Abstract
Hematopoietic stem cells (HSCs) are functionally defined as cells that upon transplantation into irradiated or otherwise immunocompromised adult organisms provide long-term reconstitution of the entire hematopoietic system. They emerge in the vertebrate conceptus around midgestation. Genetic studies have identified a number of transcription factors and signaling molecules that act at the onset of hematopoiesis, and have begun to delineate the molecular mechanisms underlying the formation of HSCs. One molecule that has been a particularly useful marker of this developmental event in multiple species is Runx1 (also known as AML1, Pebp2α). Runx1 is a sequence-specific DNA-binding protein, that along with its homologues Runx2 and Runx3 and their shared non-DNA binding subunit CBFβ, constitute a small family of transcription factors called core-binding factors (CBFs). Runx1 is famous for its role in HSC emergence, and notorious for its involvement in leukemia, as chromosomal rearrangements and inactivating mutations in the human RUNX1 gene are some of the most common events in de novo and therapy-related acute myelogenous leukemia, myelodysplastic syndrome and acute lymphocytic leukemia. Here we will review the role of Runx1 in HSC emergence in the mouse conceptus and describe some of the genetic pathways that operate upstream and downstream of this gene. Where relevant, we will include data obtained from other species and embryonic stem (ES) cell differentiation cultures.
Keywords: Runx1, HSC, hemogenic endothelium, transcription, ontogeny
Mouse developmental hematopoiesis
Hematopoiesis in the conceptus (we use the word conceptus to include all the products of conception, including the embryo and extra-embryonic tissues) is generally divided into primitive and definitive hematopoiesis. Primitive hematopoiesis is restricted to the extra-embryonic yolk sac and starts at 7.5 days post coitus (dpc) in the mouse. It gives rise to primitive erythrocytes, macrophages, and megakaryocytes (Haar and Ackerman, 1971; Moore and Metcalf, 1970; Palis et al., 1999; Tober et al., 2007). Definitive hematopoiesis occurs asynchronously at distinct locations in the mouse conceptus. These include the extra-embryonic yolk sac and placenta, the vitelline and umbilical arteries (which connect the yolk sac and placenta, respectively, to the embryo), and within the embryo proper in the area of the dorsal aorta where it is flanked by the urogenital ridges, the so-called aorta-gonad-mesonephros (AGM) region, and its predecessor the para-aortic splanchopleura (p-Sp) (Alvarez-Silva et al., 2003; Cumano et al., 1996; de Bruijn et al., 2000; Garcia-Porrero et al., 1995; Godin et al., 1995; Medvinsky and Dzierzak, 1996; Medvinsky et al., 1993; Müller et al., 1994; Palis et al., 1999). Definitive hematopoiesis gives rise to erythrocytes expressing βmajor and not embryonic globin (Brotherton et al., 1979; Palis et al., 1999; Wong et al., 1983), cells of all myeloid and lymphoid lineages and ultimately to the definitive HSCs which maintain lifelong hematopoiesis. The first definitive blood cells appear in the yolk sac and are committed progenitors of various types, whilst functional definitive HSCs are first and autonomously generated in the >34 somite pair stage (sp) AGM (Medvinsky and Dzierzak, 1996; Müller et al., 1994). For a more comprehensive description of the types of blood cell progenitors that emerge over the several day period between 7.5 and 11.0 dpc, and the types of assays used to detect them, we refer the reader to Table 1 in the review by Speck and Dzierzak (Dzierzak and Speck, 2008).
Runx1 deficiency blocks the formation of definitive hematopoietic cells
The phenotype of Runx1 deficient mice is unique, phenocopied only by knockout of the gene encoding for its heterodimeric partner CBFβ (Cbfb) (Okuda et al., 1996; Sasaki et al., 1996; Wang et al., 1996a; Wang et al., 1996b). Runx1 and CBFβ deficient embryos undergo a dramatic death, often heralded by profound hemorrhaging in relatively specific areas including the central nervous system and VII/VIII cranial nerve. Blood cells in the sites of hemorrhaging consist solely of primitive erythrocytes, while definitive blood cells (definitive hematopoietic progenitors and HSCs), are virtually absent from Runx1 deficient conceptuses, and severely reduced in CBFβ mutants (Cai et al., 2000; Li et al., 2006; Okuda et al., 1996; Sasaki et al., 1996; Wang et al., 1996a; Wang et al., 1996b). This lack of definitive hematopoietic cells is observed in all hematopoietic sites of the mouse conceptus (yolk sac, vitelline and umbilical arteries, AGM region, allantois/placenta, fetal liver, thymus) (Mukouyama et al., 2000; North et al., 1999; Okuda et al., 1996; Rhodes et al., 2008; Sasaki et al., 1996; Wang et al., 1996a; Wang et al., 1996b; Yokomizo et al., 2001; Zeigler et al., 2006). This is unlike defects in Notch signaling that affect hematopoiesis in some sites (AGM region) but not others (yolk sac) (Hadland et al., 2004; Kumano et al., 2003; Robert-Moreno et al., 2005; Robert-Moreno et al., 2008).
Of the primitive lineages, primitive macrophages and their progenitors are also absent from Runx1-deficient mouse conceptuses and Runx1-null ES cell cultures (Lacaud et al., 2002; Li et al., 2006) (K. Liddiard and MdB, unpublished observations). Although primitive megakaryocyte development in Runx1-deficient conceptuses has not been examined directly, megakaryocytes and platelets are absent (Okuda et al., 1996; Wang et al., 1996a), indicative of a block somewhere during the differentiation of this lineage. Primitive erythrocytes, in contrast, were present in both Runx1- and CBFβ-deficient mouse conceptuses, and are generated from mouse ES cells with either Runx1 or Cbfb mutations (Lacaud et al., 2002; Miller et al., 2001; Okuda et al., 1996; Sasaki et al., 1996; Wang et al., 1996a; Wang et al., 1996b). However, some defects in Runx1-deficient primitive erythrocytes (or in those expressing a dominant negative CBFβ protein) have been reported, including abnormal morphology, delayed maturation, and altered expression of cell surface markers (Ter119, CD41) and transcription factors (EKLF, KLF1, GATA1) (Castilla et al., 1996; Yokomizo et al., 2008). Knock down of Runx1 in zebrafish by morpholino injection similarly affects definitive hematopoiesis but not/marginally primitive erythropoiesis (Burns et al., 2005; Gering and Patient, 2005; Kalev-Zhylinska et al., 2002). Runx1 was the first gene that upon mutation so neatly segregated primitive erythropoiesis from definitive hematopoiesis, and for that reason it became a focal point for studying the origins of definitive (adult) blood.
A role for Runx1 in the formation of definitive blood from hemogenic endothelium in the embryo
The sites and cells in which Runx1 is expressed provided profound insights into the process by which definitive hematopoietic progenitors and HSCs are formed. Runx1 expression is found in all sites of hematopoiesis in the conceptus and precedes the emergence of definitive hematopoietic progenitors and HSCs. Quite strikingly, expression is seen in a subset of endothelial cells in the vitelline and umbilical arteries, the yolk sac, the placenta, and the ventral aspect of the dorsal aorta in the AGM region, but not in endothelial cells elsewhere (North et al., 1999; Ottersbach and Dzierzak, 2005; Rhodes et al., 2008). Runx1 is also expressed in mesenchymal cells in some of these sites, specifically those underlying the dorsal aorta in the AGM region, in the placenta, and in hematopoietic cell clusters attached to the luminal wall of the aorta and vitelline and umbilical arteries. The presence of Runx1 transcripts in the ventral aspect of the dorsal aorta is conserved in all vertebrate species that have been examined (Bollerot et al., 2005; Burns et al., 2002; Ciau-Uitz et al., 2000; Gering and Patient, 2005; Kalev-Zhylinska et al., 2002; North et al., 1999).
The observation that Runx1 was expressed in endothelial cells prior to the onset of definitive hematopoiesis, supported an articulated, but at the time, not widely accepted hypothesis that blood developed from a “hemogenic endothelium” (Jaffredo et al., 1998; Jordan, 1916; Nishikawa et al., 1998; Smith and Glomski, 1982). The hemogenic endothelium was proposed to line the lumens of certain arteries in the conceptus and give rise to clusters of hematopoietic cells on the luminal side of the artery (or in the case of chick embryos and zebrafish embryos, also in the ventral mesenchyme (Jaffredo et al., 2005b; Kissa et al., 2008)) that expressed markers such as CD45, αIIb integrin (CD41), c-myb, Mpl, and c-kit (Bernex et al., 1996; Burns et al., 2005; Corbel, 2002; Gering and Patient, 2005; Jaffredo et al., 2005a; Jaffredo et al., 1998; Labastie et al., 1998; Manaia et al., 2000; Marshall et al., 1999; Ody et al., 1999; Pardanaud et al., 1996; Petit-Cocault et al., 2007; Tavian et al., 1996; Thompson et al., 1998). The clusters were found closely associated with aortic endothelium in the ventral aspect of the dorsal aorta, and the vitelline and umbilical arteries, suggesting an intimate relationship between the two lineages (Dieterlen-Lièvre and Martin, 1981; Garcia-Porrero et al., 1995; Sabin, 1920).
The first direct experimental evidence for a hemogenic endothelial precursor for blood was provided by Dieterlen-Lievre and colleagues, who performed lineage-tracing analyses in the chick embryo using vital dyes (acetylated low density lipoprotein labeled with DiI) and retroviruses. She and her colleagues demonstrated that labeling of the entire vasculature prior to the formation of intra-arterial clusters, resulted in labeled CD45+ cells in the clusters that subsequently formed, as well as in the mesenchyme ventral to the chick dorsal aorta (Jaffredo et al., 2000; Jaffredo et al., 1998). Further support for the differentiation of blood from endothelium was provided by the Nishikawa lab, who showed that cells isolated from mouse embryos on the basis of their expression of vascular endothelial cadherin (VE-cadherin) and the absence of the hematopoietic markers Ter119 and CD45, could give rise to blood following in vitro culture (Nishikawa et al., 1998). However, there was still some doubt that blood is borne directly from endothelium, and alternative models exist. For example, it was proposed that in the mouse the immediate precursors to the intra-arterial clusters originate in the mesenchyme ventral to the dorsal aorta in so called sub-aortic patches, and migrate towards the arteries and squeeze between endothelial cells to enter the lumen (Bertrand et al., 2005; Godin and Cumano, 2002; Manaia et al., 2000; Yoon et al., 2008).
