Abstract
Congenital diaphragmatic hernia (CDH) is a serious birth defect that accounts for 8% of all major birth anomalies. Approximately 40% of cases occur in association with other anomalies. As sporadic complex CDH likely has a significant impact on reproductive fitness, we hypothesized that de novo variants would account for the etiology in a significant fraction of cases. We performed exome sequencing in 39 CDH trios and compared the frequency of de novo variants with 787 unaffected controls from the Simons Simplex Collection. We found no significant difference in overall frequency of de novo variants between cases and controls. However, among genes that are highly expressed during diaphragm development, there was a significant burden of likely gene disrupting (LGD) and predicted deleterious missense variants in cases (fold enrichment = 3.2, P-value = 0.003), and these genes are more likely to be haploinsufficient (P-value = 0.01) than the ones with benign missense or synonymous de novo variants in cases. After accounting for the frequency of de novo variants in the control population, we estimate that 15% of sporadic complex CDH patients are attributable to de novo LGD or deleterious missense variants. We identified several genes with predicted deleterious de novo variants that fall into common categories of genes related to transcription factors and cell migration that we believe are related to the pathogenesis of CDH. These data provide supportive evidence for novel genes in the pathogenesis of CDH associated with other anomalies and suggest that de novo variants play a significant role in complex CDH cases.
Introduction
Congenital diaphragmatic hernia (CDH) is a serious birth defect characterized by incomplete formation of the diaphragm, resulting in herniation of the abdominal viscera into the chest cavity. The incidence of CDH is ∼1 in 3000 live births, accounting for 8% of all major birth anomalies (1,2). CDH can occur as an isolated defect or complex defect associated with other congenital anomalies, most commonly heart, brain, renal and genitourinary malformations (3). Pulmonary hypoplasia and pulmonary hypertension are the most significant causes of morbidity and mortality for isolated CDH patients. Although recent advances in the postnatal care of infants with CDH have reduced the overall mortality to 30% (4,5), the long-term morbidity in survivors of severe CDH is significant (6). The survival rate, however, is still <50% for severe isolated CDH or complex CDH, particularly when associated with major cardiac defects (7,8). The annual medical cost for caring for CDH survivors in the United States is nearly $160 million dollars, and projected national costs exceed $250 million for all CDH care per year (9), making it the costliest non-cardiac birth defect (10).
The etiology of CDH in most cases remains unclear. Gene knockout mice associated with CDH, rare monogenetic disorders in humans, familial aggregation and association with chromosomal anomalies provide support that there is a genetic contribution in CDH (11–13). Chromosomal anomalies, in particular de novo and large events spanning multiple genes, have been identified as conferring risk for ∼10% of patients with CDH (2). Cytogenetic and array-based methods have identified many recurrent complete or partial aneuploidies in CDH such as trisomy 21, trisomy 18 and deletions of 15q26, 8p23.1 and 1q41–q42 (12). However, the majority of sporadic or familial CDH cases are not caused by detectable chromosomal anomalies, and many of the genes involved in CDH are yet to be identified.
Currently, over 60 genes have been implicated in diaphragm development through animal models and monogenic syndromes associated with CDH (11,14). Despite the abundance of candidate genes, few causative genes and variants have been identified in humans with CDH. The high mortality of CDH has made it difficult to utilize classical genetic approaches for gene identification. The early death of affected individuals, prior to reproduction, means that the number of familial cases is limited; additionally, there is a lack of biospecimens from many affected children, who died shortly after birth. Diseases with such severe effects on reproductive fitness have increasingly been shown to be in part due to de novo variants, including single-nucleotide variants (SNVs), short insertions and deletions (indels) or larger copy number variants (CNVs) (15). Massively parallel sequencing technologies, including whole-exome sequencing (WES), provide the opportunity to detect de novo variants in sporadic genetic diseases. WES of hundreds of simplex autism or schizophrenia trios with no family history has shown that de novo rare variants, especially those predicted to be severe or disruptive (nonsense, splice site and frameshift), are enriched in affected individuals compared with their unaffected siblings or other healthy controls (16–20), and we have demonstrated similar results for congenital heart disease (CHD) (21). We also identified several rare de novo variants in CDH associated with the transcriptional factors GATA4 and GATA6 using WES, providing support for using WES to identify deleterious de novo variants in CDH by analyzing parent–child trios (22,23).
We hypothesized that CDH associated with additional major malformations would be frequently associated with de novo variants and sought to identify genes with potential pleiotropic effects on the development of the diaphragm and other organs. These cases represent the most severely impacted patients, and identifying the genes in these individuals is particularly valuable. We performed a study using exome sequencing of 39 complex CDH parent–child trios to test our hypothesis that de novo variants are present in a significant fraction of sporadic complex CDH cases and represent a significant burden compared with unaffected controls.
Results
De novo variant filtering
WES was performed on a series of 39 complex CDH trios. The sequence data had a median of 93% of targeted bases covered by 15 or more reads (Supplementary Material, Table S1). Variant calling with the GATK HaplotypeCaller produced an average of 18 183 SNVs and 473 indels in coding regions per CDH case (Supplementary Material, Table S2). We used the Simons Simplex Collection (SSC) unaffected offspring with their parents as control trios, which were sequenced using the same exome capture platform. There were an average of 19 824 coding SNVs and 408 indels per SSC sample. Biological parentage was confirmed from the sequence data in all trios. The population structures of the case and control data are shown in Supplementary Material, Figure S1. Under the assumption that the rate of de novo variants is in principle independent of population, the burden analysis was not stratified.
After filtration designed to remove false positives, we identified 42 de novo variants in 39 CDH probands and 749 in 787 SSC unaffected controls. Supplementary Material, Table S3 contains a complete list of de novo variants identified in CDH cases. Of the 42 de novo variants in the CDH cases, we were able to validate 40 (95%) by Sanger sequencing. We removed these two non-validated variants from all following analysis. Among these 40 variants, 30 were missense, 2 were nonsense, 3 were frameshift deletions, 1 was a non-frameshift insertion and 4 were synonymous variants (Supplementary Material, Table S3). Nine of the missense variants were predicted to be deleterious (D-miss) by MetaSVM (24) (Supplementary Material, Table S3). Genes with confirmed de novo variants predicted to be deleterious are provided in Table 1.
Table 1.
