Abstract
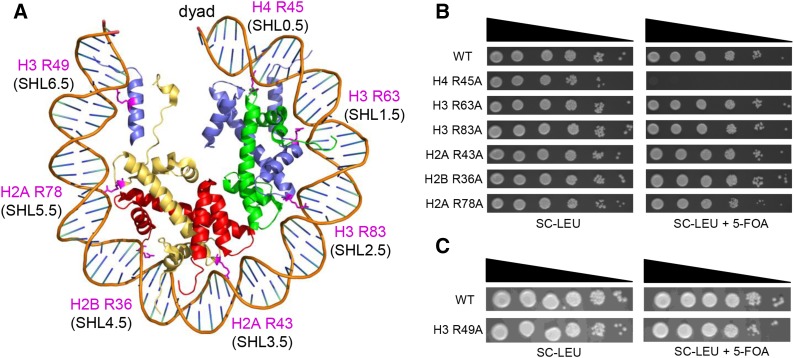
A critical feature of the intermolecular contacts that bind DNA to the histone octamer is the series of histone arginine residues that insert into the DNA minor groove at each superhelical location where the minor groove faces the histone octamer. One of these “sprocket” arginine residues, histone H4 R45, significantly affects chromatin structure in vivo and is lethal when mutated to alanine or cysteine in Saccharomyces cerevisiae (budding yeast). However, the roles of the remaining sprocket arginine residues (H3 R63, H3 R83, H2A R43, H2B R36, H2A R78, H3 R49) in chromatin structure and other cellular processes have not been well characterized. We have genetically characterized mutations in each of these histone residues when introduced either singly or in combination to yeast cells. We find that pairs of arginine residues that bind DNA adjacent to the DNA exit/entry sites in the nucleosome are lethal in yeast when mutated in combination and cause a defect in histone occupancy. Furthermore, mutations in individual residues compromise repair of UV-induced DNA lesions and affect gene expression and cryptic transcription. This study reveals simple rules for how the location and structural mode of DNA binding influence the biological function of each histone sprocket arginine residue.
Keywords: nucleotide excision repair, cryptic transcription, nucleosome, histone assembly
THE histone octamer, which is composed of two copies each of histones H2A, H2B, H3, and H4, binds with high affinity to ∼147 bp of DNA to form the nucleosome core particle. Histone-DNA binding is mediated by >100 histone main-chain and side-chain interactions with the DNA sugar-phosphate backbone and a similar number of indirect water-mediated interactions (Davey et al. 2002; Muthurajan et al. 2003). These interactions occur primarily at 14 locations in the nucleosome structure where the DNA minor groove faces the histone octamer [superhelical locations (SHL) −6.5 to 6.5] (Luger et al. 1997). At each of these locations, an arginine side chain extends into the DNA minor groove and makes extensive contacts with the DNA backbone (Figure 1A).
Figure 1.
Growth phenotypes of single sprocket arginine histone mutants. (A) Location of the sprocket arginine residues is highlighted in pink in the yeast nucleosome structure (1id3) (White et al. 2001). (B and C) Yeast strains containing a single histone arginine mutation were spotted at decreasing serial dilutions on SC-LEU and SC-LEU+5-FOA plates. The SC-LEU+5-FOA plates select against a URA3 plasmid containing wild-type histones, revealing the growth phenotype of the histone mutant. SHL, superhelical location.
We will refer to the arginine residues that insert into the DNA minor groove as “sprocket” arginines, since they engage the DNA “chain” like the teeth of a bicycle sprocket wheel. Sprocket arginine residues are highly conserved (Muthurajan et al. 2003; Sullivan and Landsman 2003), and the insertion of sprocket arginine side chains into the DNA minor groove comprises a significant fraction of the solvent accessible surface area that is buried upon histone-DNA binding (Davey et al. 2002). Studies have suggested that sprocket arginine–DNA contacts may play an important role in the rotational positioning of nucleosomes (Luger et al. 1997; Harp et al. 2000; Rohs et al. 2009; Wang et al. 2010; West et al. 2010). For example, short poly(A) stretches narrow the DNA minor groove, potentially enhancing electrostatic interactions between the DNA phosphate backbone and the sprocket arginine (Rohs et al. 2009; West et al. 2010).
While sprocket arginine residues appear to have unique and important roles in DNA binding and are highly conserved among eukaryotic species, mutational screens in yeast revealed that only one of these arginine residues, H4 R45, is essential for cell viability (Matsubara et al. 2007; Dai et al. 2008; Nakanishi et al. 2008) (Figure 1B). H4 R45 contacts the DNA at superhelical locations flanking the dyad (SHL ±0.5) and is lethal when mutated to alanine or cysteine (Fry et al. 2006; Matsubara et al. 2007; Dai et al. 2008; Nakanishi et al. 2008). It is not clear if H4 R45 is essential because it binds near the dyad, because it binds immediately adjacent SHLs (−0.5 and +0.5), or because it plays some other essential function.
Mutations in the H4 R45 residue (H4 R45C and H4 R45H) were originally isolated from a yeast genetic screen for switch-independent (sin) mutants (Kruger et al. 1995). Expression of the H4 R45C (or R45H) mutant partially alleviates the requirement for the SWI/SNF remodeling complex to activate the expression of the yeast homothallic switching endonuclease (HO) gene. H4 R45C and R45H mutants also enhance nucleotide excision repair (NER) of UV-induced cyclobutane pyrimidine dimers (CPDs) and render yeast cells more resistant to UV radiation (Nag et al. 2008). These effects on transcription and DNA repair appear to be a consequence of how the H4 R45C mutant alters the DNA accessibility, intrinsic mobility, and stability of yeast nucleosomes (Kurumizaka and Wolffe 1997; Wechser et al. 1997; Flaus et al. 2004; Muthurajan et al. 2004). The crystal structure of the H4 R45C mutant nucleosome revealed no significant distortions in the nucleosome structure (Muthurajan et al. 2004). Instead, the H4 R45C mutant eliminates stabilizing van der Waals contacts between H4 R45 and the DNA minor groove (Muthurajan et al. 2004). The H4 R45C mutant can also affect Mg2+-dependent higher-order folding of nucleosome arrays (Horn et al. 2002), indicating that the effects of this mutation may extend beyond the mononucleosome.
Another sprocket arginine residue, H3 R83, was isolated in a genetic screen for mutants that caused the loss of ribosomal DNA (rDNA) silencing (LRS) in yeast (Park et al. 2002). The H3 R83A mutant causes recessive defects in rDNA and telomere silencing and a dominant defect in HML silencing (Park et al. 2002). The H3 R83A mutant cells are marginally more sensitive to the DNA-damaging agent methyl methanesulfonate (MMS) in some strain backgrounds (Matsubara et al. 2007). However, the H3 R83A mutant does not have a sin phenotype, nor does it affect Mg2+-dependent higher-order folding of nucleosome arrays (Fry et al. 2006), indicating that it has phenotypes distinct from that of the H4 R45 mutants.
Much less is known about the function of the other sprocket arginine residues (H3 R63, H2A R43, H2B R36, H2A R78, and H3 R49). Global screens of histone mutant phenotypes in yeast found that both H3 R49A and H2A R78A mutants have increased sensitivity to hydroxyurea (HU, a DNA replication inhibitor), high temperature (39°), and the DNA-damaging agent MMS (Dai et al. 2008; Huang et al. 2009; Sakamoto et al. 2009; Dai et al. 2010). The H3 R49A mutant was also found to be more sensitive to UV radiation (Dai et al. 2008; Huang et al. 2009) and induces cryptic transcription (Hainer and Martens 2011). The H2A R78A mutant was found to be more sensitive to the DNA-damaging agent camptothecin (Dai et al. 2010). One study observed marginal HU and MMS sensitivities for the H3 R63A mutant (Sakamoto et al. 2009). In contrast, the H2A R43A and H2B R36A mutants have no apparent phenotype when mutated to alanine (Matsubara et al. 2007; Huang et al. 2009). The H2B R36 residue is part of the histone H2B repression (HBR) cassette (Parra et al. 2006). Deletion of the eight-residue HBR cassette increases the expression of a large fraction of the yeast genome, renders cells more sensitive to UV radiation and other DNA-damaging agents (Parra et al. 2006), and affects NER at certain loci (Nag et al. 2010). It is not clear whether H2B R36 or other residues in the HBR domain play a functional role in these processes.
