.Chahrour M, Jung SY, Shaw C, et al. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320(5880):1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo JU, Su Y, Shin JH, et al. Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat Neurosci. 2014;17(2):215–222. doi: 10.1038/nn.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mehler MF. Epigenetic principles and mechanisms underlying nervous system functions in health and disease. Prog Neurobiol. 2008;86(4):305–341. doi: 10.1016/j.pneurobio.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fahrner JA, Bjornsson HT. Mendelian disorders of the epigenetic machinery: Tipping the balance of chromatin states. Annu Rev Genomics Hum Genet. 2014 Sep;15 doi: 10.1146/annurev-genom-090613-094245. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maze I, Noh KM, Allis CD. Histone regulation in the CNS: basic principles of epigenetic plasticity. Neuropsychopharmacology. 2013;38(1):3–22. doi: 10.1038/npp.2012.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bjornsson HT, Brown LJ, Fallin MD, et al. Epigenetic specificity of loss of imprinting of the IGF2 gene in Wilms tumors. J Natl Cancer Inst. 2007;99(16):1270–1273. doi: 10.1093/jnci/djm069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ratnakumar K, Bernstein E. ATRX: the case of a peculiar chromatin remodeler. Epigenetics. 2013;8(1):3–9. doi: 10.4161/epi.23271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walton EL, Francastel C, Velasco G. Maintenance of DNA methylation: Dnmt3b joins the dance. Epigenetics. 2011;6(11):1373–1377. doi: 10.4161/epi.6.11.17978. [DOI] [PubMed] [Google Scholar]
- 15.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 16.Klein CJ, Botuyan MV, Wu Y, et al. Mutations in DNMT1 cause hereditary sensory neuropathy with dementia and hearing loss. Nat Genet. 2011;43(6):595–600. doi: 10.1038/ng.830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Winkelmann J, Lin L, Schormair B, et al. Mutations in DNMT1 cause autosomal dominant cerebellar ataxia, deafness and narcolepsy. Hum Mol Genet. 2012;21(10):2205–2210. doi: 10.1093/hmg/dds035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bjornsson HT, Fallin MD, Feinberg AP. An integrated epigenetic and genetic approach to common human disease. Trends Genet. 2004;20(8):350–358. doi: 10.1016/j.tig.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 19.Berdasco M, Esteller M. Genetic syndromes caused by mutations in epigenetic genes. Hum Genet. 2013;132(4):359–383. doi: 10.1007/s00439-013-1271-x. [DOI] [PubMed] [Google Scholar]
- 20.Tatton-Brown K, Seal S, Ruark E, et al. Childhood Overgrowth Consortium. Mutations in the DNA methyltransferase gene DNMT3A cause an overgrowth syndrome with intellectual disability. Nat Genet. 2014;46(4):385–388. doi: 10.1038/ng.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hagleitner MM, Lankester A, Maraschio P, et al. Clinical spectrum of immunodeficiency, centromeric instability and facial dysmorphism (ICF syndrome) J Med Genet. 2008;45(2):93–99. doi: 10.1136/jmg.2007.053397. [DOI] [PubMed] [Google Scholar]
- 22.Matarazzo MR, De Bonis ML, Vacca M, Della Ragione F, D’Esposito M. Lessons from two human chromatin diseases, ICF syndrome and Rett syndrome. Int J Biochem Cell Biol. 2009;41(1):117–126. doi: 10.1016/j.biocel.2008.07.026. [DOI] [PubMed] [Google Scholar]
- 23.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99(3):247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 24.Ueda Y, Okano M, Williams C, Chen T, Georgopoulos K, Li E. Roles for Dnmt3b in mammalian development: a mouse model for the ICF syndrome. Development. 2006;133(6):1183–1192. doi: 10.1242/dev.02293. [DOI] [PubMed] [Google Scholar]
