Abstract
Less than half of patients with suspected genetic disease receive a molecular diagnosis. We have therefore integrated next-generation sequencing (NGS), bioinformatics, and clinical data into an effective diagnostic workflow. We used variants in the 2741 established Mendelian disease genes [the disease-associated genome (DAG)] to develop a targeted enrichment DAG panel (7.1 Mb), which achieves a coverage of 20-fold or better for 98% of bases. Furthermore, we established a computational method [Phenotypic Interpretation of eXomes (PhenIX)] that evaluated and ranked variants based on pathogenicity and semantic similarity of patients’ phenotype described by Human Phenotype Ontology (HPO) terms to those of 3991 Mendelian diseases. In computer simulations, ranking genes based on the variant score put the true gene in first place less than 5% of the time; PhenIX placed the correct gene in first place more than 86% of the time. In a retrospective test of PhenIX on 52 patients with previously identified mutations and known diagnoses, the correct gene achieved a mean rank of 2.1. In a prospective study on 40 individuals without a diagnosis, PhenIX analysis enabled a diagnosis in 11 cases (28%, at a mean rank of 2.4). Thus, the NGS of the DAG followed by phenotype-driven bioinformatic analysis allows quick and effective differential diagnostics in medical genetics.
INTRODUCTION
At the time of this writing, roughly 7000 Mendelian diseases are recognized (1-3). Although these diseases are individually rare, up to 8% of the population is affected by a specific genetic disorder (4). Because of the vast number of diseases, many of which have a broad and incompletely understood phenotypic spectrum, and the high genetic heterogeneity of many clinical syndromes such as intellectual disability, the diagnostic process in medical genetics is often challenging, even for experienced and expert clinicians. The traditional medical genetics evaluation relies on recognizing a characteristic pattern of signs or symptoms to guide targeted genetic testing for confirmation of the diagnosis, with the major diagnostic methods including karyotyping, array comparative genomic hybridization (CGH), biochemical testing, and Sanger sequencing of individual genes. However, the diagnostic yield remains less than 50% even after extensive workups (5), with the costs of clinical and molecular genetic analysis for patients whose diagnosis is not clear after the first visit reaching 25,000 U.S. dollars or more (5).
The term “diagnostic odyssey” has been used to describe the experience of patients and families affected by rare diseases that cannot be diagnosed; for instance, the average time between the onset of symptoms and the correct diagnosis is currently 14 years for patients with type 2 myotonic dystrophy (6). The lack of a diagnosis can mean missed opportunities for tailored approaches to clinical management and treatment strategies, a substantial burden of guilt and uncertainty for families, and the inability to make accurate statements on recurrence risk and prognosis, not to mention the economic costs of unnecessary diagnostic procedures.
Whole-exome sequencing (WES), first used in 2010 to identify the cause of a Mendelian disease (7), is rapidly becoming attractive as a tool for diagnostic testing in general medical genetics (8). Additionally, next-generation sequencing (NGS) panel, WES, and whole-genome sequencing (WGS) approaches have been introduced for carrier screening (9), as well as in neonatal intensive care units (10). However, medical interpretation of WES results remains challenging, and the successes have, for the most part, been limited to single cases or small groups of patients (11). Identifying the one or two causative mutations among the myriad of variants present in the WES findings of an individual has been compared to finding a needle in a haystack (12). A typical exome contains more than 30,000 variants when compared to the human reference sequence, with about 10,000 of them representing nonsynonymous amino acid substitutions, alterations of conserved splice site residues, or small insertions or deletions (13, 14). Although the community has developed numerous bioinformatic tools to filter out common variants and predict their pathogenicity (15, 16), each human genome harbors about 100 genuine loss-of-function variants with ~20 genes completely inactivated (17). Therefore, purely sequence-based evaluation of genes in diagnostic WES typically identifies tens or hundreds of candidates. Although this is acceptable in a research context, in which other strategies such as genetic linkage or comparison with a study group of individuals thought to have the same disease can often reduce the search space, extensive evaluation of long lists of candidate genes does not scale well to the diagnostic setting.
Depth and uniformity of coverage have a major influence on the performance of targeted capture for NGS. For instance, at a mean on-target read depth of 20×, up to 15% of heterozygous single-nucleotide variants (SNVs) will be missed (18). Although initial WES studies aimed for a coverage of 20-fold, deeper coverage is needed for accurate detection of heterozygous variants (19), and current studies typically use a coverage of 50- to 70-fold (20, 21) or higher. This has led to debate in the community as to the relative value of various NGS approaches for diagnostics, with proponents of targeted panel sequencing (22), WES (23), and WGS (24).
Here, we explore a different approach toward the translation of NGS-based diagnostics into clinical diagnostics in a medical genetics clinic. We contend that WES is not optimal in a purely diagnostic setting because we can currently offer a confident interpretation of variants only in ~2740 known Mendelian disease genes; the identification of a potentially pathogenic variant in a gene regarded as a good candidate because of biochemical or model organism data often represents the starting point for a good research project, but is more likely to engender confusion in a diagnostic setting. Therefore, by enriching for genes known to be associated with Mendelian disease, we shift the focus from the whole exome to that part of the exome/genome that is clinically interpretable in a diagnostic setting. We refer to this portion of our genome as the disease-associated genome (DAG). A pathogenic variant in one of these genes is, in principle, interpretable in the context of the presenting clinical phenotype and our knowledge of the diseases associated with the gene in question. We have previously shown that phenotypically driven genomic data fusion (25) and comparison of human to model organism phenotypes (26) markedly improves the ability to correctly identify candidate disease-causing mutations in WES studies. Here, we use the Human Phenotype Ontology (HPO) and associated data to develop a computational procedure for differential diagnosis with the DAG panel. The HPO provides a structured, comprehensive, and well-defined set of more than 10,000 terms describing human phenotypic abnormalities. It provides annotations of nearly 7300 human hereditary syndromes that yield computable representations of the diseases, associated disease genes, as well as the signs, symptoms, laboratory findings, and other phenotypic abnormalities that characterize the diseases (3, 27). Here, we adapt our semantic similarity approach toward differential diagnosis, using terms and annotations from the HPO (28), to rank candidate genes in a diagnostic setting. Our algorithm is freely available for academic use through http://compbio.charite.de/PhenIX/.
RESULTS
Here, we present an approach to Mendelian disease diagnostics that involves the targeted sequencing of the DAG panel combined with a phenotype-driven computational analysis strategy [Phenotypic Interpretation of eXomes (PhenIX)] that ranks candidate genes based on the presence of rare, predicted pathogenic variants and the clinical relevance of the genes with associated disease phenotypes. Our algorithm first filters the variants according to rarity, target region location, and predicted pathogenicity. Next, the remaining candidate genes are evaluated for clinical relevance on the basis of the semantic similarity of the patient’s phenotypic abnormalities to the phenotypic spectrum of diseases associated with each candidate gene. In brief, our method aims to identify and rank disease genes by combining potential clinical relevance with deleterious variants found within those genes (see Materials and Methods).
Design and validation of the DAG panel
We established a comprehensive catalog of Mendelian disease genes using data from the HPO project (3), part of which is derived from information in the Online Mendelian Inheritance in Man (OMIM) (1) and Orphanet (2) resources. The HPO project, which was initiated in 2007, has grown to include more than 10,000 terms describing individual phenotypic abnormalities that have been used to generate more than 110,000 annotations to more than 7000 mainly Mendelian disease entries (3, 27). The data in the HPO thus provide a powerful curated resource for translational research by providing the means to capture, store, and exchange phenotypic information about human disease and have been used to integrate phenotypic information into computational analysis (25, 26, 28-32). We additionally included plausible candidate disease genes from recent publications describing large-scale WES studies, obtained by surveying the recent literature (8, 33-37), for a total of 2741 genes (genes and references are included in table S6).
Because our aim was to obtain nearly complete coverage of the DAG, we designed enrichment probes for the DAG using SureSelect technology (38). In total, 96 samples were sequenced (6 samples per lane of an Illumina HiSeq 1500 sequencer), resulting in an average coverage of 361.7 ± 81.6 reads (135.6 ± 10.6 after removal of duplicates), with 98% of the DAG target region being covered by at least 20 reads (fig. S1 and tables S1 and S2).
