Abstract
Germline cells segregate from the soma to maintain their totipotency, but the cellular mechanisms of this segregation are unclear. The Drosophila melanogaster embryo forms a posterior group of primordial germline cells (PGCs) by their division from the syncytial soma. Extended plasma membrane furrows enclose the PGCs in response to the germ plasm protein Germ cell-less (Gcl) and Rho1–actomyosin activity. Recently, we found that loss of the Arf-GEF Steppke (Step) leads to similar Rho1-dependent plasma membrane extensions but from pseudocleavage furrows of the soma. Here, we report that the loss of step also leads to premature formation of a large cell group at the anterior pole of the embryo . These anterior cells lacked germ plasm, but budded and formed at the same time as posterior PGCs, and then divided asynchronously as PGCs also do. With genetic analyses we found that Step normally activates Arf small G proteins and antagonizes Rho1–actomyosin pathways to inhibit anterior cell formation. A uniform distribution of step mRNA around the one-cell embryo cortex suggested that Step restricts cell formation through a global control mechanism. Thus, we examined the effect of Step on PGC formation at the posterior pole. Reducing Gcl or Rho1 levels decreased PGC numbers, but additional step RNAi restored their numbers. Reciprocally, GFP–Step overexpression induced dosage- and Arf-GEF-dependent loss of PGCs, an effect worsened by reducing Gcl or actomyosin pathway activity. We propose that a global distribution of Step normally sets an inhibitory threshold for Rho1 activity to restrict early cell formation to the posterior.
Keywords: germline segregation, cell division, Arf-GEF activity, Rho1 inhibition, Drosophila
EMBRYONIC specification of primordial germline cells (PGCs) distinguishes them from somatic cells to maintain the totipotency of the germline (Seydoux and Braun 2006; Hayashi et al. 2007; Strome and Lehmann 2007; Johnson et al. 2011). Although this segregation occurs across animals, its cellular bases remain unclear.
The early Drosophila embryo segregates the germline from the soma through an extreme form of asymmetric cell division. The very early embryo is a syncytium of dividing nuclei. At nuclear cycle 9, a group of nuclei are recruited from the subcortex to the posterior pole of the syncytium, and each induces transient, shallow, dome-like buds at the embryo surface. During nuclear cycle 10, these posterior cells bud again and then divide fully from the remaining syncytium. This asymmetric division forms the PGCs at the posterior pole of the embryo. The remaining somatic nuclei continue dividing as a syncytium until 13 rounds of nuclear division are complete, at which point they too divide into separate cells through the process of cellularization that forms the blastoderm (Foe and Alberts 1983).
The asymmetric division of PGCs from the soma is dictated by germ plasm deposited maternally at the posterior pole (Wilson and Macdonald 1993; Lehmann and Ephrussi 1994; Mahowald 2001). Specifically, the germ plasm protein Germ cell-less (Gcl) promotes activity of Rho1 and downstream actomyosin pathways to form extended plasma membrane furrows that encase single PGCs laterally and then basally (Cinalli and Lehmann 2013). Once the lateral membranes form, their basal tips expand perpendicularly to form basal membranes beneath each nucleus. These basal membranes have been termed "bud furrows" and are coated with cytoskeletal networks composed of actin, nonmuscle myosin II, and Anillin. These furrows form independently of spindles, and other than positive roles for Gcl and Rho1 (Cinalli and Lehmann 2013), it is unclear how bud furrows form and what prevents their formation elsewhere around the embryo.
We recently reported that early embryo depletion of the plasma membrane Arf-guanine nucleotide exchange factor (Arf-GEF) Steppke (Step) leads to premature basal membrane formation, but for pseudocleavage furrows that transiently separate somatic, syncytial nuclei (Lee and Harris 2013). Without Step, these abnormal basal membranes have dramatic effects at equatorial (nonpolar) regions of the embryo. They sporadically capture nuclei to form single cells, but also displace nuclei from the syncytial blastoderm into the yolk below. Despite their random and disruptive effects on equatorial somatic nuclei, we were struck by how similar these abnormal basal membranes are to PGC bud furrows: (1) they each have a similar architecture relative to lateral membrane domains, (2) they each are coated with actin, nonmuscle myosin II, and Anillin, and (3) they each form through Rho1–actomyosin activity. Thus, we hypothesized that PGC formation may not depend solely on induction by the germ plasm, but that Step’s inhibition of Rho1 additionally controls where the asymmetric division occurs. This hypothesis invokes a commonly used mechanism of pattern formation, the combination of local activation with global inhibition (Turing 1952; Gierer and Meinhardt 1972; Roussos et al. 2011; Chau et al. 2012; Fletcher et al. 2012).
Step is the sole Drosophila member of the cytohesin Arf-GEF family. Cytohesins localize to the plasma membrane and activate Arf small G proteins. In response, Arf small G proteins induce proximal signals that trigger endocytosis and other effects (D’souza-Schorey and Chavrier 2006; Gillingham and Munro 2007; Donaldson and Jackson 2011). We found that Step is enriched at the base of somatic plasma membrane furrows and regulates their structure through its Arf-GEF activity in cooperation with the clathrin adaptor complex, AP-2. These data suggested that a local, Step-dependent, endocytic pathway regulates somatic furrows, and an increase of furrow Rho1 protein levels with step loss suggested that Rho1, or an associated protein, might be the endocytic target (Lee and Harris 2013).
Here, we tested whether Step regulates PGC segregation from the soma by comparing the two poles of the embryo with step loss or gain. Strikingly, step RNAi or mutant embryos formed structurally similar cell groups at both the posterior and anterior poles. The anterior group lacked germ plasm but formed and behaved as PGCs initially do. Not only does Step inhibit this anterior cell formation, but we found that it also hinders normal PGC formation. In each case, Step seems to use its Arf-GEF activity to antagonize Rho1–actomyosin pathways. We additionally documented a uniform expression of Step across the embryo. Thus, Step seems to normally set a global inhibitory threshold for Rho1 activity, and one major effect of this threshold is the restriction of cell formation to the posterior pole.
