Abstract
Mitochondrial (mt) DNA can be classified into haplogroups representing different geographic and/or racial origins of populations. The H haplogroup is protective against age-related macular degeneration (AMD), while the J haplogroup is high risk for AMD. In the present study, we performed comparison analyses of human retinal cell cybrids, which possess identical nuclei, but mtDNA from subjects with either the H or J haplogroups, and demonstrate differences in total global methylation, and expression patterns for two genes related to acetylation and five genes related to methylation. Analyses revealed that untreated-H and -J cybrids have different expression levels for nuclear genes (CFH, EFEMP1, VEGFA and NFkB2). However, expression levels for these genes become equivalent after treatment with a methylation inhibitor, 5-aza-2′-deoxycytidine. Moreover, sequencing of the entire mtDNA suggests that differences in epigenetic status found in cybrids are likely due to single nucleotide polymorphisms (SNPs) within the haplogroup profiles rather than rare variants or private SNPs. In conclusion, our findings indicate that mtDNA variants can mediate methylation profiles and transcription for inflammation, angiogenesis and various signaling pathways, which are important in several common diseases.
Introduction
Mitochondria are unique organelles with their own maternally inherited DNA (mtDNA), containing a non-coding control region (MT-Dloop), important for replication and transcription. The mtDNA-coding region encodes for 37 genes, including 13 proteins, 22 tRNAs and 2 rRNAs, that are critical for oxidative phosphorylation (OXPHOS). The mtDNA can be classified into haplogroups, defined by an accumulation of single nucleotide polymorphisms (SNPs), which represent the different geographic origins of populations. Investigations report that the mtDNA haplogroups can be either protective or high risk for a number of diseases, including cancers, diabetes, Alzheimer's disease, Parkinson's disease and cardiomyopathies (1,2). Age-related macular degeneration (AMD) is the leading cause of vision loss in older individuals. It has been shown that the J haplogroup is associated with AMD (3–8), while the H haplogroup has a protective effect (7). Recent studies have shown that epigenetics may also play a key role in the progression of AMD (9).
Transmitochondrial cybrids (cell lines containing identical nuclei, but mtDNA from different individuals) have been used to investigate the mitochondrial–nuclear interactions for various diseases (10–22). Our previous studies have shown that cybrids containing different mtDNA haplogroups have: (i) differential expression of nuclear genes involved in cell signaling, inflammation and apoptosis pathways; and (ii) different responses to sublethal ultraviolet radiation (23–26). However, the signaling transduction mechanism(s) involved with the mitochondrial–nuclear interactions in the cybrids are not fully understood. Studies have shown that mitochondria and methylation status are interconnected. When cells were depleted of their mitochondria, the degree of DNA methylation was affected (27). Osteosarcoma cybrids with the J haplogroup mtDNA had higher levels of total methylation compared with cybrids with H haplogroup mtDNA (28). Methylation levels can be influenced by S-adenosylmethionine (SAM) formation and mitochondrial functions (29,30). Although human mtDNA has been shown to have both CpG and non-CpG methylation sites (31), the interplay between various mtDNA haplogroups associated with methylation-related nuclear gene expressions needs further elucidation.
This report compares human retinal cell cybrids possessing either the H or J haplogroup mtDNA, and demonstrates differences in: (i) total global methylation and (ii) expression patterns for two genes related to acetylation (HAT1 and HDAC1) and five genes related to methylation (MAT2B, DNMT1, DNMT3A, DNMT3B and MBD2). The untreated H versus J cybrids have different expression levels for methylation-regulated nuclear genes (CFH, EFEMP1, VEGFA and NFkB2), but after treatment with 5-aza-dC, a methylation inhibitor, the expression levels become equivalent. Our results suggest that the mtDNA haplogroup background within cells can significantly influence epigenetic profiles and create an environment capable of upregulating specific major pathways critical for disease processes.
Results
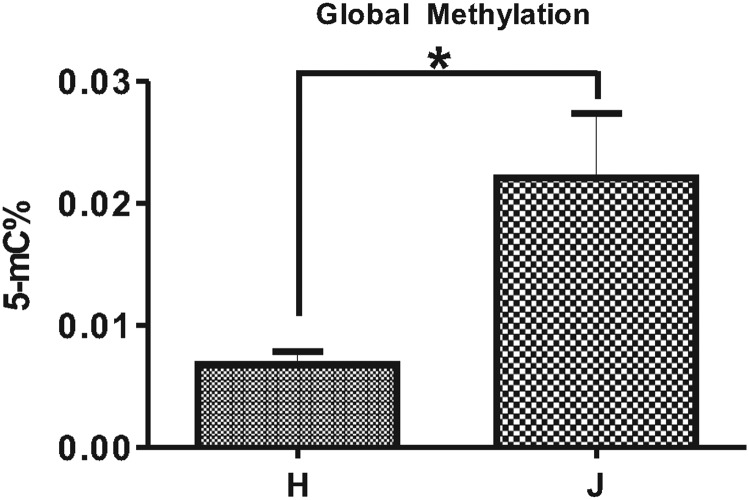
Elevated levels of global DNA methylation in J versus H cybrids
All cybrid cultures were grown under identical conditions and periods of time. Results showed the 5-mC% mean value for the H cybrids was 0.007 ± 0.001 and the 5-mC% mean value for the J cybrids was 0.022 ± 0.0053 (Fig. 1). The mean difference for the H versus the J cybrids was −0.015 ± 0.005 (P = 0.02). This indicates that the J cybrids showed higher levels of total global methylation compared with the H cybrids.
Figure 1.
J cybrids show increased levels of global DNA methylation. The levels of methylated DNA (5-mC%) were quantified in J (n = 3) and H (n = 3) cybrids, cultured under identical conditions. The H cybrids had a lower 5-mC% mean value (0.007 ± 0.001) compared with the J cybrids (0.022 ± 0.0053, P = 0.02). Samples were run in duplicate and the experiment was repeated twice. Statistical significance is denoted by *P < 0.05.
Altered expression levels of genes associated with epigenetic pathways
The gene expression levels for 11 genes related to epigenetic modification (Fig. 2) were analyzed by Q-PCR in H cybrids versus J cybrids (Tables 1 and 2). The expression levels for HAT1, a histone acetyltransferase, were significantly lower in the J cybrids compared to the H cybrids (0.63-fold, P = 0.0001). With respect to the deacetylase enzymes, the J cybrids showed a 0.68-fold decrease in expression for HDAC1 gene (P = 0.003) compared with the H cybrids, but HDAC6 (P = 0.18) and HDAC11 (P = 0.4) showed similar gene expression levels for H and J cybrids. The expression levels for SIN3A, a transcriptional regulator protein, which promotes deacetylation, were also similar in H and J cybrids (P = 0.19).
Figure 2.
Schematic of the acetylation and methylation enzymes affecting transcription. Upper panel shows the active transcription state for chromatin with unmethylated CpG sites on the DNA and acetylated histone sites. Bottom panel shows the inactive transcription state for chromatin with methylated CpG sites and non-acetylated histone H3 lysine residues. HATs, histone acetyltransferase; HDACs, histone deacetylase; DNMTs, DNA (cystosine-5) methyltransferase; MBDs, methyl-CpG binding domain protein 2; H3, histone lysine residue; M, methylated CpG site.
Table 1.
