Abstract
Genetics of gene expression (eQTLs or expression QTLs) has proved an indispensable tool for understanding biological pathways and pathomechanisms of trait-associated SNPs. However, power of most genome-wide eQTL studies is still limited. We performed a large eQTL study in peripheral blood mononuclear cells of 2112 individuals increasing the power to detect trans-effects genome-wide. Going beyond univariate SNP-transcript associations, we analyse relations of eQTLs to biological pathways, polygenetic effects of expression regulation, trans-clusters and enrichment of co-localized functional elements. We found eQTLs for about 85% of analysed genes, and 18% of genes were trans-regulated. Local eSNPs were enriched up to a distance of 5 Mb to the transcript challenging typically implemented ranges of cis-regulations. Pathway enrichment within regulated genes of GWAS-related eSNPs supported functional relevance of identified eQTLs. We demonstrate that nearest genes of GWAS-SNPs might frequently be misleading functional candidates. We identified novel trans-clusters of potential functional relevance for GWAS-SNPs of several phenotypes including obesity-related traits, HDL-cholesterol levels and haematological phenotypes. We used chromatin immunoprecipitation data for demonstrating biological effects. Yet, we show for strongly heritable transcripts that still little trans-chromosomal heritability is explained by all identified trans-eSNPs; however, our data suggest that most cis-heritability of these transcripts seems explained. Dissection of co-localized functional elements indicated a prominent role of SNPs in loci of pseudogenes and non-coding RNAs for the regulation of coding genes. In summary, our study substantially increases the catalogue of human eQTLs and improves our understanding of the complex genetic regulation of gene expression, pathways and disease-related processes.
Introduction
Expression quantitative trait loci (eQTLs) are pairs of genomic variants and genes for which there is an association of the genomic variant with the mRNA expression of the gene (1). If the genomic variant is an SNP, we call it ‘eSNP’ (2,3). Corresponding genes are referred to as ‘regulated genes'. Analysis of eQTLs is considered an important avenue for the mechanistic understanding of genotype–phenotype associations (4), especially in the context of genome-wide association studies (GWAS) analysing the genetics of complex traits. Genes regulated by a GWAS-SNP are promising candidates for follow-up functional studies and may point towards novel and relevant regulatory mechanisms (5). Observed enrichment of GWAS-SNPs within eSNPs corroborates this approach (6). Consequently, an increasing number of eQTL-studies in humans in different tissues have been performed (3,4,7–27).
Commonly, eQTLs are differentiated in cis- (i.e. local) and trans- (i.e. distant) eQTLs. Cis-eSNPs are typically defined to be located within the transcribed region of a regulated gene or within a maximum distance of 1 Mb to the transcribed region. If multiple genes are regulated by a single trans-eSNP, the term eQTL hot-spot or trans-cluster is used (12,13,21,28). Published eQTL studies have shown that there are numerous cis-eSNPs with high effect size on corresponding transcript levels. Conversely, trans-effects are usually small in size requiring larger studies for detection and confirmation.
Here, we performed a comprehensive genome-wide eQTL analysis of gene expression in peripheral blood mononuclear cells (PBMCs). We studied a large cohort of 2112 individuals allowing us to detect small effects which are common for trans-eQTLs. Exploiting the high power of our study, we also perform extensive replication analysis of published eQTLs including a large recent meta-analysis (23). From this, we find a good replication rate of previously published eQTLs in our data and conclude that about one-third of eQTLs identified in our study are novel. Going beyond pure univariate analysis of SNP-transcript pairs, we analyse pleiotropic effects of gene-expression regulation by studying eQTL hot spots where we discover novel trans-clusters of regulated genes. We demonstrate that these genes can provide meaningful mechanistic hypotheses. Additionally, we estimate the polygenetic effects of expression regulation by calculating chip-wise (CW) heritability (29). We propose to contrast these estimates with the combined correlation adjusted explained variances (30) of all significantly associated eSNPs for cis- and trans-regulation. This allows us to approximate the gap between the heritability already explained by the discovered cis- and trans-eQTLs and the heritability accessible with our SNP microarray technology in even larger studies. Finally, we perform a comprehensive analysis of annotated genomic elements including novel classes of non-protein coding loci. Our results support a prominent role for loci of non-coding RNAs (ncRNAs) and loci of pseudogenes in the regulation of expression of coding genes in humans. This result may facilitate further research regarding eQTL identification and regulatory mechanisms. Throughout the manuscript, we discuss implications for our functional understanding of gene-expression regulations and SNP–phenotype associations by contrasting our results with GWAS-SNPs or by pathway enrichment analyses.
Results
The power of the study allows detection of small genetic effects on gene expression
We studied the genetics of gene expression in PBMCs of 2112 individuals from the LIFE-Heart Study (31). We assessed the power of our study in comparison with previously published eQTL studies (Supplementary Material, Fig. S1). Exemplarily, we had 80% power to detect an eSNP that explains 1.8% variance of a trans-regulated transcript or 0.7% variance of a cis-regulated transcript. This is considerably more than the power to detect the same effects in a study comprising 1500 individuals (44.8 and 59.4%, respectively), what was the largest single study published so far (21).
Summary information of identified eQTLs
Controlling the false-discovery rate at 5% separately for cis- and trans-eQTLs, we identified a total of 1 840 232 eQTLs involving 11 410 (85.5%) genes. After pruning of eSNPs, in order to account reporting for linkage disequilibrium (LD), this number corresponds to ∼151 277 eQTLs. A genome-wide eQTL-plot displaying positions of eSNPs against those of corresponding regulated genes is shown in Supplementary Material, Figure S2. In our data, 17.6% of all genes expressed in mononuclear blood cells were associated with a trans-eSNP and 83.2% of all genes were associated with a cis-eSNP. Conversely, 779 042 (29.7%) of all SNPs were associated with a gene in cis, whereas 38 034 (1.4%) were associated with a gene in trans. After pruning, these observations corresponded to 81 148 (28.4%) cis- and 3800 (1.3%) trans-acting SNPs, respectively. Note that the smallest identified effect sizes with study-wide significance are different for cis- and trans-eQTLs (0.4 and 1.3% explained variance of gene-expression levels, respectively). All eQTLs are individually reported in Supplementary Material, Table S1 and are available as custom track for the UCSC genome browser (30) in Supplementary Material, Table S2. A summary information of all identified eQTLs is provided in Table 1.
Table 1.
Distribution of eQTLs (FDR ≤ 5%) at different significance cut-offs
| max. P-value | min. R2 | cis-eQTLs | cis-eSNPs (%) | cis-eSNPs pruned (%) | cis-regulated genes (%) | trans-eQTLs | trans-eSNPs (%) | trans-eSNPs pruned (%) | trans-regulated genes (%) |
|---|---|---|---|---|---|---|---|---|---|
| 0.00285 | >0.0042 | 1,739 991 | 779 042 (30) | 81 148 (28) | 11 098 (83) | ||||
| <10−5 | >0.0092 | 940 389 | 483 797 (18) | 40 314 (14) | 6 718 (50) | ||||
| <1.02 × 10−7 | >0.013 | 709 956 | 393 740 (15) | 30 225 (11) | 5788 (43) | 100 241 | 38 034 (1.4) | 3800 (1.3) | 2354 (18) |
| <10−10 | >0.02 | 519 833 | 311 001 (12) | 22 045 (8) | 4884 (37) | 58 072 | 23 809 (0.91) | 1356 (0.47) | 600 (4.5) |
| <10−15 | >0.03 | 360 548 | 231 440 (8.8) | 14 939 (5) | 3977 (30) | 31 660 | 15 732 (0.6) | 820 (0.28) | 374 (2.8) |
| <10−20 | >0.04 | 274 719 | 184 139 (7) | 11 093 (4) | 3366 (25) | 20 943 | 11 629 (0.44) | 553 (0.19) | 269 (2) |
| <10−50 | >0.1 | 103 318 | 77 368 (2.9) | 3705 (1.3) | 1747 (13) | 5772 | 3846 (0.15) | 131 (0.045) | 77 (0.58) |
| <10−100 | >0.19 | 41 375 | 32 415 (1.2) | 1309 (0.46) | 924 (6.9) | 1864 | 1579 (0.06) | 38 (0.013) | 28 (0.21) |
| <10−200 | >0.35 | 14 257 | 10 995 (0.42) | 420 (0.15) | 396 (3) | 955 | 869 (0.033) | 9 (0.003) | 11 (0.082) |
| <10−300 | >0.48 | 6971 | 5606 (0.21) | 221 (0.08) | 223 (1.7) | 821 | 754 (0.029) | 5 (0.002) | 7 (0.052) |
R2 corresponds to the variance of the transcription levels explained by corresponding eSNPs. Note that a gene can be both, cis- and trans-associated. After all pre-processing and filtering steps, we analysed a total of 2 625 374 autosomal SNPs and 18 738 expression probes within 2112 individuals. Pruning was done separately for cis- and trans-eQTLs.
Replication analysis and estimation of novel eQTLs
We next investigated novelty of identified eQTLs. Therefore, we compared our results with 22 published eQTL studies (3,7–27) subsequently referred to as ‘published studies' (Supplementary Material, Table S3). Sample numbers in previously published single studies ranged from N = 52 to N = 1490, the meta-study included 5311 individuals. Many of these studies were carried out in blood or blood-derived cell lines, but some studies used tissues derived from other organs such as liver, skin and brain.
About 590 228 (34.3%) of all cis-eQTLs and 46 115 (46.4%) of all trans-eQTLs detected in our study were not previously reported in these 22 studies and are further on termed ‘novel eQTLs'. After pruning, novel eQTLs corresponded to 75 790 unique cis-associations and 4375 unique trans-associations. Vice versa, 65.4% (7122) of our cis-regulated genes, and 64.7% (1494) of our trans-regulated genes were replicated/reported with the same or linked eSNP in these 22 studies.
Reported regulated genes were enriched in our results [92.0 versus 91.1%, odds ratio (OR) 1.1, 95% confidence interval (95%CI) 1.02–1.22, P = 0.02]. As expected, previously reported eQTLs had higher effect sizes than novel eQTLs in our data (reported: R2median = 1.11%, novel: R2median = 0.98%).
For a more detailed comparison, we investigated whether we can replicate eQTLs of published genome-wide studies including more than 1000 individuals as well as eQTLs of the meta-study.
In order to minimize the influence of technical differences, we limited replication analysis to SNP–gene pairs available in our study. Replication rates of cis-regulated genes were 72% for Fehrmann et al. (12), 84% for the meta-study (23) and 95% for Zeller et al. (21), while replication rate of trans-regulated genes were 26, 25 and 61%, respectively (Supplementary Material, Fig. S3). These rates are good in comparison with previously reported replication rates (12,13,21). The lowest cis-replication rate was observed for results from Fehrmann et al. (12). A reason might be that the cohort of Fehrmann et al. (12) comprised several distinct sub-cohorts with different diseases and leveraged two different gene-expression analysis platforms thereby increasing variance of gene expression. This reasoning might also be partly relevant for the meta-study. Additionally, the meta-study includes a considerable proportion of eQTLs with small effect size which reduces power for replication analysis. High trans-replication of Zeller et al. (21) in comparison to the other studies might mainly reflect higher effect sizes of trans-eQTLs due to the stricter Bonferroni-based significance level adopted there. Additionally, similar to our study, Zeller et al. (21) investigated a homogenous cohort and used as tissue a purified cell-population from blood (monocytes). Monocytes are also enriched in PBMCs, which might have positively affected replication rate of tissue-specific trans-eQTLs. Furthermore, we analysed vice-versa replication of our results (FDR ≤ 0.05) in Westra et al. (FDR ≤ 0.5). Here, we found replication rates on a similar level: we could replicate 80.5% of our cis-regulated genes and 48.5% of our trans-regulated genes for which at least one overlapping eSNP is available in Westra et al. (23).
Local eSNPs are enriched within 5 Mb to the regulated gene
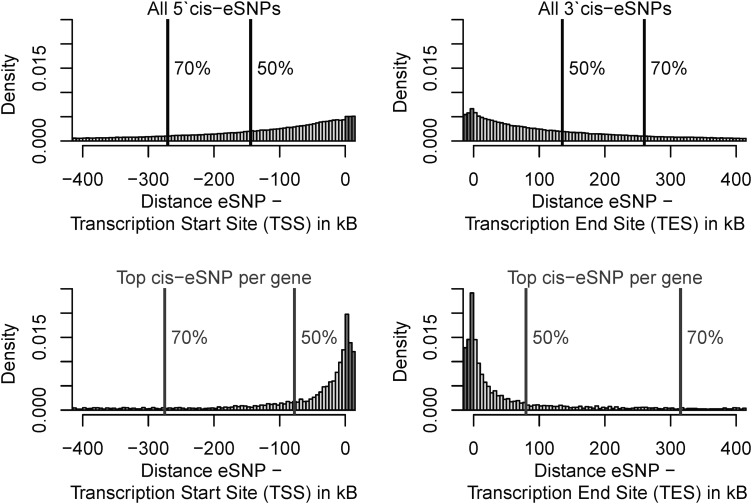
We were interested in estimating the genomic range in which local regulation is observable. On average, 85.3% of all eSNPs of a given chromosome were located outside the transcribed region of the regulated gene. More than half of all eSNPs of a regulated gene were located within a margin of 140 kb. When restricting to the strongest eSNP per gene (called ‘top eSNP’), this margin is reduced to 82 kb. Note that in our data, the top-eSNP accounted on median for about 40% of the total variance explained by all cis-eSNPs located on the same chromosome. Consistent with previous studies (32,33), we observed approximately symmetric enrichment of eSNPs at the transcript start site (TSS) and transcript end site (TES, Fig. 1). Note that the enrichment peak in Figure 1 seen at the TES most likely results from exon-specific QTLs and the known 3′ bias of Illumina probes (34). The decline of eQTL effect sizes with increasing distance to the transcript is shown in Supplementary Material, Figure S4.