Materials and Methods
Drosophila genetics and molecular reagents
Mutant alleles included: dia5 (Bloomington Drosophila Stock Center, BDSC, no. 9138); gcl∆ (gift of R. Lehmann, New York University School of Medicine, New York, NY); gclDf(2R)BSC269 and gclDf(2R)Exel7098 (BDSC nos. 23165 and 7864); rho172O and rho172F (BDSC nos. 7325 and 7326); stepK08110 and stepSH0323 (gifts of M. Hoch, Life and Medical Science Institute of Bonn, Germany); and zip1 (BDSC no. 4199). UAS constructs included: UAS–GFP-Step and UAS–GFP-StepE173K as well as corresponding RNAi-resistant forms (Lee and Harris 2013); UAS–GFP (gift of U. Tepass, University of Toronto, Toronto, Canada); UAS–step-shRNA (P[VALIUM20-TRiP.HMS00365]attP2; BDSC no. 32374); and UAS–mcherry-shRNA (P[VALIUM20-mCherry]attp2; BDSC no. 35785). UAS constructs were expressed maternally using maternal-α4-tubulin–GAL4-VP16 (gift of M. Peifer, University of North Carolina, Chapel Hill, NC) and defects were assessed in offspring. y w embryos were used as a control (gift of M. Peifer).
All complex genotypes were synthesized using standard Drosophila genetics and the presence of alleles and transgenes was confirmed after synthesis by probing for their expected phenotypes in single disruption analyses.
Embryo staining and Imaging
Embryos were fixed for 20 min in 1:1 3.7% formaldehyde in PBS:heptane and then devitellinized in methanol. Blocking and staining were in PBS containing 1% goat serum, 0.1% Triton X-100, and 1% sodium azide. Antibodies used were rabbit, Amphiphysin (1:2000; gift of G. Boulianne, Hospital for Sick Children, Toronto, Canada), Anillin (1:500; gift of J. Brill, Hospital for Sick Children, Toronto, Canada), Step (1:350; gift of M. Hoch); chicken, Vasa (1:2000; gift of K. Howard care of M. van Doren, Johns Hopkins University, USA); mouse, Discs large (Dlg) (1:100; Developmental Studies Hybridoma Bank (DSHB), Peanut (1:10; DSHB)), phospho-Histone H3 (1:2000; Cell Signaling Technology, Beverly, MA). Following a 10-min, 1:1 10% formaldehyde/PBS:heptane fixation and hand peeling devitellinization, F-actin was stained with Alexa Fluor 568-conjugated phalloidin (1:200; Life Technologies, Burlington, Canada). Secondary antibodies were conjugated to Alexa Fluor 488, Alexa Fluor 546, and Alexa Fluor 647 (Life Technologies). Embryos were mounted in Aqua Polymount (Polysciences).
Immunofluorescent images were collected by a spinning disk confocal system (Quorum Technologies, Guelph, Canada) at room temperature using 10× EC Plan-Neofluar NA 0.3, 40× Plan Neofluar NA 1.3, and 63× Plan Apochromat NA 1.4 objectives (Carl Zeiss, Toronto, Canada) with a piezo top plate and an EM CCD camera (Hamamatsu Photonics, Hamamatsu, Japan), where z-stacks had 1-µm and 300-nm step sizes, respectively. These images were analyzed with Volocity software (PerkinElmer, Waltham, MA) and Imaris 6.2 software (Bitplane, Zurich, Switzerland). Live phase-contrast imaging was performed at a single focal plane, at room temperature, with a BX51 microscope and a UPlanFl 20×/0.50 Ph1 objective (Olympus, Richmond Hill, Canada), an Evolution VF camera (Media Cybernetics, Rockville, MD), and QCapturePro software (QImaging, Surrey, Canada).
Photoshop (Adobe, Mountain View, CA) was used for figure preparation. Except where noted, input levels were adjusted so the main signal range spanned the entire output grayscale. Images were resized by bicubic interpolation without noticeable changes at normal viewing magnifications.
Fluorescent in situ hybridization
y w embryos were fixed, and hybridized as described as in Wilk et al. (2010) with the probe RE34385 for step (FlyFISH database, http://fly-fish.ccbr.utoronto.ca). Probe production was described in Lecuyer et al. (2008) and Wilk et al. (2010). Embryos were imaged as described above. There was no detectable signal in negative controls lacking the probe.
Quantification of GFP-Step expression levels
For each cellularization embryo with furrow depths of 2–7 µm, mean intensities of GFP–Step were determined in 0.8-µm circles for the top five brightest regions at similarly sized furrow canals from five distinct intact somatic cell compartments across the field of view using Image J. For each measurement, a background measurement was taken at a neighboring nuclear region in the same way and then subtracted from the furrow canal measurement. These background-corrected measurements were averaged to assign an overall metric for each embryo. The embryo metrics were then normalized relative to the highest value in the experiment, graphed, and compared.
Statistics
Comparisons were done using Student’s t-tests. Means are shown with standard deviation.