Genes with confirmed de novo nonsense variants, indels or missense variants predicted deleterious (D) by MetaSVM in CDH probands
| Proband ID | Gene | Variant | Amino acid change | MetaSVM prediction (score) | MDD expressed genes |
|---|---|---|---|---|---|
| 01–0460 | ARFGEF2 | c.G3326A | p.R1109H | D (0.332) | Expressed |
| 01–0761 | CDO1 | c.259delG | p.D87fs | N/A | Highly expressed |
| 01–0568 | CLCN4 | c.G43A | p.D15N | D (0.644) | |
| 01–0109 | DLST | c.297_298del | p.99_100del | N/A | Highly expressed |
| 01–0562 | GATA6 | c.C1366T | p.R456C | D (0.881) | Highly expressed |
| 05–0011 | INHBB | c.C1055G | p.T352R | D (0.507) | |
| 01–0057 | LONP1 | c.C1325T | p.T442M | D (1.087) | Expressed |
| 03–0001 | PPAPDC2 | c.T824A | p.V275E | D (0.427) | Expressed |
| 01–0634 | PRKACB | c.C277T | p.R93X | N/A | Highly expressed |
| 01–0215 | PTPN12 | c.C77T | p.T26M | D (0.465) | Highly expressed |
| 01–0450 | SIN3A | c.1570_1577del | p.Y524Vfs*26 | N/A | Highly expressed |
| 01–0562 | SLC5A9 | c.C172T | p.R58C | D (0.856) | Expressed |
| 01–0147 | STAG2 | c.C1840T | p.R614X | N/A | Highly expressed |
| 01–0083 | TLN1 | c.G98A | p.R33H | D (0.351) | Highly expressed |
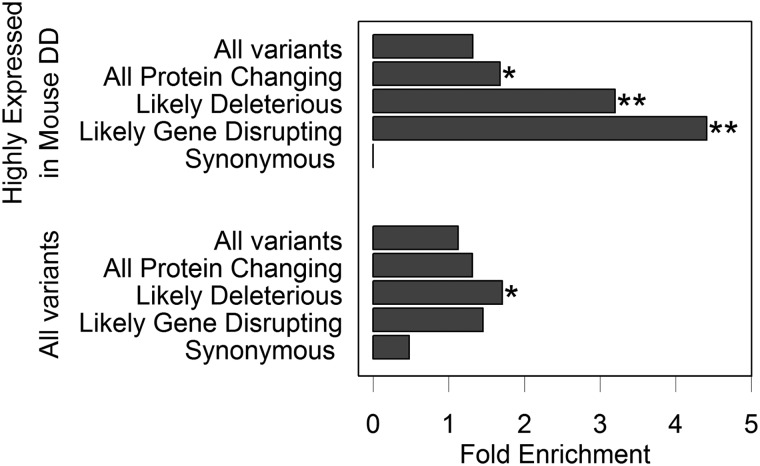
Excess of de novo variant burden in genes highly expressed in mouse developing diaphragm
The distribution of de novo variants in cases and controls both conformed closely to a Poisson distribution (Supplementary Material, Fig. S2). We hypothesized that CDH cases carry an excess burden of gene damaging de novo variants; we therefore estimated burden in three groups of non-silent variants: ‘likely gene disrupting’ (LGD) composed of nonsense, frameshift indels and splicing site variants, ‘likely deleterious’ composed of LGD variants plus D-miss variants, and all protein changing variants. When considering variants across all genes, there was no significant enrichment of de novo variants in any groups (Supplementary Material, Table S4).
We hypothesized that genes that contribute to CDH should be functionally relevant to diaphragm development. Therefore, we refined the burden analysis by partitioning the variants according to gene expression datasets from the pleuroperitoneal folds of mouse developing diaphragm (MDD) (25). In genes that were highly expressed in MDD, defined as the top 25th percentile of the robust multichip averaging (RMA)-normalized expression data (4510 genes), there were 13 de novo variants in CDH cases (31.0% of CDH variants) (Table 2) and 196 in controls (26.2% of control variants). We found a significant burden of de novo likely deleterious variants in cases compared with controls [P = 0.003, fold enrichment = 3.2, 95% confidence interval (CI) 1.64–21.18, Table 2 and Fig. 1], with comparable enrichment in LGD (P = 0.005, fold enrichment = 4.41, 95% CI 1.82–21.18). LGD variants also occurred at a rate of 5.4 times higher (P = 0.003, Table 3) than expected estimated by gene-specific background mutation rate (26) in cases, whereas no significant enrichment was observed in control relative to expectation (P = 0.6, Table 3).
Table 2.
Frequency of de novo variants by top 25% mouse RNA expression
| Category | Total number of de novo variants |
Frequency of de novo variants/subject |
Fold enrichment (95% CI)a | P-value* | ||
|---|---|---|---|---|---|---|
| CDH trios (n = 39) | Control (n = 787) | CDH trios (n = 39) | Controls (n = 787) | |||
| Synonymous | 0 | 45 | 0 | 0.06 | 0 (0–21.18) | 1.000 |
| LGD | 5 | 19 | 0.13 | 0.02 | 4.41 (1.82–21.18) | 0.005 |
| Likely deleterious | 8 | 45 | 0.21 | 0.06 | 3.20 (1.64–21.18) | 0.003 |
| All protein changing | 13 | 151 | 0.33 | 0.19 | 1.68 (1.01–21.18) | 0.048 |
| All variants | 13 | 196 | 0.33 | 0.25 | 1.32 (0.79–21.18) | 0.191 |
Bold fonts indicate significant after Bonferroni correction.
aThe fold enrichment is the ratio of variants in cases to variants in controls divided by the ratio of cases to controls.
*P-values compare the number of variants in each category between cases and controls using a two-sided binomial exact test (uncorrected).
CI, confidence interval.
Figure 1.
Fold enrichment of de novo variants in cases versus controls. The graph shows the fold enrichment of de novo variants in different variant categories for all variants, and variants in genes that were highly expressed in MDD. The fold enrichment is the ratio of variants in cases to variants in controls divided by the ratio of cases to controls. *P ≤ 0.05 and **P ≤ 0.01, as assessed by a binomial exact test.
Table 3.
Frequency of de novo variants in CDH cases and controls by top 25% mouse RNA expression compared with expected mutation rate
| Class | Cases (n = 39) |
P-value* | Controls (n = 787) |
Fold enrichmenta | P-value* | |||
|---|---|---|---|---|---|---|---|---|
| Expected | Observed | Fold enrichmenta | Expected | Observed | ||||
| All | 9.38 | 13 | 1.40 | 0.154 | 189.36 | 193 | 1.00 | 0.405 |
| Synonymous | 2.63 | 0 | 0.00 | 1.000 | 53.06 | 44 | 0.83 | 0.909 |
| Missense | 6.23 | 8 | 1.30 | 0.288 | 125.63 | 126 | 1.00 | 0.499 |
| LGD | 0.93 | 5 | 5.40 | 0.003 | 18.86 | 18 | 0.95 | 0.609 |
Bold fonts indicate significant after Bonferroni correction.
aThe fold enrichment is the ratio of observed variants to expected variants.
*P-values compare the number of observed variants to the number of expected variants using a one-tailed Poisson exact test.
