Abstract
The opportunistic pathogen Candida albicans has a large repertoire of mechanisms to generate genetic and phenotypic diversity despite the lack of meiosis in its life cycle. Its parasexual cycle enables shifts in ploidy, which in turn facilitate recombination, aneuploidy, and homozygosis of whole chromosomes to fuel rapid adaptation. Here we show that the tetraploid state potentiates ploidy variation and drives population heterogeneity. In tetraploids, the rate of losing a single heterozygous marker [loss of heterozygosity (LOH)] is elevated ∼30-fold higher than the rate in diploid cells. Furthermore, isolates recovered after selection for LOH of one, two, or three markers were highly aneuploid, with a broad range of karyotypes including strains with a combination of di-, tri-, and tetrasomic chromosomes. We followed the ploidy trajectories for these tetraploid- and aneuploid-derived isolates, using a combination of flow cytometry and double-digestion restriction-site-associated DNA analyzed with next-generation sequencing. Isolates derived from either tetraploid or aneuploid isolates predominately resolved to a stable euploid state. The majority of isolates reduced to the conventional diploid state; however, stable triploid and tetraploid states were observed in ∼30% of the isolates. Notably, aneuploid isolates were more transient than tetraploid isolates, resolving to a euploid state within a few passages. Furthermore, the likelihood that a particular isolate will resolve to the same ploidy state in replicate evolution experiments is only ∼50%, supporting the idea that the chromosome loss process of the parasexual cycle is random and does not follow trajectories involving specific combinations of chromosomes. Together, our results indicate that tetraploid progenitors can produce populations of progeny cells with a high degree of genomic diversity, from altered ploidy to homozygosis, providing an excellent source of genetic variation upon which selection can act.
Keywords: parasex, ploidy, yeast
SHIFTS in ploidy between haploid, diploid, and/or polyploid states are mediated by both sexual and asexual mechanisms and promote genetic variation within a population of cells. Such transitions not only yield altered whole-genome ploidy states, but also facilitate increased frequencies of recombination, aneuploidy, and whole-chromosome homozygosis that appear to be common features of mitotic divisions in eukaryotic pathogens, including parasites and fungi (Calo et al. 2013). Importantly, these genome changes have the potential to fuel rapid adaptation in microbial populations (Selmecki et al. 2006; Rancati et al. 2008; Yona et al. 2012). Similar nonmeiotic ploidy shifts occur in somatic mammalian cells such as hepatocytes, and it is thought that some of the aneuploidies and ploidy shifts may provide cells with a selective advantage (Berman and Hadany 2012; Duncan et al. 2012; Duncan 2013). Unusual ploidy states also are a common feature of tumor cell populations, where mitotic collapse (Gordon et al. 2012) and/or telomere defects (Davoli and De Lange 2012) yield tetraploids that ultimately produce aneuploid progeny cells, some of which outcompete the surrounding cells. Understanding how nonmeiotic ploidy reduction occurs in tetraploid and aneuploid cells will provide important insights into these unconventional, but not uncommon, processes.
Candida albicans, the most prevalent opportunistic fungal pathogen of humans, has long been considered an asexual, highly heterozygous, obligate diploid organism, but this idea has been challenged by the discovery of haploid (Hickman et al. 2013) and tetraploid isolates (Suzuki et al. 1986; Legrand et al. 2004) produced in vitro and/or in vivo. Tetraploid cells can arise through cell cycle defects when exposed to fluconazole and subsequently undergo unequal mitotic divisions with multiple spindles that give rise to aneuploid progeny (Harrison et al. 2014). Tetraploids can also arise through a diploid–tetraploid parasexual cycle (Hull et al. 2000; Magee and Magee 2000; Bennett and Johnson 2003; Forche et al. 2008; Song and Petes 2012). This parasexual cycle involves mating between diploid cells followed by ploidy reduction, or “concerted chromosome loss” processes, in which loss of markers on one chromosome is frequently accompanied by loss of markers on other chromosomes frequently resulting in diploid or near-diploid progeny (Bennett and Johnson 2003). These near-diploid parasexual progeny diverge from the parental type by whole-chromosome aneuploidy and by loss of heterozygosity (Forche et al. 2008), both of which provide population heterogeneity upon which natural selection can act (Berman and Hadany 2012).
Completion of the parasexual cycle is thought to be nonmeiotic: C. albicans has lost multiple genes required for meiosis in related yeasts, including the transcriptional regulators IME1 and SUM1 and several spore assembly genes (Wong et al. 2003; Butler et al. 2009). Furthermore, no obvious ascospores have been detected (Van Der Walt 1967). Other fungal species also undergo nonmeiotic ploidy reduction and parasexual cycles, including the closely related C. tropicalis (Seervai et al. 2013) and more distantly related Aspergillus species (Pontecorvo 1956) as well as the rice blast fungus, Magnaporthe grisea (Zeigler et al. 1997). In slime molds (e.g., Dictyostelium), ploidy shifts occur through transient aneuploidy (Sinha and Ashworth 1969). Nonmeiotic ploidy reduction is not restricted to organisms with unconventional life cycles: Saccharomyces cerevisiae, which has a conventional sexual cycle, can also undergo nonmeiotic ploidy reductions, e.g., in mutant backgrounds deficient for DNA repair (Song and Petes 2012) or for chromosome cohesion (Alabrudzinska et al. 2011) or when polyploid cells are passaged for several hundred generations (Gerstein et al. 2006).
In S. cerevisiae, tetraploid and triploid cells are less stable than diploid cells (Mayer and Aguilera 1990; Pavelka et al. 2010), triploids that undergo meiosis generate aneuploid progeny (St. Charles et al. 2012), and tetraploids are prone to chromosome missegregation events due to increased monopolar attachments (Storchova et al. 2006), which may result in aneuploid derivatives. Cells carrying aneuploidies can delay the cell cycle and decrease growth rates (Torres et al. 2007), especially when cells are propagated in laboratory conditions that optimize rapid population growth. In particular, cells with high degrees of aneuploidy (with genome contents between 1.5N and 2N) are more unstable than cells carrying a small number of aneuploidies (i.e., with genome contents close to 1N) (Zhu et al. 2012). This phenomenon is similar to observations in mammalian cells where highly aneuploid genomes are far less viable than those with small numbers of extra chromosomes (Weaver and Cleveland 2006).
In C. albicans, the stability of unconventional ploidy states (i.e., tetraploid and aneuploid) and the process by which ploidy reduction occurs are not well understood. C. albicans can tolerate and maintain chromosomal aneuploidy and large tracts of allele homozygosity (Legrand et al. 2004; Selmecki et al. 2005; Arbour et al. 2009; Bouchonville et al. 2009; Selmecki et al. 2010; Abbey et al. 2011; Andaluz et al. 2011). In addition, exposure to external stress conditions promotes genome instability and increases the rates and types of loss of heterozygosity (LOH) in diploid cells (Forche et al. 2011). Specifically, chromosome loss events are observed more frequently following exposure to fluconazole (Forche et al. 2011) or heat shock (Hilton et al. 1985; Bouchonville et al. 2009) in diploid cells. Furthermore, elevated temperatures and prolonged growth (7–10 days) on glucose-rich or on sorbose media results in approximately diploid progeny from tetraploid parents (Bennett and Johnson 2003). However, it remains unclear how rapidly tetraploid cells lose chromosomes to return to a diploid state or whether other ploidy states can be maintained.
Here we found that the C. albicans tetraploid genome undergoes higher levels of LOH than the diploid genome. Tetraploid cells underwent detectable ploidy changes over both short and extended times of experimental evolution. Selection for marker loss in tetraploid cells enriched for the rare cells that lose chromosomes, and these resulting aneuploids were intermediates in the chromosome loss phase of the parasexual cycle. By determining the chromosomal copy number of these isolates, we revealed a very broad range of ploidy states and karyotypes, including some genomes with a combination of disomic, trisomic, and tetrasomic chromosomes. Despite the relatively frequent appearance of highly aneuploid cells, these aneuploids resolved to euploid states (diploid, triploid, tetraploid, and rarely haploid) when passaged for several more generations. This study supports a model in which the ploidy-reduction phase of the parasexual cycle initiates with random chromosome loss events to produce highly unstable aneuploid derivatives that continue to have dynamic ploidy transitions until they reach a near-euploid state. Ploidy reduction in tetraploid and highly aneuploid cells is stochastic, and individual isolates rarely reduce ploidy to the same degree or in the same manner. Thus, the production of heterogeneous populations of cells is an integral property of nonmeiotic ploidy reduction.
Materials and Methods
Strain construction
Tetraploid strains (Supporting Information, Table S1) were constructed by mating opaque diploid cells of opposite mating types together and subsequently selected for mating products by plating on double selective media after 48 hr on mating media. Isolation of opaque cells and mating conditions were performed as described previously (Bennett and Johnson 2003; Alby and Bennett 2009; Hnisz et al. 2011; Hickman et al. 2013). To construct YJB12712 (one LOH marker, gal1Δ/gal1Δ/gal1Δ/GAL1) we crossed YJB12234 (MTLa gal1Δ/Δ uraΔ/Δ) with DSY919 (MTLα gal1Δ/GAL1 his1Δ/Δ leu2Δ/Δ) and tetraploid mating products were selected for on media lacking uridine and histidine. To construct YJB12651 (two LOH markers, gal1Δ/gal1Δ/GAL1/GAL1), we crossed YJB12234 (MTLa gal1Δ/Δ uraΔ/Δ ENO1-GFP) with YJB12552 (MTLα GAL1/GAL1 ade2Δ/Δ ENO1-RFP) and tetraploid mating products were selected on media lacking uridine and adenine. To construct YJB12779 (three LOH markers, gal1Δ/GAL1/GAL1/GAL1), we crossed CHY477 (MTLα GAL1/GAL1 ade2Δ/Δ) with DSY917 (MTLa gal1Δ/GAL1 his1Δ/Δ leu2Δ/Δ) and tetraploid mating products were selected on media lacking histidine and adenine. The tetraploid state of mating products was verified via flow cytometry for DNA content.
