Abstract
For heterotrophic microbes, limited availability of carbon and energy sources is one of the major nutritional factors restricting the rate of growth in most ecosystems. Physiological adaptation to this hunger state requires metabolic versatility which usually involves expression of a wide range of different catabolic pathways and of high-affinity carbon transporters; together, this allows for simultaneous utilization of mixtures of carbonaceous compounds at low concentrations. In Escherichia coli the stationary phase sigma factor RpoS and the signal molecule cAMP are the major players in the regulation of transcription under such conditions; however, their interaction is still not fully understood. Therefore, during growth of E. coli in carbon-limited chemostat culture at different dilution rates, the transcriptomes, expression of periplasmic proteins and catabolomes of strains lacking one of these global regulators, either rpoS or adenylate cyclase (cya), were compared to those of the wild-type strain. The inability to synthesize cAMP exerted a strong negative influence on the expression of alternative carbon source uptake and degradation systems. In contrast, absence of RpoS increased the transcription of genes belonging to high-affinity uptake systems and central metabolism, presumably due to reduced competition of σD with σS. Phenotypical analysis confirmed this observation: The ability to respire alternative carbon substrates and to express periplasmic high-affinity binding proteins was eliminated in cya and crp mutants, while these properties were not affected in the rpoS mutant. As expected, transcription of numerous stress defence genes was negatively affected by the rpoS knock-out mutation. Interestingly, several genes of the RpoS stress response regulon were also down-regulated in the cAMP-negative strain indicating a coordinated global regulation. The results demonstrate that cAMP is crucial for catabolic flexibility during slow, carbon-limited growth, whereas RpoS is primarily involved in the regulation of stress response systems necessary for the survival of this bacterium under hunger conditions.
Introduction
As a rule, two major factors control heterotrophic bacterial growth in most ecosystems: On the one hand it is temperature, as it controls the rate of biochemical processes (and in most ecosystems temperatures are low); on the other hand, availability of assimilable carbon and energy sources is severely restricted, as most of the potentially utilizable carbon compounds are present in polymeric forms and are not directly accessible [1,2]. This latter situation, usually referred to as oligotrophy, starvation, or (technically) carbon and energy limited conditions, has also been described physiologically as the “hunger status” [3]. Availability of organic nutrients (particularly of assimilable organic carbon, AOC) is the result of the usually slow rate of AOC generation from, e.g, decaying plant material and its consumption by the competing and interacting microbial cells present in a given environment. In addition, microbial cells are exposed very directly to potentially harmful changes in temperature, light and other physico-chemical parameters. Hence, they have to be prepared for such challenges, and when necessary, also to be able to rapidly react to environmental stresses. Microbial cells are known to be able to adjust their cellular composition to environmental needs [4] and they possess fine-tuned global regulatory mechanisms that allow coordinated gene expression in complex regulatory networks resulting from the interaction of different global regulators [5–7].
As carbon and energy availability is a major factor determining the survival and proliferation of microbes in ecosystems, a rapid response to fluctuations in nutrient concentrations is an important fitness determinant [8–9]. An increased catabolic versatility and flexibility, allowing the simultaneous uptake and utilization of multiple organic nutrients and a rapid adjustment of uptake spectrum and fluxes according to availability in the immediate environment, is—in contrast to carbon (glucose) catabolite repression—a useful physiological strategy under hunger conditions [8,10–12]. Furthermore, cells respond to hunger by expressing a variety of high-affinity uptake systems involved in carbon and energy substrate transport [8, 13–18].
Concerning the numerous adverse physical and chemical challenges a cell might face, a broadly expressed stress response is an essential requirement for successful survival. Therefore, slowly growing bacterial cells also depend on the induction of general stress response mechanisms [19]. Even more important here is to be prepared for all possible situations as there are neither sufficient resources to express stress response and defence systems in times of need, nor is there probably enough time to react [11]. Two global regulators are known to control nutrient scavenging abilities and stress response systems in the model bacterium E. coli, namely cAMP-CRP and the alternative (stress response) sigma factor RpoS. Under glucose-limited conditions intracellular concentrations of cAMP are increased and a (strain specific) correlation between specific growth rate and cAMP in glucose-limited continuous culture was found [20–23]. A similar increase was observed for RpoS levels during the transition from exponential growth phase to stationary phase in complex medium, where during slow growth RpoS reached 30–50% of the levels of the housekeeping sigma factor RpoD [24]. A direct correlation between increasing RpoS-levels and decreasing specific growth rates was also reported for exponentially growing batch cultures and carbon-limited continuous cultures of E. coli [19,25], whereas levels of RpoD remained constant [25]. When investigating and comparing gene expression and global physiology of microbes under different growth conditions it is necessary to control the cell’s environment as well as possible [4,26]. Of particular importance with respect to overall cellular composition and global response are two parameters, namely the type of the growth-limiting nutrient (whether C, N, P etc.), and the specific growth rate [4,17,27–29]. With the exception of the phase of genuinely exponential and nutritionally unrestricted growth in batch culture [17], steady-state continuous (or chemostat) cultivation is the only method that allows a precise control of the specific growth rate by a selected limiting nutrient of bacterial cells, and thus of their physiological state [4,26,30]. Regarding slow, nutrient-limited growth, chemostat culture seems to be the only well reproducible experimental approach [18,31]. Although numerous studies addressed the effect of knock-out mutations of global regulatory genes on the transcriptome of E. coli in batch culture, similar experiments under constant, carbon- and energy-limited growth conditions in chemostats are largely missing. Transcriptome studies of global regulatory mutant strains in batch culture were performed for RpoS [32–37], cAMP-CRP [38,39], Lrp [40,41], and Crl [42]. Already in 2002 a cross-talk between cAMP and RpoS was proposed [43]. Later many genes, known to be RpoS-dependent, were reported to be also regulated by either cAMP [38], or to have a cAMP-CRP binding site within the promoter region in transcriptome/genome studies [37]. However, results are far from being consistent; several operons seem to be also under the control of additional global regulators, indicating a complex mechanism governing gene expression.
In an attempt to disentangle the effects of the cAMP-CRP and RpoS global regulators and their roles for physiological adaptation to (environmentally relevant) slow growth exerted by limited availability of carbon and energy sources, we compared the transcriptome and global physiological state of wild-type versus ΔrpoS or Δcya E. coli strains during cultivation in glucose- and LB-limited chemostat culture.
Material and Methods
Bacterial strains, growth medium and growth conditions
Wild-type E. coli MG1655 and rpoS knock-out strains used in this study have been described elsewhere [18,31]. The Δcya and Δcrp strains are also derivatives of E. coli MG1655 and were obtained from A. C. Matin and M. Cashel [44]. The genotype of all strains is given in Table 1. Glucose-limited mineral medium and carbon-limited modified LB medium for batch and chemostat cultivation were described previously [15,25]. The Δcya strain exhibited a strongly reduced growth yield in modified LB medium, therefore, for chemostat cultivation tryptone and yeast extract concentrations were increased to 2 g L-1 and 1 g L-1, respectively, (which corresponds to 20% of the original strength of LB medium). Cultivation conditions were the same as in a previous study [15]. For continuous cultivation small volume glass bioreactors were used. Aeration with a fine pore glass purger and magnetic stirring was sufficient to keep oxygen saturation above 60%. For batch cultivation with modified LB medium, baffled Erlenmeyer flasks (300 ml) and a low liquid volume (50 ml) were used. Flasks were vigorously shaken as electromagnetic stirring was not sufficient for maintaining fully oxic conditions during the transition to stationary phase. To avoid adaptive mutations in the wild-type strain and compensatory mutations in knock-out strains [18], new starter cultures were prepared for each experiment. Stability and purity of chemostat cultures were checked for wild-type and ΔrpoS strains as described previously [25] and for Δcya and Δcrp strains by confirming in addition the strongly reduced number of substrates which support growth on BIOLOG AN MicroPlates.
Table 1. Effect of global regulatory mutations on growth kinetic parameters and specific hydroperoxidase activity of E. coli in glucose mineral medium cultures at 37°C.
The dilution rate in glucose-limited chemostat cultures was set at a value of approximately half the maximum specific growth rate (D = 0.3 h-1 for wild-type and ΔrpoS strains, D = 0.16 h-1 for the Δcya strain and D = 0.14 h-1 for the Δcrp strain). Values represent the average of replicate measurements (n ≥ 3), standard deviations are given in parentheses.
| strain | genotype | μmax | Ks | Yield | HPI+II a | HPII b |
|---|---|---|---|---|---|---|
| [h-1] | [mg L-1] | [g dr. wt. g glc. -1] | [μmol H2O2 min-1 mgprotein -1] | |||
| MG1655 (wt) | F - λ - rph1 | 0.63 c | 0.54 | 0.46 d | 62.9 d | 27.7 d |
| (± 0.01) | (± 0.12) | (± 0.01) | (± 8.1) | (± 3.9) | ||
| ΔrpoS | MG1655ΔrpoS::Tn10 | 0.69 | 0.29 | 0.37 | 67.0 d | 1.1 d |
| (± 0.01) | (± 0.03) | (± 0.06) | (± 3.4) | (± 0.2) | ||
| Δcya | MG1655Δcya-854 | 0.35 | 5.7 | 0.33 | 51.0 | 23.3 |
| (± 0.03) | (± 1.2) | (± 0.04) | (± 16) | (± 4.7) | ||
| Δcrp | MG1655 Δcrp-39 rpsL | 0.28 | 19.6 | 0.31 | n. d. e | n. d. e |
| (± 0.03) | (± 1.8) | (± 0.02) | ||||
Cultures were first run in the batch mode and growth was followed by determining OD546 spectrophotometrically (model Uvikon 930, Kontron, Zurich, Switzerland) at regular intervals. After 4–5 hours the culture mode was changed to continuous at a dilution rate corresponding to half μmax by switching on the medium feed pump. Steady-state with respect to optical density was reached approximately 20 hours after starting the medium feed. Samples for catabolic pathway assays, periplasmic proteins and transcriptome analysis were taken 40 hours after switching to the continuous culture mode, and each experiment was done in triplicate.
Determination of growth kinetic parameters, enzyme activities and catabolic ability
Maximum specific growth rates (μmax, h-1) were determined for growth in both mineral glucose and modified LB medium batch cultures at 37°C. To avoid lag phases, pre-warmed medium was inoculated from exponential phase cultures growing in the same medium. Specific growth rates were calculated from the linear part of logarithmic growth curves by least squares regression. The linear part of logarithmic growth curves in modified LB medium ended at an OD546 of approximately 0.2 (see [45]), higher optical densities were excluded from the calculation of μmax,LB. Growth yield was determined as described in Ihssen and Egli [25]. Steady-state glucose concentrations were determined by ion chromatography [25], the results were used for the calculation of glucose affinities (Ks,Glc, μg L-1) with a modified Monod equation including an smin of 12 μg L-1 [46].
