Abstract
MicroRNAs (miRNAs) are critical post-transcriptional regulators. Based on a previous genome-wide association (GWA) scan, we conducted a polymorphism in microRNA target sites (poly-miRTS)-centric multistage meta-analysis for lumbar spine (LS)-, total hip (HIP)- and femoral neck (FN)-bone mineral density (BMD). In stage I, 41 102 poly-miRTSs were meta-analyzed in seven cohorts with a genome-wide significance (GWS) α = 0.05/41 102 = 1.22 × 10−6. By applying α = 5 × 10−5 (suggestive significance), 11 poly-miRTSs were selected, with FGFRL1 rs4647940 and PRR5 rs3213550 as top signals for FN-BMD (P = 7.67 × 10−6 and 1.58 × 10−5) in gender-combined sample. In stage II in silico replication (two cohorts), FGFRL1 rs4647940 was the only signal marginally replicated for FN-BMD (P = 5.08 × 10−3) at α = 0.10/11 = 9.09 × 10-3. PRR5 rs3213550 was also selected based on biological significance. In stage III de novo genotyping replication (two cohorts), FGFRL1 rs4647940 was the only signal significantly replicated for FN-BMD (P = 7.55 × 10−6) at α = 0.05/2 = 0.025 in gender-combined sample. Aggregating three stages, FGFRL1 rs4647940 was the single stage I-discovered and stages II- and III-replicated signal attaining GWS for FN-BMD (P = 8.87 × 10−12). Dual-luciferase reporter assays demonstrated that FGFRL1 3′ untranslated region harboring rs4647940 appears to be hsa-miR-140-5p's target site. In a zebrafish microinjection experiment, dre-miR-140-5p is shown to exert a dramatic impact on craniofacial skeleton formation. Taken together, we provided functional evidence for a novel FGFRL1 poly-miRTS rs4647940 in a previously known 4p16.3 locus, and experimental and clinical genetics studies have shown both FGFRL1 and hsa-miR-140-5p are important for bone formation.
Introduction
Osteoporosis, a common skeletal disease characterized by low bone mineral density (BMD) and deterioration in bone microarchitecture, impaired bone strength and leads to an increased risk of fragility fractures (1). Twin and family studies of BMD have shown that 60–90% of BMD variation can be attributed to genetic factors (2–6).
MicroRNAs (miRNAs) are evolutionarily conserved, endogenous, single-stranded, non-coding RNAs of 18–24 nucleotides in length that regulate gene expression, typically by binding to their complementary sequences located at 3′ untranslated regions (UTRs) of target mRNAs by mediating target mRNA degradation and/or protein synthesis repression (7,8). There are 2578 mature human miRNA sequences currently listed in miRBase (9,10). Further, more than 45 000 conserved miRNA target sites within human 3′ UTRs have been identified, and more than 60% of human protein-coding genes are under selective pressure to maintain pairing to miRNAs (11). Each miRNA can modulate about 200 target mRNAs (12,13).
Canonical miRNA targeting is mediated by a perfect Watson–Crick pairing of miRNA's 5′ seed region, typically comprising nucleotides 2–7 at 5′-end of an miRNA (12), which determines both target site specificity (14–16) and most of energy change involved in miRNA–mRNA binding (17,18). Some 6-mer seeds also match to positions 1–6 (19,20). The miRNA–mRNA binding is determined not only by the thermodynamic property of miRNA, but also by characteristic secondary structure of the target mRNA's 3′ UTR containing the miRNA binding site (21), which can be affected by single-nucleotide polymorphisms (SNPs) either inside or outside the miRNA target site that alter miRNA accessibility (13,22).
Disruptions of miRNA binding sites by SNPs present in 3′ UTRs of mammalian genes known as polymorphisms in microRNA target sites (poly-miRTSs), have been clearly documented, e.g. (23–25). By abolishing/modifying/creating miRNA target sites, such SNPs may affect target mRNA/protein expressions, which could modulate diseases risks (23,26–28).
Although genome-wide association (GWA) studies have been successful in identifying new genes with common variants that exert modest effects on complex traits (e.g. BMD), conventional ‘hypothesis-free’ GWA approach commonly performs 1 million independent association tests, and therefore has strict requirements for controlling occurrences of false positives in such an ‘unbiased’ approach [typically, a genome-wide significance (GWS) threshold of α = 5 × 10−8 is applied], resulting in severe multiple testing corrections. True positive associations could be missed, particularly in the presence of limited replication resources (29). More than 90% of SNPs collected in the National Human Genome Research Institute (NHGRI) Genome-wide Association Study catalog (30) are located within non-coding regions (31). However, many such non-coding SNPs are simply neglected due to a lack of functional annotations. Therefore, revisiting GWA scans from the perspective of biological knowledge (e.g. miRNA target sites, promoter methylation regions) could discover new genetic variants that are experimentally testable. Because the human genome harbors a large number of poly-miRTSs that are potentially important (32,33), screening exclusively those poly-miRTSs could discover novel regulatory SNPs affecting gene expressions thanks to a less number of statistical tests based on biological meaning. Because such a functional candidate genomic region association approach may increase type I error rate (i.e. α), to minimize false positives, a multi-stage approach was applied, such that those stage I-selected poly-miRTSs based on a moderate threshold (α = 5 × 10−5) are subject to stringent stage II- and stage III-independent replications based on their respective Bonferroni-corrected significance thresholds.
Results
Basic genotype information and phenotype characteristics for the three stages of GWA meta-analysis focusing on poly-miRTSs were presented in Table 1.
Table 1.
Basic genotype information and phenotype characteristics of the study cohorts at stages I, II and IIIa
| Sample | N | Population | # Genotyped SNPs | # Imputed SNPs | Female (%) | Age (years) | LS-BMD (g/cm2) | HIP-BMD (g/cm2) | FN-BMD (g/cm2) | Bone densitometer |
|---|---|---|---|---|---|---|---|---|---|---|
| Stage Ib (GWA meta-analysis) | ||||||||||
| OOS | 987 | Caucasian | 407 634 | 6 427 198 | 49.6 | 50.3 (18.3) | 1.03 (0.16) | 0.97 (0.16) | 0.81 (0.14) | Hologic QDR 4500W |
| KCOS | 2250 | Caucasian | 743 768 | 7 085 256 | 75.9 | 51.4 (13.8) | 1.02 (0.16) | 0.97 (0.17) | 0.80 (0.15) | Hologic QDR 4500W |
| COS | 1547 | Han Chinese | 725 883 | 5 823 600 | 50.7 | 34.8 (13.4) | 0.95 (0.13) | 0.92 (0.13) | 0.81 (0.13) | Hologic QDR 4500W |
| FHS | 3747 | Caucasian | 405 948 | 6 456 277 | 57.3 | 60.3 (10.7) | 1.22 (0.23) | 0.95 (0.17) | 0.89 (0.16) | Lunar DPX-L |
| IFS | 1488 | Caucasian | 543 932 | 7 202 569 | 100.0 | 32.7 (7.2) | — | — | — | Lunar DPX-L |
| WHI-AA | 712 | African American | 799 328 | 11 862 114 | 100.0 | 60.9 (6.9) | 1.05 (0.17) | 0.95 (0.15) | 0.83 (0.14) | Hologic QDR-2000 |
| WHI-HIS | 409 | Hispanic | 647 362 | 6 916 926 | 100.0 | 60.7 (7.2) | 0.97 (0.16) | 0.86 (0.13) | 0.73 (0.12) | Hologic QDR-2000 |
| Total | 11 140 | |||||||||
| Stage IIc (in silico replication) | ||||||||||
| RS | 4904 | Caucasian | 512 349 | 6 986 425 | 57.0 | 69.4 (9.0) | 1.09 (0.18) | NA | 0.86 (0.13) | Lunar DPX-L |
| AOGC | 1955 | Caucasian | Region-wise | Region-wise | 100.0 | 69.6 (8.6) | 1.08 (0.26) | 0.89 (0.25) | 0.79 (0.21) | Multipled |
| Total | 6859 | |||||||||
| Stage III (de novo genotyping replication) | ||||||||||
| KCOSR | 3923 | Caucasian | 3 | 0 | 57.6 | 46.8 (14.5) | 1.03 (0.15) | 0.96 (0.15) | 0.80 (0.14) | Hologic QDR |
| COSR | 2740 | Han Chinese | 3 | 0 | 57.7 | 32.3 (14.4) | 0.93 (0.12) | 0.91 (0.13) | 0.80 (0.13) | Hologic QDR 4500W |
| Total | 6663 | |||||||||
aAdapted from Supplementary Material, Table S1 of Zhang et al. (34). Although the Korean Genome Epidemiology Study (KoGES) was included in stage 2 (in silico replication) of Zhang et al. (34), because none of KoGES results were included in the in silico replication of stage I-selected poly-miRTSs, the stage II (in silico replication) of the current study only included RS and AOGC, but not KoGES. Data were presented as mean (SD). N denotes sample size for each study cohort. OOS, Omaha Osteoporosis Study; KCOS, Kansas-City Osteoporosis Study; COS, China Osteoporosis Study; FHS, Framingham Heart Study; IFS, Indiana Fragility Study; WHI-AA, Women's Health Initiative African American ancestry; WHI-HIS, Women's Health Initiative Hispanic ancestry. RS, Rotterdam Study; AOGC, Australian Osteoporosis Genetics Consortium; KCOSR, KCOS Replication; COSR, COS Replication. SNP, single-nucleotide polymorphism; poly-miRTS, polymorphism in microRNA target sites; BMD, bone mineral density; LS, lumbar spine; HIP, total hip; FN, femoral neck. ‘—’, not available. λGC ranges from 0.99 to 1.06 (34).
bAs indicated in Zhang et al. (34), 5842 825 common SNPs (either genotyped or imputed) were shared across the seven stage I genome-wide association studies and only those 41 102 poly-miRTSs were included in stage I.
cAs indicated in Zhang et al. (34), for AOGC study, genotype imputations were only performed for specific regions, and only 11 stage I-selected poly-miRTSs were included in stage II of the current study.
dEither Hologic QDR or Lunar DPX-L series was used to measure BMD.