Description of genes analyzed in this study
| Symbol | Gene name | GenBank accession no. |
Function |
|---|---|---|---|
| HAT1 | Histone acetyltransferase | NM_001033085 NM_003642 | Adds acetyl group; promotes transcription. |
| HDAC1 | Histone deacetylase 1 | NM_004964 | Class I histone deacetylase involved in control of cell proliferation, differentiation, growth and apoptosis; represses transcription. |
| HDAC6 | Histone deacetylase 6 | NM_006044 | Class II histone deacetylase. |
| HDAC11 | Histone deacetylase 11 | NM_024827 | Class IV histone deacetylase localized to the nucleus; involved in regulating interleukin 10 expression. |
| SIN3A | Sin3 transcription regulator homolog A (yeast) |
NM_001145357 NM_001145358 NM_015477 |
Transcriptional regulator for STAT3; Promotes deacetylation. |
| MAT2B | Methionine adenosyltransferase II, beta | NM_013283 | Catalyzes the biosynthesis of S-adenosylmethionine, the major methyl donor; Regulatory beta subunit. |
| MBD2 | methyl-CpG binding domain protein 2 | NM_003927 | Part of nuclear protein family related by the presence of a methyl-CpG binding domain (MBD). Binds specifically to methylated DNA; represses transcription from methylated gene promoters; functions as a demethylase to activate transcription. |
| MBD4 | Methyl-CpG binding domain protein 4 | NM_003925 | Binds specifically to methylated DNA; Involved in protein interactions, and DNA repair. |
| DNMT1 | DNA (cystosine-5) methyltransferase 1 | NM_001130823 NM_001379 | Methylates CpG residues, preferentially methylates hemimethylated DNA. Associates with DNA replication sites in S phase maintaining the methylation pattern in newly synthesized strand, essential for epigenetic inheritance. |
| DNMT3A | DNA (cystosine-5) methyltransferase 3A |
NM_022552 NM_153759 NM_175629 |
Methylates de novo during development. Important for genomic imprinting. |
| DNMT3B | DNA (cystosine-5) methyltransferase 3B |
NM_001207055; NM_001207056 NM_006892 NM_175848 NM_175849 NM_175850 |
Methylates de novo during development. Important for genomic imprinting. |
| CFH | Complement factor H | NM_000186 | Essential in regulation of complement activation. |
| EFEMP1 | EGF containing fibulin-like extracellular matrix protein 1 |
NM_004105 NM_018894 NM_001039348 NM_001039349 |
Encoded protein contains tandemly repeated EGF-like repeats followed by C-terminus fibulin-like domain. |
| NFκB2 | Nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 |
NM_001077494 NM_001077493 NM_002502 |
Subunit of transcription factor complex nuclear factor-kappa-B complex which is central activator of inflammation and immune function genes. |
| VEGFA | Vascular endothelial growth factor A |
NM_001025366 NM_001025367 NM_001025368 NM_001025369 NM_001025370 NM_001033756 NM_001171622 NM_001171623 NM_001171624 NM_001171625 NM_001171626 NM_001171627 NM_001171628 NM_001171629 NM_001171630 NM_001204384 NM_001204385 NM_003376 |
Member of PDGF/VEGF growth factor family. Acts on endothelial cells. Causes increased vascular permeability, angiogenesis, vasculogenesis, endothelial cell growth, cell migration and inhibits apoptosis. Mutations in this gene have been associated with proliferative and non-proliferative diabetic retinopathy. |
| HPRT1 | Hypoxanthine phosphoribosyltransferase 1 | NM_000194 | Transferase, which catalyzes conversion of hypoxanthine to inosine monophosphate & guanine to guanosine monophosphate. Plays a central role in the generation of purine nucleotides through the purine salvage pathway. Endogenous control. |
| HMBS | Hydroxymethylbilane synthase |
NM_000190 NM_001024382 NM_001258208 NM_001258209 |
Member of the hydroxymethylbilane synthase superfamily. Third enzyme of the heme biosynthetic pathway. Catalyzes the head to tail condensation of four porphobilinogen molecules into the linear hydroxymethylbilane. Endogenous control. |
| ALAS1 | 5′-aminolevulinate synthase 1 |
NM_000688 XM_005264944 |
This gene encodes the mitochondrial enzyme which catalyzes the rate-limiting step in heme (iron-protoporphyrin) biosynthesis. The enzyme encoded by this gene is the housekeeping enzyme; the level of the mature encoded protein is regulated by heme: high levels of heme down-regulate the mature enzyme in mitochondria while low heme levels up-regulate. |
| TUBB | Tubulin, beta class I | NM_178014 NM_001293213 | This gene encodes a beta tubulin protein. This protein forms a dimer with alpha tubulin and acts as a structural component of microtubules. |
Table 2.
Differential gene expression in cybrids H versus J
| Symbol | H versus J ΔΔCT/ |
H versus J Fold | H versus J P-value |
|---|---|---|---|
| HAT1a | −0.67 ± 0.16 | 0.63 | 0.0001 |
| HDAC1b | −0.55 ± 0.16 | 0.68 | 0.003 |
| HDAC6b | −0.43 ± 0.5 | 0.74 | 0.18 |
| HDAC11b | −0.33 ± 0.38 | 0.8 | 0.4 |
| SIN3Ab | −0.41 ± 0.30 | 0.75 | 0.19 |
| MAT2Bb | 0.6 ± 0.16 | 1.51 | 0.002 |
| MBD2^^a | −1.3 ± 0.36 | 0.4 | 0.001 |
| MBD4b | 0.08 ± 0.23 | 1.06 | 0.74 |
| DNMT1a | −2 ± 0.2 | 0.24 | <0.0001 |
| DNMT3A^a | −1.73 ± 0.2 | 0.3 | <0.0001 |
| DNMT3B^a | −1.88 ± 0.2 | 0.32 | <0.0001 |
Fold values >1 indicate upregulation of the gene compared with H cybrids; Fold values <1 indicate downregulation of the gene compared with H cybrids; H cybrids are assigned a value of 1.
Measured versus HMBS; ^ versus ALAS1; ^^ versus TUBB as housekeeper; Fold = 2ΔΔCT.
an = 7 different H cybrids and 6 different J cybrids, with three values for each sample.
bn = 3 different H cybrids and 3 different J cybrids, with three values for each sample.
The expression levels for the MAT2B gene, which catalyzes the biosynthesis of SAM a major methyl donor, were significantly higher in the J cybrids compared with the H cybrids (1.51-fold, P = 0.002). The MAT1A levels were minimal in both the J and H cybrids (data not shown). DNMT1, a DNA methyltransferase, which specifically targets the mtDNA (32), was expressed at lower levels in the J cybrids compared with the H cybrids (0.24-fold, P = 0.0001). The DNMT3A and DNMT3B expression levels were also significantly lower in the J cybrids compared with the H cybrids (0.3-fold, P < 0.0001 and 0.27-fold, P < 0.001). The J cybrids showed a significant decrease in the expression levels of MBD2 compared with the H cybrids (0.4-fold, P = 0.001). There was no difference observed in the expression levels for MBD4 in the H versus J cybrids (1.06-fold, P = 0.74).
Methylation inhibitor studies comparing H versus J cybrids
Studies were performed to compare responses of the H and J cybrids to methylation inhibition. Briefly, H and J cybrids were treated with the methylation inhibitor 5-aza-dC and subsequently the expression levels of four nuclear genes associated with AMD were measured (Table 3). When untreated-J and -H cultures were compared with each other, the CFH expression levels were 0.44-fold lower in the untreated-J cybrids compared with the untreated-H cybrids (P < 0.0001). After 5-aza-dC treatment, the treated-J and -H cybrids expressed similar levels of CFH (1.07-fold, P = 0.42). When the untreated cybrids were compared, the EFEMP1 gene showed 0.71-fold lower expression levels in the untreated-J cybrids compared with the untreated-H cybrids (P = 0.015). However, there was no difference in the expression levels for EFEMP1 in the treated-H and -J cybrids (1.2-fold, P = 0.2) after treatment with 5-aza-dC. The NFkB2 gene was expressed at lower levels in the untreated-J cybrids compared with the untreated-H cybrids (0.69-fold, P = 0.03), but after the 5-aza-dC treatment, the treated-H and -J cybrids expressed similar levels of NFkB2 (1.04-fold, P = 0.71). VEGFA is a critical gene for neovascularization and is important for development and disease processes. When the untreated cybrids were compared with each other, the untreated-J cybrids expressed 0.62-fold lower levels of VEGFA compared with the untreated-H cybrids (P = 0.0006). When the cybrids are demethylated with 5-aza-dC, the treated-H and -J cybrids showed similar expression levels for the VEGFA gene (1.1-fold, P = 0.3) (Fig. 3).
Table 3.
Expression levels of genes before and after treatment with 5-aza-dC, a methylation inhibitor
| H Cybridsa versus J Cybrids (Untreated) P-value ΔΔCT/Fold | H Cybridsa versus J Cybrids (5-aza-dC Treated) P-value ΔΔCT/Fold | |
|---|---|---|
| CFHb | <0.0001 −1.17 ± 0.12/0.44 |
0.42 0.1 ± 0.13/1.07 |
| EFEMP1c | 0.015 −0.48 ± 0.2/0.71 |
0.2 0.23 ± 0.18/1.2 |
| NFkB2c | 0.03 −0.52 ± 0.22/0.69 |
0.71 0.06 ± 0.16/1.04 |
| VEGFAc | 0.0006 −0.67 ± 0.15/0.62 |
0.3 0.18 ± 0.17/1.1 |
Fold values >1 indicate upregulation of the gene.
Fold values <1 indicate downregulation of the gene.
Fold = 2ΔΔCT.
H cybrids, n = 3 different individuals; J cybrids, n = 3 different individuals. Each sample was run in triplicate. Experiment was repeated twice.
aH cybrids assigned a value of 1.
bMeasured versus HMBS as housekeeper.
cMeasured versus HPRT1 as housekeeper.