- 25.Heyn H, Vidal E, Sayols S, et al. Whole-genome bisulfite DNA sequencing of a DNMT3B mutant patient. Epigenetics. 2012;7(6):542–550. doi: 10.4161/epi.20523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin B, Tao Q, Peng J, et al. DNA methyltransferase 3B (DNMT3B) mutations in ICF syndrome lead to altered epigenetic modifications and aberrant expression of genes regulating development, neurogenesis and immune function. Hum Mol Genet. 2008;17(5):690–709. doi: 10.1093/hmg/ddm341. [DOI] [PubMed] [Google Scholar]
- 27.Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet. 2002;3(9):662–673. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- 28.Fan G, Hutnick L. Methyl-CpG binding proteins in the nervous system. Cell Res. 2005;15(4):255–261. doi: 10.1038/sj.cr.7290294. [DOI] [PubMed] [Google Scholar]
- 29.Boyes J, Bird A. DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell. 1991;64(6):1123–1134. doi: 10.1016/0092-8674(91)90267-3. [DOI] [PubMed] [Google Scholar]
- 30.Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol Cell Biol. 1998;18(11):6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sasai N, Nakao M, Defossez PA. Sequence-specific recognition of methylated DNA by human zinc-finger proteins. Nucleic Acids Res. 2010;38(15):5015–5022. doi: 10.1093/nar/gkq280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bogdanović O, Veenstra GJ. DNA methylation and methyl-CpG binding proteins: developmental requirements and function. Chromosoma. 2009;118(5):549–565. doi: 10.1007/s00412-009-0221-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mackay DJ, Callaway JL, Marks SM, et al. Hypomethylation of multiple imprinted loci in individuals with transient neonatal diabetes is associated with mutations in ZFP57. Nat Genet. 2008;40(8):949–951. doi: 10.1038/ng.187. [DOI] [PubMed] [Google Scholar]
- 34.de Greef JC, Wang J, Balog J, et al. Mutations in ZBTB24 are associated with immunodeficiency, centromeric instability, and facial anomalies syndrome type 2. Am J Hum Genet. 2011;88(6):796–804. doi: 10.1016/j.ajhg.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nitta H, Unoki M, Ichiyanagi K, et al. Three novel ZBTB24mutations identified in Japanese and Cape Verdean type 2 ICF syndrome patients. J Hum Genet. 2013;58(7):455–460. doi: 10.1038/jhg.2013.56. [DOI] [PubMed] [Google Scholar]
- 36.Weemaes CM, van Tol MJ, Wang J, et al. Heterogeneous clinical presentation in ICF syndrome: correlation with underlying gene defects. Eur J Hum Genet. 2013;21(11):1219–1225. doi: 10.1038/ejhg.2013.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laget S, Joulie M, Le Masson F, et al. The human proteins MBD5 and MBD6 associate with heterochromatin but they do not bind methylated DNA. PLoS ONE. 2010;5(8):e11982. doi: 10.1371/journal.pone.0011982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hodge JC, Mitchell E, Pillalamarri V, et al. Disruption of MBD5 contributes to a spectrum of psychopathology and neurodevelopmental abnormalities. Mol Psychiatry. 2014;19(3):368–379. doi: 10.1038/mp.2013.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mullegama S, Rosenfeld J, Orellana C, et al. MBD5 dosage affects multiple neurodevelopmental pathways in common with other genetic syndromes. Presented at the American Society of Human Genetics Meeting; Nov 6–10, 2012; San Francisco, CA. [Google Scholar]
- 40.Bonnet C, Ali Khan A, Bresso E, et al. Extended spectrum of MBD5 mutations in neurodevelopmental disorders. Eur J Hum Genet. 2013;21(12):1457–1461. doi: 10.1038/ejhg.2013.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mullegama SV, Rosenfeld JA, Orellana C, et al. Reciprocal deletion and duplication at 2q23.1 indicates a role for MBD5 in autism spectrum disorder. Eur J Hum Genet. 2014;22(1):57–63. doi: 10.1038/ejhg.2013.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weng SM, Bailey ME, Cobb SR. Rett syndrome: from bed to bench. Pediatr Neonatol. 2011;52(6):309–316. doi: 10.1016/j.pedneo.