Introduction
Toxin production by pathogenic microorganisms likely serves to (i) protect against phagocytosis by predatory cells, (ii) aid in penetrating tissue barriers, (iii) promote nutrient release, or (iv) alter cellular architecture and metabolism in ways that facilitate the establishment of a niche for colonization and replication. Useful enzymatic targets for some bacterial toxins may be similar between prokaryotic and eukaryotic environments, necessitating a need for eukaryotic-specific cofactors to regulate toxin activity. Host cell—derived factors may impart localization properties to an effector, induce folding events, provide a platform for the inhibition of cellular processes or support greater substrate promiscuity (Fig 1). The aim of this review is to describe the diversity of bacterial effectors known to require, or to be stimulated by eukaryotic cofactors and to integrate new ideas regarding the structural and functional implications of this relationship (Table 1).
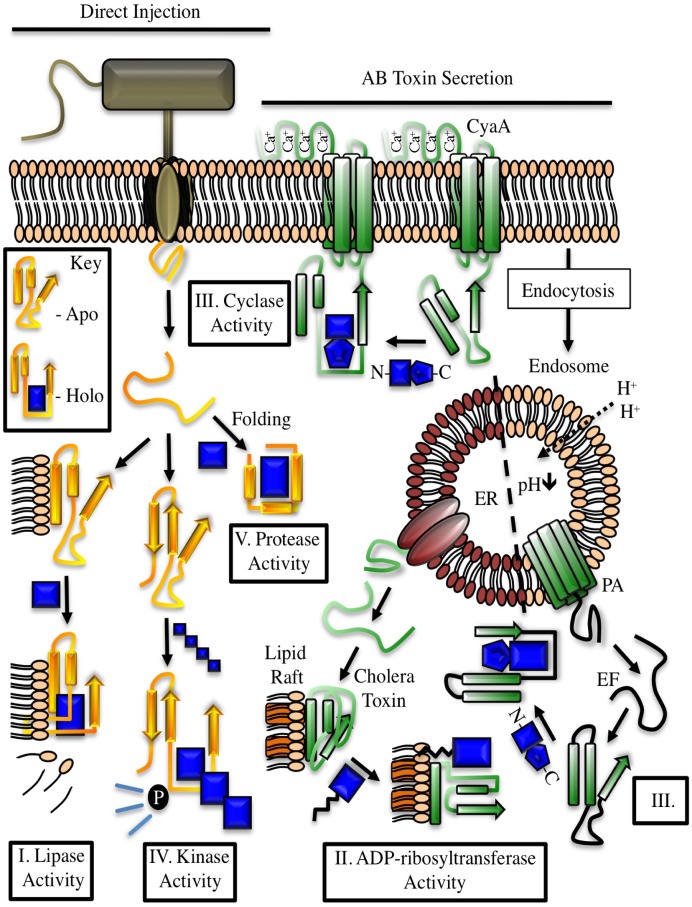
Fig 1. Examples of cofactor regulation of secreted bacterial enzymes.
Toxins delivered to their target cell by either direct injection (yellow) or cell surface binding and translocation (green) can be host cofactor activated. These enzymes generally contain dynamic structures that assume a catalytically active fold upon complex formation with a host factor. The key depicts this process through cofactor-mediated organization (blue) of an unstructured sequence. Apo, the apoenzyme catalytically-inactive state. Holo, the holoenzyme active state in which the toxin is in complex with its cofactor. CyaA, the plasma membrane-localized nucleotide cyclase toxin from Bordetella pertussis complexed to calcium ions and calmodulin. EF, edema factor from Bacillus anthracis, binds to calmodulin in a different orientation than CyaA. PA, protective antigen. The cofactor for cholera toxin (ARF) is shown in the myristolated, GTP-bound form. ER, endoplasmic reticulum, with cholera toxin peptide being secreted through ER protein channels.