Runx1 deficiency blocked the formation of the intra-arterial clusters in vivo, and the formation of hematopoietic cells from purified endothelial cells ex vivo, suggesting a role for Runx1 in the hemogenic endothelium to hematopoietic cell transition (North et al., 1999; Yokomizo et al., 2001). Indeed, arterial endothelial cells in Runx1 deficient embryos initiated reporter gene expression from endogenous Runx1 regulatory elements at the proper time of development (although expression was lost later on), indicating that the observed defect in hematopoiesis did not result from impaired migration/incorporation of endothelial progenitors into the aortic endothelium, or impaired specification of hemogenic endothelium, but rather from impaired differentiation of blood from endothelium (North et al., 1999). Morpholino knockdown of Runx1 in zebrafish, also impedes hematopoietic cluster formation without affecting the aortic endothelium (Burns et al., 2005; Gering and Patient, 2005; Kalev-Zhylinska et al., 2002), in line with Runx1 being responsible for a hemogenic endothelium to hematopoietic cell transition. Runx1 was the first gene specifically implicated to function at this step.
Although the histological evidence for Runx1 expression and activity in the endothelium of the major arteries (dorsal aorta, vitelline, umbilical) was compelling, and was observed in multiple organisms, expression in the yolk sac and placenta were less easy to interpret. The yolk sac is a source of both primitive and definitive blood progenitors, which appear in separate waves and anatomical locations (Ferkowicz et al., 2003; Ferkowicz and Yoder, 2005; Palis et al., 1999). Progenitors to primitive erythrocytes, which are CD41dim, develop in a band of prospective blood islands that encircle the proximal yolk sac at 7.5 dpc (Ferkowicz et al., 2003; Li et al., 2005). Slightly later (8.25 dpc) definitive progenitors appear as small clusters of CD41bright cells located at the proximal border of the blood islands (Ferkowicz et al., 2003; Li et al., 2005). Runx1 is expressed in the mesoderm of the prospective blood islands in mid to late primitive streak conceptuses (~7.5 dpc), and in the primitive erythrocytes as they begin to differentiate from mesoderm in the yolk sac (Lacaud et al., 2002; North et al., 1999). This “background” of Runx1 expression during the formation of the blood islands and the initial stages of primitive erythropoiesis makes identifying the emergence of Runx1+ definitive precursors in the yolk sac difficult to visualize. Nevertheless Runx1 expression was observed in yolk sac endothelial cells and putative definitive blood cells associated with them, after expression in erythrocytes is extinguished by 8.5 dpc as Ter119 levels become elevated and maturation progresses (North et al., 1999; North et al., 2004). In addition, it was noted that in Runx1 deficient conceptuses, expression of reporter gene knockins in the yolk sac blood islands was normal, but later expression in endothelial cells scattered around the yolk sac was absent, as were clusters (North et al., 1999; Yokomizo et al., 2001). Studies in the ES cell differentiation model of yolk sac hematopoiesis showed that Runx1 expression in embryoid bodies (EBs) is upregulated contemporaneously with that of Flk1, and blast colony-forming cells (BL-CFCs), thought to represent hemangioblasts that differentiated from EBs were uniformly Runx1 positive (Lacaud et al., 2002). In the absence of Runx1, the generation of definitive type hematopoietic progenitors in the blast colonies was completely blocked, with just the core of the colony, which has endothelial characteristics, remaining (Lacaud et al., 2002; Lancrin et al., 2009). Thus, similar to the situation in the dorsal aorta, Runx1 is an early marker of hematopoiesis in the yolk sac and plays a role in the generation of definitive hematopoietic cells, despite the fact that it is not required in the hemangioblast or for the onset of primitive erythropoiesis.
In the placenta, which recently was shown to harbor hematopoietic progenitors and stem cells (Alvarez-Silva et al., 2003; Gekas et al., 2005; Ottersbach and Dzierzak, 2005; Rhodes et al., 2008), Runx1 positive endothelium in the labyrinth is similarly overshadowed by a larger population of Runx1 positive mesenchymal cells (Ottersbach and Dzierzak, 2005; Rhodes et al., 2008; Zeigler et al., 2006), and intra-luminal clusters of hematopoietic cells are relatively infrequent (Ottersbach and Dzierzak, 2005; Rhodes et al., 2008). One potential explanation for the difficulty in finding Runx1+ clusters is that the Runx1-lacZ allele that many investigators have used to track expression is nonfunctional (North et al., 1999), and it is well-documented that Runx1 haploinsufficiency depresses definitive hematopoietic progenitor numbers and cluster formation (Cai et al., 2000; Mukouyama et al., 2000; Wang et al., 1996a).
Most recently, the transition of cells with an endothelial phenotype and morphology into free-floating hematopoietic cells was directly visualized by time-lapse microscopy in culture (Eilken et al., 2009), in live zebrafish embryos, and in thick sections of the mouse aorta cultured ex vivo (Bertrand et al., 2010; Boisset et al., 2010; Kissa and Herbomel, 2010). Runx1 mutants or morpholinos were used to demonstrate the specificity of the budding process, as they completely impaired hematopoietic cell formation in all systems (Boisset et al., 2010; Kissa and Herbomel, 2010; Lam et al., 2010). In a study using zebrafish embryos it was shown that very few cells budded from the endothelium of Runx1 morphants, and those that tried died immediately (Kissa and Herbomel, 2010). These data suggest that signals other than Runx1 may initiate the budding process, but that Runx1 is absolutely required for it to progress normally.
The vast majority of adult blood is derived from endothelium
Although intra-aortic clusters were clearly observed at developmental times when definitive hematopoietic progenitors and HSCs appeared, a question that remained was to what extent do the endothelial cells and intra-aortic clusters that appear so briefly in the midgestation conceptus contribute to the HSCs that are ultimately found in adult marrow? Two groups addressed this question in mice by labeling all cells expressing, or that at one time had expressed the endothelial marker VE-cadherin, by crossing VE-cadherin-Cre recombinase transgenic mice to Rosa26 reporter mice (Chen et al., 2009; Zovein et al., 2008). Both groups showed that cells in the adult bone marrow, which do not express cell surface VE-cadherin (Kim et al., 2005; Taoudi et al., 2005), were indeed derived from a precursor that had expressed VE-cadherin. The percentage of labeled cells in the adult marrow was >95% in one study (Chen et al., 2009), which, given the less than 100% efficiency of VE-cadherin/Cre excision suggests that almost all adult blood cells are born from VE-cadherin expressing cells. VE-cadherin is also transiently expressed in 7.5 dpc yolk sac mesoderm, raising the possibility that blood is not derived from VE-cadherin positive endothelium (Yokomizo et al., 2007). However one group took advantage of an estrogen regulated form of Cre expressed from the VE-cadherin promoter, and activated Cre after the transient wave of VE-cadherin expression in yolk sac mesoderm had ended. They showed that blood was labeled, and was therefore derived from VE-cadherin positive endothelium (Zovein et al., 2008). Together with the finding that deletion of Runx1 by VE-cadherin-Cre blocked the formation of intra-aortic clusters, HSCs, and definitive hematopoietic progenitors (in essence phenocopying germline Runx1 deficiency (Chen et al., 2009)), these data provide compelling in vivo evidence that definitive hematopoietic progenitors and HSCs differentiate from VE-cadherin+ cells, most of which are endothelial cells in a Runx1-dependent manner. Conversely, restricted expression of Runx1 or CBFβ only in Tie2+ cells or their progeny allowed for the formation of HSCs and/or hematopoietic progenitors, consistent with the above-mentioned results (Liakhovitskaia et al., 2009; Miller et al., 2002). However, since Tie2 is not only expressed on endothelial cells but also on some mesenchymal cells and on quiescent adult HSCs (Arai et al., 2004; Li et al., 2006), these data were not as conclusive as those generated by conditional knockout using VE-cadherin-Cre. Recent experiments in zebrafish, using an endothelial cell-associated kdrl:Cre to activate a reporter gene, are in line with an endothelial origin of most blood cells (Bertrand et al., 2010), although the spatiotemporal window during which the kdrl:Cre is active is not entirely clear (Bertrand et al., 2010; Kissa and Herbomel, 2010).
Runx1 is transiently required for hematopoietic progenitor and HSC emergence
Loss of Runx1 function in the adult causes not a reduction, but rather an expansion of phenotypic HSCs and progenitors in the bone marrow (Chen et al., 2009; Growney et al., 2005; Ichikawa et al., 2004; Putz et al., 2006; Schindler et al., 2009), thus at some point hematopoietic stem and progenitor cells no longer absolutely require Runx1. Several studies have attempted to define the temporal window during which Runx1 is absolutely required (Fig. 1). Chen et al. showed that the transition to relative Runx1 “independence” occurs in the fetus, as deletion of Runx1 with Vav1-Cre, which is active in fetal liver hematopoietic progenitors and HSCs starting at approximately 11.5 dpc, does not block HSC or progenitor formation (Chen et al., 2009). In line with this, retroviral transduction of Runx1 into cells derived from the AGM of 11.5 dpc Runx1 deficient embryos could not rescue hematopoietic progenitor formation, while transduction of cells derived from the 9.5 dpc p-Sp could do so (Goyama et al., 2004; Mukouyama et al., 2000). However, as the extent to which hematopoiesis was rescued was not addressed, it is not clear whether or not optimal progenitor and/or HSC production requires Runx1 during the entire window of its expression in the 8.5 dpc to 11.5 dpc p-Sp/AGM. Whether it is required only in the endothelium and not once the intra-arterial clusters have formed, is another outstanding question. Finally, given that hematopoiesis unfolds gradually in multiple anatomical locations, it is likely that the time at which Runx1 is required will differ in each hemogenic site.
Fig. 1. The outer boundaries of the proposed developmental window of the absolute Runx1 requirement in definitive blood cell generation (solid red bar).
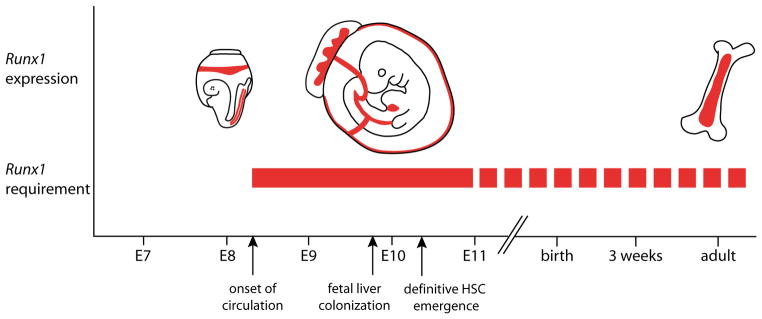
A continued role for Runx1in specific hematopoietic cell types and lineages is represented by the dotted red bar. See text for additional explanation.