To estimate the advantage of the high coverage of the DAG panel with respect to comprehensive variant calling, we randomly sampled reads from the Binary Alignment/Map (BAM) files from the DAG target region (twice over each of the 96 sequenced DAG samples) to a target average coverage of 100-fold to simulate the coverage expected from typical exome sequencing. After this, the down-sampled BAM files were processed in the same way as the original BAM files, and the distribution of called variants was compared (table S3). A substantial number of variants called from the original BAM file were not called from the files simulated to have exome or genome coverage, including an average of 5.2 ± 2.0 variants listed in the Human Gene Mutation Database (HGMD) (39).
Phenotypic interpretation of eXomes: PhenIX
We developed a computational algorithm to filter and rank candidate genes according to variant rarity and pathogenicity and potential clinical relevance of the gene harboring the variants. As input, PhenIX requires (i) a variant call format (VCF) file representing the results of sequencing the DAG target region (or an exome or a genome), and (ii) a list of HPO terms representing the clinical features of the individual being sequenced. Each variant is scored on the basis of rarity and predicted pathogenicity; after this, all variants mapping to a given gene are combined. The genes harboring predicted pathogenic variants are assigned a phenotype score by using the semantic similarity between associated disease phenotypes and the patient’s phenotype. However, the gene is down-weighted if the distribution of variants in a gene is incompatible with the mode of inheritance of the associated disease, for example, if a single heterozygous variant is observed in a gene associated with an autosomal recessively inherited disease. Finally, a rank is calculated on the basis of the combined variant and the phenotype scores.
To estimate the performance of our method, we conducted extensive computational simulations using mutation data from the HGMD. Sample data sets were simulated for a given disease and inheritance model by spiking with mutations from HGMD into a VCF file generated with the DAG panel. Appropriate HPO terms were chosen from the annotations of the corresponding disease. Several test scenarios were considered. The performance of the method was near 100% when all the HPO terms annotating the disease were used for the analysis (for example, Greig cephalopolysyndactyly syndrome is annotated with 44 HPO terms representing individual signs and symptoms of that disease). In another, more realistic, test scenario, up to five terms were chosen at random, of which two were made imprecise by exchanging them with the more general parent term, and two unrelated confounder (“noise”) terms were added at random. Here, the correct gene was ranked in first place in 86.5% of 8504 simulations, corresponding to a 32.5-fold improvement over pure variant filtering (Fig. 1 and fig. S2).
Fig. 1.
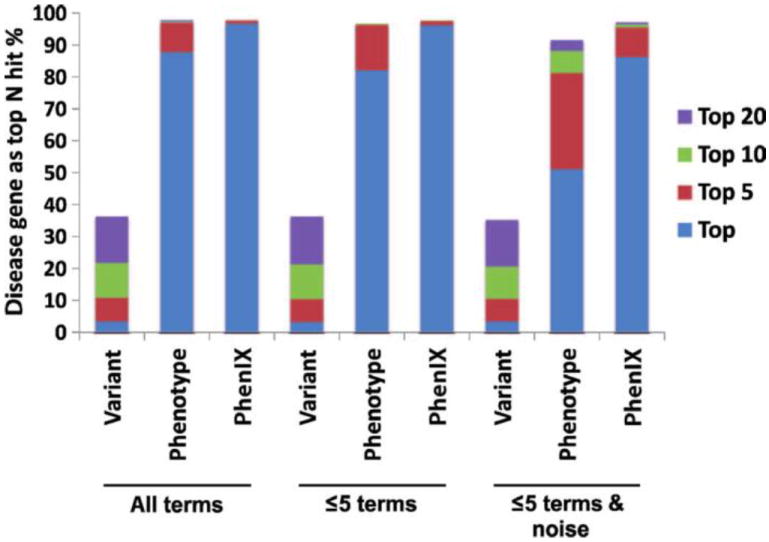
Computational evaluation of PhenIX. HGMD mutations were inserted into variant files from DAG panels from which the causative mutations had been removed and phenotypic annotations of the corresponding diseases were extracted from the HPO database. The genes were ranked with PhenIX. Results were simulated either on the entire disease set (All) or by filtering for known autosomal dominant (AD) or autosomal recessive (AR) diseases (fig. S2). A total of 8504 (All), 3471 (AD), and 5006 (AR) simulations were performed. Data are shown as the percentage of simulations in which the correct genes was ranked in Nth place. Variant, only variant scores used to rank candidate genes; All terms, all HPO terms used to annotate a disease were used for PhenIX analysis; ≤5 terms, up to five HPO terms were chosen at random from the terms used to annotate the disease; ≤5 terms & noise, up to five annotations are used, two of which are made imprecise by exchanging them with a more general parent term; additionally, two random noise terms were added. Results are shown for the correct gene being ranked as the single top hit, or being among the top 5, 10, or 20 hits for the three test scenarios.
Retrospective analysis
We then tested the performance of our method with the generated DAG data from 52 individuals with a diagnosis of Mendelian disease that had been confirmed by Sanger sequencing (Table 1). HPO terms were entered, and filtering was performed at a frequency threshold of 1%. The average rank of the correct gene among the 2741 disease genes in the DAG panel was 2.1. The mean rank of the autosomal recessive genes was 5, substantially lower than for the autosomal dominant genes (1.7). The lower rank for the recessive genes was partially related to results for an individual with eczematoid acrodermatitis enteropathica, who had a missense mutation in SLC39A4 that was correctly flagged as pathogenic, as well as a synonymous mutation that had been shown to cause a splice defect. The latter mutation was not identified as deleterious by PhenIX, resulting in a final rank of 14 for SLC39A4.
Table 1. Fifty-two control patient cases with known mutations.
The number of patients with a mutation in the given gene is indicated in parentheses.
| Mode of inheritance | Genes | Average rank |
|---|---|---|
| AD | ACVR1, ATL1, BRCA1, BRCA2, CHD7 (4), CLCN7, COL1A1, COL2A1, EXT1, FGFR2 (2), FGFR3, GDF5, KCNQ1, MLH1 (2), MLL2/KMT2D, MSH2, MSH6, MYBPC3, NF1 (6), P63, PTCH1, PTH1R (2), PTPN11 (2), SCN1A, SOS1 TRPS1, TSC1, WNT10A | 1.7 |
| AR | ATM, ATP6V0A2, CLCN1 (2), LRP5, PYCR1, SLC39A4 | 5 |
| X | EFNB1, MECP2 (2), DMD, PHF6 | 1.8 |
Prospective analysis
To further validate our methodology, we investigated 40 individuals who, after extensive clinical genetic evaluation (physical examination by medical geneticist, array CGH, and often targeted Sanger gene sequencing), remained without a diagnosis (clinical features summarized in Table 2). We designed a standard evaluation procedure in which deep phenotyping (40) with the selection of representative HPO terms (3, 27) was followed by targeted NGS of the DAG panel. Computational analysis was performed as described above to generate a ranked list of candidates based on the combined variant and clinical relevance scores. Because our computational simulations almost always placed the true disease gene in the top 10 candidates, we limited our evaluation to the top 20 ranked genes, as well as any gene with a pathogenic mutation at the same nucleotide position listed in HGMD (39) or ClinVar (41) for each patient. Initial clinical evaluation was performed by one of the authors, and a short list of the most likely candidates was presented to the entire group in clinical rounds, where up to the best two candidate genes were chosen on the basis of clinical experience. These genes were subjected to Sanger validation and cosegregation studies. If the variants in the selected genes cosegregated as expected and the clinical manifestations of the patient were sufficiently explained by a disease associated with the gene, then a positive diagnosis was made. Otherwise, the short list was reexamined for additional candidates (Fig. 2). We estimate that an experienced clinical geneticist would spend a total of 1 hour in the initial evaluation of the patient and in deciding whether to perform DAG panel sequencing and an additional 1 hour studying the list of top 20 candidates, evaluating the results of the Sanger validation and co-segregation studies before being able to decide whether a definitive diagnosis can be made.