Stage I (GWA discovery)
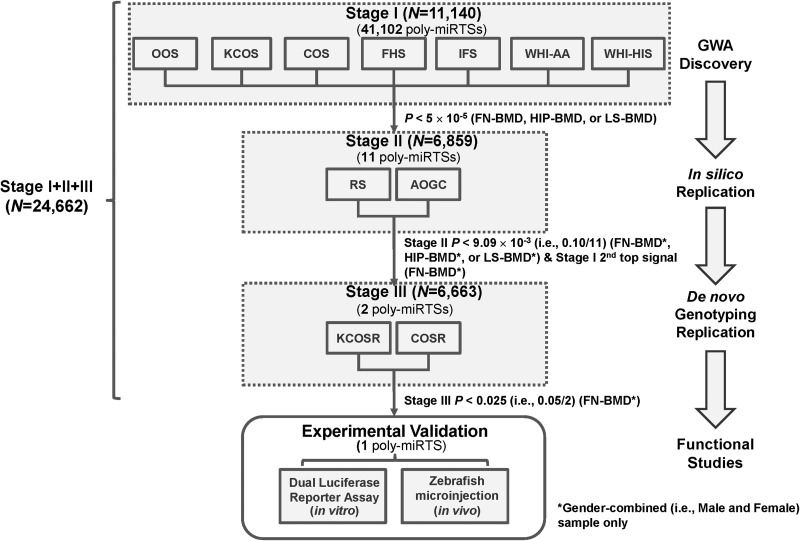
As shown in Figure 1, seven cohorts, i.e. OOS (Omaha Osteoporosis study), KCOS (Kansas City Osteoporosis study), COS (Chinese Osteoporosis), FHS (Framingham Heart Study), WHI-AA [Women's Health Initiative (WHI), African American ancestry], and WHI-HiS (WHI, Hispanic ancestry) were included in this GWA discovery stage. Through genotype imputation using 1 KG reference panels, 5 842 825 SNPs (genotyped + imputed) were imputed (34). Because the current study only focused on 41 102 poly-miRTSs of 5 842 825 SNPs, quantile–quantile (Q–Q) plots were generated exclusively for these 41 102 poly-miRTSs for gender-combined (i.e. male and female) sample and female-specific sample, respectively (Supplementary Material, Fig. S1). SNP ranks, names and P-values for five top signals of 11 stage I-selected poly-miRTS for femoral neck (FN)-BMD in gender-combined sample (i.e. rs4647940, rs3213550, rs523200, rs11581122 and rs10739677) and in female-specific sample (i.e. rs4647940, rs1599795, rs10739677, rs3213550 and rs523200) were indicated. After adjusting BMD phenotypes by five principal components, genomic control (GC) inflation factor (λGC) was computed for each of lumbar spine (LS)-, total hip (HIP)- and FN-BMD traits for stage I 41 102 poly-miRTSs in gender-combined and female-specific samples, respectively, and these λGC's were compared with those of 5 842 825 SNPs of the traditional GWA study of Zhang et al. (34). Overall, because these 41 102 poly-miRTSs were not random SNPs, but rather SNPs with biologically higher prior probabilities, λGC's were slightly increased from the range of 1.03−1.04 to the range of 1.033−1.121 (Supplementary Material, Table S1). To protect against an increase in type I error rate (i.e. α), we applied a multi-stage approach such that those stage I-selected poly-miRTSs based on a moderate threshold (α = 5 × 10−5) are subject to stage II- and stage III-independent replications based on their respective Bonferroni-corrected significance thresholds. Three poly-miRTSs of two genes attained GWS (i.e. α = 0.05/41 102 = 1.22 × 10−6): OSBPL2 rs6089342 (P = 2.70 × 10−7) and rs6121978 (P = 3.65 × 10−7) located on chromosome 20q13.33 for LS-BMD in gender-combined sample, and TMEM135 rs550773 (P = 5.93 × 10−7) located on chromosome 11q14.2 for FN-BMD in gender-combined sample. Another 37 poly-miRTSs had P-values in the range of 1.22 × 10−6 to 5 × 10−5. Of these 40 top-ranking poly-miRTSs at α = 5 × 10−5 (suggestive significance), 11 (i.e. SLC22A2 rs3127592 and rs3127593, FGFRL1 rs4647940, CD80 rs1599795, C6orf97-ESR1 rs9479085, PRR5 rs3213550, PTH2R rs1057392, FAM125B rs10739677, AMAC1 rs17547201, FAM46C rs11581122 and SF1 rs523200, located in 10 gene regions) were selected based on SNP selection criteria: (i) statistical significance threshold (P < 5 × 10−5); (ii) allelic association direction (if ≥2 traits are available, same directions for ≥6 of seven cohorts for each trait; if only one trait is available, same directions for all seven cohorts); and (iii) biological significance, [i.e. being an miRNA target site disease-associated SNP (miRTSdSNP) according to Bruno et al. (35)].
Figure 1.
A flow chart of the poly-miRTS-centric three-stage GWA meta-analysis and experimental validation. GWA, genome-wide association; SNP, single-nucleotide polymorphism; DB, database; poly-miRTSs, polymorphisms in microRNA target sites; study name abbreviations are shown in footnote of Table 1.
Stage II (in silico replication)
At stage II, these above 11 stage I-selected poly-miRTSs were further subject to in silico replication in two independent cohorts: (i) RS (Rotterdam Study; N = 4904, 57.0% women) and (ii) AOGC (Angle-Australasian Osteoporosis Genetics Consortium; N = 1955, 100.0% women). The following SNP selection criteria were applied for stage II SNP selection: (i) statistical significance threshold (Bonferroni-corrected marginal significance, i.e. P < 0.1/11 = 9.09 × 10−3) and (ii) biological significance. Only one poly-miRTS met statistical significance threshold—FGFRL1 rs4647940, with a P = 5.078 × 10−3 for FN-BMD in gender-combined sample (Table 2). At stage I, FGFRL1 rs4647940 is top signal among these stage I-selected 11 poly-miRTSs for FN-BMD in gender-combined sample. As shown in Supplementary Material, Figure S1A, for this trait, there was a bigger gap of -log10(P-value) between rs3213550 and rs523200 [Δ-log10(P-value) = −log10(1.58 × 10−5) + log10(4.76 × 10−5) = 0.4789] than between rs523200 and rs11581122 [Δ-log10(P-value) = −log10(4.76 × 10−5) + log10(6.66 × 10−5) = 0.1459], and between rs11581122 and rs10739677 [Δ-log10(P-value) = −log10(6.66 × 10−5) + log10(1.267 × 10−4) = 0.2793]. In addition, as shown in Supplementary Material, Figure S1B, second top signal for FN-BMD trait in female-specific sample, i.e. rs1599795, had a less significant P-value (i.e. 8.32 × 10−5) than the P-value of second top signal (i.e. 1.58 × 10−5) for the same trait in gender-combined sample, i.e. rs3213550. Further, besides rs4647940 (P = 7.67 × 10−6), which attained an α = 5 × 10−5 threshold in gender-combined and female-specific samples, rs3213550 (P = 1.58 × 10−5) is the only other poly-miRTS that attained an α = 5 × 10−5 threshold in gender-combined sample, and an α = 5.5 × 10−4 (a more relaxed threshold compared with α = 5 × 10−5) in female-specific sample, respectively. Taken together, at stage II, PRR5 poly-miRTS rs3213550 exhibited a relatively robust association for FN-BMD at stage I. Although this SNP did not reach stage II marginal significance threshold (P = 0.3138 > 9.09 × 10−3), it is notable that at stage I, it had consistent significant associations for FN-BMD in gender-combined and female-specific samples. PRR5 is a recently identified component of mammalian target of rapamycin complex 2 (mTORC2), which also contains four other components, i.e. mTOR, Rictor, mSin1, mLST8/GL and plays a critical role in bone development and homeostasis (36–38). PRR5 interacts with Rictor in regulating skeletal growth (39). Further, prior biological evidence suggests that rs3213550, which is located in 3′ UTR of PRR5 gene, is a disease-associated miRTSdSNP (35). Therefore, at stage II, based on biological significance of PRR5 and mTORC2 for bone growth and stage I statistical significance for FN-BMD (i.e. a runner up following rs4647940), PRR5 poly-miRTS rs3213550 was also selected for stage III de novo genotyping replication.
Table 2.
GWA meta-analysis results for FGFRL1 and PRR5 Poly-miRTSsa
| Poly-miRTS |
Stage I |
Stage II |
Stage III |
Stage I + II + III | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dbSNP ID | Locus | Gene | Phenotype | Analysis | P-value | Phenotype | Analysis | P-value | Phenotype | Analysis | P-value | P-value |
| rs3127592 | 6q25.3 | SLC22A2 | HIP-BMD | Combined | 3.24 × 10−6 | FN-BMD | Combined | 0.5034 | — | — | — | — |
| rs3127593 | 6q25.3 | SLC22A2 | HIP-BMD | Combined | 6.59 × 10−6 | FN-BMD | Combined | 0.4638 | — | — | — | — |
| rs4647940 | 4p16.3 | FGFRL1 | FN-BMD | Combined | 7.67× 10−6 | FN-BMDb | Combined | 5.078 × 10−3 | FN-BMDb | Combined | 7.55 × 10−6 | 8.87 × 10−12 |
| rs1599795 | 3q13.33 | CD80 | HIP-BMD | Combined | 1.32 × 10−5 | FN-BMD | Combined | 0.3689 | — | — | — | — |
| rs9479085 | 6q25.1 | C6orf97 | LS-BMD | Combined | 1.47 × 10−5 | LS-BMDb | Combined | 9.202 × 10−3 | — | — | — | — |
| rs3213550 | 22q13.31 | PRR5 | FN-BMD | Combined | 1.58 × 10−5 | FN-BMDb | Combined | 0.3138 | FN-BMD | Combined | 0.08208 | 1.17 × 10−5 |
| rs1057392 | 2q34 | PTH2R | HIP-BMD | Combined | 2.60 × 10−5 | FN-BMD | Combined | 0.3167 | — | — | — | — |
| rs10739677 | 9q33.3 | FAM125B | HIP-BMD | Female | 2.67 × 10−5 | FN-BMD | Combined | 0.4262 | — | — | — | — |
| rs17547201 | 17q12 | AMAC1 | HIP-BMD | Female | 2.79 × 10−5 | FN-BMD | Combined | 0.4009 | — | — | — | — |
| rs11581122 | 1p12 | FAM46C | HIP-BMD | Combined | 4.47 × 10−5 | FN-BMD | Combined | 0.6717 | — | — | — | — |
| rs523200 | 11q13.1 | SF1 | FN-BMD | Combined | 4.76 × 10−5 | FN-BMDb | Combined | 0.1879 | — | — | — | — |
aBMD, bone mineral density; FN, femoral neck; HIP, total hip; LS, lumbar spine; poly-miRTS, polymorphism in microRNA Target Sites, SNP, single nucleotide polymorphism. Combined refers to male and female. Genome-wide significance (GWS) is defined as α = 5.00 × 10−8 in the previous traditional study (34) and 5.21 × 10−6 in the current study, respectively. P-values attaining the GWS level are highlighted in bold font. The SNP reaching GWS for stage I + II + III (i.e. P < 5.21 × 10−6) was highlighted in bold font.
bData were available for two cohorts in each respective stage of stages II and III. Otherwise, data were available for only one cohort in each respective stage of stages II and III.
Stage III (de novo genotyping replication)
At stage III, the above two stage II-selected poly-miRTSs located in two respective gene regions—FGFRL1 and PRR5, were further subject to replication by de novo genotyping in two independent cohorts: (i) KCOSR (KCOS Replication; N = 3932, 57.6% women) and (ii) COSR (COS Replication; N = 2740, 57.7% women). In this stage, SNP selection criterion is solely based on Bonferroni-corrected statistical significance threshold (P < 0.05/2 = 0.025) for FN-BMD trait in gender-combined sample. Only FGFRL1 rs4647940 met this criterion for FN-BMD (P = 7.55 × 10−6), in gender-combined sample (Table 2).
Stage I+II+III meta-analysis
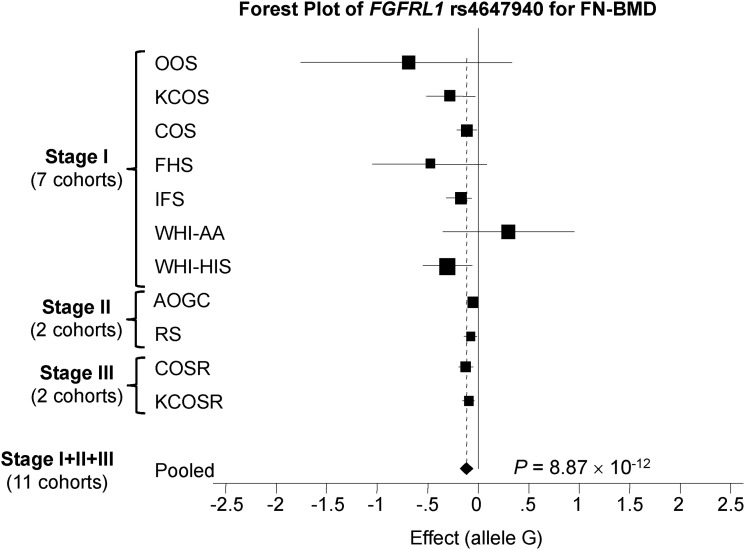
Meta-analysis results pooling all available data across stages I, II and III for two poly-miRTSs—FGFRL1 rs4647940 and PRR5 rs3213550 for FN-BMD in gender-combined sample revealed that FGFRL1 rs4647940 (P = 8.87 × 10-12) was the only poly-miRTS that attained both the current study's GWS (i.e. P < 1.22 × 10−6) and conventional GWS (i.e. P < 5 × 10−8) (Table 2). Stage I's second top signal for FN-BMD of 11 stage I-selected poly-miRTSs—PRR5 rs3213550 (P = 1.17 × 10−5) did not attain the current study's reduced GWS. Taken together, the current study has identified and robustly replicated FGFRL1 rs4647940 as the only signal associated with FN-BMD in gender-combined sample. As shown in forest plot of stage I + II + III meta-analysis of poly-miRTS, FGFRL1 rs4647940 for FN-BMD in gender-combined sample (Fig. 2), not only the overall meta-analysis across these 11 cohorts attained conventional GWS (i.e. P < 5 × 10−8), but also all replication cohorts of stage II (AOGC and RS) and stage III (COSR and KCOSR) have same directions of allelic associations as ≥ 6 out of seven stage I cohorts.