Determination of LOH rates by fluctuation analysis
To measure LOH rates, fluctuation analysis was performed essentially as described previously (Hull et al. 2000; Magee and Magee 2000; Bennett and Johnson 2003; Forche et al. 2008, 2011; Bouchonville et al. 2009). Briefly, strains were plated for single colonies on rich media (YPD: 1% yeast extract, 1% Bacto peptone, 2% dextrose, 0.04% adenine, 1.5% agar). Twelve independent single colonies per strain were inoculated into 2 ml liquid YPD or stress media (described below) and grown for 16–20 hr at 30°. Cells were pelleted, washed once with distilled water, and resuspended in 500 μl distilled water. Dilutions of each culture were spotted onto YPD for total cell count and onto 2-deoxygalactose (2-DOG) to determine the proportion of cells that lost GAL1. YPD CFUs were counted on day 2, and 2-DOGR colonies were counted on day 3, which is prior to the time that colonies would have formed adaptive mutations on the 5-FOA medium (Forche et al. 2003). GAL1 loss rates were determined using the method of the median (Lea and Coulson 1948) and standard deviations were calculated in Excel. For each strain, at least two independent fluctuation analyses were performed. Twenty-four single colonies were picked from YPD and 2-DOG plates for each strain, grown overnight in liquid YPD, and simultaneously prepared for flow cytometry or for long-term storage in 50% glycerol at −80°.
Flow cytometry for ploidy determination
Midlog-phase cells were harvested, washed, and resuspended in 50 mM Tris (pH 8):50 mM EDTA (50:50 TE) and fixed with 95% ethanol overnight at 4°. Cells were washed twice with 50:50 TE, resuspended in 1 mg/ml RNAse A, incubated 1 hr at 37° and then collected, resuspended in 5 mg/ml Proteinase K, and incubated for 30 min at 37°. Cells were washed once with 50:50 TE, resuspended in SybrGreenI (1:85 dilution in 50:50 TE), incubated overnight at 4°, collected, resuspended in 50:50 TE, briefly sonicated, and analyzed using a FACScaliber. Data were fitted with a multi-Gaussian cell cycle model to produce estimates for whole-genome ploidy, based on the assumption that the G2 peak has twice the fluorescence of the G1 peak and minimizing the contribution of S-phase cells to the error function. Ploidy values were calculated by comparing the ratio of peak location in experimental samples to diploid and tetraploid controls as published previously (Hickman et al. 2013).
Double-digestion restriction-site-associated DNA analyzed with next-generation sequencing preparation and analysis
Double-digestion restriction-site-associated DNA analyzed with next-generation sequencing (ddRAD-seq) was performed similarly to that in Tan et al. (2013), digesting genomic DNA with MbeI and then with MfoI to yield thousands of fragments with unique adjacent sequences. After each restriction digest, fragments were end ligated with multiplexing bar-coded PCR amplification primers (P1 and P2 after MbeI and MfoI digests, respectively). The fragments were size selected using gel purification (∼75–400 bp) and then PCR amplified with the P1 and P2 primers and analyzed by next-generation sequencing (NGS). The end result is high-depth sequencing of selected regions bordered by both MbeI and MboI restriction digest sites. Sequence data were analyzed for chromosomal copy number, using the yeast mapping analysis pipeline (Abbey et al. 2014). Reads for the C. albicans reference SC5314 and experimental data sets were aligned to the C. albicans SC5314 reference genome assembly 21 (http://www.candidagenome.org/) and then processed with a series of tools to generate a file containing the read depth at each genome coordinate. Usable restriction fragments were determined via in silico digestion of the reference genome and combined with the per-position read depth and an average read depth is calculated per restriction fragment. This average fragment read depth has biases related to restriction fragment length, GC content, and a position effect, which is consistent across data sets. The restriction fragment-length and GC-content biases are corrected by locally weighted scatterplot smoothing (LOWESS) normalization, where each data point is divided by the LOWESS curve fit to the data. The position-effect bias is then corrected by dividing the LOWESS-normalized experimental data set by the LOWESS-normalized reference data set. Fragments <50 bp or >1000 bp were discarded because of increased noise in average read depth for those fragments. A final copy number estimate (CNV) estimate was made by calculating the weighted average of the corrected read depths.
Passaging experiments
We inoculated 5 μl of glycerol stock for 24 non-LOH (picked from YPD plates) and 24 LOH (picked from 2-DOG plates) single colonies for DSY919 (2N, +Δ), YJB12712 (4N, +ΔΔΔ), YJB12651 (4N+, ++ΔΔΔ), and YJB12779 (4N, +++Δ) into 1 ml liquid YPD in 96-deep-well culture blocks and incubated them at 30° with shaking. Every 24 hr, 10 μl of culture was diluted in 1 ml fresh YPD (1:100 dilution) and reincubated at 30° with shaking. On days 7, 14, 21, and 28 cultures were simultaneously prepared for flow cytometry or for long-term storage in 50% glycerol at −80°. Glycerol stocks were also prepared on days 4, 10, 17, and 24. Ploidy analysis of day 4 isolates was performed after the completion of the passaging experiment from glycerol stocks and included an additional overnight growth in YPD to prepare cells for flow cytometry. We repeated the passaging experiment with YJB12779 by inoculating the same glycerol stocks of 24 non-LOH (picked from YPD plates) and 24 LOH (picked from 2-DOG plates) single colonies into 1 ml YPD, incubating the isolates under the same conditions, including preparations for flow cytometry and long-term storage at the same number days of passaging.
Growth rates
Cells were grown at 30° to saturation in liquid YPD medium, diluted 1:1000 in fresh YPD medium, and grown for 24 hr at 30° with shaking in a microplate reader (Sunrise model; Tecan). OD600 was measured every 15 min and used to calculate maximum growth rate. Each experiment included a minimum of two replicates for each isolate and P-values were determined by Mann–Whitney tests.
Results
The tetraploid genome is intrinsically unstable
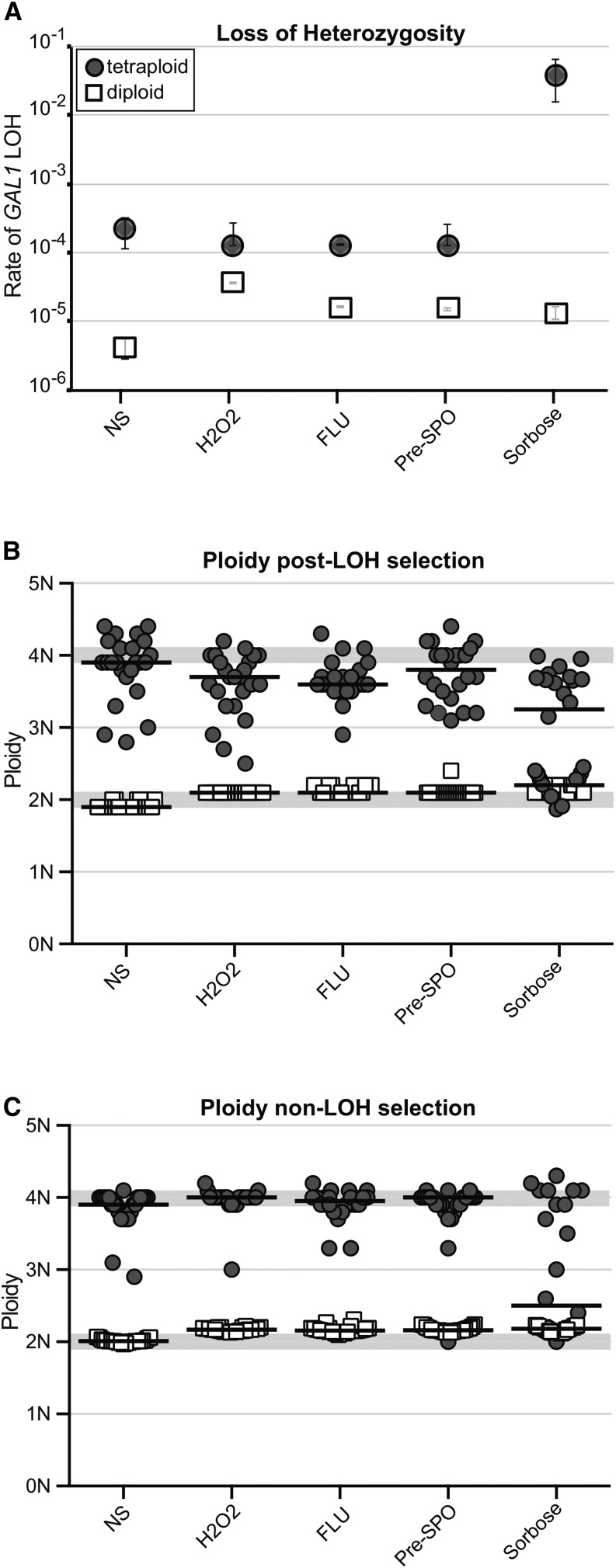
We assessed the contribution of tetraploidy to genome stability by using fluctuation analyses (Lea and Coulson 1948) to measure the rates of LOH of GAL1 [located on chromosome 1 (Chr1)]. We compared the number of cells that grew on 2-deoxygalactose, which is toxic to cells that retain GAL1 function, relative to the total number of cells [determined by plating on YPD (Legrand et al. 2007; Forche et al. 2009, 2011)]. In YPD medium [i.e., no stress (“NS”)], LOH rates for a single GAL1 locus in a tetraploid strain (shaded circles, +ΔΔΔ, YJB12712) were ∼30-fold higher (2.16 × 10−4 per cell division) than the LOH rate for a diploid strain (open squares, +Δ, DSY919; 4.18 × 10−6 per cell division) (Figure 1A, NS).
Figure 1.
Tetraploid cells are highly unstable. (A) Rate of GAL1 marker loss by fluctuation analysis for diploid (DSY919, open squares) and tetraploid (YJB12712, shaded circles) strains when grown overnight in YPD (“NS,” no stress), YPD + 5 mM H2O2 (“H2O2”), YPD + 1 μg/ml fluconazole (“FLU”), YPD with 20% glucose (“Pre-SPO,” initially described in Bennett and Johnson 2003), or media containing 2% L-sorbose (“Sorbose”) as the carbon source. At least two independent fluctuation analyses were performed per strain; error bars indicate the standard deviation. (B) Ploidy analysis of 24 single colonies following LOH selection (isolated from 2-DOG media) for the diploid and tetraploid strains in A. The ploidies for individual colonies isolated from each strain and condition are displayed as symbols and the median for each strain and condition is indicated with a solid bar. The regions with light shading indicate the ploidy range for diploid and tetraploid. (C) Ploidy analysis of 24 single colonies with no LOH selection (isolated from YPD media) for diploid and tetraploid strains. Data are displayed as in B.