Expression of catabolic pathways in carbon-limited chemostat cultures was analyzed with BIOLOG AN MicroPlate respiration assays [15]. Periplasmic proteins were extracted by chloroform shock treatment and analysed on 15% SDS-PAGE gels which were stained with Coomassie Blue [15]. Hydroperoxidase I and II specific activities were determined with a spectrophotometric assay as described previously [25].
RNA isolation and cDNA synthesis
Total RNA was isolated using standard procedures [47]. The RNA extracted was checked for purity by gel electrophoresis and was quantified by measuring absorbance at 260 nm. CyScribe First-Strand cDNA Labelling Kit (Amersham Bioscience, Little Chalfont, England) was used to reversely transcribe mRNA into fluorescently labelled cDNA, followed by hybridization onto the microarray slide as previously described [13]. Slide microarrays were purchased from MWG-Biotech AG (MWG E. coli K12 V2 array, Ebersberg, Germany); they contained gene specific oligonucletide probes representing the complete E. coli K12 genome (4,288 ORF).
Image data analysis
Microarray slides were scanned with an Affymetrix 428 Array Scanner (High Wycombe, England) and the obtained images were analysed with the Affymetrix Jaguar software version 2.0. Spot intensities and corresponding background signals were further analyzed with the program GeneSpring from Silicon Genetics (Redwood City, USA) as reported elsewhere [13]. After background correction, the mean value of three replicate spots per sample was used for calculation of induction/repression factors of individual genes. Data from the three independent experiments were combined, genes that were differently regulated ≥3 and ≤-3 (t-test, p ≤ 0.2) were defined as being statistically significant. We did not choose a stricter p value because gene expression levels of Δcya and ΔrpoS strains exhibited a large variation after normalization. Technical replicates on the same slide exhibited a signal variability below 10%, but a much higher variance was observed for gene expression ratios of biological replicates after normalization, in particular for genes with low expression in one of the two compared strains. A multitude of genes exhibited an up- or down-regulation factor between 2 and 3. We considered this as background variability and therefore set the threshold of for significant effects of the studied global regulatory mutations as ≥3 and ≤-3. Validity of this value is backed by results for genes previously reported under the control of cAMP-CRP [38,39]. Although exhibiting a high variability of expression ratios, the mean value was always above 3 when comparing the wild-type and the Δcya strain, for example, ptsG (4.0 ± 1.4, from 3.0 to 5.6), mglA (3.9 ± 2.6, from 2.2 to 6.9), mglB (4.9 ± 1.5, from 3.4 to 6.4), rbsD (3.6 ± 0.8, from 3.0 to 4.5) and mdh (4.1 ± 1.0, from 3.0 to 5.0). The same was observed for genes known to be regulated by RpoS, for example idnT (-3.0 ± 0.85, from -2-1 to -3.75), prpE (5.9 ± 2.8, from 3.5 to 9.0) and talA (8.6 ± 7.3, from 3.2 to 16.9) all surpassed the threshold of the expression ratios when comparing wild type and ΔrpoS strains. The complete set of microarray data is available on the Gene Expression Omnibus (GEO) website (http://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE25982.
Results and Discussion
Growth of heterotrophic microbes in the natural environment is most of the time restricted by the availability of carbon and energy sources [48]. This is also the case for E. coli, even in the gut periods of real abundance are probably rare and short, followed virtually immediately by limitation. The situation was described earlier as feast and famine existence [49], with famine being the dominating nutritional condition outside its primary habitat [2], because concentrations of easily usable monomeric growth substrates such as sugars, amino acids and organic acids in the environment are usually in the range of 1–100 μg L-1 [50]. Even though E. coli cannot be considered an oligotrophic microorganism, steady-state concentrations of the limiting carbon substrate (glucose) in slow-growing chemostat cultures were 50 μg L-1 and lower (D ≤ 0.1 h-1), indicating that wild-type E. coli MG1655 is capable of transporting and metabolizing minute amounts of organic nutrients [31]. Yet more, the ability to survive, grow and even compete with the natural bacterial flora in freshwater was recently confirmed also for E. coli O157 [51]. The physiological state during carbon-limited growth is fundamentally different from that of cells growing with excess carbon and energy sources; the former is characterized by an up-regulation of a multitude of high-affinity transport systems and catabolic pathways for diverse organic nutrients [13,15,18].
As both, cAMP and RpoS are considered to be the two global regulators responsible for carbon- and energy-limitation physiological phenotype (hunger phenotype), we have investigated the effect of the two regulators under comparable growth conditions in either glucose- or LB-limited chemostat culture.
cAMP-CRP is strictly required for expression of the hunger phenotype
The extent to which the hunger phenotype depends on the cAMP-CRP global regulator was investigated by comparing the isogenic wild-type strain E. coli K12 MG1655 to two knock-out mutants, deficient in either adenylate cyclase (cya) or in CRP, during growth with a defined mineral glucose medium in carbon- and energy-limited chemostat culture. Transcriptome analysis of the wild-type and the isogenic Δcya strain was performed in three replicate glucose-limited chemostat cultures operated at a dilution rate (D) corresponding to approximately half μmax,Glc, i.e. at extracellular glucose concentrations corresponding to Ks,Glc; hence, the wild-type strain was grown at a D of 0.3 h-1, whereas the Δcya strain was cultivated at a D of 0.16 h-1 (see Table 1). This choice was made because, first, wild-type E. coli exhibited the best Ks to glucose at a dilution rate of 0.3 h-1 [31] and, second, because this dilution rate equivalent to half μmax was used in earlier proteome [18], catabolome [15] and transcriptome analyses [13] performed with wild-type E. coli K12 MG1655. Due to the fact that μmax,Glc of the Δcya strain in mineral medium was reduced by 50% (Table 1), which is in agreement with a previous study [52], a D of 0.16 h-1 was chosen for cultivating this mutant (0.14 h-1 for the Δcrp strain), in order to obtain stable growth under glucose-limited continuous culture conditions. Similar levels of KatE (belonging to the RpoS regulon) activity were found in both wild-type and Δcya E. coli strain, compared to the low levels present in the ΔrpoS mutant (Table 1), which suggests minor secondary effects on RpoS levels in the Δcya strain resulting from the reduced μmax,Glc. The transcriptome results for the wild-type and Δcya strain cultivated under glucose-limited chemostat conditions are shown in Table 2 and S1 Table. Applying a rather strict cut-off factor of 3 for defining a significant difference in relative gene expression, 192 genes were affected by the adenylate cyclase knock-out mutation (Table 2 and S1 Table). The majority of these genes (152) was down-regulated, which is in agreement with the role of cAMP-CRP as a transcription activator. A large fraction of the Δcya affected genes (51) were reported to possess one or more cAMP-CRP binding sites in their promoter region [53,54] and 44 of them were previously found to be down-regulated in crp mutant strains [38,39]. This observation is in agreement with previous studies reporting that cAMP-CRP regulates more than 400 genes, making it the broadest global regulator after the housekeeping sigma factor RpoD [6].
Table 2. Genes coding for proteins involved in transport and metabolism of carbon substrate with significant up- or down-regulation in ΔrpoS and Δcya strains (difference in average signal intensity compared to wild-type E. coli K12 ≥ 3 or ≤ -3, p-value ≤ 0.2) in glucose-limited continuous culture cultivated at D = 0.3 h-1.