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 43.Nan X, Ng HH, Johnson CA, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393(6683):386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 44.Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23(2):185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 45.Neul JL, Kaufmann WE, Glaze DG, et al. RettSearch Consortium. Rett syndrome: revised diagnostic criteria and nomenclature. Ann Neurol. 2010;68(6):944–950. doi: 10.1002/ana.22124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liyanage VR, Rastegar M. Rett syndrome and MeCP2. Neuromolecular Med. 2014;16(2):231–264. doi: 10.1007/s12017-014-8295-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Naidu S, Bibat G, Kratz L, et al. Clinical variability in Rett syndrome. J Child Neurol. 2003;18(10):662–668. doi: 10.1177/08830738030180100801. [DOI] [PubMed] [Google Scholar]
- 48.Leuzzi V, Di Sabato ML, Zollino M, Montanaro ML, Seri S. Early-onset encephalopathy and cortical myoclonus in a boy with MECP2 gene mutation. Neurology. 2004;63(10):1968–1970. doi: 10.1212/01.wnl.0000144350.97844.94. [DOI] [PubMed] [Google Scholar]
- 49.Schüle B, Armstrong DD, Vogel H, Oviedo A, Francke U. Severe congenital encephalopathy caused by MECP2 null mutations in males: central hypoxia and reduced neuronal dendritic structure. Clin Genet. 2008;74(2):116–126. doi: 10.1111/j.1399-0004.2008.01005.x. [DOI] [PubMed] [Google Scholar]
- 50.Moog U, Van Roozendaal K, Smeets E, et al. MECP2 mutations are an infrequent cause of mental retardation associated with neurological problems in male patients. Brain Dev. 2006;28(5):305–310. doi: 10.1016/j.braindev.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 51.Van Esch H, Bauters M, Ignatius J, et al. Duplication of the MECP2 region is a frequent cause of severe mental retardation and progressive neurological symptoms in males. Am J Hum Genet. 2005;77(3):442–453. doi: 10.1086/444549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cuddapah VA, Pillai RB, Shekar KV, et al. Methyl-CpG-binding protein 2 (MECP2) mutation type is associated with disease severity in Rett syndrome. J Med Genet. 2014;51(3):152–158. doi: 10.1136/jmedgenet-2013-102113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cohen DR, Matarazzo V, Palmer AM, et al. Expression of MeCP2 in olfactory receptor neurons is developmentally regulated and occurs before synaptogenesis. Mol Cell Neurosci. 2003;22(4):417–429. doi: 10.1016/s1044-7431(03)00026-5. [DOI] [PubMed] [Google Scholar]
- 54.Chen RZ, Akbarian S, Tudor M, Jaenisch R. Deficiency of methyl-CpG binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nat Genet. 2001;27(3):327–331. doi: 10.1038/85906. [DOI] [PubMed] [Google Scholar]
- 55.Guy J, Hendrich B, Holmes M, Martin JE, Bird A. A mouse Mecp2- null mutation causes neurological symptoms that mimic Rett syndrome. Nat Genet. 2001;27(3):322–326. doi: 10.1038/85899. [DOI] [PubMed] [Google Scholar]
- 56.Chapleau CA, Calfa GD, Lane MC, et al. Dendritic spine pathologies in hippocampal pyramidal neurons from Rett syndrome brain and after expression of Rett-associated MECP2 mutations. Neurobiol Dis. 2009;35(2):219–233. doi: 10.1016/j.nbd.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fukuda T, Itoh M, Ichikawa T, Washiyama K, Goto Y. Delayed maturation of neuronal architecture and synaptogenesis in cerebral cortex of Mecp2-deficient mice. J Neuropathol Exp Neurol. 2005;64(6):537–544. doi: 10.1093/jnen/64.6.537. [DOI] [PubMed] [Google Scholar]