Transcriptional regulation of Runx1 expression: promoters and cis-elements
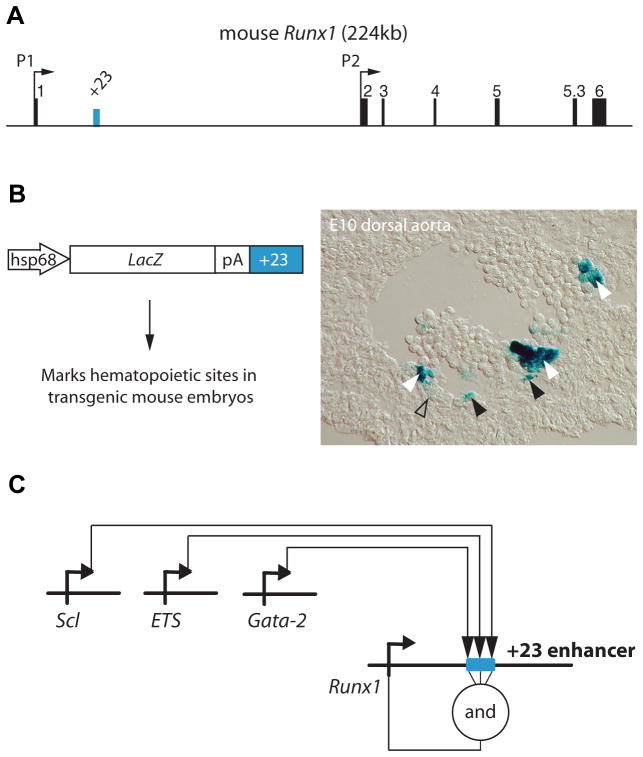
The pivotal role for Runx1 at the onset of definitive hematopoiesis raises the question how the expression of this master regulator itself is controlled. The large size of the Runx1 locus (224 Kb in the mouse) has made defining the cis-regulatory elements and trans-acting factors that govern its spatiotemporal expression a challenge. Runx1, like the other vertebrate RUNX genes, is transcribed from two alternative promoters, a distal P1 and a proximal P2 (Bee et al., 2009b; Ghozi et al., 1996; Levanon et al., 2001; Levanon and Groner, 2004; Telfer and Rothenberg, 2001) (Fig. 2A). Alternative gene promoters are frequently found in the mammalian genome, and are believed to contribute to its greater regulatory complexity (Davuluri et al., 2008). For Runx1, alternative promoter usage is known to result in the generation of a series of transcripts that differ in their untranslated regions and/or protein-coding exons, influencing mRNA stability, efficiency of translation by the use of alternative translation initiation mechanisms and micro-RNAs, and/or Runx1 protein structure (reviewed in (Bee et al., 2009b; Levanon et al., 2001; Levanon and Groner, 2004). In the mouse, both promoters are active at and required for the normal onset of definitive hematopoiesis, in a partially overlapping but non-redundant fashion (Bee et al., 2009b; Bee et al., 2010; Pozner et al., 2007). In general, transcripts from the P2 promoter are produced earlier than those from the P1 promoter, in hemogenic endothelium as well as in the very first emerging hematopoietic cells in the mouse embryo and ES cell cultures, while P1 is the dominant Runx1 promoter in fetal liver and adult HSCs (Bee et al., 2009b; Challen and Goodell, 2010; Fujita et al., 2001; Sroczynska et al., 2009; Telfer and Rothenberg, 2001).
Fig. 2. The Runx1 +23 enhancer recapitulates the hematopoietic specific expression pattern of Runx1.
(A) Schematic of the Runx1 locus. Vertebrate Runx1 is transcribed from two promoters, the P1 and the P2. A 531 bp mouse-frog conserved enhancer was identified (Nottingham et al., 2007) and is located 23 kb downstream of the ATG in exon 1. (B) This +23 enhancer targets reporter gene expression to hematopoietic sites in the developing embryos, including all emerging HSCs. A transverse section through the dorsal aorta of an E10 transient transgenic embryo shows Xgal staining in emerging hematopoietic clusters (white arrowheads), in scattered cells of the endothelial wall (black arrowheads), and in a few mesenchymal cells (open arrowhead). Identical Xgal staining is seen in established mouse lines carrying the hsp68LacZ+23 transgene (not shown). (C) Targeted mutagenesis of putative transcription factor binding sites and chromatin IP (Nottingham et al., 2007), and trans-activation assays (Landry et al., 2008) placed the Runx1 +23 enhancer directly downstream of the ETS/GATA/SCL kernel (Liu et al., 2008; Pimanda et al., 2007b) that is active at the onset of developmental hematopoiesis.
Maintenance of Runx1 expression in hemogenic endothelial cells requires continued Runx1 function, raising the possibility that Runx1 positively regulates its own expression (North et al., 1999). Negative regulation of Runx gene expression has also been observed; overexpression of Runx3 downregulated Runx1 expression in human B cell lines (Spender et al., 2005), Runx2 repressed Runx3 in tooth development (Wang et al., 2005), and Runx2 has been shown to repress its own expression in vitro (Drissi et al., 2000). Several Runx binding sites are located in/nearby the mammalian P1 and P2 core promoters suggesting that both promoters may be auto- and cross-regulated by Runx proteins. The two adjacent Runx binding sites in the P1, which are located in the P1 5′UTR, are conserved between all three Runx genes and in multiple species extending to frog; and a positive autoregulatory loop was shown to act on the P1 promoter in a myeloid progenitor cell line (Bee et al., 2009a; Levanon and Groner, 2004; Pimanda et al., 2007a). However, eliminating both of these sites did not affect the activity of a P1 promoter fragment in transgenic mice (Bee et al., 2009a). Presumably sequences at the P2, or more distal to either promoter confer autoregulation.
Despite the specific activity pattern of mouse P1 and P2 Runx1 promoters in developmental hematopoiesis, neither core promoter confers Runx1-specific expression to transgenic reporters in vivo or in vitro (Bee et al., 2009a; Ghozi et al., 1996), indicating that tissue-specific cis-regulatory elements are located elsewhere in the locus. Recently, a deeply conserved hematopoietic-specific enhancer was identified that is located in the first intron of Runx1, 23.5 kb downstream of the translation start site in exon 1 (Fig. 2 A,B). This +23 enhancer was found to act with either of the Runx1 promoters or an exogenous promoter to confer specific expression of a reporter gene in hematopoietic sites in the conceptus, in a subset of the cells in which endogenous Runx1 is expressed (Bee et al., 2009a; Nottingham et al., 2007). Specifically, the +23 enhancer was active in the intra-arterial clusters, including the emerging HSCs, but in only a subset of endothelial cells and mesenchymal cells ventral to the dorsal aorta (Fig. 2B), probably reflecting the absence of other cis-acting sequences necessary to drive the full spectrum of Runx1 expression at this site. It is of interest to note that in zebrafish, cis-acting sequences sufficient for runx1 expression in the dorsal aorta are contained within 8 kb upstream of the P2 promoter (Lam et al., 2009), whilst the +23 enhancer is not conserved in this species. Thus, there appears to be a divergence in the cis-regulation of Runx1 between mammals and fish, although upstream factors appear to be conserved (discussed below and in (Bee et al., 2009a; Bee et al., 2009b)).
Direct transcriptional regulators of Runx1 in developmental hematopoiesis
The +23 enhancer contains several conserved motifs corresponding to putative binding sites for hematopoietic transcription factors, with GATA, ETS, and RUNX motifs critical for in vitro enhancer activity (Nottingham et al., 2007). During the onset of hematopoiesis in the conceptus, enhancer activity was dependent on the GATA and ETS motifs, but not on the RUNX motif, indicating that in vivo the +23 enhancer acts during the initiation of Runx1 expression (Nottingham et al., 2007) rather than during maintenance through autoregulation (Ferjouxet al., 2007; Gajewski et al., 2007). Chromatin immunoprecipitation analyses showed specific enrichment of Gata2, the ETS factors PU.1, Fli-1 and Elf1, and the SCL complex (presumably through GATA factors) on the +23 enhancer (Landry et al., 2008; Nottingham et al., 2007). Taken together, this places Runx1 directly downstream of GATA, ETS and SCL transcription factors (Fig. 2C), which are themselves well known players in the generation of the vasculature and the hematopoietic system in the mouse and zebrafish, ((De Val and Black, 2009) and references therein).
The dorsal aorta is initially formed by endothelial precursors derived from lateral plate mesoderm (Esner et al., 2006; Pouget et al., 2006; Wasteson et al., 2008). In the dorsal lateral plate mesoderm of Xenopus embryos, cells expressing Flk1, the endothelial maker Fli1 (XFli-1, encoding an ETS family protein), Scl (encoded by Tal1) and Gata2, but not Runx1, are found before the dorsal aorta forms (Ciau-Uitz et al., 2000). A subset of these cells migrates towards a VEGF signal at the midline of the embryo that is secreted by the hypochord. Although Flk1 and XFli-1 expression are maintained as these endothelial precursors move towards the midline, Scl and Gata2 expression are extinguished in the actively migrating cells (Ciau-Uitz et al., 2000). After the endothelial precursors reach the midline and form the (unpaired) dorsal aorta they activate Runx1 expression, and reactivate the expression of Scl and Gata2 (Ciau-Uitz et al., 2000). This sequence of events suggests that a local signal in the vicinity of the ventral aspect of the dorsal aorta is responsible for activating Runx1, and for reactivating Scl and Gata2 expression.
SCL, Gata2 and Fli1 are expressed in the mouse pSp/AGM in endothelial cells and the hematopoietic clusters, and are required for normal definitive hematopoiesis (Elefanty et al., 1999; Hart et al., 2000; Khandekar et al., 2007; Kobayashi-Osaki et al., 2005; Ling et al., 2004; Melet et al., 1996; Minegishi et al., 1999; Minegishi et al., 2003; Pimanda et al., 2007b; Porcher et al., 1996; Sanchez et al., 1999; Sinclair et al., 1999; Spyropoulos et al., 2000; Tsai et al., 1994). In addition, SCL and Gata2 were shown to be expressed in and play a role in HSC function in the embryo and/or the adult (Elefanty et al., 1999; Lacombe et al., 2009; Ling et al., 2004; Rodrigues et al., 2005; Sanchez et al., 1999). In HSC emergence, Scl and its interaction partner, the LIM domain only 2 protein (Lmo2) function earlier than Runx1, prior to expression of the endothelial markers VE-Cadherin and Tie2 (Drake and Fleming, 2000; Endoh et al., 2002; Gering et al., 2003; Li et al., 2006; Patterson et al., 2007; Schlaeger et al., 2005). Scl is required in the lateral plate mesoderm for hematopoietic specification, and unlike Runx1, is no longer required once the hemogenic endothelium forms (Chen et al., 2009; Endoh et al., 2002; Lancrin et al., 2009; Li et al., 2006; Schlaeger et al., 2005). The ETS transcription factor Fli1 is at the top of the genetic hierarchy that leads to Gata2, Scl, Flk1, Tie2, and ultimately Runx1 expression (Liu et al., 2008). Morpholinos against Xenopus Fli1 dramatically decreased the expression of all of these genes. Gata2 morphants, on the other hand, expressed Fli1 normally but not Scl, Lmo2, Tie2, or Runx1, placing Gata2 downstream of Fli1 but upstream of these other genes (Liu et al., 2008). The +23 hematopoietic-specific enhancer in Runx1 contains binding sites for all of the transcription factors in this early hematopoietic “kernel” (Gata2, Scl/Lmo2, Fli-1), and thus may integrate the information coming from all three signals (Landry et al., 2008; Liu et al., 2008; Nottingham et al., 2007; Pimanda et al., 2007b) (Fig. 2C).