Figure 1.
Distances between eSNPs and regulated genes. Histogram of the distance in kilobase between eSNPs and transcription start sites (show at the left side) and between eSNPs and transcription end sites of corresponding genes (shown at the right sight). Dark grey bars represent start and end of transcribed regions. Vertical lines and adjacent numbers are percentiles of all upstream and downstream distances found within 5 Mb 3′ from TSS and 5′ from TES. The upper panels show all eSNPs at FDR ≤ 5%, the lower panels are restricted to the strongest eSNP per regulated gene.
Only 62.3% of all cis-regulated genes were immediately adjacent to the top-eSNP. This means, a considerable amount of cis-regulations bridges at least one gene not regulated by the SNP considered (Supplementary Material, Fig. S5).
To go further, we were interested in eQTL enrichment in dependence on the distance to the regulated gene. Remarkably, even when excluding known long-range LD-regions, enrichment of eQTLs was observed up to 5 Mb to the TSS /TES. At this distance, the density of local eQTLs approximately met the density of inter-chromosomal trans-eQTLs (6.9 eQTLs/Mb, Supplementary Material, Fig. S6, lower panel). When restricting this analysis to the top-eSNP in order to account for LD, enrichment was still observed up to a distance of 2 Mb to the TSS/TES (Supplementary Material, Fig. S6, upper panel). This challenges the current commonly adopted limit of 1 Mb to define cis-regulations.
Pathway analysis demonstrates relevance of identified eQTLs for GWAS-related disease phenotypes and traits
To explore relevance of identified regulated genes, we investigated enrichment of KEGG pathways (35) among genes regulated by eSNPs that are in LD with a GWAS-SNP (R2 ≥ 0.5). This was done for each GWAS-trait separately. In Table 2, we show the KEGG pathway that was strongest enriched within regulated genes found for a certain GWAS trait. This table is restricted to GWAS-traits with regulated genes outside of the HLA-locus, an extended table including all data is shown as Supplementary Material, Table S4. From these tables, the discrepancy between position-based identification of candidate genes (nearest gene to GWAS-SNP) and functional identification by eQTL analysis becomes highly apparent: only in a single GWAS-trait ('Comprehensive strength and appendicular lean mass’), position-based and eQTL-analysis based genes were identical. For all other traits, almost all genes were different. This was observed for cis- as well as trans-regulated genes. Still, identified enriched pathways are meaningful for the corresponding GWAS-trait: this includes obvious examples like ‘Bitter taste perception’ showing enrichment for pathway ‘Taste transduction’, or ‘Asthma and hay fever’ showing enrichment for ‘Cytokine–cytokine receptor interaction’. Further examples are the KEGG ‘PPAR signalling pathway’ found to be enriched in the GWAS trait ‘acute lymphoblastic leukaemia’. Involvement of this pathway in this disease was debated (36). Similarly, KEGG pathway ‘Glutathione metabolism’ was enriched within GWAS-trait ‘Stearic acid (18:0) plasma levels': evidence of binding of stearic acid (18:0) to glutathione S-transferase was described (37). Furthermore, roles of Helicobacter pylori infection in autoimmune diseases were discussed (38) providing a reasoning for the identified relation of the GWAS-trait ‘Helicobacter pylori serologic status' the KEGG-pathway ‘Rheumatoid arthritis'. The relation between ‘Mean platelet volume’ and ‘Platelet counts' with KEGG pathways ‘ECM-receptor interaction’ and ‘Focal adhesion’, respectively, is mainly driven by trans-clusters and discussed in the following section named ‘Examples of GWAS-trait-related trans-clusters provide meaningful mechanistic hypotheses'.
Table 2.
Enrichment of KEGG pathways within regulated genes of GWAS-traits
| GWAS trait | GWAS-P-value range | KEGG-Term | Found in trait (%) | Enrich-ment factor | P-value | Genes fond in pathway |
|---|---|---|---|---|---|---|
| Acute lymphoblastic leukaemia (childhood) | 6 × 10−46–9 × 10−06 | PPAR signaling pathway | 4 (8.5) | 19 | 4.1 × 10−05 | cis: ACSL3*+, LPL+, NR1H3+, PCK2*+; trans: NR1H3+ |
| Asthma and hay fever | 5 × 10−12–2 × 10−06 | Cytokine–cytokine receptor interaction | 10 (6.0) | 9 | 2.8 × 10−08 | cis: IL18R1+, IL18RAP+; trans: CCL20*+, CCL3*+, CCL3L3*+, CCL4*+, IL1A*+, IL1B*+, IL8*+, TNF*+ |
| Bitter taste perception | 2 × 10−62–3 × 10−08 | Taste transduction | 3 (13.6) | 112 | 1.3 × 10−06 | cis: TAS2R14+, TAS2R20+, TAS2R43+ |
| Blood pressure measurement (cold pressor test) | 4 × 10−09–3 × 10−06 | RNA polymerase | 2 (8.7) | 178 | 3.0 × 10−05 | cis: POLR2J+, POLR2J2+ |
| Comprehensive strength and appendicular lean mass | 2 × 10−07–8 × 10−07 | Biosynthesis of unsaturated fatty acids | 2 (10.5) | 216 | 2.0 × 10−05 | cis: FADS1, FADS2 |
| Economic and political preferences (immigration/crime) | 2 × 10−06–6 × 10−06 | Steroid hormone biosynthesis | 3 (14.3) | 195 | 1.2 × 10−07 | cis: AKR1C2+, AKR1C3+, AKR1C4*+ |
| Helicobacter pylori serologic status | 1 × 10−18–2 × 10−08 | Rheumatoid arthritis | 7 (9.9) | 18 | 4.8 × 10−08 | trans: CCL20*+, CCL3*+, CCL3L3*+, IL1A*+, IL1B*+, IL8*+, TNF*+ |
| Lipoprotein-associated phospholipase A2 activity and mass | 2 × 10−23–5 × 10−06 | Drug metabolism: cytochrome P450 | 3 (9.7) | 40 | 4.5 × 10−05 | cis: GSTM1*+, GSTM2*+, GSTM4+ |
| Mean platelet volume | 1 × 10−103–7 × 10−06 | ECM-receptor interaction | 11 (20.0) | 6 | 9.1 × 10−07 | cis: CD36; trans: COL6A3+, GP1BA+, GP1BB+, GP6+, GP9+, ITGA2B+, ITGB1+, ITGB3+, ITGB5+, VWF |
| Metabolite levels (HVA-5-HIAA Factor score) | 2 × 10−06–6 × 10−06 | Fatty acid elongation in mitochondria | 2 (28.6) | 585 | 2.5 × 10−06 | cis: HADHA+, HADHB+ |
| Platelet counts | 3 × 10−54–7 × 10−06 | Focal adhesion | 17 (12.0) | 3 | 3.9 × 10−05 | cis: PRKCB+, VASP+; trans: ACTN1+, COL6A3+, EGF+, ILK+, ITGA2B, ITGB1+, ITGB3+, ITGB5+, MYL9+, PARVB+, PTK2+, RAP1B*+, TLN1+, VCL+, VWF |
| Serum uric acid levels | 1 × 10−80–3 × 10−06 | Systemic lupus erythematosus | 10 (11.8) | 44 | 9.3 × 10−17 | cis: HIST1H2AB*+, HIST1H2AC+, HIST1H2AE*+, HIST1H2BB+, HIST1H2BD+, HIST1H4A+, HIST1H4B*+, HIST1H4C+, HIST1H4D*+, HIST1H4H+ |
| Stearic acid (18:0) plasma levels | 1 × 10−20–5 × 10−06 | Glutathione metabolism | 4 (10.0) | 37 | 2.4 × 10−06 | cis: GGT7*+, GSTM1+, GSTM2+, GSTM4+ |
| Type 1 diabetes autoantibodies | 2 × 10−111–2 × 10−06 | Sulphur metabolism | 3 (42.9) | 47 | 2.3 × 10−05 | cis: SULT1A1+, SULT1A2+, SUOX+ |
Found in trait (%): Number of genes belonging to the KEGG-Term that are also regulated by an eSNP. The percentage relates to all genes belonging to the KEGG-Term. Enrichment: Found genes versus the genes expected without any enrichment. P-value: nominal enrichment P-value. Asterisks indicate novel identified eQTLS, the ‘+’ sign indicates that the respective gene was not mentioned as ‘reported gene’ or ‘mapped gene’ in the GWAS-catalogue.
Summary information on trans-clusters
We defined trans-clusters as trans-eSNPs associated with at least two trans-regulated genes. Within our data, we identified 14 953 trans-clusters. After pruning, this number corresponded to 849 unique SNPs or ∼175 genomic loci, i.e. 11.9% of all loci that included a trans-eSNP were associated with more than one trans-regulated gene. Our data confirm previously reported large trans-clusters related to HLA-SNPs on chromosome 6 and the large trans-cluster on chromosome 3 related to rs12485738 (Supplementary Material, Fig. S2). The latter corresponds to an SNP known to be associated with mean platelet volume (39). Most of our trans-cluster loci were found on chromosomes 6 (9.7%) and chromosome 2 (9.7%).
Analysis of trans-clusters is especially appealing for eSNPs that are in LD with known GWAS-SNPs, as further hypotheses about pathomechanisms of the disease-associated SNP can be generated. Therefore, we contrasted trans-clusters where the eSNPs is in LD (R2 ≥ 0.5) with a GWAS-SNP to trans-clusters unrelated to GWAS-SNPs. Indeed, trans-cluster eSNPs that were in LD with GWAS-SNPs appeared to regulate more genes (on average 1.6 times more genes, Quasi-Poisson-fit P<10−15). They also showed stronger associations with regulated transcripts (i.e. the median of the MANOVA-log10 P-values for association was shifted by 20.9, Wilcoxon test P < 10−15). Interestingly, trans-cluster eSNPs in LD with GWAS-SNPs were less frequently associated with an additional cis-regulated transcript, (92.6 versus 95.4%, Fisher-test P < 10−15). However, if an additional cis-regulation was present, cis-effect sizes were stronger (median difference of explained variance of transcript levels was 0.11%, Wilcoxon test P < 10−15). Finally, we compared the average decrease of the correlations among trans-regulated genes when expression levels of these genes were adjusted to the trans-cluster eSNP. A decrease is supportive for a causal effect of the eSNP on expression levels of trans-genes. The decrease of the correlation was stronger for GWAS-related trans-clusters than for non-GWAS related trans-clusters (on median −11 versus −8%, Wilcoxon test P < 10−15).
Examples of GWAS-trait-related trans-clusters provide meaningful mechanistic hypotheses
Outstanding trans-clusters with implication for GWAS-SNPs are shown in Table 3. This table is restricted to trans-clusters that are in LD with GWAS-SNPs and that have at least three trans-regulated genes from which at least 75% had to be novel. Additionally, correlation of expression levels of regulated genes was required to decrease when adjusting expression levels on the corresponding trans-cluster eSNP.
Table 3.