Figure 2.
A forest plot of FGFRL1 rs4647940 for FN-BMD trait for stage I + II + III for gender-combined (i.e. male and female) cohorts. BMD, bone mineral density; FN, femoral neck.
Regional association plots and linkage disequilibrium r2 plots
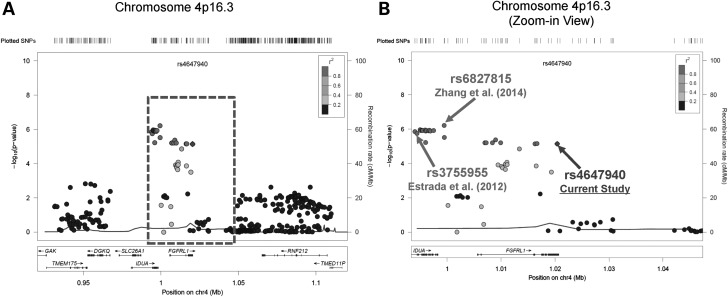
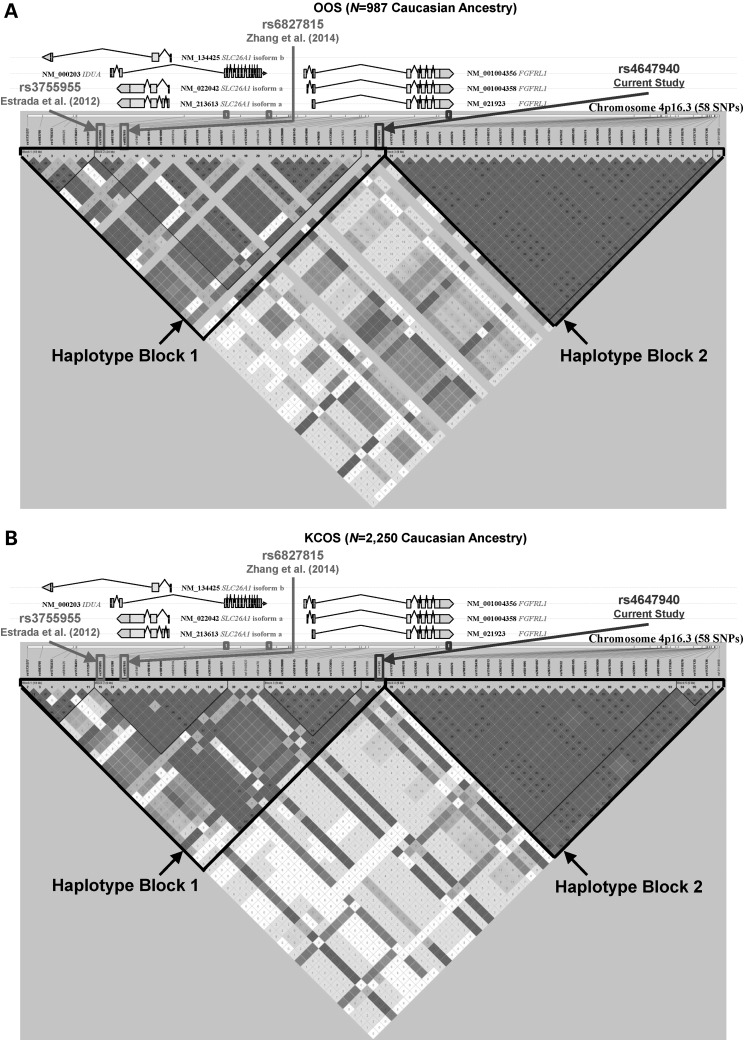
A regional association plot of traditional GWA meta-analysis results for FN-BMD trait of FGFRL1 poly-miRTS rs4647940 and those stage I SNPs in the ± 100-kb flanking regions, particularly intergenic rs6827815 (currently considered as located within IDUA gene region; http://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?rs=6827815) of Zhang et al. (34) and IDUA rs3755955 of Estrada et al. (40), were shown in Figure 3A, with a zoom-in view of the middle part (i.e. ± 27-kb flanking regions) spanning these three SNPs shown in Figure 3B. In order to delineate a high-resolution linkage disequilibrium (LD) structure for 4p16.3 locus encompassing these three SNPs, an LD r2 plot for 58 SNPs (from rs11731377 to rs13114658 spanning 80.744-kb) was generated for OOS (Fig. 4A) and KCOS (Fig. 4B) cohorts, respectively, both of which are of Caucasian ancestry. This entire region is partitioned into left and right haplotype blocks: block 1 (left) extends from rs11731377 to rs4647940 (30 SNPs) and block 2 (right) extends from rs13137819 and rs13114658 (28 SNPs) in each cohort. All these three SNPs (i.e. rs3755955, rs6827815 and rs4647940) are located within block 1 in both cohorts, such that rs4647940 is in a moderately high LD with both rs3755955 [r2 = 0.649 and 0.842 for OOS (N = 987) and KCOS (N = 2250), respectively] and rs6827815 (r2 = 0.702 and 0.857 in OOS and KCOS, respectively).
Figure 3.
Regional association plots for chromosome 4p16.3 loci FGFRL1 rs4647940, IDUA rs6827815 and IDUA rs3755955 for FN-BMD trait (based on hg19/1000 Genomes Mar 2012 EUR). (A) The region encompassing rs4647940 with ±100-kb flanking regions, and (B) a zoom-in view of the center region, [i.e. the dashed rectangle in (A)], encompassing rs4647940 with ±27-kb regions. BMD, bone mineral density; EUR, European; FN, femoral neck.
Figure 4.
The physical locations and linkage disequilibrium pairwise r2 plot generated by Haploview (41,42) for the 80.744-kb human FGFRL1 region (total 58 SNPs) encompassing FGFRL1 rs4647940 (current study), rs6827815 [Zhang et al. (34)] and rs3755955 [Estrada et al. (40)] for (A) OOS and (B) KCOS, respectively. Haplotype blocks 1 and 2 are shown by dashed lines. OOS, Omaha Osteoporosis Study; KCOS, Kansas-City Osteoporosis Study; SNP, single-nucleotide polymorphism.
Conditional meta-analysis
In rs3755955-rs6827815-rs4647940 region, because rs6827815 (distance: 20.946 kb) is physically closer to rs4647940 than rs3755955 (distance: 25.977 kb), two conditional analyses were performed: (i) rs4647940 conditional on rs6827815: under a fixed-effects model, P = 1.35 × 10−3 for FN-BMD in stage I gender-combined sample, which remains to be significant at α = 5.00 × 10−3, suggesting rs4647940 and rs6827815 represent distinct signals at this chromosomal locus and (ii) rs4647940 conditional on both rs3755955 and rs6827815: under a fixed-effects model, P = 1.33 × 10−3 for FN-BMD in stage I gender-combined sample.
Predicted RNA secondary structures of FGFRL1 3′ UTR harboring rs4647940
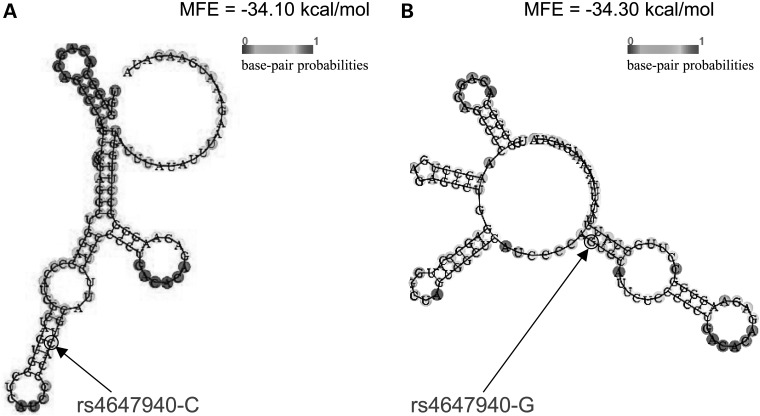
The secondary structure of a 121-bp (i.e. rs4647940 with ± 60-bp flanking regions) FGFRL1 3′ UTR segment carrying either wild-type (WT) (i.e. ‘C’) or mutant (MUT) (i.e. ‘G’) allele of rs4647940 was predicted by RNAfold. Difference of minimum free energies (MFEs) between folding patterns for MUT and WT alleles is ΔMFE = MFEMUT – MFEWT = (−34.30) − (−34.10) = −0.20 kcal/mol. Therefore, the secondary structure of WT allele such that the SNP is located in a stem connected to a terminal loop and an internal loop (Fig. 5A) is less stable than that of MUT allele such that the SNP is located in a stem connected to two internal loops (Fig. 5B). Therefore, this SNP causes a change of RNA secondary structure of FGFRL1 3′ UTR, potentially affecting miRNA target site accessibility.
Figure 5.
Predicted secondary structure of FGFRL1 3′ UTR 121-bp mRNA sequence carrying either (A) rs4647940-C allele or (B) rs4647940-G allele by RNAfold. MFE, minimum free energy.
Evolutionary conservation of FGFRL1 3′ UTR harboring rs4647940
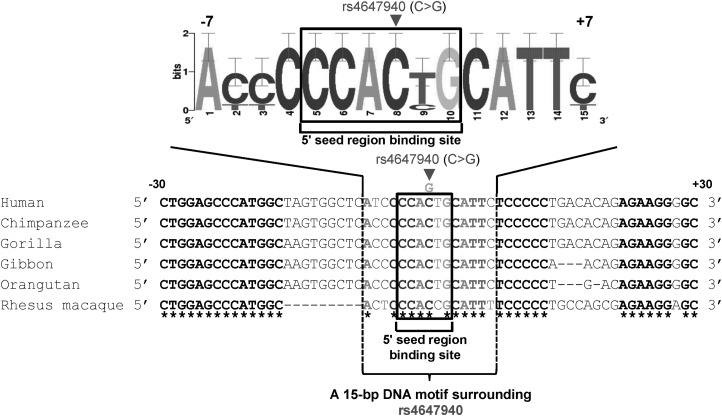
Multiple sequence alignment of orthologous sequences of FGFRL1 3′ UTR segment encompassing rs4647940 from human, chimpanzee, gorilla, gibbon, orangutan and rhesus macaque species showed that this SNP corresponds to a highly conserved nucleotide (Fig. 6), which could be under high selective pressures during evolution, and therefore is likely functional.
Figure 6.
Cross-species comparison of DNA sequences of FGFRL1 3′ UTR encompassing rs4647940 in human (GenBank accession number NM_001004356.2), chimpanzee (GenBank accession number AC194569.3), gibbon (GenBank accession number NW_003501383.2), gorilla (GenBank accession number NW_004002168.1), orangutan (GenBank accession number NW_002878038.1) and rhesus macaque (GenBank accession number NW_001111081.1). A 15-bp consensus motif surrounding FGFRL1 rs4647940 is represented by a WebLogo. The box (a 6-bp sequence) denotes a 5′ seed region binding site for hsa-miR-140-5p, which includes FGFRL1 rs4647940 (i.e. at fourth position). Multiple DNA sequence alignment was performed by ClustalX version 1.81. The asterisks denote nucleotides conserved across all the species that were compared.
Predicted miRNAs that potentially bind to FGFRL1 rs4647940
Based on predictions of four in silico tools (i.e. MicroSNiPer, MirSNP, RegRNA 2.0 and TargetScan), 13 miRNAs could target FGFRL1 rs4647940 (Table 3).
Table 3.