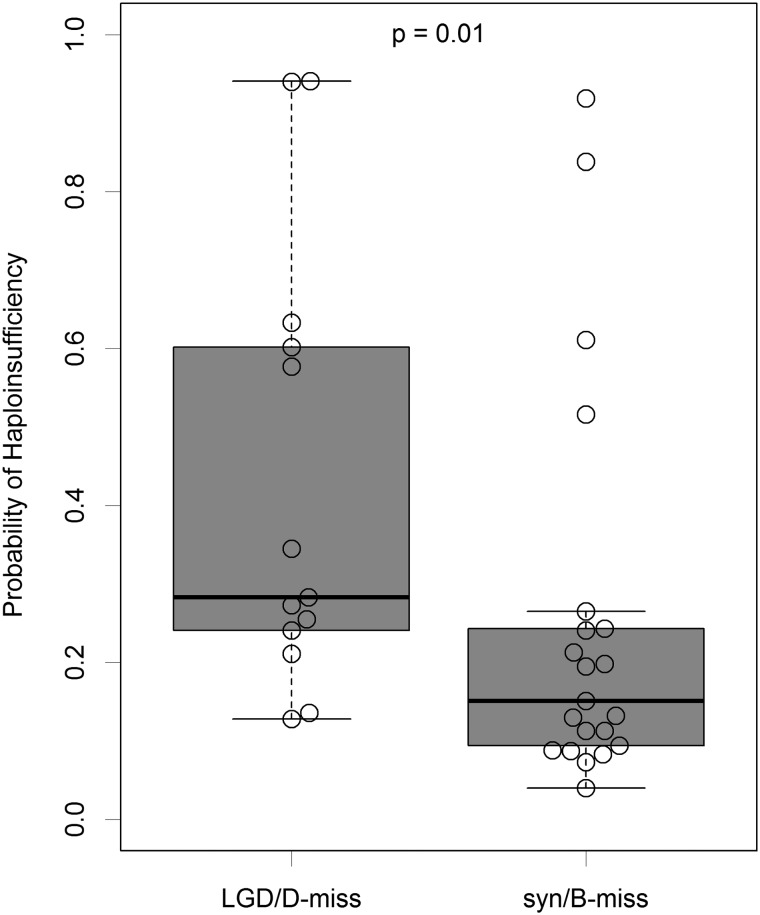
In addition to burden, we hypothesize that genes with LGD or D-miss de novo variants in CDH cases are likely to be haploinsufficient. We compared predicted haploinsufficiency probability (27) between genes with LGD or D-miss variants and the ones with benign missense or synonymous variants among CDH cases and found that genes with LGD or D-miss variants are ranked significantly higher (Mann–Whitney U-test P-value = 0.01, Fig. 2). This does not apply to controls (P-value = 0.31).
Figure 2.
Haploinsufficiency in genes with different types of de novo variants in CDH cases. The boxplot shows the genes with likely gene damaging (LGD) de novo variants (nonsense, splicing and frameshift indels) or damaging missense (D-miss) de novo variants in CDH cases have higher probability of haploinsufficiency than the ones with synonymous (syn) or benign missense (B-miss) variants. P-value was calculated by Mann–Whitney U-test. The beeswarm plot embedded in the boxplot was made by R package ‘beeswarm’.
Genes with de novo variants in cases are functionally similar to candidate genes from mouse models
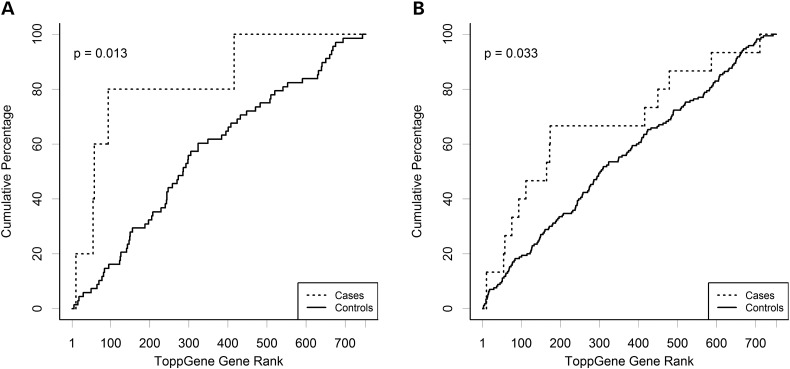
We hypothesized that de novo variants observed in CDH cases would be found in genes that were more functionally relevant to CDH than those carrying de novo variants in controls. Thus, we employed a targeted approach similar to that described by Longoni et al. (28). We selected 61 genes from mouse models with diaphragmatic hernia or thin diaphragm muscle as our seed genes (Supplementary Material, Table S5). There were a total of 748 genes carrying de novo variants in both cases and controls. These were ranked using ToppGene (29) based on functional similarity to the seed set, and the ranks for CDH and control variants were compared using a Wilcoxon rank sum test. The ToppGene rank for genes with de novo variants in CDH are shown in Table 4. Two genes FBN1 and MUSK, which carried non-synonymous de novo variants in controls, were not ranked by ToppGene as they occurred in the seed set; these genes were given a nominal rank of 0 for the purposes of the Mann–Whitney–Wilcoxon test. We found that genes with LGD variants in cases are significantly (P = 0.013) ranked higher than the ones in controls (Fig. 3), and the same trend holds when adding D-miss variants, especially among genes highly expressed in MDD (P = 0.01, Supplementary Material, Fig. S3).
Table 4.
Rank of genes with deleterious de novo variants based on ToppGene functional similarity
| Gene | Rank | Variant type |
|---|---|---|
| GATA6 | 1 | nonsynonymousSNV |
| SIN3A | 11 | frameshiftdeletion |
| HSPG2 | 29 | nonsynonymousSNV |
| STAG2 | 55 | stopgainSNV |
| CDO1 | 58 | frameshiftdeletion |
| PTPN12 | 76 | nonsynonymousSNV |
| PRKACB | 94 | stopgainSNV |
| TRIB2 | 100 | nonsynonymousSNV |
| INHBB | 112 | nonsynonymousSNV |
| FAT3 | 152 | nonsynonymousSNV |
| PPL | 165 | nonsynonymousSNV |
| MYBBP1A | 168 | nonsynonymousSNV |
| TLN1 | 173 | nonsynonymousSNV |
| ARFGEF2 | 174 | nonsynonymousSNV |
| HIST1H3C | 182 | nonsynonymousSNV |
| KMT2B | 215 | nonsynonymousSNV |
| ORC1 | 224 | nonsynonymousSNV |
| SPAM1 | 266 | nonsynonymousSNV |
| PFKL | 308 | synonymousSNV |
| ZNF25 | 317 | nonsynonymousSNV |
| CCDC80 | 320 | nonsynonymousSNV |
| LSS | 336 | nonsynonymousSNV |
| IGSF9B | 342 | nonsynonymousSNV |
| TGM6 | 364 | nonsynonymousSNV |
| KIAA1161 | 400 | nonsynonymousSNV |
| DLST | 416 | frameshiftdeletion |
| WDHD1 | 447 | nonsynonymousSNV |
| LONP1 | 450 | nonsynonymousSNV |
| ATP7B | 453 | synonymousSNV |
| PEAR1 | 454 | nonsynonymousSNV |
| EME1 | 469 | nonsynonymousSNV |
| FAM109A | 473 | synonymousSNV |
| PPAPDC2 | 479 | nonsynonymousSNV |
| ALDH9A1 | 493 | synonymousSNV |
| RASSF10 | 551 | nonsynonymousSNV |
| PXMP4 | 552 | nonsynonymousSNV |
| CLCN4 | 587 | nonsynonymousSNV |
| DMXL2 | 610 | nonsynonymousSNV |
| SLC5A9 | 711 | nonsynonymousSNV |
| PRR14 | 712 | nonsynonymousSNV |
| CCDC173 | 738 | nonsynonymousSNV |
| SLC26A8 | 747 | nonframeshiftinsertion |
Figure 3.