Table 2. Summary of clinical signs and symptoms in 40 patients with unknown diagnosis.
| Clinical presentation | n |
|---|---|
| Intellectual disability + multiple congenital anomalies (more than two other organ systems affected) | 13 |
| Intellectual disability + other neuropsychological features | 7 |
| Intellectual disability + musculoskeletal abnormalities | 5 |
| Intellectual disability + eye abnormalities | 1 |
| Intellectual disability + dysmorphic features | 1 |
| Multiple congenital anomalies (more than two organ systems affected) without intellectual disability | 6 |
| Skeletal phenotype | 5 |
| Eye and/or ear phenotype | 2 |
Fig. 2.
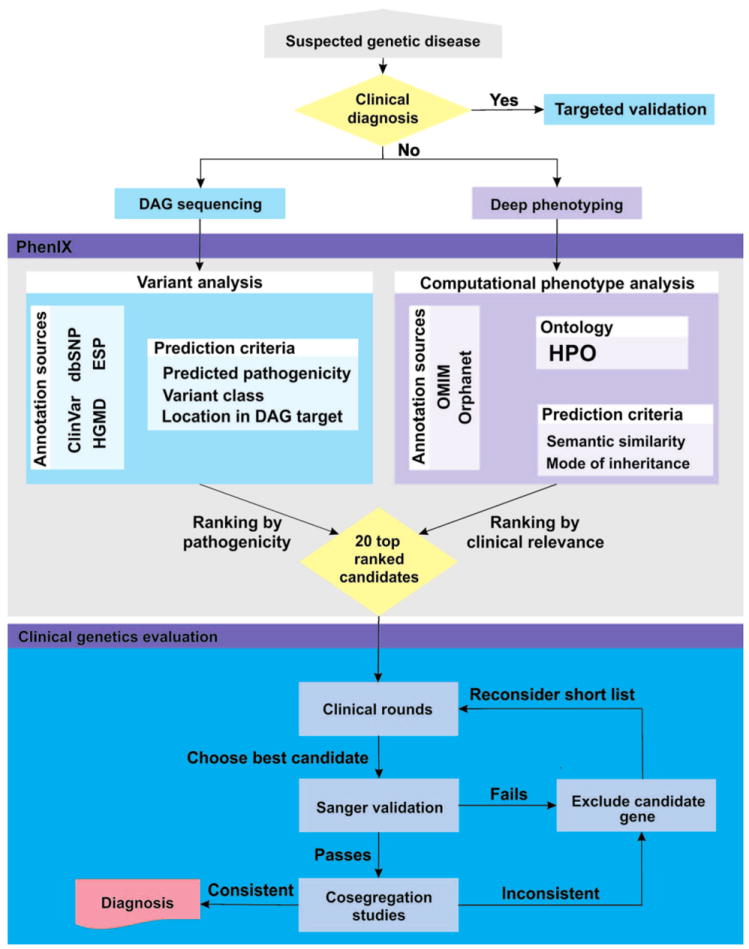
PhenIX workflow, showing the clinical and bioinformatic analysis steps. After initial clinical evaluation, a decision is made to perform PhenIX analysis if no clinical diagnosis can be found. After sequencing and computational analysis, clinical evaluation of the top 20 gene candidates identifies genes for validation by Sanger sequencing and cosegregation studies.
By applying this procedure to 40 individuals, we identified a definitive diagnosis in 11 (28%) cases. Table 2 shows a clinical summary of these cases, and tables S4 and S5 include a full list of HPO terms used to search in PhenIX. PhenIX analysis was performed according to the flow chart in Fig. 2, and the top 20 genes were inspected. Discussion at clinical genetics rounds flagged one (n = 16 only one) or two (n = 6) genes as being likely candidates. These genes were then subjected to Sanger validation, cosegregation studies, and close examination. This led to definitive diagnoses being made in 11 of 40 patients (28%) (Table 3).
Table 3. Clinical category and final diagnoses of 11 patients whose diagnoses were identified by PhenIX analysis.
Additional information, including complete lists of HPO terms used to describe the phenotypic abnormalities seen in these patients, is available in table S4. Patients P6 and P10 were referred from external centers. Rank shows the rank after PhenIX analysis before clinical evaluation. The average rank for all 11 cases was 2.5. MoI, mode of inheritance; AD, autosomal dominant; AR, autosomal recessive; XR, X-linked recessive.
| ID | Age, sex | Presentation | Gene | Rank | Diagnosis | MoI |
|---|---|---|---|---|---|---|
| P1 | 3 years (female) | Intellectual disability + multiple congenital anomalies | MLL | 2 | Wiedemann-Steiner syndrome (54) | AD |
| P2 | 5 years (female) | Intellectual disability + multiple congenital anomalies | SYNGAP1 | 4 | Mental retardation, MRD5 (55) | AD |
| P3 | 6 years (female) | Skeletal phenotype | FGFR2 | 1 | Pfeiffer syndrome (56) | AD |
| P4 | Death at 5.5 months (female) | Multiple congenital anomalies without intellectual disability | SH3PXD2B | 6 | Frank-ter Haar syndrome (57) | AR |
| P5 | 6 months (female) | Intellectual disability + neurological abnormalities | SLC6A3 | 1 | Parkinsonism-dystonia (58) | AR |
| P6 | Fetus (male), death at 22 weeks of gestation | Skeletal phenotype | ALPL | 2 | Infantile hypophosphatasia (59) | AR |
| P7 | 7 years (male) | Eye phenotype | NHS | 2 | Nance-Horan syndrome/cataract 40, X-linked (60) | XR |
| P8 | 14 years (male) | Intellectual disability + multiple congenital anomalies | MLL | 1 | Wiedemann-Steiner syndrome (54) | AD |
| P9 | 6 years (female) | Intellectual disability + multiple congenital anomalies | DYRK1A | 4 | Mental retardation, MRD7 (61) | AD |
| P10 | 4 children between 1½ and 7 years | Intellectual disability + multiple congenital anomalies | MCOLN1 | 1 | Type IV mucolipidosis (62) | AR |
| P11 | 3 years (male) | Intellectual disability + multiple congenital anomalies | RBM10 | 3 | TARP syndrome (63) | XR |
DISCUSSION
Genomic medicine, including WES and WGS, is poised to transform clinical practice in many fields (42). Here, we present a phenotype-driven computational and clinical workflow for the efficient diagnosis of rare Mendelian diseases. Our approach uses the results of clinical analysis to substantially improve the ranking of candidate genes, and provides a clear pathway to integrate the results of bioinformatics analysis into the clinical workflow by clinical evaluation of phenotypic matching among the best candidates.
Here, we have shown how to use a computable representation of clinical phenotypes to prioritize candidate genes in diagnostic sequencing with a target panel of 2741 known Mendelian disease genes. Our workflow represents a tight integration of clinical and bioinformatic analysis (Fig. 2). Clinical expertise is required to perform deep phenotyping and choose representative HPO terms to describe the clinical features of the patient being investigated. Experience is necessary to realize whether a given phenotypic abnormality is likely to be characteristic of a disease or an incidental finding, for example, a feature such as low-grade myopia may not be related to the genetic disease being sought and adding this feature to PhenIX analysis may lower the score of the actual disease-causing gene. After sequencing, alignment, and variant calling, PhenIX analysis is used to generate a list of the top 20 candidates. Additional candidates can be listed if desired. Clinical expertise is required to examine this list for promising candidates based on additional information from original publications and databases, such as OMIM. To assist with this process, the PhenIX Web page provides links to a number of useful resources including OMIM, the University of California Santa Cruz (UCSC) Genome Browser, ClinVar, and HGMD. We suggest that a presentation of the case together with a description of the best PhenIX candidates at clinical genetics rounds should be performed, followed by validation of the most plausible candidate(s) by Sanger sequencing and cosegregation studies. In our experience, discussions on the differential diagnosis proceed quickly when organized in this fashion and fit well into a typical clinical workflow. We chose to limit our NGS analysis only to the sample from the affected individual, because in the diagnostic setting, family samples (trios) may not be available initially. In addition, the cost of sequencing may be a factor. However, trio sequencing could easily be adapted into our workflow.