- 58.Li Y, Wang H, Muffat J, et al. Global transcriptional and translational repression in human-embryonic-stem-cell-derived Rett syndrome neurons. Cell Stem Cell. 2013;13(4):446–458. doi: 10.1016/j.stem.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou Z, Hong EJ, Cohen S, et al. Brain-specific phosphorylation of MeCP2 regulates activity-dependent Bdnf transcription, dendritic growth, and spine maturation. Neuron. 2006;52(2):255–269. doi: 10.1016/j.neuron.2006.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen WG, Chang Q, Lin Y, et al. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science. 2003;302(5646):885–889. doi: 10.1126/science.1086446. [DOI] [PubMed] [Google Scholar]
- 61.Asaka Y, Jugloff DG, Zhang L, Eubanks JH, Fitzsimonds RM. Hippocampal synaptic plasticity is impaired in the Mecp2-null mouse model of Rett syndrome. Neurobiol Dis. 2006;21(1):217–227. doi: 10.1016/j.nbd.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 62.Chao HT, Zoghbi HY, Rosenmund C. MeCP2 controls excitatory synaptic strength by regulating glutamatergic synapse number. Neuron. 2007;56(1):58–65. doi: 10.1016/j.neuron.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Blue ME, Naidu S, Johnston MV. Altered development of glutamate and GABA receptors in the basal ganglia of girls with Rett syndrome. Exp Neurol. 1999;156(2):345–352. doi: 10.1006/exnr.1999.7030. [DOI] [PubMed] [Google Scholar]
- 64.Blue ME, Naidu S, Johnston MV. Development of amino acid receptors in frontal cortex from girls with Rett syndrome. Ann Neurol. 1999;45(4):541–545. doi: 10.1002/1531-8249(199904)45:4<541::aid-ana21>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 65.Blue ME, Kaufmann WE, Bressler J, et al. Temporal and regional alterations in NMDA receptor expression in Mecp2-nullmice. Anat Rec (Hoboken) 2011;294(10):1624–1634. doi: 10.1002/ar.21380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lim DH, Maher ER. Human imprinting syndromes. Epigenomics. 2009;1(2):347–369. doi: 10.2217/epi.09.24. [DOI] [PubMed] [Google Scholar]
- 67.Bartolomei MS. Genomic imprinting: employing and avoiding epigenetic processes. Genes Dev. 2009;23(18):2124–2133. doi: 10.1101/gad.1841409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chamberlain SJ. RNAs of the human chromosome 15q11–q13 imprinted region. Wiley Interdiscip Rev RNA. 2013;4(2):155–166. doi: 10.1002/wrna.1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Buiting K, Gross S, Lich C, Gillessen-Kaesbach G, el-Maarri O, Horsthemke B. Epimutations in Prader-Willi and Angelman syndromes: a molecular study of 136 patients with an imprinting defect. Am J Hum Genet. 2003;72(3):571–577. doi: 10.1086/367926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Williams CA, Driscoll DJ, Dagli AI. Clinical and genetic aspects of Angelman syndrome. Genet Med. 2010;12(7):385–395. doi: 10.1097/GIM.0b013e3181def138. [DOI] [PubMed] [Google Scholar]
- 71.Thibert RL, Larson AM, Hsieh DT, Raby AR, Thiele EA. Neurologic manifestations of Angelman syndrome. Pediatr Neurol. 2013;48(4):271–279. doi: 10.1016/j.pediatrneurol.2012.09.015. [DOI] [PubMed] [Google Scholar]
- 72.Leyser M, Penna PS, de Almeida AC, Vasconcelos MM, Nascimento OJ. Revisiting epilepsy and the electroencephalogram patterns in Angelman syndrome. Neurol Sci. 2014;35(5):701–705. doi: 10.1007/s10072-013-1586-3. [DOI] [PubMed] [Google Scholar]
- 73.Kishino T, Lalande M, Wagstaff J. UBE3A/E6-AP mutations cause Angelman syndrome. Nat Genet. 1997;15(1):70–73. doi: 10.1038/ng0197-70. [DOI] [PubMed] [Google Scholar]
- 74.Nawaz Z, Lonard DM, Smith CL, et al. The Angelman syndrome-associated protein, E6-AP, is a coactivator for the nuclear hormone receptor superfamily. Mol Cell Biol. 1999;19(2):1182–1189. doi: 10.1128/mcb.19.2.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dindot SV, Antalffy BA, Bhattacharjee MB, Beaudet AL. The Angelman syndrome ubiquitin ligase localizes to the synapse and nucleus, and maternal deficiency results in abnormal dendritic spine morphology. Hum Mol Genet. 2008;17(1):111–118. doi: 10.1093/hmg/ddm288. [DOI] [PubMed] [Google Scholar]