ToppGene ranking in cases and controls. The graph shows the ranking of genes by ToppGene for (A) likely gene damaging (LGD) variants (nonsense, splicing and frameshift indels) and (B) likely deleterious (LGD plus deleterious non-synonymous). P-values are the result of a Mann–Whitney U-test.
Estimation of percentage of CDH patients with risk-associated variants
As likely deleterious de novo variants in genes that were very highly expressed in MDD were significantly increased in our CDH patients, we estimated the percentage of these de novo variants that are associated with CDH risk in our patients. There were 8 of these de novo variants in the 39 CDH patients (33.0% of CDH variants) and 45 in the 787 controls (8.8% of control variants) (Supplementary Material, Table S6). An estimated 72% (95% CI: 22–86%) of these variants are associated with CDH risk.
For the same group of variants, there were 8 of 39 CDH patients and 41 of 787 controls who had at least 1 variant. We estimated the percentage of CDH patients carrying CDH risk-associated de novo variants to be 15% (95% CI: 4.3–29%) (Supplementary Material, Table S6).
Discussion
The morbidity and mortality of CDH are high, and patients with complex CDH have substantially higher morbidity and mortality when compared with patients with isolated CDH. In this study, we performed WES on 39 complex CDH parent–child trios to try to determine the etiology of the CDH in these patients. We identified and validated 40 de novo single-nucleotide and indel variants (Supplementary Material, Table S3), 15 of which (38.5%) were nonsense, indels or missense variants that were predicted to be deleterious by MetaSVM.
When considering all genes carrying de novo variants, we did not find significant enrichment in any type of genetic variant in our CDH patients relative to controls. Neither cases nor controls showed any significant deviation from the number of expected de novo variants, either overall or by variant type. However, when we considered genes that are expressed in the pleuroperitoneal fold of the MDD (25) and those genes that are intolerant of genetic variation (24), we found that all LGD variants in our CDH cases were in genes that are expressed in MDD and that are intolerant to functional genetic variants. Considering genes that are intolerant of genetic variation alone did not yield any significant increase in burden (Supplementary Notes and Supplementary Material, Tables S8–S10). The frequency of LGD variants in our CDH patients was higher than expected (fold enrichment 4.3×, P = 0.007), and there was no increased frequency of LGDs observed in the controls. Additionally, in CHD patients, the genes with LGD or D-miss de novo variants are significantly ranked higher in haploinsufficiency probability than the ones with benign missense or synonymous variants.
Normal formation of the pleuroperitoneal fold is important for the development of the diaphragm (30). By limiting the analysis to the genes that are highly expressed in the MDD (top quartile of expression), we found that protein changing de novo variants were more frequent in CDH cases than in controls, consistent with previous studies in CHD (19,21). This result was most significant for LGD variants (P = 0.003) and occurred at a rate of 5.4 times higher than expected in cases, but not in controls.
We ranked all the genes with de novo variants in cases and controls based on the similarity to the genes implicated in abnormal diaphragm in murine models and found that genes with LGD variants were ranked significantly higher in our CDH patients than those in controls. In fact, 4 of 5 genes with LGD variants in cases were ranked in the top 12%, and 6 out of the 10 D-miss variants were ranked in the top 22%. These results suggest that genes with de novo LGD variants identified in our CDH patients are relevant to the disease pathogenesis based on prior biological knowledge.
All the results indicate an excess of LGD variants in our CDH patients. Haploinsufficiency and dosage sensitivity of single genes can alter developmental processes. LGD mutations, such as R112X in ZFPM2 (28,31,32) and G238X and V358Cfs34* in GATA6 (23), have been reported in humans and mouse models of CDH. In our sample of 39 cases, we identified five LGD variants in the genes SIN3A, STAG2, PRKACB, DLST and CDO1. These genes are highly expressed in the murine developing diaphragm. SIN3A is a corepressor that plays a role in the regulation of gene transcription through interactions with retinoic acid receptors (RARs) (33). Retinoic acid signaling has been previously implicated in CDH (34). Double mutants of RARs produced a diaphragmatic hernia in mice (35). The protein encoded by STAG2 is part of the cohesin complex, which is crucial to regulate the separation of sister chromatids during cell division. Haploinsufficiency of cohesin genes is associated with human multisystem developmental disorders such as Cornelia de Lange Syndrome (CdLS, OMIM 122470) (36). A duplication CNV of Xq25 that includes STAG2 was reported in a patient with a learning disability and microcephaly (37). Our CDH patient has microcephaly, a sacral dimple, scoliosis and developmental delay. These data may suggest a role for STAG2 in brain and diaphragm development.
CNVs have been previously reported in association with CDH. To identify the causative CDH gene within the set of contiguous genes in CNVs, we determined that seven potentially deleterious de novo variants we identified map to CDH CNVs (Supplementary Material, Table S3). Four of these seven were consistently predicted to be deleterious (STAG2, PTPN12, SIN3A and PPAPDC2). Variants in these four genes are either LGD variants (p.R614X in STAG2 and p.Y524Vfs*26 in SIN3A) or conserved missense variants that are predicted to be pathogenic (c.C77T, p.T26M in PTPN12 and c.T824A, p.V275E in PPAPDC2). The CNV data add to the weight of the evidence supporting the role of these genes in CDH.
We estimate that at least 15% of sporadic complex CDH patients carry LGD or miss-D CDH associated de novo variants in genes highly expressed during mouse diaphragm development. The rate is slightly lower than that in autism (21%) (38) and schizophrenia (17.6%) (20) and slightly higher than that in CHD (10%) (21). We also estimate that ∼72% of LGD and predicted deleterious missense de novo variants are likely associated with CDH, a rate comparable with a recent autism study (38). We note that owing to our small sample size, our estimate has a wide confidence interval (95% CI is 4.3–29% for the percentage of patients explained, and 22–86% for contributing de novo LGD and predicted deleterious missense variants). Our estimates could be low because the result we present is limited to those genes highly expressed in MDD.