Putative human miRNAs that bind to FGFRL1 Poly-miRTS rs4647940
| miR-Name | # Programs | MicroSNiPer | MirSNP Score | RegRNA2.0 Score | TargetScan Context Score Percentile |
|---|---|---|---|---|---|
| hsa-miR-17-3p | 1 | Seed length: 7 (Yes) | — (No) | <150 (No) | — (No) |
| hsa-miR-30c-2-3p | 1 | Seed length: 7 (Yes) | — (No) | <150 (No) | — (No) |
| hsa-miR-34c-5p | 1 | Seed length: 6 (Yes) | — (No) | <150 (No) | — (No) |
| hsa-miR-92a-2-5p | 1 | Seed length: 8 (Yes) | — (No) | <150 (No) | — (No) |
| hsa-miR-106b-3p | 1 | Seed length: 6 (Yes) | — (No) | <150 (No) | — (No) |
| hsa-miR-140-5p | 1 | Seed length: 6 (Yes) | — (No) | <150 (No) | — (No) |
| hsa-miR-143-5p | 1 | Seed length: 6 (Yes) | — (No) | <150 (No) | — (No) |
| hsa-miR-4667-5p | 2 | Seed length: 8 (Yes) | 144.00 (Yes) | <150 (No) | — (No) |
| hsa-miR-4700-5p | 2 | Seed length: 11 (Yes) | 161.00 (Yes) | <150 (No) | — (No) |
| hsa-miR-4723-5p | 2 | Seed length: — (No) | 144.00 (Yes) | <150 (No) | 77 (Yes) |
| hsa-miR-5698 | 2 | Seed length: — (No) | 162.00 (Yes) | ≥150 (Yes) | — (No) |
| hsa-miR-3175 | 3 | Seed length: — (No) | 159.00 (Yes) | ≥150 (Yes) | 86 (Yes) |
| hsa-miR-491-5p | 4 | Seed length: 8 (Yes) | 148.00 (Yes) | ≥150 (Yes) | 76 (Yes) |
GWA, genome-wide association; poly-miRTS, polymorphism in microRNA target sites; ‘# Programs’, total number of programs that gave the prediction, denoted by ‘Yes’ in parentheses (otherwise, ‘No’ is shown in parentheses); ‘—’, not available. Seven human miRNAs that correspond to mouse miRNAs significantly differentially expressed in bone between different time points [He et al. (43)] were in italic font, which includes hsa-miR-140-5p (in bold font). The miRNAs were first sorted by the ‘# Programs’ (lowest to highest), and then sorted by miR number (smallest to largest).
Differential expressions of mouse miRNA orthologs in bone during skeletal development
Based on miRNA-seq study of a mouse model for age-related osteoporosis (43), i.e. mmu-miR-140-5p, mmu-miR-143-5p, mmu-miR-34c-5p, mmu-miR-106b-3p, mmu-miR-30c-2-3p, mmu-miR-92a-2-5p and mmu-miR-17-3p, were differentially expressed between 25-m versus 2-m, 8-m versus 2-m and 25-m versus 8-m time points in bone (Table 4), with their corresponding human orthologs highlighted in Table 3. Two of them, i.e. mmu-miR-34c-5p and mmu-miR-140-5p, not only were significantly differentially expressed across all three comparisons, but also had same directions of fold changes (FCs) (i.e. consistently down-regulated from 2 to 8 and to 25 months). Most prominently, mmu-miR-140-5p had greatest FCs (5.88-, 4- and 1.47-folds of down-regulation) among these seven miRNAs, which attained highest statistical significance levels for 25-m versus 2-m (P = 9.15 × 10−91) and 8-m versus 2-m (P = 9.63 × 10−82) comparisons. Because hsa-miR-140-5p is highly conserved during evolution across a wide spectrum of species (Supplementary Material, Fig. S2), such that both mmu-miR-140-5p and dre-miR-140-5p have same sequences as hsa-miR-140-5p, only hsa-miR-140-5p was chosen for experimental validation.
Table 4.
Mouse miRNAs that show differential expressions in bone among three time points determined by miRNA-Seq
| miR-Name | 25-m versus. 2-m |
8-m versus. 2-m |
25-m versus. 8-m |
|||
|---|---|---|---|---|---|---|
| FC | P-value | FC | P-value | FC | P-value | |
| mmu-miR-140-5p | 0.17 | 9.15 × 10−91 | 0.25 | 9.63 × 10−82 | 0.68 | 1.79 × 10−3 |
| mmu-miR-143-5p | 0.45 | 1.91 × 10−27 | 0.34 | 1.22 × 10−51 | 1.34 | 1.90 × 10−3 |
| mmu-miR-34c-5p | 0.47 | 7.33 × 10−24 | 0.72 | 8.95 × 10−8 | 0.66 | 1.85 × 10−7 |
| mmu-miR-106b-3p | 1.34 | 1.10 × 10−4 | 1.00 | 0.96 | 1.33 | 1.43 × 10−4 |
| mmu-miR-30c-2-3p | 0.91 | 0.048 | 0.88 | 2.80 × 10−3 | 1.04 | 0.44 |
| mmu-miR-92a-2-5p | 0.66 | 0.1134 | 1.45 | 0.061 | 0.45 | 1.11 × 10−3 |
| mmu-miR-17-3p | 0.91 | 0.17 | 1.90 | 2.90 × 10−32 | 0.47 | 1.20 × 10−33 |
2-m, 2-month; 8-m, 8-month; 25-m, 25-month; FC, fold change. Data were obtained from He et al. (43). Three mouse miRNAs with significant differences (i.e. P < 0.05/3 = 0.0167) of their expressions across three time points (i.e. 2-m, 8-m and 25-m) were shown in italic font, which includes -miR-140-5p (in bold font). The miRNAs were sorted by their levels of statistical significance for 25-m versus. 2-m comparison (highest to lowest).
In vitro dual-luciferase reporter assay results
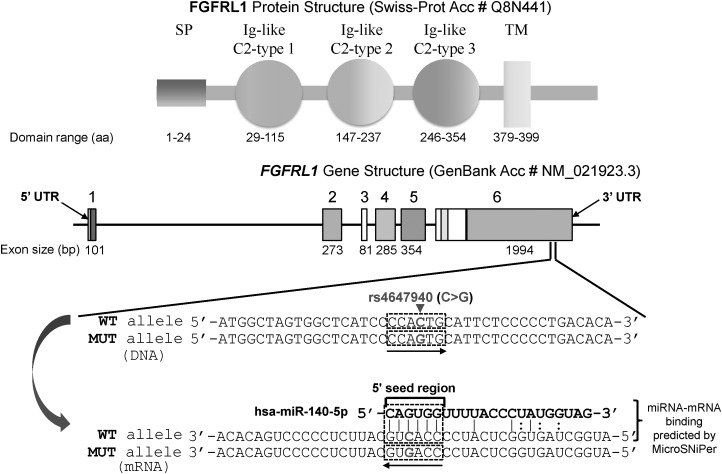
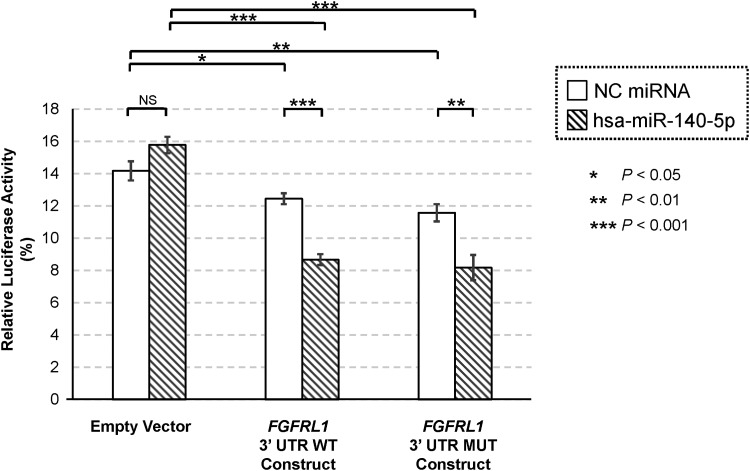
Human FGFRL1 gene, which resides in chromosome 4p16.3, consists of six exons. Exon 1 contains 5′ UTR followed by a coding sequence for signal peptide (SP). Exons 2, 4 and 5 encode immunoglobulin (Ig)-like C2-type 1, Ig-like C2-type 2 and Ig-like C2-type 3 domains, respectively. Exon 6 encodes transmembrane (TM) domain, cytoplasmic tail and 3′ UTR. Exon–intron structure of FGFRL1 is highly conserved based on previous phylogenetic analysis (44), and rs4647940 is located in the 3′ UTR, which is a part of exon 6 (Fig. 7). FGFRL1 rs4647940 WT allele (i.e. ‘C’) is complementary with the third position (underlined) of hsa-miR-140-5p 5′ seed region (i.e. 5′-CAGUGG-3′), whereas MUT allele (i.e. ‘G’) causes a mismatch. In dual-luciferase reporter assay, a negative control (NC) miRNA mimic was co-transfected with an empty vector, or a vector carrying FGFRL1 3′ UTR WT insert, or a vector carrying FGFRL1 3′ UTR MUT insert. In human embryonic kidney (HEK) 293 cells, for NC miRNA co-transfection, compared with an empty vector as baseline, a vector carrying either FGFRL1 3′ UTR WT insert (−12.15 ± 4.83%, P = 0.031) or FGFRL1 3′ UTR MUT insert (−18.33 ± 5.66%, P = 8.88 × 10−3) demonstrated significant inhibitions of respective relative luciferase activities, but no statistically significant difference was observed between 3′ UTR WT and MUT inserts (P = 0.20) (Fig. 8). For hsa-miR-140-5p co-transfection, compared with an empty vector, a vector carrying either FGFRL1 3′ UTR WT insert (−45.07 ± 3.90%, P = 4.19 × 10−7) or FGFRL1 3′ UTR MUT insert (−48.20 ± 5.95%, P = 1.06 × 10−5) exhibited significant inhibitions of relative luciferase activities. No statistically significant difference was detected between 3′ UTR WT and MUT inserts (P = 0.58). Nonetheless, relative luciferase activities differed significantly between NC miRNA and hsa-miR-140-5p for both 3′ UTR WT insert (P = 1.33 × 10−5) and 3′ UTR MUT insert (P = 0.0051), indicating that the FGFRL1 3′ UTR insert bearing rs4647940 is a potential target site for hsa-miR-140-5p.
Figure 7.
FGFRL1 gene structure, rs4647940 physical location and the potential binding of FGFRL1 mRNA with hsa-miR-140-5p. aa, amino acid; bp, base pair; Acc #, accession number; Ig, immunoglobulin; SP, signal peptide; TM, transmembrane; UTR, untranslated region; WT, wild-type; MUT, mutant. ‘|’ denotes perfect match; ‘:’ denotes G:U wobble.
Figure 8.
Dual-luciferase report assay results for FGFRL1 rs4647940. The relative luciferase activities for six different conditions (three reporter constructs: Empty/WT/MUT vectors, two miRNA types: NC miRNA/hsa-miR-140-5p miRNA). On the y-axis, relative luciferase activity (in percentage) was computed as the ratio of firefly luciferase activity to Renilla luciferase activity is given. On the x-axis, different transfection groups are shown. Data are presented as mean ± SEM from six independent replicates. miRNA, microRNA; MUT, mutant; NC, negative control; NS, not significant; SEM, standard error of the mean.