Signaling pathways acting upstream of Runx1
Hedgehog signaling is required at multiple stages of blood cell formation in zebrafish (Gering and Patient, 2005). It is required first for the migration of endothelial precursors from the dorsal lateral plate towards the midline, and slightly later for arterial specification. Inhibition of hedgehog signaling at either of these two steps severely reduces the number of Runx1+ cells in the dorsal aorta. Hedgehog signaling, however, is not thought to activate Runx1 expression directly (Wilkinson et al., 2009). VEGF and Notch signaling are also upstream of Runx1 expression (Burns et al., 2005; Gering and Patient, 2005). Inhibition of either signaling pathway prevents arterial specification (monitored by the expression of ephrinB2a) and the formation of Runx1+ cells. The Notch signaling hematopoietic defect can be overcome in both zebrafish and mice by ectopic expression of Runx1, indicating that Runx1 is genetically downstream of Notch (Burns et al., 2005; Nakagawa et al., 2006). However, hematopoietic markers including Runx1, are detectable in the dorsal aorta prior to the requirement for Notch activity (Burns et al., 2005; Gering and Patient, 2005; Kalev-Zhylinska et al., 2002; Wilkinson et al., 2009), suggesting that Notch may be necessary to maintain but not to initiate Runx1 expression. Whether Runx1 is a direct or indirect target of Notch signaling, and whether it also works in parallel with Notch is not known. No binding sites for the CSL transcription factor (CBF1; recombinant binding protein-J kappa (RBPjÍ); Suppressor of Hairless (Su[H]); Lag-1), which transmits the Notch signal to downstream target genes, have been reported within the Runx1 gene. It has been shown that Hes1, a downstream effector of Notch1, augments the transcriptional activity of Runx1 (McLarren et al., 2000), and thus the two pathways may work in parallel, as they do during the early stages of T cell development (Guo et al., 2008; Nakagawa et al., 2006).
One mode by which Notch signaling regulates cell fate is through lateral inhibition, in which a cell expressing Notch ligands activates signaling in adjacent cells, and the cells delivering and receiving the signal adopt different identities (Bray, 2006). Since not all Runx1+ endothelial cells appear to give rise to hematopoietic clusters, Notch signaling could potentially be involved in selecting the Runx1+ cells that do form clusters. Notch signaling in both zebrafish and mice is required only for the formation of definitive hematopoietic cells in the AGM region, but surprisingly not in the yolk sac (Burns et al., 2005; Gering and Patient, 2005; Hadland et al., 2004; Kumano et al., 2003; Robert-Moreno et al., 2005; Robert-Moreno et al., 2008). Differences in signaling pathways like these could underlie the differences in definitive blood cell types in the various anatomical sites of blood cell formation.
The signaling pathway activated by bone morphogenic protein 4 (Bmp4) is also essential for blood cell formation, and may be one of the local signals that induce Runx1 expression in the ventral aspect of the dorsal aorta. Bmp4 expression in mesenchyme underlying intra-arterial clusters in the dorsal aorta has been documented in multiple species (Durand et al., 2007; Marshall et al., 2000; Pimanda et al., 2007a; Suonpaa et al., 2005), and inhibition of Bmp4 signaling in zebrafish blocked both the initiation and maintenance of Runx1 expression (McReynolds et al., 2007; Wilkinson et al., 2009). Bmp4 appears to act later than Hedgehog and Notch, in that specification of arterial endothelium, which requires the latter two signals, does not require Bmp4 signaling (Wilkinson et al., 2009). Runx1 may be a direct target of Bmp4 signaling, as one of its downstream effectors, Smad1, was shown to occupy the distal P1 promoter of Runx1 in a hematopoietic cell line in vivo (Pimanda et al., 2006). A schematic representing the main steps in HSC formation as discussed above is shown in Fig. 3.
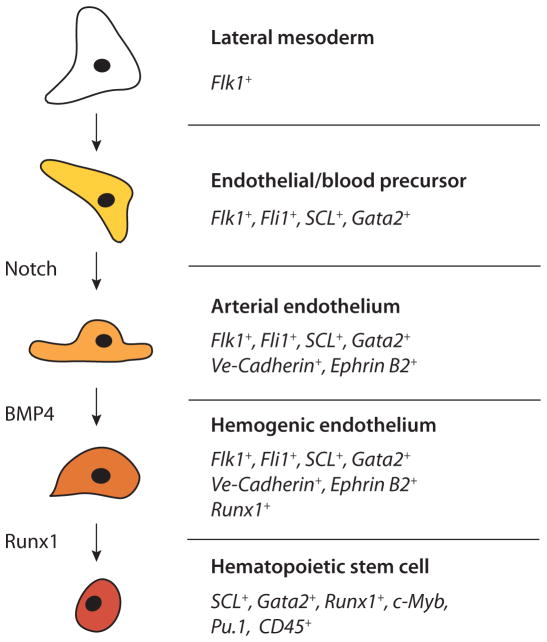
Fig. 3. Steps in hematopoietic stem cell formation, based on the model in which definitive blood is derived from so-called hemogenic endothelium.
Arrows indicate the order of events only and should not be taken to represent direct transitions in all instances. Transcription factors, signaling pathways, and cell type specific genes active during this process are shown. This list is not exhaustive and is based on data available on mRNA and/or gene enhancer-mediated expression in mouse, chick, zebrafish and/or Xenopus embryos. In some instances there is a discrepancy in mRNA and protein expression. For example, VE-Cadherin protein is expressed on the cell membrane of mouse HSCs isolated from the AGM region, while in the chicken embryo aortic clusters do not express VE-Cadherin mRNA). This may be caused by the perdurance of protein following loss of mRNA synthesis. Pu.1 and c-Myb are expressed in pSp/AGM hematopoietic cells and play a role in fetal liver and adult bone marrow HSCs (Garcia et al., 2009; Iwasaki et al., 2005, Kim et al., 2004, Lieu and Reddy, 2009; Mukouyama et al., 1999; Sandberg et al., 2005). Thus, these transcription factors are likely to be expressed in AGM HSCs, although to our knowledge this was not formally shown.
Runx1 expression has been used as a convenient marker to screen for genes, compound libraries, or conditions that affect its expression and thus definitive hematopoiesis (Burns et al., 2009; North et al., 2009; North et al., 2007). North et al. (North et al., 2007) used a combination of probes for Runx1 and c-Myb expression to monitor the effects of small molecules on AGM hematopoiesis in zebrafish (North et al., 2007). This resulted in the identification of compounds that modulate the prostaglandin E2 pathway as regulators of HSC formation in the embryo. This finding was quickly translated into a clinical trial (http://clinicaltrials.gov/ct2/show/NCT00890500) designed to test the ability of a stabilized derivative of prostaglandin E2 1(6, 16 Dimethyl-Prostaglandin E2) to improve the engraftment of patients receiving allogeneic umbilical cord blood stem cell transplants for the treatment of their leukemia.
The same screen for Runx1/c-Myb expression in zebrafish found that a group of compounds that regulate blood flow also affect blood cell formation in the AGM region (North et al., 2009). Compounds that increased Runx1/c-Myb expression and blood formation included antagonists of the adrenergic signaling pathway, the Ca2+-channel blocker nifedipine, and compounds that increased the production of nitric oxide (NO). All of these compounds increased vasodilation and subsequently the volume of blood flowing through the dorsal aorta. Genes associated with the three affected pathways (adra2b, adra2, da, adra2c; ace2, agtrl1a, adt; nos1) were expressed in endothelial and hematopoietic (Lmo2+ and/or CD41+) cells of zebrafish embryos, and the expression of several were upregulated at the onset of definitive hematopoiesis. The importance of blood flow in regulating adult blood cell formation from endothelium was confirmed genetically by demonstrating that zebrafish lacking a heartbeat due to a mutation in the gene encoding cardiac troponin T (sih) had dramatically decreased Runx1/c-myb expression in the AGM region.
Since HSC formation occurs subsequent to the establishment of the embryonic circulation, the mechanical stimulation of NO production, modulated by sheer stress and alterations in blood flow (Fukumura et al., 2001), could play an important role in inducing HSC and intra-arterial cluster formation. Activation of NO signaling prior to the establishment of the circulation enhanced Runx1/c-myb expression and blood formation, and inhibition of NO signaling had the opposite effect in both zebrafish and mouse embryos (North et al., 2009). Interestingly, chemicals that enhance NO signaling (S-nitroso-N-acetyl-penicillamine, or SNAP) could rescue Runx1/c-myb expression in zebrafish with defective Notch signaling, and correspondingly could block the enhanced expression of ephrinB2a and Runx1/c-myb in transgenic zebrafish expressing the activated intracellular component of the Notch receptor. These data indicate that the NO pathway, and by extension blood flowing through the vasculature, functions downstream of Notch to specify arterial identity and promote blood cell formation.
The effect of flow on blood cell formation from mouse embryonic stem cells was also monitored using Runx1 and c-Myb as markers (Adamo et al., 2009). Using an apparatus that reproduced the biomechanical sheer stress experienced in the embryonic dorsal aorta from the flow of blood, Adamo et al. demonstrated that Runx1 and c-Myb expression, as well as the number committed hematopoietic progenitor numbers were upregulated by approximately 2–3 fold by sheer stress. Similar increases were also observed in explants of para-aortic splanchnopleuras or AGM regions exposed to the sheer stress caused by flow. Runx1 expression was reduced in the para-aortic splanchnopleuras of mice deficient for the Na+/Ca+ exchanger encoded by Ncx1 (Slc8a), which lack a heartbeat (Koushik et al., 2001), providing genetic confirmation for the ex vivo effects that were observed (Adamo et al., 2009).