Trans-clusters that are correlated with GWAS SNPs
| eSNP | Chr | GWAS phenotype | GWAS SNP | R2 | GWAS reported genes | P-value | trans- regulated genes novel, % | n trans- regulated genes | trans-regulated genes | cis-regulated genes | Mean correl. Change, % |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs34856868 | 1 | Obesity-related traits | rs34856868 | 1 | BTBD8 | 1.1 × 10−45 | 100 | 7 | CDC42BPB*, ZAK*, KCNK13*, C6orf192*, PGA5*, FOLR2*, ACOX2* | FAM69A* | −4.4 |
| rs17616434 | 4 | Alcohol consumption, Allergic sensitization, Asthma and hay fever, Helicobacter pylori serologic status, Self-reported allergy | rs10004195, rs17616434, rs2101521, rs4543123, rs4833095 | 0.88–1 | FAM114A1, Intergenic, KLF3, MIR574, TLR1, TLR10, TLR6 | 9.8 × 10−59 | 100 | 38 | CCL3*, NFKBIA*, TNF*, CCL3L3*, CCL20*, IL1B*, SLC25A24*, IL1A*, MAFF*, CCL4*, CYP4B1*, IER2*, ZC3H12A*, IL8*, NFKBIZ*, CD83*, PPP1R15A*, PNRC1*, GADD45B*, FFAR2*, G0S2*, CDKN1A*, LOC338758*, FTH1*, ZFP36*, IER5*, TNFAIP3*, PIM3*, KLF10*, RAD1*, OTUD1*, BTG2*, JUNB*, IGFBPL1* | TLR1, KLHL5*, C4orf34*, KLHL5 | −4.8 |
| rs9275698 | 6 | Asthma | rs9275698 | 1 | HLA-DQA2 | 4.3 × 10−25 | 80 | 5 | BTN3A2, HLA-G*, VARS2*, DEF8*, KPNA2* | HLA-DPB1, HLA-DRB1, PSMB9, HLA-DQA1, RDBP*, SKIV2L*, HLADMA, HLA-DOB, HLA-DOA*, C2* | −6.3 |
| rs3132468 | 6 | Dengue shock syndrome | rs3132468 | 1 | MICB | 3.1 × 10−52 | 80 | 6 | HLA-DRB1, LIMS1*, HLA-A*, XRCC6*, TMEM154* | ATP6V1G2, HLA-C, LST1, HSPA1B, DDAH2, SKIV2L, ABCF1*, AIF1, AIF1*, LY6G5C, TUBB* | −11 |
| rs2858870 | 6 | Nodular sclerosis Hodgkin lymphoma | rs204999, rs2858870, rs6903608, rs9268528, rs9268542 | 1 | HLA-DQB1, HLA-DRB1 | 3.8 × 10−122 | 87.5 | 9 | EXOC1*, ZNF672*, TMEM154*, SSRP1*, HLA-C*, HLA-C, TRIM56*, XRCC6*, ARHGAP24* | HLA-DRB1, SKIV2L, HLA-DQA1, HSPA1L, PSMB9*, TAP2*, AGPAT1, HLA-DOB*, HLADRA, FKBPL, C6orf48* | −2.3 |
| rs2293889 | 8 | HDL cholesterol | rs2293889 | 1 | TRPS1 | 7.1 × 10−28 | 100 | 5 | EMR1*, EMR3*, MBOAT7*, MYB*, ADAM8* | TRPS1* | −5.2 |
| rs5016282 | 11 | Attention-deficit hyperactivity disorder | rs5016282 | 1 | GRM5 | 4.1 × 10−18 | 100 | 3 | ACP2*, NR1H3*, DDB2* | −5.2 | |
| rs10876864 | 12 | Alopecia areata, Asthma, Polycystic ovary syndrome, Rheumatoid arthritis, Type 1 diabetes and autoantibodies, Vitiligo | rs10876864, rs11171739, rs1701704, rs2292239, rs2456973, rs705702, rs773125 | 0.6–1 | CDK2, DGKA, ERBB3, IKZF4, PA2G4, PMEL, RAB5B, RPS26, SUOX, ZNFN1A4 | <10−220 | 76.9 | 13 | IP6K2*, STX1B*, LAP3*, DPF2, KCTD11, LDHC*, PRIC285, PTBP1*, ARPC5*, TRAM1*, C20orf24*, SEC61B*, BTF3* | RPS26, SUOX, SMARCC2*, GDF11, ESYT1*, ATP5B*, RDH5, DGKA | −42 |
| rs10512472 | 17 | Mean platelet volume, Platelet counts | rs10512472, rs16971217 | 1 | AP2B1, SNORD7 | 2.6 × 10−09 | 77.8 | 9 | ANKRD9, C7orf41*, TSPAN9, ITGA2B*, MYL9*, CTDSPL*, MARCH2*, PGRMC1*, ITGB3* | −0.4 | |
| rs3027234 | 17 | Parkinson's disease Telomere length | rs3027234, rs3027247 | 0.67–1 | C17orf68, CTC1 | 7.5 × 10−37 | 100 | 3 | HSD17B7*, SASS6*, RNF187* | CTC1, AURKB, PFAS | −19 |
Shown are trans-clusters with >70% novel trans-regulated genes correlated with a GWAS-SNP. Regulated genes are ordered according to explained variance. Asterisk (*): novel regulated genes R2: linkage disequilibrium between eSNP and GWAS SNP, p-value: MANOVA-p-value of eSNP and all trans-regulated transcripts, mean correl. change: relative change of the correlation between trans-regulated transcripts when adjusting expression levels on the eSNP considered.
The first trans-cluster in Table 3 is rs34856868 on chromosome 1. This SNP is also associated with obesity-related traits. Identified trans-regulated gene ACOX2 (Acyl-CoA Oxidase 2, Branched Chain) is functionally plausible as it has a prominent role in lipid metabolism (Supplementary Material, Fig. S7). Consistently, this gene is also included in the GO-Term ‘fatty acid beta-oxidation using acyl-CoA oxidase’ (Supplementary Material, Table S5).
The second trans-cluster rs17616434 on chromosome 4 is associated with several GWAS-phenotypes, mainly immunity and Helicobacter-related phenotypes. In line with this, many novel identified trans-regulated genes can be found in plausible GO- and KEGG pathways, e.g. ‘Cytokine–cytokine receptor interaction’, ‘response to molecule of bacterial origin' or ‘Toll-like receptor signaling pathway’ (Supplementary Material, Table S5).
Plausible trans-regulated genes were also found for the trans-clusters on chromosome 6. Exemplarily, for rs2858870 associated with GWAS-trait ‘Nodular sclerosis Hodgkin lymphoma’, we found novel trans-regulated gene SSRP1 located on chromosome 11. This gene is known to be part of the heterodimer FACT that is critically involved in the anticancer mechanism of cisplatin (40). For trans-cluster rs9275698 associated with GWAS-trait ‘Asthma’, novel trans-regulated gene KPNA2 located on chromosome 17 was reported to be related to V(D)J recombination (41), thereby providing a link to immunity.
The trans-cluster rs10876864 on chromosome 12 is linked with type 1 diabetes and vitiligo. For this SNP, we also confirm the previously reported cis-regulated gene RPS26. Importantly, RPS26 was excluded as causal gene involved in type I diabetes (42,43). Our novel trans-regulated genes provide alternative pathomechanistic hypotheses to understand downstream effect of this SNP. Gene-Ontology categories that include novel trans-regulated genes show possibly hints to proteins targeting to membrane and purine nucleoside triphosphate biosynthesis (Supplementary Material, Table S5). Note that correlations among genes of this trans-cluster changed on average for −42% when expression levels were adjusted to rs10876864, which was the largest value for trans-clusters reported in Table 3.
For trans-cluster rs11651199 on chromosome 17 related to Parkinson's disease, the GO-terms ‘proteasomal protein catabolic process' and ‘ubiquitin-dependent protein catabolic process' include novel trans-regulated gene RNF187 and the confirmed reported cis-regulated gene AURKB (Supplementary Material, Table S5). Fittingly, involvement of the ubiquitin proteasome system is known to be related to Parkinson's disease. Therefore, our findings may be useful to improve understanding of this system as cause or consequence of early pathological alterations in Parkinson's disease (44). Note that, in this example, a cis- as well as a trans-regulated gene are included in the same enriched pathway, which further supports a functional relevance (45).
For trans-cluster rs10512472 on chromosome 17 related to the GWAS-traits ‘platelet count’ and ‘mean platelet volume’, we found several relevant GO-terms as well as KEGG-terms nominally enriched (Supplementary Material, Table S5). This includes the KEGG-term ‘Hematopoietic cell lineage’ which includes novel trans-regulated genes ITGA2B and ITGB3. As shown in Supplementary Material, Figure S8, both genes are implicated in formation of platelets thereby providing a clear link to the GWAS-trait ‘platelet count’. Similarly, Supplementary Material, Figure S9 shows KEGG-pathway ‘Regulation of actin cytoskeleton’ that includes novel trans-regulated genes ITGA2B, ITGB3 and MYL9. According to this pathway, these genes are involved in actomyosin assembly contraction which is a reasonable functional link with GWAS-trait ‘mean platelet volume’.
Evidence for a mechanistic relevance of novel trans-regulated genes can also be derived from previously reported SNP-association studies. Exemplarily, for trans-cluster rs2293889 on chromosome 8 related to HDL cholesterol (46), we found a novel cis-regulated gene TRPS1 and five novel trans-regulated genes. For most of these five novel trans-regulated genes, genetic association studies with conceptually related phenotypes can be found: SNPs in ADAM8 were reported to be involved in advanced atherosclerotic lesion areas and myocardial infarction (47), for SNPs in EMR1 and EMR3 nominal associations for heart failure and blood pressure determination, respectively, were reported in the Phenotype–Genotype Integrator (rs3895916 in EMR1 in dbGaP:phs000226 and rs45508602 in EMR3 in dbGaP:ph00221 as well as dbGaP:phs000501, accessed 11/19/13) (48). Variants in MYB were associated with coronary artery disease (27) and levels of ghrelin (49), a peptide hormone that stimulates food intake and growth-hormone release. Ghrelin in turn is known to interact with HDL (50). As it is known that TRPS1 is a transcriptional repressor with GATA-type zinc finger binding sites (51), we analysed chromatin immunoprecipitation data (52) for enrichment of binding of transcription factors near the five trans-regulated genes. Indeed, GATA1 binding sites were the most prominently enriched human binding sites in the genes of the cluster [due to genes EMR1, EMR3 and MYB (P = 0.004)]. A trend towards enrichment of GATA3 binding sites was found for MBOAT7 (P = 0.12). This is in line with observed expression levels: when we grouped individuals according to genotypes of rs2293889, we observed that expression levels of the trans-regulated genes increased (i.e. highest for genotype TT, lowest for genotype GG), whereas for the same genotypes, expression levels of TRPS1 decreased (i.e. lowest for genotype TT, highest for genotype GG, Supplementary Material, Fig. S10). This opposing behaviour is unlikely due to chance (P < 0.05) and supportive for a GATA-mediated control of the trans-regulated genes. A possible mechanism might be an effect of rs2293889 on RNA stability: RNA immunoprecipitation data reveal binding of ELAVL1 at the same chromosomal location where rs2293889 is located (53). ELAVL1 is an RNA-binding protein that can stabilize RNA in order to counteract RNA degradation.
The majority of common cis-eSNPs of strongly heritable genes seems to be identified, but the majority of common trans-acting eSNPs remains to be discovered
We were interested in polygenetic effects on gene expression. More precisely, we examined the CW heritability (29,54), which corresponds to the variance of gene-expression levels explained by all SNPs present on the chip. In our study, CW-heritability can be used to estimate how much of the genetics of gene expression is attributable to common genetic variants covered by our technology.
We contrasted the CW-heritability with the variance of gene-expression levels explained by all eQTLs identified in this study. To quantify the latter, we combined the explained variances of all identified eSNPs (FDR ≤ 5%) for each transcript using correlation adjusted scores. This approach fully accounts for the LD-structure between SNPs (30). We refer to this measure as the ‘explained variance of combined eSNPs'. The observed differences between CW-heritabilities and ‘explained variance of combined eSNPs' allows a rough quantification of how many novel eQTLs can be still discovered in even larger studies or meta-analyses applying similar technology.
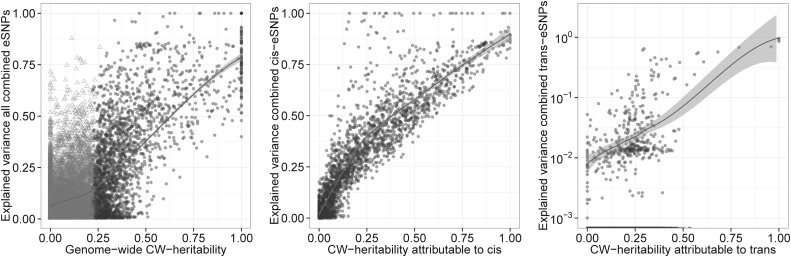
Within our data, 2556 (14.2%) of all autosomal transcripts corresponding to 2260 (17.6%) of all genes showed a CW-heritability significantly greater than zero. These genes are further on termed ‘strongly heritable genes' and the analysis further on is restricted to those genes. Smallest CW-heritability of these genes was 21.3%. Respective standard errors ranged from 0.094 to 0.155 (median 0.143). Still, we found correlation of our estimates with reported twin-based heritability of 0.5 or larger (P = 0.004, r = 0.39, 95%CI 0.13–0.60, see also Supplementary Material, Fig. S11). On median, all SNPs located on the chromosome where the regulated gene is located contributed for 36.9% of the genome-wide CW-heritability. We call this the ‘cis-attributable component of CW-heritability’. All SNPs located on different chromosomes contributed on median to 65.2% of the genome-wide CW heritability (Supplementary Material, Fig. S12). We call this the ‘trans-attributable component of CW-heritability’. These numbers suggest that for strongly heritable genes, most common variant-related heritability is hidden in trans-regulations.