Results
Without Step an anterior cell group forms independently of germ plasm
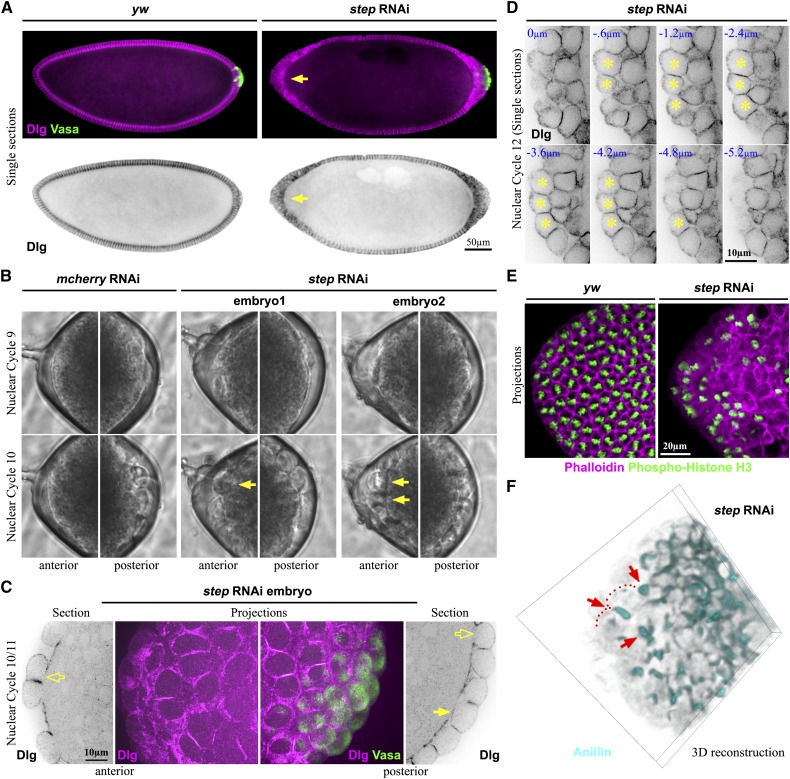
We previously reported that depletion of maternal step gene product leads to abnormal expansion of basal membranes in equatorial (nonpolar) regions of the soma during the syncytial divisions of the early Drosophila embryo (Lee and Harris 2013). In these regions, the expanded basal membranes displaced nuclei into the yolk below or toward the embryo surface—occasionally forming single, isolated, nucleated cells (Lee and Harris 2013). In analyzing step RNAi and mutant embryos further, we noted a striking abnormality at the anterior pole. Here, in contrast to controls, a large group of cells formed in nearly all of the embryos (Figure 1A and Figure 2C). Since a large group of PGCs normally forms at the posterior pole of the embryo under control of the germ plasm, we tested whether the germ plasm marker Vasa accumulated anteriorly with the loss of Step. However, the abnormal group of anterior cells was devoid of Vasa, in contrast to the posterior of control and step RNAi embryos (Figure 1A). Comparing the posterior poles further revealed Vasa-negative cells surrounding the PGCs with step loss, but not in control embryos (Figure 1A). Thus, Step plays an important role in preventing large groups of cells from forming at the poles of the syncytial Drosophila embryo, regardless of the presence or absence of germ plasm.
Figure 1.
A cell group forms abnormally at the anterior pole of step RNAi embryos. (A) Staining for the plasma membrane marker Discs large (Dlg) shows a cell group formed at the anterior of a step RNAi embryo in contrast to control (arrow) at blastoderm cellularization. The germ plasm marker Vasa shows that this abnormal cell group lacks germ plasm in contrast to the posterior PGCs. For each embryo, three separate images were stitched together, and signal intensities were adjusted to be equal across the composite image. (B) Live phase-contrast imaging showing the budding and formation of abnormal anterior cells in step RNAi embryos with the same timing as PGCs in step RNAi and control embryos. The anterior (left) and posterior (right) ends of the same embryos at nuclear cycles 9 and 10 are shown (frames from File S1 and File S2). (C) Imaging of a step RNAi embryo showing that the abnormal anterior cells lack full basal membranes at nuclear cycle 10 (hollow arrow), similar to some cells at the posterior (hollow arrow) although other posterior cells are fully separated from the soma (solid arrow). (D) Confocal sectioning shows that by nuclear cycle 12 the abnormal anterior cells of step RNAi embryos are fully enclosed by plasma membrane (stained with Dlg). (E) Phosphohistone H3 staining of mitotic chromosomes shows the asynchronous cell cycling of the abnormal anterior cells of step RNAi embryos, in contrast to the synchronous divisions of wild-type syncytial somatic nuclei, left. (F) A 3D rendering of Anillin staining shows that the abnormal anterior cells of step RNAi embryos form individual cytokinetic rings (arrows). The cortex on either side of the Anillin-positive ring is indicated with dots for one dividing cell.
Figure 2.
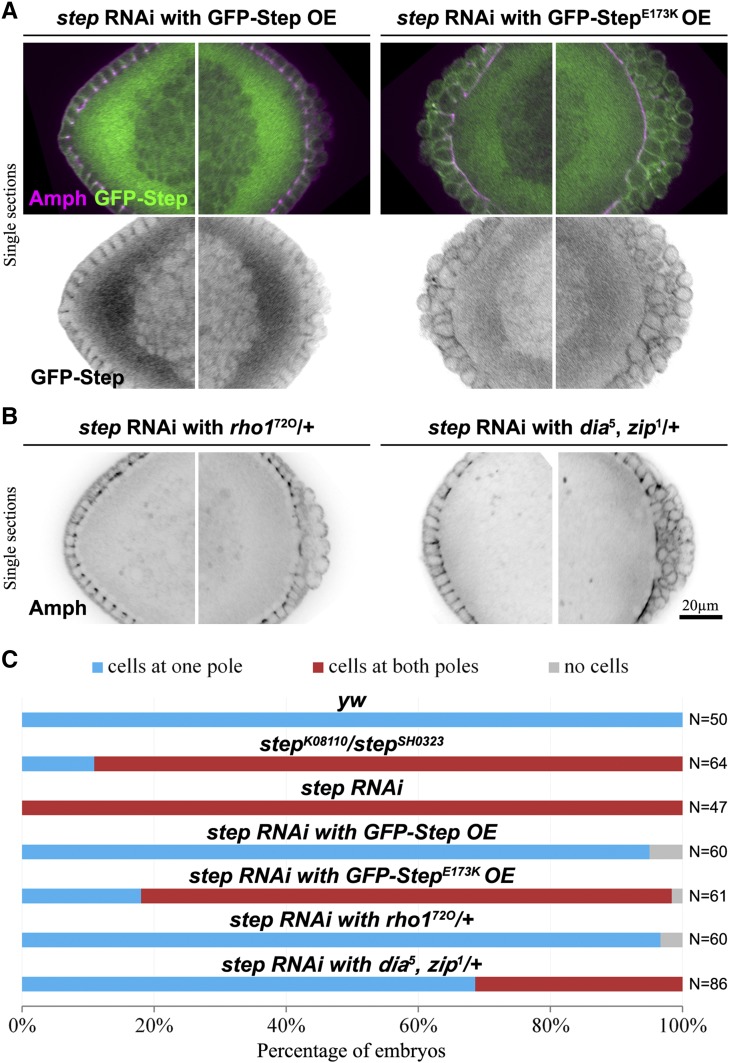
Step prevents anterior cell formation by activating Arf G proteins and antagonizing Rho1. (A) Embryos in which the step RNAi abnormal anterior cell formation is rescued by a GFP–Step construct but not by a GEF–dead construct (StepE173K). Amphiphysin (Amph) staining marks plasma membranes and the plasma membrane localization of the Step constructs is shown with their GFP tags. The anterior (left) and posterior (right) ends of the same embryos are shown at early blastoderm cellularization. (B) Embryos in which the step RNAi abnormal anterior cell formation is suppressed by reducing the levels of Rho1 or Dia and Zip. The anterior (left) and posterior (right) ends of the same embryos are shown. (C) Quantifications of the abnormal anterior cell formation for control, step mutant, step RNAi, rescue attempts, and suppression experiments. Embryos at nuclear cycle 13 or early blastoderm cellularization with depth of 2–7 µm were counted.