In this study, we investigated the role of histone sprocket arginine residues in gene expression, DNA repair, and cell viability. We found that mutants in sprocket arginine residues that contact the distal ends of nucleosomal DNA (i.e., H2A R78 and H3 R49) show enhanced sensitivity to UV radiation, which appears to be due to a defect in NER of UV-induced DNA lesions. In contrast, while these mutants also show enhanced sensitivity to the DNA alkylating agent MMS, we did not observe a corresponding defect in base excision repair (BER). Moreover, our data indicate that the H3 R49A mutant has a unique SIN phenotype, while the H2A R78A mutant derepresses the expression of many genes and causes a defect in telomeric silencing. Additionally, multiple sprocket arginine mutants show enhanced levels of cryptic transcription. Finally, we find that the double mutants of adjacent sprocket arginines that contact the distal ends of the nucleosomal DNA (i.e., H2A R78A-H3 R49A or H2A R78A-R36A) are lethal in yeast.
Materials and Methods
Yeast strains and plasmids
The yeast strains and plasmids used in this study are described in Supporting Information, Table S1 and Table S2. These strains were derived from strain JHY205 (Ahn et al. 2005), which has the chromosomal copies of each canonical histone gene deleted. The sprocket arginine histone mutants were constructed in plasmid pQQ18 (Ahn et al. 2005), which contains genes for each of the canonical histones (i.e., HTA1-HTB1HHT2-HHF2). Mutants were constructed by site-directed mutagenesis using a modified version of the QuikChange method and verified by DNA sequencing. The HO-lacZ reporter was constructed by first replacing a portion of the HO gene with a URA3 marker, followed by replacement of the URA3 marker with the HO-lacZ construct, which is inserted at position +58 in the HO open reading frame. The FLO8-HIS3 reporter was constructed using the CRISPR/Cas9 system, which will be described in more detail elsewhere. Primer sequences are available upon request. Standard growth conditions, media, and yeast (and Escherichia coli) methods were used in strain construction and cell growth.
Yeast growth curves
Growth curves were conducted as previously described (Jin et al. 2007). Yeast strains were grown to an optical density at 600 nm (OD600) of 0.01 in selective media containing galactose, permitting the expression of wild-type histones. Cells were then washed with water and resuspended in glucose media, repressing the expression of wild-type histones. Time “0” is defined as the point at which the cells were switched to glucose. OD600 measurements were taken at the indicated time points. Growth curves shown are representative of at least three independent replicates.
Yeast spotting
Logarithmically growing yeast strains were spotted as 1:10 serial dilutions onto the indicated media with 2% glucose. For MMS experiments, yeast strains were spotted onto synthetic complete (SC) media containing the indicated doses of MMS (Sigma). Images were taken after 4 days incubation at 30°. For UV experiments, plates were irradiated with the indicated dose of UV light (50, 100, or 150 J/m2) after spotting onto SC media. Images were taken after 4 days incubation in the dark (i.e., plates wrapped in aluminum foil) at 30°.
Quantitative UV sensitivity assays
To assess UV sensitivity, yeast strains were grown overnight in yeast extract peptone dextrose (YPD) medium to logarithmic phase. Serial dilutions were plated onto YPD plates and irradiated with the indicated dose of UV light (50, 100, or 150 J/m2). Irradiated plates were wrapped in aluminum foil and incubated at 30° for 2–3 days, after which point the aluminum foil was removed and an initial colony count was made. Plates were allowed to incubate at 30° for up to 2 weeks, and subsequent counts were made every few days, with the majority of colonies appearing by the fourth day. A minimum of three replicates was performed for each strain. Survival was determined by dividing the relative colony-forming units (CFUs) on the irradiated plates by the relative CFUs of plates that were not irradiated.
LacZ assays
LacZ assays were performed as described in the Clontech Yeast Protocols Handbook. Briefly, cells were grown in YPD medium to an OD600 of 0.4–0.9. The cells were pelleted, washed, and then resuspended with Z buffer (60 mM Na2HPO4·7H2O, 40 mM NaH2PO4·H2O, 10 mM KCL, 1 mM MgSO4·7H2O), and the supernatants were removed. Cells were lysed by repeated freeze/thaw cycles using liquid nitrogen, and BMEZ buffer (Z buffer + 0.27% β-mercaptoethanol) was added. Reactions were started with the addition of ο-nitrophenyl-β-d-galactoside dissolved in Z buffer and incubated at 30° for the duration of the experiment. Reactions were stopped by the addition of Na2CO3 and absorbance readings at OD420 were measured and used to calculate β-galactosidase units. A minimum of four replicates was performed for each strain.
Nucleotide excision repair assays
Yeast cells were grown in YPD liquid media to an OD600 of ∼0.6. After removing a “no UV” control aliquot, the YPD medium was removed and the cells were resuspended in water and subsequently irradiated with 100 J/m2 of UV light (time “0”). Cells were resuspended in YPD and incubated at 30° with continuous shaking. Aliquots were spun down and placed in the −80° freezer at the indicated time points. Cells were lysed by bead beating, and DNA was extracted as described (Rose et al. 1990). DNA was resuspended in T4 endonuclease reaction buffer to a final concentration of 4 mM Tris (pH 7.4), 2 mM EDTA, 25 mM NaCl, and 25 µg/mL BSA in the presence or absence of T4 endonuclease V (Epicentre) for a minimum of 2 hr at 37°. Afterward, single-strand DNA was resolved on a 1.2% alkaline gel and stained with SYBR Gold (Invitrogen). The gel was scanned by Typhoon FLA 7000 (GE Healthcare), and quantifications were performed using ImageQuant 5.2 (Molecular Dynamics) to determine the number of CPDs per kilobase as previously described (Bespalov et al. 2001).
Base excision repair assays
Repair assays were conducted as previously described (Czaja et al. 2010). Briefly, yeast cells were grown in YPD to an OD600 of ∼0.6. After removing a “no MMS” control aliquot, cultures were treated with 0.2% MMS (Sigma) for 10 min. Cells were washed with water and resuspended in YPD (time “0”). Cells were allowed to repair for indicated periods of time at 30° with continuous shaking, harvested, and frozen at −80°. Cells were lysed by bead beating and DNA was extracted as described (Rose et al. 1990). DNA was resuspended in MOPS buffer (70 mM MOPS pH 7.5, 1 mM DTT, 1 mM EDTA, 5% glycerol) with or without alkyladenine DNA glycosylase (AAG, from Leona Samson, Massachusetts Institute of Technology) and APE1 endonculease (NEB) for a minimum of 2 hr. These and undigested controls were run on a 1.2% alkaline agarose gel to resolve single-stranded DNA and stained with SYBR Gold (Invitrogen). The gel was scanned by Typhoon FLA 7000 (GE Healthcare), and removal of lesions was quantified using ImageQuant 5.2 (Molecular Dynamics) and analyzed using previously published quantification methods. Data are given as the percentage of lesions removed compared to the “0” time point for three independent replicate experiments.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was conducted and analyzed using a modified version of previously published protocols (Kyriss et al. 2010; Zheng et al. 2010). Briefly, yeast cells were grown to an OD600 of 0.6–0.8 and cross-linked using 1% formaldehyde (v/v). Cells were lysed by bead beating at 4°, and chromatin was sonicated using a Bioruptor (Diagenode) to create chromatin fragments averaging ∼500 bp. Chromatin extract was immunoprecipitated using an anti-FLAG antibody (Sigma) bound to magnetic beads. The immunoprecipitated and input chromatin were proteinase K (Thermo) digested, cross-links were reversed, and DNA was phenol-chloroform-extracted and ethanol-precipitated. After treating with RNAse A (Thermo), the DNA was analyzed by quantitative PCR (qPCR). qPCR experiments were conducted using EvaGreen qPCR master mix (Biotium) and the ABI 7500 Fast Real-Time PCR system using region specific primers. Data are given as the percentage immunoprecipitated compared to input whole-cell extract, representing at least three replicates (except for Figure S6; see legend). Primer sequences are available upon request.