Table 1. Cross kingdom enzyme stimulators for secreted bacterial toxins.
| Strain | Enzyme | Stimulated Activity | Activator |
|---|---|---|---|
| Pseudomonas aeruginosa | ExoU | PLA2/Lyso-PLA | Ubiquitin |
| Pseudomonas fluorescens | ExoU-like | PLA2 | Ubiquitin |
| Photorhabdus asymbiotica | LopU | PLA2 | Ubiquitin |
| Burkholderia thailandensis | ExoU-like | PLA2 | Ubiquitin |
| Rickettsia prowazekii | RP534 | PLA1/PLA2/Lyso-PLA2 | bSOD(ubiquitin) |
| Rickettsia typhi | RT0590/Pat1 | PLA2 | bSOD(ubiquitin) |
| Rickettsia typhi | RT0522/Pat2 | PLA2 | bSOD(ubiquitin) |
| Legionella pneumophila | VipD | PLA1 | Rab5 |
| Salmonella typhimurium | SseJ | Esterase | GTP-RhoA |
| Bordetella pertussis | CyaA | Nucleotidyl cyclase | Calmodulin |
| Bacillus anthracis | Edema factor | Nucleotidyl cyclase | Calmodulin |
| Pseudomonas aeruginosa | ExoY | Nucleotidyl cyclase | Actin? |
| Aeromonas hydrophila | AexU | ADP-ribosyltransferase | 14-3-3 |
| Aeromonas spp. | AexT | ADP-ribosyltransferase | 14-3-3 |
| Pseudomonas aeruginosa | ExoS | ADP-ribosyltransferase | 14-3-3 |
| Pseudomonas aeruginosa | ExoT | ADP-ribosyltransferase | 14-3-3 |
| Vibrio parahaemolyticus | VopT | ADP-ribosyltransferase | 14-3-3 |
| Vibrio cholerae | Cholera toxin | ADP-ribosyltransferase | ARF |
| Escherichia coli | Heat labile toxin | ADP-ribosyltransferase | ARF |
| Shigella flexneri | OspG | Kinase | E2-Ubiquitin |
| Yersinia pestis | YopO/YpkA | Kinase | G-actin |
| Pseudomonas syringae | HopZ1 | Acetyl transferase | IP6 |
| Yersinia spp. | YopJ | Acetyl transferase | IP6 |
| Salmonella typhimurium | AvrA | Acetyl transferase | IP6 |
| Vibrio parahaemolyticus | VPA1380 | Cysteine protease | IP6 |
| Pseudomonas syringae | AvrRpt2 | Cysteine protease | Cyclophilin |
| Totals | 26 | 9 | 12 |
I. Phospholipases
Many bacterial toxins contain catalytic domains with homology to plant patatins, which are lipid acyl hydrolases found in potato tubers. Cofactor activation of phospholipase activity is best characterized for ExoU, a lipid membrane—hydrolyzing protein encoded by the opportunistic pathogen P. aeruginosa. Monoubiquitin, ubiquitin polymers, and ubiquitylated proteins are capable of activating ExoU [1]. Bioinformatic analyses identified at least 17 additional bacterial patatin-like phospholipases that fit the criteria for ubiquitin-mediated activation [2]. Functional studies of a selected subset of enzymes demonstrated that ubiquitin activates phospholipases from P. asymbiotica, B. thailandensis, and P. fluorescens [2].
ExoU orthologs are also found in frank pathogens within the Rickettsiae, Legionellae, and Salmonellae. R. typhus encodes ExoU homologs, Pat1 and Pat2 [3,4]. Both proteins are activated by preparations of bovine SOD1 (bSOD1 [3,4]), which may provide a source of ubiquitin [1]. Additionally, R. prowazekii, the etiologic agent of typhus, displays enhanced enzymatic activity in the presence of bSOD1 (ubiquitin, [5]). Rickettsial-derived PLA enzymes function in cellular entry [6], phagosomal escape, and noncytolytic free fatty acid release [7].