| Gene | b no. | Gene product | Ratio cAMP | Ratio RpoS |
|---|---|---|---|---|
| Transport | ||||
| nhaA | b0019 | Na+/H antiporter, pH dependent | 4.1 | |
| yajR | b0427 | putative transport protein | -3.3** | |
| nmpC b , e , f , g | b0553 | outer membrane porin protein, locus of qsr prophage | 6.3 | -3.2* |
| cusB | b0574 | outer membrane transport protein involved in copper tolerance | -3.1* | |
| fepE | b0587 | ferric enterobactin (enterochelin) transport | -3.0** | |
| cstA f , i | b0598 | carbon starvation protein | 7.6 | |
| ybeJ | b0655 | glutamate/aspartate periplasmic binding transport protein | 5.4 | |
| artI g | b0863 | arginine 3rd transport system periplasmic binding protein | 4.1 | |
| ycaD | b0898 | putative transport protein | 4.4 | |
| ompF f , i | b0929 | outer membrane protein 1a | 27.4* | |
| ompA f , g | b0957 | outer membrane protein 3a | 4.0 | |
| ycdG | b1006 | putative transport protein | 3.6 | |
| ptsG c , f , g , i | b1101 | PTS system, glucose-specific IIBC component | 4.0 | |
| nhaB | b1186 | Na+/H+ antiporter, pH independent | -4.5** | -5.4** |
| ydcS e | b1440 | putative ABC transporter periplasmic binding protein | 8.9 | 9.8 |
| ydcU | b1442 | putative transport system permease protein | 11.9 | 7.3 |
| xasA e , h | b1492 | acid sensitivity protein, putative transporter | 34.1* | |
| ego | b1513 | putative ATP-binding component of a transport system | 15.2 | 14.3 |
| manX c , f , g , i | b1817 | PTS enzyme IIAB, mannose-specific | 8.0 | |
| manY c , f , g | b1818 | PTS enzyme IIC, mannose-specific | 15.1 | |
| manZ f , g , i | b1819 | PTS enzyme IID, mannose-specific | 5.8 | |
| gatC c , i | b2092 | PTS system galactitol-specific enzyme IIC | 35.3* | |
| gatB c , i | b2093 | galactitol-specific enzyme IIB of phosphotransferase system | 60.6* | |
| gatA c | b2094 | galactitol-specific enzyme IIA of phosphotransferase system | 55.9* | |
| yohC e | b2135 | putative transport protein | 10.1 | 7.8 |
| mglA c , f , g , i | b2149 | ATP-binding component of methyl-galactoside transport | 3.9 | |
| mglB c , f , g , i | b2150 | galactose-binding transport protein | 4.9 | |
| exbD | b3005 | uptake of enterochelin tonB-dependent uptake of B colicins | -3.0 | |
| mscL | b3291 | mechanosensitive channel | 6.0 | |
| feoA | b3408 | ferrous iron transport protein A | -4.9** | |
| hdeD b , e | b3511 | putative transporter protein | 11.5 | |
| dppA | b3544 | dipeptide transport protein | 11.9 | |
| rbsD g , i | b3748 | D-ribose high-affinity transport system, membrane-associated protein | 3.6 | |
| rbsB g | b3751 | D-ribose periplasmic binding protein | 9.4 | |
| glpF c , f , i | b3927 | facilitated diffusion of glycerol | 9.6 | |
| yjbB i | b4020 | putative alpha helix protein | -3.6 | |
| malG f , g , i | b4032 | part of maltose permease, inner membrane | 5.9 | |
| malF f , g , i | b4033 | part of maltose permease, periplasmic | 6.9 | |
| malE c , f , g , i | b4034 | periplasmic maltose-binding protein | 7.5 | |
| malK c , f , g , i | b4035 | ATP-binding component of transport system for maltose | 4.2 | |
| lamB c , f , g , i | b4036 | phage lambda receptor protein, maltose high-affinity receptor | 8.1 | |
| malM c , f , g , i | b4037 | periplasmic protein of mal regulon | 12.5 | |
| actP f | b4067 | acetate permease | 10.7 | |
| alsB | b4088 | D-allose-binding periplasmic protein | -3.2** | |
| cycA | b4208 | transport of D-alanine, D-serine, and glycine | 4.0 | |
| ytfQ | b4227 | putative D-ribose transport protein, ABC superfamily | 3.6 | |
| ytfR | b4228 | putative ATP-binding component of a transport system | 5.2 | |
| idnT f , g | b4265 | L-idonate transporter | -3.3* | -3.0 |
| sgcX | b4305 | putative PTS transport system | 4.4 | |
| Carbon & Energy metabolism | ||||
| acnB b , f | b0118 | aconitate hydrase B | -3.0** | |
| prpB b , f | b0331 | putative carboxyphosphoenolpyruvate mutase | 17.0 | 15.8 |
| prpC f | b0333 | citrate synthase, propionate metabolism | 30.8* | 17.6 |
| prpD f | b0334 | 2-methyl citrate dehydratase | 5.6 | 7.6 |
| prpE f | b0335 | putative propionyl-CoA synthetase | 5.9 | |
| yajO | b0419 | putative NAD(P)H-dependent xylose reductase | 3.7 | |
| thiJ | b0424 | 4-methyl-5(beta-hydroxyethyl)-thiazole monophosphate synthesis | 6.1 | |
| gltA c , f , g | b0720 | citrate synthase | 6.9 | |
| sdhD a , b , i | b0722 | succinate dehydrogenase, hydrophobic subunit | -3.1 | |
| sdhB f , g , i | b0724 | succinate dehydrogenase, iron sulfur protein | 7.2 | |
| sucA c , g , i | b0726 | 2-oxoglutarate dehydrogenase (decarboxylase component) | 3.7 | |
| sucC c , g | b0728 | succinyl-CoA synthetase, beta subunit | 4.4 | |
| moa | b0781 | molybdopterin biosynthesis, protein A | -6.2** | -7.4** |
| dmsB | b0895 | anaerobic dimethyl sulfoxide reductase subunit B | -7.4** | -3.0** |
| mgsA | b0963 | methylglyoxal synthase | 5.5 | |
| nark | b1223 | nitrite extrusion protein | 4.7 | |
| aldA c , f , g | b1415 | aldehyde dehydrogenase, NAD-linked | 6.5 | |
| gapC_1 b | b1417 | glyceraldehyde 3-phosphate dehydrogenase C, interrupted | 5.6 | |
| astA | b1747 | arginine succinyltransferase | 7.0 | |
| gapA g | b1779 | glyceraldehyde-3-phosphate dehydrogenase A | 3.5 | |
| gatD c , f , i | b2091 | galactitol-1-phosphate dehydrogenase | 11.2 | |
| gatZ c , f | b2095 | putative tagatose 6-phosphate kinase 1 | 54.4 | |
| gatY f | b2096 | tagatose-bisphosphate aldolase 1 | 50.5** | |
| fbaB b , d , e , h | b2097 | fructose-biphosphate aldolase | 4.0 | 5.6 |
| menB | b2262 | dihydroxynaphtoic acid synthetase | -3.0* | |
| nuoH | b2282 | NADH dehydrogenase I chain H | 3.7 | |
| nuoE | b2285 | NADH dehydrogenase I chain E | 4.3 | |
| talA a , b , d , e , h | b2464 | transaldolase A | 8.6 | |
| eno | b2779 | Enolase | 3.6 | |
| glcB | b2976 | malate synthase G | 4.1 | |
| glcF | b2978 | glycolate oxidase iron-sulfur subunit | 7.1 | |
| glcD | b2979 | glycolate oxidase subunit D | 7.0 | |
| mdh f , g , i | b3236 | malate dehydrogenase | 4.1 | |
| pckA g | b3403 | phosphoenolpyruvate carboxykinase | 8.7 | |
| atpD g | b3732 | membrane-bound ATP synthase, F1 sector, beta-subunit | 4.7 | |
| atpH g | b3735 | membrane-bound ATP synthase, F1 sector, delta-subunit | 4.0 | |
| atpE g | b3737 | membrane-bound ATP synthase, F0 sector, subunit c | 4.6 | |
| atpB g | b3738 | membrane-bound ATP synthase subunit a | 5.3 | |
| aceB c , f | b4014 | malate synthase A | 4.1 | |
| aceA c , f , i | b4015 | isocitrate lyase | 5.1 | |
| acs f , g , i | b4069 | acetyl-CoA synthetase | 19.5 | |
| frdD | b4151 | fumarate reductase, anaerobic, membrane anchor polypeptide | 4.1 | |
| frdB b | b4153 | fumarate reductase, anaerobic, iron-sulfur protein subunit | -3.1** | -3.2** |
Most of the genes down-regulated in the Δcya strain belong to operons involed in the uptake of carbon sources (Table 2): glucose (ptsG), maltose (malEFG and malK-lamB-malM), galactose (gatABCD and mglBA), ribose (rbsDB and ytfQR), mannose (manXYZ) and acetate (actP). Genes for the uptake of amino acids were also affected, e.g., those for glutamate and aspartate (ybeJ), arginine (artI), alanine, serine and glycine (cycA), peptides (cstA) and dipeptides (dppA). Several outer membrane porins (nmpC, ompF, ompA and ompC), a mechanosensitive channel (mscL), and an antiporter (nhaA) were also down-regulated in the Δcya strain, whereas another antiporter (nhaB) was up-regulated. The down-regulation of several genes encoding proteins involved in the transport of carbon sources in the absence of cAMP is in agreement with the results of previous studies performed in batch cultures [38,39]. Among the cAMP-dependent transporter genes found in this study, the mal operon, mgl operon and ompF have previously been shown to be important for high-affinity uptake of glucose under carbon-limited conditions and to be under the transcriptional control of cAMP [18,55–57]. The low expression of genes encoding ABC-type high-affinity uptake systems and the PTS transporter for glucose is in agreement with the 10 to 20-fold poorer affinity (Ks,Glc) of both Δcya and Δcrp strains for glucose as determined from steady-state concentrations in chemostat cultures (Table 1). In contrast, the growth yield for glucose was only marginally affected by these global regulatory mutations, indicating that biosynthetic pathways are not part of the cAMP-CRP and RpoS regulons (Table 1). The transcriptome data and the increased Ks,Glc are consistent with the missing over-expression of high-affinity periplasmic binding proteins for carbon substrates in the Δcya and Δcrp strains under carbon- and energy-limited growth conditions (Fig 1). Expression patterns for periplasmic proteins in Δcya and Δcrp strains were similar, although minor differences existed. Interestingly, such minor differences in transcriptome analyses between Δcya and Δcrp strains were also reported for Vibrio cholerae [58].
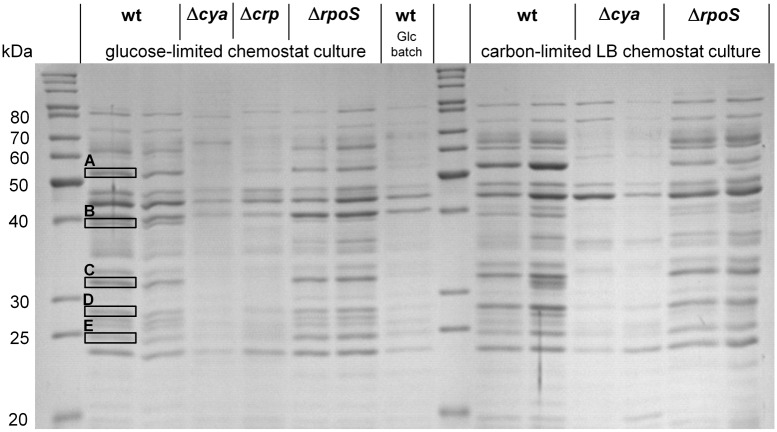
Fig 1. Expression of periplasmic proteins in carbon-limited chemostat cultures of wild-type (wt),Δcrp, Δcya and ΔrpoS E. coli strains.
Carbon-limited LB chemostat cultures were set at a dilution rate of 0.3 h-1, while the dilution rate of glucose-limited chemostat cultures was operated at approximately half the maximum specific growth rate (D = 0.3 h-1 for wild-type and rpoS strains, D = 0.16 h-1 for thecya strain and D = 0.14 h-1 for thecrp strain). The protein bands marked in the figure were identified in a previous study [15] as (A) dipeptide-binding protein DppA, (B) maltose-binding protein MalE, (C) galactose/glucose-binding protein MglB, (D) ribose-binding protein RbsB and (E) glutamine-binding protein GlnH. When two lanes per strain for a similar condition are shown, chloroform shock extracts from two separate chemostat cultures were loaded onto the gel. Loading volumes were normalized to the optical density of the culture samples. Protein size markers are shown in the first and ninth lane from the left.