Western blot analysis of yeast extracts
Yeast strains were grown to mid-log phase and proteins were extracted using 0.1% NaOH. Blots were probed using 1:3000 dilution of anti-H2A (ActiveMotif catalog no. 39235), 1:3000 dilution of anti-H2B (ActiveMotif catalog no. 39237), 1:1000 dilution of anti-H3 (ActiveMotif catalog no. 39163), or 1:1000 dilution of anti-H3K36me3 (Abcam catalog no. ab9050) primary antibody followed by 1:3000 dilution of anti-rabbit (Bio-Rad catalog no. 172-1019) secondary. Anti-GAPDH antibody (Thermo Scientific catalog no. MA5-15738) was used as a loading control using an anti-mouse (Bio-Rad catalog no. 170-6516) secondary. Blots were imaged using a Typhoon FLA 7000 (GE Healthcare) and analyzed using ImageQuant 5.2 software.
Microarray data and analysis
Microarray experiments were performed as previously described (Kyriss et al. 2010) with slight modifications. Triplicate yeast cultures for the wild-type and H2A R78A mutant strains were grown to mid-log phase in YPD media at 30° and harvested. Total RNA was isolated by hot acid phenol–chloroform extraction and used for microarray analysis following standard Affymetrix protocols for Yeast Genome 2.0 GeneChip arrays.
Microarray data were processed using the MAS 5.0 algorithm implemented in the Bioconductor R package (Gentleman et al. 2004) after removing data from probes for Schizosaccharomyces pombe transcripts using published methods (Gillespie et al. 2010). Differentially expressed genes were identified using the limma R package (Smyth 2005), using a Benjamini–Hochberg adjusted P-value threshold of 0.05 and fold-change threshold of 1.7 (up or down), as previously described (Van Wageningen et al. 2010; Lenstra et al. 2011). Genes up-regulated in the H2A R78A mutant were analyzed for enriched chromatin features using the ChromatinDB database (O’Connor and Wyrick 2007).
Results
Effects of histone sprocket arginine mutants on cell growth, gene repression, and cryptic transcription
To characterize the biological functions of sprocket arginine residues, we constructed alanine mutations in each of the histone arginine residues that insert into the DNA minor groove (i.e., H4 R45A, H3 R63A, H3 R83A, H2A R43A, H2B R36A, H2A R78A, and H3 R49A; see Figure 1A). Each sprocket arginine mutant was constructed in the same plasmid (pQQ18) (Ahn et al. 2005), which contains genes coding for each of the four core histone proteins (HTA1, HTB1, HHT2, HHF2) under the control of their native promoters. Using this strategy, we were able to introduce each histone mutant into a common yeast strain background (JHY205) (Ahn et al. 2005) and thus directly compare phenotypes for mutants in each of the four core histone genes. While the H4 R45A mutant was lethal, in accordance with previous reports (Matsubara et al. 2007; Dai et al. 2008; Nakanishi et al. 2008), the other sprocket arginine mutants were viable (Figure 1, B and C). Additionally, the viable histone sprocket arginine mutants were expressed at roughly wild-type levels based on Western blot analysis of yeast whole-cell extracts (Figure S1).
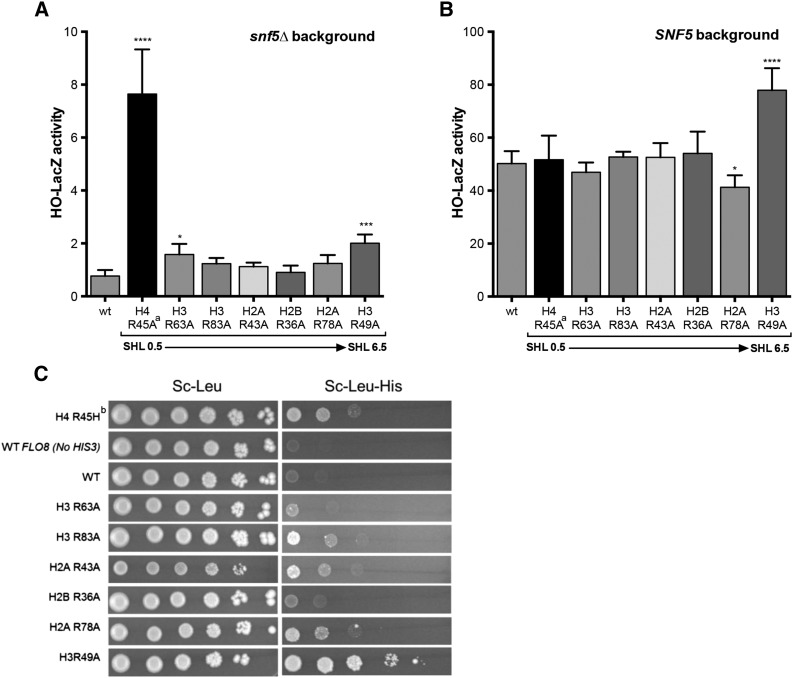
We first examined the role of sprocket arginine residues in regulating yeast gene expression. Mutations in H4 R45 were originally isolated in a genetic screen for mutants that enabled expression of an HO-lacZ reporter gene in the absence of a functional SWI/SNF complex (Kruger et al. 1995). In our strain background, we confirmed that the H4 R45A mutant (in the presence of wild-type H2A; see legend for Figure 2) significantly induced expression of the HO-lacZ reporter in the snf5∆ mutant background (Figure 2A), as judged by LacZ activity assays. Significant increases in LacZ expression were also detected for the H3 R63A and H3 R49A mutants, although the magnitude of the increase was considerably smaller in these mutants (Figure 2A). Intriguingly, the H3 R49A mutant also significantly increased expression of the HO-lacZ reporter in the SNF5 wild-type background (Figure 2B). In contrast, the H4 R45A mutant had no effect on HO-lacZ expression in SNF5 wild-type cells, indicating that these two sprocket arginine mutants likely affect HO-lacZ expression via distinct mechanisms.
Figure 2.
Effects of histone sprocket arginine mutants on expression of the HO-lacZ reporter and cryptic transcription of the FLO8-HIS3 reporter. (A) LacZ activity was measured for extracts from each histone mutant strain in a snf5∆ genetic background. Mean and standard deviation of LacZ activity from at least three independent experiments is depicted for each strain. The SHLs of nucleosomal DNA bound by each arginine residue are indicated. P-values were corrected for multiple hypothesis testing. ****P < 0.0001; ***P < 0.001; **P < 0.01; *P < 0.05. ªThe H4 R45A mutant strain also contains the wild-type (wt) histone H4 gene. (B) Same as in A, except the strains are wild type for SNF5. (C) Yeast strains containing the FLO8-HIS3 reporter (excepting the wild-type FLO8 control) were spotted at decreasing serial dilutions on SC-LEU and SC-LEU-HIS plates. Growth on SC-LEU-HIS plates indicates activation of the FLO8 cryptic transcript containing the HIS3 reporter. bThe H4 R45H mutant strain also contains the wild type (wt) histone H4 gene.