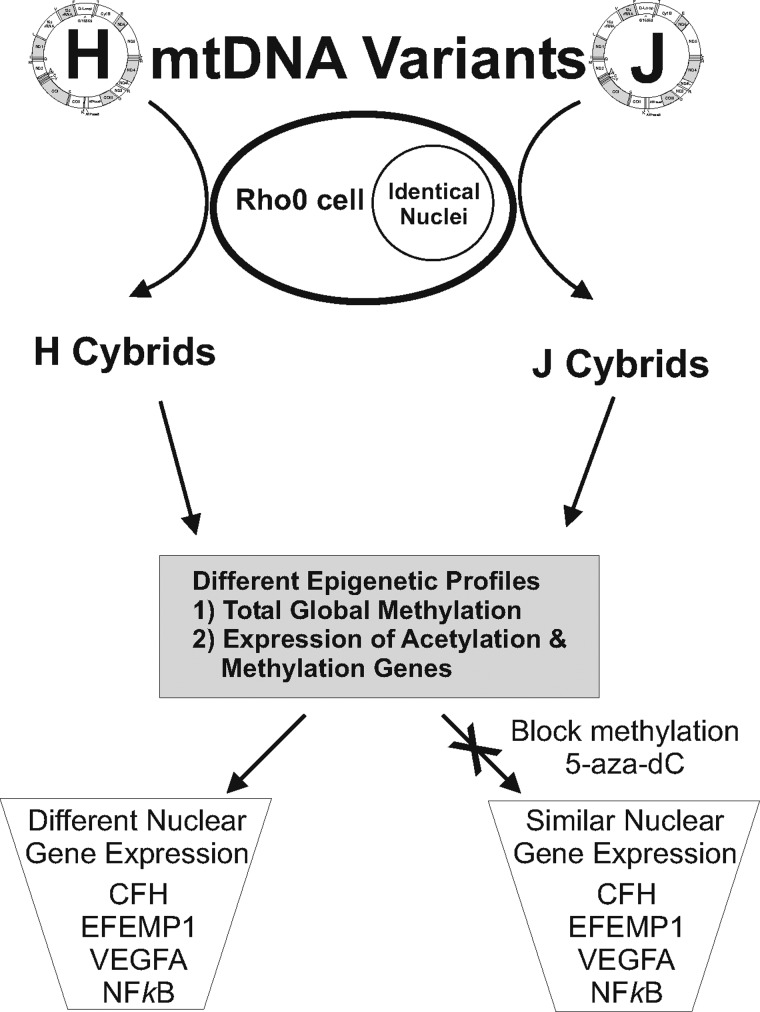
Figure 3.
Schematic summarizing epigenetic profiles of H versus J cybrids. The H and J cybrids have different levels of total global methylation and expression of acetylation and methylation-related genes. Untreated H and J cybrids show significantly different transcription levels for CFH (P < 0.0001), EFEMP1 (P = 0.015), NFkB2 (P = 0.03) and VEGFA (P = 0.0006). After 48 h treatment with 5-aza-dC, a methylation inhibitor, then the gene expression levels are equivalent in the H and J cybrid cultures; CFH (P < 0.42, EFEMP1 (P = 0.2), NFkB2 (P = 0.71) and VEGFA (P = 0.3). Rho0, lacking mtDNA; 5-aza-dC, 5-aza-2′-deoxycytidine; CFH, Complement factor H; EFEMP1, EGF containing fibulin-like extracellular matrix protein1; VEGFA, vascular endothelial growth factor A; NFkB2, nuclear factor of kappa light polypeptide gene enhancer in B-cells 2.
In the 5-aza-dC experiments, the HPRT1 gene was used as the housekeeper for EFEMP1, NFkB2 and VEGFA. The HMBS gene was the housekeeper for CHF. To determine if the changes in the demethylation experiments were due to methylation regulation of the housekeeper genes, the expression level of HMBS as a target gene and HPRT1 as the housekeeper gene was measured and found to be similar before and after treatment with 5-aza-dC (0.97-fold, P = 0.77). As a second analysis, the HPRT1 gene (target) was measured against TUBB gene (housekeeper) and similarly, there was no change in expression after treatment with 5-aza-dC (1.22-fold, P = 0.15). If the 5-aza-dC were causing demethylation, then the values for the genes would have varied.
Sequences comparisons of the H and J cybrids for the MT-Dloop
The entire control regions for H (n = 7) and J (n = 6) cybrids were sequenced and analyzed. Sequencing profiles for the MT-Dloops of H and J cybrids were similar to the Cambridge Reference Sequence (Table 4). The areas of greatest SNP variations were from nucleotides (np) 263–461, a region which contains the mtDNA loci for Conserved Sequence Block 2 (np 299–315), H Strand Origin (np 110–441) and Hypervariable Segment 2 (np 57–372). There was also high variability with C insertions at the np 310–321 region. The H and J cybrids had similar total numbers of CpG and non-CpG methylation sites in the MT-Dloop, as represented by ACA, CCA, TCA, GCA and ACT (33) (Table 5).
Table 4.
Representative sequences showing the CpG and non-CpG sites (as represented by ACA,  ,
,  ,
,  , and
, and  ) within the MT-Dloop from J Cybrid (J) and H Cybrid (H) compared to the revised Cambridge Reference Standard (R)
) within the MT-Dloop from J Cybrid (J) and H Cybrid (H) compared to the revised Cambridge Reference Standard (R)
 |
Table 5.
Potential methylation sites in the MT-Dloop for J and H Cybrids
| CpG Sites | Non-CpG Sites |
Total | |||||
|---|---|---|---|---|---|---|---|
| CpG | ACA | CCA | TCA | GCA | ACT | ||
| rCRS | 23 | 26 | 23 | 15 | 12 | 15 | 114 |
| J | 23 | 26 | 20 | 17 | 12 | 14 | 112 |
| H | 23 | 26 | 23 | 14 | 12 | 14 | 112 |
rCRS, revised Cambridge Reference Standard (based upon GenBank number NC_012920).
(From http://www.mitomap.org).
Sequence variations of the H and J cybrid mtDNA
The original three H and J cybrids have been sequenced and reported to be J1d1a, J1c1 and J1c7 along with H, H5a and H66a (24). In this investigation, additional cybrids were used and the entire mtDNA from each of the cybrids was sequenced (Tables 6 and 7) and compared with the Cambridge Reference Sequence. SNPs we define as unique are not listed in www.MitoMap.org. Private SNPs are those that do not define the haplogroup. The rs# numbers for the mtDNA SNPs were obtained from rCRS GenBank number NC_012920. The rs# numbers for 31 SNPs for the J cybrids have been listed. However, 11 of the mtDNA SNPs have not been assigned rs# numbers. For the H cybrids, there are 24 SNPs that had rs# numbers, but 17 SNPs lack the rs# identification.
Table 6.
Sequencing results for the entire mtDNA in J Cybrids
| Loci: MT- | AA Change | rs# | J 13-74 | J1c1a | J 13-107 | J1b | J 12-43 | J1b1b1 |
|---|---|---|---|---|---|---|---|---|
| RNR1 | 2 853 518 | 750A>G | a | 750A>G | a | 750A>G | a | |
| RNR1 | 2 001 030 | 1438A>G | a | 1438A>G | a | 1438A>G | a | |
| RNR2 | 3 928 306 | 3010G>A | J1 | 3010G>A | J1 | 3010G>A | J1 | |
| RNR2 | Unique | na | 3847T>C | J1c17 | ||||
| ND1 | Y-H | 41 460 449 | 3394T>C | J1c1 | ||||
| ND1 | Y-H | 1 599 988 | 4216T>C | JT | 4216T>C | JT | 4216T>C | JT |
| TI | Unique | na | 4322C>T | b | 4322C>T | b | ||
| ND2 | SYN | na | 4664C>T | b | ||||
| ND2 | SYN | 3 021 086 | 4769A>G | a | 4769A>G | a | 4769A>G | a |
| ND2 | A-T | 3 021 088 | 5460G>A | J1b1 | ||||
| CO2 | V-I | na | 7805G>A | b | ||||
| CO2 | SYN | na | 8200T>C | b | ||||
| CO2 | Non-Coding | 8896 | 8269G>A | J1b | 8269G>A | J1b | ||
| ATP6 | T-A | 2 001 031 | 8860A>G | a | 8860A>G | a | 8860A>G | a |
| CO3 | SYN | na | 9635A>C | J1c1a | ||||
| ND3 | T-A | 2 853 826 | 10398A>G | J | 10398A>G | J | 10398A>G | J |
| TR | 2 00 478 835 | 10410T>A | J1b1b1 | |||||
| ND4 | SYN | 3 915 952 | 11251A>G | JT | 11251A>G | JT | 11251A>G | JT |
| ND4 | SYN | na | 11623C>T | J1c1a | ||||
| ND5 | SYN | 28 359 172 | 12612A>G | J | 12612A>G | J | 12612A>G | J |
| ND5 | A-I | 28 359 178 | 13708G>A | J | 13708G>A | J | 13708G>A | J |
| ND5 | S-P | 2 00 380 057 | 13879T>C | J1b1 | ||||
| ND5 | SYN | 3 70 031 192 | 13899T>C | J1c1a | ||||
| ND5 | unique | na | 14028A>G | b | ||||
| CYB | F-L | 28 357 681 | 14798T>C | J1c | ||||
| CYB | T-A | 2 853 508 | 15326A>G | a | 15326A>G | a | 15326A>G | a |
| CYB | L-I | 5 27 236 209 | 15452C>A | JT | 15452C>A | JT | 15452C>A | JT |
| Dloop | Non-Coding | 1 47 903 261 | 16069C>T | J | 16069C>T | J | 16069C>T | J |
| Dloop | Non-Coding | 1 47 029 798 | 16126T>C | JT | 16126T>C | JT | 16126T>C | JT |
| Dloop | Non-Coding | 41 419 246 | 16145G>A | J1b | 16145G>A | J1b | ||
| Dloop | Non-Coding | na | 16222C>T | J1b | ||||
| Dloop | Non-Coding | 1 38 126 107 | 16261C>T | J1b | 16261C>T | J1b | ||
| Dloop | Non-Coding | 62 581 312 | 150C>T | J1b17 | ||||
| Dloop | Non-Coding | 3 72 099 630 | 200A>G | b | ||||
| Dloop | Non-Coding | 41 323 649 | 228G>A | J1c | ||||
| Dloop | Non-Coding | 2 853 515 | 263A>G | a | 263A>G | a | ||
| Dloop | Non-Coding | na | 271C>T | J1b1b | ||||
| Dloop | Non-Coding | 41 528 348 | 295C>T | J | 295C>T | J | 295C>T | J |
| Dloop | Non-Coding | 41 402 146 | 462C>T | J1 | 462C>T | J1 | 462C>T | J1 |
| Dloop | Non-Coding | na | 482T> | J1c1 | ||||
| Dloop | Non-Coding | 28 625 645 | 489T>C | J | 489T>C | J | 489T>C | J |
| Dloop | Non-Coding | 1 13 683 159 | 508A>G | b |
aPrivate: non-haplogroup defining SNP found in both H and J cybrids.
bPrivate: non-haplogroup defining SNP found in single H or J cybrid.
na, rs# not assigned. Unique: SNP not listed on www.mitomap.org.