Genes previously identified in CDH have been transcription factors (GATA4, NR2F2, ZFPM2, WT1), proteins involved in cell migration or components of the extracellular matrix (SLIT3, ROBO1) (39). Among the genes with likely pathogenic missense variants in our study, i.e. those which were predicted to be deleterious by MetaSVM (Table 1), GATA6 is a transcription factor. GATA6 has been identified as a candidate CDH gene in our previous CDH study (23). PTPN12, TLN1 and ARFGEF2 are involved in cell migration. PTPN12 is a member of the protein tyrosine phosphatase (PTP) family, which is a critical regulator of cell adhesion, migration and cell–matrix interactions (40). PTPN12 was reported to play an essential role in early embryogenesis, and embryos with PTP-PEST (−/−) had severe mesenchyme deficiency and morphological abnormalities, resulting in early embryonic lethality (41). TLN1 is a cytoskeleton protein involved in cell adhesion by regulation of integrin activation (42). Skeletal muscle development and function are dependent on β1 integrins (43). The TLN1 missense variant we identified, p.R33H, is located in a F0 subdomain of the Talin N-terminus and may affect the activation of integrin (44), resulting in abnormal cell spreading. ARFGEF2 plays an important role in intracellular vesicular trafficking and neural proliferation and migration (45). Mutations in ARFGEF2 are associated with a wide range of movement disorders and neuronal migration disorders associated with microcephaly (46). ARFGEF2 is essential for early embryonic development. Homozygous knockout mice are embryonic lethal (47).
All the patients in this series were complex CDH cases. Some of the patients showed developmental delay when we completed the 2-year and 5-year developmental assessments (Supplementary Material, Table S7). The genes we identified (ARFGEF2, GATA6, HSPG2 and ORC1) overlap with the genes implicated in the Deciphering Developmental Disorder study (48). ARFGEF2 is a probable developmental disorder gene, and GATA6, HSPG2 and ORC1 are confirmed developmental disorder genes, suggesting that the genes identified in our CDH study may also contribute to other developmental disorders. Our results in the complex CDH cases may or may not be generalizable to patients with isolated CDH. Owing to the limited sample size of this study, we did not identify genes with recurrent mutations.
In conclusion, we identified a significantly higher frequency of protein damaging de novo variants, including LGD and deleterious missense variants, in functionally relevant genes in our complex CDH patients, and such de novo variants may explain ∼24% of complex CDH cases. Genes that are involved in transcriptional regulation or cell migration are enriched in the genes with de novo variants. This relatively high yield of de novo predicted pathogenic variants suggests that WES has clinical utility in complex CDH cases and has significant implications for genetic counseling because these cases will have a low risk of recurrence for the parents.
Materials and Methods
Patients
All patients were recruited as part of the DHREAMS (Diaphragmatic Hernia Research & Exploration; Advancing Molecular Science) study (http://www.cdhgenetics.com/, accessed 1 June 2015) (49). All participants provided informed consent/assent for participation in this study, which was approved by the Institutional Review Boards of Columbia University, University of Pittsburgh, Omaha Children's Hospital/University of Nebraska and University of Michigan/CS Mott Children's Hospital.
Thirty-nine patients with complex CDH and their unaffected parents were analyzed by WES. Patients had at least one additional of the following birth defects: CHD, central nervous system defect, pyloric stenosis, omphalocele, polysplenia, asplenia, Hirschsprung's disease, intestinal malrotation, situs inversus, genital urinary defect, skeletal anomalies, cleft lip/palate, abnormal hearing, microtia, coloboma, dysmorphic features, club foot, limb anomalies, congenital cystic adenomatoid malformation/congenital pulmonary airway malformation (CCAM/CPAM) or bronchopulmonary sequestration. A complete family history including history of diaphragm defects and major malformations was collected on all patients by a single genetic counselor, and no patients had a family history of CDH or fit diagnostic criteria for any recognizable syndrome (Supplementary Material, Table S7). A blood, saliva and/or skin/diaphragm tissue sample was collected from the affected patient and both parents. All probands had normal chromosome microarray results without large de novo deletions or duplications. Totally, 787 unaffected control trios were included in our analysis. The control group was the unaffected siblings of children with sporadic autism from two publicly available SSC datasets (50): Wigler data (17) and State data (19).
Exome sequencing
Genomic DNA from whole blood or tissue was processed with the Agilent SureSelect V2 or V4 exome capture reagent and TruSeq DNA Sample Prep Kits (Illumina), followed by 100-bp paired-end sequencing reads on IlluminaHiSeq 2000 platform according to the manufacturer's instructions (Illumina, Inc., San Diego, California, USA).
We applied a uniform analytical pipeline based on BWA-mem/Picard/GATK to process sequencing reads of both case and control trios. BWA-mem (51) was used for mapping sequencing reads to the human reference genome (hg19), followed by GATK (52) for local multiple realignment, recalibration of base quality scores and calling variants. Variant calling was carried out jointly with all CDH cases, and with each of the two SSC datasets using GATK (version 3) HaplotypeCaller.
We annotated all variants using ANNOVAR (http://www.openbioinformatics.org/annovar/, accessed 1 June 2015) (53) to obtain information about protein coding changes, conservation, functional prediction [PolyPhen-2 (54), SIFT (55)], dbSNP status (dbSNP137) and allele frequency in the 1000 genome project (www.1000genomes.org/, accessed 1 June 2015) and the NHLBI GO Exome Sequencing Project (ESP) (http://evs.gs.washington.edu/EVS/, accessed 1 June 2015).
Principal component analysis of population structure
Principal component analysis of common variants was carried out using Eigenstrat (56) to determine the population structure of both cases and controls. Common variants were identified as those with an alternate allele frequency of >5% in both the 1000 genome project and NHLBI GO ESP. The HapMap 3 sample collection data (57) were downloaded as a reference. Variants that were missing in >5% of the dataset were removed, and the combined data were analyzed with Eigenstrat.
Identification and confirmation of de novo variants
We started from candidate de novo variant sites at which the genotype was identified as heterozygous in the child and homozygous reference in both parents. We then applied a filtration procedure to remove potential false positives. Specifically, we removed a candidate site if it did not meet any of the following criteria: (a) allele frequency in 1000 Genomes and ESP of <0.1%, (b) fraction of the alternative allele in both parents of <0.02, (c) fraction of the alternative allele in proband of >0.2 for SNVs and of >0.3 for indels, (d) alternative allele depth of >5 in proband, (e) total read depth ≥10 in parents and (f) genotype quality of >30 in parents and >70 in proband. We validated the resulting candidate de novo variants in CDH cases by Sanger dideoxynucleotide sequencing. Primer3 was used to design the oligonucleotides for the amplification of regions that include the variants.