In diploid C. albicans cells, mild, physiologically relevant stresses cause increased rates of LOH (Forche et al. 2011). We tested whether external stress conditions exacerbated the degree of genome instability observed in tetraploid cells by measuring rates of LOH after preincubation under stress conditions, including oxidative (5 mM H2O2) and antifungal drug [1 μg/ml fluconazole (FLU)] stresses, both of which had been previously measured in diploid cells (Forche et al. 2011). In addition, we measured LOH rates after pregrowth in high concentrations of glucose (pre-sporulation, i.e. pre-SPO) or with an alternative carbon source (Sorbose), conditions that have been documented to promote ploidy reduction and marker loss in tetraploid cells at elevated temperatures (Bennett and Johnson 2003). Interestingly, unlike those in diploid strains, LOH rates did not increase when tetraploid strains were exposed to oxidative, antifungal, and high-glucose conditions (Figure 1A). The elevated basal rate of LOH in tetraploids and the failure of physiologically relevant stressors to increase the basal LOH rate suggest that tetraploidy itself may act as an intrinsic genome stress.
The one exceptional condition with dramatically increased LOH rates was L-sorbose, an alternative carbon source, which caused approximately two orders of magnitude increase in the rate of LOH. Sorbose induces ploidy reduction in tetraploids following parasexual mating in C. albicans and other Candida species (Forche et al. 2008; Chakraborty et al. 2013; Seervai et al. 2013) and has been used to select for Chr5 monosomy in diploid cells (Rustchenko et al. 1994; Ahmad et al. 2012). We measured the DNA content of 24 initially tetraploid isolates that had undergone an LOH event (selected for 2-DOGR) following overnight growth in sorbose (Figure 1B, “Sorbose”). Ploidy reduction was evident in ∼90% (21/24) of initially tetraploid isolates, whereas all initially diploid isolates remained diploid following exposure to sorbose. While ploidy reduction was most prevalent in tetraploids exposed to sorbose, exposure to all other conditions also resulted in isolates with reduced ploidy among the 2-DOGR isolates. In contrast, we never detected a ploidy shift among the initially diploid strains selected for LOH (Figure 1B, open squares). Thus, LOH in tetraploid cells is frequently associated with reduction of DNA content and this phenomenon is exacerbated considerably by growth in L-sorbose.
We hypothesized that selection for LOH at GAL1 in tetraploid backgrounds enriches for the rare cells that have begun the chromosome loss process, and thus overall DNA contents should be reduced. Furthermore, if tetraploidy per se acts as an intrinsic genomic stress, then we expect isolates to undergo chromosome loss events in the absence of stress or without selection for GAL1 LOH (i.e., plated on YPD rather than 2-DOG). Consistent with this hypothesis, a subset of isolates displayed reduced DNA content (<3.8, 20/120 over all conditions) (Figure 1C). Exposure to sorbose yielded a particularly high fraction on nontetraploid derivatives (16/24) as well as a high frequency of cell death (data not shown). This suggests that sorbose is a severe stress that destabilizes chromosomes in tetraploid cells, rapidly yielding derivatives with reduced ploidy. By contrast, the diploid strain (open squares, DSY919) did not diverge from the 2N DNA content under any stress conditions or under LOH selection. Together, these results indicate that the tetraploid state exhibits chromosome instability and that mild stresses do not affect the rate of chromosome instability, while the severe stress of growth on a poorly metabolized carbon source does.
Dramatic ploidy changes accompany double and triple LOH events
If the tetraploid state is intrinsically unstable, then multiple LOH events on independent chromosomes should be relatively common. We tested this hypothesis by measuring the rates of double or triple LOH events, using three additional strains. Two of the strains, RBY18 and YJB12651, carried two wild-type GAL1 alleles but differed in that RBY18 was euploid (GAL1 genotype: ++ΔΔ) while YJB12651 was aneuploid, carrying an extra copy of both Chr1 and Chr5 (++ΔΔΔ). The third tetraploid strain, YJB12779, carried three wild-type GAL1 alleles and one gal1Δ allele (+++Δ). Rates of double LOH for strains losing two GAL1 alleles (++ΔΔ, RBY18 and ++ΔΔΔ, YJB12651; 1.4 × 10−5 and 5.1 × 10−6 per cell division, respectively) were slightly lower than the rate of LOH at a single locus (2.16 × 10−4 per cell division). Furthermore, the extra copy of Chr1 did not alter the LOH rates. Nonetheless, LOH rates for double events were two to three orders of magnitude higher than expected if LOH for each chromosome harboring GAL1 was entirely random (predicted to be 4.67 × 10−8 if loss events are independent). Remarkably, the rate of loss for three GAL1 alleles (+++Δ, YJB12779: 8.7 × 10−8) was detectable and much higher than that expected for three random events (predicted to be 1.01 × 10−11 if loss events are independent). Thus, tetraploids undergo double and triple LOH events at rates that suggest that loss of a single chromosome is likely to be accompanied by the loss of other chromosomes as well.
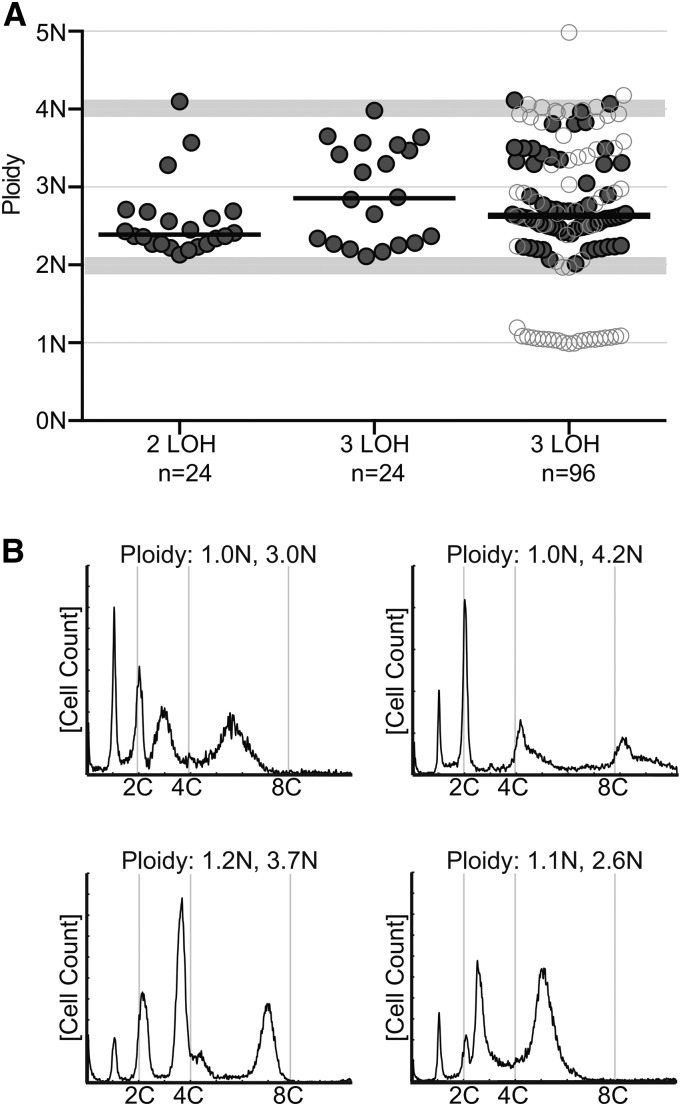
We next asked how double or triple LOH events at GAL1 affect overall DNA content, using 24 independent isolates randomly picked after LOH selection (Figure 2A). Strikingly, the ploidy distributions for the different LOH selections were distinct. In contrast to selection for loss of a single GAL1 allele, which yielded mostly subtetraploid derivatives (parent, +ΔΔΔ, YJB12712; LOH-derived median ploidy = 3.8 ± 0.3, Figure 1B, NS), selection for loss of two GAL1 alleles yielded a majority of near-diploid derivatives (parents, ++ΔΔ, RBY18 and ++ΔΔΔ, YJB12651; LOH-derived medians = 2.6 ± 0.4 and 2.4 ± 0.5, respectively). This is consistent with previous studies that selected only for tetraploids that lost two GAL1 alleles (Bennett and Johnson 2003; Forche et al. 2008), although the time frame and growth conditions for double GAL1 LOH are different in this study (overnight growth in liquid YPD at 30° compared to 8 days on solid pre-SPO/sorbose media at 37°). Importantly, this result suggests that chromosome loss can occur rapidly in tetraploid cells. Selection for loss of three GAL1 alleles yielded isolates with a broad range of DNA content (parent, +++Δ, YJB12779; LOH-derived median = 2.9 ± 0.6, range 2.1–4.0; Figure 2A, center, n = 24).
Figure 2.
Ploidy reduction in double and triple LOH isolates. (A) Ploidy analysis of 24 single colonies following double (2 LOH; n = 24, YJB12651) or triple LOH (3 LOH; n = 24, YJB12779) or 96 single colonies following triple LOH (3 LOH; n = 96, YJB12779) selection (isolated from 2-DOG media). Data are displayed as in Figure 1. Isolates with a single ploidy are displayed as shaded circles and isolates with multiple-ploidy subpopulations are displayed as open circles. The median for each strain and experiment is indicated with the solid bar. The regions with light shading indicate the ploidy range for diploid and tetraploid. (B) Selected representative flow cytometry profiles of isolates with haploid subpopulations from A (3 LOH, n = 96).