On the basis of our results, we suggest that targeting all known disease genes, that is, a DAG, rather than the whole exome or genome, is advantageous in terms of target coverage, cost per sample, and the ability to provide quick and accurate clinical interpretation of the variants. Cases that remain unsolved after PhenIX analysis of the DAG panel can be considered for more time-intensive clinical research WES/WGS studies, because these approaches are able to search for potential mutations in previously undescribed disease genes.
There are several areas in which our approach can be improved and extended. The phenotypic analysis based on semantic similarity depends on an annotated corpus of information about the phenotypic features that characterize various diseases. The HPO currently has more than 110,000 annotations to more than 7000 diseases listed in OMIM (3). Increasing the depth of annotation to these diseases would improve the performance (43). A number of challenges remain in the ontological modeling of certain classes of diseases and phenotypes in areas such as neurobehavioral abnormalities (44). The DAG panel, as presented here, currently contains baits only for protein coding genes. However, other medically relevant sequences of the genome could be captured in a similar way, such as enhancers of the sonic hedgehog gene, in which point mutations can cause characteristic skeletal malformations (45). Hand in hand with this, future bioinformatics research will be required to confidently identify medically relevant variants in noncoding sequences, as well as presumptive synonymous variants that actually lead to a deleterious effect such as defective splicing in the case of the “silent” SLC39A4 mutation mentioned above. Our approach concentrates on known disease genes and is thus not designed or intended to identify new disease genes; other computational tools such as the Exomiser (26) and eXtasy (25) have been presented for this purpose.
In summary, we have presented a diagnostic tool for genetics professionals that combines targeted enrichment and NGS of a comprehensive panel of genes known to be associated with Mendelian disease; bioinformatics analysis of sequencing results is tightly coupled to the expertise and workflow of genetics professionals, allowing a complete workup of NGS results in roughly 2 hours per patient. A recent study on the use of diagnostic exome sequencing of 250 unselected, consecutive cases achieved a diagnostic yield of 25% (8), and another larger-scale exome-based study on persons with intellectual disability reached a diagnostic yield of 16% (46). Although it is hard to compare the diagnostic yield between different studies, the results presented here are competitive, with an average rank of the correct gene of 2.1 in a retrospective study on representative diseases and a yield of 28% in a prospective study with cases chosen for the fact that a diagnosis could not be achieved. Additionally, our method requires less sequencing than high coverage WES or WGS, which may translate into cost benefits. Our bioinformatic and clinical workflow could be completed in roughly 2 hours per patient, and PhenIX analysis is easy to use, requiring only a VCF file and a list of HPO terms. Our method thus provides the means for quick and effective differential diagnostics in medical genetics.
MATERIALS AND METHODS
Consent
This study was approved by the Institutional Review Board of the Charité Universitätsmedizin Berlin. Informed written consent was obtained from adult subjects and parents of children.
Case selection
The control group consisted of 52 individuals with suspected genetic diagnoses seen at the Institute of Medical Genetics and Human Genetics of the Charité university hospital between 2010 and 2013, and who received an etiological diagnosis based on clinical findings and the identification of mutations in the genes indicated in Table 1. In addition, 38 patients seen during this time frame who remained without an etiological diagnosis were investigated in this study. Patients were chosen on the basis of availability of DNA samples from parents (for validation of cosegregation by Sanger sequencing), consent for research, and the inability to identify a genetic diagnosis despite a high index of suspicion of an underlying genetic cause. Two additional cases were referred from external clinics and were not seen in our department (P6 and P10 in Table 3).
Capture of the targeted disease-related genome and NGS
A SureSelectXT Automation Custom Capture Library (Agilent) target enrichment panel was generated using the coordinates given in table S6. The enrichment panel comprised all coding exons of 2741 genes associated with at least one Mendelian disease, as well as 133 control genes. Capture was performed according to the manufacturer’s instructions using an NGS Workstation Option B (Agilent) for automated library preparation starting with 3 mg of DNA per sample. Then, sequencing of 100–base pair paired-end reads was carried out on a HiSeq 1500 (Illumina). Sequence reads were mapped to the haploid human reference genome (hg19) with Novoalign (Novocraft Technologies). SNVs and short insertions and deletions (indels) were called using GATK version 2.8 (47). Variant annotation was performed with Jannovar (48). In total, 96 samples were sequenced on two HiSeq 1500 flow cells.
PhenIX: Bioinformatic ranking of candidate genes
Ranking of candidate genes was performed in two steps. First, off-target and synonymous variants were removed, and the remaining variants were analyzed with respect to population frequency by using data from dbSNP (49) and from the Exome Variant Server [National Heart, Lung, and Blood Institute (NHLBI) GO Exome Sequencing Project 2014, http://evs.gs.washington.edu/EVS/]. For the purposes of analysis, we assumed the minor allele frequency of each variant to be the maximum frequency reported by dbSNP or that of the African American or European American populations represented in the Exome Variant Server. A frequency score is calculated as max(0,1 – 0.13533e100*f), and variants with no frequency data (f = 0) were assigned a score of 1.0, resulting in values between 1.0 and 0.0 for variants with frequencies of up to 2%. Predicted pathogenicity of missense variants was derived from dbNSFP version 2.4 (50) using the fields for MutationTaster (16), polyphen-2 (15), and SIFT (51). Scores from these three prediction tools were normalized to be between 0.0 (benign) and 1.0 (pathogenic), and the single most pathogenic score was taken for each variant. For classes of variants other than missense mutations, a pathogenicity score was calculated as described (26). Finally, the overall variant score was calculated as the product of the frequency and pathogenicity score. A clinical relevance score was calculated using the semantic similarity between phenotypic abnormalities entered by the user and 2741 disease genes in our database. The phenotypic abnormalities of all diseases associated with a given gene were assigned to the gene, because our method ranks candidate genes rather than individual diseases. For instance, the FBN1 gene is mutated in Marfan syndrome, acromicric dysplasia, and a number of other diseases, and the phenotypic abnormalities of each of those diseases were assigned to FBN1. Then, the semantic similarity score of the Phenomizer algorithm (28) was calculated for each of the genes. The maximum score was set to 1.0, and the other scores were normalized accordingly. The final score was calculated as the average of the variant and the gene relevance score. However, if the variant distribution for a gene was not compatible with the mode of inheritance of the associated diseases (for example, a gene has only a single heterozygous mutation but the associated disease is autosomal recessive, or the gene has only a single homozygous mutation but the disease is autosomal dominant), then the gene relevance score was divided by 2 before calculating the final score. The final score was calculated as the mean of the variant score and the gene relevance score. The major distinctions between PhenIX and our previously published algorithm PHIVE, which is implemented in the Exomiser (26), are, thus, the restriction of the analysis to variants in clinically interpretable disease genes using only human phenotype information rather than model organism phenotype data, the analysis of sequencing results for previously reported mutations in ClinVar and the public version of HGMD, and the use of prioritization based on the modes of inheritance of diseases associated with candidate genes compared with the distribution of sequenced variants.
Computational evaluation of PhenIX prioritization
To test the performance of PhenIX prioritization with DAG panel sequencing, we used a simulation approach based on known disease-causing mutations from the HGMD. A total of 28,516 mutations were selected on the basis of being assigned as disease-causing, single-nucleotide mutations (including indels) by HGMD and with HPO annotations available for the disease in question. For the simulations, 10,000 variants were randomly selected from this set. We first removed the causative mutations from the 52 VCF files generated from the retrospective cohort with known mutations. Then, we added an additional mutation to one of these files. For autosomal dominant diseases, one heterozygous mutation was added, and for autosomal recessive diseases, either one homozygous mutation or two heterozygous mutations were added. The phenotypic (HPO) annotations for the corresponding disease were then compared to the HPO annotations associated with the 2741 disease genes (if a disease gene was associated with multiple diseases, all annotations were merged). There were three test scenarios. In the first case, all HPO annotations for the disease in question were used. To simulate incomplete phenotyping, we performed the simulations with up to five HPO terms chosen at random from the annotations of the disease. Finally, to simulate the effects of noise, we randomly chose two of the five terms and promoted them to their less-specific parent terms, and finally, two new terms were chosen randomly from the whole of HPO and added to the annotations.