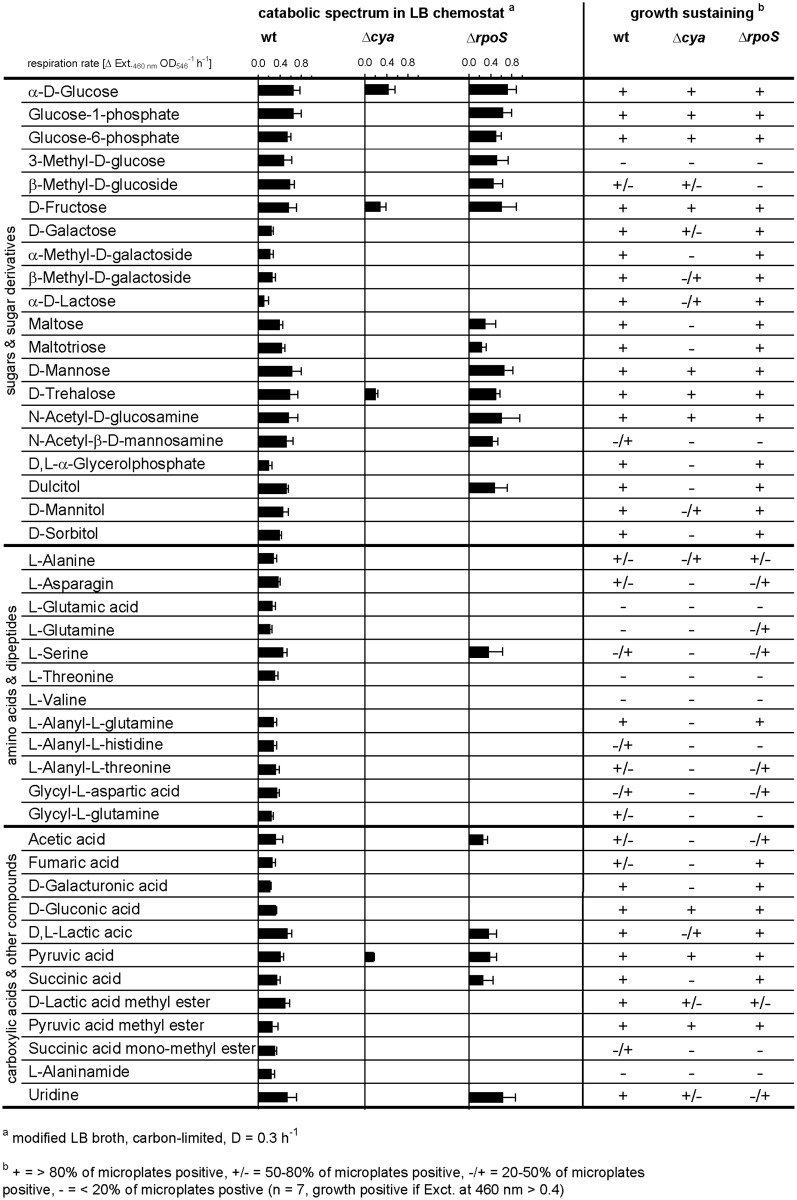
In the wild-type strain strong induction of high-affinity binding proteins for e.g. maltose, ribose, galactose, dipeptides and glutamine occurred in response to carbon and energy limitation. This has repeatedly been observed in previous studies for both laboratory strains and natural isolates of E. coli [13,15,18]. In addition to the negative effect of the absence of cAMP on the expression of transport systems for alternative carbon substrates, many genes encoding enzymes of central catabolic pathways were down-regulated, e.g. components of the tricarboxylic acid cycle (TCA), the glyoxylate bypass, glycolysis, gluconeogenesis, the pentose phosphate pathway, the methylglyoxyl pathway, propionate metabolism and galactitol degradation (Table 2). Genes encoding subunits of ATP synthase were also negatively affected. Among the most down-regulated genes in the Δcya strain were those for the galactitol degradation pathway (gatD, gatZ and gatY), propionate metabolism (prpB, prpC and prpD) and acetyl-CoA synthetase (acs). The genes belonging to the propionate metabolism are known to be cAMP-dependent, in addition σN is required for their transcription [59]. Acetyl-CoA synthetase (acs) was reported by others to be under the control of cAMP, Fnr and the flux of carbon through the acetate pathway [60,61]. Genes encoding enzymes of the tricarboxylic acid (TCA) cycle and glyoxylate bypass (AceA, AceB and GlcB) were also transcribed at a lower level in the absence of adenylate cyclase. A significant reduced activity of the glyoxylate bypass for Δcya and Δcrp strains has been previously reported under glucose limitation [23,60,62]. The reduced expression of genes encoding components of the TCA cycle and the glycolysis pathway is in agreement with the significantly reduced max,Glc of Δcya and Δcrp strains in glucose-excess batch culture (Table 1). At the physiological level the mandatory presence of cAMP-CRP for induction of transport systems and catabolic pathways for alternative carbon substrates was reflected in a drastically reduced capability to respire and grow on carbon substrates other than glucose (Figs 2 and 3). The high catabolic flexibility of glucose-limited growing E. coli cultures reported previously [15] was abolished in strains without functional cAMP-CRP global regulation (Fig 2). The limited catabolic flexibility of these strains in glucose-limited chemostats resembled the pattern observed for wild-type E. coli cultivated under conditions of excess glucose [15]. Similarly, the expression of an enormous number of alternative catabolic pathways in wild-type E. coli observed in complex medium under carbon- and energy-limited conditions was completely abolished in the absence of functional adenylate cyclase (Fig 3). With the exception of L-serine, which was not oxidized by Δcya cells, the BIOLOG respiration pattern of the Δcya strain cultivated in a carbon-limited LB chemostat was comparable to that of fast-growing cells of the wild-type strain in LB batch cultures [15]. In agreement with the missing expression of catabolic pathways for alternative organic substrates, the growth yield of the Δcya strain in modified LB medium was strongly reduced, both in batch and chemostat culture (Fig 4 and Table 3). By contrast, the maximal specific growth rate in the same medium (determined at low cell densities) was only slightly reduced in cAMP-CRP knock-out strains (Table 3), again suggesting that the expression of biosynthetic pathways is not influenced by this global regulatory mechanism.
Fig 2. Substrate respiration rates of wild-type, Δcrp, Δcya and ΔrpoS strains of E. coli in glucose-limited chemostat cultures.
Respiration rates were determined with chloramphenicol-inhibited cells on BIOLOG AN MicroPlates and normalized to OD546 of the used cell suspension. Bars give average rates of ≥ 4 BIOLOG plates, for each strain cells were taken from two separate chemostat cultures. Error bars represent standard deviations.
Fig 3. Substrate respiration rates of wild-type, Δcrp, Δcya and ΔrpoS strains of E. coli in carbon-limited LB chemostat cultures and substrates that supported growth in BIOLOG plates.
Respiration rates were determined as described in legend to Fig 2. For growth, all substrates leading to significant colour formation in any strain are shown. L-valine was included in the table as negative control.
Fig 4. Growth of wild-type (■, □), ΔrpoS (●, ○), and Δcya (▲, Δ) strains of E. coli in batch cultures with modified LB medium buffered at pH 7.
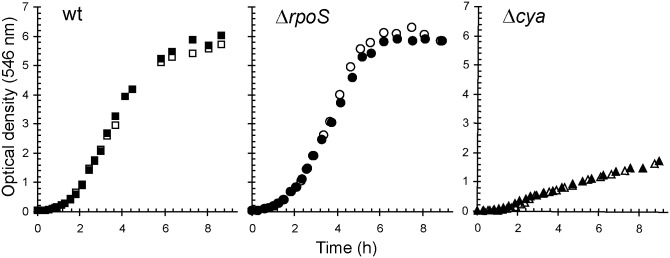
Data from two individual experiments (black and white symbols).
Table 3. Effect of global regulatory mutations on growth kinetic parameters and specific hydroperoxidase activity of E. coli MG1655 in modified LB complex medium cultures at 37°C.
Growth yield and specific hydroperoxidase activities were determined in carbon-limited LB chemostat cultures operated at a dilution rate of 0.3 h-1.
| strain | μmax | Yield | HPI+II b | HPII c |
|---|---|---|---|---|
| [h-1] | [g dr. wt. g LB -1] a | [μmol H2O2 min-1 mgprotein -1] | ||
| MG1655 (wt) | 2.13 | 0.111 | 104 | 48.0 |
| (± 0.11) | (± 0.03) | (± 11) | (± 6.8) | |
| ΔrpoS | 2.04 | 0.092 | 50.7 | 0.7 |
| (± 0.04) | (± 0.011) | (± 13.0) | (± 0.3) | |
| Δcya | 1.64 | 0.015 | 66.5 | 57.5 |
| (± 0.08) | (± 0.001) | (± 11) | (± 22) | |
a biomass yield per amount of tryptone and yeast extract (2:1) present in the feed medium of carbon-limited chemostat cultures
b combined activities of heat labile hydroperoxidase I (KatG) and heat stable hydroperoxidase II (KatE)
c heat stable hydroperoxidase II activity (KatE)
An interesting exception to the general rule of down-regulation of transport systems in the Δcya strain are metal transport systems, which were found to be up-regulated: cusB (involved in copper transport), feoA (involved in ferrous ion transport) and fepE (involved in ferric enterobactin transport) transcript levels were all 3-5-fold enhanced in the mutant (Table 2).
RpoS is not required for the physiological response (hunger-response) of E. coli to carbon and energy limitation
Similarly to cAMP, the levels of the alternative sigma factor RpoS increase with decreasing specific growth rate in carbon- and energy-limited chemostat cultures and during the transition to stationary phase in batch culture [22,25]. This is the reason why RpoS was also called starvation or stationary phase sigma factor [63].
We compared the transcriptome of a ΔrpoS mutant to that of the wild-type strain under identical growth conditions in glucose-limited chemostat culture at a D of 0.3 h-1 (doubling time: 2.3 h). Genes with significant differential expression ratio are listed in Table 2 and S1 Table (cut-off: 3-fold difference of expression signal). The number of affected genes was lower than in the case of cAMP, 100 genes were found to be affected by the absence of RpoS, of which 52 were down-regulated and 48 up-regulated. Among the genes down-regulated many (42 of 52) were reported to be RpoS-dependent in previous transcriptome studies carried out in batch culture [32,33,35–37]. However, only one out of the 48 up-regulated genes had been reported to be up-regulated in the absence of RpoS in an earlier transcriptome study under batch stationary phase conditions [36]. Interestingly, the major part of genes detected at higher transcription levels in the ΔrpoS strain had a low expression ratio (between 3 and 5), which may be explained by the loss of competition between σD and σS, resulting in an enhanced transcription of σD-dependent genes. RpoS competes with RpoD for free RNA-polymerases and the loss of RpoS has an impact on gene expression of genes transcribed by σD due to the fact that more RNA-polymerases are bound with σD [34,64–66]. Only few transport systems were more than 3-fold up-regulated in the ΔrpoS strain, e.g., the allose uptake system (alsB) and the idonate/gluconate uptake system (idnT). Genes encoding components of transport systems were generally not down-regulated in the ΔrpoS strain, with the notable exceptions of xasA, which encodes an acid sensitive transport protein, hdeD, which has been suggested to be involved in acid stress resistance [67] and genes encoding ABC-type transporters of unknown specifity (ydcS, ydcU, ego and yohC). Some hypothetical genes (13) that were highly down-regulated in the ΔrpoS strain were also affected by cAMP, suggesting a co-regulation of the two global regulators.