VipD lipase from L. pneumophila displays a Rab5-dependent PLA1 activity [8] that targets the enzyme to endosomes resulting in inhibition of phagosome maturation. SseJ, a glycerophospholipid-cholesterol acyltransfersase (GCAT)-like enzyme from Salmonella, is activated by GTP-RhoA [9] likely for the temporal regulation of cholesterol metabolism in infected cells [10]. The Y. enterocolitica homolog to SseJ, YspM, may have a similar function [11].
II. Transferases
Most cofactor-stimulated transferases catalyze the transfer of an ADP-ribose moiety from nicotinamide adenine dinucleotide to a target protein. Exoenzyme S from P. aeruginosa was one of the first identified bacterial ADP-ribosyltransferases requiring a cofactor for activation [12]. Members of the 14-3-3 family of eukaryotic scaffolding proteins stimulate ADP-ribosylation of a variety of targets causing disruptions in cytoskeletal integrity and vesicular trafficking [13]. Homologous enzymes are present in A. hydrophila [14] and V. parahaemolyticus [15].
Another enzyme from P. aeruginosa, ExoT, is similar (76% identity) to ExoS and requires 14-3-3 proteins as cofactors, but has limited target recognition and is not overtly cytotoxic [16]. CrkI and CrkII are the only proteins modified by ExoT [17]. ADP-ribosylation uncouples integrin signaling and alters the cytoskeleton. A. hydrophila and A. salmonicida encode enzymes similar to ExoT/S termed AexT [18].
Bacterial effectors with acetyltransferase activity include YopJ, a type III secreted enzyme from Yersinia that inhibits immune responses through acetylation of several residues in the kinases involved in the MAPK and NFκB pathways [19]. Acetyltransferase activity is stimulated by a highly abundant eukaryotic signaling carbohydrate, inositol hexakisphosphate (IP6, [20]). P. syringae secretes an IP6-activated acetyltransferase, HopZ1, which interferes with plant cell cytoskeletal networks and downstream cell wall—mediated immune responses by acetylating microtubules [21]. The binding of IP6 to S. typhimurium AvrA suggests that stimulation of acetyltransferase activity in this family of enzymes likely involves allosteric conformational changes [20]. Structural investigations of V. cholerae RTX toxin [22] and Clostridium difficile toxin A [23] suggest that IP6 interactions with bacterial enzymes, and their subsequent allosteric conformational changes, may be a conserved strategy.
ADP-ribosylation factors (ARFs) have been identified as stimulators of type I and type II heat labile toxin (LT) activity from enteric bacteria. Cholera toxin from V. cholerae, LT-I and LT-II from E. coli, are the most studied enzymes of this family to date; each are highly homologous in sequence and structure. Cholera toxin A1 subunit was shown to fold on host cell lipid rafts [24] before interacting with ARF proteins in their GTP-bound state [25]. Activated enzymes ADP-ribosylate the alpha subunit of the heterotrimeric G stimulatory protein (Gs), locking it into the GTP-bound form. This stimulates adenylyl cyclase activity and generates supraphysiological amounts of cAMP, deregulating the chloride balance in the intestinal lumen to cause diarrhea.
III. Nucleotide Cyclases
Cofactor-stimulated nucleotide cyclases include the adenylyl cyclases edema factor (EF) of anthrax toxin and CyaA of B. pertussis. These enzymes contain structurally similar components but differ in affinity and binding mechanism for their eukaryotic activator, calmodulin (CaM). Edema factor has three domains: a protective antigen-binding domain, the adenylyl cyclase core domain, and an alpha-helical domain. The helical domain interacts with the core domain to maintain an unstructured activation loop. The N-terminal domain of CaM interacts with the EF helical domain, causing a conformational switch between the helical and core domains [26]. The C-terminal domain of CaM then docks into a crevice between the helical and core domains, stabilizing the catalytic loop [27].
In contrast to soluble EF, CyaA localizes to the cytosolic interface at the plasma membrane. The CaM binding domain involves completely different sequences with an affinity that is roughly 100-fold stronger as compared to EF. N- and C-terminal domains of CaM participate in binding, stabilization, and activation of CyaA. The C-terminal domain stabilizes a catalytic loop, and the N-terminal domain is postulated to bind a β-hairpin motif of CyaA to promote substrate (ATP) binding within the catalytic chamber [28].