We also examined the effects of the sprocket arginine residues on cryptic intragenic transcription. Previous studies have discovered that the H3 R49A sprocket arginine mutant activates transcription initiation from a cryptic promoter within the FLO8-coding sequence due to a defect in Set2-dependent H3 K36 methylation (Hainer and Martens 2011). We tested the effects of this and other sprocket arginine mutants on FLO8 cryptic transcription using a FLO8-HIS3 reporter (Cheung et al. 2008) and scoring for growth on media lacking histidine (SC-Leu-His; see Figure 2C). We confirmed that the H3 R49A mutant induced cryptic transcription from the FLO8-HIS3 reporter, enabling the strain to grow in media lacking histidine. In contrast, essentially no growth was detected on media lacking histidine for the wild-type strain (Figure 2C). We found that multiple sprocket arginine mutants induced FLO8 cryptic transcription, namely H3 R83A, H2A R43A, H2A R78A, and H4 R45H (Figure 2C), although apparently to a lesser extent than the H3 R49A mutant. Unlike H3 R49A, however, these other sprocket arginine mutants do not affect H3 K36 trimethylation (Figure S2), indicating that they regulate cryptic transcription through a distinct pathway. In summary, the H4 R45 and H3 R49 residues regulate HO repression but through distinct mechanisms, and multiple sprocket arginine residues are required to repress cryptic transcription.
H3 R49 and H2A R78 sprocket arginine residues are required for efficient nucleotide excision repair of UV-induced DNA lesions
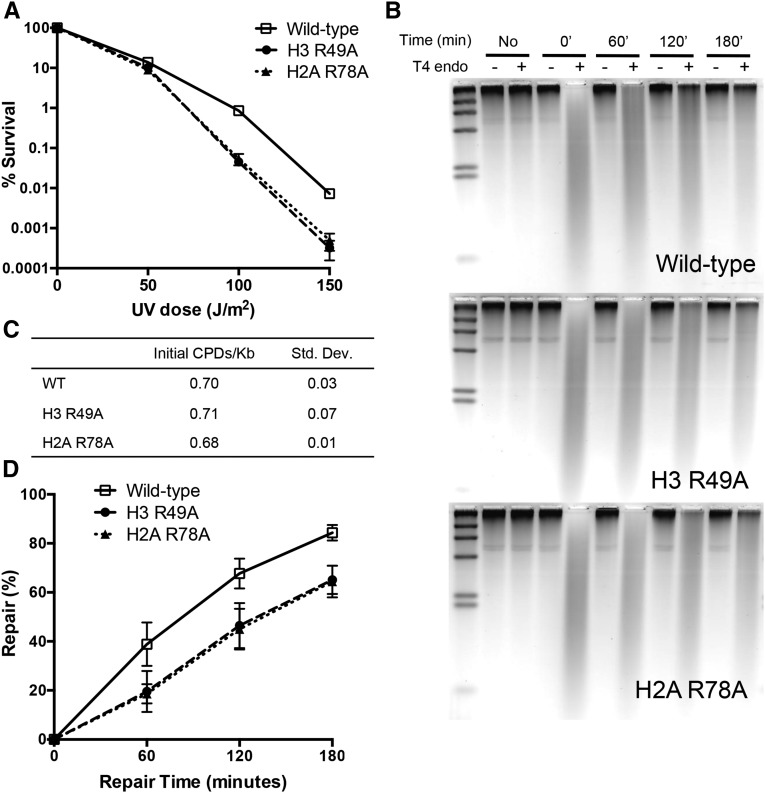
We next examined the role of sprocket arginine residues in DNA repair. Yeast cells with mutations in the H4 R45 sprocket arginine have the unusual phenotype of being more resistant to UV radiation, likely because the cells more efficiently repair UV-induced DNA lesions (Nag et al. 2008). We found that none of the other sprocket arginine mutants were more resistant to killing by UV radiation (Figure S3); however, the H2A R78A and H3 R49A sprocket arginine mutants were significantly more sensitive to UV radiation than wild-type cells in our strain background (Figure S3 and Figure 3A).
Figure 3.
H3 R49 and H2A R78 histone residues are required for survival and efficient NER of UV-induced DNA lesions. (A) UV survival assays of histone mutant strains. (B) Representative alkaline gels from genomic NER assays of histone mutants. Genomic DNA was isolated from yeast cells before UV exposure or at the indicated time points following UV radiation (100 J/m2). Isolated genomic DNA was treated with or without T4 endonuclease, which makes single-stranded DNA (ssDNA) nicks at sites of UV-induced CPDs. (C) Quantification of initial frequency of CPDs per kilobase of genomic DNA. (D) Quantification of the percentage of repair of CPDs from alkaline gels. Mean and standard deviation of the percentage of repair of CPDs from three independent experiments is depicted.
We tested whether the increased UV sensitivity in these mutants was due to a defect in NER by measuring the repair of CPDs, a major class of UV-induced DNA lesions that is repaired by the NER pathway, in the histone mutant cells following UV exposure. We observed a significant global defect in the repair of CPDs in the H2A R78A and H3 R49A mutants relative to wild type (Figure 3, B and D). Both mutants had a very similar defect in NER efficiency (and UV sensitivity), but neither mutant significantly altered the initial CPD damage levels following UV exposure (Figure 3C). These data indicate that sprocket arginine residues that bind near the ends of the nucleosomal DNA function in NER.
H2A R78A sprocket arginine residue does not affect basal expression of NER genes, but is required for gene repression
One possible explanation for the observed NER defects is that the histone mutants may reduce the expression of one or more key NER genes. We tested this possibility by measuring genome-wide expression changes in the H2A R78A mutant relative to wild type using Affymetrix microarrays. Importantly, we observed no significant decreases in the expression of any NER genes under normal growth conditions (Table S3).
Overall, 290 genes showed an increase in messenger RNA (mRNA) levels, and only 8 genes showed a decrease in mRNA levels in the H2A R78A mutant strain. Gene ontology analysis using the FunSpec software (Robinson et al. 2002) revealed that the up-regulated genes were enriched for the sporulation (P = 3.8 × 10−10), oxidation-reduction (P = 2.2. × 10−8), and carbohydrate metabolism (P = 5.2 × 10−7) functional categories. Because the H2A R78A mutant induced cryptic transcription of the FLO8-HIS3 reporter gene (see Figure 2C), it is possible that some of the gene expression changes could reflect activation of cryptic transcripts. However, we did not observe a significant enrichment of yeast genes with known cryptic transcripts (Cheung et al. 2008). Analysis of histone occupancy ChIP-chip data in ChromatinDB (O’Connor and Wyrick 2007) indicated that the 290 up-regulated genes had higher levels of histone occupancy in their promoter regions in wild-type cells and were depleted for permissive chromatin marks (Figure S4). Taken together, these data suggest that the H2A R78 sprocket arginine is required for chromatin-mediated repression of many yeast genes.
Histone sprocket arginine mutants are sensitive to MMS but do not affect base excision repair
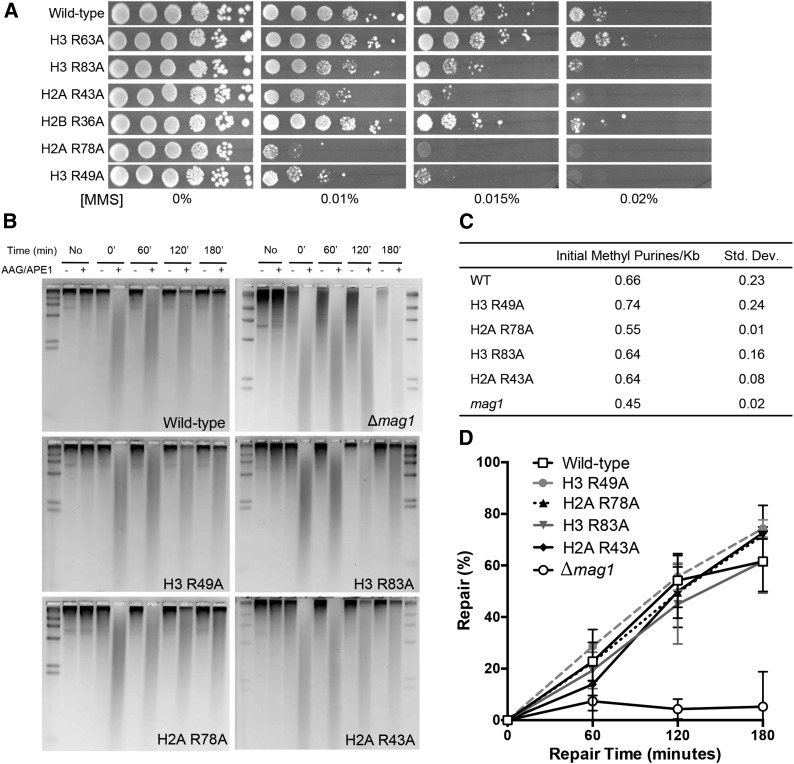
Many of the sprocket arginine mutants are sensitive to MMS, a DNA-damaging agent that generates alkylated base lesions that are primarily repaired by the BER pathway. We confirmed the MMS sensitivity of the H3 R83A, H2A R43A, H2A R78A, and H3 R49A mutants in our strain background (Figure 4A). As two of these mutants showed an NER defect, we tested each MMS-sensitive strain for a possible defect in BER by measuring the repair of the two major MMS-induced DNA lesions, 3-methyladenine and 7-methylguanine. However, we found that none of these histone mutants caused a global defect in BER (Figure 4, B–D), indicating that the MMS sensitivity is likely not due to a global deficiency in repairing these base lesions.