In vivo zebrafish microinjection experiment results
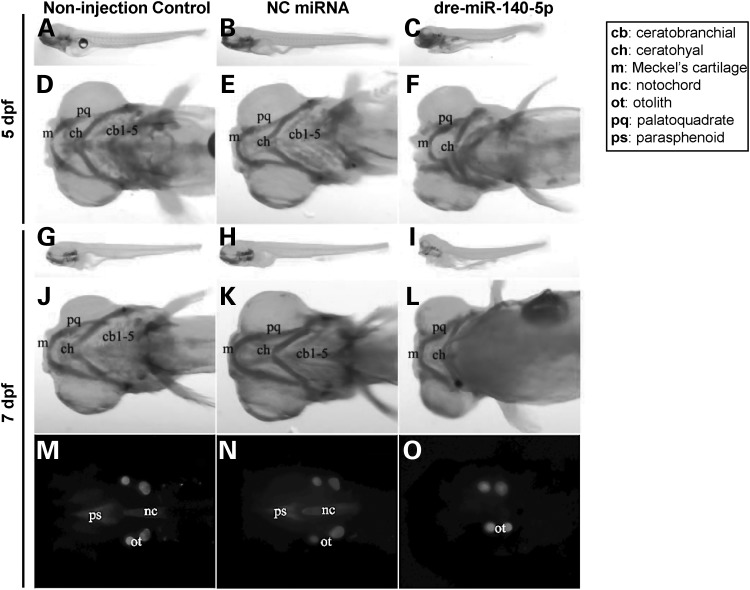
Three independent miRNA microinjection experiments were performed, with similar morphological phenotype results obtained. Data from a representative experiment of them were shown in Figure 9. N = 11, 14 and 14 zebrafish embryos were, respectively, included in non-injection control (left panel), NC miRNA-injected (middle panel), and dre-miR-140-5p-injected (right panel) groups. Morphological phenotypes were obtained at two time points—5 days post-fertilization (dpf) and 7 dpf, respectively. Figure 9A–F compared zebrafish skeletal morphologies across non-injection control, NC miRNA-injected and dre-miR-140-5p-injected groups at 5 dpf, whereas Figure 9G–O compared skeletal morphologies across them at 7 dpf, respectively. At 5 dpf, the body length of dre-miR-140-5p-injected larva appeared shorter (in example shown, 22.61%) compared with either non-injection control, or NC miRNA-injected larva, which had same lengths (Fig. 9A–C). A similar pattern of body length comparisons was observed at 7 dpf (Fig. 9G–I). Further, compared with non-injection control and NC miRNA-injected larvae at 5 dpf (Fig. 9D and J) and 7 dpf (Fig. 9E and K), dre-miR-140-5p-injected larvae at 5 dpf (Fig. 9F) and 7 dpf (Fig. 9L) showed somewhat reduced Meckel's cartilage (denoted by ‘m’) and almost absent ceratobranchial cartilages (denoted by ‘cb1-5’). Significant skeletal morphological differences were detected between non-injection control/NC miRNA-injected and dre-miR-140-5p-injected larvae at 7 dpf (Fig. 9M–O). Florescent images demonstrated that notochord (denoted by ‘nc’) and parasphenoid (denoted by ‘ps’) were present in non-injection control and NC miRNA-injected groups (Fig. 9M and N), but were absent in dre-miR-140-5p-injected group (Fig. 9O). Overall, skeletal malformations were observed in 0/14 (0.00%), 2/14 (14.29%) and 5/11 (45.45%) for non-injection control, NC miRNA-injected, and dre-miR-140-5p-injected groups, respectively. Although the percentage of dre-miR-140-5p-injected group (45.45%) was not statistically different from that of NC miRNA-injected group (14.29%) [P-value (two-sided) = 0.18], the percentage of non-injection group (0.00%) was significantly different from that of dre-miR-140-5p-injected group (45.45%) with a P-value (two-sided) = 0.0087. Further, non-injection control + NC miRNA-injected groups compared with dre-miR-140-5p-injected group gives a significant P-value (two-sided) = 0.012.
Figure 9.
Craniofacial and overall morphological phenotypes of zebrafish larvae in microRNA microinjection experiments. (A-O) Left, middle and right panels depict images corresponding to non-injection control, NC miRNA-injected and dre-miR-140-5p-injected groups at 5 dpf and 7 dpf larval stages, respectively. (A-E) 5 dpf stage of zebrafish larvae after microinjection with respective miRNAs were stained with Alcian blue 8 GX to observe overall morphological phenotypes (A-C) or craniofacial cartilage structures (D-E). (G-O) 7 dpf stage of zebrafish larvae after microinjection with respective miRNAs were stained with both Alcian blue 8 GX (G-L) and Alizarin red S (M-O) to observe overall morphological phenotypes (G-I) or craniofacial cartilage structures (J-L) or mineralized bone (MO). The RNA sequence of dre-miR-140-5p is exactly the same as that of hsa-miR-140-5p, indicating cross-species conservation (Supplementary Material, Fig. S1). Three independent experiments comparing non-injection control, NC miRNA-injected and dre-miR-140-5p-injected groups were performed, and consistent morphological phenotypes were obtained for each of these three groups across these experiments. Images presented are from one representative experiment. NC, negative control; dpf, days post-fertilization.
Discussion
A traditional GWA meta-analysis approach has been widely applied for discovering novel genetic loci for common human diseases. To date, such an approach has identified a large number of genetic loci underlying disease susceptibilities (45–47). However, in traditional GWA studies, the significantly associated variants are seldom those that play functional roles in phenotypic determination, but are instead in high LD with a causal variant. Consequently, the identification of causal variants is important in understanding molecular mechanisms underlying disease pathogenesis, such that additional well-focused intensive studies (e.g. fine mapping) are required to complement traditional GWA studies to identify disease-causing alleles. Because a functional candidate genomic region association approach focusing exclusively on SNPs with higher prior probabilities for biological functions may increase the type I error rate (i.e. α), the best strategy to differentiate true positives from false positives is to perform replication studies in independent samples. Therefore, focusing exclusively on those poly-miRTSs, a multi-stage approach, which typically includes a GWA discovery stage followed by in silico and de novo genotyping replication stages, is a conservative strategy to identify novel functional SNPs located in non-coding 3′ UTRs potentially undetected by previous traditional GWA meta-analysis studies due to an overly strict statistical significance threshold. Such a ‘hybrid’ approach combines strengths of both a multi-stage GWA meta-analysis approach and a hypothesis-driven approach. Johnson et al. (29) employed a similar multi-stage strategy of the current study for blood pressure and hypertension traits focusing exclusively on 6683 SNPs located within 60-kb of each of 30 candidate gene regions encoding known anti-hypertensive drug targets in stage I. Two novel SNPs, i.e. ADRB1 rs1801253 [for systolic blood pressure (SBP) and diastolic blood pressure (DBP)], and AGT rs2004776 (for DBP) were identified and replicated, which were missed by their previous traditional GWA scan (i.e. CHARGE Consortium) (48). Another study by Xu et al. (49) also applied a functional candidate gene association approach for coronary artery disease (CAD) that revealed two novel regulatory SNPs (rs7842 and rs4400266) located, respectively, in C3AR1 and C6 genes to be associated with CAD, which were missed by previous traditional GWA scans.
By interrogating exclusively 41 102 poly-miRTSs collected from Poly-miRTS database (50,51), the probability of detecting functional poly-miRTSs causally linked with BMD traits has significantly increased. A total of 11 stage I-selected poly-miRTSs (corresponding to 10 chromosomal loci) were selected based on (i) statistical significance threshold, (ii) allelic association direction and (iii) biological significance. In stage II in silico replication, FGFRL1 rs4647940 (P = 5.078 × 10−3) met a relatively stringent statistical significance threshold (i.e. P < 9.09 × 10−3) for FN-BMD in gender-combined sample. Because FGFRL1 rs4647940 is top signal among these stage I-selected 11 poly-miRTSs for FN-BMD in gender-combined sample, in order to avoid missing potentially functional poly-miRTSs for FN-BMD trait, among these 11 stage I-selected poly-miRTSs, we also included stage I's second top signal for FN-BMD in gender-combined sample, i.e. PRR5 poly-miRTS rs3213550 (P-value: 1.58 × 10−5), which is a disease-associated miRTSdSNP, although this SNP did not reach stage II significance threshold. In stage III, based on Bonferroni-corrected statistical significance threshold (P < 0.05/2 = 0.025) for FN-BMD trait in gender-combined sample, only FGFRL1 rs4647940 attained statistical significance (Fig. 1). In Zhang et al. (34), 33 stage 1-selected SNPs, [i.e. 10 SNPs reaching GWS (i.e. α = 5.00 × 10−8) and 23 SNPs reaching borderline significance (i.e. α = 1.00 × 10−6)] corresponding to 33 chromosomal loci were selected and subjected to stage 2 in silico analysis. None of the current study's 11 stage I-selected poly-miRTSs overlap with these 33 stage 1-selected SNPs of previous traditional study. Four chromosomal loci {i.e. 2q33 (PTH2R), 4p16.3 [intergenic (currently IDUA)], 6q25.1 (C6orf67–ESR1) and 20q13.33 (OSBPL2)} were shared in common, but were represented by totally different SNPs. Of these 33 stage 1-selected chromosomal loci, 13 previously reported and 2 novel loci reached conventional GWS (34). Of them, only chromosome 4p16.3 [intergenic (currently IDUA)] was commonly shared with the current study, but was represented by a totally different gene, i.e. FGFRL1. Estrada et al. (40) performed a traditional multi-stage GWA scan that studied ∼2.5 million autosomal + chromosome × SNPs in 17 cohorts (N = 32 961) in stage 1 (GWA discovery). Based on SNP selection criteria: (i) P < 5 × 10−6 (both unconditional and conditional analysis) for autosomal SNPs and (ii) P < 5 × 10−5 for chromosome × SNPs, 96 SNPs (87 autosomal SNPs and 9 chromosome × SNPs) corresponding to 83 chromosomal loci were subject to stage 2 de novo + in silico replication in 34 cohorts (N = 50 933). None of the current study's 11 stage I-selected poly-miRTSs overlap with these 96 stage 1-selected SNPs of Estrada et al. (40), but three chromosomal loci [4p16.3 (IDUA), 6q25.1 (C6orf67–ESR1) and 11q13.1 (SCYL1)] were shared in common, which were represented by totally different SNPs, and at 4p16.3 and 11q13.1, were represented by totally different genes. At stage 2 of Estrada et al. (40), 64 [54 chromosomal loci containing 56 gene loci (40)] of these 96 stage 1-selected SNPs attained conventional GWS (i.e. P < 5 × 10−8, stages 1 + 2-only), 16 (14 chromosomal loci) of which were further replicated in stage 3 for association with fractures (P < 5 × 10−4, stage 3-only) in 50 fracture cohorts (N = 133 460). Of these 54 BMD-associated chromosomal loci that attained conventional GWS, only 4p16.3 (IDUA) was commonly shared with the current study, but was represented by a totally different gene, i.e. FGFRL1. None of those 16 BMD- and fracture-associated loci were shared with the current study. Taken together, FGFRL1 rs4647940 is a novel poly-miRTS for FN-BMD that has been discovered in stage I and subsequently replicated in stages II and III of the current study, respectively. If not focusing exclusively on poly-miRTSs in stage I, this FGFRL1 rs4647940 (P = 7.67 × 10−6 for FN-BMD at stage I) would not have been discovered at all—e.g. this potentially functional SNP did not reach stage 1’s borderline significance threshold α = 1.0 × 10−6 in Zhang et al. (34), and therefore did not survive to be further validated in stages 2 and 3. Therefore, by using a more relaxed statistical significance threshold (i.e. 5 × 10−5, because only 41 102 poly-miRTSs were examined), this SNP was detected in stage I of the current study, and upon replications in subsequent stages II and III, reached GWS (P = 8.87 × 10−12) in stage I + II + III (N = 24 662). A meta-analysis of rs4647940 conditional on rs3755955 remained suggestively significant (P = 1.35 × 10−3) for FN-BMD in stage I gender-combined sample at α = 5.00 × 10−3, whereas a meta-analysis of rs4647940 conditional on both rs3755955 and rs6827815 was also significant (P = 1.33 × 10−3) for FN-BMD in stage 1 gender-combined sample.
For FGFRL1 rs4647940, based on predictions by four in silico programs, 13 human miRNAs could potentially bind to this poly-miRTS site (Table 3). Of them, according to miRNA-seq data (43), mouse orthologs of hsa-miR-17-3p, mmu-miR-30c-2-3p, hsa-miR-34c-5p, hsa-miR-92a-2-5p, hsa-miR-106b-3p, hsa-miR-140-5p and hsa-miR-143-5p appear to be differentially expressed at different time points during development in a mouse model of age-related osteoporosis (Table 4). Of them, mmu-miR-140-5p (a mouse ortholog of hsa-miR-140-5p) appeared to exhibit greatest magnitudes of differential expressions among comparisons of three time points (i.e. 2-m, 8-m and 25-m), and highest levels of statistical significance for 25-m versus 2-m and 8-m versus 2-m comparisons (Table 4). To experimentally test the potential biological role of FGFRL1 poly-miRTS rs4647940, both in vitro and in vivo functional studies were conducted. First, luciferase activity assay results showed that, when co-transfected with hsa-miR-140-5p, reporter construct containing either FGFRL1 3′ UTR WT insert or FGFRL1 3′ UTR MUT insert repressed luciferase activity to similar extents (Fig. 8). These experimental findings are concordant with a regulatory role of hsa-miR-140-5p in modulating FGFRL1 expression by binding to this 3′ UTR segment harboring rs4647940, causing a pronounced inhibition of luciferase activity. As indicated in both Figure 7 and Supplementary Material, Figure S2, hsa-miR-140-5p contains a 6-mer 5′ seed region (predicted by MicroSNiPer) that is complementary to FGFRL1 rs4647940 SNP site. Because miRNAs with 6-mer seed regions typically modestly down-regulate their respective mRNA targets (52), these luciferase activity assay results are in agreement with the notion that FGFRL1 poly-miRTS rs4647940 is located in 5′ seed region binding site of hsa-miR-140-5p. Because an miRNA's 5′ seed sequence plays an important role in target recognition and binding, a 3′ UTR polymorphism located in an miRNA's 5′ seed region binding site, e.g. FGFRL1 rs4647940, may have a higher probability of functional importance, compared with a 3′ UTR polymorphism located in that miRNA's 3′ mismatch tolerant region (MTR) binding site.(53) We further tested whether hsa-miR-140-5p plays a significant role in skeletal formation in vivo. Nakamura et al. (54) demonstrated that Mir140−/− mice had shortened endochondral bones. In another Mir140−/− mouse model, a mild skeletal phenotype was observed postnatally, with reduced stature and lower body weight, as well as craniofacial deformities characterized by shorter snout and domed skull (55). In a previous zebrafish model, Mir140 is found expressed in cartilage of pharyngeal arches and in head skeleton (56). Further, Eberhart et al. (57) observed that dre-miR-140-5p-injected zebrafish exhibited craniofacial skeletal defects. The current study's in vivo zebrafish miRNA microinjection experiments gave rise to similar results, demonstrating that dre-miR-140-5p-injected group had a significantly higher percentage of abnormal craniofacial skeletal formation compared with non-injected control + NC miRNA-injected groups.