De novo burden analysis
We used a two-sided binomial exact test to test the null hypothesis that the average number of de novo variants per proband in complex CDH cases is the same as controls. We grouped de novo variants into four functional classes based on predicted impact: ‘LGD’ composed of nonsense, frameshift indels and splicing site variants; ‘likely deleterious’ composed of LGD variants and missense variants predicted to be deleterious by MetaSVM (24); all protein changing variants; and synonymous variants. Analyses were then carried out on each category of variant in addition to testing total variants. To account for multiple testing outcomes were considered significant at P ≤ 0.01 and nominally significant at P < 0.05.
We used a two-sided Poisson exact test to test the null hypothesis that the number of observed de novo variants was not greater than that the number of expected de novo variants. To calculate the number of expected de novo variants for CDH cases and controls, we used the gene-specific probabilities of mutation calculated by Samocha et al. (24) for all variants classes, synonymous variants, missense variants and LGD variants, where LGD comprised splicing, frameshift indels and nonsense variants.
We hypothesized that genes that contribute to CDH should be expressed in the developing diaphragm and, furthermore, that genes that contribute to CDH should also be more intolerant to functional genetic variation (58). Thus, to further investigate the burden of de novo variants, we stratified our analysis using gene expression datasets from E11.5 MDD (25) and residual variation intolerance score (58). Genes above the median of RMA-normalized hybridization intensities for probes were interpreted as expressed (25).
We performed all analyses with two false positives removed from the cases. As it was not feasible to identify and remove false-positive variants in the controls, the analyses would produce conservative estimate of burden.
Percentage of risk-associated variants and CDH patients with those variants
We estimated the percentage of CDH risk-associated variants and percentage of CDH patients with risk-associated variants according to the deleterious de novo variants identified in cases and controls. The formula for the percentage of risk-associated variants is
where N1 is the number of deleterious de novo variants in the 39 CDH patients, and N2 is the number of deleterious de novo variants in the 787 controls. To obtain 95% CI, we simulated de novo mutation counts for CDH patients and controls and based on the Poisson distribution with parameters fitted from the data (19). We performed 100 000 simulations to estimate 5 and 95% quartiles.
The formula for the percentage of CDH patients with risk-associated variants is as follows:
where n1 is the number of CDH patients who have at least one deleterious variant, and n2 is the number of controls who have at least one deleterious variant. To obtain 95% CI, we simulated a number of cases and controls that have at least one deleterious variant by binomial distributions, with success rates estimated from the study using maximum likelihood method. We performed 100 000 simulations to estimate 5 and 95% quartiles.
Gene prioritization by functional similarity
Genes with de novo variants in cases and controls were ranked using ToppGene candidate gene prioritization based on functional similarity to training gene list (https://toppgene.cchmc.org/, accessed 1 June 2015) (29). To generate a training gene list, we searched the literature for genes associated with abnormal diaphragmatic phenotypes in mouse models. We then compared the ranks of CDH and control genes using a Mann–Whitney U-test.
Supplementary Material
Funding
This work was supported by National Institute of Health grant (HD057036) and was supported in part by Columbia University's Clinical and Translational Science Award (CTSA), grant (UL1 RR024156) from National Center for Advancing Translational Sciences/National Institutes of Health (NCATS-NCRR/NIH), a grant from CHERUBS, a grant from the National Greek Orthodox Ladies Philoptochos Society, Inc. and generous donations from the Wheeler foundation, Vanech Family Foundation, Larsen Family, Wilke Family and many other families.
Supplementary Material
Acknowledgements
We are thankful to all the families for their generous contributions. We are grateful for the technical assistance provided by Patricia Lanzano, Jiancheng Guo, Liyong Deng, Badri Vardarajan and Jing He from Columbia University. We also thank Jeannie Kreutzman from University of Michigan, Sheila Horak from University of Nebraska and Laurie Luther and Min Shi from University of Pittsburgh. Study data were collected and managed using Research Electronic Data Capture (REDCap) electronic data capture tools hosted at Columbia University. REDCap is a secure, web-based application designed to support data capture for research studies. We are grateful to all of the families at the participating SFARI Simplex Collection (SSC) sites, as well as the principal investigators (A. Beaudet, R. Bernier, J. Constantino, E. Cook, E. Fombonne, D. Geschwind, D. Grice, A. Klin, D. Ledbetter, C. Lord, C. Martin, D. Martin, R. Maxim, J. Miles, O. Ousley, B. Pelphrey, B. Peterson, J. Piggot, C. Saulnier, M. State, W. Stone, J. Sutcliffe, C. Walsh and E. Wijsman).
Conflict of Interest statement. None declared.