A rank was determined for the original disease gene after PhenIX analysis. In all the analysis, an ordinal ranking method was used in which equal scoring genes are resolved arbitrarily but consistently by assigning a unique rank to each of the ties. In our case, we sorted the equally scored genes alphabetically and assign the ranks. We recorded the number of times the correct disease gene was ranked in first place, as well as the total recall (correct gene listed at any rank). For each simulation, 1 of the 52 DAG panel VCF files was chosen.
Clinical evaluation and validation of NGS results
We clinically evaluated the NGS results using the PhenIX server, which implements the algorithm described above. PhenIX presents a ranked gene list together with links to various other resources such as the UCSC browser (52), Entrez Gene (53), OMIM (1), Orphanet (2), ClinVar (41), MutationTaster (16), and HGMD (39). Evaluation was performed by trained genetics professionals. For each unsolved case, the top 20 ranked candidates were examined by comparison with the above-mentioned data sources and as appropriate with the original literature. An initial assessment of these 20 candidates was possible in about 2 hours and resulted in a short list of candidates thought to be potential matches. These were discussed at clinical rounds by a team of clinicians and researchers including L.M., T.Z., L.G.N., S.D., N.E., M.S., N.C.Ø., M.R.S., R.F., U.K., P.K., P.N.R., S.M., and D.H. A consensus decision was reached on candidates to be validated by Sanger sequencing and cosegregation studies. We considered a case to be solved after clinical analysis and cosegregation studies if a degree of certainty was reached that led to reporting of the mutation and diagnosis in our clinical setting.
Supplementary Material
www.sciencetranslationalmedicine.org/cgi/content/full/6/252/252ra123/DC1 Fig S1. Distribution of the coverage fraction for all sequenced 96 samples. Fig. S2. Computational evaluation of PhenIX. Table S1. Percentage of target bases that exceed coverages of 10, 20, …, 100 reads. Table S2. Read alignment and coverage summary statistics.
Table S3. Average number of variants called only from the original BAM files from the DAG panels. Table S4. Detailed clinical and molecular findings for the 11 individuals in whom a previously unknown diagnosis was clarified by PhenIX analysis. Table S5. Clinical presentation of 29 patients for whom PhenIX analysis failed to reveal a molecular diagnosis.
Table S6. List of genes (with references) present in the DAG panel. References (64-70)
Acknowledgments
We thank the patients and their families for taking part in this study. Funding: The study was supported by grants from the Bundesministerium für Bildung und Forschung (BMBF project numbers 0313911, 0316065E, and 0316190A), core infrastructure funding from the Wellcome Trust, NIH 1R24OD011883-02, and by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under contract no. DE-AC02-05CH11231, the Volkswagenstiftung (Lichtenberg Program to M.R.S.), and a grant to S.M. by the Max Planck Foundation. Agilent supplied the SureSelect kits at no charge.
Footnotes
Author contributions: T.Z., S.M., N.C.Ø., D.H., and P.N.R. participated in drafting/or revising the manuscript; T.Z., D.H., D.S., S.M., and P.N.R. designed the study; T.Z., S.K., L.M., M.J., J.H., P.K., L.G.N., S.D., N.E., M.S., N.C.Ø., M.R.S., U. Krüger, G.F., B.F., U. Kornak, R.F., A.A., Y.M., S.E.L., M.H., D.S., D.H., S.M., and P.N.R. participated in the acquisition and/or analysis of data; and T.Z., D.H., S.M., P.N.R., and G.F. provided administrative, technical, or supervisory support.
Competing interests: Y.M. is an equity holder of Cartagenia NV, and S.M. is a paid consultant for Agilent. The other authors declare that they have no competing interests.
Data and materials availability: The PhenIX server is freely available for academic use at http://compbio.charite.de/PhenIX/. The HPO is freely available for all users at http://www.human-phenotype-ontology.org.
REFERENCES AND NOTES
- 1.Amberger J, Bocchini C, Hamosh A. A new face and new challenges for Online Mendelian Inheritance in Man (OMIM®) Hum Mutat. 2011;32:564–567. doi: 10.1002/humu.21466. [DOI] [PubMed] [Google Scholar]
- 2.Rath A, Olry A, Dhombres F, Brandt MM, Urbero B, Ayme S. Representation of rare diseases in health information systems: The Orphanet approach to serve a wide range of end users. Hum Mutat. 2012;33:803–808. doi: 10.1002/humu.22078. [DOI] [PubMed] [Google Scholar]
- 3.Köhler S, Doelken SC, Mungall CJ, Bauer S, Firth HV, Bailleul-Forestier I, Black GC, Brown DL, Brudno M, Campbell J, FitzPatrick DR, Eppig JT, Jackson AP, Freson K, Girdea M, Helbig I, Hurst JA, Jähn J, Jackson LG, Kelly AM, Ledbetter DH, Mansour S, Martin CL, Moss C, Mumford A, Ouwehand WH, Park SM, Riggs ER, Scott RH, Sisodiya S, Van Vooren S, Wapner RJ, Wilkie AO, Wright CF, Vulto-van Silfhout AT, de Leeuw N, de Vries BB, Washingthon NL, Smith CL, Westerfield M, Schofield P, Ruef BJ, Gkoutos GV, Haendel M, Smedley D, Lewis SE, Robinson PN. The Human Phenotype Ontology project: Linking molecular biology and disease through phenotype data. Nucleic Acids Res. 2014;42:D966–D974. doi: 10.1093/nar/gkt1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baird PA, Anderson TW, Newcombe HB, Lowry RB. Genetic disorders in children and young adults: A population study. Am J Hum Genet. 1988;42:677–693. [PMC free article] [PubMed] [Google Scholar]
- 5.Shashi V, McConkie-Rosell A, Rosell B, Schoch K, Vellore K, McDonald M, Jiang YH, Xie P, Need A, Goldstein DB. The utility of the traditional medical genetics diagnostic evaluation in the context of next-generation sequencing for undiagnosed genetic disorders. Genet Med. 2014;16:176–182. doi: 10.1038/gim.2013.99. [DOI] [PubMed] [Google Scholar]
- 6.Hilbert JE, Ashizawa T, Day JW, Luebbe EA, Martens WB, McDermott MP, Tawil R, Thornton CA, Moxley RT., III Diagnostic odyssey of patients with myotonic dystrophy. J Neurol. 2013;260:2497–2504. doi: 10.1007/s00415-013-6993-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ng SB, Buckingham KJ, Lee C, Bigham AW, Tabor HK, Dent KM, Huff CD, Shannon PT, Jabs EW, Nickerson DA, Shendure J, Bamshad MJ. Exome sequencing identifies the cause of a mendelian disorder. Nat Genet. 2010;42:30–35. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Y, Muzny DM, Reid JG, Bainbridge MN, Willis A, Ward PA, Braxton A, Beuten J, Xia F, Niu Z, Hardison M, Person R, Bekheirnia MR, Leduc MS, Kirby A, Pham P, Scull J, Wang M, Ding Y, Plon SE, Lupski JR, Beaudet AL, Gibbs RA, Eng CM. Clinical whole-exome sequencing for the diagnosis of Mendelian disorders. N Engl J Med. 2013;369:1502–1511. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bell CJ, Dinwiddie DL, Miller NA, Hateley SL, Ganusova EE, Mudge J, Langley RJ, Zhang L, Lee CC, Schilkey FD, Sheth V, Woodward JE, Peckham HE, Schroth GP, Kim RW, Kingsmore SF. Carrier testing for severe childhood recessive diseases by next-generation sequencing. Sci Transl Med. 2011;3:65ra4. doi: 10.1126/scitranslmed.3001756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saunders CJ, Miller NA, Soden SE, Dinwiddie DL, Noll A, Alnadi NA, Andraws N, Patterson ML, Krivohlavek LA, Fellis J, Humphray S, Saffrey P, Kingsbury Z, Weir JC, Betley J, Grocock RJ, Margulies EH, Farrow EG, Artman M, Safina NP, Petrikin JE, Hall KP, Kingsmore SF. Rapid whole-genome sequencing for genetic disease diagnosis in neonatal intensive care units. Sci Transl Med. 2012;4:154ra135. doi: 10.1126/scitranslmed.3004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johansen Taber KA, Dickinson BD, Wilson M. The promise and challenges of next-generation genome sequencing for clinical care. JAMA Intern Med. 2014;174:275–280. doi: 10.1001/jamainternmed.2013.12048. [DOI] [PubMed] [Google Scholar]