Table 7.
Sequencing results for the entire mtDNA in H Cybrids
| Loci: MT- | AA Change | rs# | H 11-10 | H4a1a | H 11-23 | H11a2a2 | H 11-35 | H1 | H 13-49 | H1j |
|---|---|---|---|---|---|---|---|---|---|---|
| RNR1 | 2 853 518 | 750A>G | a | 750A>G | a | 750A>G | a | 750A>G | a | |
| RNR1 | 3 888 511 | 961T>G | H11a | |||||||
| RNR1 | 2 001 030 | 1438A>G | a | 1438A>G | a | 1438A>G | a | 1438A>G | a | |
| RNR2 | 2 854 128 | 2706A>G | H | 2706A>G | H | 2706A>G | H | 2706A>G | H | |
| RNR2 | 3 928 306 | 3010G>A | H1 | 3010G>A | H1 | |||||
| ND1 | T-M | 41 402 945 | 3992C>T | H4 | ||||||
| ND1 | T-A | 41 504 646 | 4024A>G | H4a | ||||||
| TI | Unique | na | 4322C>T | b | ||||||
| ND2 | SYN | na | 4733T>C | H1j | ||||||
| ND2 | SYN | 3 021 086 | 4769A>G | H2a1dc | 4769A>G | H2a1dc | 4769A>G | H2a1dc | 4769A>G | H2a1dc |
| ND2 | SYN | 41 419 549 | 5004T>C | H4 | ||||||
| NC3 | Non-Coding | na | 5585G>A | H11a2a2 | ||||||
| CO1 | SYN | na | 6951G>A | H4a1a4b2 | ||||||
| CO1 | SYN | 2 015 062 | 7028T>C | H | 7028T>C | H | 7028T>C | H | 7028T>C | H |
| CO2 | SYN | 8896 | 8269G>A | H4a1a | ||||||
| ATP8 | M-T | na | 8448T>C | H11 | ||||||
| ATP6 | T-A | 2 001 031 | 8860A>G | a | 8860A>G | a | ||||
| ATP6 | SYN | 28 358 270 | 9123G>A | H4 | ||||||
| ND4 | Unique | na | 11587C>T | b | ||||||
| ND4 | SYN | 2 853 495 | 11719G>A | H | 11719G>A | H | 11719G>A | H | 11719G>A | H |
| ND4 | SYN | na | 12130T>C | H45b | ||||||
| ND5 | A-T | na | 13759G>A | H11 | ||||||
| ND5 | C-Y | na | 13889G>A | H4a1a3 | ||||||
| ND5 | SYN | na | 13911A>G | b | ||||||
| ND5 | SYN | na | 14025T>C | b | ||||||
| ND6 | SYN | 2 853 815 | 14365C>T | H4a1 | ||||||
| ND6 | V-A | 41 354 845 | 14582A>G | H4a | ||||||
| ND6 | SYN | na | 14587A>G | H11a2 | ||||||
| CYB | T-I | na | 14766T>C | H | 14766T>C | H | 14766T>C | H | 14766T>C | H |
| CYB | T-A | 2 853 508 | 15326A>G | a | 15326A>G | a | 15326A>G | a | 15326A>G | a |
| CYB | SYN | na | 15670T>C | H11a2a2 | ||||||
| Dloop | Non-Coding | 3 134 562 | 16140T>C | H11a2a | ||||||
| Dloop | Non-Coding | 55 749 223 | 16189T>C | c | ||||||
| Dloop | Non-Coding | na | 16265A>G | H11a2a2 | ||||||
| Dloop | Non-Coding | na | 16289A>G | b | ||||||
| Dloop | Non-Coding | na | 16293A>G | H11a | ||||||
| Dloop | Non-Coding | 34 799 580 | 16311T>C | H11c | ||||||
| Dloop | Non-Coding | 3 087 742 | 73 G>A | H | 73 G>A | H | 73 G>A | H | 73 G>A | H |
| Dloop | Non-Coding | 62 581 312 | 150C>T | |||||||
| Dloop | Non-Coding | 2 857 291 | 195T>C | c | ||||||
| Dloop | Non-Coding | 2 853 515 | 263A>G | a | 263A>G | a | 263A>G | a | 263A>G | a |
aPrivate: non-haplogroup defining SNP found in both H and J cybrids.
bPrivate: non-haplogroup defining SNP found in single H or J cybrid.
cBack mutation to ancestral state; na, rs# not assigned.
Unique: SNP not listed on www.mitomap.org.
The new J cybrids were classified into J1c1a (#J13-74), J1b (#J13-107) and J1b1b1 (#J12-43). There were a total of eight private SNPs in the J cybrids: J1c1a cybrid with m.4322C>T (unique, MT-TI); J1b cybrid with m.4322C>T (unique, MT-TI), m.8200T>C (syn, MT-CO2) and m.200A>G (non-coding); and J1b1b1 cybrid with m.4664C>T (syn, MT-ND2), m.7805G>A (V-I, MT-CO2), m.14028A>G (unique, MT-ND5) and m.508A>G (non-coding). The private SNPs were found in different mitochondrial loci (MT-TI, MT-CO2, MT-ND2, and non-coding). There were seven non-synonymous, J-defining haplogroup SNPs: m.3394T>C (Y-H, MT-ND1), m.4216T>C (Y-H, MT-ND1*), m.10398A>G (T-A, MT-ND3*), m.13708G>A (A-I, MT-ND5*), m.13879T>C (S-P, MT-ND5), m.14798T>C (F-L, MT-CYB) and m.15452C>A (L-I, MT-CYB*). The four SNPs with asterisks are J-defining SNPs found in all three J cybrids. The non-coding region of the J cybrids possessed 15 SNPs, 12 of which defined the J haplogroup. It is important to note that the three J cybrids listed in this paper and the previously published J cybrid sequences (24) did not have private or unique SNPs found in all samples, supporting the idea that it is the background mtDNA haplogroup profile which causes the differences detected in the J versus H cybrids.
Based upon the sequences, the additional H cybrids were classified into H4a1a (#H11-10), H11a2a2 (#H11-23), H1 (#H11-35) and H1j (#H13-49). The H-defining SNPs were m.73G>A, m.2706G>A, m.7028T>C, m.11719A>G and m.14766T>C. There were a total of five private SNPs in the H cybrids. The H4a1a cybrid had no private SNPs, but four non-synonymous H-defining SNPs: m.3992T>C (M-T, MT-ND1), m.4024A>G (T-A, MT-ND1), m.13889G>A (C-Y, MT-ND5) and m.14582A>G (V-A, MT-ND6). H11a2a2 had two private SNPs: 4322C>T (unique, MT-TI and 11587C>T (unique, MT-ND4) and two non-synonymous H-defining SNPs: 8448T>C, (M-T, MT-ATP8) and 13759G>A (A-T, MT-ND5). The H1 cybrid had one private SNP, 16289A>G (non-coding, MT-Dloop). The H1j cybrid had two private SNPs: 13911A>G (syn, MT-ND5) and 14025T>C (syn, MT-ND5). The H1 and H1j cybrids did not have any non-synonymous H-defining SNPs.
Discussion
Our present studies illustrate that cybrids containing the J haplogroup mtDNA have significantly higher levels of total global methylation compared with H cybrids, a finding similar to that found in osteosarcoma cybrids (28). Since the total methylation was varied, the expression levels of methylation- and acetylation-specific genes were also examined and found to be different in H versus J cybrids. In mammalian cells, SAM is the universal methyl donor to cytosines in CpG sites in promoters. We found that the MAT2B gene, which catalyzes the biosynthesis of SAM from ATP and methionine, was expressed at a higher level in J cybrids compared with H cybrids. In cancer cells, increased MAT2B levels lead to lower SAM, which in turn facilitates the growth of hepatocellular carcinoma cells (34). The MAT2B levels may be influencing growth in cybrids as studies have shown that J cybrids grow more rapidly than H cybrids, which in turn grow faster than Uk cybrids (23,24,35). In J haplogroup osteosarcoma cybrids, elevated levels of MAT1A were expressed (28). However, our H and J RPE cybrids expressed only very low levels of MAT1A (the hepatic isoform) or MAT2A (non-hepatic isoform). This disparity may be due to differences in the underlying host cells (human RPE cells versus osteosarcoma cells). Our findings herein suggest that the MAT2B is the major methionine adenosyltransferase in human RPE cells in vitro and its expression can be modulated by the mtDNA variants within cells.