For 2115 (93.6%) of all strongly heritable genes, one or more eSNPs was identified in our study (Fig. 2). For 99% of genes with a significant cis-attributable component of CW-heritability, we found one or more cis-eQTLs in our study. Only for 23% of these genes, the explained variance of combined cis-eSNPs was smaller than the corresponding cis-CW-heritability. Therefore, our data suggest that for strongly heritable genes, the vast majority of common cis-eSNPs accessible with our technology seems to be identified.
Figure 2.
Estimating the gap between explained and predicted heritability of gene expression. To estimate the gap between explained and predicted heritability of gene expression, we compared the explained variance of gene expression of combined eSNPs versus the genetic variance of gene-expression levels resulting from all imputed SNPs (CW-heritability). This is shown at the left side for all SNPs, in the middle for all SNPs found on the chromosome, where the regulated transcript is located (cis-regulation), and at the right side for all SNPs found on all chromosomes, where the regulated transcript is not located (trans-regulation). Triangles indicate transcripts with significant genome-wide CW-heritability (P ≤ 0.05). For each graph, a loess-estimator including confidence bounds is shown. Note that, for convenience, the ordinate in (C) is log10-transformed. Transcripts with an explained variance of combined trans-eSNPs of zero are shown at the bottom of the graph.
In contrast, for only 19% of genes with a significant trans-attributable component of CW-heritability, we found one or more trans-eQTLs. On median, the explained variance of combined trans-eSNPs explained only 7% of the corresponding trans-CW-heritability (Fig. 2, right). Therefore, we conclude that most common cis-eSNPs are discovered, but the majority of common trans-acting eSNPs remains to be identified.
Note that explained variance of combined trans-eSNPs tends to be large only when explained variance of combined cis-eSNPs of the gene is small (Supplementary Material, Fig. S13).
Loci of pseudogenes and multiple classes of non-coding RNAs are enriched at sites of eSNPs
Finally, we were interested in enrichment of eSNPs within functionally annotated genomic loci. Knowledge of enrichment can facilitate the search for yet undetected eQTLs and provides hypotheses about molecular mechanisms of the regulation of gene expression. Table 4 and Supplementary Material, Table S6 show results of this analyses: we could confirm enrichment of eSNPs within previously reported functionally annotated genomic loci. This includes loci of certain classes of histones, transcribed coding genes, CpG islands, transcription factors and miRNA target sites (21,51,52). We also found enrichment within yet unreported annotated loci. This includes loci of ncRNAs and loci of pseudogenes, loci of transcripts of uncertain coding potential (55) and predicted open-reading frames in intergenic space (56). Classes of enriched ncRNA loci were loci of large intergenic RNAs (lincRNAs), ncRNA loci relevant in transcriptome regulation (i.e. bona fide ncRNAs regulated by TP53-mediated apoptosis or in the cell cycle (57) and ncRNA-loci displaying conserved secondary structures. Those ncRNA loci were found in intergenic regions as well as in introns of coding transcripts. Importantly, eSNPs were only enriched in exons of lincRNAs, but not introns of lincRNAs (Supplementary Material, Table S6). This corroborates a functional relevance of these lincRNA-related eSNPs.
Table 4.
Enrichment of functionally annotated genomic loci co-localizing with eSNPs
| Annotation | OR | P-value | Overlap |
|---|---|---|---|
| Exons of long non-coding RNAs | |||
| Cabili et al. (55) | 1.13 (1.08–1.18) | 4.3 × 10−8 | 4041 |
| Introns of long non-coding RNAs | |||
| Gencode v13 | 0.77 (0.75–0.8) | 1.9 × 10−74 | 8688 |
| Cabili et al. (55) | 0.83 (0.81–0.84) | 2.5 × 10−75 | 17 311 |
| Bona fide non-coding RNAs regulated in cell cycle, TP53 pathway or STAT3 pathway | |||
| Cell cycle (transcript located in introns of protein-coding genes) | 1.48 (1.33–1.65) | 3.4 × 10−13 | 783 |
| TP53 (transcript located in intergenic space) | 1.51 (1.2–1.9) | 3.0 × 10−4 | 181 |
| TP53 (transcript located in introns of protein-coding genes) | 1.61 (1.5–1.74) | 9.8 × 10−38 | 1714 |
| Bona fide non-coding genomic regions predicted to contain conserved secondary structure motifs | |||
| SISSIz (motif located in intron of protein-coding gene) | 1.35 (1.31–1.39) | 4.2 × 10−94 | 10 658 |
| RNAz (motif located in intergenic space) | 1.16 (1.12–1.22) | 1.1 × 10−12 | 4459 |
| RNAz (motif located in intron of protein-coding gene) | 1.58 (1.53–1.65) | 2.3 × 10−128 | 6555 |
| miRNA target sites | |||
| TsmiRNA (conserved miRNA target sites–UCSC track) | 1.66 (1.37–2) | 6.5 × 10−8 | 277 |
| Novel transcripts with putative coding function | |||
| Exons of transcripts of uncertain coding potential (TUCP) | 1.26 (1.19–1.34) | 7.2 × 10−15 | 2414 |
| Predicted ORF in Intergenic space (RNAcode) | 1.27 (1.19–1.35) | 1.7 × 10−12 | 1897 |
| Pseudogenes | |||
| Gencode v13 | 1.35 (1.32–1.39) | 2.1 × 10−125 | 13 933 |
| Protein-coding gene annotation (Gencode v13) | |||
| 5′UTRs | 1.57 (1.52–1.62) | 7.2 × 10−188 | 10 084 |
| Coding exons | 1.43 (1.4–1.45) | 4.0 × 10−288 | 24 729 |
| 3′UTRs | 1.54 (1.51–1.57) | <1 × 10−220 | 24 828 |
| Intergenic space | 0.9 (0.9–0.91) | 4.3 × 10−93 | 174 239 |
| Intron | 1.33 (1.32–1.34) | <1 × 10−220 | 180 540 |
| Regulatory sites | |||
| CpG islands (UCSC track) | 1.54 (1.5–1.59) | 2.5 × 10−186 | 10 779 |
| Most conserved sequences (MCS, UCSC track) | 1.21 (1.19–1.22) | 1.2 × 10−140 | 42 451 |
| Open source for Regulatory Annotation (OregAnno, UCSC track) | 1.26 (1.23–1.3) | 2.4 × 10−52 | 9074 |
| Promoter regions (2 kb upstream of 5′UTR) | 1.51 (1.49–1.53) | <1 × 10−220 | 46 383 |
| Promoter regions (5 kb upstream of 5′UTR) | 1.5 (1.48–1.51) | <1 × 10−220 | 69 202 |
| Pol-II binding sites (ENCODE) | 1.44 (1.43–1.46) | <1 × 10−220 | 85 931 |
| Transcription factor binding sites (ENCODE) | 1.27 (1.25–1.28) | <1 × 10−220 | 102 616 |
| Transcription factors from Transfac database | 1.17 (1.15–1.19) | 2.6 × 10−66 | 26 801 |
| DNaseI hypersensitivity sites (ENCODE) | 1.24 (1.23–1.25) | <1 × 10−220 | 115 291 |
| Chromatin marks associated with enhancer or promoter sites (ENCODE) | |||
| H3K4 monomethylation | 1.25 (1.24–1.27) | <1 × 10−220 | 220 210 |
| H3K4 trimethylation | 1.4 (1.38–1.41) | <1 × 10−220 | 108 138 |
| H3K27 acetylation | 1.36 (1.34–1.37) | <1 × 10−220 | 135 114 |
| Chromatin marks associated with active regions of POL-II transcripts (ENCODE) | |||
| H3K36 trimethylation | 1.55 (1.54–1.57) | <1 × 10−220 | 213 995 |
| Chromatin marks associated with repressed regions of POL-II transcripts (ENCODE) | |||
| H3K27 trimethylation | 0.81 (0.8–0.82) | <1 × 10−220 | 251 569 |
OR, odds ratios; P-value, P-value of Fisher's Exact Test; Overlap, number of eSNPs overlapping with an annotation. A non-coding transcript is bona fide non-coding if it does not exhibit any evidence for open-reading frames or any sequence similarity to known amino acid coding sequences. Within this analysis, 787 378 unique eSNPs were included. In this table, enriched or depleted categories are reported if significance level was smaller than 0.05 after Bonferroni correction for 42 categories considered.
When investigating pseudogene-loci in more detail, we found systematic differences between pseudogene loci containing an eSNP and those not: pseudogene loci with co-localized eSNPs were more often transcribed and showed in general more often evidence for transcription-related activities. These loci were also less likely to result from retro-transposition of mRNA but more likely to result from processes like gene duplication or gene inactivation (Supplementary Material, Table S7). A special class of pseudogenes comprehends those regulating their parent gene. A pseudogene's parent gene is defined as a coding gene, from which the pseudogene originates, e.g. via gene duplication. We searched for pseudogenes loci that contain an eSNP regulating the parent gene of the pseudogene. Within our data, we identified 44 such pairs. Of those, 16 pseudogenes were reported to be transcribed. Six of these included a pseudogene locus where the co-located eSNPs was in LD with GWAS-SNPs (Table 5).
Table 5.
Examples of pseudogenes co-located with an eSNP that regulates the pseudogene's parent gene
| Pseudogene ID | Pseudogene position | Pseudogene biotype | eSNPs | Regulated gene = pseudogene parent gene | Regulated gene position | Corresponding GWAS trait |
|---|---|---|---|---|---|---|
| ENST00000427240.1 | chr1: 39 997 510–40 024 379 (−) | Unprocessed | rs2746050 (0.006) | PPIE | chr1: 40 204 517–40 229 585 (+) | C-reactive protein (rs12037222–rs2746050, R2 = 0.55) HDL cholesterol (rs4660293–rs2746050, R2 = 0.50) Red blood cell traits (rs3916164–rs2746050, R2 = 0.62) |
| ENST00000428767.1 | chr2: 73 898 157–73 912 212 (+) | Unprocessed | rs1052162 (0.027), rs10206899 (0.022) | ALMS1 | chr2: 73 612 886–73 837 046 (+) | Chronic kidney disease (rs13538–rs10206899, R2 = 1.00) Creatinine levels (rs10206899–rs10206899, R2 = 1.00) Glomerular filtration rate (rs10206899–rs10206899, R2 = 1.00) Metabolic traits (rs13391552–rs1052162, R2 = 0.96) Metabolite levels (rs9309473–rs10206899, R2 = 1.00) Metabolite levels (X-11787) (rs13538–rs10206899, R2 = 1.00) |
| ENST00000475455.1 | chr3: 133 407 036–133 431 646 (+) | Unprocessed | rs1006097 (0.005) | TF | chr3: 133 464 977–133 497 849 (+) | Iron status biomarkers (rs2718812–rs1006097, R2 = 0.90) |
| ENST00000377662.2 | chr6: 26 422 347–26 431 843 (+) | Processed | rs6456723 (0.005) | BTN2A1 | chr6: 26 458 189–26 469 865 (+) | Iron levels (rs17342717–rs6456723, R2 = 0.32) Iron status biomarkers (rs17342717–rs6456723, R2 = 0.32) Red blood cell traits (rs17342717–rs6456723, R2 = 0.32) |
| ENST00000435769.1 | chr7: 72 040 483–72 298 654 (−) | Unprocessed | rs3015844 (0.105), rs13238203 (0.005) | TYW1 | chr7: 66 461 817–66 704 496 (+) | Subcutaneous adipose tissue (rs2058059–rs3015844, R2 = 0.75) Triglycerides (rs13238203–rs13238203, R2 = 1.00) |
| ENST00000415709.1 | chr22: 25 851 679–25 855 648 (+) | Unprocessed | rs6423498 (0.373) | CRYBB2 | chr22: 25 615 612–25 627 836 (+) | Bipolar disorder (mood-incongruent) (rs1930961–rs6423498, R2 = 0.94) |
Pseudogenes were restricted to those reported to be transcribed (58), additionally, a corresponding GWAS trait had to exist. Pseudogene biotype: ‘processed’, pseudogene originates from retrotransposition; ‘unprocessed’, pseudogene originates from gene-duplication (58); eSNPs, all co-localized eSNPs that also are associated with expression levels of the pseudogene's parent gene. Values following SNP-Ids show explained variance of the regulated gene's expression level, corresponding GWAS phenotype, GWAS phenotype with a GWAS SNP in LD with the eSNPs. LD between GWAS-SNPs and eSNPs is shown in hyphens.
We wanted to provide additional hypotheses for ncRNA- and pseudogene-related regulatory processes in GWAS-traits. Therefore, we first filtered all published GWAS-SNPs to those GWAS-SNPs that were in LD (R2 ≥ 0.8) with an eSNP identified in our study. We searched for GWAS-traits enriched for eSNPs located in loci of pseudogene and loci of ncRNAs. We found 17 GWAS-traits nominally enriched for eSNPs located in loci of ncRNAs (Supplementary Material, Table S8) and 14 GWAS-traits nominally enriched for eSNPs located in loci of pseudogenes (Supplementary Material, Table S9). This information may provide a starting point for further exploration of possibly ncRNA- or pseudogene-related mechanisms of expression modulation in these traits.