We initially hypothesized that selection for loss of three GAL1 alleles might yield rare haploids among the progeny of initially tetraploid isolates, given that double LOH selection resulted in near-diploid progeny. To test this hypothesis, we analyzed ploidy in an additional 96 individual colonies from parent strain YJB12779 (+++Δ) (Figure 2A, right, “96 isolates”). The ploidy distributions between the two experiments were not significantly different (initial analysis, n = 24, median = 2.9N; repeated analysis, n = 96, median = 2.6N; P = 0.45, Mann–Whitney test). Importantly, of the 96 new isolates analyzed, 30 had multiple ploidy subpopulations and 21 of these contained a detectable haploid subpopulation (Figure 2B), supporting the hypothesis that haploids appear frequently, albeit transiently, under conditions selecting for loss of three allelic markers. We propose that LOH selection enriches for rare derivatives of tetraploid cells that have initiated the chromosome loss process, that selection for loss of two alleles enriches for near-diploid progeny, and that selection for loss of three alleles promotes the appearance of transient and/or unstable haploids.
Parasexual ploidy reduction initiates with random chromosome loss
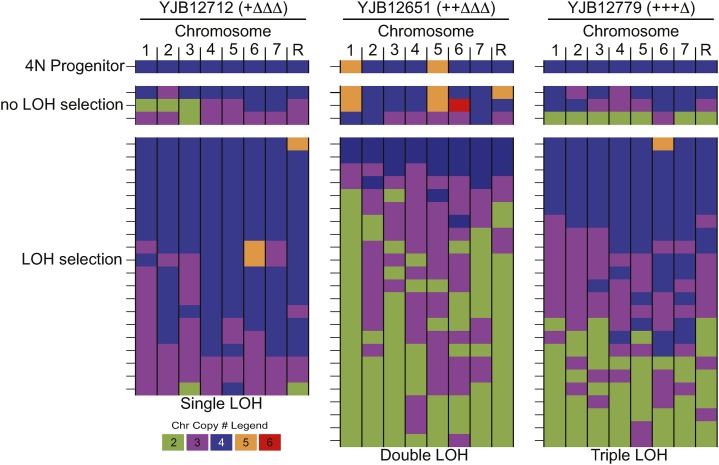
Chromosome loss from tetraploids could proceed via frequent loss of specific chromosomes or via random loss of any chromosome. To better understand the changes in ploidy observed in LOH-derived isolates, we assessed the specific chromosomes lost in individual isolates. To determine the karyotype for the large number of presumably aneuploid C. albicans isolates, we adapted ddRAD-seq, a cost-effective and high-throughput approach (Baird et al. 2008), to determine relative chromosomal copy number in C. albicans. Data from 68 LOH-derived isolates, including 20 from YJB12712 (+∆∆∆, no stress), 24 from YJB12651 (++∆∆∆), and 24 from YJB12779 (+++∆), were analyzed and displayed using YMAP (Abbey et al. 2014). As controls, we also included a small number of aneuploid isolates that had not been selected for LOH (“no LOH”) (9 total, 3 from each background) as well as each tetraploid parental strain (Figure 3). Importantly, ploidy determined by calculating relative chromosomal copy number with RAD-seq was strongly correlated with ploidy determined using flow cytometry analysis of DNA content (Figure S1). The types and degree of chromosome loss observed in non-LOH-derived isolates were similar to the chromosome loss events detected in the LOH-derived isolates (Figure 3, middle compared to bottom), indicating that LOH selection enriches for cells undergoing chromosome loss and that growth on 2-DOG media does not induce chromosome loss events.
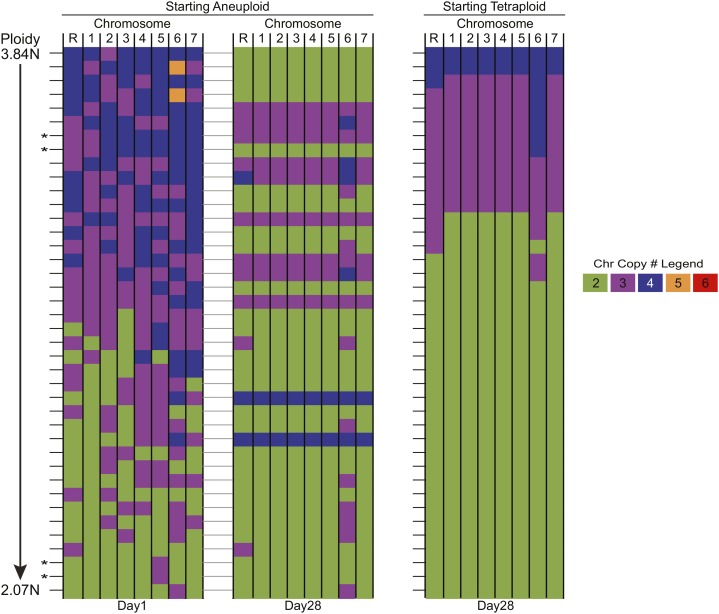
Figure 3.
Karyotype analysis following LOH selection. Shown is a summary of ddRAD-seq analysis of chromosomal copy number for the progenitor tetraploid strains (left, YJB12712; center, YJB12651; right, YJB12779), non-LOH isolates (n = 3 for every strain background), and the complete collection of LOH isolates (left, YJB12712, n = 20; center, YJB12651, n = 24; right, YJB12712, n = 24) from each of these strains. Every line indicates a unique derived isolate where the eight Candida chromosomes are aligned horizontally and copy number is colored as indicated in the legend.
As expected, among the LOH-derived isolates, Chr1 (which harbors the GAL1 locus) was lost frequently: 73% of isolates had only two or three copies of Chr1, and only a single isolate had a segmental aneuploidy on Chr1 (Figure S5), suggesting that chromosome loss was the major mechanism of LOH in tetraploid cells. Loss of Chr1 often was accompanied by the loss of multiple other chromosomes, generating a diverse set of aneuploid progeny whose overall DNA content ranged from 2.1N to 4.1N (Figure 3) with no clear pattern of chromosome imbalance (Figure S2). Notably, three isolates were perfectly triploid. Furthermore, a significant proportion of individual isolates carried interesting combinations of disomic, trisomic, and tetrasomic chromosome copy numbers [9%, (6/68) for LOH-derived isolates and 11% (1/9) for non-LOH isolates]. Furthermore, we detected strains that had gained or lost any one of the eight C. albicans chromosomes. Thus, it appears that few, if any, chromosomes are lost or gained in a coordinated manner. Rather, the initial process appears to be random, because viable nondiploid progeny with extra copies of many different combinations of the eight C. albicans chromosomes were recovered.
Tetraploid and highly aneuploid cells converge to stable euploid states
If tetraploids go through an aneuploid intermediate state during the ploidy reduction process, then we would expect aneuploids to return to the diploid state more rapidly than tetraploids. To test this hypothesis, we compared the long-term ploidy dynamics of a large set of tetraploid isolates (n = 50; 13 LOH and 37 non-LOH) and compared it to the set of aneuploid isolates characterized by RAD-seq (n = 45; 36 LOH and 9 non-LOH) as well as to the three triploid LOH isolates (Figure 4 and Figure S3).
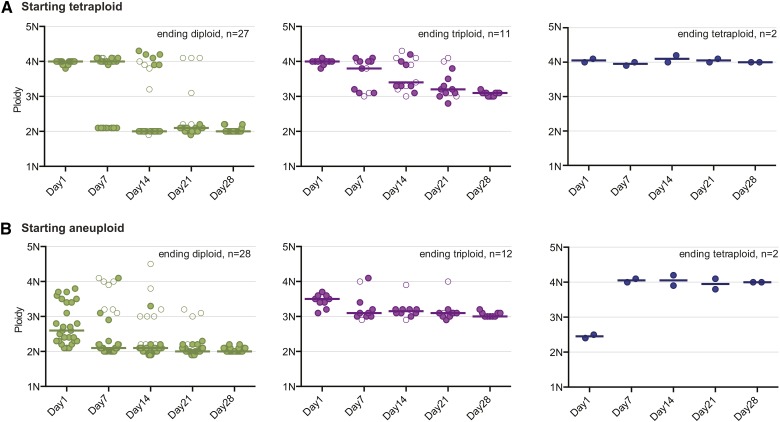
Figure 4.
Long-term ploidy dynamics of tetraploid and aneuploid isolates. Isolates that were initially (A) tetraploid or (B) aneuploid were serially transferred daily in liquid YPD for 28 days. Flow cytometry for DNA content was performed for each isolate on days 7, 14, 21, and 28. Ploidy values for day 1 are the same data presented in Figure 1 and Figure 2 (now organized as either starting tetraploid or aneuploid). Ploidy plots are divided into three classes: ending diploid (green), ending triploid (purple), or ending tetraploid (blue). As in Figure 2, isolates with a single ploidy are displayed as colored solid circles and isolates with multiple-ploidy subpopulations are displayed as colored open circles. The median for each class at every time point is indicated with a bar.
To measure ploidy dynamics, we propagated cultures by daily serial transfer for 28 days under standard laboratory conditions. Predictably, the majority of isolates returned to a diploid state regardless of whether they were initially tetraploid (54%; Figure 4A, “ending diploid”) or initially aneuploid (62%; Figure 4B, ending diploid). A significant proportion of the passaged cultures had not resolved to a single ploidy by day 28 and instead had multiple ploidy subpopulations in the culture (22% initially tetraploid, 16% initially aneuploidy; Figure S4), identified by multiple G1/G2 peaks in their flow cytometry profile (for examples see Figure 2B and description in Hickman et al. 2013). Surprisingly, not all ending states were diploid. Rather, several became triploid (22% initially tetraploid, 18% initially aneuploid; Figure 4, A and B, “ending triploid”). A few remained tetraploid (4%) or returned to the tetraploid state by gaining chromosomes (4%) (Figure 4, A and B, “ending tetraploid”). Of the three isolates that began the passaging experiment perfectly triploid, two ended as diploid and the other as tetraploid (Figure S3). In contrast, a collection of diploid isolates (DSY919; n = 24 LOH derived) did not undergo any major ploidy changes over the 28 days of passaging (Figure S3).
A striking difference between the initially tetraploid and initially aneuploid collections of isolates was the timing of ploidy resolution (defined as reaching and/or maintaining a single ploidy state). Initially aneuploid isolates rapidly underwent changes in ploidy: by day 7 >60% (n = 28) of aneuploids and <25% (n = 12) of tetraploids had reached and maintained their final ploidy state (Figure S5A). Furthermore, aneuploid isolates that reached ploidy resolution did so extremely rapidly: by day 4 (only 96 hr of growth), the collection of isolates that were diploid on day 28 had reached a median ploidy of 2.2N; isolates ending triploid had reached a median ploidy of 3.3N, and one of the two isolates ending tetraploid was already tetraploid (Figure S5B). In contrast, the median ploidy of initially tetraploid isolates was 4.0N by day 4, regardless of ending ploidy state on day 28. Together, these results suggest that chromosome loss in tetraploids occurs infrequently, but once tetraploids lose one or more chromosomes (thus becoming aneuploid), they are much more unstable and ploidy change is expedited.