Mechanisms of Runx1 action
Although several target genes of Runx1 have been reported ((Otto et al., 2003) and references therein, (Hug et al., 2004; Michaud et al., 2008; Sakai et al., 2009; Wotton et al., 2008)), critical direct targets at the onset of hematopoiesis remain to be identified. Given that Runx1 is a transcription factor required for an endothelial to hematopoietic cell transition, logic dictates that it should repress an endothelial program, while activating genes specifically expressed in blood. Indeed, expression of several endothelial markers, including Flk1 and VE-cadherin, are downregulated in cells after Runx1 is expressed, and before intra-arterial clusters expressing hematopoietic markers are apparent (Hirai et al., 2003; Jaffredo et al., 2005a; Jaffredo et al., 1998). Corresponding changes in endothelial and hematopoietic gene expression were observed in ES cell differentiation cultures in which Runx1 cDNA was conditionally expressed (Sakai et al., 2009). The dynamic expression of hematopoietic and endothelial markers has been most carefully described in chick, Xenopus, and zebrafish embryos. Expression of Runx1 in the ventral endothelium of the dorsal aorta in chick embryos is followed by the progressive loss of Flk1 and VE-cadherin transcripts, and the upregulation of the hematopoietic markers Scl, Lmo2, Gata2, and Gata3 (Jaffredo et al., 2005a). Intra-aortic clusters are conspicuously VE-cadherin and Flk1 negative (although in the mouse cell surface VE-Cadherin perdures on HSCs in the clusters (North et al., 2002; Taoudi et al., 2005)), and express additional hematopoietic markers including Pu.1, c-myb, CD41, and CD45 (Jaffredo et al., 2005a; Jaffredo et al., 1998; Manaia et al., 2000; Ody et al., 1999; Pardanaud et al., 1996). Some of these genes may be Runx1 targets (Okada et al., 1998). Runx1 was shown to down-regulate the expression of Flk1, but whether Flk1 is a direct or indirect target is not known (Hirai et al., 2005). Pu.1 is a bona fide downstream target of Runx1 that contains an upstream regulatory element (URE) that is occupied by Runx1 in vivo (Hoogenkamp et al., 2009; Huang et al., 2008).
Runx1 is expressed in the hemangioblast, and although Runx1 deficient ES cells can generate hemangioblasts expressing Flk1, Scl, and Fli1, they cannot express the Runx1 downstream target Pu.1 (Hoogenkamp et al., 2009). Runx1 binding to its target sites (the 3′ URE and promoter) in the Pu.1 locus in hemangioblasts was weak and unstable, but nonetheless sufficient to induce local chromatin unfolding, as defined by increased sensitivity to DNaseI treatment and demethylation of CpGs. A twelve-hour exposure to Runx1 appeared sufficient to sustain Pu.1 expression and chromatin occupancy by other transcription factors (C/EBP, ETS proteins) on the 3′ URE and promoter, even after Runx1 was withdrawn (Hoogenkamp et al., 2009). No ATP-dependent chromatin-remodeling proteins that associate with Runx1 have been identified, so the mechanistic details by which Runx1 promotes this early chromatin unfolding is not known.
The Runx proteins are well-documented activators and repres-sors of transcription, and domains mediating those activities have been mapped to sequences N- and C-terminal to the DNA-binding Runt domain (Kanno et al., 1998; Liu et al., 2006). Transcriptional activation seems to be the predominant function during HSC emergence, as mutant forms of Runx1 lacking the C-terminal transactivation domain are unable to rescue hematopoiesis in Runx1 deficient ES cells (Nishimura et al., 2004; Okuda et al., 2000), or when reintroduced into cells derived from the p-Sp of Runx1 deficient embryos (Goyama et al., 2004). Deletion of repressive sequences, on the other hand, did not affect definitive progenitor cell emergence, although it did seem to impair the contribution of ES cells carrying those mutations to the fetal liver and thymus of chimeric mice following injection into blastocysts (Goyama et al., 2004; Nishimura et al., 2004).
Several co-factors and co-activators that interact with the transactivation domain of Runx1 could potentially mediate its activity during HSC emergence. The C-terminal transactivation domain, which was mapped between amino acids 291–371 (Kanno et al., 1998), interacts with the transcriptional co-activators MOZ (KAT6a, a member of the MYST family), p300, and the CREB-binding protein (CBP), all three of which have histone acetyltransferase activity (Kitabayashi et al., 2001; Kitabayashi et al., 1998). All three proteins co-purified with Runx1 as a complex that also contained the homeodomain interacting protein kinase 2 (HIPK2), and the promyelocytic leukemia protein (PML) (Kitabayashi et al., 2001; Kitabayashi et al., 1998). Germline deletion of MOZ caused a 5–10 fold decrease in the number of functional and phenotypic hematopoietic progenitors, and the fetal livers contained no transplantable HSCs (Katsumoto et al., 2006). Deletion of p300 or CBP did not affect the emergence of HSCs in the fetus, although CBP was essential for HSC self-renewal (Rebel et al., 2002). Mice doubly deficient for HIPK2, a nuclear serine/threonine kinase that phosphorylates Runx1, p300, and MOZ (Aikawa et al., 2006), and its paralog HIPK1 (Hipk1−/−Hipk2−/−) had a phenotype very similar to that caused by p300 or CBP deficiency (Isono et al., 2006). The fact that progenitors or HSCs emerged at all would indicate that the function of Runx1 in HSC emergence is not entirely dependent on MOZ, p300, CBP, or HIPK1/2. Either there is functional redundancy between these proteins, or other co-activator proteins must be involved.
Conclusions and future directions
The current interest in stem cell-based therapies has emphasized the importance of understanding how tissue-specific stem cells are specified in development. Studies on the expression and role of Runx1 at the onset of definitive hematopoiesis have firmly established this transcription factor as a pivotal player in HSC emergence. The next challenge will be to establish the gene regulatory network in which Runx1 functions. This will provide an important mechanistic framework of HSC emergence and will open the way to future studies into how to modulate this network to promote stem cell maintenance and/or de novo generation. In addition, given the link between deregulation of Runx1 function and leukemia, such a framework will form a basis to explore the changes in the network associated with disease.
Abbreviations used in this paper
- AGM
aorta-gonad-mesonephros region
- dpc
days post coitus
- ES
embryonic stem cell
- HSC
hematopoietic stem cell
- pSp
para-aortic splanchn opleura
References
- ADAMO L, NAVEIRAS O, WENZEL PL, MCKINNEY-FREEMAN S, MACK PJ, GRACIA-SANCHO J, SUCHY-DICEY A, YOSHIMOTO M, LENSCH MW, YODER MC, et al. Biomechanical forces promote embryonic haematopoiesis. Nature. 2009;459:1131–1135. doi: 10.1038/nature08073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AIKAWA Y, NGUYEN LA, ISONO K, TAKAKURA N, TAGATA Y, SCHMITZ ML, KOSEKI H, KITABAYASHI I. Roles of HIPK1 and HIPK2 in AML1- and p300-dependent transcription, hematopoiesis and blood vessel formation. EMBO J. 2006;25:3955–3965. doi: 10.1038/sj.emboj.7601273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ALVAREZ-SILVA M, BELO-DIABANGOUAYA P, SALAUN J, DIETERLEN-LIEVRE F. Mouse placenta is a major hematopoietic organ. Development. 2003;130:5437–5444. doi: 10.1242/dev.00755. [DOI] [PubMed] [Google Scholar]
- ARAI F, HIRAO A, OHMURA M, SATO H, MATSUOKA S, TAKUBO K, ITO K, KOH GY, SUDA T. Tie2/angiopoietin-1 signaling regulates hematopoietic stem cell quiescence in the bone marrow niche. Cell. 2004;118:149–161. doi: 10.1016/j.cell.2004.07.004. [DOI] [PubMed] [Google Scholar]
- BEE T, ASHLEY EL, BICKLEY SR, JARRATT A, LI PS, SLOANE-STANLEY J, GOTTGENS B, DE BRUIJN MF. The mouse Runx1 +23 hematopoietic stem cell enhancer confers hematopoietic specificity to both Runx1 promoters. Blood. 2009a;113:5121–5124. doi: 10.1182/blood-2008-12-193003. [DOI] [PubMed] [Google Scholar]
- BEE T, LIDDIARD K, SWIERS G, BICKLEY SR, VINK CS, JARRATT A, HUGHES JR, MEDVINSKY A, DE BRUIJN MF. Alternative Runx1 promoter usage in mouse developmental hematopoiesis. Blood Cells Mol Dis. 2009b;43:35–42. doi: 10.1016/j.bcmd.2009.03.011. [DOI] [PubMed] [Google Scholar]
- BEE T, SWIERS G, MUROI S, POZNER A, NOTTINGHAM W, SANTOS AC, LI PS, TANIUCHI I, DE BRUIJN MF. Non-redundant roles for Runx1 alternative promoters reflect their activity at discrete stages of developmental hematopoiesis. Blood. 2010;115:3042–3050. doi: 10.1182/blood-2009-08-238626. [DOI] [PubMed] [Google Scholar]
- BERNEX F, DE SEPULVEDA P, KRESS C, ELBAZ C, DELOUIS C, PANTHIER JJ. Spatial and temporal patterns of c-kit-expressing cells in WlacZ/+ and WlacZ/WlacZ mouse embryos. Development. 1996;122:3023–3033. doi: 10.1242/dev.122.10.3023. [DOI] [PubMed] [Google Scholar]