Exoenzyme Y is a type III-secreted, cofactor-stimulated adenylyl cyclase from P. aeruginosa [29]. Functionally, ExoY cyclase activity disrupts the actin cytoskeleton and is correlated with Tau hyperphosphorylation, which leads to microtubule breakdown, endothelial cell gap formation, and increased tissue permeability [30]. The cofactor for ExoY remains to be established, but preliminary work suggests that host cell actin stimulates cyclase activity [31]. Sensitive detection methods demonstrate that ExoY, EF, and CyaA have broad nucleotidyl cyclase activity and catalyze the formation of cCMP and cUMP (or cGMP and cUMP with ExoY) in addition to cAMP [32,33]. The production of cCMP and cUMP during in vivo infection may provide an experimental system to explore the roles of these cyclic nucleotides as new second messengers [34].
IV. Kinases
Yersinia species inject actin-stimulated effector kinases into host cells [35]. YopO or YpkA binds to actin in a manner that does not interfere with the association of other actin-targeting proteins, while simultaneously blocking actin incorporation into a filament. Thus, actin facilitates catalysis by stabilizing the enzyme’s catalytic loop while serving as a platform for phosphorylation of other actin-binding proteins [36].
Another kinase characterized from Shigella species, but also present in strains of Yersinia and enterohemorrhagic E. coli, is OspG [37–39]. OspG is involved in attenuating NFκB signaling by interfering with IκBα degradation [40]. Ubiquitin and ubiquitin chains were first identified as stimulators of OspG kinase activity in vitro [39]. Two groups subsequently solved structures of OspG in complex with E2 ubiquitin-conjugating enzymes covalently linked to monoubiquitin, UbcH5a~Ub [38] and UbcH7~Ub [37]. At least 10 different E2~Ub conjugates appear to be able to activate OspG, suggesting that there may not be a strict preference towards a particular E2.
V. Proteases
V. parahaemolyticus has recently been shown to secrete an IP6-activated enzyme, VPA1380, with homology to the cysteine protease domains of multifunctional-autoprocessing RTX (MARTX) domains from Vibrio and large clostridial toxins A and B [41]. The mechanism of activation, toxicity, and target substrates are yet to be elucidated. P. syringae secretes a protease, AvrRpt2, which is homologous to the staphopain cysteine proteases that are activated through interactions with cyclophilins [42]. AvrRpt2 is predominantly in an unstructured coil conformation until binding to a host cyclophilin, which induces folding into a stable conformer [43]. Analysis of protease activity supports a model in which AvrRpt2 must be in complex with cyclophilin cofactor for maximal enzymatic activity [44].
Conclusions
Numerous secreted bacterial enzymes interact with host cell factors to ensure enzymatic activation in the correct environment, and in some cases, trafficking to a specific location within the host. Common properties of the activators are that they are ubiquitously distributed throughout the eukaryotic kingdom, are present in high concentration, and play roles in cell signaling. The prevailing theme of a high cellular concentration may facilitate the required conditions for toxins to establish a cross kingdom binding partner as they evolved [45]. In this way, highly regulated and flexible proteins that are easily translocated across lipid barriers could be allosterically reorganized to a stable and active conformation. Importantly, bacterial utilization of host cell machinery to ensure maximal toxin activity in the correct environment minimizes the maintenance of bacterial genetic information.