Fibroblast growth factor (FGF) signaling pathway is pivotal in a variety of developmental processes, including cartilage formation (58) and bone growth (59). FGFRL1 is FGFR gene family's fifth member, and mutations of FGFR1-FGFR4 cause skeletal disorders (e.g. craniosynostosis syndromes and chondrodysplasias) (60). Interestingly, FGFRL1 orthologous genes in species ranging from cnidarians to bilaterians are linked with other FGF signaling pathway members within a physical distance <10 Mb (44). For instance, in sea urchin, Fgfrl1 is closely linked to Fgfr1. Human FGFRL1 is located in close proximity to FGFR3 (44). The fact that FGFRL1 is physically linked to other FGF signaling pathway genes throughout evolution is suggestive of FGFRL1’s functional importance in this biological pathway, and likely, also in bone formation: (i) FGFRL1 is expressed in skeletal tissues shown by northern blotting and in situ hybridization (61); (ii) a depletion of zebrafish fgfrl1a and fgfrl1b results in a craniofacial phenotype (62); (iii) Fgfrl1−/− mice exhibited hypoplasia of all skeletal elements, including shortened axial and appendicular skeletons, malformed vertebrae, small pelvic girdle and small rib cage (63); and (iv) in humans, as in zebrafish and mice, FGFRL1 could be implicated in craniofacial formation during embryogenesis, such that FGFRL1 deletions or frameshift mutation result in craniofacial malformations (64–66).
The current study's in vitro and in vivo experimental validation results, combined with findings of previous functional studies of FGFRL1 and hsa-miR-140-5p as well as clinical genetic studies of FGFRL1 gene mutations, provided strong evidence that FGFRL1 and hsa-miR-140-5p, which are evolutionarily conserved, play significant biological roles during skeletogenesis. FGFRL1 rs4647940, attaining conventional GWS for FN-BMD in the current study, could change hsa-miR-140-5p's 5′ seed region binding site located in FGFRL1’s 3′ UTR, which could then modulate FGFRL1’s mRNA expression level that can affect bone formation.
Our study has several limitations. First, the luciferase activity assay's results demonstrated that FGFRL1 3′ UTR WT and MUT inserts could both bind to hsa-miR-140-5p rather than NC miRNA, but MUT allele did not appear to significantly de-repress luciferase activity in comparison with WT allele. However, rs4647940 may only have a relatively moderate effect on FGFRL1 mRNA-hsa-miR-140-5p binding, because 5′ seed region of hsa-miR-140-5p is a 6-mer. Further, multiple factors could affect results of luciferase activity assay, such as insert length, cell line, and transfection conditions. Besides hsa-miR-140-5p, FGFRL1 rs4647940 could affect other bone metabolism-related miRNAs' binding sites, e.g. hsa-miR-17-3p and hsa-miR-92a-2-5p (Table 3) predicted by RNAfold. Secondly, in vivo experiment in a zebrafish model showed that dre-miR-140-5p microinjection results in a dramatic effect on bone development (Fig. 9), but whether such an effect was mediated by its binding with a target site located on 3′ UTR of either fgfrl1a or fgfrl1b in zebrafish remains to be elucidated. Nevertheless, results of mouse miRNA-seq (Table 4) and zebrafish microinjection (Fig. 9) experiments unequivocally demonstrated a critical role of miR-140-5p in bone growth. Given that FGFRL1 rs4647940 is possibly located in a 5′ seed region binding site of miR-140-5p based on in vitro luciferase activity assay results (Fig. 7), FGFRL1 rs4647940 is likely a regulatory variant causally linked with BMD.
Taken together, through a combination of a three-stage, poly-miRTS-centric GWA meta-analysis and functional experiments, FGFRL1 poly-miRTS rs4647940 is significantly associated with FN-BMD trait, attaining conventional GWS. Based on abundant data accumulated from in vitro and in vivo experimental and clinical genetics studies, FGFRL1 rs4647940 is likely to play an important role in modulating the expression of FGFRL1 during skeletogenesis, and further functional characterizations of this SNP in bone metabolism is deserved.
Materials and Methods
A flow chart outlining steps of the current study is depicted in Figure 1. Detailed information on study participants, phenotype measurement and modeling, DNA genotyping, quality control (QC) and genotype imputation for the three-stage GWA meta-analysis is provided in Supplementary material, Materials and Methods.
Association analysis
In the current study, only at stage I (seven GWA cohorts), gender-specific (i.e. male- and female-specific) and gender-combined (i.e. male and female) analyses were performed. Nonetheless, only four stage I's GWA cohorts (i.e. OOS, KCOS, COS and FHS) contain males (Table 1), such that male-specific sample size was much smaller than female-specific sample size, giving rise to less statistical power. Therefore, same as in the traditional GWA scan (34), Q–Q plot was generated only for female-specific and gender-combined samples, respectively. At stages II and III, because there were two cohorts at each stage, only gender-combined analyses were performed. Association tests at each stage are elaborated as follows. In stage I, associations of 41 102 directly typed or imputed poly-miRTSs with LS-, HIP- and FN-BMD traits were tested under an additive genetic model for each GWA cohort. Specifically, for five population-based samples—OOS, KCOS, COS, WHI-AA and WHI-HIS, associations were examined by fitting a linear regression model by MACH2QTL (http://www.sph.umich.edu/csg/abecasis/MACH/download/), in which an allele dosage was used as a predictor of phenotype. For two family-based samples—FHS and IFS, a mixed linear model was applied that took into consideration the effect of genetic relatedness within each pedigree (67). In stage II, association analyses in RS sample were performed by MACH2QTL implemented by GRIMP (68). Imputation and association analyses in KoGES were performed by IMPUTE v2 (69) and PLINK (70). Detailed statistical methods for association analyses of HIP-, LS- and FN-BMD traits in AOGC sample were presented previously by Duncan et al. (71). Briefly, for HIP-BMD, association analysis for imputed SNPs was performed using an additive genetic model including four EIGENSTRAT eigenvectors by MACH2DAT, accounting for uncertainties in imputed genotype data by weighting genotypes by their posterior probabilities. For LS- and FN-BMD, Z-transformed residual BMD scores (in g/cm2) were generated for the entire AOGC cohort after adjusting for age, age2, weight and center of BMD measurement. An association test developed by Kung et al. (72) was applied to account for selections of extreme samples. In stage III, association tests were carried out using PLINK (70) for each study.
Meta-analysis
As described by Zhang et al. (34), a P-value based meta-analysis was used in METAL (http://www.sph.umich.edu/csg/abecasis/metal/) (73), weighted by square root of sample size. Cochran's Q statistic and I2 were computed as measures of between-study heterogeneity. Random-effect meta-analyses were performed for those poly-miRTSs with Q statistic P < 0.05 or I2 value > 50% by applying the ‘rmeta’ package (http://cran.r-project.org/web/packages/rmeta/index.html). In stage 1, meta-analysis of a given SNP conditional on one or two other SNPs in LD was also performed by GCTA version 1.20 (http://www.complextraitgenomics.com/software/gcta/massoc.html) (74).
Regional association plot and LD r2 plot
Regional association plots were generated by applying LocusZoom (http://csg.sph.umich.edu/locuszoom/) (75). LD r2 Plots were generated by Haploview (41,42) for 58 SNPs at chromosome 4p16.3 for OOS and KCOS cohorts, respectively.
Poly-miRTSs in miRNA target sites
The poly-miRTS-centric GWA meta-analysis focuses exclusively on 41 102 poly-miRTSs in stage I, in contrast to 5 842 825 SNPs in stage 1 of previous traditional GWA scan (34). Detailed information on poly-miRTS selection is presented in Supplementary Material, Text S1.
Computational predictions of RNA secondary structures
The miRNA–mRNA hybridization is a two-step process such that an miRNA first binds to a short accessible region of its target mRNA, and then, the target mRNA's secondary structure unfolds as the miRNA completes its binding to target site (76). The secondary structure of an mRNA can influence its accessibility to miRNA (77). In order to assess potential impacts of WT and MUT alleles on the secondary structure of the 3′ UTR of FGFRL1, a 121-bp FGFRL1 3′ UTR segment carrying either WT or MUT allele of rs4647940 were predicted by RNAfold from the Vienna RNA package (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi) (78,79) based on MFE algorithm with default parameters.
Multiple sequence alignment and sequence logo
Polymorphisms located at evolutionarily conserved nucleotides are likely to be of greater functional significance than those located at evolutionarily divergent nucleotides. To further bioinformatically assess functional significance for FGFRL1 rs4647940, cross-species comparisons of orthologous sequences of FGFRL1 3′ UTR segment encompassing rs4647940 from six species, i.e. human, chimpanzee, gorilla, gibbon, orangutan and rhesus macaque, was performed by ClustalX version 1.81 (80) with default parameters. A Sequence Logo corresponding to a middle 15-bp DNA segment was generated using WebLogo online program (81).
In silico predictions of miRNAs that bind to a poly-miRTS
Human miRNAs that could bind to FGFRL1 rs4647940 were predicted by four online tools: (i) MicroSNiPer (http://cbdb.nimh.nih.gov/microsniper) (82), (ii) MirSNP (http://cmbi.bjmu.edu.cn/mirsnp) (83), (iii) RegRNA 2.0 (http://regrna2.mbc.nctu.edu.tw/) (84,85) and (iv) TargetScan (http://www.targetscan.org/) 6.2 (11,16,86).
In vitro dual-luciferase reporter assays
To assess whether FGFRL1 3′ UTR segment containing either WT or MUT allele of rs4647940 could bind to hsa-miR-140-5p in vitro, a pmirGLO dual-luciferase miRNA target expression vector harboring FGFRL1 3′ UTR WT or MUT inset was constructed, which was subsequently transfected into HEK 293 cells. Details about cloning and transfection, as well as luciferase activity assays are presented in Supplementary Material, Text S1.
In vivo zebrafish microinjection experiments
To assess an in vivo role of hsa-miR-140-5p in bone formation, zebrafish microinjection experiments were performed, with each experiment consisting of three groups: (i) a non-injection control group, (ii) an NC miRNA microinjection group and (iii) a dre-miR-140-5p microinjection group, respectively. Detailed experimental procedures were described in Supplementary Material, Text S1. These procedures were repeated three times, and microscope image data of morphological phenotypes from a representative experiment at 5 dpf and 7 dpf were obtained from each respective group.