- BERTRAND JY, CHI NC, SANTOSO B, TENG S, STAINIER DY, TRAVER D. Haematopoietic stem cells derive directly from aortic endothelium during development. Nature. 2010;464:108–111. doi: 10.1038/nature08738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BERTRAND JY, GIROUX S, GOLUB R, KLAINE M, JALIL A, BOUCONTET L, GODIN I, CUMANO A. Characterization of purified intraembryonic hematopoietic stem cells as a tool to define their site of origin. Proc Natl Acad Sci USA. 2005;102:134–139. doi: 10.1073/pnas.0402270102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BOISSET JC, VAN CAPPELLEN W, ANDRIEU-SOLER C, GALJART N, DZIERZAK E, ROBIN C. In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature. 2010;464:116–120. doi: 10.1038/nature08764. [DOI] [PubMed] [Google Scholar]
- BOLLEROT K, ROMERO S, DUNON D, JAFFREDO T. Core binding factor in the early avian embryo: cloning of Cbfbeta and combinatorial expression patterns with Runx1. Gene Expr Patterns. 2005;6:29–39. doi: 10.1016/j.modgep.2005.05.003. [DOI] [PubMed] [Google Scholar]
- BRAY SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- BROTHERTON TW, CHUI DH, GAULDIE J, PATTERSON M. Hemoglobin ontogeny during normal mouse fetal development. Proc Natl Acad Sci USA. 1979;76:2853–2857. doi: 10.1073/pnas.76.6.2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BURNS CE, DEBLASIO T, ZHOU Y, ZHANG J, ZON L, NIMER SD. Isolation and characterization of runxa and runxb, zebrafish members of the runt family of transcriptional regulators. Exp Hematol. 2002;30:1381–1389. doi: 10.1016/s0301-472x(02)00955-4. [DOI] [PubMed] [Google Scholar]
- BURNS CE, GALLOWAY JL, SMITH AC, KEEFE MD, CASHMAN TJ, PAIK EJ, MAYHALL EA, AMSTERDAM AH, ZON LI. A genetic screen in zebrafish defines a hierarchical network of pathways required for hematopoietic stem cell emergence. Blood. 2009;113:5776–5782. doi: 10.1182/blood-2008-12-193607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BURNS CE, TRAVER D, MAYHALL E, SHEPARD JL, ZON LI. Hematopoietic stem cell fate is established by the Notch-Runx pathway. Genes Dev. 2005;19:2331–2342. doi: 10.1101/gad.1337005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CAI Z, DE BRUIJN MFTR, MA X, DORTLAND B, LUTEIJN T, DOWNING JR, DZIERZAK E. Haploinsufficiency of AML1/CBFA2 affects the embryonic generation of mouse hematopoietic stem cells. Immunity. 2000;13:423–431. doi: 10.1016/s1074-7613(00)00042-x. [DOI] [PubMed] [Google Scholar]
- CASTILLA LH, WIJMENGA C, WANG Q, STACY T, SPECK NA, ECKHAUS M, MARÍN-PADILLA M, COLLINS FS, WYNSHAW-BORIS A, LIU PP. Defects of embryonic hematopoiesis and lethal hemorrhaging in mouse embryos heterozygous for a knocked-in leukemia gene CBFB-MYH11. Cell. 1996;87:687–696. doi: 10.1016/s0092-8674(00)81388-4. [DOI] [PubMed] [Google Scholar]
- CHALLEN GA, GOODELL MA. Runx1 Isoforms Show Differential Expression Patterns During Hematopoietic Development But Have Similar Functional Effects in Adult Hematopoietic Stem Cells. Exp Hematol. 2010;38:403–416. doi: 10.1016/j.exphem.2010.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHEN MJ, YOKOMIZO T, ZEIGLER BM, DZIERZAK E, SPECK NA. Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature. 2009;457:889–891. doi: 10.1038/nature07619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CIAU-UITZ A, WALMSLEY M, PATIENT R. Distinct origins of adult and embryonic blood in Xenopus. Cell. 2000;102:787–796. doi: 10.1016/s0092-8674(00)00067-2. [DOI] [PubMed] [Google Scholar]
- CORBEL C. Expression of alphaVbeta3 integrin in the chick embryo aortic endothelium. Int J Dev Biol. 2002;46:827–830. [PubMed] [Google Scholar]
- CUMANO A, DIETERLEN-LIÈVRE F, GODIN I. Lymphoid potential, probed before circulation in mouse, is restricted to caudal intraembryonic splanchnopleura. Cell. 1996;86:907–916. doi: 10.1016/s0092-8674(00)80166-x. [DOI] [PubMed] [Google Scholar]
- DAVULURI RV, SUZUKI Y, SUGANO S, PLASS C, HUANG TH. The functional consequences of alternative promoter use in mammalian genomes. Trends Genet. 2008;24:167–177. doi: 10.1016/j.tig.2008.01.008. [DOI] [PubMed] [Google Scholar]
- DE BRUIJN MF, SPECK NA, PEETERS MC, DZIERZAK E. Definitive hematopoietic stem cells first develop within the major arterial regions of the mouse embryo. EMBO J. 2000;19:2465–2474. doi: 10.1093/emboj/19.11.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DE VAL S, BLACK BL. Transcriptional control of endothelial cell development. Dev Cell. 2009;16:180–195. doi: 10.1016/j.devcel.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DIETERLEN-LIÈVRE F, MARTIN C. Diffuse intraembryonic hemato-poiesis in normal and chimeric avian development. Dev Biol. 1981;88:180–191. doi: 10.1016/0012-1606(81)90228-1. [DOI] [PubMed] [Google Scholar]
- DRAKE CJ, FLEMING PA. Vasculogenesis in the day 6.5 to 9.5 mouse embryo. Blood. 2000;95:1671–1679. [PubMed] [Google Scholar]
- DRISSI H, LUC Q, SHAKOORI R, CHUVA DE SOUSA LOPES S, CHOI JY, TERRY A, HU M, JONES S, NEIL JC, LIAN JB, et al. Transcriptional autoregulation of the bone related CBFA1/RUNX2 gene. J Cell Physiol. 2000;184:341–350. doi: 10.1002/1097-4652(200009)184:3<341::AID-JCP8>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- DURAND C, ROBIN C, BOLLEROT K, BARON MH, OTTERSBACH K, DZIERZAK E. Embryonic stromal clones reveal developmental regulators of definitive hematopoietic stem cells. Proc Natl Acad Sci USA. 2007;104:20838–20843. doi: 10.1073/pnas.0706923105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DZIERZAK E, SPECK NA. Of lineage and legacy: the development of mammalian hematopoietic stem cells. Nat Immunol. 2008;9:129–136. doi: 10.1038/ni1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EILKEN HM, NISHIKAWA S, SCHROEDER T. Continuous single-cell imaging of blood generation from haemogenic endothelium. Nature. 2009;457:896–900. doi: 10.1038/nature07760. [DOI] [PubMed] [Google Scholar]
- ELEFANTY AG, BEGLEY CG, HARTLEY L, PAPAEVANGELIOU B, ROBB L. SCL expression in the mouse embryo detected with a targeted lacZ reporter gene demonstrates its localization to hematopoietic, vascular, and neural tissues. Blood. 1999;94:3754–3763. [PubMed] [Google Scholar]
- ENDOH M, OGAWA M, ORKIN S, NISHIKAWA S. SCL/tal-1-dependent process determines a competence to select the definitive hemato-poietic lineage prior to endothelial differentiation. EMBO J. 2002;21:6700–6708. doi: 10.1093/emboj/cdf674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ESNER M, MEILHAC SM, RELAIX F, NICOLAS JF, COSSU G, BUCKINGHAM ME. Smooth muscle of the dorsal aorta shares a common clonal origin with skeletal muscle of the myotome. Development. 2006;133:737–749. doi: 10.1242/dev.02226. [DOI] [PubMed] [Google Scholar]
- FERJOUX G, AUGE B, BOYER K, HAENLIN M, WALTZER L. A GATA/RUNX cis-regulatory module couples Drosophila blood cell commitment and differentiation into crystal cells. Dev Biol. 2007;305:726–734. doi: 10.1016/j.ydbio.2007.03.010. [DOI] [PubMed] [Google Scholar]
- FERKOWICZ MJ, STARR M, XIE X, LI W, JOHNSON SA, SHELLEY WC, MORRISON PR, YODER MC. CD41 expression defines the onset of primitive and definitive hematopoiesis in the murine embryo. Development. 2003;130:4393–4403. doi: 10.1242/dev.00632. [DOI] [PubMed] [Google Scholar]
- FERKOWICZ MJ, YODER MC. Blood island formation: longstanding observations and modern interpretations. Exp Hematol. 2005;33:1041–1047. doi: 10.1016/j.exphem.2005.06.006. [DOI] [PubMed] [Google Scholar]
- FUJITA Y, NISHIMURA M, TANIWAKI M, ABE T, OKUDA T. Identification of an alternatively spliced form of the mouse AML1/RUNX1 gene transcript AML1c and its expression in early hematopoietic development. Biochem Biophys Res Commun. 2001;281:1248–1255. doi: 10.1006/bbrc.2001.4513. [DOI] [PubMed] [Google Scholar]
- FUKUMURA D, GOHONGI T, KADAMBI A, IZUMI Y, ANG J, YUN CO, BUERK DG, HUANG PL, JAIN RK. Predominant role of endothelial nitric oxide synthase in vascular endothelial growth factor-induced angiogenesis and vascular permeability. Proc Natl Acad Sci USA. 2001;98:2604–2609. doi: 10.1073/pnas.041359198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GAJEWSKI KM, SORRENTINO RP, LEE JH, ZHANG Q, RUSSELL M, SCHULZ RA. Identification of a crystal cell-specific enhancer of the black cells prophenoloxidase gene in Drosophila. Genesis. 2007;45:200–207. doi: 10.1002/dvg.20285. [DOI] [PubMed] [Google Scholar]
- GARCIA P, CLARKE M, VEGIOPOULOS A, BERLANGA O, CAMELO A, LORVELLEC M, FRAMPTON J. Reduced c-Myb activity compromises HSCs and leads to a myeloproliferation with a novel stem cell basis. EMBO J. 2009;28:1492–1504. doi: 10.1038/emboj.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GARCIA-PORRERO JA, GODIN IE, DIETERLEN-LIÈVRE F. Potential intraembryonic hemogenic sites at pre-liver stages in the mouse. Anat Embryol. 1995;192:425–435. doi: 10.1007/BF00240375. [DOI] [PubMed] [Google Scholar]
- GEKAS C, DIETERLEN-LIEVRE F, ORKIN SH, MIKKOLA HK. The placenta is a niche for hematopoietic stem cells. Dev Cell. 2005;8:365–375. doi: 10.1016/j.devcel.2004.12.016. [DOI] [PubMed] [Google Scholar]