Funding Statement
This work was supported by National Institutes of Health / National Institute of Allergy and Infectious Disease grant 1R01 AI104922 to DWF. and National Institutes of Health / General Medicine grant 1R01 GM114234 to JBF. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Anderson DM, Schmalzer KM, Sato H, Casey M, Terhune SS, Haas AL, et al. Ubiquitin and ubiquitin-modified proteins activate the Pseudomonas aeruginosa T3SS cytotoxin, ExoU. Molecular microbiology. 2011;82(6):1454–67. 10.1111/j.1365-2958.2011.07904.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anderson DM, Sato H, Dirck AT, Feix JB, Frank DW. Ubiquitin activates patatin-like phospholipases from multiple bacterial species. Journal of bacteriology. 2015;197(3):529–41. 10.1128/JB.02402-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rahman MS, Ammerman NC, Sears KT, Ceraul SM, Azad AF. Functional characterization of a phospholipase A(2) homolog from Rickettsia typhi . Journal of bacteriology. 2010;192(13):3294–303. 10.1128/JB.00155-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rahman MS, Gillespie JJ, Kaur SJ, Sears KT, Ceraul SM, Beier-Sexton M, et al. Rickettsia typhi possesses phospholipase A2 enzymes that are involved in infection of host cells. PLoS pathogens. 2013;9(6):e1003399 10.1371/journal.ppat.1003399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Housley NA, Winkler HH, Audia JP. The Rickettsia prowazekii ExoU homologue possesses phospholipase A1 (PLA1), PLA2, and lyso-PLA2 activities and can function in the absence of any eukaryotic cofactors in vitro. Journal of bacteriology. 2011;193(18):4634–42. 10.1128/JB.00141-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Silverman DJ, Santucci LA, Meyers N, Sekeyova Z. Penetration of host cells by Rickettsia rickettsii appears to be mediated by a phospholipase of rickettsial origin. Infection and immunity. 1992;60(7):2733–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Winkler HH, Daugherty RM. Phospholipase A activity associated with the growth of Rickettsia prowazekii in L929 cells. Infection and immunity. 1989;57(1):36–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gaspar AH, Machner MP. VipD is a Rab5-activated phospholipase A1 that protects Legionella pneumophila from endosomal fusion. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(12):4560–5. 10.1073/pnas.1316376111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Christen M, Coye LH, Hontz JS, LaRock DL, Pfuetzner RA, Megha, et al. Activation of a bacterial virulence protein by the GTPase RhoA. Science signaling. 2009;2(95):ra71 10.1126/scisignal.2000430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kolodziejek AM, Miller SI. Salmonella modulation of the phagosome membrane, role of SseJ. Cellular microbiology. 2015;17(3):333–41. 10.1111/cmi.12420 [DOI] [PubMed] [Google Scholar]
- 11. Witowski SE, Walker KA, Miller VL. YspM, a newly identified Ysa type III secreted protein of Yersinia enterocolitica . Journal of bacteriology. 2008;190(22):7315–25. 10.1128/JB.00861-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Coburn J, Kane AV, Feig L, Gill DM. Pseudomonas aeruginosa exoenzyme S requires a eukaryotic protein for ADP-ribosyltransferase activity. The Journal of biological chemistry. 1991;266(10):6438–46. [PubMed] [Google Scholar]
- 13. Fu H, Coburn J, Collier RJ. The eukaryotic host factor that activates exoenzyme S of Pseudomonas aeruginosa is a member of the 14-3-3 protein family. Proceedings of the National Academy of Sciences of the United States of America. 1993;90(6):2320–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sha J, Wang SF, Suarez G, Sierra JC, Fadl AA, Erova TE, et al. Further characterization of a type III secretion system (T3SS) and of a new effector protein from a clinical isolate of Aeromonas hydrophila—part I. Microbial pathogenesis. 2007;43(4):127–46. [DOI] [PubMed] [Google Scholar]