The abnormal anterior cells form and behave as posterior PGCs normally do
To determine whether the mechanism underlying the abnormal anterior cell formation was similar to that of normal PGC formation, we compared their initial formation by phase contrast, time-lapse microscopy. In 5/5 control embryos, our results were consistent with previous observations of posterior pole buds forming at nuclear cycle 9 and PGCs forming at nuclear cycle 10, with no observable cells formed at the anterior pole at these stages (Figure 1B and Supporting Information, File S1; Foe and Alberts 1983). Strikingly, in 2/5 step RNAi embryos, formation of shallow buds at nuclear cycle 9 and then round cells at nuclear cycle 10 was apparent at both the anterior and posterior poles with the same timing (Figure 1B, arrows indicate the ectopic cells, and File S2). For the remaining 3/5 step RNAi embryos, anterior buds and round cells formed during later nuclear cycles but still prior to normal blastoderm cellularization (data not shown). To examine the early abnormal cells at higher resolution, we stained step RNAi embryos with the plasma membrane marker Dlg and performed confocal imaging. These analyses revealed that the cycle 10, round, anterior cells had not yet formed full basal membranes (Figure 1C, hollow arrow), similar to some cells at the posterior (Figure 1C, hollow arrow; compare with fully formed PGCs, solid arrow). These data suggest an early action of Step to prevent initial stages of cell formation at nuclear cycles 9 and 10.
To test if the abnormal anterior cell compartments became functional, single cells, we analyzed their subsequent structure and behavior. Using Dlg as a plasma membrane marker, we found that the abnormal anterior cells became fully membrane enclosed by nuclear cycle 12 (Figure 1D). With full membrane enclosure, a loss of syncytial behavior would be expected. By labeling with the mitotic chromosomal marker phosphohistone H3, we found that, in contrast to the synchronous nuclear cycling of the wild-type syncytial soma, asynchronous nuclear cycling occurred in the abnormal anterior cell group (Figure 1E). Finally, by labeling cytoskeletal markers (Anillin and the Drosophila septin, Peanut) we found evidence for individual cytokinetic rings scattered within the anterior cell groups (Figure 1F and Figure S1), suggestive of single cell divisions, as occurs for PGCs after they normally form (Technau and Campos-Ortega 1986). Overall, these results indicate that Step normally prevents a group of single, functional cells from forming at the anterior pole of the early embryo.
Step activates Arf small G proteins to prevent anterior cell formation
Since Step is a member of the cytohesin Arf-GEF family, which activates Arf small G proteins at the plasma membrane (D’souza-Schorey and Chavrier 2006; Gillingham and Munro 2007; Donaldson and Jackson 2011), we hypothesized that Step prevents anterior cell formation through Arf G proteins. To test this possibility, we expressed RNAi-resistant GFP-Step constructs in step RNAi embryos and analyzed them during blastoderm cellularization when step RNAi embryos can be most accurately staged. Expressing an active form of the GFP-Step construct restored the embryo morphology of step RNAi embryos (95% of embryos displayed a cell group only at one pole; Figure 2, A and C). In contrast, expression of RNAi-resistant GFP-Step with a mutation inactivating its Arf-GEF domain (GFP-StepE173K; Cherfils et al. 1998; Mossessova et al. 1998) failed to prevent the abnormal anterior cell formation with step RNAi (80% of embryos displayed cell groups at both poles; Figure 2, A and C). Significantly, both constructs localized to the plasma membrane of the cell compartments involved (Figure 2A). Thus, Step appears to act at the plasma membrane to activate Arf small G proteins for the inhibition of abnormal cell formation at the anterior pole.
Step antagonizes Rho1 to prevent anterior cell formation
Previously, we found that Rho1 pathways were responsible for the abnormal expansion of basal membranes in equatorial regions of step loss-of-function embryos (Lee and Harris 2013). Since the Rho1–actomyosin pathway also drives normal PGC formation (Cinalli and Lehmann 2013), we hypothesized that Rho1–actomyosin pathways might also be responsible for the abnormal anterior cell formation with step loss. Thus, we pursued suppression experiments. Maternal heterozygosity for rho172O dramatically suppressed the abnormal cell formation of step RNAi embryos (97% of these embryos displayed a cell group only at one pole; Figure 2, B and C). Combined heterozygosity for mutant alleles of the Drosophila formin diaphanous (dia) and non-muscle myosin heavy chain, zipper (zip), encoding downstream effectors of Rho1, had a similar but milder effect (Figure 2, B and C). Together, these data argue that Step antagonizes Rho1–actomyosin pathways to restrict early cell formation to the embryo posterior.
Step is expressed uniformly across the syncytial soma and posterior pole
Since Step inhibits cell formation and Rho1 promotes cell formation, we hypothesized that a regional difference in their relative activities could dictate normal PGC formation. We considered three models. At one extreme, Step inhibition of Rho1 could be high in the soma and absent at the posterior pole, with equivalent Rho1 activation across the embryo. At the other extreme, Step inhibition of Rho1 could be uniform across the embryo, with greater Rho1 activation at the posterior pole. A third model would be a combination of both differences. In all three cases, the result would be greater Rho1–actomyosin pathway output at the furrows of presumptive PGCs and thus asymmetric division of the syncytial Drosophila embryo.
To begin to test these models, we pursued the expression pattern of step gene products. An antibody shown to specifically detect Step in larval tissues (Hahn et al. 2013) and the later embryo (our unpublished observations) detected no signal above background in the syncytial embryo (data not shown), suggesting that Step protein levels are relatively low at this stage. Thus, we examined the localization of step mRNA by fluorescent in situ hybridization. Focusing specifically on the developmental stages when PGCs bud and first form, we detected no differences in step mRNA localization around the full embryo periphery. Higher magnification confocal imaging of the embryo posterior verified that there was no detectable difference in step mRNA levels or localization between forming PGCs and neighboring, peripheral, somatic regions (Figure S2). These data suggested a model in which Step inhibition of Rho1 is equivalent across the soma and the posterior pole.