References
- 1.Doyle N.M., Lally K.P. (2004) The CDH Study Group and advances in the clinical care of the patient with congenital diaphragmatic hernia. Semin. Perinatol., 28, 174–184. [DOI] [PubMed] [Google Scholar]
- 2.Pober B.R., Russell M.K., Ackerman K.G. (1993) Congenital diaphragmatic hernia overview. In Pagon R.A., Adam M.P., Ardinger H.H., Wallace S.E., Amemiya A., Bean L.J.H., Bird T.D., Dolan C.R., Fong C.T., Smith R.J.H., Stephens K. (eds), In GeneReviews(R). Seattle, WA: Available from: http://www.ncbi.nlm.nih.gov/books/NBK1359/ (date last accessed, 1 June 2015). [Google Scholar]
- 3.Pober B.R. (2007) Overview of epidemiology, genetics, birth defects, and chromosome abnormalities associated with CDH. Am. J. Med. Genet. C Semin. Med. Genet., 145C, 158–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Langham M.R., Jr, Kays D.W., Ledbetter D.J., Frentzen B., Sanford L.L., Richards D.S. (1996) Congenital diaphragmatic hernia. Epidemiology and outcome. Clin. Perinatol., 23, 671–688. [PubMed] [Google Scholar]
- 5.Downard C.D., Jaksic T., Garza J.J., Dzakovic A., Nemes L., Jennings R.W., Wilson J.M. (2003) Analysis of an improved survival rate for congenital diaphragmatic hernia. J. Pediatr. Surg., 38, 729–732. [DOI] [PubMed] [Google Scholar]
- 6.Skari H., Bjornland K., Haugen G., Egeland T., Emblem R. (2000) Congenital diaphragmatic hernia: a meta-analysis of mortality factors. J. Pediatr. Surg., 35, 1187–1197. [DOI] [PubMed] [Google Scholar]
- 7.Zaiss I., Kehl S., Link K., Neff W., Schaible T., Sutterlin M., Siemer J. (2011) Associated malformations in congenital diaphragmatic hernia. Am. J. Perinatol., 28, 211–217. [DOI] [PubMed] [Google Scholar]
- 8.Lally K.P., Lasky R.E., Lally P.A., Bagolan P., Davis C.F., Frenckner B.P., Hirschl R.M., Langham M.R., Buchmiller T.L., Usui N., et al. (2013) Standardized reporting for congenital diaphragmatic hernia—an international consensus. J. Pediatr. Surg., 48, 2408–2415. [DOI] [PubMed] [Google Scholar]
- 9.Raval M.V., Wang X., Reynolds M., Fischer A.C. (2011) Costs of congenital diaphragmatic hernia repair in the United States-extracorporeal membrane oxygenation foots the bill. J. Pediatr. Surg., 46, 617–624. [DOI] [PubMed] [Google Scholar]
- 10.Metkus A.P., Esserman L., Sola A., Harrison M.R., Adzick N.S. (1995) Cost per anomaly—what does a diaphragmatic-hernia cost. J. Pediatr. Surg., 30, 226–230. [DOI] [PubMed] [Google Scholar]
- 11.Brady P.D., Srisupundit K., Devriendt K., Fryns J.P., Deprest J.A., Vermeesch J.R. (2011) Recent developments in the genetic factors underlying congenital diaphragmatic hernia. Fetal Diagn. Ther., 29, 25–39. [DOI] [PubMed] [Google Scholar]
- 12.Holder A.M., Klaassens M., Tibboel D., de Klein A., Lee B., Scott D.A. (2007) Genetic factors in congenital diaphragmatic hernia. Am. J. Hum. Genet., 80, 825–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Veenma D.C., de Klein A., Tibboel D. (2012) Developmental and genetic aspects of congenital diaphragmatic hernia. Pediatr. Pulmonol., 47, 534–545. [DOI] [PubMed] [Google Scholar]
- 14.Slavotinek A.M. (2007) Single gene disorders associated with congenital diaphragmatic hernia. Am. J. Med. Genet. C Semin. Med. Genet., 145C, 172–183. [DOI] [PubMed] [Google Scholar]
- 15.Veltman J.A., Brunner H.G. (2012) De novo mutations in human genetic disease. Nat. Rev. Genet., 13, 565–575. [DOI] [PubMed] [Google Scholar]
- 16.Fromer M., Pocklington A.J., Kavanagh D.H., Williams H.J., Dwyer S., Gormley P., Georgieva L., Rees E., Palta P., Ruderfer D.M., et al. (2014) De novo mutations in schizophrenia implicate synaptic networks. Nature, 506, 179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iossifov I., Ronemus M., Levy D., Wang Z., Hakker I., Rosenbaum J., Yamrom B., Lee Y.H., Narzisi G., Leotta A., et al. (2012) De novo gene disruptions in children on the autistic spectrum. Neuron, 74, 285–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Roak B.J., Vives L., Girirajan S., Karakoc E., Krumm N., Coe B.P., Levy R., Ko A., Lee C., Smith J.D., et al. (2012) Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature, 485, 246–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanders S.J., Murtha M.T., Gupta A.R., Murdoch J.D., Raubeson M.J., Willsey A.J., Ercan-Sencicek A.G., DiLullo N.M., Parikshak N.N., Stein J.L., et al. (2012) De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature, 485, 237–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu B., Ionita-Laza I., Roos J.L., Boone B., Woodrick S., Sun Y., Levy S., Gogos J.A., Karayiorgou M. (2012) De novo gene mutations highlight patterns of genetic and neural complexity in schizophrenia. Nat. Genet., 44, 1365–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zaidi S., Choi M., Wakimoto H., Ma L., Jiang J., Overton J.D., Romano-Adesman A., Bjornson R.D., Breitbart R.E., Brown K.K., et al. (2013) De novo mutations in histone-modifying genes in congenital heart disease. Nature, 498, 220–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu L., Wynn J., Cheung Y.H., Shen Y., Mychaliska G.B., Crombleholme T.M., Azarow K.S., Lim F.Y., Chung D.H., Potoka D., et al. (2013) Variants in GATA4 are a rare cause of familial and sporadic congenital diaphragmatic hernia. Hum. Genet., 132, 285–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu L., Bennett J.T., Wynn J., Carvill G.L., Cheung Y.H., Shen Y., Mychaliska G.B., Azarow K.S., Crombleholme T.M., Chung D.H., et al. (2014) Whole exome sequencing identifies de novo mutations in GATA6 associated with congenital diaphragmatic hernia. J. Med. Genet., 51, 197–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dong C., Wei P., Jian X., Gibbs R., Boerwinkle E., Wang K., Liu X. (2015) Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum. Mol. Genet., 24, 2125–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Russell M.K., Longoni M., Wells J., Maalouf F.I., Tracy A.A., Loscertales M., Ackerman K.G., Pober B.R., Lage K., Bult C.J., et al. (2012) Congenital diaphragmatic hernia candidate genes derived from embryonic transcriptomes. Proc. Natl Acad. Sci. USA, 109, 2978–2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Samocha K.E., Robinson E.B., Sanders S.J., Stevens C., Sabo A., McGrath L.M., Kosmicki J.A., Rehnstrom K., Mallick S., Kirby A., et al. (2014) A framework for the interpretation of de novo mutation in human disease. Nat. Genet., 46, 944–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang N., Lee I., Marcotte E.M., Hurles M.E. (2010) Characterising and predicting haploinsufficiency in the human genome. PLoS Genet., 6, e1001154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Longoni M., Russell M.K., High F.A., Darvishi K., Maalouf F.I., Kashani A., Tracy A.A., Coletti C.M., Loscertales M., Lage K., et al. (2015) Prevalence and penetrance of ZFPM2 mutations and deletions causing congenital diaphragmatic hernia. Clin. Genet., 87, 362–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen J., Bardes E.E., Aronow B.J., Jegga A.G. (2009) ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucl. Acids Res., 37, W305–W311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clugston R.D., Zhang W., Greer J.J. (2010) Early development of the primordial mammalian diaphragm and cellular mechanisms of nitrofen-induced congenital diaphragmatic hernia. Birth Defects Res. Part A: Clin. Mol. Teratol., 88, 15–24. [DOI] [PubMed] [Google Scholar]