Our Q-PCR studies showed significantly lower expression levels for DNMT1, DNMT3A and DNMT3B in the J cybrids compared with the H cybrids. Shock et al.(32) identified DNMT1 inside mitochondria, while others have found DNMT3A associated with mitochondrial fractions (36). While the degree of mtDNA methylation may provide a biomarker and diagnostic tool to identify abnormalities of mitochondrial metabolism (37), it is necessary to learn more about its significance before it might have a correlative value to diseases.
The MBD genes encode for a family of proteins that bind to the methylated DNA, block transcription, act as demethylases to activate transcription and are associated with various cancers (38). In our study, the J cybrids showed significantly lower levels of MBD2 expression compared with the H cybrids. In contrast, the levels for MBD4 gene expression were similar in the H and J RPE cybrids, which is consistent with findings in the osteosarcoma cybrids (28). Both in vitro and in vivo studies have shown that MBD2 can bind to methylated DNA and silence IL-4, a major signaling intermediate molecule for IFN-gamma and the immune system (39,40). Differential expression of MBD2 by the J and H cybrids could potentially lead to different methylation status and transcription levels for nuclear genes in complement and inflammation pathways, both of which are activated in AMD.
Epigenetics involves not only modification of DNA methylation, but also acetylation and deacetylation of histones. To our knowledge, we are the first group to demonstrate that the mtDNA variants can modulate the expression levels of acetylation-related genes. Recent evidence has shown that mitochondrial signaling is likely regulated through acetylation, with >1000 mitochondrial-related proteins having >4500 acetylation sites, indicating this form of regulation significantly impacts cellular homeostasis (41). Therefore, some of the acetylation-related genes in our cybrids were examined using Q-PCR. The J cybrids had significantly lower levels of HAT1, an acetyltransferase which adds acetyl groups to histones and promotes transcription. The inhibition of acetylation can lower inflammation and may be a possible therapy to block progression of diabetic retinopathy (42). Deregulation of acetylation and deacetylation is also associated with various cancers. Current studies are underway to identify acetylation modifiers for use as therapeutic agents (43–45).
In J cybrids, we detected significantly decreased expression levels of HDAC1 compared with H cybrids. HDAC1 is part of a complex responsible for deacetylation of histones, which represses transcription. When examined by GeneChip analyses, the H and J cybrids did not show differences of expression for another group of deacetylase enzymes, the sirtuins (SIRT3, SIRT4, SIRT5), which are actually found within mitochondria and are responsible for major protein modifications (46). In particular, the mitochondrial SIRT3 acts by deacetylation of various substrates to regulate reactive oxygen species (ROS) production and detoxification (47). This suggests that the mtDNA variants are influencing acetylation through the HDACs rather than SIRTs, but additional studies are required to understand this intricate process.
Our initial experiments showed that H and J cybrids have different levels for total global methylation and expression of acetylation and methylation genes. However, we also wanted to investigate the influences that H mtDNA versus J mtDNA might have on genes known to be regulated by methylation. CFH is a major susceptibility gene which confers significant risk for development of AMD (48–50). EFEMP1 is a high-risk gene for AMD and other retinal degenerations (51,52) and misfolded EFEMP1 causes altered RPE cell function and inflammation (53). VEGFA is associated with increased neovascularization found in AMD patients and is the target for anti-VEGF therapies commonly used to control the choroidal neovascularization associated with AMD. NFkB2 is a major signaling molecule for inflammation and the activated form is associated with AMD fibrovascular membranes (54).
The H and J cybrids were cultured with and without 5-aza-dC, a DNA methyltransferase inhibitor. Although the untreated-J cybrids showed lower expression levels for CFH (P < 0.0001), EFEMP1 (P = 0.015), NFkB2 (P = 0.03) and VEGFA (P < 0.0006) compared with the untreated-H cybrids, once the cells were demethylated, the expression levels for these were similar in J-treated and H-treated cybrids. These results suggest that methylation status may be regulated in part by mtDNA variants, because when methylation is present, the J cybrids express lower levels of these genes, but once demethylated, the levels are similar.
Methylation status of tissues is extremely important in diseases. In gastric tumor cells, demethylation with 5-aza-dC increased microRNA-195 and microRNA-378 levels, which in turn down-regulated VEGF levels (55). In hepatocellular carcinoma cells, Yang et al. (56) reported an inverse relationship between the degree of methylation at CpG sites and CFH expression levels. This is similar to what we found in the J cybrids, which also have higher total methylation, but lower expressions of CFH (23). Expression levels of CFH can correlate with metastasis, mortality risk, insulin resistance in adipose tissues and possibly tumor suppression (56–58). The degree of methylation in the EFEMP1 promoter plays a key role in tumor metastases and disease outcomes (59–69). Therefore, the fact that mtDNA variants (J versus H haplogroups) can affect significantly, via the methylation profile, the expression levels of CFH, EFEMP1 and VEGFA, suggests that targeting the methylation pathways may serve as a therapeutic target/approach in future studies.
It has been shown that 2–5% of human mtDNA has methylation at the CCGG regions (70). Normally the methylation sites for human nuclear DNA occur through the CpG sites while plants and fungi utilize non-CpG methylation sites; however, reports have shown that mtDNA actually contain both types of methylation sites, CpG and non-CpG sites (31). The MT-Dloop contains the control region possessing the Light-Strand Promoter, responsible for transcription of the MT-ND6 gene and eight of the MT-tRNAs, and the Heavy-Strand Promoter, which controls the transcription for the other mtDNA encoded genes. Previous studies have shown that J cybrids express significantly lower MT-RNA levels than H cybrids (24,71). Based upon reported differences in MT-RNA expression levels, we speculated that mtDNA defining J haplogroups may have different numbers of CpG and non-CpG sites within the MT-Dloop. However, the numbers of CpG and non-CpG sites in the Light-Strand Promoter and Heavy-Strand Promoter for the H and J cybrids were similar, indicating that altered MT-RNA expression levels were likely not a result of SNP substitutions within those regions. Our sequencing data suggest that although the J haplogroup mtDNA have numerous SNP variants, there was no disruption of the SNP pattern within the MT-Dloop causing increased numbers of potential CpG or non-CpG methylation sites. Future studies will focus on understanding the mechanism(s) underlying differential methylation levels associated with different mtDNA variants.
Sequencing of the entire mtDNA for the cybrids showed that the majority of SNPs defined either the H or J haplogroups. There were no private or unique SNPs that were found uniformly in all of the H or J cybrids. This supports the concept that the differences in gene expression are not due to a single mutation, private or unique SNP, but rather: (i) the accumulation of multiple SNPs in the J or H haplogroups or (ii) a particular, as of yet, unidentified SNP, which defines the haplogroup, but has properties to modulate nuclear gene expression via a retrograde signaling mechanism. In addition, all of the J cybrids belonged in the haplogroup J1 family, with none being part of the J2 haplogroup, the other main branch of the J haplogroup. This may be significant because J1 and J2 branches are quite different, especially when mutations in complex III are considered. The SNP m.195T>C found in H11a2a2 cybrid has been found to be associated with bipolar disorder (72) (reported in www.MitoMap.org). None of the other SNPs in either the H or J cybrids were associated with diseases when compared with the ‘Reported Mitochondria DNA Base Substitution in Diseases' or the ‘Coding and Control Region Point Mutations' sections. Research studies need to be completed before we fully comprehend how the mtDNA variants: (i) mediate total global methylation levels; (ii) affect expression levels of epigenetic-related genes or (iii) influence the transcription of non-energy-related nuclear genes. Further investigations aimed at elucidating the mechanisms of these mtDNA variants will help in paving the way to personalized therapeutic advancements and interventions.
Materials and Methods
Cybrid cultures and culture conditions
Institutional review board (#2003-3131) approval was obtained from the University of California-Irvine. For DNA analyses, 20 ml of peripheral blood was collected by venipuncture in tubes containing sodium citrate buffer. DNA was isolated with a DNA extraction kit (PUREGENE, Qiagen, Valencia, CA). Platelets were isolated by a series of centrifugation steps and final pellets were suspended in Tris-buffered saline (TBS). The ARPE-19 cells deficient in mtDNA (Rho0) were created by serial passage in low-dose ethidium bromide as previously described (73). Cybrids were created by polyethylene glycol fusion of platelets with Rho0 ARPE-19 cells according to modified procedures of Chomyn (74). Cybrids were cultured until confluent in DMEM-F12 containing 10% dialyzed fetal bovine serum, 100 unit/ml penicillin and 100 µg/ml streptomycin, 2.5 μg/ml fungizone, 50 µg/ml gentamycin and 17.5 mm glucose as described previously (23). The H and J cybrids used in this set of experiments were previously shown to have similar mtDNA copy numbers (23). To eliminate potential technical variability, all experiments were performed under exactly the same controlled conditions. For each experiment, all H and J cybrids were at passage 5 and cultured side-by-side in an incubator using identical media conditions.