Figure 4.
Histone sprocket arginine mutants are sensitive to MMS, but do not affect BER efficiency of MMS-induced DNA lesions. (A) Sprocket arginine mutant strains were spotted at decreasing serial dilutions on plates containing the indicated doses of MMS. (B) Representative alkaline gels from genomic BER assays of histone mutants and a Δmag1 control strain. DNA was isolated from yeast cells before MMS exposure or at the indicated time points following MMS treatment (0.2% for 10 min). Isolated genomic DNA was treated with (or without) AAG/APE1 to induce ssDNA nicks at sites of 3-methyladenine and 7-methylguanine lesions. (C) Quantification of initial frequency 3-methyladenine and 7-methylguanine lesions per kilobase of genomic DNA. (D) Quantification of the percentage of repair of MMS-induced DNA damage from alkaline gels. Mean and standard deviation of the percentage of repair from three independent experiments is depicted.
Double mutants of adjacent sprocket arginine residues near the DNA exit/entry sites are lethal
While our data clearly indicate that multiple sprocket arginine residues have important roles in transcription and DNA repair (particularly H2A R78 and H3 R49), only the H4 R45 sprocket arginine residue is essential for cell viability (Figure 1B). We hypothesized that the H4 R45A mutant is lethal because it is the only sprocket arginine mutant that affects DNA interactions at adjacent superhelical locations (SHL +0.5 and SHL −0.5). To test our hypothesis, we constructed double mutants of nonessential sprocket arginine residues that contacted adjacent DNA superhelical locations [e.g., SHL ±1.5 (H3 R63) and SHL ±2.5 (H3 R83)]. While a number of double-mutant combinations are viable in yeast, the H2A R78A-H2B R36A and H2A R78A-H3 R49A double mutants are lethal (Figure 5A). Intriguingly, these sprocket arginine residues contact the DNA at superhelical locations near the DNA exit/entry sites in the nucleosome (Figure 1A). In contrast, double mutants of sprocket arginine residues that contact the DNA at superhelical locations distal from the exit/entry sites (i.e., SHL ±1.5, SHL ±2.5, SHL ±3.5) are viable (Figure 5A), although the H3 R83A-H2A R43A and H2A R43A-H2B R36A double mutants show increased sensitivity to MMS (data not shown).
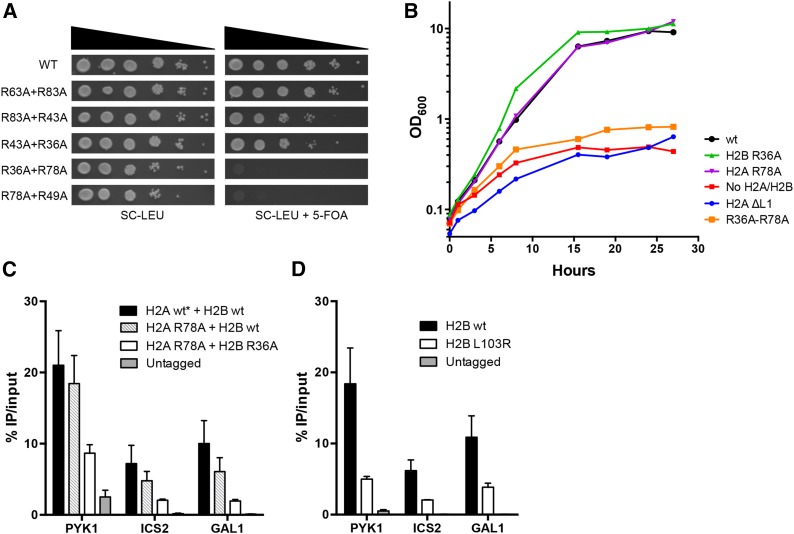
Figure 5.
The histone H2A R78A-H2B R36A double mutant causes a rapid cessation of cell growth and a defect in histone occupancy. (A) Yeast strains containing the indicated double mutants were spotted as described in Figure 1B. (B) Growth rate of histone mutants upon repression of wild-type histone H2A and H2B genes by shifting to glucose media (“0” hour). The H2A ∆L1 strain has a deletion in the essential L1 loop (residues 39–42). (C) Histone occupancy of the FLAG-tagged wild-type or mutant histone H2B in the indicated genetic background (see text for more details) was measured by ChIP at select genomic regions. *Strain contained wild-type H2A and H2A R78A mutant. (D) Same as in C using a FLAG-tagged wild-type H2B or mutant H2B L103R.
To investigate the mechanism for lethality, we constructed a yeast strain expressing the H2A R78A-H2B R36A double mutant under the control of the endogenous histone promoter and wild-type H2A and H2B under the control of the heterologous GAL1 promoter. This strain was viable in galactose media, as the wild-type H2A and H2B genes were expressed at high levels. Wild-type H2A and H2B expression was shut off when shifted to glucose media, permitting us to better characterize the lethal phenotype of the H2A R78A-H2B R36A double mutant.
We first used this experimental system to test the growth rates of various control histone mutants following the shutoff of wild-type H2A and H2B expression in glucose media. Strains containing the H2A R78A or H2B R36A single mutants grew similar to wild type after the shift to glucose media, eventually reaching stationary phase (Figure 5B). In contrast, an H2A L1 loop deletion (H2A ∆39-42) mutant, which presumably cannot fold or assemble into the nucleosome, arrested in growth about as rapidly as the “no H2A/H2B” control. The H2A R78A-H2B R36A double mutant typically arrested in growth after the shift to glucose media with kinetics similar to both the H2A L1 loop deletion and the “no H2A/H2B” controls (Figure 5B). These data suggest that the double mutant may not be able to properly assemble into nucleosomes.
H2A R78A-H2B R36A double mutant causes a defect in histone occupancy in vivo
To test this hypothesis directly, we used ChIP to measure the incorporation of the H2A R78A-H2B R36A double-mutant dimer in chromatin. Since the double mutant is lethal, we constructed a strain that is homozygous for the H2A R78A mutation and expresses both a FLAG-tagged H2B R36A mutant gene and an untagged wild-type H2B gene (strain YAH013; see Table S1). This strain was viable due to the presence of the untagged H2B wild-type gene, and all H2B-FLAG-containing dimers contained the H2A R78A-H2B R36A double mutation. Western blot analysis indicated that the FLAG-tagged H2B protein was expressed at only slightly reduced levels as compared to untagged wild-type H2B (Figure S5A). We performed ChIP-qPCR experiments on this strain using an anti-FLAG antibody to measure the incorporation and occupancy of the double-mutant dimer (i.e., H2A R78A-H2B R36A) at four well-characterized genomic loci: the promoter of ICS2, the 5′-coding regions of PYK1 and GAL1, and the 3′-coding region of PMA1. The PMA1 primer set had high background signal in a few of the untagged control experiments, so we focused on the first three genomic regions for our analysis.