Flow cytometry does not have the resolution to distinguish gains or losses of the smallest C. albicans chromosomes. To determine whether isolates presumed di-, tri-, or tetraploid after 28 days of passaging were euploid or continued to carry aneuploidies, we performed ddRAD-seq analysis on all isolates with a single ploidy population on day 28 (Figure 5, Day28). The majority of all isolates (66%; n = 49) were completely euploid, yet a significant number (∼34%; n = 25) carried additional chromosomes, irrespective of whether they were initially aneuploid (Figure 5, Day28, n = 14) or initially tetraploid (Figure 5, n = 11). Notably, unlike the remarkable diversity in chromosomal copy number observed in initially aneuploid isolates (day 1), only two chromosomes, Chr6 and ChrR, remained aneuploid by day 28. These two specific aneuploidies were also observed in initially tetraploid isolates (Figure 5), either alone (n = 18 for Chr6; n = 2 for ChrR) or in combination (n = 5). These results support the premise that, while tetraploid and highly aneuploid isolates are unstable and resolve toward a euploid state (Figure 4 and Bennett and Johnson 2003; Forche et al. 2008), aneuploidy of specific chromosomes can be tolerated and even maintained for prolonged periods of time.
Figure 5.
Karyotype analysis after long-term passaging. Shown is a summary of ddRAD-seq chromosome copy number analysis for (left and center) individual isolates starting aneuploid (n = 37) on day 1 (left, same as in Figure 3, now ordered by decreasing ploidy) and those same isolates on day 28 (center, connected by dashed line) and for (right) isolates starting tetraploid on day 28 (n = 37) of passaging in YPD. Tetraploid isolates on day 1 are not represented here since they were all euploid and similar in chromosomal copy number. Data are presented as in Figure 3.
To determine whether specific combinations of aneuploid chromosomes are likely to resolve to a specific euploid state, we compared the day 28 and day 1 chromosome composition profiles of the initially aneuploid isolates (Figure 5). Predictably, all isolates that already were very close to diploid (i.e., 2N + 1, 2, or 3 extra chromosomes) resolved to an approximately diploid state (50% 2N, 50% 2N + 1). However, isolates with more DNA content (i.e., 2N + ≥4 extra chromosomes or 3N + extra chromosomes) resolved to either the approximately diploid (60%) or the approximately triploid (40%) state. Interestingly, dramatic gains in DNA content, as well as ploidy resolution to the tetraploid state, were detected exclusively among the isolates with “complex” aneuploid chromosome combinations (di-, tri-, and tetrasomic chromosomes in a single isolate). Further examination of the individual ploidy dynamics over time for each of these complex aneuploids (n = 6) reveals transient DNA content gains in three additional isolates (Figure S6). This suggests that highly aneuploid genomes are particularly unstable and prone to genome duplication events.
In our collection of initially aneuploid isolates, only two pairs had identical karyotypes (Figure 5, marked with *): one pair with nearly diploid DNA content (2N + Chr5) and one pair with higher DNA content (3N + ChrR + Chr1). In the 2N + Chr5 pair of isolates, both isolates resolved to the diploid state, suggesting that having a single extra chromosome limited the range of potential ploidy states that they could assume. In contrast, for the 3N + ChrR + Chr1 pair of isolates, one isolate resolved to diploid and the other resolved to triploid. These results, together with the diverse ploidy resolutions observed from the three triploids (Figure S3) and in the collection of tetraploid isolates (Figure 4B), strongly support the idea that cells with high DNA content generate progeny with diverse ploidy outcomes. Thus, we propose that cells with high DNA content represent a very high potential to resolve and reduce ploidy randomly. Furthermore, once the majority of extra chromosomes have been lost, the potential range of ploidy fates is more limited.
Consequences of chromosome loss and growth rate
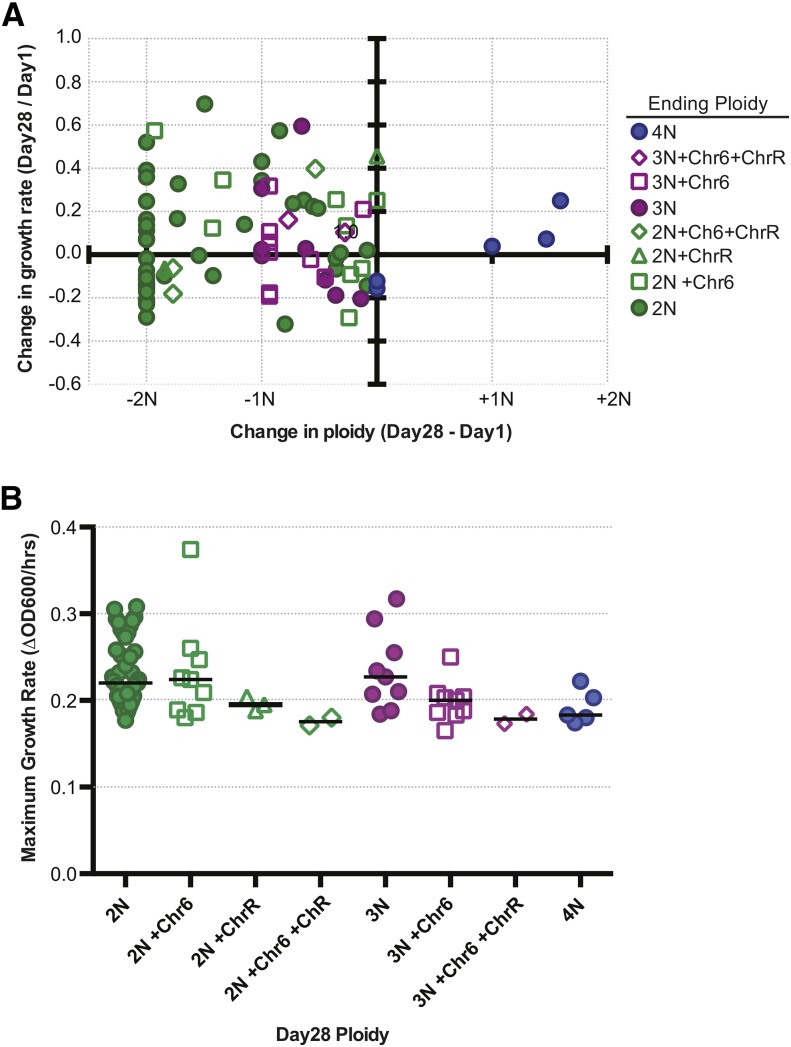
Under nonselective conditions, aneuploidy often incurs a fitness cost, usually due to altered gene dosage and stoichiometry of protein complex components (Torres et al. 2007, 2010; Tang et al. 2011; Oromendia et al. 2012). Given the high degree of aneuploidy that we observed on day 1 and the subsequent convergence toward euploidy with serial passaging, one prediction is that growth rate should increase with the loss of excess DNA content. We assessed how growth rate changed in relationship to ploidy change at day 28 (Figure 6A) relative to day 1 (Figure S7). While growth rate increased as DNA content decreased in a large number of isolates (Figure 6A, top left quadrant), growth rate was reduced in 44% of the diploids (Figure 6A, solid green circles, bottom left quadrant) and other isolates with reduced DNA content. Reduced growth rates are likely the consequence of retaining homozygous chromosomes (i.e., loss of allelic variation), an established outcome of parasexual ploidy reduction (Forche et al. 2008).
Figure 6.
Fitness consequences of ploidy reduction and aneuploidy for growth in YPD. (A) XY scatterplot of the change in ploidy (x-axis, calculated by subtracting day 28 from day 1) and the corresponding change in growth rate (y-axis, day 28 divided by day 1). The color and shape of the symbols indicate the final ploidy state on day 28 as described in the key. (B) Growth rates of day 28 isolates grouped by final karyotype. Each symbol represents a single isolate and the bar indicates the median value.
To investigate the fitness consequences of the isolates that maintained Chr6 and/or ChrR aneuploidy on day 28, we compared the growth rate of aneuploid isolates to that of those that were perfectly euploid on day 28 (Figure 6B). The growth rate for diploids (median = 0.220 hr−1, n = 43) and that for diploids carrying an additional Chr6 (median = 0.224 hr−1, n = 9) are not statistically distinct (P = 0.6728, Mann–Whitney test). However, the growth rates for euploid triploids (median = 0.227 hr−1, n = 9) are significantly different from triploids carrying an additional Chr6 (median = 0.200 hr−1, n = 9) (P = 0.0422, Mann–Whitney test), suggesting that there is a fitness cost for additional chromosomes in nondiploid backgrounds.