Lack of RpoS reduced the transcription of only few metabolic genes, these are involved in propionate metabolism, the pentose phosphate pathway, glycolysis and biotin synthesis (Table 2). Surprisingly, the entire propionate operon (prpBCDE) was strongly down-regulated in the ΔrpoS strain, which confirms a previous transcriptome study [33]. In contrast to the Δcya strain where genes encoding components of the TCA cycle were generally down-regulated, aconitate hydrase B (acnB), fumarate reductase (frdB) and succinate dehydrogenase (sdhD) were up-regulated in the ΔrpoS strain, which is in agreement with the reported down-regulation of sdhD at high RpoS levels [68].
In general, the ΔrpoS mutation had no negative effect on the transcription of the majority of transport systems and catabolic enzymes under glucose-limited growth conditions, excluding RpoS as important global regulator for the adaptation of E. coli to a restricted supply of carbon and energy sources.
The transciptome data recorded for the ΔrpoS strain matched results of the physiological characterization. The slight up-regulation of genes involved in the citric acid cycle by the loss of RpoS was in agreement with the slightly improved max,Glc in glucose-excess batch cultures (Table 1). The limited effect of the absence of RpoS on the transcription of catabolic genes for alternative carbon substrates coincided with the results from BIOLOG respiration assays for glucose-limited growing ΔrpoS cells, which were to a large extent similar to wild-type cells grown under the same conditions (Figs 2 and 3). Exceptions were trehalose, mannitol, sorbitol and D-malate, which were oxidized at high rates by wild-type cells, but were either not oxidized or utilized only at reduced rates by the ΔrpoS mutant (Fig 2). The number of catabolic pathways expressed by the ΔrpoS strain in carbon-limited chemostat cultures with modified LB medium was considerably larger than that found in Δcya cultures, although the capacity to oxidise many substrates was negatively affected, e.g., for D-sorbitol, L-alanine, L-alanyl-L-glutamine, gluconate and fumarate (Fig 3). For the ΔrpoS strain the spectrum of growth-sustaining organic substrates assessed on BIOLOG plates was almost identical to that of the wild-type strain (Fig 3), indicating that most uptake systems and catabolic enzymes can be expressed properly in the absence of functional RpoS.
Contrary to the observed negative effect of the Δcya mutation on the expression of high-affinity binding proteins in response to carbon and energy limitation, loss of functional RpoS had no such effect (Fig 1). Also in contrast to the Δcya and Δcrp mutants, the RpoS knock-out strain exhibited a similar or even increased fitness under carbon-limited growth conditions compared to the wild-type strain. Maximal specific growth rate (μmax), affinity for glucose (Ks,Glc) and growth yields of the ΔrpoS strain were either similar or enhanced, both when growing in glucose mineral medium and in complex medium (Table 1 and Table 3, Fig 4). The observed improved affinity for glucose is in agreement with previous reports on positive effects of rpoS mutations on high-affinity transport of glucose in glucose-limited chemostat cultures [31,69,70].
Effect of cAMP and RpoS on expression of stress defence genes under carbon- and energy-limited conditions
RpoS is known to play a crucial role in the response of E. coli to adverse physico-chemical conditions such as acidic pH, high osmolarity, changes in temperature and more, offering an explanation for the increased stress tolerance of stationary phase cells [63]. Here we discuss the observed differences in the transcriptomes between wild-type E. coli K12 MG1655, and RpoS- and cAMP/CRP-deficient isogenic mutants under balanced (steady-state) growth conditions (glucose-limited continuous cultures), and compare the results to those reported in the literature.
Surprisingly, only few stress defence genes (6) were found to be significantly negatively affected in cells of the ΔrpoS mutant strain during slow growth in glucose-limited chemostat culture (Table in S1 Table). Genes involved in acid resistance were affected, e.g. gadB encoding the glutamate decarboxylase isozyme and two periplasmic proteins located in the same operon (hdeA and hdeB). This down-regulation of genes involved in acid resistance is in agreement with transcriptome data obtained with a ΔrpoS strain in batch culture [33,36,37]. Two additional genes down-regulated in the ΔrpoS strain in our experiments are known to belong to the σS-regulon (OsmE, a stress inducible lipoprotein, and Bfr, a bacterioferrin). However, both genes were also down-regulated in the absence of cAMP, although the effect of cAMP on bfr expression was less pronounced than that of RpoS (Table in S1 Table). Furthermore, the RpoS-dependent gene marC, involved in antibiotic resistance, was highly down-regulated and not affected significantly by cAMP. In contrast, genes necessary for resistance against antibiotics (fsr and creD) and phages (dcrB and pspE) were affected by both cAMP and RpoS.
Cyclic AMP was observed to regulate transcription of cold shock proteins in different ways. While mRNA levels of CspD and CspC were reduced in the Δcya strain, transcription of CspF was up-regulated; this result is in agreement with a previous report [38]. Also one enzyme necessary for DNA repair (umuC) was found to be up-regulated in the absence of cAMP, which is in agreement with a previous study [39].
Unexpectedly, genes encoding two universal stress proteins, UspA and UspB, were down-regulated in the Δcya strain but not in the ΔrpoS strain, suggesting the existence of a stress regulon under the control of cAMP-CRP. The expression of uspB was previously reported to be under the RpoS-regulon in some studies [33,37], while others could not find any difference in the expression of this gene under similar conditions [32,35,36]. These results challenge the assumed dominant role of RpoS for the survival of E. coli under different stress conditions and suggest that also the cAMP-CRP regulon might contribute significantly to stress resistance. Nevertheless, RpoS might influence the cellular levels of UspA and UspB by controlling the expression of other genes that control translation, folding and stability of these stress-defence proteins.
Activity of heat-stable hydroperoxidase II, encoded by katE, was suggested to be an indicator for RpoS-dependent gene expression [25,69]. Although the negative effect of the ΔrpoS mutation on hydroperoxidase II activity was again observed in our study in carbon- and energy-limited chemostat cultures (in both glucose mineral and LB medium, Table 1), DNA microarray data gave no indication of significantly decreased transcription of katE (Table 3). The lacking negative effect of the rpoS knock-out mutation on the transcriptional level is in agreement with the results of other studies [32,33,35,36]. Possibly, RpoS exerts an indirect effect on the stability on KatE activity, e.g. by controlling the expression of genes that affect either translation, folding, activity or half-life of KatE. Expression of this gene was also not regulated by cAMP. Neither the transcription level of katE nor hydroperoxidase II activity was affected in the Δcya and Δcrp strains (Table 1 and Table 3).
Effect of cAMP and RpoS on expression of biosynthetic genes under carbon- and energy-limited conditions
The effect of either Δcya or ΔrpoS global regulatory mutations on the expression of biosynthetic pathways was limited; only few of these genes were down-regulated in the absence of one or the other of the two global regulators (Table in S1 Table).
An exception was the gene rpsV, coding for a stationary-phase ribosome-associated protein that might control protein synthesis and prevent mistranslation occurring in stationary phase [71]. This gene was activated in an RpoS-dependent manner in glucose-limited continuous culture, whereas the absence of cAMP led to a strong down-regulation (Table in S1 Table).
Transcription of a gene encoding another ribosomal protein, RpmJ, was also down-regulated in the Δcya strain; rpmJ belongs to the spc operon encoding 11 ribosomal proteins [72]. Furthermore, in the absence of cAMP mRNA levels of SecY were also reduced; this protein is a component of the type II secretory pathway for proteins [73] and plays an important role in the secretion of binding proteins to the periplasm [74]. This result is consistent with the observed reduced expression of most genes encoding periplasmic binding proteins in the absence of cAMP (Fig 1, Table 2); presumably expression and secretion are tightly coordinated.
Furthermore, cAMP seems to be involved in the regulation of the biosynthesis of histidine, as both hisG and hisC were down-regulated in the Δcya strain.
A heterogeneous pattern was observed for transcription of genes encoding enzymes involved in nucleotide biosynthesis. Whereas several genes were transcribed at higher levels in both mutants (Table in S1 Table), transcription of 2-deoxyribose-5-phosphate aldolase (DeoC) was down-regulated in the absence of cAMP. deoC is member of the deoCABD operon, which is predicted to be regulated by cAMP [53]. Interestingly, deoD, a member of this operon, was reported in other studies to be not only cAMP—but also RpoS-dependent [35,38].
Effect of cAMP and RpoS knock-out mutations on transcription of regulatory proteins
Global regulators often act via regulatory cascades by affecting the expression of other regulatory proteins that are specific for particular operons. In this sense, when grown in glucose-limited chemostat culture the two E. coli strains studied, carrying either a Δcya or a ΔrpoS mutation, were affected in the transcription of 31 regulatory genes (Table in S1 Table). In this group of genes, the majority of those affected more than 3-fold were down-regulated in the absence of cAMP (23 out of 26), whereas 9 out of 13 were up-regulated when cells were lacking RpoS.
In the Δcya strain, the genes down-regulated code for regulators controlling the transcription of operons and genes with functions ascribed to metabolic pathways, cell structures, DNA structure, DNA modification, sigma factors, sensor proteins, stress regulation and DNA-binding proteins. Only 4 such genes were down-regulated in the ΔrpoS strain and they are considered to be responsible for the regulation of functions assigned to cell structure and stress responses. Genes found to be up-regulated (9) are known to be involved in the regulation of proteins used in different functions, such as RNA modification, the glc operon for sugar transport, cell structure and sensory proteins.
Transcription of the four genes down-regulated in the ΔrpoS strain was also affected in the strain lacking cAMP; whereas bolA and csrA were down-regulated to comparable levels, yiaG and dps were considerably more affected by the lack of RpoS. BolA, the transcription of which is reported to be under the control of σS [75], regulates the morphology of cells [76] and seems to be involved in biofilm formation [77]. CsrA is a regulator of carbohydrate metabolism and affects glycogen degradation [78], glycogen biosynthesis, gluconeogenesis, and glycolysis [79,80]. CsrA has also a regulatory role in biofilm formation [81], and also plays a role in motility via post-transcriptional activation of the expression of the flhDC flagellar regulatory genes [82]. YiaG is a putative transcriptional regulator and was also found to be down-regulated in an ΔrpoS mutant in batch culture [35–37].
Interestingly, transcription of the dps gene was reduced in both mutant strains. Dps is an abundant protein in stationary phase cells and is involved in stress response [83], its main function is assumed to be the chelation of intracellular iron in order to prevent DNA damage [84]. Dps expression was reported to be regulated by RpoS and repressed by cAMP-CRP [85], which is in contrast to our results. The repression observed earlier in a Δcrp strain was suggested to occur indirectly and it was speculated that this de-repression was caused by an up-regulation of RpoS [86]. In our experiments we did not observe an up-regulation of RpoS in the Δcya mutant and, therefore, the mechanism proposed by Joeng and colleagues [86] is unlikely. We assume that either other regulatory proteins or additional factors are involved in the regulation of dps as recently proposed [87,88].