Step antagonizes Rho1 pathways during posterior PGC formation
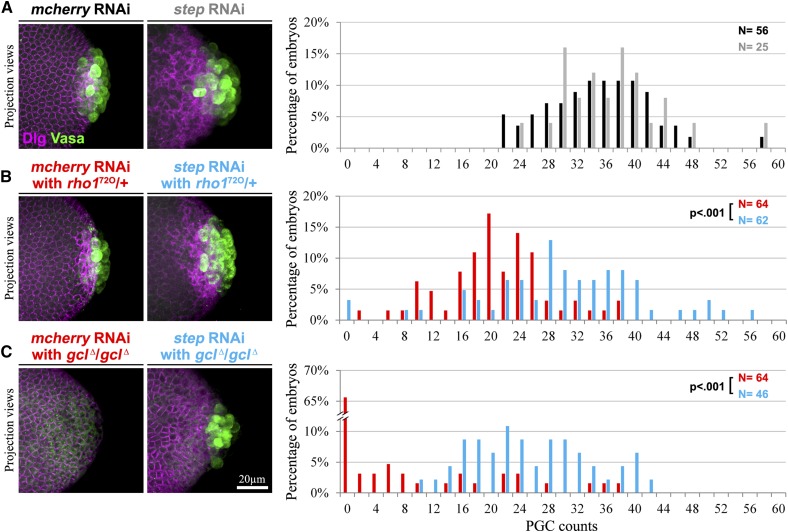
To test whether Step activity affects the posterior pole, we examined genetic interactions between step and rho1 or gcl, the two genes known to promote the bud furrows of PGCs (Cinalli and Lehmann 2013). For rho1, we noted that maternal heterozygosity for either of two null alleles (rho172O and rho172F) led to a substantial reduction of PGC numbers, as marked by Vasa, vs. control embryos (Figure 3, A and B, and Figure S3A). We found that these PGC numbers were reduced both at blastoderm cellularization (Figure 3, A and B) and at nuclear cycles 11 and 12 (Figure S4), suggesting that the rho1 heterozygotes had fewer PGCs due to defective PGC formation, rather than defects in later PGC division or maintenance. If Step antagonizes Rho1 during normal PGC formation, then two factors might contribute to the reduced ability of Rho1 to promote PGC formation in these heterozygotes: the genetic reduction of rho1 gene product and inhibition from Step. To test the effect of Step, we reduced its levels by RNAi in the rho1 heterozygotes and found a restoration of PGC numbers vs. controls at both blastoderm cellularization and at nuclear cycles 11 and 12 (Figure 3, A and B, and Figure S4). Similarly, we found that step RNAi restored PGC numbers in homozygous gcl null mutants observed at blastoderm cellularization (Figure 3, A and C). Notably, rho1 heterozygotes at nuclear cycle 11 (Figure S4), and most gcl mutants at blastoderm cellularization (Figure 3C; Robertson et al. 1999), had no PGCs, and thus the recovery of PGC numbers with the reduction of Step further confirms a restoration of PGC formation in each case. Together, these data indicate that Step hinders Rho1–actomyosin activity during normal PGC formation.
Figure 3.
Endogenous Step antagonizes Rho1–actomyosin pathways during PGC formation. (A–C) PGCs (marked with Vasa) are shown and quantified during early blastoderm cellularization. Dlg indicates plasma membranes. (A) Similar numbers of PGCs are found in control RNAi (solid) and step RNAi (shaded) embryos. (B) Reduced PGC numbers in rho1 heterozygotes (red) are restored with step RNAi (blue). (C) Reduced or absent PGC numbers in gcl homozygotes (red) are restored with step RNAi (blue).
Elevated global Step Arf-GEF activity blocks PGC formation
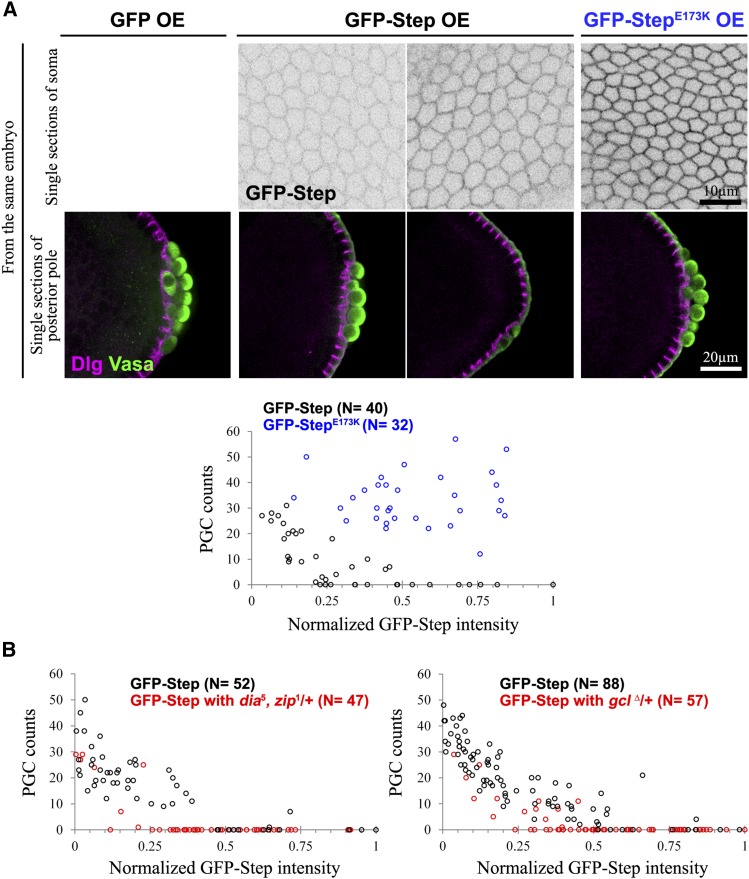
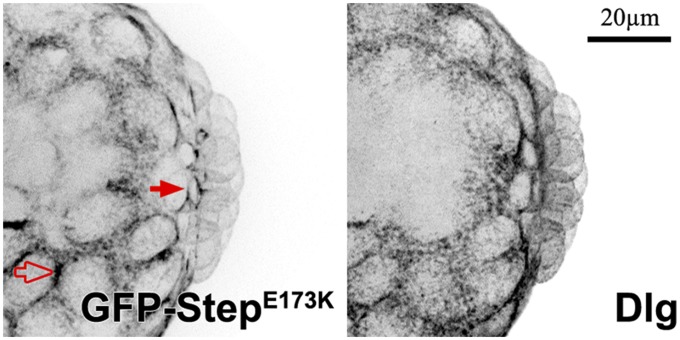
Our loss-of-function and step expression studies suggested a model in which global Step activity sets an inhibitory threshold for the Rho1 activity needed for PGC formation. To test this model, we overexpressed GFP–Step globally and examined effects on PGC numbers. This GFP–Step overexpression inhibited PGC formation, whereas GEF–dead GFP–StepE173K overexpression did not (Figure 4A). To test the responsiveness of the system to Step overexpression, we compared GFP–Step plasma membrane levels and PGC numbers embryo-by-embryo. This analysis revealed that the effect of GFP–Step on PGC numbers was dose dependent, whereas the same range of GFP–StepE173K expression had no effect (Figure 4A). To determine where the constructs acted within PGCs during their formation we imaged the nondisruptive GFP–StepE173K. In forming PGCs, this protein was strongly enriched at the basal tips of lateral plasma membrane furrows (Figure 5), where actomyosin activity normally promotes bud furrow formation (Cinalli and Lehmann 2013). Together, these observations indicate that Step acts through Arf small G proteins at the plasma membrane to restrict PGC formation. It also seems that factors for the localization and activity of these Step constructs are not limiting in the PGCs.