ChIP analyses indicated that the H2A R78A-H2B R36A double-mutant dimer was incorporated into chromatin as its occupancy was significantly greater than the untagged control at each of the three genomic regions tested (Figure 5C and Figure S5B). However, the occupancy of the H2A R78A-H2B R36A double-mutant dimer was decreased compared to wild type or the H2A R78A mutant alone (Figure 5C). In contrast, the H2A R78A mutant alone caused a marginal decrease in H2B occupancy that was not significantly different from wild type (Figure 5C; P > 0.05). Similar results were obtained using FLAG-tagged H2A strains for the ChIP experiments (Figure S6A) although the decrease in H2A occupancy in the double mutant was statistically significant only at the GAL1 5′-coding region (P < 0.05). A caveat is the H2A-FLAG protein did not express well in our strain background (Figure S6B).
The ChIP data indicate that the H2A R78A-H2B R36A double mutant can be assembled into chromatin in vivo, but this assembly is less efficient (or less stable) than wild-type H2A-H2B. For comparison, we used ChIP to measure the decrease in H2B occupancy in the H2B L103R mutant, which partially disrupts the binding of the H2A-H2B dimer to the nucleosome (Jamai et al. 2007). Interestingly, the H2B L103R mutant caused a similar decrease in histone occupancy as the H2A R78A-H2B R36A double mutant (Figure 5D), yet the H2B L103R mutant is viable (Jamai et al. 2007 and data not shown). In summary, these data show that the H2A R78A-H2B R36A double-mutant dimer has a defect in chromatin incorporation, but the relative magnitude of this defect suggests that it is not the primary cause of lethality.
Discussion
In this study, we have characterized the biological functions of histone sprocket arginine residues in the model eukaryote Saccharomyces cerevisiae. Unlike previous studies (Matsubara et al. 2007; Nakanishi et al. 2008; Sakamoto et al. 2009), our study employed a common strain background for assaying the phenotypes of both histone H2A/H2B and H3/H4 mutants, which enabled us to directly compare these histone mutant phenotypes. The H4 R45 sprocket arginine residue is known to affect HO transcription, UV resistance, NER efficiency, and cell viability in yeast. As detailed below, we have identified novel functions for the remaining six sprocket arginines in these and other biological pathways. Our systematic analysis reveals simple rules for how the location and structural mode of DNA binding influence the biological function of each histone sprocket arginine residue.
The H4 R45 residue, which inserts into adjacent DNA minor grooves immediately flanking the dyad, is the only sprocket arginine required for cell viability. We hypothesized that the H4 R45A mutant is lethal because it is the only sprocket arginine mutant that affects DNA interactions at adjacent superhelical locations (SHL +0.5 and SHL −0.5). However, we found that double sprocket arginine mutants that affect binding to adjacent superhelical locations were in many cases viable (i.e., H3 R63A-H3 R83A, H3 R83A-H2A R43A, and H2A R43A-H2B R36A). Only double mutants that affect adjacent superhelical locations near the DNA ends were lethal (i.e., H2A R78A-H2B R36A and H2A R78A-H3 R49A).
We focused on investigating the mechanism of lethality in the H2A R78A-H2B R36A, since this double mutant was more amenable to study. We hypothesized that the H2A R78A-H2B R36A double mutant is lethal due to a defect in nucleosome assembly. This hypothesis is supported by data from time-course experiments that revealed that the H2A R78A-H2B R36A double mutant arrests in growth fairly rapidly after the expression of wild-type H2A-H2B is shut off. Consistent with the growth kinetics, ChIP experiments indicate that the H2A R78A-H2B R36A double mutant can be assembled into chromatin but at a significantly reduced efficiency (or stability) compared to wild-type H2A-H2B or the H2A R78A mutant alone. While this decrease in histone occupancy could potentially explain the lethal phenotype, we do not favor this hypothesis because we also found that the viable H2B L103R mutant has a similar decrease in histone H2B occupancy. Instead, our current working hypothesis is that incorporation of one double-mutant dimer (i.e., a “heterozygous” nucleosome) is tolerated, but incorporation of two double-mutant dimers within a single nucleosome is destabilizing, thus leading to lethality. It is not clear if a related mechanism may be responsible for the lethality of the H2A R78A-H3 R49A double mutant.
The H4 R45 sprocket arginine residue was originally characterized in a screen for sin mutations that induced HO expression in the absence of the SWI/SNF chromatin-remodeling complex (Kruger et al. 1995). None of the other sprocket arginine residues had the same magnitude of effect on HO expression as the H4 R45 mutant, but we did observe significant changes in HO expression in a few other sprocket arginine mutants, particularly H3 R49A. A previous study did not detect a sin phenotype for the H3 R49A mutant in the presence of the wild-type H3 gene (Hainer and Martens 2011), indicating that the H3 R49A sin phenotype is likely to be recessive, unlike the original SIN histone mutants. The H3 R49A sin phenotype is also distinct in that it increases HO expression even when the wild-type SWI/SNF complex is present. Based on these data, we hypothesize that the H3 R49A mutant increases HO expression through a different pathway than SWI/SNF-dependent chromatin remodeling at the HO promoter. H3 R49 binds near the nucleosomal DNA exit/entry site and has been shown to affect nucleosomal DNA unwrapping in vitro (Ferreira et al. 2007). Our current model is that the H3 R49A mutant causes increased DNA unwrapping from HO promoter nucleosomes, which stimulates HO transcription.
An alternative hypothesis is that the H3 R49A mutant increases HO-lacZ expression by inducing the expression of cryptic transcripts within the HO locus that encode a functional LacZ protein. A previous study discovered that the H3 R49A mutant activates cryptic transcription at multiple loci (e.g., FLO8, STE11, and SYF1) due to a defect in H3 K36 methylation (Hainer and Martens 2011). We confirmed that H3 R49A induced cryptic transcription using a FLO8-HIS3 reporter (Cheung et al. 2008). Furthermore, we identified additional sprocket arginine mutants (i.e., H4 R45H, H3 R83A, H2A R43A, and H2A R78A) that also activated cryptic transcription of this reporter, although to a lesser extent than was observed for the H3 R49A mutant and through a pathway distinct from H3 K36 methylation.
Mutations in the H4 R45 sprocket arginine residue also enhance UV resistance and increase NER efficiency in yeast cells, presumably by rendering the nucleosomal DNA more accessible to NER enzymes (Nag et al. 2008; Nag and Smerdon 2009). While none of the other sprocket arginine mutants significantly enhance UV resistance, we discovered that the H2A R78A and H3 R49A mutants are more sensitive to UV damage and have a significant defect in NER. While the H4 R45 residue contacts DNA near the dyad center, the H3 R49 and H2A R78 residues bind near the ends of the nucleosomal DNA. These observations suggest that the translational position of the nucleosomal DNA bound by the sprocket arginine residue (i.e., near dyad vs. near exit/entry) influences its role in UV resistance and repair.
The mechanism underlying the UV sensitivity and repair phenotypes is currently unclear. The H2A R78A and H3 R49A mutants presumably decrease the stability of DNA binding within the nucleosome, particularly near the ends of the nucleosomal DNA. A decrease in the stability of DNA binding would be expected to enhance access by NER enzymes and thus increase NER efficiency, but the opposite phenotype was observed. Moreover, there were no differences in initial damage levels in either histone mutant strain, and we further showed that the H2A R78A mutant did not decrease the basal expression of any NER genes. Recently, a number of studies have found that histone chaperones such as Spt16 and Nap1 bind to the histone surface that normally interacts with the nucleosomal DNA (D’Arcy et al. 2013; Hondele and Ladurner 2013; Hondele et al. 2013). For example, the H2A R78 sprocket arginine is part of the binding interface with the Spt16 Middle domain (Hondele et al. 2013). Hence, our current working hypothesis is that these histone mutants may be poor substrates for histone chaperones and thus may inhibit chromatin remodeling necessary for efficient NER.
Multiple sprocket arginine mutants, particularly H2A R78A and H3 R49A, were sensitive to the DNA alkylating agent MMS as anticipated from previous studies (Matsubara et al. 2007; Dai et al. 2008, 2010; Huang et al. 2009; Sakamoto et al. 2009). However, none of these mutants affected the efficiency of BER in repairing MMS-induced DNA lesions. Instead, we suspect that the MMS sensitivity may be due to a defect in DNA damage tolerance or signaling. Future studies will be needed to investigate these hypotheses.