- 15. Kodama T, Rokuda M, Park KS, Cantarelli VV, Matsuda S, Iida T, et al. Identification and characterization of VopT, a novel ADP-ribosyltransferase effector protein secreted via the Vibrio parahaemolyticus type III secretion system 2. Cellular microbiology. 2007;9(11):2598–609. [DOI] [PubMed] [Google Scholar]
- 16. Yahr TL, Barbieri JT, Frank DW. Genetic relationship between the 53- and 49-kilodalton forms of exoenzyme S from Pseudomonas aeruginosa . Journal of bacteriology. 1996;178(5):1412–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sun J, Barbieri JT. Pseudomonas aeruginosa ExoT ADP-ribosylates CT10 regulator of kinase (Crk) proteins. The Journal of biological chemistry. 2003;278(35):32794–800. [DOI] [PubMed] [Google Scholar]
- 18. Vilches S, Wilhelms M, Yu HB, Leung KY, Tomas JM, Merino S. Aeromonas hydrophila AH-3 AexT is an ADP-ribosylating toxin secreted through the type III secretion system. Microbial pathogenesis. 2008;44(1):1–12. [DOI] [PubMed] [Google Scholar]
- 19. Orth K, Palmer LE, Bao ZQ, Stewart S, Rudolph AE, Bliska JB, et al. Inhibition of the mitogen-activated protein kinase kinase superfamily by a Yersinia effector. Science. 1999;285(5435):1920–3. [DOI] [PubMed] [Google Scholar]
- 20. Mittal R, Peak-Chew SY, Sade RS, Vallis Y, McMahon HT. The acetyltransferase activity of the bacterial toxin YopJ of Yersinia is activated by eukaryotic host cell inositol hexakisphosphate. The Journal of biological chemistry. 2010;285(26):19927–34. 10.1074/jbc.M110.126581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee AH, Hurley B, Felsensteiner C, Yea C, Ckurshumova W, Bartetzko V, et al. A bacterial acetyltransferase destroys plant microtubule networks and blocks secretion. PLoS pathogens. 2012;8(2):e1002523 10.1371/journal.ppat.1002523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lupardus PJ, Shen A, Bogyo M, Garcia KC. Small molecule-induced allosteric activation of the Vibrio cholerae RTX cysteine protease domain. Science. 2008;322(5899):265–8. 10.1126/science.1162403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pruitt RN, Chagot B, Cover M, Chazin WJ, Spiller B, Lacy DB. Structure-function analysis of inositol hexakisphosphate-induced autoprocessing in Clostridium difficile toxin A. The Journal of biological chemistry. 2009;284(33):21934–40. 10.1074/jbc.M109.018929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ray S, Taylor M, Banerjee T, Tatulian SA, Teter K. Lipid rafts alter the stability and activity of the cholera toxin A1 subunit. The Journal of biological chemistry. 2012;287(36):30395–405. 10.1074/jbc.M112.385575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kahn RA, Gilman AG. The protein cofactor necessary for ADP-ribosylation of Gs by cholera toxin is itself a GTP binding protein. The Journal of biological chemistry. 1986;261(17):7906–11. [PubMed] [Google Scholar]
- 26. Drum CL, Yan SZ, Bard J, Shen YQ, Lu D, Soelaiman S, et al. Structural basis for the activation of anthrax adenylyl cyclase exotoxin by calmodulin. Nature. 2002;415(6870):396–402. [DOI] [PubMed] [Google Scholar]
- 27. Tang WJ, Guo Q. The adenylyl cyclase activity of anthrax edema factor. Molecular aspects of medicine. 2009;30(6):423–30. 10.1016/j.mam.2009.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Guo Q, Jureller JE, Warren JT, Solomaha E, Florian J, Tang WJ. Protein-protein docking and analysis reveal that two homologous bacterial adenylyl cyclase toxins interact with calmodulin differently. The Journal of biological chemistry. 2008;283(35):23836–45. 10.1074/jbc.M802168200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yahr TL, Vallis AJ, Hancock MK, Barbieri JT, Frank DW. ExoY, an adenylate cyclase secreted by the Pseudomonas aeruginosa type III system. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(23):13899–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ochoa CD, Alexeyev M, Pastukh V, Balczon R, Stevens T. Pseudomonas aeruginosa exotoxin Y is a promiscuous cyclase that increases endothelial tau phosphorylation and permeability. The Journal of biological chemistry. 2012;287(30):25407–18. 10.1074/jbc.M111.301440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schneider EH, Seifert R. Report on the Third Symposium "cCMP and cUMP as New Second Messengers". Naunyn-Schmiedeberg's archives of pharmacology. 2015;388(1):1–3. 10.1007/s00210-014-1072-3 [DOI] [PubMed] [Google Scholar]