Figure 4.
Excess Step can block PGC formation dose-dependently by activating Arf G proteins. (A and B) PGCs (marked with Vasa) are shown and quantified during early blastoderm cellularization. Dlg indicates plasma membranes. (A) Increasing GFP–Step levels (imaged in equatorial regions of the soma, top) correlates with decreased PGC numbers, but the same range of GEF–dead GFP–StepE173K had no effect (graphed below; the PGC counts were significantly different (P < 0.001)) (B) Reducing Rho1–actomyosin pathway activity through heterozygosity for dia and zip, or gcl, enhanced the inhibition of PGC numbers by GFP–Step (in each case the reductions in PGC counts were significant; P < 0.001).
Figure 5.
Evidence for Step localization to the bud furrows of PGCs. GFP–StepE173K localizes to the bud furrows of forming PGCs (solid arrows) at a local level similar to that of surrounding somatic plasma membranes (open arrows) and lower than that of other regions of PGC plasma membranes. Dlg is shown as a separate plasma membrane marker.
To test whether the inhibitory effects of Step overexpression are related to Rho1–actomyosin pathways, we evaluated the effects of Step overexpression in two contexts in which Rho1–actomyosin pathways were weakened but had no effects on PGC numbers on their own. Specifically, maternal heterozygosity for mutant alleles of both dia and zip, or single maternal heterozygosity for null alleles of gcl, had minimal effect on PGC numbers alone (Figure S3, B and C). However, these perturbations enhanced the inhibitory effects of GFP–Step overexpression on PGC numbers (Figure 4B and Figure S5). These data further indicate that Step controls cell formation by setting a threshold for Rho1–actomyosin activity.
Discussion
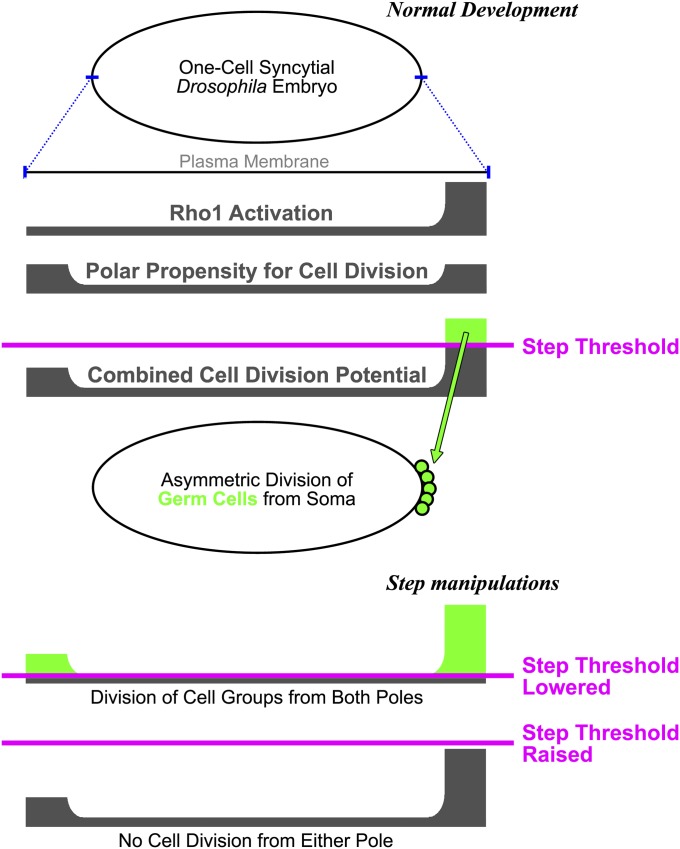
To contribute fully to the next generation, PGCs segregate from the influences of somatic cells. In the early Drosophila embryo, PGC division from the soma is dictated by inductive signals from the germ plasm, previously the only mechanism known to control germline formation. Our data show that a global inhibitory mechanism also exists in the early embryo. This mechanism is based on Arf-GEF inhibition of actomyosin activity and restricts the formation of large groups of cells within the syncytial embryo. Thus, germline separation is controlled by a combination of local activation and global inhibition, a combination that creates robust patterns in many different biological contexts. Specifically, our data frame a model in which a uniform distribution of Step sets an inhibitory threshold for Rho1 activity to control the division of PGCs from the syncytial soma (Figure 6). Normally, Rho1 activity overcomes the threshold only at the posterior pole, through its upstream activation by the germ plasm (Cinalli and Lehmann 2013).