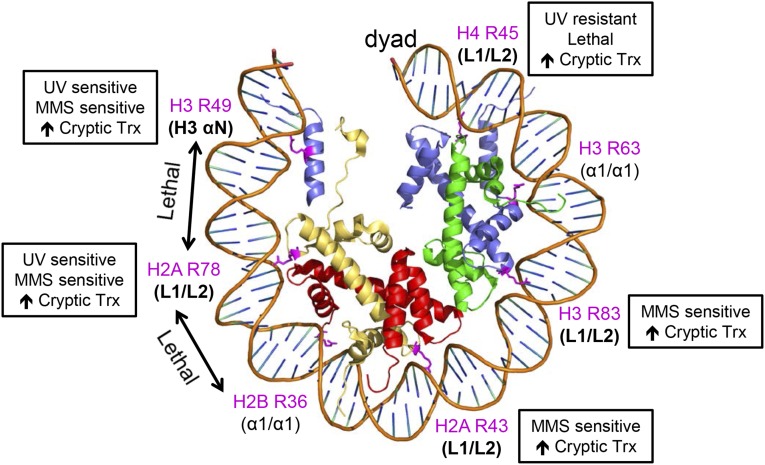
Within the nucleosome, each histone heterodimer binds to ∼27–28 bp of DNA via a central α1/α1-binding motif and two flanking Loop 1–Loop 2 (L1/L2)-binding motifs (Luger et al. 1997). Intriguingly, our phenotypic analysis indicates that the sprocket arginine residues located in L1/L2-binding motifs (i.e., H4 R45, H3 R83, H2A R43, and H2A R78) have important roles in cryptic transcription and MMS sensitivity among other phenotypes (Figure 6). In contrast, sprocket arginine residues located in α1/α1-binding motifs (i.e., H3 R63 and H2B R36) are not required for these biological processes (Figure 6) and appear to be largely dispensable.
Figure 6.
Translational positions of sprocket arginine residues, their associated DNA-binding motifs, and their mutant phenotypes, as identified by this study. The yeast nucleosome structure (1id3) (White et al. 2001) was displayed using PyMOL.
Analysis of published data sets of histone mutant phenotypes (Matsubara et al. 2007; Dai et al. 2008; Huang et al. 2009; Sakamoto et al. 2009) largely confirms these conclusions, with two exceptions: (i) a previous study (Matsubara et al. 2007) did not report an MMS sensitivity for the H2A R43A mutant and (ii) the same group reported that the H3 R63A mutant was MMS sensitive (and also had HU and Spt− phenotypes) (Sakamoto et al. 2009). These discrepancies could be due to differences in genetic background or in the construction of the mutant strains. While we used the same genetic background for all of our sprocket arginine mutants, the other group used different strain backgrounds for the histone H2A/H2B mutants vs. H3/H4 mutants. Moreover, these strains likely had different relative histone dosages since either H2A-H2B or H3-H4 were expressed from a plasmid, while the remaining histones were expressed from the chromosomal loci. In contrast, all of the histone genes were expressed from the same plasmid in our strains. Interestingly, Jef Boeke’s group reported results in agreement with ours (Dai et al. 2008; Huang et al. 2009), namely that the H3 R63A mutant was not sensitive to MMS (or HU) nor had any other reported phenotypes (e.g., telomeric silencing, etc.). Their study used an H3 R63A mutant integrated at its chromosomal locus and thus was presumably expressed at normal levels.
In summary, our data suggest that sprocket arginine residues located in L1/L2-binding motifs tend to have more important biological functions in yeast than sprocket arginines in α1/α1-binding motifs. Moreover, L1/L2 sprocket arginine residues located near the dyad (i.e., H4 R45) or near the DNA exit/entry sites (i.e., H2A R78) have more severe phenotypes when mutated to alanine. It is possible that α1/α1 sprocket arginine residues have important biological functions, but these functions are masked by redundancy with other nearby histone residues. The H3 R49 sprocket arginine residue does not readily fit into this model as it is located in the H3 αN helix. H3 R49 shares many phenotypes with the adjacent H2A R78 sprocket arginine (e.g., UV sensitivity, NER deficiency, MMS sensitivity, etc.), perhaps because both sprocket arginines contact the DNA near the nucleosomal DNA exit/entry sites. Hence, our data suggest that the translational position of the sprocket arginine within the nucleosome, as well as its associated binding motif, appear to strongly influence its biological function.
Supplementary Material
Acknowledgments
We thank David Allis and Linda Breeden for providing yeast strains or plasmids; Michael Smerdon and Peng Mao for helpful comments and suggestions on the manuscript; and Katie Adolphsen, Susan Kim, Anna Lindsay, Travis Ostbye, and Russell Wilson for expert technical assistance. This work was supported by grant no. ES002614 from the National Institute of Environmental Health Sciences (NIEHS). Isaura Gallegos was supported by National Institutes of Health/National Institute of General Medical Sciences through institutional training grant award T32-GM008336. The contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIEHS, NIGMS, or NIH. Microarray data for this study have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus database under accession no. GSE68159.
Footnotes
Communicating editor: M. Hampsey
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.175885/-/DC1.
Literature Cited
- Ahn S. H., Cheung W. L., Hsu J. Y., Diaz R. L., Smith M. M., et al. , 2005. Sterile 20 kinase phosphorylates histone H2B at serine 10 during hydrogen peroxide-induced apoptosis in S. cerevisiae. Cell 120: 25–36. [DOI] [PubMed] [Google Scholar]
- Bespalov V. A., Conconi A., Zhang X., Fahy D., Smerdon M. J., 2001. Improved method for measuring the ensemble average of strand breaks in genomic DNA. Environ. Mol. Mutagen. 38: 166–174. [DOI] [PubMed] [Google Scholar]
- Cheung V., Chua G., Batada N. N., Landry C. R., Michnick S. W., et al. , 2008. Chromatin- and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome. PLoS Biol. 6: e277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaja W., Bespalov V. A., Hinz J. M., Smerdon M. J., 2010. Proficient repair in chromatin remodeling defective ino80 mutants of Saccharomyces cerevisiae highlights replication defects as the main contributor to DNA damage sensitivity. DNA Repair (Amst.) 9: 976–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J., Hyland E. M., Yuan D. S., Huang H., Bader J. S., et al. , 2008. Probing nucleosome function: a highly versatile library of synthetic histone H3 and H4 mutants. Cell 134: 1066–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J., Hyland E. M., Norris A., Boeke J. D., 2010. Yin and Yang of histone H2B roles in silencing and longevity: a tale of two arginines. Genetics 186: 813–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Arcy S., Martin K. W., Panchenko T., Chen X., Bergeron S., et al. , 2013. Chaperone Nap1 shields histone surfaces used in a nucleosome and can put H2A–H2B in an unconventional tetrameric form. Mol. Cell 51: 662–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey C. A., Sargent D. F., Luger K., Maeder A. W., Richmond T. J., 2002. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. J. Mol. Biol. 319: 1097–1113. [DOI] [PubMed] [Google Scholar]
- Ferreira H., Somers J., Webster R., Flaus A., Owen-Hughes T., 2007. Histone tails and the H3 alphaN helix regulate nucleosome mobility and stability. Mol. Cell. Biol. 27: 4037–4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaus A., Rencurel C., Ferreira H., Wiechens N., Owen-Hughes T., 2004. Sin mutations alter inherent nucleosome mobility. EMBO J. 23: 343–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry C. J., Norris A., Cosgrove M., Boeke J. D., Peterson C. L., 2006. The LRS and SIN domains: two structurally equivalent but functionally distinct nucleosomal surfaces required for transcriptional silencing. Mol. Cell. Biol. 26: 9045–9059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman R. C., Carey V. J., Bates D. M., Bolstad B., Dettling M., et al. , 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie C. S., Lei G., Boys R. J., Greenall A., Wilkinson D. J., 2010. Analysing time course microarray data using Bioconductor: a case study using yeast2 Affymetrix arrays. BMC Res. Notes 3: 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hainer S. J., Martens J. A., 2011. Identification of histone mutants that are defective for transcription-coupled nucleosome occupancy. Mol. Cell. Biol. 31: 3557–3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harp J. M., Hanson B. L., Timm D. E., Bunick G. J., 2000. Asymmetries in the nucleosome core particle at 2.5 A resolution. Acta Crystallogr. D Biol. Crystallogr. 56: 1513–1534. [DOI] [PubMed] [Google Scholar]