Inhibition of methylation in cybrid cultures
These experiments were designed to determine if the different mtDNA variants (H versus J haplogroups) influence the RNA expression for nuclear genes with known methylation sites in their promoter regions (EFEMP1 and VEGFA) or key genes associated with signaling transduction or complement pathways (NFkB2 and CFH). H and J cybrids (n = 3 for each cybrid, each from different individuals) were plated for 24 h, media were removed and replaced with the same media containing a final concentration of 250 nm 5-aza-2′-deoxycytidine (5-aza-dC, Sigma-Alrich, St Louis, MO, USA). Cells were treated with 5-aza-dC for 48 h, the culture media being replaced after 24 h with fresh media containing the compound. Cells were pelleted, RNA isolated and cDNA synthesized as described in what follows. Q-PCR was performed with primers for CFH, EFEMP1, NFkB2 and VEGFA.
Identification of cybrid haplogroups
Cybrid DNA was extracted from cell pellets using a spin column kit (DNeasy Blood and Tissue Kit, Qiagen, Valencia, CA, USA) as described previously (24). The mitochondrial haplogroups were identified by PCR along with restriction enzyme digestions, allelic discrimination and sequencing (5,24). The major defining SNP for the H haplogroup is T7028C. The major SNP defining the J haplogroup is G13708A. Allelic discrimination was performed at the GenoSeq UCLA Genotyping and Sequencing Core. Data were analyzed with Sequence Detection Systems software (ABI7900HT). The mtDNA sequences were compared with the classification from www.phylotree.org and www.MitoMap.com. In an earlier publication, the entire mtDNA sequences for H cybrids (n = 3) were categorized as H, H5a and H66a, and the J haplogroup cybrids (n = 3) were classified into J1d1a, J1c1 and J1c7 (24). These cybrids were used for the global methylation and inhibitor studies. Additional H (H4a1a, H11a2a2, H1 and H1j; total of n = 7) and J (J1c1a, J1b, J1b1b1, total of n = 6) cybrids were also sequenced and used for the Q-PCR analyses.
Global DNA methylation assay
The global DNA methylation status was detected using the MethylFlash Methylated DNA Quantification Kit (Colormetric) (EpiGenTek, Farmingdale, NY, USA) according to the manufacturer's protocol. The amount of DNA used in the assay was 100 ng. Briefly, the H (n = 3) and J (n = 3) cybrids were cultured until confluent and DNA isolated as described earlier. The DNA was bound to strip wells that were specifically treated by the manufacturer to have a high DNA affinity. The methylated fraction of DNA was detected using capture and detection antibodies and then quantified through an ELISA-like reaction by reading the absorbance in a microplate spectrophotometer at 450 nm. The amount of methylated DNA (5-mC%) was proportional to the OD intensity measured with an absorbance plate reader (BioTek, Winooski, VT, USA), calculated according to the kit's formulas for relative methylation status of two different DNAs. Samples were run in duplicate and the experiment was repeated twice.
RNA extraction, amplification of cDNA and quantitative PCR (Q-PCR) analyses
Cells from cybrid cultures were pelleted and RNA isolated using the RNeasy Mini-Extraction kit (Qiagen, Inc., Valencia, CA, USA) as described previously (24). This study compares relative differences in gene expression levels between the H and J cybrids. For Q-PCR analyses, 100 ng of individual RNA samples were reverse transcribed into cDNA using the QuantiTect Reverse Transcription Kit (Qiagen, Inc.). Q-PCR was performed in triplicate using 10 different primers (QuantiTect Primer Assay, Qiagen, Inc.) for genes associated with various aspects of acetylation (HAT1, HDAC1, HDAC6, HDAC11, SIN3A) or methylation (MAT2B, MBD4, DNMT1, DNMT3A and DNMT3B). The Q-PCR was performed on individual samples using a QuantiFast SYBR Green PCR Kit (Qiagen) on a Bio-Rad iCycler iQ 500 detection system. For the various target genes, housekeeping genes that had comparable amplification efficiencies to the genes of interest were chosen in order to maximize the accuracy of our ΔΔCT values. The housekeeper genes were either HPRT1 (hypoxanthine phosphorbosyltransferase 1), HMBS (hydroxymethylbilane synthase), ALAS1 (aminolevulinate, delta-synthase) or TUBB (Tubulin, beta class 1).
Statistical analyses
Statistical analyses of the data were performed by ANOVA (GraphPad Prism, version 5.0). Newman–Keuls multiple-comparison or the two-tailed t-tests were used to compare the data within each experiment. P < 0.05 was considered statistically significant. Error bars in the graphs represent standard error of the mean (SEM).
Funding
This work was supported by Discovery Eye Foundation, Guenther Foundation, Beckman Initiative for Macular Research, Polly and Michael Smith Foundation, Max Factor Family Foundation, Iris and B. Gerald Cantor Foundation, and the National Institute on Aging (S.M.J., R37 AG006168). J.C.d.C. is the David & Julianna Pyott Pan-American-Retina Research fellow. D.M. was the recipient of the David & Julianna Pyott Pan-American-Retina Research Fellowship (2012–2014).
Acknowledgements
We wish to thank the subjects who participated in this study.
Conflict of Interest statement. None declared.
References
- 1.Czarnecka A.M., Bartnik E. (2011) The role of the mitochondrial genome in ageing and carcinogenesis. J. Aging Res., 2011, 136435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wallace D.C., Lott M.T., Procaccio V. (2007) Mitochondrial genes in degenerative diseases, cancer and aging. In: Rimoin C., Pyeritz K. (eds), Emery and Rimoin's Principles and Practices of Medical Genetics. Churchill Livingstone Elsevier, Philadelphia, PA. [Google Scholar]
- 3.Jones M.M., Manwaring N., Wang J.J., Rochtchina E., Mitchell P., Sue C.M. (2007) Mitochondrial DNA haplogroups and age-related maculopathy. Arch. Ophthalmol., 125, 1235–1240. [DOI] [PubMed] [Google Scholar]
- 4.Canter J.A., Olson L.M., Spencer K., Schnetz-Boutaud N., Anderson B., Hauser M.A., Schmidt S., Postel E.A., Agarwal A., Pericak-Vance M.A., et al. (2008) Mitochondrial DNA polymorphism A4917G is independently associated with age-related macular degeneration. PLoS ONE, 3, e2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Udar N., Atilano S.R., Memarzadeh M., Boyer D., Chwa M., Lu S., Maguen B., Langberg J., Coskun P., Wallace D.C., et al. (2009) Mitochondrial DNA haplogroups associated with age-related macular degeneration. Invest. Ophthalmol. Vis. Sci., 50, 2966–2974. [DOI] [PubMed] [Google Scholar]
- 6.SanGiovanni J.P., Arking D.E., Iyengar S.K., Elashoff M., Clemons T.E., Reed G.F., Henning A.K., Sivakumaran T.A., Xu X., DeWan A., et al. (2009) Mitochondrial DNA variants of respiratory complex I that uniquely characterize haplogroup T2 are associated with increased risk of age-related macular degeneration. PLoS ONE, 4, e5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mueller E.E., Schaier E., Brunner S.M., Eder W., Mayr J.A., Egger S.F., Nischler C., Oberkofler H., Reitsamer H.A., Patsch W., et al. (2012) Mitochondrial haplogroups and control region polymorphisms in age-related macular degeneration: a case-control study. PLoS ONE, 7, e30874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kenney M.C., Hertzog D., Chak G., Atilano S.R., Khatibi N., Soe K., Nobe A., Yang E., Chwa M., Zhu F., et al. (2013) Mitochondrial DNA haplogroups confer differences in risk for age-related macular degeneration: A case control study. BMC Med. Genet., 14, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu M.M., Chan C.C., Tuo J. (2012) Genetic mechanisms and age-related macular degeneration: common variants, rare variants, copy number variations, epigenetics, and mitochondrial genetics. Hum. Genomics, 6, 13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carelli V., Vergani L., Bernazzi B., Zampieron C., Bucchi L., Valentino M., Rengo C., Torroni A., Martinuzzi A. (2002) Respiratory function in cybrid cell lines carrying European mtDNA haplogroups: implications for Leber's hereditary optic neuropathy. Biochim. Biophys. Acta, 1588, 7–14. [DOI] [PubMed] [Google Scholar]