Reproducibility of specific ploidy reduction processes
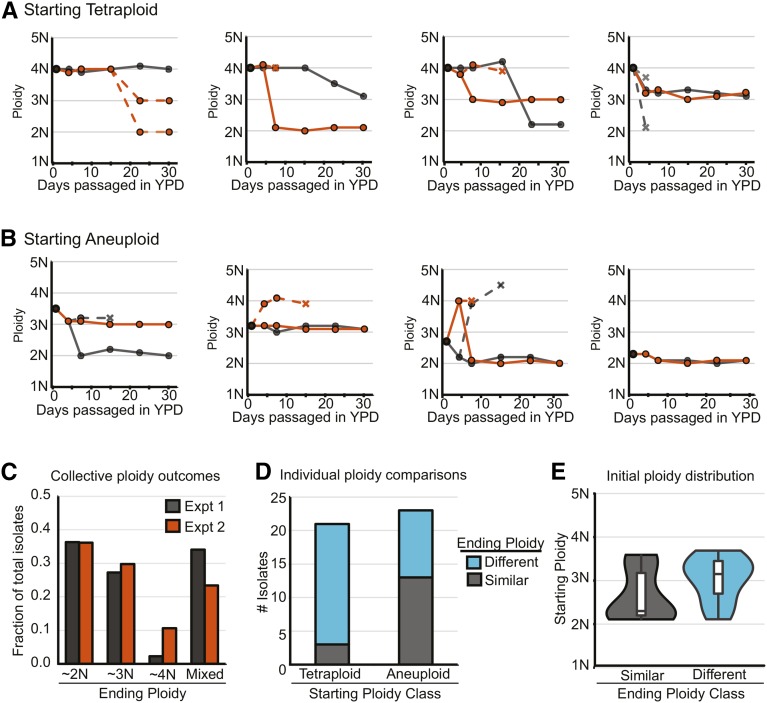
To specifically assess the ploidy “fate” of individual isolates, we repeated the passaging experiment, using the complete set of YJB12779-derived isolates including 23 isolates that were initially aneuploid (Figure 7A and Figure S8) and 21 that were initially tetraploid (Figure 7B and Figure S9). Overall, isolates converged toward a euploid state in both experiments (Figure 7C). However, there was a striking difference between initially tetraploid and initially aneuploidy isolates when the specific outcomes for the same individual were compared between the two experiments (illustrated in Figure 7, A and B, Figure S8, and Figure S9). Only 14% (3/21) of the initially tetraploid isolates resolved to the same ploidy in the two parallel experiments (Figure 7D). In contrast, for initially aneuploid isolates, >50% (13/23) resolved to the same ploidy in both experiments. Notably, the specific isolates that were initially aneuploid and resolved to a similar ploidy level had lower initial DNA content (median = 2.3N, n = 13) than those that resolved to different ploidy states in the two passaging experiments (median = 3.2N, n = 10) (Figure 7E). Thus, isolates with higher DNA content (tetraploid and highly aneuploid), are more likely to produce derivatives with a range of different ploidy states. Presumably these high-DNA content isolates retain more potential chromosome loss trajectories than aneuploid isolates that are already very close to the diploid state (i.e., plus one or two chromosomes).
Figure 7.
Ploidy potential of tetraploid and aneuploid cells. (A and B) Examples of individual ploidy trajectories for (A) tetraploid and (B) aneuploid isolates that were repassaged for 28 days (black, “experiment 1”; orange, “experiment 2”). Dashed lines indicate time points with multiple ploidy subpopulations and X’s represent ploidy subpopulations that became extinct (were not observed in subsequent passages of the same individual culture). (C) The percentage of total isolates (including both the starting tetraploid and aneuploid collections) that on day 28 are near diploid, triploid, or tetraploid or have multiple-ploidy subpopulations (“Mixed”). (D) The number of isolates that when compared individually between experiments 1 and 2 either have the same ploidy resolution (gray, “Similar”) or do not (blue, “Different”) for the tetraploid and aneuploid collections. (E) Violin and box plots of the initial ploidy for aneuploid isolates that had similar (gray) or different (teal) ending ploidy resolutions in experiments 1 and 2. The median ploidy for each class is displayed as a black bar.
Discussion
Here we show that the tetraploid state in C. albicans is highly unstable and undergoes the parasexual ploidy reduction process that initiates with stochastic chromosome loss to produce highly aneuploid derivatives that rapidly resolve to a euploid state. Tetraploid cells have higher genomic instability relative to diploid cells, resulting in chromosome loss and ploidy reduction regardless of external stressors (Figure 1 and Figure 2). Selection for LOH events enriches for rare chromosome loss events, but such events also occur even in the absence of selection. Furthermore, the combination of chromosomes that are lost varies considerably and we explicitly show that parasexual ploidy reduction is random and generates transient aneuploid intermediates, with some genomes having complex aneuploidies including the simultaneous presence of disomic, trisomic, and tetrasomic chromosomes (Figure 3). By assessing the long-term ploidy dynamics of tetraploid and aneuploid cells, we found that euploidy is preferred and the diploid state is predominant, although a significant proportion of the euploid progeny are triploid or tetraploid (Figure 4). Aneuploidy, especially of Chr6 and/or ChrR, was also a common outcome (Figure 5). Finally, isolates with high DNA content, whether tetraploid or highly aneuploid, are likely to follow different chromosome loss trajectories in parallel experiments (Figure 6). Thus, the nonmeiotic parasexual ploidy reduction in C. albicans potentiates the production of diverse progeny.
This work has important implications for our understanding of the C. albicans life cycle. Historically, C. albicans had been classified as an asexual, diploid organism and early phylogenetic analyses initially indicated that the species was exclusively clonal (Pujol et al. 1993). However, a diploid–tetraploid parasexual cycle has been elucidated (Hull et al. 2000; Magee and Magee 2000; Miller and Johnson 2002; Bennett and Johnson 2003; Forche et al. 2008) and a small number of reports show that ploidy variation occurs in clinical isolates (Suzuki et al. 1982, 1986, 1989). Furthermore, subsequent experimental characterization of the diploid–tetraploid parasexual cycle suggests that cryptic mating events may occur in the host environment (Lachke et al. 2003; Ramírez-Zavala et al. 2008; Huang et al. 2010). These studies have informed the interpretation of more recent population genetic reports to now include the possibility of rare mating events (Tavanti et al. 2004, 2005; Odds et al. 2007; Bougnoux et al. 2008). An important insight from the work presented here is that polyploid states are intrinsically unstable, may exist only transiently, and thus may be difficult to detect in natural populations.
While C. albicans is most frequently isolated as a diploid, other ploidy states are viable, including haploid strains, which have been isolated under a variety of conditions (Hickman et al. 2013), as well as tetraploid and aneuploid isolates from clinical samples (Selmecki et al. 2005, 2006; Hirakawa et al. 2014; Ford et al. 2015), suggesting that unconventional ploidy states can be found in vivo as well as in vitro. Not only did haploid subpopulations arise in this study (Figure 2), but also triploid isolates appeared (Figure 3), consistent with previous chromosome loss studies (Bennett and Johnson 2003). Even in S. cerevisiae, stable, unconventional ploidy states like triploidy have been observed (Gerstein et al. 2008). Triploidy cannot be maintained in organisms that routinely go through meiosis, yet there is no a priori reason that it cannot be maintained in organisms that reproduce primarily through mitosis.
The viability and mitotic stability of nondiploid states suggest there may be unknown benefits to maintaining ploidy variation, at least over moderate time frames (e.g., 28 days of serial passaging). In budding yeasts, ploidy level correlates with cellular size and surface area-to-volume ratios (Mortimer 1958; Hickman et al. 2013) and may be important for proliferating under stress or nutrient-limited conditions. While the fitness consequences of different whole-genome ploidy states have been studied (Zörgö et al. 2013), the results do not suggest that one ploidy state is “better” than another or even that the more fit state will predominate the microbial population (Gerstein and Otto 2011). As such, the ability to switch between ploidy states, via sexual or less conventional mechanisms, may be an important bet-hedging strategy for microbial fitness, particularly in dynamic or fluctuating environments. Thus, we propose that whole-genome shifts in ploidy potentially facilitate adaptation.
Aneuploidy also has well-documented roles in adaptation, both in the laboratory and in natural populations. In S. cerevisiae, particular aneuploid chromosomes facilitate multicellular phenotypic switching in natural isolates (Tan et al. 2013) and can confer fitness advantages in laboratory strains under a variety of stresses (Pavelka et al. 2010). Such changes may provide a transient intermediate in adaptation to unfavorable environmental conditions (Yona et al. 2012) and genetic backgrounds (Rancati et al. 2008). In pathogenic species, aneuploidies that provide additional copies of specific genes on Chr5 are responsible for the acquisition of resistance to azole antifungals in C. albicans (Selmecki et al. 2006, 2008, 2009) and Chr5 aneuploidy often appears, albeit transiently, during the acquisition of azole resistance in individual patients (Ford et al. 2015). Similar genes are amplified with Chr1 disomy, which is associated with azole resistance in the basidiomycete budding yeast Cryptococcus neoformans (Sionov et al. 2010).
In S. cerevisiae, the presence of a single aneuploid chromosome often carries a fitness cost thought to be due to imbalances in the corresponding encoded proteins. (Torres et al. 2007, 2010; Tang et al. 2011; Oromendia et al. 2012). In addition, haploids selected for a single aneuploidy often, but not always, exhibit increased genome instability (Sheltzer et al. 2011). Here, highly aneuploid isolates rapidly underwent ploidy reduction to a euploid state, suggesting that chromosomal imbalances in general are detrimental to the cells under standard laboratory conditions. Nonetheless, specific aneuploidies persisted over the course of the 28-day passaging experiments. Chr6 aneuploidy was most common in this study (Figure 5) and was also the most common aneuploidy in haploid C. albicans isolates (Hickman et al. 2013). Chr6 is one of the smallest chromosomes in the genome and would be expected to cause fewer dosage problems. However, ChrR is one of the largest chromosomes and it is also frequently found in aneuploidy alone or together with Chr6. Thus, the presence of extra DNA or protein-coding genes, per se, is not sufficient to explain the likelihood that extra copies of some chromosomes are well tolerated. We suggest an alternative hypothesis, that specific genes or gene families on the aneuploid chromosome (e.g., the ALS and SAP genes on Chr6) provide a selective advantage for growth and/or may be limiting for growth such that amplified genes provided by the additional chromosome overcome a growth limitation. This also implies that any growth advantage conferred by the extra gene copies is larger than the cost incurred by increased stress on the protein quality-control system.
An important insight from this work is that ploidy level per se is not sufficient to predict fitness. Diploidy is likely to be the “most fit” of the ploidy states, given that the majority of isolates returned to the diploid state. Importantly, the range of growth rates exhibited by diploids relative to triploids was not significantly different (P = 0.8725, Mann–Whitney test) nor was the range of growth rates exhibited by euploid diploids relative to diploid isolates carrying an extra copy of Chr6 (P = 0.6728, Mann–Whitney test). In contrast, tetraploid cells and aneuploid isolates carrying extra copies of larger (i.e., ChrR) or multiple chromosomes generally had slower growth rates. It is important to note that diploid isolates exhibited a broad range of growth rates (0.177–0.308 hr−1, n = 43). We presume this is because ploidy reduction can result in whole-chromosome homozygosis and we posit that not only ploidy, but also the degree of allelic heterozygosity, contributes to fitness in C. albicans. To this end, we previously found that completely homozygous diploid genomes grow slower in vitro and are outcompeted in vivo compared to heterozygous diploids (Hickman et al. 2013).