- 31.Ackerman K.G., Herron B.J., Vargas S.O., Huang H.L., Tevosian S.G., Kochilas L., Rao C., Pober B.R., Babiuk R.P., Epstein J.A., et al. (2005) Fog2 is required for normal diaphragm and lung development in mice and humans. PLoS Genet., 1, 58–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bleyl S.B., Moshrefi A., Shaw G.M., Saijoh Y., Schoenwolf G.C., Pennacchio L.A., Slavotinek A.M. (2007) Candidate genes for congenital diaphragmatic hernia from animal models: sequencing of FOG2 and PDGFRalpha reveals rare variants in diaphragmatic hernia patients. Eur. J. Hum. Genet., 15, 950–958. [DOI] [PubMed] [Google Scholar]
- 33.Hong S.H., David G., Wong C.W., Dejean A., Privalsky M.L. (1997) SMRT corepressor interacts with PLZF and with the PML-retinoic acid receptor alpha (RARalpha) and PLZF-RARalpha oncoproteins associated with acute promyelocytic leukemia. Proc. Natl Acad. Sci. USA, 94, 9028–9033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kling D.E., Schnitzer J.J. (2007) Vitamin A deficiency (VAD), teratogenic, and surgical models of congenital diaphragmatic hernia (CDH). Am. J. Med. Genet. C Semin. Med. Genet., 145C, 139–157. [DOI] [PubMed] [Google Scholar]
- 35.Mendelsohn C., Lohnes D., Decimo D., Lufkin T., LeMeur M., Chambon P., Mark M. (1994) Function of the retinoic acid receptors (RARs) during development (II). Multiple abnormalities at various stages of organogenesis in RAR double mutants. Development, 120, 2749–2771. [DOI] [PubMed] [Google Scholar]
- 36.Pistocchi A., Fazio G., Cereda A., Ferrari L., Bettini L.R., Messina G., Cotelli F., Biondi A., Selicorni A., Massa V. (2013) Cornelia de Lange Syndrome: NIPBL haploinsufficiency downregulates canonical Wnt pathway in zebrafish embryos and patients fibroblasts. Cell Death Dis., 4, e866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roberts J.L., Hovanes K., Dasouki M., Manzardo A.M., Butler M.G. (2014) Chromosomal microarray analysis of consecutive individuals with autism spectrum disorders or learning disability presenting for genetic services. Gene, 535, 70–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iossifov I., O'Roak B.J., Sanders S.J., Ronemus M., Krumm N., Levy D., Stessman H.A., Witherspoon K.T., Vives L., Patterson K.E., et al. (2014) The contribution of de novo coding mutations to autism spectrum disorder. Nature, 515, 216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bielinska M., Jay P.Y., Erlich J.M., Mannisto S., Urban Z., Heikinheimo M., Wilson D.B. (2007) Molecular genetics of congenital diaphragmatic defects. Ann. Med., 39, 261–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng Y., Yang W., Xia Y., Hawke D., Liu D.X., Lu Z. (2011) Ras-induced and extracellular signal-regulated kinase 1 and 2 phosphorylation-dependent isomerization of protein tyrosine phosphatase (PTP)-PEST by PIN1 promotes FAK dephosphorylation by PTP-PEST. Mol. Cell. Biol., 31, 4258–4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sirois J., Cote J.F., Charest A., Uetani N., Bourdeau A., Duncan S.A., Daniels E., Tremblay M.L. (2006) Essential function of PTP-PEST during mouse embryonic vascularization, mesenchyme formation, neurogenesis and early liver development. Mech. Dev., 123, 869–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bouaouina M., Lad Y., Calderwood D.A. (2008) The N-terminal domains of talin cooperate with the phosphotyrosine binding-like domain to activate beta1 and beta3 integrins. J. Biol. Chem., 283, 6118–6125. [DOI] [PubMed] [Google Scholar]
- 43.Conti F.J., Felder A., Monkley S., Schwander M., Wood M.R., Lieber R., Critchley D., Mueller U. (2008) Progressive myopathy and defects in the maintenance of myotendinous junctions in mice that lack talin 1 in skeletal muscle. Development, 135, 2043–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Domadia P.N., Li Y.F., Bhunia A., Mohanram H., Tan S.M., Bhattacharjya S. (2010) Functional and structural characterization of the talin F0F1 domain. Biochem. Biophys. Res. Commun., 391, 159–165. [DOI] [PubMed] [Google Scholar]
- 45.Sheen V.L., Ganesh V.S., Topcu M., Sebire G., Bodell A., Hill R.S., Grant P.E., Shugart Y.Y., Imitola J., Khoury S.J., et al. (2004) Mutations in ARFGEF2 implicate vesicle trafficking in neural progenitor proliferation and migration in the human cerebral cortex. Nat. Genet., 36, 69–76. [DOI] [PubMed] [Google Scholar]
- 46.de Wit M.C.Y., de Coo I.F.M., Halley D.J.J., Lequin M.H., Mancini G.M.S. (2009) Movement disorder and neuronal migration disorder due to ARFGEF2 mutation. Neurogenetics, 10, 333–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grzmil P., Enkhbaatar Z., Gundsambuu B., Oidovsambuu O., Yalcin S., Wolf S., Engel W., Neesen J. (2010) Early embryonic lethality in gene trap mice with disruption of the Arfgef2 gene. Int. J. Dev. Biol., 54, 1259–1266. [DOI] [PubMed] [Google Scholar]
- 48.Wright C.F., Fitzgerald T.W., Jones W.D., Clayton S., McRae J.F., van Kogelenberg M., King D.A., Ambridge K., Barrett D.M., Bayzetinova T., et al. (2015) Genetic diagnosis of developmental disorders in the DDD study: a scalable analysis of genome-wide research data. Lancet, 385, 1305–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu L., Wynn J., Ma L., Guha S., Mychaliska G.B., Crombleholme T.M., Azarow K.S., Lim F.Y., Chung D.H., Potoka D., et al. (2012) De novo copy number variants are associated with congenital diaphragmatic hernia. J. Med. Genet., 49, 650–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fischbach G.D., Lord C. (2010) The Simons Simplex Collection: a resource for identification of autism genetic risk factors. Neuron, 68, 192–195. [DOI] [PubMed] [Google Scholar]
- 51.Li H., Durbin R. (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics, 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., et al. (2010) The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res., 20, 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang K., Li M.Y., Hakonarson H. (2010) ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucl. Acids Res., 38, e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. (2010) A method and server for predicting damaging missense mutations. Nat. Methods, 7, 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kumar P., Henikoff S., Ng P.C. (2009) Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc., 4, 1073–1082. [DOI] [PubMed] [Google Scholar]
- 56.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet., 38, 904–909. [DOI] [PubMed] [Google Scholar]
- 57.Altshuler D.M., Gibbs R.A., Peltonen L., Dermitzakis E., Schaffner S.F., Yu F.L., Bonnen P.E., de Bakker P.I.W., Deloukas P., Gabriel S.B., et al. (2010) Integrating common and rare genetic variation in diverse human populations. Nature, 467, 52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Petrovski S., Wang Q., Heinzen E.L., Allen A.S., Goldstein D.B. (2013) Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet., 9, e1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.