Discussion
In this work, we performed an eQTL-study designed to detect small effects as we analysed the largest single eQTL-cohort available so far. This allows us to validate published variants, to detect several new variants and to dissect cis- and trans-effects. As we analysed cis- and trans-eQTLs in a genome-wide manner, our data are a valuable resource to explore the relevance of trait associated SNPs identified in current as well as future GWAS studies. Going beyond univariate association analyses, we derived insights into the gap between estimated heritability and discovered eQTLs and provided potential functional explanation of genotype–phenotype associations and their relation to functional classes of the genome.
In our analysis, we found cis-regulation in about 83% of tested genes. This is higher than the percentage of cis-regulated gene reported in a recent meta-study (44%). The explanation is that (i) more SNPs survived the quality filter steps in our study (2.63 Mio SNPs versus 1.96 Mio. SNPs in the meta-analysis); (ii) we considered an eQTL to be cis-regulated in a much larger genomic region: in our cis-analysis, we included SNPs located within 1 Mb of either side of the transcription start or end site or within the gene body. In contrast, the meta-analysis considered only the probe binding site ± 250 kb. In consequence, many of our identified cis-eQTLs were small in effect size, as the effect size of cis-eQTL decreases as distance to the regulated gene increases: the effect size of the strongest eSNP in 34.9% of all regulated genes explained at most 1% of the variance of gene expression; (iii) we analysed a less diverse sample as we studied a single cohort using purified PBMCs as tissue. In contrast, the meta-analysis included many studies and focussed on whole blood. A relation between the number of detected eQTLs and study homogeneity was demonstrated previously (2). Also note that a high number of cis-regulated genes is in line with previous assumption, that most, if not all genes may have allele-specific expression differences (59).
By comparing our results with the literature, we could replicate 72–95% of reported cis-regulated genes and 25–61% of reported trans-regulated genes. Vice versa, we could replicate at Westra's FDR ≤ 0.5 level 5174 (80.5%) of our cis-regulated genes for which at least one overlapping eSNP is available in Westra et al. and 228 (47.8%) of our trans-regulated genes. Therefore, replication rate can be considered good in comparison with previously reported replication rates (12,13,21). We estimated that about one-third of our eQTLs are novel in respect to 22 published eQTL studies including the meta-analysis (Supplementary Material, Table S3). Although such comparisons are always limited due to differences in design and methodology of different eQTL studies, this number illustrates the increased power of our study.
Concerning cis-regulated eQTLs, we found most eSNPs within <150 kb of the gene's start and end site. When we analysed the density of eQTLs, we found enrichments even at a distance of 5 Mb away from the transcribed region of a gene. This suggests that cis-eQTL analysis might benefit from a more liberal definition of the maximum distance from eSNP to gene and further illustrates that the concept of cis- and trans-eQTLs is limited. In this sense, we believe that the usually applied concept of calculating cis- and trans-specific FDRs is not optimal. A Bayesian approach including chances of true positives in dependence on the distance of an eSNP to its regulated genes might be a future improvement to be developed.
More than one-third of all top-eSNPs were not located directly adjacent to the regulated gene (Supplementary Material, Fig. S5). Furthermore, when looking at enriched pathways within regulated genes related to GWAS-phenotypes (Table 2), immediately neighbouring genes of an eSNP almost never matched the regulated gene identified via eQTL-mapping. This illustrates the relevance of functional studies in order to assign phenotype-associated SNPs to causal genes.
This is even more relevant for trans-regulated genes. In Table 3, we show trans-clusters related to GWAS phenotypes thereby providing novel candidate genes for future studies on various traits and diseases (5). We underline potential relevance of these trans-clusters by showing that regulated genes are related to conceptually linked biological annotations and pathways. At the example of a trans-cluster related to HDL-levels, we also use data from chromatin-immunoprecipitation experiments to demonstrate how further functional evidence can be added to the discovered associations.
Although the high number of novel eQTLs detected by our study implies a considerable progress, it is important to describe the gap between identified eQTLs and those still to be discovered. Here, we estimated this gap by comparing the combined contribution of all identified eSNPs with a global heritability measure summarizing the effect of all imputed SNPs (CW-heritability) (29,54). This accounts for LD (30,60) and allows separate analysis of the cis- and trans-component.
Thereby, we like to acknowledge that our CW-heritability estimates are still imprecise with standard errors ranging from 0.094 to 0.155 (median 0.143) for genome-wide CW-heritability and from 0.012 to 0.054 (median 0.0346) for cis-attributable component of CW heritability. Therefore, conclusion from these results are limited to strongly heritable genes and this analysis could benefit from even larger sample sizes than ours. Still, results drastically differed between cis- and trans-regulated genes: for almost all strongly heritable genes with a significant cis-attributable heritability component, we could identify one or more eSNPs. Combined cis-eSNPs explained most of the cis-attributable component of CW-heritability. In fact, due to winner's curse, the combined contribution of all identified eSNPs was often even larger than the cis-attributable CW heritability as the latter is less affected by winner's curse. Although an exact quantification of these effects is difficult, these results suggest that for strongly heritable genes, the vast majority of common cis-eQTLs seems to be identified (given our tissue and expression microarray technique). A strong contribution of known cis-eSNPs to the CW-heritability is in line with previous findings (13,61).
In contrast, only for 19% of genes with a significant trans-attributable CW-heritability, we could identify one or more eSNPs, on median all trans-eSNPs accounted for <10% of the trans-attributable heritability component (Fig. 2). This is especially important as we found that on median, most of the total CW-heritability result from trans (Supplementary Material, Fig. S12). These findings are also in line with other studies reporting that the trans-component has a stronger influence on the heritability of the transcriptome and that the number of yet-identified responsible trans-eSNPs is still very limited (61). Note, that the architecture of the genetics of gene expression is reported to be in the main additive, thereby closely resembling those of common diseases (62). Therefore, these results warrant that a comprehensive identification of trans-eQTLs requires even larger eQTL studies and respective meta-analyses with sample sizes comparable to those required in GWAS of common diseases. Additionally, this points to the relevance of improved methodological approaches for trans-eQTL detection.
EQTL detection could benefit from the identification of functional elements that co-localize with eSNPs. Knowledge of such functional classes can be used to better identify and predict eQTLs (63,64) and improve understanding of the regulatory architecture of the genome. For the first time, we report that loci of eSNPs are enriched at genomic sites of distinct classes of ncRNAs and pseudogenes (Table 4 and Supplementary Material, Table S6). This is reasonable, as ncRNAs are known to regulate expression of protein-coding genes in cis (65) and in trans (66,67). Previously, 108 cis-regulated ncRNAs were directly described (68). Our study further extends this finding, as our enrichment analysis comprised many additional ncRNA loci.
Our observation of enrichment of eQTLs in loci of pseudogenes hints towards a more general regulatory relevance of pseudogenes. This is supported by the reported enrichment of GWAS-SNPs—which are themselves enriched for eSNPs (6)—within genomic loci of pseudogenes (69). Examples for mechanisms on how a pseudogene can influence expression levels of other genes include miRNA-related interaction, influence of RNA stability and antisense regulation (70). Many of these reported mechanisms include interaction with the pseudogene's parent gene (i.e. the gene from which the pseudogene originates). Accordingly, we found 44 pseudogenes with a co-located eSNP that appears to regulate the pseudogene's parent gene. Table 5 shows examples of such pairs with potential relevance for GWAS-traits. To provide further starting points for the exploration of possibly ncRNA- or pseudogene-related mechanisms of gene-expression modulation, we report GWAS-traits showing nominal enrichment of eSNPs located at genomic loci of ncRNAs and pseudogenes (Supplementary Material, Tables S8 and S9).
In summary, our study substantially increases the catalogue of human eQTLs and improves our understanding of the complex genetic regulation of gene-expression, pathways and disease-related processes hereon. By numerous examples, we demonstrated how our study can support the identification of biologically plausible and testable hypotheses facilitating further research to understand the mechanisms of genotype–phenotype associations.
Materials and Methods
Ethics statement
The study meets the ethical standards of the Declaration of Helsinki. It has been approved by the Ethics Committee of the Medical Faculty of the University Leipzig, Germany (registration number 276–2005) and is registered at ClinicalTrials.gov (NCT00497887). Written informed consent including agreement with genetic analyses was obtained from all participants enrolled in the study.
Description of the cohort
Samples were derived from the ongoing Leipzig LIFE Heart Study which is an observational study designed to analyse molecular-genetic modifiers of atherosclerosis risk and related phenotypes. Individuals comprise either patients with suspected coronary artery disease due to clinical symptoms/non-invasive testing or with stable left main coronary artery disease. Details of the study can be found elsewhere (31,71). Patients with acute myocardial infarction were excluded from this analysis.
Measurement of gene expression
PBMC isolation (N = 2580) was performed using Cell Preparation Tubes (CPT, Becton Dickinson) as described (71). Total RNA was extracted using TRIzol reagent (Invitrogen) and quantified with an UV-Vis spectrophotometer (NanoDrop, Thermo Fisher). A total of 500 ng RNA per sample were ethanol precipitated with GlycoBlue (Invitrogen) as carrier and dissolved at a concentration of 50–300 ng/µl prior to probe synthesis. N = 79 samples were not further processed due to low RNA concentrations. N = 2501 samples were hybridized to Illumina HT-12 v4 Expression BeadChips (Illumina, San Diego, CA, USA) in batches of 48 and scanned on the Illumina iScan instrument according to the manufacturer's specifications (67). Documentation of sample processing included batch information at any processing step to allow adjustment in subsequent data analysis.
Raw data of all 47 323 probes were extracted by Illumina GenomeStudio, 47 308 probes could be successfully imputed in all samples. Data were further processed within R 2.13.1/Bioconductor. A total of 123 (4.9%) individuals having an extreme number of expressed genes [<7505 genes, defined as median ± 3 interquartile ranges (IQR) of the cohort's values] were excluded. Transcripts that were not found to be expressed according to Illumina's internal cut-off as implemented in Bioconductor package ‘lumi’ P ≤ 0.05 in at least 5% of all samples were not further considered in the analysis. Expression values were quantile normalized and log2-transformed (72). For further outlier detection, we calculated the Euclidian distance between all individuals and an artificial individual having average expression levels of all transcripts. Sixty-nine (2.9%) of the remaining individuals with a distance larger than median + 3 IQR were excluded. Furthermore, we defined for each individual a combined quantitative measure combining quality control features available for HT-12 v4 (i.e. perfect-match and miss-match control probes, control probes present at different concentrations, mean of negative control probes, mean of house-keeping genes, Euclidian distances of expression values, number of expressed genes, mean signal strength of biotin-control-probes). We calculated Mahalanobis-distance between all individuals and an artificial individual having average values for these quality control features. Thirty-one (1.3%) of the remaining individuals with a distance larger than median + 3 IQR were excluded. Transcript levels were adjusted for the known batch Sentrix barcode (i.e. expression chip-ID) using an empirical Bayes method as described (73). The empirical Bayes method required that at least two individuals for each batch are provided. This excluded two individuals. Success of adjustment was checked using ANOVA for both, the Sentrix barcode as well as the processing batch (in a processing batch, several expression chips were jointly processed, in consequence, within a processing-batch, several Sentrix barcodes are completely nested). The multivariate model included age, sex, monocyte counts and lymphocyte counts as covariates. A QQ-plot showing the distribution of ANOVA P-values before and after adjustment is shown in Supplementary Material, Figure S14. A total of 625 (2.2%) expression probes still over-inflated following Bonferroni-correction were excluded and 28 295 probes residualized for age, sex, monocyte counts and lymphocyte counts remained in analysis. Due to incomplete data of these covariates, 20 (0.9%) of the remaining individuals were excluded. Additionally, we calculated principal components of the expression data residualized for its first five principal components to account for unmeasured batch effects as outlined elsewhere (12). Using this number of PCAs, we found no evidence that trans-eQTL detection was compromised and still observed increasing numbers and effect sizes of detected eQTLs due to the adjustment. Probes were assigned to genes using Entrez-gene IDs via the R add-on package from Bioconductor illuminaHumanv4.db_1.14.0 that relates to NCBI data dated on 7 March 2012 (74). Entrez-gene IDs were used to retrieve information for the abbreviated gene names (HGNC identifier) (8) and transcription start site and transcription end site of corresponding genes via Bioconductor package ‘org.Hs.eg.db_2.7.1’. This package is based on hg19 coordinates retrieved from Golden Path data provided by UCSC Genome Bioinformatics at ftp://hgdownload.cse.ucsc.edu/goldenPath/hg19 with a date stamp of 22 March 2010. Remapping information (74) was only accepted if distance between chromosomal coordinates of expression probes and TSS/TSE was smaller than 1 Mb. The initial pre-processing resulted in 28 295 expression probes corresponding to 15 217 genes. Of those, 18 738 probes corresponding to 13 338 genes mapped uniquely within the human genome and had a probe annotation quality score (74) of at least ‘good’. Throughout the manuscript, corresponding gene names are provided as HGNC identifiers. For 2112 (93.5%) of the remaining individuals, valid genotype data were available allowing eQTL analysis. Raw and pre-processed gene-expression data are available from GEO (https://submit.ncbi.nlm.nih.gov/geo, GEO accession no. GSE65907).