Figure 6.
A three-factor model for the control of asymmetric PGC division from the syncytial Drosophila soma. Two factors contribute to the potential for cell division (an elevation of Rho1 activity at the posterior pole and a propensity for cell division at both poles), and a global threshold of inhibitory Step activity controls where the division occurs (normally at the posterior pole). See Discussion for further details.
The uniformity of Step activity was evident from several pieces of data. First, our analyses of endogenous step mRNA localization revealed a uniform pattern around the embryo cortex, and our inability to detect endogenous Step protein suggested that its levels are low relative to later developmental stages. Second, our functional analyses showed endogenous Step activity at both the soma and the posterior pole where PGCs form. Activity in the soma was evident from the abnormal formation of the anterior cell group with step loss. Activity at the posterior pole was evident from the suppression of PGC formation defects of rho1 heterozygous mutants, or gcl homozygous mutants, by step RNAi. The posterior analyses indicated that PGC formation was impaired in rho1 or gcl mutants not simply because of reduced Rho1 activity, but because the experimentally reduced Rho1 activity was unable to surmount the inhibitory effects of endogenous Step. Third, GFP–Step constructs, overexpressed globally, localized similarly to the plasma membranes of somatic cell compartments and forming PGCs, and construct activity was apparently not limited at the posterior pole (it is possible though that the localization or activity of the lower levels of endogenous Step might be limited at the posterior). Notably, protein levels of the Rho–GEF Pebble and nonmuscle myosin II (Zip) are higher in forming PGCs than surrounding somatic regions (Young et al. 1991; Prokopenko et al. 2000). Moreover, a survey of the FlyFISH database revealed six actin-related mRNA transcripts enriched in the pole plasm at embryonic stages 1–3 (the early syncytial divsions): pebble, spire, Septin 4, Arp2/3 complex subunit 2, Tropomyosin 1, and Ankyrin. Overall, these data suggest that Step inhibition of Rho1 is similar across the soma and presumptive PGCs and that greater Rho1 activation, and possibly other actin cytoskeleton induction, occurs at the PGCs.
The Step threshold for Rho1 activity is not trivial. Specifically, step loss alone is sufficient for robust, ectopic formation of a large group of cells (Figure 1). Strikingly, this precocious cell division occurred without germ plasm, arguing against a longstanding view that PGC formation occurs solely by induction from the germ plasm (Wilson and Macdonald 1993; Lehmann and Ephrussi 1994; Mahowald 2001; Cinalli and Lehmann 2013). PGC formation also seems to be controlled by an inhibitory threshold set by Step. If the Step threshold for Rho1 activity is lowered, then the need for Rho1 for PGC formation is also lowered, and, strikingly, the need for the germ plasm component Gcl is eliminated, since PGC formation defects of gcl null mutants can be rescued by step RNAi (Figure 3). If the threshold is increased by Step overexpression, then PGC cell formation is inhibited in a dose-dependent manner (Figure 4). In wild-type embryos, the threshold seems to be specifically tuned for PGC formation at the embryo posterior.
Although step loss leads to the abnormal anterior cell group, it is intriguing that widespread cell formation does not occur at equatorial regions of step loss-of-function embryos. We documented previously that Step activity is needed to restrain actomyosin networks at equatorial pseudocleavage furrows, but in this context the regulation seems to coordinate syncytial nuclear divisions (without Step, sporadic cell formation and nuclear loss occurs; Lee and Harris 2013). This same global Step activity appears to have a related, but distinct, impact at the poles, hindering uniform formation of large cell groups. Thus, Step’s inhibition of Rho1 seems to have distinct cellular effects in different parts of the embryo. The difference between the polar and equatorial regions suggests an additional cue for cell formation at the poles. There are two main differences between the regions: the poles have greater curvature, and the poles are regulated by the terminal patterning system. The greater curvature of the poles could promote cell formation by directing the basal tips of furrows closer to one another than they are at the flatter equatorial regions of the embryo. However, germ plasm positioned ectopically to equatorial regions can induce formation of small groups of cells (Smith et al. 1992), although the relative effectiveness of this cell division is unclear. Regulation by terminal genes could conceivably make the poles more susceptible to the effects of step loss by increasing signals for Rho1 activation or decreasing factors for Step activity. To our knowledge, such effects have not been reported, and, in fact, terminal gene activity seems to be incompatible with normal PGC formation (Martinho et al. 2004; de Las Heras et al. 2009), but perhaps genes specific to the equatorial region could help distinguish the poles. Regardless of the specific mechanism, our data indicate that an additional property of the poles promotes the formation of cell groups. With the Step threshold in place, this property may normally act with germ plasm to promote posterior cell formation. Without Step, this property elicits the formation of cell groups at both poles, but presumably only cells capturing the germ plasm would contribute to the germline (an idea that is difficult to test because of the general disruption of step loss-of-function embryos).
Overall, our data support a three-factor model for the control of PGC segregation from the soma (Figure 6). A potential for cell division is generated by two factors—Rho1 activation by the posterior germ plasm plus a propensity for cell division at both poles—and this potential must overcome an inhibitory threshold set by Step Arf-GEF activity. In this model, Step matches the physical behavior of the early embryo plasma membrane with the maternal positioning of the germ plasm so that the first cells formed in the embryo are the germ cells.
Supplementary Material
Acknowledgments
We thank D. Godt for helpful critiques of the manuscript and G. Boulianne, J. Brill, M. Hoch, K. Howard, R Lehmann, M. Peifer, U. Tepass, and M. van Doren for reagents. The work was supported by a Canadian Institutes of Health Research (CIHR) operating grant to T. Harris (MOP82829) and an Ontario Graduate Scholarship to D. Lee. T. Harris also holds a Tier 2 Canada Research Chair.
Footnotes
Communicating editor: L. Cooley
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.176867/-/DC1.