- 11.Bellizzi D., Cavalcante P., Taverna D., Rose G., Passarino G., Salvioli S., Franceschi C., De Benedictis G. (2006) Gene expression of cytokines and cytokine receptors is modulated by the common variability of the mitochondrial DNA in cybrid cell lines. Genes Cells, 11, 883–891. [DOI] [PubMed] [Google Scholar]
- 12.Bellizzi D., Taverna D., D'Aquila P., De Blasi S., De Benedictis G. (2009) Mitochondrial DNA variability modulates mRNA and intra-mitochondrial protein levels of HSP60 and HSP75: Experimental evidence from cybrid lines. Cell Stress Chaperones, 14, 265–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.D'Aquila P., Rose G., Panno M.L., Passarino G., Bellizzi D. (2012) SIRT3 gene expression: a link between inherited mitochondrial DNA variants and oxidative stress. Gene, 497, 323–329. [DOI] [PubMed] [Google Scholar]
- 14.Ghosh S.S., Swerdlow R.H., Miller S.W., Sheeman B., Parker W.D., Jr., Davis R.E. (1999) Use of cytoplasmic hybrid cell lines for elucidating the role of mitochondrial dysfunction in Alzheimer's disease and Parkinson's disease. Ann. NY Acad. Sci., 893, 176–191. [DOI] [PubMed] [Google Scholar]
- 15.Hwang S., Kwak S.H., Bhak J., Kang H.S., Lee Y.R., Koo B.K., Park K.S., Lee H.K., Cho Y.M. (2011) Gene expression pattern in transmitochondrial cytoplasmic hybrid cells harboring type 2 diabetes-associated mitochondrial DNA haplogroups. PLoS ONE, 6, e22116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jun A.S., Trounce I.A., Brown M.D., Shoffner J.M., Wallace D.C. (1996) Use of transmitochondrial cybrids to assign a complex I defect to the mitochondrial DNA-encoded NADH dehydrogenase subunit 6 gene mutation at nucleotide pair 14459 that causes Leber hereditary optic neuropathy and dystonia. Mol. Cell Biol., 16, 771–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaipparettu B.A., Ma Y., Park J.H., Lee T.L., Zhang Y., Yotnda P., Creighton C.J., Chan W.Y., Wong L.J. (2013) Crosstalk from non-cancerous mitochondria can inhibit tumor properties of metastatic cells by suppressing oncogenic pathways. PLoS ONE, 8, e61747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koga Y., Davidson M., Schon E.A., King M.P. (1995) Analysis of cybrids harboring MELAS mutations in the mitochondrial tRNA (Leu(UUR)) gene. Muscle Nerve Suppl. 3, S119–S123. [DOI] [PubMed] [Google Scholar]
- 19.Pacheu-Grau D., Gomez-Duran A., Iglesias E., Lopez-Gallardo E., Montoya J., Ruiz-Pesini E. (2013) Mitochondrial antibiograms in personalized medicine. Hum. Mol. Genet., 22, 1132–1139. [DOI] [PubMed] [Google Scholar]
- 20.Tu Y.F., Kaipparettu B.A., Ma Y., Wong L.J. (2011) Mitochondria of highly metastatic breast cancer cell line MDA-MB-231 exhibits increased autophagic properties. Biochim. Biophys. Acta, 1807, 1125–1132. [DOI] [PubMed] [Google Scholar]
- 21.Vithayathil S.A., Ma Y., Kaipparettu B.A. (2012) Transmitochondrial cybrids: Tools for functional studies of mutant mitochondria. Methods Mol. Biol., 837, 219–230. [DOI] [PubMed] [Google Scholar]
- 22.Weng S.W., Kuo H.M., Chuang J.H., Lin T.K., Huang H.L., Lin H.Y., Liou C.W., Wang P.W. (2013) Study of insulin resistance in cybrid cells harboring diabetes-susceptible and diabetes-protective mitochondrial haplogroups. Mitochondrion, 13, 888–897. [DOI] [PubMed] [Google Scholar]
- 23.Kenney M.C., Chwa M., Atilano S.R., Pavlis J.M., Falatoonzadeh P., Ramirez C., Malik D., Hsu T., Woo G., Soe K., et al. (2013) Mitochondrial DNA variants mediate energy production and expression levels for CFH, C3 and EFEMP1 genes: implications for age-related macular degeneration. PLoS ONE, 8, e54339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kenney M.C., Chwa M., Atilano S.R., Falatoonzadeh P., Ramirez C., Malik D., Tarek M., Caceres Del Carpio J., Nesburn A.B., Boyer D.S., et al. (2014) Inherited mitochondrial DNA variants can affect complement, inflammation, and apoptosis pathways: insights into mitochondrial-nuclear interactions. Hum. Mol. Genet., 23, 3537–3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kenney M.C., Chwa M., Atilano S.R., Falatoonzadeh P., Ramirez C., Malik D., Tarek M., Del Carpio J.C., Nesburn A.B., Boyer D.S., et al. (2014) Molecular and bioenergetic differences between cells with African versus European inherited mitochondrial DNA haplogroups: implications for population susceptibility to diseases. Biochim. Biophys. Acta, 1842, 208–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malik D., Hsu T., Falatoonzadeh P., Caceres-Del-Carpio J., Tarek M., Chwa M., Atilano S.R., Ramirez C., Nesburn A.B., Boyer D.S., et al. (2014) Human retinal transmitochondrial cybrids with J or H mtDNA haplogroups respond differently to ultraviolet radiation: implications for retinal diseases. PLoS ONE, 9, e99003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smiraglia D.J., Kulawiec M., Bistulfi G.L., Gupta S.G., Singh K.K. (2008) A novel role for mitochondria in regulating epigenetic modification in the nucleus. Cancer Biol. Therapy, 7, 1182–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bellizzi D., D'Aquila P., Giordano M., Montesanto A., Passarino G. (2012) Global DNA methylation levels are modulated by mitochondrial DNA variants. Epigenomics, 4, 17–27. [DOI] [PubMed] [Google Scholar]
- 29.Naviaux R.K. (2008) Mitochondrial control of epigenetics. Cancer Biol. Therapy, 7, 1191–1193. [DOI] [PubMed] [Google Scholar]
- 30.Wallace D.C., Fan W. (2010) Energetics, epigenetics, mitochondrial genetics. Mitochondrion, 10, 12–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bellizzi D., D'Aquila P., Scafone T., Giordano M., Riso V., Riccio A., Passarino G. (2013) The control region of mitochondrial DNA shows an unusual CpG and non-CpG methylation pattern. DNA Res.: An International Journal for Rapid Publication of Reports on Genes and Genomes, 20, 537–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shock L.S., Thakkar P.V., Peterson E.J., Moran R.G., Taylor S.M. (2011) DNA methyltransferase 1, cytosine methylation, and cytosine hydroxymethylation in mammalian mitochondria. Proc. Natl. Acad. Sci. USA, 108, 3630–3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo W., Chung W.Y., Qian M., Pellegrini M., Zhang M.Q. (2014) Characterizing the strand-specific distribution of non-CpG methylation in human pluripotent cells. Nucleic Acids Res., 42, 3009–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lo T.F., Tsai W.C., Chen S.T. (2013) MicroRNA-21–3p, a berberine-induced miRNA, directly down-regulates human methionine adenosyltransferases 2A and 2B and inhibits hepatoma cell growth. PLoS ONE, 8, e75628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gomez-Duran A., Pacheu-Grau D., Lopez-Gallardo E., Diez-Sanchez C., Montoya J., Lopez-Perez M.J., Ruiz-Pesini E. (2010) Unmasking the causes of multifactorial disorders: OXPHOS differences between mitochondrial haplogroups. Hum. Mol. Genet., 19, 3343–3353. [DOI] [PubMed] [Google Scholar]
- 36.Chestnut B.A., Chang Q., Price A., Lesuisse C., Wong M., Martin L.J. (2011) Epigenetic regulation of motor neuron cell death through DNA methylation. J. Neurosci., 31, 16619–16636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iacobazzi V., Castegna A., Infantino V., Andria G. (2013) Mitochondrial DNA methylation as a next-generation biomarker and diagnostic tool. Mol. Genet. Metab., 110, 25–34. [DOI] [PubMed] [Google Scholar]
- 38.Parry L., Clarke A.R. (2011) The roles of the Methyl-CpG binding proteins in cancer. Genes Cancer, 2, 618–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bhattacharya S.K., Ramchandani S., Cervoni N., Szyf M. (1999) A mammalian protein with specific demethylase activity for mCpG DNA. Nature, 397, 579–583. [DOI] [PubMed] [Google Scholar]
- 40.Hutchins A.S., Artis D., Hendrich B.D., Bird A.P., Scott P., Reiner S.L. (2005) Cutting edge: a critical role for gene silencing in preventing excessive type 1 immunity. J. Immunol., 175, 5606–5610. [DOI] [PubMed] [Google Scholar]
- 41.Amado F.M., Barros A., Azevedo A.L., Vitorino R., Ferreira R. (2014) An integrated perspective and functional impact of the mitochondrial acetylome. Expert Rev. Proteomics, 11, 383–394. [DOI] [PubMed] [Google Scholar]
- 42.Kadiyala C.S., Zheng L., Du Y., Yohannes E., Kao H.Y., Miyagi M., Kern T.S. (2012) Acetylation of retinal histones in diabetes increases inflammatory proteins: Effects of minocycline and manipulation of histone acetyltransferase (HAT) and histone deacetylase (HDAC). J. Biol. Chem., 287, 25869–25880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cress W.D., Seto E. (2000) Histone deacetylases, transcriptional control, and cancer. J. Cell Physiol., 184, 1–16. [DOI] [PubMed] [Google Scholar]