Supplementary Material
Funding
The study was partially supported by Shanghai Leading Academic Discipline Project (S30501) and start-up fund from Shanghai University of Science and Technology. The investigators of this work were also supported by grants from NIH (P50AR055081, R01AG026564, R01AR050496, RC2DE020756, R01AR057049, R03TW008221 and R01GM109068) and the Franklin D. Dickson/Missouri Endowment and the Edward G. Schlieder Endowment. The Framingham Heart Study (FHS) is conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with Boston University (Contract Number N01-HC-25195). This manuscript was not prepared in collaboration with investigators of FHS and does not necessarily reflect the opinions or views of FHS, Boston University or NHLBI. Funding for SHARe genotyping was provided by NHLBI Contract N02-HL-64278. SHARe Illumina genotyping was provided under an agreement between Illumina and Boston University. Funding support for the Framingham Bone Mineral Density data sets was provided by NIH grants R01 AR/AG 41398, R01 AR050066 and R03 AG20321. Funding support for the Genetic Determinants of Bone Fragility was provided through the National Institute of Aging (NIA) Division of Geriatrics and Clinical Gerontology. Genetic Determinants of Bone Fragility is a genome-wide association studies funded as part of the NIA Division of Geriatrics and Clinical Gerontology. Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, was provided by the NIA Division of Geriatrics and Clinical Gerontology and the NIA Division of Aging Biology. Support for the collection of datasets and samples were provided by the parent grant, Genetic Determinants of Bone Fragility (P01-AG018397). Funding support for the genotyping which was performed at the Johns Hopkins University Center for Inherited Diseases Research was provided by the NIH NIA. The Women's Health Initiative (WHI) program is funded by the NHLBI, National Institutes of Health, US Department of Health and Human Services through contracts N01WH22110, 24152, 32100-2, 32105-6, 32108-9, 32111–13, 32115, 32118-32119, 32122, 42107–26, 42129–32 and 44221. This manuscript was not prepared in collaboration with investigators of the WHI, has not been reviewed and/or approved by WHI, and does not necessarily reflect the opinions of the WHI investigators or the NHLBI. WHI Population Architecture Using Genomics and Epidemiology (PAGE) is funded through the NHGRI PAGE network (U01 HG004790). Assistance with phenotype harmonization, SNP selection, data cleaning, meta-analyses, data management and dissemination and general study coordination, was provided by the PAGE Coordinating Center (U01HG004801-01). The FHS datasets used for the analyses described in this manuscript were obtained from dbGaP at http://www.ncbi.nlm.nih.gov/sites/entrez?db:gap through dbGaP accession phs000007.v14.p6. Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, of Genetic Determinants of Bone Fragility study was provided by the NIA Division of Geriatrics and Clinical Gerontology and the NIA Division of Aging Biology. The lndiana Fragility Study (IFS) datasets used for the analyses described in this manuscript were obtained from dbGaP at http://www.ncbi.nlm.nih.gov/sites/entrez?db:gap through dbGaP accession phs000138.v2.p1. The WHI datasets used for the analyses described in this manuscript were obtained from dbGaP at http://www.ncbi.nlm.nih.gov/sites/entrez?db:gap through dbGaP accession phs000200.v6.p2. This work was supported in part by the Netherlands Organization of Scientific Research NWO Investments (175.010.2005.011 and 911-03-012 to K.E., F.R. and A.G.U.); the Research Institute for Diseases in the Elderly (014-93-015 and RIDE2 to K.E., F.R. and A.G.U.); the Netherlands Genomics Initiative (NGI)/Netherlands Organization for Scientific Research (050-060-810 to K.E., F.R. and A.G.U.); the National Genome Research Institute, Korean Center for Disease Control and Prevention (2001-2003-348- 6111-221, 2004-347-6111-213 and 2005-347-2400-2440-215 to H.J.C., J.Y.L., B.G.H., C.S.S. and N.H.C.); the National Health and Medical Research Council Project Grant (511132 to AOGC) and the National Health and Medical Research Council (Australia) Career Development Award (569807 to E.L.D.). Funding of AOGC was also received from the Australian Cancer Research Foundation and Rebecca Cooper Foundation (Australia). The Rotterdam study is funded by Erasmus Medical Center and Erasmus University, Rotterdam, Netherlands Organization for the Health Research and Development (ZonMw), the Research Institute for Diseases in the Elderly (RIDE), the Ministry of Education, Culture and Science, the Ministry for Health, Welfare and Sports, the European Commission (DG XII) and the Municipality of Rotterdam. T.N. is supported in part by a start-up fund of Tulane University. L.Z. is partially supported by National Natural Science Foundation of China project (31100902). M.A.B. is funded by a National Health and Medical Research Council (Australia) Senior Principal Research Fellowship. Y.-P. W. is funded by the US National Institutes of Health and National Science Foundation. H.-W. D. is partially supported by the Franklin D. Dickson/Missouri Endowment and is supported by the Edward G. Schlieder Endowment.
Supplementary Material
Acknowledgements
We thank Dr Karol Estrada and Dr Fernando Rivadeneira (both from Department of Internal Medicine and Department of Epidemiology, Erasmus Medical Center, Rotterdam, The Netherlands) for their help with analyzing and delivering the Rotterdam Study data, and Tian Hu (Department of Epidemiology, Tulane University School of Public Health and Tropical Medicine) for assistance in generating the meta-analysis figure. We are grateful for Dr Jesse D. Ziebarth and Dr Yan Cui (both from University of Tennessee Health Science Center) for helpful comments on application of Poly-miRTS Databases. The AOGC thanks the research nurses involved in this study [Ms Barbara Mason and Ms Amanda Horne (Auckland), Ms Linda Bradbury and Ms Kate Lowings (Brisbane), Ms Katherine Kolk and Ms Rumbidzai Tichawangana (Geelong); Ms Helen Steane (Hobart); Ms Jemma Christie (Melbourne); and Ms Janelle Rampellini (Perth)]. The AOGC also acknowledges gratefully technical support from Ms Kathryn Addison, Ms Marieke-Brugmans, Ms Catherine Cremen, Ms Johanna Hadler and Ms Karena Pryce.
Conflict of Interest statement. None declared.
References
- 1.Kanis J.A., Melton L.J., III, Christiansen C., Johnston C.C., Khaltaev N. (1994) The diagnosis of osteoporosis. J. Bone Miner. Res., 9, 1137–1141. [DOI] [PubMed] [Google Scholar]
- 2.Cardon L.R., Garner C., Bennett S.T., Mackay I.J., Edwards R.M., Cornish J., Hegde M., Murray M.A., Reid I.R., Cundy T. (2000) Evidence for a major gene for bone mineral density in idiopathic osteoporotic families. J. Bone Miner. Res., 15, 1132–1137. [DOI] [PubMed] [Google Scholar]
- 3.Howard G.M., Nguyen T.V., Harris M., Kelly P.J., Eisman J.A. (1998) Genetic and environmental contributions to the association between quantitative ultrasound and bone mineral density measurements: a twin study. J. Bone Miner. Res., 13, 1318–1327. [DOI] [PubMed] [Google Scholar]
- 4.Nguyen T.V., Howard G.M., Kelly P.J., Eisman J.A. (1998) Bone mass, lean mass, and fat mass: same genes or same environments? Am. J. Epidemiol., 147, 3–16. [DOI] [PubMed] [Google Scholar]
- 5.Peacock M., Turner C.H., Econs M.J., Foroud T. (2002) Genetics of osteoporosis. Endocr. Rev., 23, 303–326. [DOI] [PubMed] [Google Scholar]
- 6.Yang T.L., Zhao L.J., Liu Y.J., Liu J.F., Recker R.R., Deng H.W. (2006) Genetic and environmental correlations of bone mineral density at different skeletal sites in females and males. Calcif. Tissue Int., 78, 212–217. [DOI] [PubMed] [Google Scholar]
- 7.Chang T.C., Mendell J.T. (2007) microRNAs in vertebrate physiology and human disease. Annu. Rev. Genomics Hum. Genet., 8, 215–239. [DOI] [PubMed] [Google Scholar]
- 8.Gu S., Kay M.A. (2010) How do miRNAs mediate translational repression? Science, 1, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kozomara A., Griffiths-Jones S. (2014) miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res., 42, D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomas L.F., Saetrom P. (2014) Circular RNAs are depleted of polymorphisms at microRNA binding sites. Bioinformatics, 30, 2243–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedman R.C., Farh K.K., Burge C.B., Bartel D.P. (2009) Most mammalian mRNAs are conserved targets of microRNAs. Genome Res., 19, 92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartel D.P. (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell, 116, 281–297. [DOI] [PubMed] [Google Scholar]
- 13.Krek A., Grun D., Poy M.N., Wolf R., Rosenberg L., Epstein E.J., MacMenamin P., da Piedade I., Gunsalus K.C., Stoffel M., et al. (2005) Combinatorial microRNA target predictions. Nat. Genet., 37, 495–500. [DOI] [PubMed] [Google Scholar]
- 14.Doench J.G., Sharp P.A. (2004) Specificity of microRNA target selection in translational repression. Genes Dev., 18, 504–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brennecke J., Stark A., Russell R.B., Cohen S.M. (2005) Principles of microRNA-target recognition. PLoS Biol., 3, e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewis B.P., Burge C.B., Bartel D.P. (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell, 120, 15–20. [DOI] [PubMed] [Google Scholar]
- 17.Ameres S.L., Martinez J., Schroeder R. (2007) Molecular basis for target RNA recognition and cleavage by human RISC. Cell, 130, 101–112. [DOI] [PubMed] [Google Scholar]
- 18.Haley B., Zamore P.D. (2004) Kinetic analysis of the RNAi enzyme complex. Nat. Struct. Mol. Biol., 11, 599–606. [DOI] [PubMed] [Google Scholar]
- 19.Maragkakis M., Alexiou P., Papadopoulos G.L., Reczko M., Dalamagas T., Giannopoulos G., Goumas G., Koukis E., Kourtis K., Simossis V.A., et al. (2009) Accurate microRNA target prediction correlates with protein repression levels. BMC Bioinformatics, 10, 295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chi S.W., Hannon G.J., Darnell R.B. (2012) An alternative mode of microRNA target recognition. Nat. Struct. Mol. Biol., 19, 321–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haas U., Sczakiel G., Laufer S.D. (2012) MicroRNA-mediated regulation of gene expression is affected by disease-associated SNPs within the 3′-UTR via altered RNA structure. RNA Biol., 9, 924–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen K., Rajewsky N. (2006) Natural selection on human microRNA binding sites inferred from SNP data. Nat. Genet., 38, 1452–1456. [DOI] [PubMed] [Google Scholar]
- 23.Chin L.J., Ratner E., Leng S., Zhai R., Nallur S., Babar I., Muller R.U., Straka E., Su L., Burki E.A., et al. (2008) A SNP in a let-7 microRNA complementary site in the KRAS 3′ untranslated region increases non-small cell lung cancer risk. Cancer Res., 68, 8535–8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mishra P.J., Mishra P.J., Banerjee D., Bertino J.R. (2008) MiRSNPs or MiR-polymorphisms, new players in microRNA mediated regulation of the cell: Introducing microRNA pharmacogenomics. Cell Cycle, 7, 853–858. [DOI] [PubMed] [Google Scholar]
- 25.Abelson J.F., Kwan K.Y., O'Roak B.J., Baek D.Y., Stillman A.A., Morgan T.M., Mathews C.A., Pauls D.L., Rasin M.R., Gunel M., et al. (2005) Sequence variants in SLITRK1 are associated with Tourette's syndrome. Science, 310, 317–320. [DOI] [PubMed] [Google Scholar]
- 26.Dickson D.W., Baker M., Rademakers R. (2010) Common variant in GRN is a genetic risk factor for hippocampal sclerosis in the elderly. Neurodegener. Dis., 7, 170–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang G., van der Walt J.M., Mayhew G., Li Y.J., Zuchner S., Scott W.K., Martin E.R., Vance J.M. (2008) Variation in the miRNA-433 binding site of FGF20 confers risk for Parkinson disease by overexpression of alpha-synuclein. Am. J. Hum. Genet., 82, 283–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Landi D., Gemignani F., Barale R., Landi S. (2008) A catalog of polymorphisms falling in microRNA-binding regions of cancer genes. DNA Cell Biol., 27, 35–43. [DOI] [PubMed] [Google Scholar]