- 12.Cooper GM, Shendure J. Needles in stacks of needles: Finding disease-causal variants in a wealth of genomic data. Nat Rev Genet. 2011;12:628–640. doi: 10.1038/nrg3046. [DOI] [PubMed] [Google Scholar]
- 13.Pelak K, Shianna KV, Ge D, Maia JM, Zhu M, Smith JP, Cirulli ET, Fellay J, Dickson SP, Gumbs CE, Heinzen EL, Need AC, Ruzzo EK, Singh A, Campbell CR, Hong LK, Lornsen KA, McKenzie AM, Sobreira NL, Hoover-Fong JE, Milner JD, Ottman R, Haynes BF, Goedert JJ, Goldstein DB. The characterization of twenty sequenced human genomes. PLOS Genet. 2010;6:e1001111. doi: 10.1371/journal.pgen.1001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li MX, Kwan JS, Bao SY, Yang W, Ho SL, Song YQ, Sham PC. Predicting Mendelian disease-causing non-synonymous single nucleotide variants in exome sequencing studies. PLOS Genet. 2013;9:e1003143. doi: 10.1371/journal.pgen.1003143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwarz JM, Rödelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 17.MacArthur DG, Balasubramanian S, Frankish A, Huang N, Morris J, Walter K, Jostins L, Habegger L, Pickrell JK, Montgomery SB, Albers CA, Zhang ZD, Conrad DF, Lunter G, Zheng H, Ayub Q, DePristo MA, Banks E, Hu M, Handsaker RE, Rosenfeld JA, Fromer M, Jin M, Mu XJ, Khurana E, Ye K, Kay M, Saunders GI, Suner MM, Hunt T, Barnes IH, Amid C, Carvalho-Silva DR, Bignell AH, Snow C, Yngvadottir B, Bumpstead S, Cooper DN, Xue Y, Romero IG, Wang J, Li Y, Gibbs RA, McCarroll SA, Dermitzakis ET, Pritchard JK, Barrett JC, Harrow J, Hurles ME, Gerstein MB, Tyler-Smith C 1000 Genomes Project Consortium. A systematic survey of loss-of-function variants in human protein-coding genes. Science. 2012;335:823–828. doi: 10.1126/science.1215040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meynert AM, Bicknell LS, Hurles ME, Jackson AP, Taylor MS. Quantifying single nucleotide variant detection sensitivity in exome sequencing. BMC Bioinformatics. 2013;14:195. doi: 10.1186/1471-2105-14-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ajay SS, Parker SC, Abaan HO, Fajardo KV, Margulies EH. Accurate and comprehensive sequencing of personal genomes. Genome Res. 2011;21:1498–1505. doi: 10.1101/gr.123638.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lohmueller KE, Sparsø T, Li Q, Andersson E, Korneliussen T, Albrechtsen A, Banasik K, Grarup N, Hallgrimsdottir I, Kiil K, Kilpeläinen TO, Krarup NT, Pers TH, Sanchez G, Hu Y, Degiorgio M, Jørgensen T, Sandbæk A, Lauritzen T, Brunak S, Kristiansen K, Li Y, Hansen T, Wang J, Nielsen R, Pedersen O. Whole-exome sequencing of 2,000 Danish individuals and the role of rare coding variants in type 2 diabetes. Am J Hum Genet. 2013;93:1072–1086. doi: 10.1016/j.ajhg.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams FM, Scollen S, Cao D, Memari Y, Hyde CL, Zhang B, Sidders B, Ziemek D, Shi Y, Harris J, Harrow I, Dougherty B, Malarstig A, McEwen R, Stephens JC, Patel K, Menni C, Shin SY, Hodgkiss D, Surdulescu G, He W, Jin X, McMahon SB, Soranzo N, John S, Wang J, Spector TD. Genes contributing to pain sensitivity in the normal population: An exome sequencing study. PLOS Genet. 2012;8:e1003095. doi: 10.1371/journal.pgen.1003095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rehm HL. Disease-targeted sequencing: A cornerstone in the clinic. Nat Rev Genet. 2013;14:295–300. doi: 10.1038/nrg3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi BO, Koo SK, Park MH, Rhee H, Yang SJ, Choi KG, Jung SC, Kim HS, Hyun YS, Nakhro K, Lee HJ, Woo HM, Chung KW. Exome sequencing is an efficient tool for genetic screening of Charcot–Marie–Tooth disease. Hum Mutat. 2012;33:1610–1615. doi: 10.1002/humu.22143. [DOI] [PubMed] [Google Scholar]
- 24.Ball MP, Thakuria JV, Zaranek AW, Clegg T, Rosenbaum AM, Wu X, Angrist M, Bhak J, Bobe J, Callow MJ, Cano C, Chou MF, Chung WK, Douglas SM, Estep PW, Gore A, Hulick P, Labarga A, Lee JH, Lunshof JE, Kim BC, Kim JI, Li Z, Murray MF, Nilsen GB, Peters BA, Raman AM, Rienhoff HY, Robasky K, Wheeler MT, Vandewege W, Vorhaus DB, Yang JL, Yang L, Aach J, Ashley EA, Drmanac R, Kim SJ, Li JB, Peshkin L, Seidman CE, Seo JS, Zhang K, Rehm HL, Church GM. A public resource facilitating clinical use of genomes. Proc Natl Acad Sci U S A. 2012;109:11920–11927. doi: 10.1073/pnas.1201904109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sifrim A, Popovic D, Tranchevent LC, Ardeshirdavani A, Sakai R, Konings P, Vermeesch JR, Aerts J, De Moor B, Moreau Y. eXtasy: Variant prioritization by genomic data fusion. Nat Methods. 2013;10:1083–1084. doi: 10.1038/nmeth.2656. [DOI] [PubMed] [Google Scholar]
- 26.Robinson PN, Köhler S, Oellrich S, Wang K, Mungall CJ, Lewis SE, Washington N, Bauer S, Seelow D, Krawitz P, Gilissen C, Haendel M, Smedley D Sanger Mouse Genetics Project. Improved exome prioritization of disease genes through cross-species phenotype comparison. Genome Res. 2014;24:340–348. doi: 10.1101/gr.160325.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robinson PN, Köhler S, Bauer S, Seelow D, Horn D, Mundlos S. The Human Phenotype Ontology: A tool for annotating and analyzing human hereditary disease. Am J Hum Genet. 2008;83:610–615. doi: 10.1016/j.ajhg.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Köhler S, Schulz MH, Krawitz P, Bauer S, Dölken S, Ott CE, Mundlos C, Horn D, Mundlos S, Robinson PN. Clinical diagnostics in human genetics with semantic similarity searches in ontologies. Am J Hum Genet. 2009;85:457–464. doi: 10.1016/j.ajhg.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bauer S, Köhler S, Schulz MH, Robinson PN. Bayesian ontology querying for accurate and noise-tolerant semantic searches. Bioinformatics. 