Literature Cited
- Chau A. H., Walter J. M., Gerardin J., Tang C., Lim W. A., 2012. Designing synthetic regulatory networks capable of self-organizing cell polarization. Cell 151: 320–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherfils J., Menetrey J., Mathieu M., Le Bras G., Robineau S., et al. , 1998. Structure of the Sec7 domain of the Arf exchange factor ARNO. Nature 392: 101–105. [DOI] [PubMed] [Google Scholar]
- Cinalli R. M., Lehmann R., 2013. A spindle-independent cleavage pathway controls germ cell formation in Drosophila. Nat. Cell Biol. 15: 839–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza-Schorey C., Chavrier P., 2006. ARF proteins: roles in membrane traffic and beyond. Nat. Rev. Mol. Cell Biol. 7: 347–358. [DOI] [PubMed] [Google Scholar]
- de Las Heras J. M., Martinho R. G., Lehmann R., Casanova J., 2009. A functional antagonism between the pgc germline repressor and torso in the development of somatic cells. EMBO Rep. 10: 1059–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson J. G., Jackson C. L., 2011. ARF family G proteins and their regulators: roles in membrane transport, development and disease. Nat. Rev. Mol. Cell Biol. 12: 362–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher G. C., Lucas E. P., Brain R., Tournier A., Thompson B. J., 2012. Positive feedback and mutual antagonism combine to polarize Crumbs in the Drosophila follicle cell epithelium. Curr. Biol. 22: 1116–1122. [DOI] [PubMed] [Google Scholar]
- Foe V. E., Alberts B. M., 1983. Studies of nuclear and cytoplasmic behaviour during the five mitotic cycles that precede gastrulation in Drosophila embryogenesis. J. Cell Sci. 61: 31–70. [DOI] [PubMed] [Google Scholar]
- Gierer A., Meinhardt H., 1972. A theory of biological pattern formation. Kybernetik 12: 30–39. [DOI] [PubMed] [Google Scholar]
- Gillingham A. K., Munro S., 2007. The small G proteins of the Arf family and their regulators. Annu. Rev. Cell Dev. Biol. 23: 579–611. [DOI] [PubMed] [Google Scholar]
- Hahn I., Fuss B., Peters A., Werner T., Sieberg A., et al. , 2013. The Drosophila Arf GEF Steppke controls MAPK activation in EGFR signaling. J. Cell Sci. 126: 2470–2479. [DOI] [PubMed] [Google Scholar]
- Hayashi K., de Sousa Lopes S. M., Surani M. A., 2007. Germ cell specification in mice. Science 316: 394–396. [DOI] [PubMed] [Google Scholar]
- Johnson A. D., Richardson E., Bachvarova R. F., Crother B. I., 2011. Evolution of the germ line-soma relationship in vertebrate embryos. Reproduction 141: 291–300. [DOI] [PubMed] [Google Scholar]
- Lecuyer, E., A. S. Necakov, L. Caceres and H. M. Krause, 2008 High-resolution fluorescent in situ hybridization of Drosophila embryos and tissues. CSH Protoc. 2008: pdb prot5019. [DOI] [PubMed]
- Lee D. M., Harris T. J., 2013. An Arf-GEF regulates antagonism between endocytosis and the cytoskeleton for Drosophila blastoderm development. Curr. Biol. 23: 2110–2120. [DOI] [PubMed] [Google Scholar]
- Lehmann R., Ephrussi A., 1994. Germ plasm formation and germ cell determination in Drosophila. Ciba Found. Symp. 182: 282–300. [DOI] [PubMed] [Google Scholar]
- Mahowald A. P., 2001. Assembly of the Drosophila germ plasm. Int. Rev. Cytol. 203: 187–213. [DOI] [PubMed] [Google Scholar]
- Martinho R. G., Kunwar P. S., Casanova J., Lehmann R., 2004. A noncoding RNA is required for the repression of RNApolII-dependent transcription in primordial germ cells. Curr. Biol. 14: 159–165. [DOI] [PubMed] [Google Scholar]
- Mossessova E., Gulbis J. M., Goldberg J., 1998. Structure of the guanine nucleotide exchange factor Sec7 domain of human arno and analysis of the interaction with ARF GTPase. Cell 92: 415–423. [DOI] [PubMed] [Google Scholar]
- Prokopenko S. N., Saint R., Bellen H. J., 2000. Tissue distribution of PEBBLE RNA and pebble protein during Drosophila embryonic development. Mech. Dev. 90: 269–273. [DOI] [PubMed] [Google Scholar]
- Robertson S. E., Dockendorff T. C., Leatherman J. L., Faulkner D. L., Jongens T. A., 1999. germ cell-less is required only during the establishment of the germ cell lineage of Drosophila and has activities which are dependent and independent of its localization to the nuclear envelope. Dev. Biol. 215: 288–297. [DOI] [PubMed] [Google Scholar]
- Roussos E. T., Condeelis J. S., Patsialou A., 2011. Chemotaxis in cancer. Nat. Rev. Cancer 11: 573–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydoux G., Braun R. E., 2006. Pathway to totipotency: lessons from germ cells. Cell 127: 891–904. [DOI] [PubMed] [Google Scholar]
- Smith J. L., Wilson J. E., Macdonald P. M., 1992. Overexpression of oskar directs ectopic activation of nanos and presumptive pole cell formation in Drosophila embryos. Cell 70: 849–859. [DOI] [PubMed] [Google Scholar]
- Strome S., Lehmann R., 2007. Germ vs. soma decisions: lessons from flies and worms. Science 316: 392–393. [DOI] [PubMed] [Google Scholar]
- Technau G. M., Campos-Ortega J. A., 1986. Lineage analysis in embryos of Drosophila melanogaster. II. Commitment and proliferative capabilities of neural and epidermal cell progenitors. Rouxs Arch. Dev. Biol. 195: 445–454. [DOI] [PubMed] [Google Scholar]
- Turing A. M., 1952. The chemical basis of morphogenesis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 237: 37–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilk R., Murthy S. U. M., Yan H., Krause H. M., 2010. In Situ Hybridization: Fruit Fly Embryos and Tissues. Wiley, New York. [Google Scholar]
- Wilson J. E., Macdonald P. M., 1993. Formation of germ cells in Drosophila. Curr. Opin. Genet. Dev. 3: 562–565. [DOI] [PubMed] [Google Scholar]
- Young P. E., Pesacreta T. C., Kiehart D. P., 1991. Dynamic changes in the distribution of cytoplasmic myosin during Drosophila embryogenesis. Development 111: 1–14. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.