Here we found that C. albicans is able to generate a variety of ploidy states, including atypical euploidy (i.e., triploidy) and diverse aneuploid combinations, by utilizing a stochastic chromosome loss mechanism in its parasexual cycle rather than a conventional meiotic program to achieve ploidy reduction. Meiosis and/or highly coordinated reductional division are tightly regulated and subject to multiple checkpoints to prevent such asymmetrical divisions from occurring (Murakami and Nurse 2000). C. albicans may employ imprecise mitotic divisions as a response to highly stressful environments. For example, C. albicans exposed to antifungal drugs rapidly forms unstable tetraploids through an asexual mechanism involving a trimera intermediate (Harrison et al. 2014). Soon after their formation, trimera-derived tetraploids undergo mitotic missegregation events involving multiple mitotic spindles. Such missegregation events frequently produce aneuploid derivatives, a proportion of which can proliferate in the presence of the antifungal drug (Harrison et al. 2014). These unusual mitoses are not unique to C. albicans; this phenomenon is reminiscent of multispindle divisions seen in cancer cells (Gordon et al. 2012). In addition, healthy human and mouse hepatocytes become polyploid and undergo multispindle mitoses (Duncan et al. 2010, 2012; Duncan 2013) to generate transient polyploid cell states and drive population heterogeneity. Furthermore, even species with fully functional sexual cycles utilize alternative pathways to promote ploidy variation. In addition to canonical bipolar mating (Kwon-Chung et al. 1992), C. neoformans can form high-DNA content titan cells (Okagaki et al. 2010) and can produce aneuploid progeny through unisexual mating (Ni et al. 2013).
We propose that tetraploidy, in C. albicans and in other types of single- and multicellular eukaryotes, provides an efficient mechanism to very rapidly generate a broad range of ploidy states, resulting in a broad range of fitness diversity in a single population. We posit that the intrinsic genomic instability of tetraploids and their high potential for different ploidy trajectories provide them with the ability to generate a high level of genetic and genomic diversity. Selective pressure, whether it be growth in vivo, growth in drug, or other conditions, can then act upon this diverse population to increase the frequency of those isolates with improved relative fitness.
Supplementary Material
Acknowledgments
We thank Darren Abbey, Gareth Cromie, Sophia Hirsch, and Eric Jeffrey for technical and analytical assistance; Richard Bennett for providing strains; and Laura Burrack and Aleeza Gerstein for comments on the manuscript. This work is supported by the National Institute of Allergy and Infectious Diseases (AI0624273) and by the European Research Council under the European Community’s Seventh Framework Programme (FP7/2007-2013)/ERC grant 340087 and Marie Curie grant 303635. M.A.H. was supported by a National Research Service Award postdoctoral fellowship (F32GM096536-02). A.D. is supported through a strategic partnership between the Institute for Systems Biology and the University of Luxembourg.
Footnotes
Communicating editor: M. Johnston
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.178020/-/DC1.
Literature Cited
- Abbey D., Hickman M., Gresham D., Berman J., 2011. A high resolution hapmap based on SNP/CGH combined microarrays reveal that loss of heterozygosity as well as aneuploidy accumulate in C. albicans laboratory strains. G3 1: 523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbey D. A., Funt J., Lurie-Weinberger M. N., Thompson D. A., Regev A., et al. , 2014. YMAP: a pipeline for visualization of copy number variation and loss of heterozygosity in eukaryotic pathogens. Genome Med. 6: 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad A., Kravets A., Rustchenko E., 2012. Transcriptional regulatory circuitries in the human pathogen Candida albicans involving sense–antisense interactions. Genetics 190: 537–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alabrudzinska M., Skoneczny M., Skoneczna A., 2011. Diploid-specific genome stability genes of S. cerevisiae: genomic screen reveals haploidization as an escape from persisting DNA rearrangement stress. PLoS ONE 6: e21124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alby K., Bennett R. J., 2009. Stress-induced phenotypic switching in Candida albicans. Mol. Biol. Cell 20: 3178–3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andaluz E., Bellido A., Gómez-Raja J., Selmecki A., Bouchonville K., et al. , 2011. Rad52 function prevents chromosome loss and truncation in Candida albicans. Mol. Microbiol. 79: 1462–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbour M., Epp E., Hogues H., Sellam A., Lacroix C., et al. , 2009. Widespread occurrence of chromosomal aneuploidy following the routine production of Candida albicans mutants. FEMS Yeast Res. 9: 1070–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird N. A., Etter P. D., Atwood T. S., Currey M. C., Shiver A. L., et al. , 2008. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS ONE 3: e3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett R. J., Johnson A. D., 2003. Completion of a parasexual cycle in Candida albicans by induced chromosome loss in tetraploid strains. EMBO J. 22: 2505–2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman J., Hadany L., 2012. Does stress induce (para)sex? Implications for Candida albicans evolution. Trends Genet. 28: 197–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchonville K., Forche A., Tang K. E. S., Selmecki A., Berman J., 2009. Aneuploid chromosomes are highly unstable during DNA transformation of Candida albicans. Eukaryot. Cell 8: 1554–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bougnoux M.-E., Pujol C., Diogo D., Bouchier C., Soll D. R., et al. , 2008. Mating is rare within as well as between clades of the human pathogen Candida albicans. Fungal Genet. Biol. 45: 221–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler G., Rasmussen M. D., Lin M. F., Santos M. A. S., Sakthikumar S., et al. , 2009. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 459: 657–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calo S., Billmyre R. B., Heitman J., 2013. Generators of phenotypic diversity in the evolution of pathogenic microorganisms. PLoS Pathog. 9: e1003181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty U., Mohamed A., Kakade P., Mugasimangalam R. C., Sadhale P. P., et al. , 2013. A stable hybrid containing haploid genomes of two obligate diploid Candida species. Eukaryot. Cell 12: 1061–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoli T., de Lange T., 2012. Telomere-driven tetraploidization occurs in human cells undergoing crisis and promotes transformation of mouse cells. Cancer Cell 21: 765–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan A. W., 2013. Aneuploidy, polyploidy and ploidy reversal in the liver. Semin. Cell Dev. Biol. 24: 347–356. [DOI] [PubMed] [Google Scholar]
- Duncan A. W., Taylor M. H., Hickey R. D., Hanlon Newell A. E., Lenzi M. L., et al. , 2010. The ploidy conveyor of mature hepatocytes as a source of genetic variation. Nature 467: 707–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan A. W., Hanlon Newell A. E., Bi W., Finegold M. J., Olson S. B., et al. , 2012. Aneuploidy as a mechanism for stress-induced liver adaptation. J. Clin. Invest. 122: 3307–3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forche A., May G., Beckerman J., Kauffman S., Becker J., et al. , 2003. A system for studying genetic changes in Candida albicans during infection. Fungal Genet. Biol. 39: 38–50. [DOI] [PubMed] [Google Scholar]
- Forche A., Alby K., Schaefer D., Johnson A. D., Berman J., et al. , 2008. The parasexual cycle in Candida albicans provides an alternative pathway to meiosis for the formation of recombinant strains. PLoS Biol. 6: e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forche A., Magee P. T., Selmecki A., Berman J., May G., 2009. Evolution in Candida albicans populations during a single passage through a mouse host. Genetics 182: 799–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forche A., Abbey D., Pisithkul T., Weinzierl M. A., Ringstrom T., et al. , 2011. Stress alters rates and types of loss of heterozygosity in Candida albicans. MBio 2: e00129-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford, C. B., J. M. Funt, D. Abbey, L. Issi, C. Guiducci et al, 2015 The evolution of drug resistance in clinical isolates of Candida albicans. eLife Sci. 4: e00662. [DOI] [PMC free article] [PubMed]
- Gerstein A. C., Otto S. P., 2011. Cryptic fitness advantage: diploids invade haploid populations despite lacking any apparent advantage as measured by standard fitness assays. PLoS ONE 6: e26599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein, A. C., H.-J. E. Chun, A. Grant, and S. P. Otto, 2006 Genomic convergence toward diploidy in Saccharomyces cerevisiae. PLoS Genet. 2: e145. [DOI] [PMC free article] [PubMed]
- Gerstein A. C., McBride R. M., Otto S. P., 2008. Ploidy reduction in Saccharomyces cerevisiae. Biol. Lett. 4: 91–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D. J., Resio B., Pellman D., 2012. Causes and consequences of aneuploidy in cancer. Nat. Rev. Genet. 13: 189–203. [DOI] [PubMed] [Google Scholar]
- Harrison B. D., Hashemi J., Bibi M., Pulver R., Bavli D., et al. , 2014. A tetraploid intermediate precedes aneuploid formation in yeasts exposed to fluconazole. PLoS Biol. 12: e1001815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman M. A., Zeng G., Forche A., Hirakawa M. P., Abbey D., et al. , 2013. The “obligate diploid” Candida albicans forms mating-competent haploids. Nature 494: 55–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton C., Markie D., Corner B., Rikkerink E., Poulter R., 1985. Heat shock induces chromosome loss in the yeast Candida albicans. Mol. Gen. Genet. 200: 162–168. [DOI] [PubMed] [Google Scholar]
- Hirakawa, M. P., D. A. Martinez, S. Sakthikumar, M. Z. Anderson, A. Berlin et al, 2014 Genetic and phenotypic intra-species variation in Candida albicans. Genome Res. 25: 413–425. [DOI] [PMC free article] [PubMed]
- Hnisz D., Tscherner M., Kuchler K., 2011. Morphological and molecular genetic analysis of epigenetic switching of the human fungal pathogen Candida albicans. Methods Mol. Biol. 734: 303–315. [DOI] [PubMed] [Google Scholar]