2012;28:2502–2508. doi: 10.1093/bioinformatics/bts471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Doelken SC, Köhler S, Mungall CJ, Gkoutos GV, Ruef BJ, Smith C, Smedley D, Bauer S, Klopocki E, Schofield PN, Westerfield M, Robinson PN, Lewis SE. Phenotypic overlap in the contribution of individual genes to CNV pathogenicity revealed by cross-species computational analysis of single-gene mutations in humans, mice and zebrafish. Dis Model Mech. 2013;6:358–372. doi: 10.1242/dmm.010322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoehndorf R, Schofield PN, Gkoutos GV. PhenomeNET: A whole-phenome approach to disease gene discovery. Nucleic Acids Res. 2011;39:e119. doi: 10.1093/nar/gkr538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hwang T, Atluri G, Xie M, Dey S, Hong C, Kumar V, Kuang R. Co-clustering phenome–genome for phenotype classification and disease gene discovery. Nucleic Acids Res. 2012;40:e146. doi: 10.1093/nar/gks615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tarpey PS, Smith R, Pleasance E, Whibley A, Edkins S, Hardy C, O’Meara S, Latimer C, Dicks E, Menzies A, Stephens P, Blow M, Greenman C, Xue Y, Tyler-Smith C, Thompson D, Gray K, Andrews J, Barthorpe S, Buck G, Cole J, Dunmore R, Jones D, Maddison M, Mironenko T, Turner R, Turrell K, Varian J, West S, Widaa S, Wray P, Teague J, Butler A, Jenkinson A, Jia M, Richardson D, Shepherd R, Wooster R, Tejada MI, Martinez F, Carvill G, Goliath R, de Brouwer AP, van Bokhoven H, Van Esch H, Chelly J, Raynaud M, Ropers HH, Abidi FE, Srivastava AK, Cox J, Luo Y, Mallya U, Moon J, Parnau J, Mohammed S, Tolmie JL, Shoubridge C, Corbett M, Gardner A, Haan E, Rujirabanjerd S, Shaw M, Vandeleur L, Fullston T, Easton DF, Boyle J, Partington M, Hackett A, Field M, Skinner C, Stevenson RE, Bobrow M, Turner G, Schwartz CE, Gecz J, Raymond FL, Futreal PA, Stratton MR. A systematic, large-scale resequencing screen of X-chromosome coding exons in mental retardation. Nat Genet. 2009;41:535–543. doi: 10.1038/ng.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cho G, Lim Y, Golden JA. XLMR candidate mouse gene, Zcchc12 (Sizn1) is a novel marker of Cajal–Retzius cells. Gene Expr Patterns. 2011;11:216–220. doi: 10.1016/j.gep.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Najmabadi H, Hu H, Garshasbi M, Zemojtel T, Abedini SS, Chen W, Hosseini M, Behjati F, Haas S, Jamali P, Zecha A, Mohseni M, Püttmann L, Vahid LN, Jensen C, Moheb LA, Bienek M, Larti F, Mueller I, Weissmann R, Darvish H, Wrogemann K, Hadavi V, Lipkowitz B, Esmaeeli-Nieh S, Wieczorek D, Kariminejad R, Firouzabadi SG, Cohen M, Fattahi Z, Rost I, Mojahedi F, Hertzberg C, Dehghan A, Rajab A, Banavandi MJ, Hoffer J, Falah M, Musante L, Kalscheuer V, Ullmann R, Kuss AW, Tzschach A, Kahrizi K, Ropers HH. Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature. 2011;478:57–63. doi: 10.1038/nature10423. [DOI] [PubMed] [Google Scholar]
- 36.Kapetanaki MG, Guerrero-Santoro J, Bisi DC, Hsieh CL, Rapić-Otrin V, Levine AS. The DDB1-CUL4ADDB2 ubiquitin ligase is deficient in xeroderma pigmentosum group E and targets histone H2A at UV-damaged DNA sites. Proc Natl Acad Sci U S A. 2006;103:2588–2593. doi: 10.1073/pnas.0511160103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rauch A, Wieczorek D, Graf E, Wieland T, Endele S, Schwarzmayr T, Albrecht B, Bartholdi D, Beygo J, Di Donato N, Dufke A, Cremer K, Hempel M, Horn D, Hoyer J, Joset P, Röpke A, Moog U, Riess A, Thiel CT, Tzschach A, Wiesener A, Wohlleber E, Zweier C, Ekici AB, Zink AM, Rump A, Meisinger C, Grallert H, Sticht H, Schenck A, Engels H, Rappold G, Schröck E, Wieacker P, Riess O, Meitinger T, Reis A, Strom TM. Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: An exome sequencing study. Lancet. 2012;380:1674–1682. doi: 10.1016/S0140-6736(12)61480-9. [DOI] [PubMed] [Google Scholar]
- 38.Gnirke A, Melnikov A, Maguire J, Rogov P, LeProust EM, Brockman W, Fennell T, Giannoukos G, Fisher S, Russ C, Gabriel S, Jaffe DB, Lander ES, Nusbaum C. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol. 2009;27:182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stenson PD, Mort M, Ball EV, Shaw K, Phillips A, Cooper DN. The Human Gene Mutation Database: Building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum Genet. 2014;133:1–9. doi: 10.1007/s00439-013-1358-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robinson PN. Deep phenotyping for precision medicine. Hum Mutat. 2012;33:777–780. doi: 10.1002/humu.22080. [DOI] [PubMed] [Google Scholar]
- 41.Landrum MJ, Lee JM, Riley GR, Jang W, Rubinstein WS, Church DM, Maglott DR. ClinVar: Public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42:D980–D985. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kingsmore SF, Saunders CJ. Deep sequencing of patient genomes for disease diagnosis: When will it become routine? Sci Transl Med. 2011;3:87ps23. doi: 10.1126/scitranslmed.3002695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oti M, Huynen MA, Brunner HG. The biological coherence of human phenome databases. Am J Hum Genet. 2009;85:801–808. doi: 10.1016/j.ajhg.2009.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robinson PN, Webber C. Phenotype ontologies and cross-species analysis for translational research. PLOS Genet. 2014;10:e1004268. doi: 10.1371/journal.pgen.1004268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Al-Qattan MM, Al Abdulkareem I, Al Haidan Y, Al Balwi M. A novel mutation in the SHH long-range regulator (ZRS) is associated with preaxial polydactyly, triphalangeal thumb, and severe radial ray deficiency. Am J Med Genet A. 2012;158A:2610–2615. doi: 10.1002/ajmg.a.35584. [DOI] [PubMed] [Google Scholar]
- 46.de Ligt J, Willemsen MH, van Bon BW, Kleefstra T, Yntema HG, Kroes T, Vultovan Silfhout AT, Koolen DA, de Vries P, Gilissen C, del Rosario M, Hoischen A, Scheffer H, de Vries BB, Brunner HG, Veltman JA, Vissers LE. Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med. 2012;367:1921–1929. doi: 10.1056/NEJMoa1206524. [DOI] [PubMed] [Google Scholar]
- 47.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jäger M, Wang K, Bauer S, Smedley D, Krawitz P, Robinson PN. Jannovar: A Java library for exome annotation. Hum Mutat. 2014;35:548–555. doi: 10.1002/humu.22531. [DOI] [PubMed] [Google Scholar]