- 32. Beckert U, Wolter S, Hartwig C, Bahre H, Kaever V, Ladant D, et al. ExoY from Pseudomonas aeruginosa is a nucleotidyl cyclase with preference for cGMP and cUMP formation. Biochemical and biophysical research communications. 2014;450(1):870–4. 10.1016/j.bbrc.2014.06.088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gottle M, Dove S, Kees F, Schlossmann J, Geduhn J, Konig B, et al. Cytidylyl and uridylyl cyclase activity of Bacillus anthracis edema factor and Bordetella pertussis CyaA. Biochemistry. 2010;49(26):5494–503. 10.1021/bi100684g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bahre H, Hartwig C, Munder A, Wolter S, Stelzer T, Schirmer B, et al. cCMP and cUMP occur in vivo. Biochemical and biophysical research communications. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Trasak C, Zenner G, Vogel A, Yuksekdag G, Rost R, Haase I, et al. Yersinia protein kinase YopO is activated by a novel G-actin binding process. The Journal of biological chemistry. 2007;282(4):2268–77. [DOI] [PubMed] [Google Scholar]
- 36. Lee WL, Grimes JM, Robinson RC. Yersinia effector YopO uses actin as bait to phosphorylate proteins that regulate actin polymerization. Nature structural & molecular biology. 2015;22(3):248–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Grishin AM, Condos TE, Barber KR, Campbell-Valois FX, Parsot C, Shaw GS, et al. Structural basis for the inhibition of host protein ubiquitination by Shigella effector kinase OspG. Structure. 2014;22(6):878–88. 10.1016/j.str.2014.04.010 [DOI] [PubMed] [Google Scholar]
- 38. Pruneda JN, Smith FD, Daurie A, Swaney DL, Villen J, Scott JD, et al. E2~Ub conjugates regulate the kinase activity of Shigella effector OspG during pathogenesis. The EMBO journal. 2014;33(5):437–49. 10.1002/embj.201386386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhou Y, Dong N, Hu L, Shao F. The Shigella type three secretion system effector OspG directly and specifically binds to host ubiquitin for activation. PloS one. 2013;8(2):e57558 10.1371/journal.pone.0057558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kim DW, Lenzen G, Page AL, Legrain P, Sansonetti PJ, Parsot C. The Shigella flexneri effector OspG interferes with innate immune responses by targeting ubiquitin-conjugating enzymes. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(39):14046–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Calder T, Kinch LN, Fernandez J, Salomon D, Grishin NV, Orth K. Vibrio type III effector VPA1380 is related to the cysteine protease domain of large bacterial toxins. PloS one. 2014;9(8):e104387 10.1371/journal.pone.0104387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Coaker G, Falick A, Staskawicz B. Activation of a phytopathogenic bacterial effector protein by a eukaryotic cyclophilin. Science. 2005;308(5721):548–50. [DOI] [PubMed] [Google Scholar]
- 43. Coaker G, Zhu G, Ding Z, Van Doren SR, Staskawicz B. Eukaryotic cyclophilin as a molecular switch for effector activation. Molecular microbiology. 2006;61(6):1485–96. [DOI] [PubMed] [Google Scholar]
- 44. Aumuller T, Jahreis G, Fischer G, Schiene-Fischer C. Role of prolyl cis/trans isomers in cyclophilin-assisted Pseudomonas syringae AvrRpt2 protease activation. Biochemistry. 2010;49(5):1042–52. 10.1021/bi901813e [DOI] [PubMed] [Google Scholar]
- 45. Spitzer J, Poolman B. The role of biomacromolecular crowding, ionic strength, and physicochemical gradients in the complexities of life's emergence. Microbiology and molecular biology reviews: MMBR. 2009;73(2):371–88. 10.1128/MMBR.00010-09 [DOI] [PMC free article] [PubMed] [Google Scholar]