- 76.Gustin RM, Bichell TJ, Bubser M, et al. Tissue-specific variation of Ube3a protein expression in rodents and in a mouse model of Angelman syndrome. Neurobiol Dis. 2010;39(3):283–291. doi: 10.1016/j.nbd.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jana NR. Understanding the pathogenesis of Angelman syndrome through animal models. Neural Plast. 2012;2012:710943. doi: 10.1155/2012/710943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Weeber EJ, Jiang YH, Elgersma Y, et al. Derangements of hippocampal calcium/calmodulin-dependent protein kinase II in a mouse model for Angelman mental retardation syndrome. J Neurosci. 2003;23(7):2634–2644. doi: 10.1523/JNEUROSCI.23-07-02634.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mabb AM, Judson MC, Zylka MJ, Philpot BD. Angelman syndrome: insights into genomic imprinting and neurodevelopmental phenotypes. Trends Neurosci. 2011;34(6):293–303. doi: 10.1016/j.tins.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Makedonski K, Abuhatzira L, Kaufman Y, Razin A, Shemer R. MeCP2 deficiency in Rett syndrome causes epigenetic aberrations at the PWS/AS imprinting center that affects UBE3A expression. Hum Mol Genet. 2005;14(8):1049–1058. doi: 10.1093/hmg/ddi097. [DOI] [PubMed] [Google Scholar]
- 81.Gentile JK, Tan WH, Horowitz LT, et al. A neurodevelopmental survey of Angelman syndrome with genotype-phenotype correlations. J Dev Behav Pediatr. 2010;31(7):592–601. doi: 10.1097/DBP.0b013e3181ee408e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lossie AC, Whitney MM, Amidon D, et al. Distinct phenotypes distinguish the molecular classes of Angelman syndrome. J Med Genet. 2001;38(12):834–845. doi: 10.1136/jmg.38.12.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Varela MC, Kok F, Otto PA, Koiffmann CP. Phenotypic variability in Angelman syndrome: comparison among different deletion classes and between deletion and UPD subjects. Eur J Hum Genet. 2004;12(12):987–992. doi: 10.1038/sj.ejhg.5201264. [DOI] [PubMed] [Google Scholar]
- 84.Eroglu A, Layman LC. Role of ART in imprinting disorders. Semin Reprod Med. 2012;30(2):92–104. doi: 10.1055/s-0032-1307417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Eggermann T, Leisten I, Binder G, Begemann M, Spengler S. Disturbed methylation at multiple imprinted loci: an increasing observation in imprinting disorders. Epigenomics. 2011;3(5):625–637. doi: 10.2217/epi.11.84. [DOI] [PubMed] [Google Scholar]
- 86.Azzi S, Rossignol S, Steunou V, et al. Multilocus methylation analysis in a large cohort of 11p15-related foetal growth disorders (Russell Silver and Beckwith Wiedemann syndromes) reveals simultaneous loss of methylation at paternal and maternal imprinted loci. Hum Mol Genet. 2009;18(24):4724–4733. doi: 10.1093/hmg/ddp435. [DOI] [PubMed] [Google Scholar]
- 87.Turner CL, Mackay DM, Callaway JL, et al. Methylation analysis of 79 patients with growth restriction reveals novel patterns of methylation change at imprinted loci. Eur J Hum Genet. 2010;18(6):648–655. doi: 10.1038/ejhg.2009.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Poole RL, Docherty LE, Al Sayegh A, et al. International Clinical Imprinting Consortium. Targeted methylation testing of a patient cohort broadens the epigenetic and clinical description of imprinting disorders. Am J Med Genet A. 2013;161(9):2174–2182. doi: 10.1002/ajmg.a.36049. [DOI] [PubMed] [Google Scholar]
- 89.Garg SK, Lioy DT, Cheval H, et al. Systemic delivery of MeCP2 rescues behavioral and cellular deficits in female mouse models of Rett syndrome. J Neurosci. 2013;33(34):13612–13620. doi: 10.1523/JNEUROSCI.1854-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Huang HS, Allen JA, Mabb AM, et al. Topoisomerase inhibitors unsilence the dormant allele of Ube3a in neurons. Nature. 2012;481(7380):185–189. doi: 10.1038/nature10726. [DOI] [PMC free article] [PubMed] [Google Scholar]