- GERING M, PATIENT R. Hedgehog signaling is required for adult blood stem cell formation in zebrafish embryos. Dev Cell. 2005;8:389–400. doi: 10.1016/j.devcel.2005.01.010. [DOI] [PubMed] [Google Scholar]
- GERING M, YAMADA Y, RABBITTS TH, PATIENT RK. Lmo2 and Scl/Tal1 convert non-axial mesoderm into haemangioblasts which differentiate into endothelial cells in the absence of Gata1. Development. 2003;130:6187–6199. doi: 10.1242/dev.00875. [DOI] [PubMed] [Google Scholar]
- GHOZI MC, BERNSTEIN Y, NEGREANU V, LEVANON D, GRONER Y. Expression of the human acute myeloid leukemia gene AML1 is regulated by two promoter regions. Proc Natl Acad Sci USA. 1996;93:1935–1940. doi: 10.1073/pnas.93.5.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GODIN I, CUMANO A. The hare and the tortoise: an embryonic haematopoietic race. Nat Rev Immunol. 2002;2:593–604. doi: 10.1038/nri857. [DOI] [PubMed] [Google Scholar]
- GODIN I, DIETERLEN-LIÈVRE F, CUMANO A. Emergence of multipotent hematopoietic cells in the yolk sac and paraaortic splanchnopleura of 8.5 dpc mouse embryos. Proc Natl Acad Sci USA. 1995;92:773–777. doi: 10.1073/pnas.92.3.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GOYAMA S, YAMAGUCHI Y, IMAI Y, KAWAZU M, NAKAGAWA M, ASAI T, KUMANO K, MITANI K, OGAWA S, CHIBA S, et al. The transcriptionally active form of AML1 is required for hematopoietic rescue of the AML1-deficient embryonic para-aortic splanchnopleural (P-Sp) region. Blood. 2004;104:3558–3564. doi: 10.1182/blood-2004-04-1535. [DOI] [PubMed] [Google Scholar]
- GROWNEY JD, SHIGEMATSU H, LI Z, LEE BH, ADELSPERGER J, ROWAN R, CURLEY DP, KUTOK JL, AKASHI K, WILLIAMS IR, et al. Loss of Runx1 perturbs adult hematopoiesis and is associated with a myeloproliferative phenotype. Blood. 2005;106:494–504. doi: 10.1182/blood-2004-08-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GUO Y, MAILLARD I, CHAKRABORTI S, ROTHENBERG EV, SPECK NA. Core binding factors are necessary for natural killer cell development, and cooperate with Notch signaling during T cell specification. Blood. 2008;112:480–492. doi: 10.1182/blood-2007-10-120261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HAAR JL, ACKERMAN GA. A phase and electron microscopic study of vasculogenesis and erythropoiesis in the yolk sac of the mouse. Anat Rec. 1971;170:199–223. doi: 10.1002/ar.1091700206. [DOI] [PubMed] [Google Scholar]
- HADLAND BK, HUPPERT SS, KANUNGO J, XUE Y, JIANG R, GRIDLEY T, CONLON RA, CHENG AM, KOPAN R, LONGMORE GD. A requirement for Notch1 distinguishes 2 phases of definitive hematopoiesis during development. Blood. 2004;104:3097–3105. doi: 10.1182/blood-2004-03-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HART A, MELET F, GROSSFELD P, CHIEN K, JONES C, TUNNACLIFFE A, FAVIER R, BERNSTEIN A. Fli-1 is required for murine vascular and megakaryocytic development and is hemizygously deleted in patients with thrombocytopenia. Immunity. 2000;13:167–177. doi: 10.1016/s1074-7613(00)00017-0. [DOI] [PubMed] [Google Scholar]
- HIRAI H, OGAWA M, SUZUKI N, YAMAMOTO M, BREIER G, MAZDA O, IMANISHI J, NISHIKAWA S. Hemogenic and nonhemogenic endothelium can be distinguished by the activity of fetal liver kinase (Flk)-1 promoter/enhancer during mouse embryogenesis. Blood. 2003;101:886–893. doi: 10.1182/blood-2002-02-0655. [DOI] [PubMed] [Google Scholar]
- HIRAI H, SAMOKHVALOV IM, FUJIMOTO T, NISHIKAWA S, IMANISHI J, NISHIKAWA S. Involvement of Runx1 in the down-regulation of fetal liver kinase-1 expression during transition of endothelial cells to hemato-poietic cells. Blood. 2005;106:1948–1955. doi: 10.1182/blood-2004-12-4872. [DOI] [PubMed] [Google Scholar]
- HOOGENKAMP M, LICHTINGER M, KRYSINSKA H, LANCRIN C, CLARKE D, WILLIAMSON A, MAZZARELLA L, INGRAM R, JORGENSEN H, FISHER A, et al. Early chromatin unfolding by RUNX1 - a molecular explanation for differential requirements during specification versus maintenance of the hematopoietic gene expression program. Blood. 2009;114:299–309. doi: 10.1182/blood-2008-11-191890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUANG G, ZHANG P, HIRAI H, ELF S, YAN X, CHEN Z, KOSCHMIEDER S, OKUNO Y, DAYARAM T, GROWNEY JD, et al. PU.1 is a major downstream target of AML1 (RUNX1) in adult mouse hematopoiesis. Nat Genet. 2008;40:51–60. doi: 10.1038/ng.2007.7. [DOI] [PubMed] [Google Scholar]
- HUG BA, AHMED N, ROBBINS JA, LAZAR MA. A chromatin immunoprecipitation screen reveals protein kinase Cbeta as a direct RUNX1 target gene. J Biol Chem. 2004;279:825–830. doi: 10.1074/jbc.M309524200. [DOI] [PubMed] [Google Scholar]
- ICHIKAWA M, ASAI T, SAITO T, YAMAMOTO G, SEO S, YAMAZAKI I, YAMAGATA T, MITANI K, CHIBA S, OGAWA S, et al. AML-1 is required for megakaryocytic maturation and lymphocytic differentiation, but not for maintenance of hematopoietic stem cells in adult hematopoiesis. Nat Med. 2004;10:299–304. doi: 10.1038/nm997. [DOI] [PubMed] [Google Scholar]
- ISONO K, NEMOTO K, LI Y, TAKADA Y, SUZUKI R, KATSUKI M, NAKAGAWARA A, KOSEKI H. Overlapping roles for homeodomain-interacting protein kinases hipk1 and hipk2 in the mediation of cell growth in response to morphogenetic and genotoxic signals. Mol Cell Biol. 2006;26:2758–2771. doi: 10.1128/MCB.26.7.2758-2771.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IWASAKI H, SOMOZA C, SHIGEMATSU H, DUPREZ EA, IWASAKI-ARAI J, MIZUNO S, ARINOBU Y, GEARY K, ZHANG P, DAYARAM T, et al. Distinctive and indispensable roles of PU.1 in maintenance of hemato-poietic stem cells and their differentiation. Blood. 2005;106:1590–1600. doi: 10.1182/blood-2005-03-0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JAFFREDO T, BOLLEROT K, SUGIYAMA D, GAUTIER R, DREVON C. Tracing the hemangioblast during embryogenesis: developmental relationships between endothelial and hematopoietic cells. Int J Dev Biol. 2005a;49:269–277. doi: 10.1387/ijdb.041948tj. [DOI] [PubMed] [Google Scholar]
- JAFFREDO T, GAUTIER R, BRAJEUL V, DIETERLEN-LIÈVRE F. Tracing the progeny of the aortic hemangioblast in the avian embryo. Dev Biol. 2000;224:204–214. doi: 10.1006/dbio.2000.9799. [DOI] [PubMed] [Google Scholar]
- JAFFREDO T, GAUTIER R, EICHMANN A, DIETERLEN-LIÈVRE F. Intraaortic hemopoietic cells are derived from endothelial cells during ontogeny. Development. 1998;125:4575–4583. doi: 10.1242/dev.125.22.4575. [DOI] [PubMed] [Google Scholar]
- JAFFREDO T, NOTTINGHAM W, LIDDIARD K, BOLLEROT K, POUGET C, DE BRUIJN M. From hemangioblast to hematopoietic stem cell: an endothelial connection? Exp Hematol. 2005b;33:1029–1040. doi: 10.1016/j.exphem.2005.06.005. [DOI] [PubMed] [Google Scholar]
- JORDAN HE. Evidence of hemogenic capacity of endothelium. Anat Rec. 1916;10:417–420. [Google Scholar]
- KALEV-ZHYLINSKA M, HORSFIELD JA, FLORES MVC, POSTLETHWAIT JH, VITAS MR, BAAS AM, CROSIER PS, CROSIER KE. Runx1 is required for zebrafish blood and vessel development and expression of a human RUNX1-CBF2T1 transgene advances a model for studies of leukemogenesis. Development. 2002;129:2015–2030. doi: 10.1242/dev.129.8.2015. [DOI] [PubMed] [Google Scholar]
- KANNO T, KANNO Y, CHEN LF, OGAWA E, KIM WY, ITO Y. Intrinsic transcriptional activation-inhibition domains of the polyomavirus enhancer binding protein 2/core binding factor a subunit revealed in the presence of the b subunit. Mol Cell Biol. 1998;18:2444–2454. doi: 10.1128/mcb.18.5.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KATSUMOTO T, AIKAWA Y, IWAMA A, UEDA S, ICHIKAWA H, OCHIYA T, KITABAYASHI I. MOZ is essential for maintenance of hematopoietic stem cells. Genes Dev. 2006;20:1321–1330. doi: 10.1101/gad.1393106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KHANDEKAR M, BRANDT W, ZHOU Y, DAGENAIS S, GLOVER TW, SUZUKI N, SHIMIZU R, YAMAMOTO M, LIM KC, ENGEL JD. A Gata2 intronic enhancer confers its pan-endothelia-specific regulation. Development. 2007;134:1703–1712. doi: 10.1242/dev.001297. [DOI] [PubMed] [Google Scholar]
- KIM HG, DE GUZMAN CG, SWINDLE CS, COTTA CV, GARTLAND L, SCOTT EW, KLUG CA. The ETS family transcription factor PU.1 is necessary for the maintenance of fetal liver hematopoietic stem cells. Blood. 2004;104:3894–3900. doi: 10.1182/blood-2002-08-2425. [DOI] [PubMed] [Google Scholar]
- KIM I, YILMAZ OH, MORRISON SJ. CD144 (VE-cadherin) is transiently expressed by fetal liver hematopoietic stem cells. Blood. 2005;106:903–905. doi: 10.1182/blood-2004-12-4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KISSA K, HERBOMEL P. Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature. 2010;464:112–115. doi: 10.1038/nature08761. [DOI] [PubMed] [Google Scholar]
- KISSA K, MURAYAMA E, ZAPATA A, CORTES A, PERRET E, MACHU C, HERBOMEL P. Live imaging of emerging hematopoietic stem cells and early thymus colonization. Blood. 2008;111:1147–1156. doi: 10.1182/blood-2007-07-099499. [DOI] [PubMed] [Google Scholar]