SNP genotyping, pre-processing and imputation
DNA was extracted from peripheral blood using the Invisorb Spin Blood Maxi Kit (Stratec) as described (71). Genotyping was performed with the Affymetrix Axiom Technology using the custom option. Axiom CEU comprising M = 541 621 autosomal markers served as a backbone of our custom array. A total of 62 471 autosomal markers enriched in 44 genomic regions associated with cardiovascular disorders were additionally placed on the array. Genotyping was performed on Affymetrix facilities. From N = 3036 DNA samples originally sent to Affymetrix facilities, N = 2925 samples were successfully genotyped. Cell files of all successfully genotyped samples were combined and genotypes were called by Affymetrix Power Tools version 1.12. We required an individual-wise call rate to be 97% or better. For N = 604 092 autosomal SNPs, we estimated allele frequency and minimal call rate with respect to plates. We tested asymptotically for Hardy–Weinberg equilibrium and for association of allele frequency with plates. For all samples, we recalculated the call rate in a CW manner and we assessed the mean squared difference of the individual's genotype and expected genotype incorporating all autosomal SNPs with non-missing genotype. Further, we estimated pair-wise relatedness (75) using N = 184 108 autosomal SNPs after filtering for minimal call rate (call rate<98%), Hardy–Weinberg equilibrium (P < 10−6) and SNPs which are associated with plates (P < 10−7). Adopting these criteria, 2857 individuals remained in analysis. Within these individuals, SNP quality was re-estimated and filtered for the following criteria: minimum of all plate-wise call rates had to be ≥90% (these criteria imply that the conventional overall SNP call rate is >94.2% and its 10th percentile is >99.2%), a Hardy–Weinberg equilibrium with P ≥ 10−6 and association of SNP frequencies with plates with a P ≥ 10−7). A total of 566 359 autosomal SNPs fulfilled these criteria and were used for imputation with IMPUTE v2.1.2. (http://mathgen.stats.ox.ac.uk/impute/impute_v2.html). HapMap2 CEU, Release 24, dbSNP-build 126, NCBI 36 was applied as reference panel comprising 3 974 237 autosomal SNPs. 555 911 of our measured SNPs could be mapped to the reference. Following imputation, we removed 19 individuals being outliers according to the commonly adopted six-standard deviation criterion in EIGENSTRAT (76). For this step, we used 213 540 SNPs fulfilling following quality criteria: call rate per plate ≥ 98%, HWE P ≥ 10−6, batch association with genotyping plates P ≥ 10−7. A graphical representation of the distribution of the first two eigenvalues of the remaining individuals together with estimated relatedness is shown in Supplementary Material, Figure S15. Furthermore, SNPs with low quality based on minor allele frequency (MAF) <1% or with IMPUTE-info score ≤0.3 were excluded. Here, MAF related to those 2112 individuals that had also valid transcriptome data. SNPs not included in imputed data but having high-quality genotypes on chip were added to the data set, quality-control criteria were call rate ≥97%, MAF ≥ 1% and Hardy–Weinberg equilibrium P ≥ 10−6. This resulted in a total number of 2 627 381 SNPs. These SNPs were lifted from hg18 dbSNP130 (given by the manufacturer) to hg19 applying the public available tool ‘liftOver’ from UCSC (http://genome.ucsc.edu/cgi-bin/hgLiftOver). From this procedure, 2 625 374 autosomal SNPs resulted for eQTL analysis.
eQTL-association analysis
For eQTL association analysis and FDR calculation, we used the Matrix eQTL software (77) in the environment of Revolution R Enterprise 5.0.1. We created a genome- and transcriptome-wide QQ plot to investigate the distribution of our test statistics using the same R-package. No evidence of inflation of our test statistic was observed (Supplementary Material, Fig. S16). In our data, the FDR at 5% for cis-eQTL corresponds to a P-value threshold of 0.0028 and the FDR at 5% for trans-eQTLs corresponds to a P-value threshold of 1.02 × 10−7. Given the threshold for trans-effects and our sample size, we performed power analysis in dependence on the explained variance of an eSNP. Note that this measure of effect size is independent of SNP-allele frequencies. Calculation was done using the R-package ‘pwr’. Calculation of the number of eQTLs when considering only one eSNPs per locus was done by counting all eSNPs per 1 Mb only once for each regulated gene. Assignment of cis and trans was done based on smallest physical distance between mapping of the SNP and transcription start and end site of the corresponding gene. Genes were considered cis-regulated, if the distance between the eSNP and the transcribed regions of the gene was at most 1 Mb, or if the eSNP was found within the transcribed region of the gene. Calculation of the number of pruned SNPs was done using PLINK 1.9. Here, we applied parameters clump_r2 = 0.3 and clump_kb = 5000. Pruning was done separately for cis- and trans-associated eSNPs resulting in a total of 285 362 and 288 875 unique SNPs corresponding to the 2 625 374 autosomal SNPs in analysis, respectively.
To analyse potential reasons for false-positive cis-eQTLs, we investigated whether the putative eSNP was correlated with another SNP located at the same position where the corresponding expression probe binds and therefore might artificially disturb gene-expression measurement (12). For this purpose, we analysed whether any SNP reported in the 1000 genomes project (release 20 110 521 version 3 f, restricted to SNPs with a MAF ≥ 1%) co-locates with the binding region of transcript probes (78). If LD between the putative eSNP and the 1000 genomes SNP was present (R² > 0.1) or was unknown, cis-regulated eQTLs were marked as potential false positive. For LD calculation, we used HapMap Data (Release #28, lifted over to GRCh37/hg19) as well as the 1000 genomes data as reference applying Plink v1.07 (79).
For trans-eQTLs, a reason for false positive is cross-hybridization of the corresponding expression probe in proximity to the putative eSNP site. If this is the case, the putative trans-eQTL might be in fact a cis-eQTL. Trans-eQTLs were marked as potential false positive, if cross-hybridization of expression probes of the putatively regulated gene was found within 1 Mb of the putative eSNP. Cross-hybridization information resulted from a previous extensive remapping approach (74). Within our data, we found 2.7% of all cis-eQTLs and 8.7% of all trans-eQTLs to have an increased chance to be false positive. Since our criteria do not necessarily result in false-positive eQTLs (80) and since the overall rate of these events was moderate, we did not filter these results in general, but marked them in Supplementary Material, Table S1 and excluded them in certain distinct analyses as described. For additional details of potential false-positive SNPs, see Supplementary Material, Tables S10, S11 and Figure S17.
To get information about the distribution of eQTLs under the null hypothesis, we performed 100 additional genome-wide eQTL studies by permuting per individual labels of SNP data and expression data. These data are referred to as ‘permuted eQTL data’. We used permutated eQTL data to verify the Benjamini–Hochberg based P-value thresholds corresponding to the FDR of 5%. For cis-eQTLs, empirical FDR was on average 0.0501 ranging from 0.0478 to 0.0521, for trans-eQTL, empirical FDR was on average 0.0515, ranging from 0.0459 to 0.0596. For hypergeometric enrichment analysis of genes within Gene Ontology and KEGG pathways, we used the R-package ‘GOstats'. Results with enrichment P-values of <0.05 were reported. We used all 13 338 genes that were included in eQTL analysis as background. When using Gene Ontology, we used the implemented adjustment option to correct significance of enriched pathways according to the redundant nature of the hierarchical annotation system.
We compared our results with results of the GWAS catalogue (81), accessed on 14 August 2014. Thereby, we used as reference both, HapMap Data (Release #28, lifted over to GRCh37/hg19) and 1000 genomes data release 20 110 521 version 3 from EUR population, and applied PLINK (79) to identify correlated SNPs.
Comparison with known eQTLs
To identify novel eQTLs, we compared our results with publicly available results of 22 studies (3,7–27) (Supplementary Material, Table S3). Data of some of these studies were summarized and available from seeQTL (82) and the Chicago eQTL browser (http://eqtl.uchicago.edu). Significance level α of reported eQTLs of those studies was required to be always <0.005. We matched regulated genes on transcription-probe information (Ensemble gene ID, RefSeq ID, Entrez-gene ID, Probe-ID, and/or HGNC ID) as available. Putative and/or badly characterized genes defined as genes starting with letters ‘KIAA’, ‘FLJ’, ‘HS.’, ‘C.*ORF’ ‘MGC’ and ‘LOC’ were excluded from summary statistics when counting novel eQTLs. eSNPs were matched between our study and databases based on their dbSNP identifiers. An eQTL was regarded novel if for a certain regulated gene, eSNPs of our study were found on different chromosomes compared with published eSNPs, or if LD between our eSNPs and published eSNPs could be calculated and was found to be lower than R2 = 0.3 or if distances between our eSNPs and published eSNPs were >5 Mb.
To identify replication rates for eQTLs in our study, we performed detailed comparison with genome-wide studies including more than 1000 individuals (12,21,23). Two of these studies adopted a cis-and trans-specific significance level at an FDR of 5% (12,23), and one a global Bonferroni-criterion (21). Reported eQTLs of these three studies were included in replication analysis if eQTLs were autosomal and if the regulated gene as well as the reported eSNP was analysed in our study. Adopting these criteria, 80, 85, 81% of reported cis-eQTLs and 79, 77, 73% of reported trans-eQTLs of Fehrmann et al., Westra et al. and Zeller et al. were available for replication analysis, respectively. Cis- and trans-classification was used as defined in the original reports. A certain gene was considered replicated if one or more of the reported eSNPs were associated with the same gene in our study at an FDR of 5%. When analysing vice-versa replication of our regulated genes in Westra et al., we restricted our data in the cis-specific comparison to any SNP and gene reported in results of Westra et al. (FDR ≤ 0.5). For the trans-specific comparison, we additionally restricted our SNPs to those included in the GWAS catalogue as done by Westra et al. to allow a direct comparison between the studies.
Analysis of eSNP densities flanking transcribed regions
To estimate ranges of cis-effects, we analysed the distribution of the physical distances between eSNPs and corresponding regulated genes if found on the same chromosome. In order to avoid bias, genomic regions of long-range LD (83) were excluded for this analysis, thereby filtering out 559 genes. Additionally, potential false positives were excluded from this analysis. If a certain SNP was associated with multiple probes of the same gene, the corresponding SNP-gene distance was counted only once per gene. Cis-eQTLs with effect sizes smaller than the minimal observed effect size of trans-eQTLs were excluded from this analysis. This was to avoid bias as the study-wide significance level was different for cis- and trans-eQTLs. In order to estimate the maximum distance, where more eSNPs are observed than expected under the null hypothesis, we compared the local eSNP density with the average eSNP density from interchromosomal trans-eQTLs. A scatter-plot was applied to estimate the distance for which the frequency of eQTLs in our data dropped below the density of inter-chromosomal eQTLs (Supplementary Material, Fig. S6). When estimating the total variance explained by all cis-eSNPs located on the same chromosome, we combined respective eQTLs using correlation adjusted scores as implemented in the R-package ‘care’ (30,60).
Trans-cluster analysis
We defined trans-clusters as trans-eSNPs associated with at least two trans-regulated genes. When counting trans-cluster loci, we considered only one trans-cluster eSNP per 1 Mb. To score significance across trans-clusters thereby accounting for correlation between regulated genes, we applied a standard framework of a MANOVA. We analysed the mean change of correlation between expression levels of trans-regulated probes when adjusting on the eSNP. For this purpose, we calculated pair-wise absolute Pearson product-moment correlation coefficients of all transcripts of a trans-cluster before and after adjusting expression levels in a linear model on the regulating trans-eSNP. A negative change indicated a decreased correlation after adjustment.
Reported novel trans-clusters in Table 3 resulted from pruning applying R2 ≤ 0.3 in order to report independent effects. Trans-cluster shown in this table were required to have at least three trans-regulated genes, 75% of those had to be novel. LD with a GWAS-SNP had to be at least R2 ≥ 0.5, additionally, at least one GWAS-SNP was required to have an R2 ≥ 0.8 with the trans-cluster eSNP. Potentially false-positive eQTLs were excluded from this analysis.
To identify enrichment of KEGG and GO terms within cis- and trans-regulated genes, we used the R-package ‘GOstats'. To visualize enriched KEGG pathways, we used the R-package ‘pathview’ (84).