- Huang G., Yi S., Sahni N., Daniels K. J., Srikantha T., et al. , 2010. N-acetylglucosamine induces white to opaque switching, a mating prerequisite in Candida albicans. PLoS Pathog. 6: e1000806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull C. M., Raisner R. M., Johnson A. D., 2000. Evidence for mating of the “asexual” yeast Candida albicans in a mammalian host. Science 289: 307–310. [DOI] [PubMed] [Google Scholar]
- Kwon-Chung K. J., Edman J. C., Wickes B. L., 1992. Genetic association of mating types and virulence in Cryptococcus neoformans. Infect. Immun. 60: 602–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachke S. A., Lockhart S. R., Daniels K. J., Soll D. R., 2003. Skin facilitates Candida albicans mating. Infect. Immun. 71: 4970–4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea D. E., Coulson C. A., 1948. The distribution of the numbers of mutants in bacterial populations. J. Genet. 49: 264–284. [DOI] [PubMed] [Google Scholar]
- Legrand M., Lephart P., Forche A., Mueller F.-M. C., Walsh T., et al. , 2004. Homozygosity at the MTL locus in clinical strains of Candida albicans: karyotypic rearrangements and tetraploid formation. Mol. Microbiol. 52: 1451–1462. [DOI] [PubMed] [Google Scholar]
- Legrand M., Chan C. L., Jauert P. A., Kirkpatrick D. T., 2007. Role of DNA mismatch repair and double-strand break repair in genome stability and antifungal drug resistance in Candida albicans. Eukaryot. Cell 6: 2194–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee B. B., Magee P. T., 2000. Induction of mating in Candida albicans by construction of MTLa and MTLalpha strains. Science 289: 310–313. [DOI] [PubMed] [Google Scholar]
- Mayer V. W., Aguilera A., 1990. High levels of chromosome instability in polyploids of Saccharomyces cerevisiae. Mutat. Res. 231: 177–186. [DOI] [PubMed] [Google Scholar]
- Miller M. G., Johnson A. D., 2002. White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell 110: 293–302. [DOI] [PubMed] [Google Scholar]
- Mortimer R. K., 1958. Radiobiological and genetic studies on a polyploid series (haploid to hexaploid) of Saccharomyces cerevisiae. Radiat. Res. 9: 312–326. [PubMed] [Google Scholar]
- Murakami H., Nurse P., 2000. DNA replication and damage checkpoints and meiotic cell cycle controls in the fission and budding yeasts. Biochem. J. 349: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M., Feretzaki M., Li W., Floyd-Averette A., Mieczkowski P., et al. , 2013. Unisexual and heterosexual meiotic reproduction generate aneuploidy and phenotypic diversity de novo in the yeast Cryptococcus neoformans. PLoS Biol. 11: e1001653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odds F. C., Bougnoux M.-E., Shaw D. J., Bain J. M., Davidson A. D., et al. , 2007. Molecular phylogenetics of Candida albicans. Eukaryot. Cell 6: 1041–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okagaki L. H., Strain A. K., Nielsen J. N., Charlier C., Baltes N. J., et al. , 2010. Cryptococcal cell morphology affects host cell interactions and pathogenicity. PLoS Pathog. 6: e1000953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oromendia A. B., Dodgson S. E., Amon A., 2012. Aneuploidy causes proteotoxic stress in yeast. Genes Dev. 26: 2696–2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavelka N., Rancati G., Zhu J., Bradford W. D., Saraf A., et al. , 2010. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature 468: 321–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontecorvo G., 1956. The parasexual cycle in fungi. Annu. Rev. Microbiol. 10: 393–400. [DOI] [PubMed] [Google Scholar]
- Pujol C., Reynes J., Renaud F., Raymond M., Tibayrenc M., et al. , 1993. The yeast Candida albicans has a clonal mode of reproduction in a population of infected human immunodeficiency virus-positive patients. Proc. Natl. Acad. Sci. USA 90: 9456–9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez-Zavala B., Reuss O., Park Y.-N., Ohlsen K., Morschhäuser J., 2008. Environmental induction of white–opaque switching in Candida albicans. PLoS Pathog. 4: e1000089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rancati G., Pavelka N., Fleharty B., Noll A., Trimble R., et al. , 2008. Aneuploidy underlies rapid adaptive evolution of yeast cells deprived of a conserved cytokinesis motor. Cell 135: 879–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustchenko E. P., Howard D. H., Sherman F., 1994. Chromosomal alterations of Candida albicans are associated with the gain and loss of assimilating functions. J. Bacteriol. 176: 3231–3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seervai R. N. H., Jones S. K., Hirakawa M. P., Porman A. M., Bennett R. J., 2013. Parasexuality and ploidy change in Candida tropicalis. Eukaryot. Cell 12: 1629–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmecki A., Bergmann S., Berman J., 2005. Comparative genome hybridization reveals widespread aneuploidy in Candida albicans laboratory strains. Mol. Microbiol. 55: 1553–1565. [DOI] [PubMed] [Google Scholar]
- Selmecki A., Forche A., Berman J., 2006. Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science 313: 367–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmecki A., Gerami-Nejad M., Paulson C., Forche A., Berman J., 2008. An isochromosome confers drug resistance in vivo by amplification of two genes, ERG11 and TAC1. Mol. Microbiol. 68: 624–641. [DOI] [PubMed] [Google Scholar]
- Selmecki, A. M., K. Dulmage, L. E. Cowen, J. B. Anderson, and J. Berman, 2009 Acquisition of aneuploidy provides increased fitness during the evolution of antifungal drug resistance. PLoS Genet. 5: e1000705. [DOI] [PMC free article] [PubMed]
- Selmecki A., Forche A., Berman J., 2010. Genomic plasticity of the human fungal pathogen Candida albicans. Eukaryot. Cell 9: 991–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheltzer J. M., Blank H. M., Pfau S. J., Tange Y., George B. M., et al. , 2011. Aneuploidy drives genomic instability in yeast. Science 333: 1026–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha U., Ashworth J. M., 1969. Evidence for the existence of elements of a parasexual cycle in the cellular slime mould, Dictyostelium discoideum. Proc. Biol. Sci. 173: 531–540. [Google Scholar]
- Sionov E., Lee H., Chang Y. C., Kwon-Chung K. J., 2010. Cryptococcus neoformans overcomes stress of azole drugs by formation of disomy in specific multiple chromosomes. PLoS Pathog. 6: e1000848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W., Petes T. D., 2012. Haploidization in Saccharomyces cerevisiae induced by a deficiency in homologous recombination. Genetics 191: 279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St. Charles J., Hazkani-Covo E., Yin Y., Andersen S. L., Dietrich F. S., et al. , 2012. High-resolution genome-wide analysis of irradiated (UV and γ-rays) diploid yeast cells reveals a high frequency of genomic loss of heterozygosity (LOH) events. Genetics 190: 1267–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storchova Z., Breneman A., Cande J., Dunn J., Burbank K., et al. , 2006. Genome-wide genetic analysis of polyploidy in yeast. Nature 443: 541–547. [DOI] [PubMed] [Google Scholar]
- Suzuki T., Nishibayashi S., Kuroiwa T., Kanbe T., Tanaka K., 1982. Variance of ploidy in Candida albicans. J. Bacteriol. 152: 893–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Kanbe T., Kuroiwa T., Tanaka K., 1986. Occurrence of ploidy shift in a strain of the imperfect yeast Candida albicans. J. Gen. Microbiol. 132: 443–453. [DOI] [PubMed] [Google Scholar]
- Suzuki T., Kobayashi I., Kanbe T., Tanaka K., 1989. High frequency variation of colony morphology and chromosome reorganization in the pathogenic yeast Candida albicans. J. Gen. Microbiol. 135: 425–434. [DOI] [PubMed] [Google Scholar]
- Tan Z., Hays M., Cromie G. A., Jeffery E. W., Scott A. C., et al. , 2013. Aneuploidy underlies a multicellular phenotypic switch. Proc. Natl. Acad. Sci. USA 110: 12367–12372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y.-C., Williams B. R., Siegel J. J., Amon A., 2011. Identification of aneuploidy-selective antiproliferation compounds. Cell 144: 499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavanti A., Gow N. A. R., Maiden M. C. J., Odds F. C., Shaw D. J., 2004. Genetic evidence for recombination in Candida albicans based on haplotype analysis. Fungal Genet. Biol. 41: 553–562. [DOI] [PubMed] [Google Scholar]
- Tavanti A., Davidson A. D., Fordyce M. J., Gow N. A. R., Maiden M. C. J., et al. , 2005. Population structure and properties of Candida albicans, as determined by multilocus sequence typing. J. Clin. Microbiol. 43: 5601–5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres E. M., Sokolsky T., Tucker C. M., Chan L. Y., Boselli M., et al. , 2007. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science 317: 916–924. [DOI] [PubMed] [Google Scholar]
- Torres E. M., Dephoure N., Panneerselvam A., Tucker C. M., Whittaker C. A., et al. , 2010. Identification of aneuploidy-tolerating mutations. Cell 143: 71–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Walt J. P., 1967. Sexually active strains of Candida albicans and Cryptococcus albidus. Antonie van Leeuwenhoek 33: 246–256. [DOI] [PubMed] [Google Scholar]
- Weaver B. A. A., Cleveland D. W., 2006. Does aneuploidy cause cancer? Curr. Opin. Cell Biol. 18: 658–667. [DOI] [PubMed] [Google Scholar]
- Wong S., Fares M. A., Zimmermann W., Butler G., Wolfe K. H., 2003. Evidence from comparative genomics for a complete sexual cycle in the “asexual” pathogenic yeast Candida glabrata. Genome Biol. 4: R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yona A. H., Manor Y. S., Herbst R. H., Romano G. H., Mitchell A., et al. , 2012. Chromosomal duplication is a transient evolutionary solution to stress. Proc. Natl. Acad. Sci. USA 109: 21010–21015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeigler R. S., R. P. Scott, H. Leung, A. A. Bordeos, J. Kumar et al, 1997. Evidence of parasexual exchange of DNA in the rice blast fungus challenges its exclusive clonality. Phytopathology 87: 284–294. [DOI] [PubMed] [Google Scholar]
- Zhu, J., N. Pavelka, W. D. Bradford, G. Rancati, and R. Li, 2012 Karyotypic determinants of chromosome instability in aneuploid budding yeast. PLoS Genet. 8: e1002719. [DOI] [PMC free article] [PubMed]
- Zörgö, E., K. Chwialkowska, A. B. Gjuvsland, E. Garré, P. Sunnerhagen et al., 2013 Ancient evolutionary trade-offs between yeast ploidy states. PLoS Genet. 9: e1003388. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.