- Hondele M., Ladurner A. G., 2013. Catch me if you can: how the histone chaperone FACT capitalizes on nucleosome breathing. Nucleus 4: 443–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hondele M., Stuwe T., Hassler M., Halbach F., Bowman A., et al. , 2013. Structural basis of histone H2A–H2B recognition by the essential chaperone FACT. Nature 499: 111–114. [DOI] [PubMed] [Google Scholar]
- Horn P. J., Crowley K. A., Carruthers L. M., Hansen J. C., Peterson C. L., 2002. The SIN domain of the histone octamer is essential for intramolecular folding of nucleosomal arrays. Nat. Struct. Biol. 9: 167–171. [DOI] [PubMed] [Google Scholar]
- Huang H., Maertens A. M., Hyland E. M., Dai J., Norris A., et al. , 2009. HistoneHits: a database for histone mutations and their phenotypes. Genome Res. 19: 674–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamai A., Imoberdorf R. M., Strubin M., 2007. Continuous histone H2B and transcription-dependent histone H3 exchange in yeast cells outside of replication. Mol. Cell 25: 345–355. [DOI] [PubMed] [Google Scholar]
- Jin Y., Rodriguez A. M., Stanton J. D., Kitazono A. A., Wyrick J. J., 2007. Simultaneous mutation of methylated lysine residues in histone H3 causes enhanced gene silencing, cell cycle defects, and cell lethality in Saccharomyces cerevisiae. Mol. Cell. Biol. 27: 6832–6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger W., Peterson C. L., Sil A., Coburn C., Arents G., et al. , 1995. Amino acid substitutions in the structured domains of histones H3 and H4 partially relieve the requirement of the yeast SWI/SNF complex for transcription. Genes Dev. 9: 2770–2779. [DOI] [PubMed] [Google Scholar]
- Kurumizaka H., Wolffe A. P., 1997. Sin mutations of histone H3: influence on nucleosome core structure and function. Mol. Cell. Biol. 17: 6953–6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriss M. N., Jin Y., Gallegos I. J., Sanford J. A., Wyrick J. J., 2010. Novel functional residues in the core domain of histone H2B regulate yeast gene expression and silencing and affect the response to DNA damage. Mol. Cell. Biol. 30: 3503–3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenstra T. L., Benschop J. J., Kim T., Schulze J. M., Brabers N. A., et al. , 2011. The specificity and topology of chromatin interaction pathways in yeast. Mol. Cell 42: 536–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K., Mader A. W., Richmond R. K., Sargent D. F., Richmond T. J., 1997. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389: 251–260. [DOI] [PubMed] [Google Scholar]
- Matsubara K., Sano N., Umehara T., Horikoshi M., 2007. Global analysis of functional surfaces of core histones with comprehensive point mutants. Genes Cells 12: 13–33. [DOI] [PubMed] [Google Scholar]
- Muthurajan U. M., Park Y. J., Edayathumangalam R. S., Suto R. K., Chakravarthy S., et al. , 2003. Structure and dynamics of nucleosomal DNA. Biopolymers 68: 547–556. [DOI] [PubMed] [Google Scholar]
- Muthurajan U. M., Bao Y., Forsberg L. J., Edayathumangalam R. S., Dyer P. N., et al. , 2004. Crystal structures of histone Sin mutant nucleosomes reveal altered protein-DNA interactions. EMBO J. 23: 260–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag R., Smerdon M. J., 2009. Altering the chromatin landscape for nucleotide excision repair. Mutat. Res. 682: 13–20. [DOI] [PubMed] [Google Scholar]
- Nag R., Gong F., Fahy D., Smerdon M. J., 2008. A single amino acid change in histone H4 enhances UV survival and DNA repair in yeast. Nucleic Acids Res. 36: 3857–3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag R., Kyriss M., Smerdon J. W., Wyrick J. J., Smerdon M. J., 2010. A cassette of N-terminal amino acids of histone H2B are required for efficient cell survival, DNA repair and Swi/Snf binding in UV irradiated yeast. Nucleic Acids Res. 38: 1450–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi S., Sanderson B. W., Delventhal K. M., Bradford W. D., Staehling-Hampton K., et al. , 2008. A comprehensive library of histone mutants identifies nucleosomal residues required for H3K4 methylation. Nat. Struct. Mol. Biol. 15: 881–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor T. R., Wyrick J. J., 2007. ChromatinDB: a database of genome-wide histone modification patterns for Saccharomyces cerevisiae. Bioinformatics 23: 1828–1830. [DOI] [PubMed] [Google Scholar]
- Park J. H., Cosgrove M. S., Youngman E., Wolberger C., Boeke J. D., 2002. A core nucleosome surface crucial for transcriptional silencing. Nat. Genet. 32: 273–279. [DOI] [PubMed] [Google Scholar]
- Parra M. A., Kerr D., Fahy D., Pouchnik D. J., Wyrick J. J., 2006. Deciphering the roles of the histone H2B N-terminal domain in genome-wide transcription. Mol. Cell. Biol. 26: 3842–3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. D., Grigull J., Mohammad N., Hughes T. R., 2002. FunSpec: a web-based cluster interpreter for yeast. BMC Bioinformatics 3: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohs R., West S. M., Sosinsky A., Liu P., Mann R. S., et al. , 2009. The role of DNA shape in protein-DNA recognition. Nature 461: 1248–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, M. D., F. Winston, and P. Hieter, 1990 Methods in Yeast Genetics: A Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Sakamoto M., Noguchi S., Kawashima S., Okada Y., Enomoto T., et al. , 2009. Global analysis of mutual interaction surfaces of nucleosomes with comprehensive point mutants. Genes Cells 14: 1271–1330. [DOI] [PubMed] [Google Scholar]
- Smyth, G. K., 2005 Limma: linear models for microarray data, pp. 397–420 in Bioinformatics and Computational Biology Solutions Using R and Bioconductor, edited by R. Gentleman, V. Carey, S. Dut, R. Irizarry, and W. Huber, Springer, New York. [Google Scholar]
- Sullivan S. A., Landsman D., 2003. Characterization of sequence variability in nucleosome core histone folds. Proteins 52: 454–465. [DOI] [PubMed] [Google Scholar]
- van Wageningen S., Kemmeren P., Lijnzaad P., Margaritis T., Benschop J. J., et al. , 2010. Functional overlap and regulatory links shape genetic interactions between signaling pathways. Cell 143: 991–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Ulyanov N. B., Zhurkin V. B., 2010. Sequence-dependent Kink-and-Slide deformations of nucleosomal DNA facilitated by histone arginines bound in the minor groove. J. Biomol. Struct. Dyn. 27: 843–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechser M. A., Kladde M. P., Alfieri J. A., Peterson C. L., 1997. Effects of Sin- versions of histone H4 on yeast chromatin structure and function. EMBO J. 16: 2086–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S. M., Rohs R., Mann R. S., Honig B., 2010. Electrostatic interactions between arginines and the minor groove in the nucleosome. J. Biomol. Struct. Dyn. 27: 861–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White C. L., Suto R. K., Luger K., 2001. Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. EMBO J. 20: 5207–5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S., Wyrick J. J., Reese J. C., 2010. Novel trans-tail regulation of H2B ubiquitylation and H3K4 methylation by the N terminus of histone H2A. Mol. Cell. Biol. 30: 3635–3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.