- 29.Johnson A.D., Newton-Cheh C., Chasman D.I., Ehret G.B., Johnson T., Rose L., Rice K., Verwoert G.C., Launer L.J., Gudnason V., et al. (2011) Association of hypertension drug target genes with blood pressure and hypertension in 86,588 individuals. Hypertension, 57, 903–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Welter D., MacArthur J., Morales J., Burdett T., Hall P., Junkins H., Klemm A., Flicek P., Manolio T., Hindorff L., et al. (2014) The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res., 42, D1001–D1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar V., Westra H.J., Karjalainen J., Zhernakova D.V., Esko T., Hrdlickova B., Almeida R., Zhernakova A., Reinmaa E., Vosa U., et al. (2013) Human disease-associated genetic variation impacts large intergenic non-coding RNA expression. PLoS Genet., 9, e1003201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen K., Song F., Calin G.A., Wei Q., Hao X., Zhang W. (2008) Polymorphisms in microRNA targets: a gold mine for molecular epidemiology. Carcinogenesis, 29, 1306–1311. [DOI] [PubMed] [Google Scholar]
- 33.Sethupathy P., Collins F.S. (2008) MicroRNA target site polymorphisms and human disease. Trends in Genetics: Trends Genet., 24, 489–497. [DOI] [PubMed] [Google Scholar]
- 34.Zhang L., Choi H.J., Estrada K., Leo P.J., Li J., Pei Y.F., Zhang Y., Lin Y., Shen H., Liu Y.Z., et al. (2014) Multistage genome-wide association meta-analyses identified two new loci for bone mineral density. Hum. Mol. Genet., 23, 1923–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bruno A.E., Li L., Kalabus J.L., Pan Y., Yu A., Hu Z. (2012) miRdSNP: a database of disease-associated SNPs and microRNA target sites on 3′UTRs of human genes. BMC Genomics, 13, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xian L., Wu X., Pang L., Lou M., Rosen C.J., Qiu T., Crane J., Frassica F., Zhang L., Rodriguez J.P., et al. (2012) Matrix IGF-1 maintains bone mass by activation of mTOR in mesenchymal stem cells. Nat. Med., 18, 1095–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Long F. (2012) Building strong bones: molecular regulation of the osteoblast lineage. Nat. Rev. Mol. Cell Biol., 13, 27–38. [DOI] [PubMed] [Google Scholar]
- 38.Fulzele K., Riddle R.C., DiGirolamo D.J., Cao X., Wan C., Chen D., Faugere M.C., Aja S., Hussain M.A., Bruning J.C., et al. (2010) Insulin receptor signaling in osteoblasts regulates postnatal bone acquisition and body composition. Cell, 142, 309–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Woo S.Y., Kim D.H., Jun C.B., Kim Y.M., Haar E.V., Lee S.I., Hegg J.W., Bandhakavi S., Griffin T.J., Kim D.H. (2007) PRR5, a novel component of mTOR complex 2, regulates platelet-derived growth factor receptor beta expression and signaling. J. Biol. Chem., 282, 25604–25612. [DOI] [PubMed] [Google Scholar]
- 40.Estrada K., Styrkarsdottir U., Evangelou E., Hsu Y.H., Duncan E.L., Ntzani E.E., Oei L., Albagha O.M., Amin N., Kemp J.P., et al. (2012) Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat. Genet., 44, 491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barrett J.C., Fry B., Maller J., Daly M.J. (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics, 21, 263–265. [DOI] [PubMed] [Google Scholar]
- 42.Barrett J.C. (2009) Haploview: Visualization and analysis of SNP genotype data. Cold Spring Harb. Protoc., 4, 1–5. [DOI] [PubMed] [Google Scholar]
- 43.He X., Zhang W., Liao L., Fu X., Yu Q., Jin Y. (2013) Identification and characterization of microRNAs by high through-put sequencing in mesenchymal stem cells and bone tissue from mice of age-related osteoporosis. PLoS One, 8, e71895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bertrand S., Somorjai I., Garcia-Fernandez J., Lamonerie T., Escriva H. (2009) FGFRL1 is a neglected putative actor of the FGF signalling pathway present in all major metazoan phyla. BMC Evol. Biol., 9, 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Evangelou E., Ioannidis J.P. (2013) Meta-analysis methods for genome-wide association studies and beyond. Nat. Rev. Genet., 14, 379–389. [DOI] [PubMed] [Google Scholar]
- 46.Panagiotou O.A., Willer C.J., Hirschhorn J.N., Ioannidis J.P. (2013) The power of meta-analysis in genome-wide association studies. Annu. Rev. Genomics Hum. Genet., 14, 441–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zeggini E., Ioannidis J.P. (2009) Meta-analysis in genome-wide association studies. Pharmacogenomics, 10, 191–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Levy D., Ehret G.B., Rice K., Verwoert G.C., Launer L.J., Dehghan A., Glazer N.L., Morrison A.C., Johnson A.D., Aspelund T., et al. (2009) Genome-wide association study of blood pressure and hypertension. Nat. Genet., 41, 677–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu C., Yang Q., Xiong H., Wang L., Cai J., Wang F., Li S., Chen J., Wang C., Wang D., et al. (2014) Candidate pathway-based genome-wide association studies identify novel associations of genomic variants in the complement system associated with coronary artery disease. Circ. Cardiovasc. Genet., 7, 887–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bao L., Zhou M., Wu L., Lu L., Goldowitz D., Williams R.W., Cui Y. (2007) PolymiRTS Database: linking polymorphisms in microRNA target sites with complex traits. Nucleic Acids Res., 35, D51–D54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ziebarth J.D., Bhattacharya A., Chen A., Cui Y. (2012) PolymiRTS Database 2.0: linking polymorphisms in microRNA target sites with human diseases and complex traits. Nucleic Acids Res., 40, D216–D221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Witkos T.M., Koscianska E., Krzyzosiak W.J. (2011) Practical aspects of microRNA target prediction. Curr. Mol. Med., 11, 93–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mishra P.J., Bertino J.R. (2009) MicroRNA polymorphisms: the future of pharmacogenomics, molecular epidemiology and individualized medicine. Pharmacogenomics, 10, 399–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nakamura Y., Inloes J.B., Katagiri T., Kobayashi T. (2011) Chondrocyte-specific microRNA-140 regulates endochondral bone development and targets Dnpep to modulate bone morphogenetic protein signaling. Mol. Cell Biol., 31, 3019–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miyaki S., Sato T., Inoue A., Otsuki S., Ito Y., Yokoyama S., Kato Y., Takemoto F., Nakasa T., Yamashita S., et al. (2010) MicroRNA-140 plays dual roles in both cartilage development and homeostasis. Genes Dev., 24, 1173–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wienholds E., Kloosterman W.P., Miska E., Alvarez-Saavedra E., Berezikov E., de Bruijn E., Horvitz H.R., Kauppinen S., Plasterk R.H. (2005) MicroRNA expression in zebrafish embryonic development. Science, 309, 310–311. [DOI] [PubMed] [Google Scholar]
- 57.Eberhart J.K., He X., Swartz M.E., Yan Y.L., Song H., Boling T.C., Kunerth A.K., Walker M.B., Kimmel C.B., Postlethwait J.H. (2008) MicroRNA Mirn140 modulates Pdgf signaling during palatogenesis. Nat.Genet., 40, 290–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Walshe J., Mason I. (2003) Fgf signalling is required for formation of cartilage in the head. Dev. Biol., 264, 522–536. [DOI] [PubMed] [Google Scholar]
- 59.Kronenberg H.M. (2003) Developmental regulation of the growth plate. Nature, 423, 332–336. [DOI] [PubMed] [Google Scholar]
- 60.Trueb B. (2011) Biology of FGFRL1, the fifth fibroblast growth factor receptor. Cell Mol. Life Sci., 68, 951–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Trueb B., Zhuang L., Taeschler S., Wiedemann M. (2003) Characterization of FGFRL1, a novel fibroblast growth factor (FGF) receptor preferentially expressed in skeletal tissues. J. Biol. Chem., 278, 33857–33865. [DOI] [PubMed] [Google Scholar]
- 62.Hall C., Flores M.V., Murison G., Crosier K., Crosier P. (2006) An essential role for zebrafish Fgfrl1 during gill cartilage development. Mech. Dev., 123, 925–940. [DOI] [PubMed] [Google Scholar]
- 63.Catela C., Bilbao-Cortes D., Slonimsky E., Kratsios P., Rosenthal N., Te Welscher P. (2009) Multiple congenital malformations of Wolf-Hirschhorn syndrome are recapitulated in Fgfrl1 null mice. Dis. Model Mech., 2, 283–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Engbers H., van der Smagt J.J., van 't Slot R., Vermeesch J.R., Hochstenbach R., Poot M. (2009) Wolf-Hirschhorn syndrome facial dysmorphic features in a patient with a terminal 4p16.3 deletion telomeric to the WHSCR and WHSCR 2 regions. Eur. J. Hum. Genet., 17, 129–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rieckmann T., Zhuang L., Fluck C.E., Trueb B. (2009) Characterization of the first FGFRL1 mutation identified in a craniosynostosis patient. Biochim. Biophys. Acta., 1792, 112–121. [DOI] [PubMed] [Google Scholar]
- 66.Matoso E., Ramos F., Ferrao J., Pires L.M., Mascarenhas A., Melo J.B., Carreira I.M. (2014) Interstitial 287 kb deletion of 4p16.3 including FGFRL1 gene associated with language impairment and overgrowth. Mol. Cytogenet., 7, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang L., Li J., Pei Y.F., Liu Y., Deng H.W. (2009) Tests of association for quantitative traits in nuclear families using principal components to correct for population stratification. Ann. Hum. Genet., 73, 601–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Estrada K., Abuseiris A., Grosveld F.G., Uitterlinden A.G., Knoch T.A., Rivadeneira F. (2009) GRIMP: a web- and grid-based tool for high-speed analysis of large-scale genome-wide association using imputed data. Bioinformatics, 25, 2750–2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Howie B.N., Donnelly P., Marchini J. (2009) A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet., 5, e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet., 81, 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li Y., Willer C., Sanna S., Abecasis G. (2009) Genotype imputation. Annu. Rev. Genomics Hum. Genet., 10, 387–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kung A.W., Xiao S.M., Cherny S., Li G.H., Gao Y., Tso G., Lau K.S., Luk K.D., Liu J.M., Cui B., et al. (2010) Association of JAG1 with bone mineral density and osteoporotic fractures: a genome-wide association study and follow-up replication studies. Am. J. Hum. Genet., 86, 229–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Willer C.J., Li Y., Abecasis G.R. (2010) METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics, 26, 2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang J., Lee S.H., Goddard M.E., Visscher P.M. (2011) GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet., 88, 76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pruim R.J., Welch R.P., Sanna S., Teslovich T.M., Chines P.S., Gliedt T.P., Boehnke M., Abecasis G.R., Willer C.J. (2010) LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics, 26, 2336–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Long D., Lee R., Williams P., Chan C.Y., Ambros V., Ding Y. (2007) Potent effect of target structure on microRNA function. Nat. Struct. Mol. Biol., 14, 287–294. [DOI] [PubMed] [Google Scholar]
- 77.Mahen E.M., Watson P.Y., Cottrell J.W., Fedor M.J. (2010) mRNA secondary structures fold sequentially but exchange rapidly in vivo. PLoS Biol., 8, e1000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gruber A.R., Lorenz R., Bernhart S.H., Neubock R., Hofacker I.L. (2008) The Vienna RNA websuite. Nucleic Acids Res., 36, W70–W74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hofacker I.L. (2003) Vienna RNA secondary structure server. Nucleic Acids Res., 31, 3429–3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jeanmougin F., Thompson J.D., Gouy M., Higgins D.G., Gibson T.J. (1998) Multiple sequence alignment with Clustal X. Trends Biochem. Sci., 23, 403–405. [DOI] [PubMed] [Google Scholar]
- 81.Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. (2004) WebLogo: a sequence logo generator. Genome Res., 14, 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Barenboim M., Zoltick B.J., Guo Y., Weinberger D.R. (2010) MicroSNiPer: a web tool for prediction of SNP effects on putative microRNA targets. Hum. Mutat., 31, 1223–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liu C., Zhang F., Li T., Lu M., Wang L., Yue W., Zhang D. (2012) MirSNP, a database of polymorphisms altering miRNA target sites, identifies miRNA-related SNPs in GWAS SNPs and eQTLs. BMC Genomics, 13, 661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Huang H.Y., Chien C.H., Jen K.H., Huang H.D. (2006) RegRNA: an integrated web server for identifying regulatory RNA motifs and elements. Nucleic Acids Res., 34, W429–W434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chang T.H., Huang H.Y., Hsu J.B., Weng S.L., Horng J.T., Huang H.D. (2013) An enhanced computational pl.atform for investigating the roles of regulatory RNA and for identifying functional RNA motifs. BMC Bioinformatics, 14(Suppl. 2), S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Grimson A., Farh K.K., Johnston W.K., Garrett-Engele P., Lim L.P., Bartel D.P. (2007) MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell, 27, 91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.