Surprisingly, the lack of cAMP affected transcription of one sigma factor, RpoE, which encodes a sigma factor involved in the response to heat shock and envelope stress [89], and its negative regulatory protein (rseA). Activity of RpoE is increased upon the entry into the stationary phase and it has been reported to be negatively regulated by RseA and positively by ppGpp [90]. The down-regulation of RpoE strongly suggests that in the absence of cAMP the strain might not survive when exposed to different stresses such as heat shock and envelope stress. This observation might be related to the fact that the combined volume of the outer membrane and the periplasm of strains lacking CRP is larger than that of wild-type strains [91].
Regulatory proteins of several catabolic operons were also down-regulated in the Δcya strain, e.g., SfsA controlling genes for maltose utilization, the repressor for galactitol utilization (GatR), and YehH which regulates molybdate metabolism. Transcription of genes encoding DNA-binding proteins H-NS and HU was also reduced. Interestingly, the mRNA levels of the soxS gene, coding for a regulatory protein involved in the response to oxidative stress, were found to be up-regulated in both mutants.
Effect of cAMP and RpoS knock-out mutations on other cellular functions
In addition to the observed effects of the absence of cAMP and RpoS on catabolic functions and stress defence, few other cellular functions were affected in cells growing in glucose-limited chemostat culture at a dilution rate corresponding to half μmax.
In the Δcya strain a variety of genes necessary for the synthesis of structural elements were down-regulated, e.g. genes coding for lipoproteins (ybjP, lpp, nlpD and blc), flagellum (flgD, belonging to class II), membrane biosynthesis enzymes (ddpX, wbbL, yfcX and pssA) and a putative sporulation protein (ycgB). Important is the finding that the major lipoprotein Lpp was down-regulated in the Δcya strain, this lipoprotein is necessary for the stabilization and integrity of the bacterial cell envelope [92]. These unexpected and so far unknown results strongly suggest that in the absence of cAMP the cellular envelope is affected by the down-regulation of many genes and, hence, enzymes involved in the synthesis of membrane components, resulting in a weak incorporation of transporters and a, consequently, a poor functioning of the cell membrane.
In the absence of cAMP some genes were up-regulated; these genes are involved in membrane biosynthesis (sfa), one putative lipoprotein (yehR) and fimbrial proteins (ycbQ and genes encoded by the fimAICDFGH operon). The up-regulation of genes involved in motility in the absence of the cAMP-CRP complex was also reported in other studies performed with E. coli [38] and V. cholerae [58]. Interestingly, ycbQ and members of the fimAICDFGH operon were also up-regulated in the ΔrpoS strain, which is in agreement with a previous observation [33]. The latter operon encodes fimbriae of type 1 pili that are mannose-sensitive [93], these pili can bind to mannose residues to colonize surfaces.
Structural genes were less affected in the ΔrpoS strain. Genes for membrane biosynthesis (kdsA) and cell surface antigens (uppS and rfe) were up-regulated while only two genes were found to be down-regulated, ycgB and blc. The ycgB gene is also regulated by RpoS in Salmonella enterica serovar Typhimurium [94] and was reported to be under the control of RpoS in other transcriptome studies with E. coli [36,37]. Consistent with the down-regulation observed for the Δcya strain in this study, the ycgB gene contains a predicted cAMP-CRP binding site in its promoter region [53]. In agreement with our transcriptome results for the ΔrpoS strain, transcription of the blc gene is induced at the beginning of the stationary phase in wild-type E. coli [95]. Its product might be involved in membrane repair, lipid storage or lipid transport [96].
Interestingly, genes involved in autoinducer 2 transport (ego or lsrA) and in biofilm production (ycdS and ycdU) were down-regulated both in the ΔrpoS and the Δcya strains to similar levels, which indicates a regulatory cooperation of both global regulators as suggested by others [81,97].
Conclusions
In our study, we have provided a comprehensive analysis of role of the global regulators cAMP and RpoS in E. coli under well-defined carbon- and energy-limited growth conditions. The lack of rpoS resulted in an enhanced fitness both under glucose-excess and glucose-limited conditions, whereas the lack of cAMP had a drastic negative effect on growth performance (Tables 1 and 3). The transcriptome data were confirmed by catabolome analysis, expression of periplasmic proteins and phenotypical analysis. The results obtained with the different methods were consistent. Thus, the picture obtained strongly suggests that the expression of a broad range of high-affinity uptake systems and of enzymes involved in the central metabolism (citric acid cycle and of glycolysis) were severely repressed in the absence of cAMP. Furthermore, the presented results are in agreement with flux analysis [62] and a transcriptome study performed in complex medium [38,39]. Hence, one can predict that the Δcya strain must exhibit a worse affinity than the wild-type strain not only for glucose but also for a broad range of other carbon sources. On the contrary, absence or down-regulation of RpoS would result in increased competitiveness of E. coli in the environment. These results are in agreement with the trade-off between self-preservation and nutritional competence (SPANC [98]). High variability in RpoS expression from several E. coli strains was reported, where high RpoS expression resulted in high resistance at the cost of lower competitiveness, e.g., reduced catabolic flexibility [98,99].
The alarmone ppGpp, which is involved in the expression/stability of RpoS [100,101] and is necessary for the transcription of several genes belonging to the RpoS-regulon [102], has also been suggested to be involved in the SPANC balance [103]. The lacking negative effect of the rpoS knockout mutation on transcription of certain stress resistance genes might be due to direct regulation by ppGpp which is, like RpoS, present at high concentrations at low specific growth rates [104].
In this study we have demonstrated that the absence of cAMP affects also different classes of genes that are necessary for surviving and growing in the environment. Interestingly, several genes found to be regulated by RpoS were also affected in the Δcya mutant, these genes belonged to all functional groups and numerous genes regulated by both global regulators in our experiments were previously reported to be under exclusive transcriptional control of RpoS. Our data confirm and extend recent suggestions that some genes are regulated by both regulators [38]. Weber and co-workers [37] have shown in a promoter analysis of ΔrpoS transcriptome data that about one half of all RpoS-regulated genes also possess a hypothetical cAMP-CRP binding motif in the promoter region. The transcriptome data reported here can serve as a useful starting point for further analysis of the involvement of cAMP in the regulation of different genes.
In summary we conclude that global regulation by cAMP-CRP, but not by RpoS is essential for growth and survival of E. coli in its natural habitats which also explains why rpoS but not cya/crp mutant strains can be isolated from the environment.
Supporting Information
(DOCX)
Data Availability
The microarray data have been submitted to Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE25982.
Funding Statement
The authors gratefully acknowledge the financial support for this research, which was provided for AGF by the Swiss National Science Foundation (SNF grant 31-63466.00) and for JI by Eawag, Swiss Federal Institute of Aquatic Science and Technology. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Morita RY. Bioavailability of energy and the starvation state In: Kjelleberg S.. Starvation in bacteria, New York: Plenum Press; 1993. pp. 1–23. [Google Scholar]
- 2. Savageau MA. Escherichia coli habitats, cell types, and molecular mechanisms of gene control. Am Nat 1983; 122: 732–744. [Google Scholar]
- 3. Ferenci T. Hungry bacteria—definition and properties of a nutritional state. Environ Microbiol 2001; 3: 605–611. [DOI] [PubMed] [Google Scholar]
- 4. Herbert D. The chemical composition of micro-organisms as a function of their environment. Symp Soc Gen Microbiol 1961; 11: 391–416. [Google Scholar]
- 5. Neidhardt FC, Savageau MA. Regulation beyond the operon In: Neidhardt F. C.. Escherichia coli and Salmonella typhimurium, cellular and molecular biology, 2nd edition, Washington DC: American Society for Microbiology; 1996. pp. 1310–1324. [Google Scholar]
- 6. Martinez-Antonio A, Janga SC, Thieffry D. Functional organisation of Escherichia coli transcriptional regulatory network. J Mol Biol 2008; 381: 238–247. 10.1016/j.jmb.2008.05.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Klumpp S, Hwa T. Bacterial growth: global effects on gene expression, growth feedback and proteome partition. Curr Opin Biotechnol 2014; 28: 96–102. 10.1016/j.copbio.2014.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Harder W, Dijkhuizen L. Strategies of mixed substrate utilization in microorganisms. Philos T Roy Soc B 1982; 297: 459–480. [DOI] [PubMed] [Google Scholar]
- 9. Nystrom T. To be or not to be: the ultimate decision of the growth-arrested bacterial cell. Fems Microbiol Rev 1998; 21: 283–290. [Google Scholar]
- 10. Egli T. The ecological and physiological significance of the growth of heterotrophic microorganisms with mixtures of substrates. Adv Microb Ecol 1995; 14: 305–386. [Google Scholar]
- 11. Egli T. How to live at very low substrate concentration. Water Res 2010; 44: 4826–4837. 10.1016/j.watres.2010.07.023 [DOI] [PubMed] [Google Scholar]
- 12. Matin A. Microbial regulatory mechanisms at low nutrient concentrations as studied in a chemostat In: Shilo M. Strategies of Microbial Life in Extreme Environments, Berlin: Dahlem Konferenzen, Berlin; 1979. [Google Scholar]
- 13. Franchini AG, Egli T. Global gene expression in Escherichia coli K-12 during short-term and long-term adaptation to glucose-limited continuous culture conditions. Microbiology 2006; 152: 2111–2127. [DOI] [PubMed] [Google Scholar]
- 14. Harder W, Kuenen JG, Matin A. Microbial selection in continuous culture. J Appl Bacteriol 1977; 43: 1–24. [DOI] [PubMed] [Google Scholar]