- 44.Mahlknecht U., Hoelzer D. (2000) Histone acetylation modifiers in the pathogenesis of malignant disease. Mol. Med., 6, 623–644. [PMC free article] [PubMed] [Google Scholar]
- 45.Timmermann S., Lehrmann H., Polesskaya A., Harel-Bellan A. (2001) Histone acetylation and disease. Cell Mol. Life Sci., 58, 728–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Osborne B., Cooney G.J., Turner N. (2014) Are sirtuin deacylase enzymes important modulators of mitochondrial energy metabolism? Biochim . Biophys. Acta, 1840, 1295–1302. [DOI] [PubMed] [Google Scholar]
- 47.Bause A.S., Haigis M.C. (2013) SIRT3 regulation of mitochondrial oxidative stress. Exp. Gerontol., 48, 634–639. [DOI] [PubMed] [Google Scholar]
- 48.Montezuma S.R., Sobrin L., Seddon J.M. (2007) Review of genetics in age-related macular degeneration. Semin. Ophthalmol., 22, 229–240. [DOI] [PubMed] [Google Scholar]
- 49.Katta S., Kaur I., Chakrabarti S. (2009) The molecular genetic basis of age-related macular degeneration: an overview. J. Genet., 88, 425–449. [DOI] [PubMed] [Google Scholar]
- 50.Ratnapriya R., Chew E.Y. (2013) Age-related macular degeneration-clinical review and genetics update. Clin. Genet., 84, 160–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stone E.M., Lotery A.J., Munier F.L., Heon E., Piguet B., Guymer R.H., Vandenburgh K., Cousin P., Nishimura D., Swiderski R.E., et al. (1999) A single EFEMP1 mutation associated with both Malattia Leventinese and Doyne honeycomb retinal dystrophy. Nat. Genet., 22, 199–202. [DOI] [PubMed] [Google Scholar]
- 52.Marmorstein L.Y., Munier F.L., Arsenijevic Y., Schorderet D.F., McLaughlin P.J., Chung D., Traboulsi E., Marmorstein A.D. (2002) Aberrant accumulation of EFEMP1 underlies drusen formation in Malattia Leventinese and age-related macular degeneration. Proc. Natl Acad. Sci. USA, 99, 13067–13072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roybal C.N., Marmorstein L.Y., Vander Jagt D.L., Abcouwer S.F. (2005) Aberrant accumulation of fibulin-3 in the endoplasmic reticulum leads to activation of the unfolded protein response and VEGF expression. Invest. Ophthalmol. Vis. Sci., 46, 3973–3979. [DOI] [PubMed] [Google Scholar]
- 54.Hammes H.P., Hoerauf H., Alt A., Schleicher E., Clausen J.T., Bretzel R.G., Laqua H. (1999) N(epsilon)(carboxymethyl)lysin and the AGE receptor RAGE colocalize in age-related macular degeneration. Invest. Ophthalmol. Vis. Sci., 40, 1855–1859. [PubMed] [Google Scholar]
- 55.Deng H., Guo Y., Song H., Xiao B., Sun W., Liu Z., Yu X., Xia T., Cui L., Guo J. (2013) MicroRNA-195 and microRNA-378 mediate tumor growth suppression by epigenetical regulation in gastric cancer. Gene, 518, 351–359. [DOI] [PubMed] [Google Scholar]
- 56.Yang J.D., Seol S.Y., Leem S.H., Kim Y.H., Sun Z., Lee J.S., Thorgeirsson S.S., Chu I.S., Roberts L.R., Kang K.J. (2011) Genes associated with recurrence of hepatocellular carcinoma: integrated analysis by gene expression and methylation profiling. J. Korean Med. Sci., 26, 1428–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Linton K.M., Hey Y., Saunders E., Jeziorska M., Denton J., Wilson C.L., Swindell R., Dibben S., Miller C.J., Pepper S.D., et al. (2008) Acquisition of biologically relevant gene expression data by Affymetrix microarray analysis of archival formalin-fixed paraffin-embedded tumours. Br. J. Cancer, 98, 1403–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moreno-Navarrete J.M., Martinez-Barricarte R., Catalan V., Sabater M., Gomez-Ambrosi J., Ortega F.J., Ricart W., Bluher M., Fruhbeck G., Rodriguez de Cordoba S., et al. (2010) Complement factor H is expressed in adipose tissue in association with insulin resistance. Diabetes, 59, 200–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yue W., Dacic S., Sun Q., Landreneau R., Guo M., Zhou W., Siegfried J.M., Yu J., Zhang L. (2007) Frequent inactivation of RAMP2, EFEMP1 and Dutt1 in lung cancer by promoter hypermethylation. Clin. Cancer Res., 13, 4336–4344. [DOI] [PubMed] [Google Scholar]
- 60.Nomoto S., Kanda M., Okamura Y., Nishikawa Y., Qiyong L., Fujii T., Sugimoto H., Takeda S., Nakao A. (2010) Epidermal growth factor-containing fibulin-like extracellular matrix protein 1, EFEMP1, a novel tumor-suppressor gene detected in hepatocellular carcinoma using double combination array analysis. Annal. Surg. Oncol., 17, 923–932. [DOI] [PubMed] [Google Scholar]
- 61.Hwang C.F., Chien C.Y., Huang S.C., Yin Y.F., Huang C.C., Fang F.M., Tsai H.T., Su L.J., Chen C.H. (2010) Fibulin-3 is associated with tumour progression and a poor prognosis in nasopharyngeal carcinomas and inhibits cell migration and invasion via suppressed AKT activity. J. Pathol., 222, 367–379. [DOI] [PubMed] [Google Scholar]
- 62.Zhang Y., Wang R., Song H., Huang G., Yi J., Zheng Y., Wang J., Chen L. (2011) Methylation of multiple genes as a candidate biomarker in non-small cell lung cancer. Cancer Lett., 303, 21–28. [DOI] [PubMed] [Google Scholar]
- 63.Sadr-Nabavi A., Ramser J., Volkmann J., Naehrig J., Wiesmann F., Betz B., Hellebrand H., Engert S., Seitz S., Kreutzfeld R., et al. (2009) Decreased expression of angiogenesis antagonist EFEMP1 in sporadic breast cancer is caused by aberrant promoter methylation and points to an impact of EFEMP1 as molecular biomarker. Int. J. Cancer, 124, 1727–1735. [DOI] [PubMed] [Google Scholar]
- 64.Kim Y.J., Yoon H.Y., Kim S.K., Kim Y.W., Kim E.J., Kim I.Y., Kim W.J. (2011) EFEMP1 as a novel DNA methylation marker for prostate cancer: array-based DNA methylation and expression profiling. Clin. Cancer Res., 17, 4523–4530. [DOI] [PubMed] [Google Scholar]
- 65.Song E.L., Hou Y.P., Yu S.P., Chen S.G., Huang J.T., Luo T., Kong L.P., Xu J., Wang H.Q. (2011) EFEMP1 expression promotes angiogenesis and accelerates the growth of cervical cancer in vivo. Gynecologic Oncol., 121, 174–180. [DOI] [PubMed] [Google Scholar]
- 66.En-lin S., Sheng-guo C., Hua-qiao W. (2010) The expression of EFEMP1 in cervical carcinoma and its relationship with prognosis. Gynecol. Oncol., 117, 417–422. [DOI] [PubMed] [Google Scholar]
- 67.Hu Y., Pioli P.D., Siegel E., Zhang Q., Nelson J., Chaturbedi A., Mathews M.S., Ro D.I., Alkafeef S., Hsu N., et al. (2011) EFEMP1 suppresses malignant glioma growth and exerts its action within the tumor extracellular compartment. Mol. Cancer, 10, 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Albig A.R., Neil J.R., Schiemann W.P. (2006) Fibulins 3 and 5 antagonize tumor angiogenesis in vivo. Cancer Res., 66, 2621–2629. [DOI] [PubMed] [Google Scholar]
- 69.Yang T., Qiu H., Bao W., Li B., Lu C., Du G., Luo X., Wang L., Wan X. (2013) Epigenetic inactivation of EFEMP1 is associated with tumor suppressive function in endometrial carcinoma. PLoS ONE, 8, e67458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shmookler Reis R.J., Goldstein S. (1983) Mitochondrial DNA in mortal and immortal human cells. Genome number, integrity, and methylation. J. Biol. Chem., 258, 9078–9085. [PubMed] [Google Scholar]
- 71.Gomez-Duran A., Pacheu-Grau D., Martinez-Romero I., Lopez-Gallardo E., Lopez-Perez M.J., Montoya J., Ruiz-Pesini E. (2012) Oxidative phosphorylation differences between mitochondrial DNA haplogroups modify the risk of Leber's hereditary optic neuropathy. Biochim. Biophys. Acta, 1822, 1216–1222. [DOI] [PubMed] [Google Scholar]
- 72.Rollins B., Martin M.V., Sequeira P.A., Moon E.A., Morgan L.Z., Watson S.J., Schatzberg A., Akil H., Myers R.M., Jones E.G., et al. (2009) Mitochondrial variants in schizophrenia, bipolar disorder, and major depressive disorder. PLoS ONE, 4, e4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Miceli M.V., Jazwinski S.M. (2005) Nuclear gene expression changes due to mitochondrial dysfunction in ARPE-19 cells, Implications for age-related macular degeneration. Invest. Ophthalmol. Vis. Sci., 46, 1765–1773. [DOI] [PubMed] [Google Scholar]
- 74.Chomyn A. (1996) Platelet-mediated Transformation of Human Mitochondrial DNA-Less Cells. Academic Press, Inc., Salt Lake City, UT. [DOI] [PubMed] [Google Scholar]