- 49.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu X, Jian X, Boerwinkle E. dbNSFP v2.0: A database of human non-synonymous SNVs and their functional predictions and annotations. Hum Mutat. 2013;34:E2393–E2402. doi: 10.1002/humu.22376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Karolchik D, Barber GP, Casper J, Clawson H, Cline MS, Diekhans M, Dreszer TR, Fujita PA, Guruvadoo L, Haeussler M, Harte RA, Heitner S, Hinrichs AS, Learned K, Lee BT, Li CH, Raney BJ, Rhead B, Rosenbloom KR, Sloan CA, Speir ML, Zweig AS, Haussler D, Kuhn RM, Kent WJ. The UCSC Genome Browser database: 2014 update. Nucleic Acids Res. 2014;42:D764–D770. doi: 10.1093/nar/gkt1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.NCBI Resource Coordinators. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2014;42:D7–D17. doi: 10.1093/nar/gkt1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jones WD, Dafou D, McEntagart M, Woollard WJ, Elmslie FV, Holder-Espinasse M, Irving M, Saggar AK, Smithson S, Trembath RC, Deshpande C, Simpson MA. De novo mutations in MLL cause Wiedemann-Steiner syndrome. Am J Hum Genet. 2012;91:358–364. doi: 10.1016/j.ajhg.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Berryer MH, Hamdan FF, Klitten LL, Møller RS, Carmant L, Schwartzentruber J, Patry L, Dobrzeniecka S, Rochefort D, Neugnot-Cerioli M, Lacaille JC, Niu Z, Eng CM, Yang Y, Palardy S, Belhumeur C, Rouleau GA, Tommerup N, Immken L, Beauchamp MH, Patel GS, Majewski J, Tarnopolsky MA, Scheffzek K, Hjalgrim H, Michaud JL, Di Cristo G. Mutations in SYNGAP1 cause intellectual disability, autism, and a specific form of epilepsy by inducing haploinsufficiency. Hum Mutat. 2013;34:385–394. doi: 10.1002/humu.22248. [DOI] [PubMed] [Google Scholar]
- 56.Jay S, Wiberg A, Swan M, Lester T, Williams LJ, Taylor IB, Johnson D, Wilkie AO. The fibroblast growth factor receptor 2 p.Ala172Phe mutation in Pfeiffer syndrome—History repeating itself. Am J Med Genet A. 2013;161A:1158–1163. doi: 10.1002/ajmg.a.35842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Iqbal Z, Cejudo-Martin P, de Brouwer A, van der Zwaag B, Ruiz-Lozano P, Scimia MC, Lindsey JD, Weinreb R, Albrecht B, Megarbane A, Alanay Y, Ben-Neriah Z, Amenduni M, Artuso R, Veltman JA, van Beusekom E, Oudakker A, Millán JL, Hennekam R, Hamel B, Courtneidge SA, van Bokhoven H. Disruption of the podosome adaptor protein TKS4 (SH3PXD2B) causes the skeletal dysplasia, eye, and cardiac abnormalities of Frank-Ter Haar syndrome. Am J Hum Genet. 2010;86:254–261. doi: 10.1016/j.ajhg.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kurian MA, Zhen J, Cheng SY, Li Y, Mordekar SR, Jardine P, Morgan NV, Meyer E, Tee L, Pasha S, Wassmer E, Heales SJ, Gissen P, Reith ME, Maher ER. Homozygous loss-of-function mutations in the gene encoding the dopamine transporter are associated with infantile parkinsonism-dystonia. J Clin Invest. 2009;119:1595–1603. doi: 10.1172/JCI39060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weiss MJ, Cole DE, Ray K, Whyte MP, Lafferty MA, Mulivor RA, Harris H. A missense mutation in the human liver/bone/kidney alkaline phosphatase gene causing a lethal form of hypophosphatasia. Proc Natl Acad Sci U S A. 1988;85:7666–7669. doi: 10.1073/pnas.85.20.7666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brooks SP, Ebenezer ND, Poopalasundaram S, Lehmann OJ, Moore AT, Hardcastle AJ. Identification of the gene for Nance-Horan syndrome (NHS) J Med Genet. 2004;41:768–771. doi: 10.1136/jmg.2004.022517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Courcet JB, Faivre L, Malzac P, Masurel-Paulet A, Lopez E, Callier P, Lambert L, Lemesle M, Thevenon J, Gigot N, Duplomb L, Ragon C, Marle N, Mosca-Boidron AL, Huet F, Philippe C, Moncla A, Thauvin-Robinet C. The DYRK1A gene is a cause of syndromic intellectual disability with severe microcephaly and epilepsy. J Med Genet. 2012;49:731–736. doi: 10.1136/jmedgenet-2012-101251. [DOI] [PubMed] [Google Scholar]
- 62.Sun M, Goldin E, Stahl S, Falardeau JL, Kennedy JC, Acierno JS, Jr, Bove C, Kaneski CR, Nagle J, Bromley MC, Colman M, Schiffmann R, Slaugenhaupt SA. Mucolipidosis type IV is caused by mutations in a gene encoding a novel transient receptor potential channel. Hum Mol Genet. 2000;9:2471–2478. doi: 10.1093/hmg/9.17.2471. [DOI] [PubMed] [Google Scholar]
- 63.Johnston JJ, Teer JK, Cherukuri PF, Hansen NF, Loftus SK, Chong K, Mullikin JC, Biesecker LG NIH Intramural Sequencing Center (NISC) Massively parallel sequencing of exons on the X chromosome identifies RBM10 as the gene that causes a syndromic form of cleft palate. Am J Hum Genet. 2010;86:743–748. doi: 10.1016/j.ajhg.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hamdan FF, Gauthier J, Spiegelman D, Noreau A, Yang Y, Pellerin S, Dobrzeniecka S, Côté M, Perreau-Linck E, Carmant L, D’Anjou G, Fombonne E, Addington AM, Rapoport JL, Delisi LE, Krebs MO, Mouaffak F, Joober R, Mottron L, Drapeau P, Marineau C, Lafrenière RG, Lacaille JC, Rouleau GA, Michaud JL. Synapse to Disease Group, Mutations in SYNGAP1 in autosomal nonsyndromic mental retardation. N Engl J Med. 2009;360:599–605. doi: 10.1056/NEJMoa0805392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Draguet C, Gillerot Y, Mornet E. Childhood hypophosphatasia: A case report due to a novel mutation. Arch Pediatr. 2004;11:440–443. doi: 10.1016/j.arcped.2004.02.018. [DOI] [PubMed] [Google Scholar]
- 66.Spentchian M, Brun-Heath I, Taillandier A, Fauvert D, Serre JL, Simon-Bouy B, Carvalho F, Grochova I, Mehta SG, Müller G, Oberstein SL, Ogur G, Sharif S, Mornet E. Characterization of missense mutations and large deletions in the ALPL gene by sequencing and quantitative multiplex PCR of short fragments. Genet Test. 2006;10:252–257. doi: 10.1089/gte.2006.10.252. [DOI] [PubMed] [Google Scholar]
- 67.Brun-Heath I, Lia-Baldini AS, Maillard S, Taillandier A, Utsch B, Nunes ME, Serre JL, Mornet E. Delayed transport of tissue-nonspecific alkaline phosphatase with missense mutations causing hypophosphatasia. Eur J Med Genet. 2007;50:367–378. doi: 10.1016/j.ejmg.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 68.Raychowdhury MK, González-Perrett S, Montalbetti N, Timpanaro GA, Chasan B, Goldmann WH, Stahl S, Cooney A, Goldin E, Cantiello HF. Molecular pathophysiology of mucolipidosis type IV: pH dysregulation of the mucolipin-1 cation channel. Hum Mol Genet. 2004;13:617–627. doi: 10.1093/hmg/ddh067. [DOI] [PubMed] [Google Scholar]
- 69.Gripp KW, Hopkins E, Johnston JJ, Krause C, Dobyns WB, Biesecker LG. Long-term survival in TARP syndrome and confirmation of RBM10 as the disease-causing gene. Am J Med Genet A. 2011;155A:2516–2520. doi: 10.1002/ajmg.a.34190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Reese MG, Eeckman FH, Kulp D, Haussler D. Improved splice site detection in Genie. J Comput Biol. 1997;4:311–323. doi: 10.1089/cmb.1997.4.311. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
www.sciencetranslationalmedicine.org/cgi/content/full/6/252/252ra123/DC1 Fig S1. Distribution of the coverage fraction for all sequenced 96 samples. Fig. S2. Computational evaluation of PhenIX. Table S1. Percentage of target bases that exceed coverages of 10, 20, …, 100 reads. Table S2. Read alignment and coverage summary statistics.
Table S3. Average number of variants called only from the original BAM files from the DAG panels. Table S4. Detailed clinical and molecular findings for the 11 individuals in whom a previously unknown diagnosis was clarified by PhenIX analysis. Table S5. Clinical presentation of 29 patients for whom PhenIX analysis failed to reveal a molecular diagnosis.
Table S6. List of genes (with references) present in the DAG panel. References (64-70)