- 15. Ihssen J, Egli T. Global physiological analysis of carbon- and energy-limited growing Escherichia coli confirms a high degree of catabolic flexibility and preparedness for mixed substrate utilization. Environ Microbiol 2005; 7: 1568–1581. [DOI] [PubMed] [Google Scholar]
- 16. Nahku R, Valgepea K, Lahtvee P-J, Erm S, Abner K, Adamberg K, et al. Specific growth rate dependent transcriptome profiling of Escherichia coli K12 MG1655 in accelerostat cultures. J Biotechnol 2010; 145: 60–65. 10.1016/j.jbiotec.2009.10.007 [DOI] [PubMed] [Google Scholar]
- 17. Wanner U, Egli T. Dynamics of microbial growth and cell composition in batch culture. Fems Microbiol Rev 1990; 75:19–44. [DOI] [PubMed] [Google Scholar]
- 18. Wick LM, Quadroni M, Egli T. Short- and long-term changes in proteome composition and kinetic properties in a culture of Escherichia coli during transition from glucose-excess to glucose-limited growth conditions in continuous culture and vice versa. Environ Microbiol 2001; 3: 588–599. [DOI] [PubMed] [Google Scholar]
- 19. Notley L, Ferenci T. Induction of RpoS-dependent functions in glucose-limited continuous culture: What level of nutrient limitation induces the stationary phase of Escherichia coli? J Bacteriol 1996; 178: 1465–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Berthoumieux S, de Jong H, Baptist G, Pinel C, Ranquet C, Ropers D, et al. Shared control of gene expression in bacteria by transcription factors and global physiology of the cell. Mol Syst Biol 2013; 9: 634 10.1038/msb.2012.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Matin A, Matin MK. Cellular levels, excretion, and synthesis rates of cyclic AMP in Escherichia coli grown in continuous culture. J Bacteriol 1982; 149: 801–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Notley-McRobb L, Death A, Ferenci T. The relationship between external glucose concentration and cAMP levels inside Escherichia coli: Implications for models of phosphotransferase-mediated regulation of adenylate cyclase. Microbiology 1997; 143: 1909–1918. [DOI] [PubMed] [Google Scholar]
- 23. Valgepea K, Adamberg K, Nahku R, Lahtvee P-J, Arike L, Vilu R. Systems biology approach reveals that overflow metabolism of acetate in Escherichia coli is triggered by carbon catablite repression of acetyl-CoA synthetase. BMC Syst Biol 2010; 4: 166 10.1186/1752-0509-4-166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jishage M, Iwata A, Ueda S, Ishihama A. Regulation of RNA polymerase sigma subunit synthesis in Escherichia coli: Intracellular levels of four species of sigma subunit under various growth conditions. J Bacteriol 1996; 178: 5447–5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ihssen J, Egli T. Specific growth rate and not cell density controls the general stress response in Escherichia coli . Microbiology 2004; 150: 1637–1648. [DOI] [PubMed] [Google Scholar]
- 26. Egli T. Microbial growth and physiology: A call for better craftsmanship. In: Roeling W, Bodegom P, eds. 50 Years after Pirt: from microbial physiology and ecology to quantitative biogeochemistry. Front Microbiol 2015; 6:287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chubukov V, Sauer U. Environmental dependence of stationary-phase metabolism in Bacillus subtilis and Escherichia coli . Appl Environ Microbiol 2014; 80: 2901–2909. 10.1128/AEM.00061-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Egli T. Nutrition, microbial In: Schaechter M. Desk Encyclopedia of Microbiology, Oxford, UK: Elsevier Academic Press; 2009: 788–804. [Google Scholar]
- 29. Ingraham JL, Maaløe O, Neidhardt FC. Growth of the Bacterial Cell. Sunderland, MA: Sinauer Assoc; 2009. [Google Scholar]
- 30. Pirt SJ. Principles of Microbe and Cell Cultivation. Oxford, London, Edinburgh, Melbourne: Blackwell Scientific Publications; 1975. [Google Scholar]
- 31. Wick LM, Weilenmann H, Egli T. The apparent clock-like evolution of Escherichia coli in glucose-limited chemostats is reproducible at large but not at small population sizes and can be explained with Monod kinetics. Microbiology 2002; 148: 2889–2902. [DOI] [PubMed] [Google Scholar]
- 32. Dong T, Kirchhof MG, Schellhorn HE. RpoS regulation of gene expression during exponential growth of Escherichia coli K12. Mol Genet Genomics 2008; 279: 267–277. [DOI] [PubMed] [Google Scholar]
- 33. Dong T, Schellhorn HE. Control of RpoS in global gene expression of Escherichia coli in minimal media. Mol Genet Genomics 2009; 281: 19–33. 10.1007/s00438-008-0389-3 [DOI] [PubMed] [Google Scholar]
- 34. Dong T, Yu R, Schellhorn HE. Antagonistic regulation of motility and transcriptome expression by RpoN and RpoS in Escherichia coli . Mol Microbiol 2011; 79: 375–386 10.1111/j.1365-2958.2010.07449.x [DOI] [PubMed] [Google Scholar]
- 35. Lacour S, Landini P. σS-dependent gene expression at the onset of stationary phase in Escherichia coli: Function of σS-dependent genes and identification of their promoter sequences. J Bacteriol 2004; 186: 7186–7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Patten CL, Kirchhof MG, Schertzberg MR, Morton RA, Schellhorn HE. Microarray analysis of RpoS-mediated gene expression in Escherichia coli K-12. Mol Genet Genomics 2004; 272: 580–591. [DOI] [PubMed] [Google Scholar]
- 37. Weber H, Polen T, Heuveling J, Wendisch VF, Hengge R. Genome-wide analysis of the general stress response network in Escherichia coli: σS-dependent genes, promoters, and sigma factor selectivity. J Bacteriol 2005; 187: 1591–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gosset G, Zhang ZG, Nayyar SN, Cuevas WA, Saier MH. Transcriptome analysis of Crp-dependent catabolite control of gene expression in Escherichia coli . J Bacteriol 2004; 186: 3516–3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Khankal R, Chin JW, Gosh D, Cirino PC. Transcriptional effects of CRP* expression in Escherichia coli . J Biol Eng 2009; 3:13 10.1186/1754-1611-3-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hung SP, Baldi P, Hatfield GW. Global gene expression profiling in Escherichia coli K12—The effects of leucine-responsive regulatory protein. J Biol Chem 2002; 277: 40309–40323. [DOI] [PubMed] [Google Scholar]
- 41. Tani TH, Khodursky A, Blumenthal RM, Brown PO, Matthews RG. Adaptation to famine: A family of stationary-phase genes revealed by microarray analysis. P Natl Acad Sci USA 2002; 99: 13471–13476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Typas A, Barembruch C, Possling A, Hengge R. Stationary phase reorganisation of the Escherichia coli transcription machinery by Crl protein, a fine-tuner of σS activity and levels. Embo J 2007; 26: 1569–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hengge-Aronis R. Stationary phase gene regulation: what makes an Escherichia coli promoter σS-selective? Curr Opin Microbiol 2002; 5: 591–595. [DOI] [PubMed] [Google Scholar]
- 44. Schultz JE, Latter GI, Matin A. Differential regulation by cyclic-AMP of starvation protein synthesis in Escherichia coli . J Bacteriol 1988; 170: 3903–3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Berney M, Weilenmann H-U, Ihssen J, Bassin C, Egli T. Specific growth rate determines the sensitivity of Escherichia coli to thermal, UVA, and solar disinfection. Appl Environ Microbiol 2006; 72: 2586–2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kovarova K, Zehnder AJB, Egli T. Temperature-dependent growth kinetics of Escherichia coli ML 30 in glucose-limited continuous culture. J Bacteriol 1996; 178: 4530–4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. New York: Cold Spring Harbour Press; 1989. [Google Scholar]
- 48. Morita RY. Bioavailability of energy and its relationship to growth and starvation survival in nature. Can J Microbiol 1988; 34: 436–441. [Google Scholar]
- 49. Koch AL. The adaptive responses of Escherichia coli to a feast and famine existence. Adv Microb Physiol 1971; 6: 147–217. [DOI] [PubMed] [Google Scholar]
- 50. Münster U. Concentrations and fluxes of organic carbon substrates in the aquatic environment. Antonie van Leeuwenhoek 1993; 63: 243–274. [DOI] [PubMed] [Google Scholar]
- 51. Vital M, Hammes F, Egli T. Competition of Escherichia coli O157 with a drinking water bacterial community at low nutrient concentrations. Water Res 2012; 46: 6279–6290. 10.1016/j.watres.2012.08.043 [DOI] [PubMed] [Google Scholar]
- 52. Perrenoud A, Sauer U. Impact of global transcriptional regulation by ArcA, ArcB, Cra, Crp, Cya, Fnr, and Mlc on glucose catabolism in Escherichia coli . J Bacteriol 2005; 187: 3171–3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Salgado H, Peralta-Gil M, Gama-Castro S, Santos-Zavaleta A, Muniz-Rascado L, Garcia-Sotelo JS, et al. RegulonDB v8.0: omics data sets, evolutionary conservation, regulatory phrases, cross-validated gold standards and more. Nucleic Acids Res 2013; 41: D203–D213. 10.1093/nar/gks1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tan K, Moreno-Hagelsieb G, Collado-Vides J, Stormo GD. A comparative genomics approach to prediction of new members of regulons. Genome Res 2001; 11: 566–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ferenci T. Adaptation to life at micromolar nutrient levels: regulation of Escherichia coli glucosetransport by endoinduction and cAMP. FEMS Microbiol Rev 1996; 18: 301–317. [DOI] [PubMed] [Google Scholar]
- 56. Notley-McRobb L, Ferenci T. Adaptive mgl-regulatory mutations and genetic diversity evolving in glucose-limited Escherichia coli populations. Environ Microbiol 1999; 1: 33–43. [DOI] [PubMed] [Google Scholar]
- 57. Notley-McRobb L, Ferenci T. The generation of multiple coexisting mal-regulatory mutations through polygenic evolution in glucose-limited populations of Escherichia coli . Environ Microbiol 1999; 1: 45–52. [DOI] [PubMed] [Google Scholar]
- 58. Fong JCN, Yildiz FH. Interplay between cyclic AMP-Cyclic AMP receptor protein and cyclic di-GMP signaling in Vibrio cholerae biofilm formation. J Bacteriol 2008; 190: 6646–6659. 10.1128/JB.00466-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lee SK, Newman JD, Keasling JD. Catabolite repression of the propionate catabolic genes in Escherichia coli and Salmonella enterica: Evidence for involvement of the cyclic AMP receptor protein. J Bacteriol 2005; 187: 2793–2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kumari S, Beatty CM, Browning DF, Busby SJW, Simel EJ, Hovel-Miner G, et al. Regulation of acetyl coenzyme A synthetase in Escherichia coli . J Bacteriol 2000; 182: 4173–4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Renilla S, Bernal V, Fuhrer T, Castaño-Cerezo S, Pastor JM, Iborra JL, et al. Acetate scavenging activity in Escherichia coli: interplay of acetyl–CoA synthetase and the PEP–glyoxylate cycle in chemostat cultures. Appl Micriobiol Biotechnol 2012; 93: 2109–2124. [DOI] [PubMed] [Google Scholar]