Estimation of CW heritability and correlation adjusted scores
To estimate CW heritability (CW-heritability) of transcripts, we used the software GCTA as previously described including all SNPs included in eQTL association analysis (54). We restricted this analysis to autosomal transcripts. CW-heritability is estimated based on mixed-model analysis of background relatedness between samples. Due to our chip-design, HapMap-based imputation and the non-family-based design of our cohort, this estimates the contribution of common variants to heritability only. This is different to usual family-based studies of the heritability of gene expression (2,11,13,62,85,86). When restricting analysis to the cis-attributable component of CW-heritability, we included all SNPs of the chromosome where the regulated gene is located. Vice versa, when restricting analysis to the trans-attributable component of CW-heritability, we excluded all SNPs of the chromosome where the regulated gene is located. We limited analysis to transcripts with CW-heritability significantly different from zero at the level of α = 0.05.
For contrasting CW-heritabilities with explained variances by identified eQTLs, we used correlation adjusted scores as implemented in the R-package ‘care’ (30,60). For each transcript, all associated eSNPs (FDR ≤ 5%) where summarized. We restricted analysis to observations without missing data (if SNP was from non-imputed data). The correlation shrinkage intensity lambda was 0 or if necessary to avoid singularity of the correlation matrix at most 10−9, i.e. we basically used the empirical correlation structure to estimate the genetic covariance. To quantify the explained variance assignable to cis- and trans-eSNPs separately, we summarized squared car-scores separately for all cis- and all trans-acting eSNPs of a certain transcript, respectively. For graphical presentations, summarized car-scores smaller than 10−3 were set to zero. When plotting ratios between summarized car-scores and CW-heritabilities, ratios larger than one were set to one. For reasons of improved comparability between summarized car-scores and cis-/trans-CW-heritability, all eSNPs located on the same chromosome as the regulated gene were regarded as cis-acting in this analysis.
Enrichment of eSNPs in functional elements
To compute the enrichment of eSNPs that harbour a genome annotation, we adapted the approach proposed by Hindorff et al. (81). Considering all SNPs in strong LD with the eSNP, the overlap with a genome annotation was computed with a selection of annotation sets of the human genome (version GRCh37/hg19). Significance of the observed overlap was inferred by Fisher's Exact Test. The expected number of overlaps was estimated from permuted eQTL data. For each eSNP, an interval (LD-block) that contains all SNPs in strong LD with that eSNP was generated. The LD-block was defined by the left and right most SNP which appears to be highly correlated with that eSNP (R² > 0.9) within a distance of 200 kb. Therefore, the maximum size an LD-block can have is 400 kb. LD data were obtained from HapMap (Release #27, NCBI 36, CEU) and lifted over to GRCh37/hg19. To avoid regional bias, replicates of LD-blocks were removed, so that the final set used in the analysis contains only unique LD-blocks. An LD-block was counted if at least one SNP within it overlaps the annotation. The LD-block is counted only once regardless of how often the overlap occurred with the annotation. We performed the same steps to calculate the overlap within permuted eQTL data and annotation sets providing expected distributions of SNPs under the null hypothesis.
A non-coding transcript was called bona fide non-coding if it does not exhibited any evidence for open-reading frames as predicted by RNAcode (56) nor any sequence similarity to known amino acid sequences in RefSeq database (version 7 March 2012). Thereby, similarity was assessed using tblastn with parameters -word-size 3 and an e-value < 0.05. A detailed listing and description of all included annotation sets is provided in Supplementary Material, Table S12. To characterize pseudogenes in detail, we used information described in psiDR version 1.0.0 (58).
Supplementary Material
Funding
This work was supported by the Initiative and Networking Fund of the Helmholtz Association [VH-NG738 to J.H. and K.R.]; Genome Quebec, Genome Canada, the Canadian Institutes of Health Research (CIHR) [for creation of data in dbGaP accession number phs000225.v1.p1 that was used in this study, see also Grundberg et al. (24) for additional sources]; and LIFE–Leipzig Research Center for Civilization Diseases, University of Leipzig [to H.K., A.G., F.B., P.A., K.H., R.B., M.S. and for the LIFE-Heart study]. LIFE is funded by means of the European Union, by the European Regional Development Fund (ERDF) and by means of the Free State of Saxony within the framework of the excellence initiative. Funding to pay the Open Access publication charges for this article was provided by the University of Leipzig.
Supplementary Material
Acknowledgements
We express our appreciation to all participants of the LIFE-Heart study. The data used for the analyses described in this paper includes data obtained from the database of Genotypes and Phenotypes (dbGaP), at http://www.ncbi.nlm.nih.gov/gap. Genotype and phenotype data for Expression QTL (eQTL) Analysis in Human Primary Cells (dbGaP accession number phs000225.v1.p1) were provided by Tomi Pastinen and Elin Grundberg, McGill University, Montreal, Canada.
Conflict of Interest statement. None declared.
References
- 1.Schadt E.E., Monks S.A., Drake T.A., Lusis A.J., Che N., Colinayo V., Ruff T.G., Milligan S.B., Lamb J.R., Cavet G., et al. (2003) Genetics of gene expression surveyed in maize, mouse and man. Nature, 422, 297–302. [DOI] [PubMed] [Google Scholar]
- 2.Emilsson V., Thorleifsson G., Zhang B., Leonardson A.S., Zink F., Zhu J., Carlson S., Helgason A., Walters G.B., Gunnarsdottir S., et al. (2008) Genetics of gene expression and its effect on disease. Nature, 452, 423–428. [DOI] [PubMed] [Google Scholar]
- 3.Schadt E.E., Molony C., Chudin E., Hao K., Yang X., Lum P.Y., Kasarskis A., Zhang B., Wang S., Suver C., et al. (2008) Mapping the genetic architecture of gene expression in human liver. PLoS Biol., 6, e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Consortium T.G. (2013) The Genotype-Tissue Expression (GTEx) project. Nat. Genet., 45, 580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cookson W., Liang L., Abecasis G., Moffatt M., Lathrop M. (2009) Mapping complex disease traits with global gene expression. Nat. Rev. Genet., 10, 184–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nicolae D.L., Gamazon E., Zhang W., Duan S., Dolan M.E., Cox N.J. (2010) Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet., 6, e1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Veyrieras J.-B., Kudaravalli S., Kim S.Y., Dermitzakis E.T., Gilad Y., Stephens M., Pritchard J.K. (2008) High-resolution mapping of expression-QTLs yields insight into human gene regulation. PLoS Genet., 4, e1000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choy E., Yelensky R., Bonakdar S., Plenge R.M., Saxena R., De Jager P.L., Shaw S.Y., Wolfish C.S., Slavik J.M., Cotsapas C., et al. (2008) Genetic analysis of human traits in vitro: drug response and gene expression in lymphoblastoid cell lines. PLoS Genet., 4, e1000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dimas A.S., Deutsch S., Stranger B.E., Montgomery S.B., Borel C., Attar-Cohen H., Ingle C., Beazley C., Gutierrez Arcelus M., Sekowska M., et al. (2009) Common regulatory variation impacts gene expression in a cell type-dependent manner. Science, 325, 1246–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding J., Gudjonsson J.E., Liang L., Stuart P.E., Li Y., Chen W., Weichenthal M., Ellinghaus E., Franke A., Cookson W., et al. (2010) Gene expression in skin and lymphoblastoid cells: Refined statistical method reveals extensive overlap in cis-eQTL signals. Am. J. Hum. Genet., 87, 779–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dixon A.L., Liang L., Moffatt M.F., Chen W., Heath S., Wong K.C.C., Taylor J., Burnett E., Gut I., Farrall M., et al. (2007) A genome-wide association study of global gene expression. Nat. Genet., 39, 1202–1207. [DOI] [PubMed] [Google Scholar]
- 12.Fehrmann R.S.N., Jansen R.C., Veldink J.H., Westra H.-J., Arends D., Bonder M.J., Fu J., Deelen P., Groen H.J.M., Smolonska A., et al. (2011) Trans-eQTLs reveal that independent genetic variants associated with a complex phenotype converge on intermediate genes, with a major role for the HLA. PLoS Genet., 7, e1002197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grundberg E., Small K.S., Hedman Å.K., Nica A.C., Buil A., Keildson S., Bell J.T., Yang T.-P., Meduri E., Barrett A., et al. (2012) Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat. Genet., 44, 1084–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Innocenti F., Cooper G.M., Stanaway I.B., Gamazon E.R., Smith J.D., Mirkov S., Ramirez J., Liu W., Lin Y.S., Moloney C., et al. (2011) Identification, replication, and functional fine-mapping of expression quantitative trait loci in primary human liver tissue. PLoS Genet., 7, e1002078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Montgomery S.B., Sammeth M., Gutierrez-Arcelus M., Lach R.P., Ingle C., Nisbett J., Guigo R., Dermitzakis E.T. (2010) Transcriptome genetics using second generation sequencing in a Caucasian population. Nature, 464, 773–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Myers A.J., Gibbs J.R., Webster J.A., Rohrer K., Zhao A., Marlowe L., Kaleem M., Leung D., Bryden L., Nath P., et al. (2007) A survey of genetic human cortical gene expression. Nat. Genet., 39, 1494–1499. [DOI] [PubMed] [Google Scholar]
- 17.Pickrell J.K., Marioni J.C., Pai A.A., Degner J.F., Engelhardt B.E., Nkadori E., Veyrieras J.-B., Stephens M., Gilad Y., Pritchard J.K. (2010) Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature, 464, 768–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Price A.L., Patterson N., Hancks D.C., Myers S., Reich D., Cheung V.G., Spielman R.S. (2008) Effects of cis and trans genetic ancestry on gene expression in African Americans. PLoS Genet., 4, e1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spielman R.S., Bastone L.A., Burdick J.T., Morley M., Ewens W.J., Cheung V.G. (2007) Common genetic variants account for differences in gene expression among ethnic groups. Nat. Genet., 39, 226–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stranger B.E., Forrest M.S., Dunning M., Ingle C.E., Beazley C., Thorne N., Redon R., Bird C.P., de Grassi A., Lee C., et al. (2007) Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science, 315, 848–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeller T., Wild P., Szymczak S., Rotival M., Schillert A., Castagne R., Maouche S., Germain M., Lackner K., Rossmann H., et al. (2010) Genetics and beyond--the transcriptome of human monocytes and disease susceptibility. PloS One, 5, e10693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greenawalt D.M., Dobrin R., Chudin E., Hatoum I.J., Suver C., Beaulaurier J., Zhang B., Castro V., Zhu J., Sieberts S.K., et al. (2011) A survey of the genetics of stomach, liver, and adipose gene expression from a morbidly obese cohort. Genome Res., 21, 1008–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Westra H.-J., Peters M.J., Esko T., Yaghootkar H., Schurmann C., Kettunen J., Christiansen M.W., Fairfax B.P., Schramm K., Powell J.E., et al. (2013) Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet., 45, 1238–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grundberg E., Kwan T., Ge B., Lam K.C.L., Koka V., Kindmark A., Mallmin H., Dias J., Verlaan D.J., Ouimet M., et al. (2009) Population genomics in a disease targeted primary cell model. Genome Res., 19, 1942–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mehta D., Heim K., Herder C., Carstensen M., Eckstein G., Schurmann C., Homuth G., Nauck M., Völker U., Roden M., et al. (2013) Impact of common regulatory single-nucleotide variants on gene expression profiles in whole blood. Eur. J. Hum. Genet., 21, 48–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schröder A., Klein K., Winter S., Schwab M., Bonin M., Zell A., Zanger U.M. (2013) Genomics of ADME gene expression: mapping expression quantitative trait loci relevant for absorption, distribution, metabolism and excretion of drugs in human liver. Pharmacogenomics J., 13, 12–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang X., Johnson A.D., Hendricks A.E., Hwang S.-J., Tanriverdi K., Ganesh S.K., Smith N.L., Peyser P.A., Freedman J.E., O'Donnell C.J. (2014) Genetic associations with expression for genes implicated in GWAS studies for atherosclerotic cardiovascular disease and blood phenotypes. Hum. Mol. Genet., 23, 782–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheung V.G., Spielman R.S. (2009) Genetics of human gene expression: mapping DNA variants that influence gene expression. Nat. Rev. Genet., 10, 595–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang J., Benyamin B., McEvoy B.P., Gordon S., Henders A.K., Nyholt D.R., Madden P.A., Heath A.C., Martin N.G., Montgomery G.W., et al. (2010) Common SNPs explain a large proportion of the heritability for human height. Nat. Genet., 42, 565–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zuber V., Strimmer K. (2011) High-dimensional regression and variable selection using CAR scores. Stat. Appl. Genet. Mol. Biol., 10, 1–27. [Google Scholar]