- 62. Nanchen A, Schicker A, Revelles O, Sauer U. Cyclic AMP-dependent catabolite repression is the dominant control mechanism of metabolic fluxes under glucose limitation in Escherichia coli . J Bacteriol 2008; 190: 2323–2330. 10.1128/JB.01353-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hengge-Aronis R. (1996). Regulation of gene expression during entry into the stationary phase In Neidhardt F. C.. Escherichia coli and Salmonella typhimurium, cellular and molecular biology, 2nd edition, Washington DC: American Society for Microbiology; 1996. pp. 1497–1512. [Google Scholar]
- 64. Farewell A, Kvint K, Nystrom T. Negative regulation by RpoS: a case of sigma factor competition. Mol Microbiol 1998; 29: 1039–1051. [DOI] [PubMed] [Google Scholar]
- 65. Maeda H, Fujita N, Ishihama A. Competition among seven Escherichia coli σ subunits: relative binding affinities to the core RNA polymerase. Nucleic Acids Res 2000; 28: 3497–3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Nystrom T. Growth versus maintenance: a trade-off dictated by RNA polymerase availability and sigma factor competition? Mol Microbiol 2004; 54: 855–862. [DOI] [PubMed] [Google Scholar]
- 67. Masuda N, Church GM. Regulatory network of acid resistance genes in Escherichia coli . Mol Microbiol 2003; 48: 699–712. [DOI] [PubMed] [Google Scholar]
- 68. Cunningham L, Gruer MJ, Guest JR. Transcriptional regulation of the aconitase genes (acnA and acnB) of Escherichia coli . Microbiology 1997; 143: 3795–3805. [DOI] [PubMed] [Google Scholar]
- 69. Notley-McRobb L, King T, Ferenci T. rpoS mutations and loss of general stress resistance in Escherichia coli populations as a consequence of conflict between competing stress responses. J Bacteriol 2002; 184: 806–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Visick JE, Clarke S. RpoS- and OxyR-independent induction of HPI catalase at stationary phase in Escherichia coli and identification of rpoS mutations in common laboratory strains. J Bacteriol 1997; 179: 4158–4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Izutsu K, Wada C, Komine Y, Sako T, Ueguchi C, Nakura S, et al. Escherichia coli ribosome-associated protein SRA, whose copy number incrreases during stationary phase. J Bacteriol 2001; 183: 2765–2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Komine Y, Adachi T, Inokuchi H, Ozeki H. Genomic organization and physical mapping of the transfer RNA genes in Escherichia coli K12. J Mol Biol 1990; 212: 579–598. [DOI] [PubMed] [Google Scholar]
- 73. Nishiyama K, Mizushima S, Tokuda H. A novel membrane protein involved in protein translocation across the cytoplasmic membrane of Escherichia coli. Embo J 1993; 12: 3409–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ikegami A, Nishiyama K, Matsuyama S, Tokuda H. Disruption of rpmJ encoding ribosomal protein L36 decreases the expression of secY upstream of the spc operon and inhibits protein translocation in Escherichia coli . Biosci Biotechnol Biochem 2005; 69: 1595–1602. [DOI] [PubMed] [Google Scholar]
- 75. Lange R, Hengge-Aronis R. Growth phase-regulated expression of bolA and morphology of stationary-phase Escherichia coli cells are controlled by the novel sigma factor σS . J Bacteriol 1991; 173: 4474–4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Aldea M, Hernandezchico C, Delacampa AG, Kushner SR, Vicente M. Identification, cloning, and expression of bolA, an ftsZ-dependent morphogene of Escherichia coli . J Bacteriol 1988; 170: 5169–5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Vieira HLA, Freire P, Arraiano CM. Effect of Escherichia coli morphogene bolA on biofilms. Appl Environ Microb 2004; 70: 5682–5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Yang HH, Liu MY, Romeo T. Coordinate genetic regulation of glycogen catabolism and biosynthesis in Escherichia coli via the CsrA gene product. J Bacteriol 1996; 178: 1012–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Sabnis NA, Yang HH, Romeo T. Pleiotropic regulation of central carbohydrate metabolism in Escherichia coli via the gene csrA . J Biol Chem 1995; 270: 29096–29104. [DOI] [PubMed] [Google Scholar]
- 80. Wei B, Shin S, LaPorte D, Wolfe AJ, Romeo T. Global regulatory mutations in csrA and rpoS cause severe central carbon stress in Escherichia coli in the presence of acetate. Journal of Bacteriology 2000; 182: 1632–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Jackson DW, Simecka JW, Romeo T. Catabolite repression of Escherichia coli biofilm formation. J Bacteriol 2002; 184: 3406–3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Wei BDL, Brun-Zinkernagel AM, Simecka JW, Pruss BM, Babitzke P, Romeo T. Positive regulation of motility and flhDC expression by the RNA-binding protein CsrA of Escherichia coli . Mol Microbiol 2001; 40: 245–256. [DOI] [PubMed] [Google Scholar]
- 83. Almiron M, Link AJ, Furlong D, Kolter R. A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli . Gene Dev 1992; 6: 2646–2654. [DOI] [PubMed] [Google Scholar]
- 84. Halsey TA, Vazquez-Torres A, Gravdahl DJ, Fang FC, Libby SJ. The ferritin-like Dps protein is required for Salmonella enterica serovar Typhimurium oxidative stress resistance and virulence. Infect Immun 2004; 72: 1155–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Altuvia S, Almiron M, Huisman G, Kolter R, Storz G. The Dps promoter is activated by OxyR during growth and by Ihf and a Sigma(S) in Stationary-Phase. Mol Microbiol 1994; 13: 265–272. [DOI] [PubMed] [Google Scholar]
- 86. Jeong KC, Baumler DJ, Kaspar CW. dps expression in Escherichia coli O157: H7 requires an extended—10 region and is affected by the cAMP receptor protein. Bba-Gene Struct Expr 2006; 1759: 51–59. [DOI] [PubMed] [Google Scholar]
- 87. Maciag A, Peano C, Pietrelli A, Egli T, De Bellis G, Landini P. In vitro transcription profiling of the σS subunit of bacterial RNA polymerase: re-definition of the σS regulon and identification of σS-specific promoter sequence elements. Nucleic Acids Res 2011; 39: 5338–5355. 10.1093/nar/gkr129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Traxler MF, Zacharia VM, Marquardt S, Summers SM, Nguyen HT, Stark SE, et al. Discretely calibrated regulatory loops controlled by ppGpp partition gene induction across the ‘feast to famine’ gradient in Escherichia coli . Mol Microbiol 2011; 79: 830–845. 10.1111/j.1365-2958.2010.07498.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Gross CA. Function and regulation of the heat shock protein In: Neidhardt F. C.. Escherichia coli and Salmonella typhimurium, cellular and molecular biology, 2nd edition, Washington DC: American Society for Microbiology; 1996. pp. 1382–1399. [Google Scholar]
- 90. Costanzo A, Ades SE. Growth phase-dependent regulation of the extracytoplasmic stress factor, σE, by guanosine 3 ',5 '-bispyrophosphate (ppGpp). J Bacteriol 2006; 188: 4627–4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Barth E, Gora KV, Gebendorfer KM, Settele F, Jakob U, Winter J. Interplay of cellular cAMP levels, σS activity and oxidative stress resistance in Escherichia coli . Microbiology 2009; 155: 1680–1689. 10.1099/mic.0.026021-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Cascales E, Bernadac A, Gavioli M, Lazzaroni JC, Lloubes R. Pal lipoprotein of Escherichia coli plays a major role in outer membrane integrity. J Bacteriol 2002; 184: 754–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Hahn E, Wild P, Hermanns U, Sebbel P, Glockshuber R, Häner M, et al. Exploring the 3D molecular architecture of Escherichia coli type 1 pili. J Mol Biol 2002; 323: 845–857. [DOI] [PubMed] [Google Scholar]
- 94. Ibanez-Ruiz M, Robbe-Saule V, Hermant D, Labrude S, Norel F. Identification of RpoS (σS)-regulated genes in Salmonella enterica serovar Typhimurium. J Bacteriol 2000; 182: 5749–5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Bishop RE, Penfold SS, Frost LS, Holtje JV, Weiner JH. Stationary-phase expression of a novel Escherichia coli outer membrane lipoprotein and its relationship with mammalian apolipoprotein-D—Implications for the origin of lipocalins. J Biol Chem 1995; 270: 23097–23103. [DOI] [PubMed] [Google Scholar]
- 96. Campanacci V, Nurizzo D, Spinelli S, Valencia C, Tegoni M, Cambillau C. The crystal structure of the Escherichia coli lipocalin Blc suggests a possible role in phospholipid binding. Febs Lett 2004; 562: 183–188. [DOI] [PubMed] [Google Scholar]
- 97. Wang L, Hashimoto Y, Tsao CY, Valdes JJ, Bentley WE. Cyclic AMP (cAMP) and cAMP receptor protein influence both synthesis and uptake of extracellular autoinducer 2 in Escherichia coli . J Bacteriol 2005; 187: 2066–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Ferenci T. Maintaining a healthy SPANC balance through regulatory and mutational adaptation. Mol Microbiol 2005; 57: 1–8. [DOI] [PubMed] [Google Scholar]
- 99. Phan K, Ferenci T. A design-constraint trade-off underpins the diversity in ecologically important traits in species Escherichia coli . ISME J 2013; 7: 2034–2043. 10.1038/ismej.2013.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Lange R, Fischer D, Hengge-Aronis R. Identification of transcriptional start sites and the role of ppGpp in the expression of rpoS, the structural gene for the sigmaS subunit of RNA polymerase in Escherichia coli. J Bacteriol 1995; 177: 4676–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Bougdour A, Gottesman S. ppGpp regulation of RpoS degradation via anti-adaptor protein IraP. Proc Natl Acad Sci USA 2007; 104: 12896–12901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Traxler MF, Zacharia VM, Marquardt S, Summers SM, Nguyen HT, Stark SE, et al. Discretely calibrated regulatory loops controlled by ppGpp partition gene induction across the ‘feast to famine’ gradient in Escherichia coli . Mol Microbiol 2011; 79: 830–845. 10.1111/j.1365-2958.2010.07498.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Ferenci T, Galbiati HF, Betteridge T, Phan K, Spira B. The constancy of global regulation across aspecies: the concentrations of ppGpp and RpoS are strain-specific in Escherichia coli . BMC Microbiology 2011; 11: 62 10.1186/1471-2180-11-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Teich A, Meyer S, Lin HY, Andersson L, Enfors SO, Neubauer P. Growth rate related concentration changes of the starvation response regulators σS and ppGpp in glucose-limited fed-batch and continuous cultures of Escherichia coli . Biotechnol Prog 1999; 15: 123–129. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Data Availability Statement
The microarray data have been submitted to Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE25982.