- 31.Beutner F., Teupser D., Gielen S., Holdt L.M., Scholz M., Boudriot E., Schuler G., Thiery J. (2011) Rationale and design of the Leipzig (LIFE) Heart Study: phenotyping and cardiovascular characteristics of patients with coronary artery disease. PloS One, 6, e29070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nica A.C., Montgomery S.B., Dimas A.S., Stranger B.E., Beazley C., Barroso I., Dermitzakis E.T. (2010) Candidate causal regulatory effects by integration of expression QTLs with complex trait genetic associations. PLoS Genet., 6, e1000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fu J., Wolfs M.G.M., Deelen P., Westra H.-J., Fehrmann R.S.N., Te Meerman G.J., Buurman W.A., Rensen S.S.M., Groen H.J.M., Weersma R.K., et al. (2012) Unraveling the regulatory mechanisms underlying tissue-dependent genetic variation of gene expression. PLoS Genet., 8, e1002431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Veyrieras J.-B., Gaffney D.J., Pickrell J.K., Gilad Y., Stephens M., Pritchard J.K. (2012) Exon-specific QTLs skew the inferred distribution of expression QTLs detected using gene expression array data. PloS One, 7, e30629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kanehisa M., Goto S. (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res., 28, 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garcia-Bates T.M., Lehmann G.M., Simpson-Haidaris P.J., Bernstein S.H., Sime P.J., Phipps R.P. (2008) Role of peroxisome proliferator-activated receptor gamma and its ligands in the treatment of hematological malignancies. PPAR Res., 2008, 834612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nishihira J., Ishibashi T., Sakai M., Nishi S., Kondo H., Makita A. (1992) Identification of the fatty acid binding site on glutathione S-transferase P. Biochem. Biophys. Res. Commun., 189, 197–205. [DOI] [PubMed] [Google Scholar]
- 38.Hasni S.A. (2012) Role of Helicobacter pylori infection in autoimmune diseases. Curr. Opin. Rheumatol., 24, 429–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meisinger C., Prokisch H., Gieger C., Soranzo N., Mehta D., Rosskopf D., Lichtner P., Klopp N., Stephens J., Watkins N.A., et al. (2009) A genome-wide association study identifies three loci associated with mean platelet volume. Am. J. Hum. Genet., 84, 66–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sand-Dejmek J., Adelmant G., Sobhian B., Calkins A.S., Marto J., Iglehart D.J., Lazaro J.-B. (2011) Concordant and opposite roles of DNA-PK and the ‘facilitator of chromatin transcription’ (FACT) in DNA repair, apoptosis and necrosis after cisplatin. Mol. Cancer, 10, 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cuomo C.A., Kirch S.A., Gyuris J., Brent R., Oettinger M.A. (1994) Rch1, a protein that specifically interacts with the RAG-1 recombination-activating protein. Proc. Natl Acad. Sci. USA, 91, 6156–6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Plagnol V., Smyth D.J., Todd J.A., Clayton D.G. (2009) Statistical independence of the colocalized association signals for type 1 diabetes and RPS26 gene expression on chromosome 12q13. Biostat. Oxf. Engl., 10, 327–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wallace C., Rotival M., Cooper J.D., Rice C.M., Yang J.H.M., McNeill M., Smyth D.J., Niblett D., Cambien F., Tiret L., et al. (2012) Statistical colocalization of monocyte gene expression and genetic risk variants for type 1 diabetes. Hum. Mol. Genet., 21, 2815–2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ebrahimi-Fakhari D., Wahlster L., McLean P.J. (2012) Protein degradation pathways in Parkinson's disease: curse or blessing. Acta Neuropathol. (Berl.), 124, 153–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Breitling R., Li Y., Tesson B.M., Fu J., Wu C., Wiltshire T., Gerrits A., Bystrykh L.V., de Haan G., Su A.I., et al. (2008) Genetical genomics: spotlight on QTL hotspots. PLoS Genet., 4, e1000232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Teslovich T.M., Musunuru K., Smith A.V., Edmondson A.C., Stylianou I.M., Koseki M., Pirruccello J.P., Ripatti S., Chasman D.I., Willer C.J., et al. (2010) Biological, clinical and population relevance of 95 loci for blood lipids. Nature, 466, 707–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Levula M., Airla N., Oksala N., Hernesniemi J.A., Pelto-Huikko M., Salenius J.-P., Zeitlin R., Järvinen O., Huovila A.-P.J., Nikkari S.T., et al. (2009) ADAM8 and its single nucleotide polymorphism 2662T/G are associated with advanced atherosclerosis and fatal myocardial infarction: Tampere vascular study. Ann. Med., 41, 497–507. [DOI] [PubMed] [Google Scholar]
- 48.Ramos E.M., Hoffman D., Junkins H.A., Maglott D., Phan L., Sherry S.T., Feolo M., Hindorff L.A. (2014) Phenotype-Genotype Integrator (PheGenI): synthesizing genome-wide association study (GWAS) data with existing genomic resources. Eur. J. Hum. Genet., 22, 144–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Comuzzie A.G., Cole S.A., Laston S.L., Voruganti V.S., Haack K., Gibbs R.A., Butte N.F. (2012) Novel genetic loci identified for the pathophysiology of childhood obesity in the Hispanic population. PloS One, 7, e51954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Beaumont N.J., Skinner V.O., Tan T.M.-M., Ramesh B.S., Byrne D.J., MacColl G.S., Keen J.N., Bouloux P.M., Mikhailidis D.P., Bruckdorfer K.R., et al. (2003) Ghrelin can bind to a species of high density lipoprotein associated with paraoxonase. J. Biol. Chem., 278, 8877–8880. [DOI] [PubMed] [Google Scholar]
- 51.Malik T.H., von Stechow D., Bronson R.T., Shivdasani R.A. (2002) Deletion of the GATA Domain of TRPS1 Causes an Absence of Facial Hair and Provides New Insights into the Bone Disorder in Inherited Tricho-Rhino-Phalangeal Syndromes. Mol. Cell. Biol., 22, 8592–8600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., Ma'ayan A. (2013) Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics, 14, 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guo L., Du Y., Chang S., Zhang K., Wang J. (2014) rSNPBase: a database for curated regulatory SNPs. Nucleic Acids Res., 42, D1033–D1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang J., Lee S.H., Goddard M.E., Visscher P.M. (2011) GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet., 88, 76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cabili M.N., Trapnell C., Goff L., Koziol M., Tazon-Vega B., Regev A., Rinn J.L. (2011) Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev., 25, 1915–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Washietl S., Findeiss S., Müller S.A., Kalkhof S., von Bergen M., Hofacker I.L., Stadler P.F., Goldman N. (2011) RNAcode: robust discrimination of coding and noncoding regions in comparative sequence data. RNA N. Y. N, 17, 578–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hackermüller J., Reiche K., Otto C., Hösler N., Blumert C., Brocke-Heidrich K., Böhlig L., Nitsche A., Kasack K., Ahnert P., et al. (2014) Cell cycle, oncogenic and tumor suppressor pathways regulate numerous long and macro non-protein-coding RNAs. Genome Biol., 15, R48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pei B., Sisu C., Frankish A., Howald C., Habegger L., Mu X.J., Harte R., Balasubramanian S., Tanzer A., Diekhans M., et al. (2012) The GENCODE pseudogene resource. Genome Biol., 13, R51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Buckland P.R. (2004) Allele-specific gene expression differences in humans. Hum. Mol. Genet., 13(Spec. no. 2), R255–R260. [DOI] [PubMed] [Google Scholar]
- 60.Zuber V., Duarte Silva A.P., Strimmer K. (2012) A novel algorithm for simultaneous SNP selection in high-dimensional genome-wide association studies. BMC Bioinformatics, 13, 284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wright F.A., Sullivan P.F., Brooks A.I., Zou F., Sun W., Xia K., Madar V., Jansen R., Chung W., Zhou Y.-H., et al. (2014) Heritability and genomics of gene expression in peripheral blood. Nat. Genet., 46, 430–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Powell J.E., Henders A.K., McRae A.F., Kim J., Hemani G., Martin N.G., Dermitzakis E.T., Gibson G., Montgomery G.W., Visscher P.M. (2013) Congruence of additive and non-additive effects on gene expression estimated from pedigree and SNP data. PLoS Genet., 9, e1003502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang D., Rendon A., Wernisch L. (2013) Transcription factor and chromatin features predict genes associated with eQTLs. Nucleic Acids Res., 41, 1450–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gaffney D.J., Veyrieras J.-B., Degner J.F., Pique-Regi R., Pai A.A., Crawford G.E., Stephens M., Gilad Y., Pritchard J.K. (2012) Dissecting the regulatory architecture of gene expression QTLs. Genome Biol., 13, R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ørom U.A., Derrien T., Guigo R., Shiekhattar R. (2010) Long noncoding RNAs as enhancers of gene expression. Cold Spring Harb. Symp. Quant. Biol., 75, 325–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guttman M., Donaghey J., Carey B.W., Garber M., Grenier J.K., Munson G., Young G., Lucas A.B., Ach R., Bruhn L., et al. (2011) lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature, 477, 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Holdt L.M., Hoffmann S., Sass K., Langenberger D., Scholz M., Krohn K., Finstermeier K., Stahringer A., Wilfert W., Beutner F., et al. (2013) Alu elements in ANRIL non-coding RNA at chromosome 9p21 modulate atherogenic cell functions through trans-regulation of gene networks. PLoS Genet., 9, e1003588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kumar V., Westra H.-J., Karjalainen J., Zhernakova D.V., Esko T., Hrdlickova B., Almeida R., Zhernakova A., Reinmaa E., Võsa U., et al. (2013) Human disease-associated genetic variation impacts large intergenic non-coding RNA expression. PLoS Genet., 9, e1003201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kindt A.S.D., Navarro P., Semple C.A., Haley C.S. (2013) The genomic signature of trait-associated variants. BMC Genomics, 14, 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li W., Yang W., Wang X.-J. (2013) Pseudogenes: pseudo or real functional elements? J. Genet. Genomics Yi Chuan Xue Bao, 40, 171–177. [DOI] [PubMed] [Google Scholar]
- 71.Holdt L.M., Beutner F., Scholz M., Gielen S., Gäbel G., Bergert H., Schuler G., Thiery J., Teupser D. (2010) ANRIL expression is associated with atherosclerosis risk at chromosome 9p21. Arterioscler. Thromb. Vasc. Biol., 30, 620–627. [DOI] [PubMed] [Google Scholar]
- 72.Schmid R., Baum P., Ittrich C., Fundel-Clemens K., Huber W., Brors B., Eils R., Weith A., Mennerich D., Quast K. (2010) Comparison of normalization methods for Illumina BeadChip HumanHT-12 v3. BMC Genomics, 11, 349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Johnson W.E., Li C., Rabinovic A. (2007) Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostat. Oxf. Engl., 8, 118–127. [DOI] [PubMed] [Google Scholar]
- 74.Barbosa-Morais N.L., Dunning M.J., Samarajiwa S.A., Darot J.F.J., Ritchie M.E., Lynch A.G., Tavaré S. (2010) A re-annotation pipeline for Illumina BeadArrays: improving the interpretation of gene expression data. Nucleic Acids Res., 38, e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang J. (2002) An estimator for pairwise relatedness using molecular markers. Genetics, 160, 1203–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet., 38, 904–909. [DOI] [PubMed] [Google Scholar]
- 77.Shabalin A.A. (2012) Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinforma. Oxf. Engl., 28, 1353–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.1000 Genomes Project Consortium Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A. (2012) An integrated map of genetic variation from 1,092 human genomes. Nature, 491, 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., de Bakker P.I.W., Daly M.J., et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet., 81, 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Alberts R., Terpstra P., Li Y., Breitling R., Nap J.-P., Jansen R.C. (2007) Sequence polymorphisms cause many false cis eQTLs. PloS One, 2, e622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hindorff L.A., Sethupathy P., Junkins H.A., Ramos E.M., Mehta J.P., Collins F.S., Manolio T.A. (2009) Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl Acad. Sci. USA, 106, 9362–9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xia K., Shabalin A.A., Huang S., Madar V., Zhou Y.-H., Wang W., Zou F., Sun W., Sullivan P.F., Wright F.A. (2012) seeQTL: a searchable database for human eQTLs. Bioinforma. Oxf. Engl., 28, 451–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Price A.L., Weale M.E., Patterson N., Myers S.R., Need A.C., Shianna K.V., Ge D., Rotter J.I., Torres E., Taylor K.D., et al. (2008) Long-range LD can confound genome scans in admixed populations. Am. J. Hum. Genet., 83, 132–135. Author reply 135–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Luo W., Brouwer C. (2013) Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinforma. Oxf. Engl., 29, 1830–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Göring H.H.H., Curran J.E., Johnson M.P., Dyer T.D., Charlesworth J., Cole S.A., Jowett J.B.M., Abraham L.J., Rainwater D.L., Comuzzie A.G., et al. (2007) Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat. Genet., 39, 1208–1216. [DOI] [PubMed] [Google Scholar]
- 86.Min J.L., Taylor J.M., Richards J.B., Watts T., Pettersson F.H., Broxholme J., Ahmadi K.R., Surdulescu G.L., Lowy E., Gieger C., et al. (2011) The use of genome-wide eQTL associations in lymphoblastoid cell lines to identify novel genetic pathways involved in complex traits. PloS One, 6, e22070. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.