- KITABAYASHI I, AIKAWA Y, NGUYEN LA, YOKOYAMA A, OHKI M. Activation of AML1-mediated transcription by MOZ and inhibition by the MOZ-CBP fusion protein. EMBO J. 2001;20:7184–7196. doi: 10.1093/emboj/20.24.7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KITABAYASHI I, YOKOYAMA A, SHIMIZU K, OHKI M. Interaction and functional cooperation of the leukemia-associated factors AML1 and p300 in myeloid cell differentiation. EMBO J. 1998;17:2294–3004. doi: 10.1093/emboj/17.11.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KOBAYASHI-OSAKI M, OHNEDA O, SUZUKI N, MINEGISHI N, YOKOMIZO T, TAKAHASHI S, LIM KC, ENGEL JD, YAMAMOTO M. GATA motifs regulate early hematopoietic lineage-specific expression of the Gata2 gene. Mol Cell Biol. 2005;25:7005–7020. doi: 10.1128/MCB.25.16.7005-7020.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KOUSHIK SV, WANG J, ROGERS R, MOSKOPHIDIS D, LAMBERT NA, CREAZZO TL, CONWAY SJ. Targeted inactivation of the sodium-calcium exchanger (Ncx1) results in the lack of a heartbeat and abnormal myofibrillar organization. FASEB J. 2001;15:1209–1211. doi: 10.1096/fj.00-0696fje. [DOI] [PubMed] [Google Scholar]
- KUMANO K, CHIBA S, KUNISATO A, SATA M, SAITO T, NAKAGAMI-YAMAGUCHI E, YAMAGUCHI T, MASUDA S, SHIMIZU K, TAKAHASHI T, et al. Notch1 but not Notch2 is essential for generating hematopoietic stem cells from endothelial cells. Immunity. 2003;18:699–711. doi: 10.1016/s1074-7613(03)00117-1. [DOI] [PubMed] [Google Scholar]
- LABASTIE MC, CORTÉS F, ROMÉO PH, DULAC C, PÉAULT B. Molecular identity of hematopoietic precursor cells emerging in the human embryo. Blood. 1998;92:3624–3635. [PubMed] [Google Scholar]
- LACAUD G, GORE L, KENNEDY M, KOUSKOFF V, KINGSLEY P, HOGAN C, CARLSSON L, SPECK N, PALIS J, KELLER G. Runx1 is essential for hematopoietic commitment at the hemangioblast stage of development in vitro. Blood. 2002;100:458–466. doi: 10.1182/blood-2001-12-0321. [DOI] [PubMed] [Google Scholar]
- LACOMBE J, HERBLOT S, ROJAS-SUTTERLIN S, HAMAN A, BARAKAT S, ISCOVE NN, SAUVAGEAU G, HOANG T. Scl regulates the quiescence and the long-term competence of hematopoietic stem cells. Blood. 2009;115:792–803. doi: 10.1182/blood-2009-01-201384. [DOI] [PubMed] [Google Scholar]
- LAM EY, CHAU JY, KALEV-ZYLINSKA ML, FOUNTAINE TM, MEAD RS, HALL CJ, CROSIER PS, CROSIER KE, FLORES MV. Zebrafish runx1 promoter-EGFP transgenics mark discrete sites of definitive blood progenitors. Blood. 2009;113:1241–1249. doi: 10.1182/blood-2008-04-149898. [DOI] [PubMed] [Google Scholar]
- LAM EY, HALL CJ, CROSIER PS, CROSIER KE, FLORES MV. Live imaging of Runx1 expression in the dorsal aorta tracks the emergence of blood progenitors from endothelial cells. Blood. 2010 May 7; doi: 10.1182/blood-2010-01-264382. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- LANCRIN C, SROCZYNSKA P, STEPHENSON C, ALLEN T, KOUSKOFF V, LACAUD G. The haemangioblast generates haematopoietic cells through a haemogenic endothelium stage. Nature. 2009;457:892–895. doi: 10.1038/nature07679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LANDRY JR, KINSTON S, KNEZEVIC K, DE BRUIJN MF, WILSON N, NOTTINGHAM WT, PEITZ M, EDENHOFER F, PIMANDA JE, OTTERSBACH K, et al. Runx genes are direct targets of Scl/Tal1 in the yolk sac and fetal liver. Blood. 2008;111:3005–3014. doi: 10.1182/blood-2007-07-098830. [DOI] [PubMed] [Google Scholar]
- LEVANON D, GLUSMAN G, BANGSOW T, BEN-ASHER E, MALE DA, AVIDAN N, BANGSOW C, HATTORI M, TAYLOR TD, TAUDIEN S, et al. Architecture and anatomy of the genomic locus encoding the human leukemia-associated transcription factor RUNX1/AML1. Gene. 2001;262:23–33. doi: 10.1016/s0378-1119(00)00532-1. [DOI] [PubMed] [Google Scholar]
- LEVANON D, GRONER Y. Structure and regulated expression of mammalian RUNX genes. Oncogene. 2004;23:4211–4219. doi: 10.1038/sj.onc.1207670. [DOI] [PubMed] [Google Scholar]
- LI W, FERKOWICZ MJ, JOHNSON SA, SHELLEY WC, YODER MC. Endothelial cells in the early murine yolk sac give rise to CD41-expressing hematopoietic cells. Stem Cells Dev. 2005;14:44–54. doi: 10.1089/scd.2005.14.44. [DOI] [PubMed] [Google Scholar]
- LI Z, CHEN MJ, STACY T, SPECK NA. Runx1 function in hematopoiesis is required in cells that express Tek. Blood. 2006;107:106–110. doi: 10.1182/blood-2005-05-1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIAKHOVITSKAIA A, GRIBI R, STAMATERIS E, VILLAIN G, JAFFREDO T, WILKIE R, GILCHRIST D, YANG J, URE J, MEDVINSKY A. Restoration of Runx1 expression in the Tie2 cell compartment rescues definitive hematopoietic stem cells and extends life of Runx1 knockout animals until birth. Stem Cells. 2009;27:1616–1624. doi: 10.1002/stem.71. [DOI] [PubMed] [Google Scholar]
- LIEU YK, REDDY EP. Conditional c-myb knockout in adult hemato-poietic stem cells leads to loss of self-renewal due to impaired proliferation and accelerated differentiation. Proc Natl Acad Sci USA. 2009;106:21689–21694. doi: 10.1073/pnas.0907623106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LING KW, OTTERSBACH K, VAN HAMBURG JP, OZIEMLAK A, TSAI FY, ORKIN SH, PLOEMACHER R, HENDRIKS RW, DZIERZAK E. GATA-2 plays two functionally distinct roles during the ontogeny of hematopoietic stem cells. J Exp Med. 2004;200:871–882. doi: 10.1084/jem.20031556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIU F, WALMSLEY M, RODAWAY A, PATIENT R. Fli1 acts at the top of the transcriptional network driving blood and endothelial development. Curr Biol. 2008;18:1234–1240. doi: 10.1016/j.cub.2008.07.048. [DOI] [PubMed] [Google Scholar]
- LIU H, CARLSSON L, GRUNDSTROM T. Identification of an N-terminal transactivation domain of Runx1 that separates molecular function from global differentiation function. J Biol Chem. 2006;281:25659–25669. doi: 10.1074/jbc.M603249200. [DOI] [PubMed] [Google Scholar]
- MANAIA A, LEMARCHANDEL V, KLAINE M, MAX-AUDIT I, ROMEO P, DIETERLEN-LIEVRE F, GODIN I. Lmo2 and GATA-3 associated expression in intraembryonic hemogenic sites. Development. 2000;127:643–653. doi: 10.1242/dev.127.3.643. [DOI] [PubMed] [Google Scholar]
- MARSHALL CJ, KINNON C, THRASHER AJ. Polarized expression of bone morphogenetic protein-4 in the human aorta-gonad-mesonephros region. Blood. 2000;96:1591–1593. [PubMed] [Google Scholar]
- MARSHALL CJ, MOORE RL, THOROGOOD P, BRICKELL PM, KINNON C, THRASHER AJ. Detailed characterization of the human aorta-gonad-mesonephros region reveals morphological polarity resembling a hematopoietic stromal layer. Dev Dyn. 1999;215:139–147. doi: 10.1002/(SICI)1097-0177(199906)215:2<139::AID-DVDY6>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- MCLARREN KW, LO R, GRBAVEC D, THIRUNAVUKKARASU K, KARSENTY G, STIFANI S. The mammalian basic helix loop helix protein HES-1 binds to and modulates the transactivating function of the runt-related factor Cbfa1. J Biol Chem. 2000;275:530–538. doi: 10.1074/jbc.275.1.530. [DOI] [PubMed] [Google Scholar]
- MCREYNOLDS LJ, GUPTA S, FIGUEROA ME, MULLINS MC, EVANS T. Smad1 and Smad5 differentially regulate embryonic hematopoiesis. Blood. 2007;110:3881–3890. doi: 10.1182/blood-2007-04-085753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MEDVINSKY A, DZIERZAK E. Definitive hematopoiesis is autonomously initiated by the AGM region. Cell. 1996;86:897–906. doi: 10.1016/s0092-8674(00)80165-8. [DOI] [PubMed] [Google Scholar]
- MEDVINSKY AL, SAMOYLINA NL, MÜLLER AM, DZIERZAK EA. An early pre-liver intra-embryonic source of CFU-S in the developing mouse. Nature. 1993;364:64–66. doi: 10.1038/364064a0. [DOI] [PubMed] [Google Scholar]
- MELET F, MOTRO B, ROSSI DJ, ZHANG L, BERNSTEIN A. Generation of a novel Fli-1 protein by gene targeting leads to a defect in thymus development and a delay in Friend virus-induced erythroleukemia. Mol Cell Biol. 1996;16:2708–2718. doi: 10.1128/mcb.16.6.2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MICHAUD J, SIMPSON KM, ESCHER R, BUCHET-POYAU K, BEISSBARTH T, CARMICHAEL C, RITCHIE ME, SCHUTZ F, CANNON P, LIU M, et al. Integrative analysis of RUNX1 downstream pathways and target genes. BMC Genomics. 2008;9:363–369. doi: 10.1186/1471-2164-9-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MILLER J, HORNER A, STACY T, LOWREY C, LIAN JB, STEIN G, NUCKOLLS GH, SPECK NA. The core-binding factor b subunit is required for bone formation and hematopoietic maturation. Nat Genet. 2002;32:645–649. doi: 10.1038/ng1049. [DOI] [PubMed] [Google Scholar]
- MILLER JD, STACY T, LIU PP, SPECK NA. CBFb, but not CBFb-SMMHC, rescues definitive hematopoiesis in CBFb deficient embryonic stem cells. Blood. 2001;97:2248–2256. doi: 10.1182/blood.v97.8.2248. [DOI] [PubMed] [Google Scholar]
- MINEGISHI N, OHTA J, YAMAGIWA H, SUZUKI N, KAWAUCHI S, ZHOU Y, TAKAHASHI S, HAYASHI N, ENGEL JD, YAMAMOTO M. The mouse GATA-2 gene is expressed in the para-aortic splanchnopleura and aorta-gonads and mesonephros region. Blood. 1999;93:4196–4207. [PubMed] [Google Scholar]
- MINEGISHI N, SUZUKI N, YOKOMIZO T, PAN X, FUJIMOTO T, TAKAHASHI S, HARA T, MIYAJIMA A, NISHIKAWA S, YAMAMOTO M. Expression and domain-specific function of GATA-2 during differentiation of the hematopoietic precursor cells in midgestation mouse embryos. Blood. 2003;102:896–905. doi: 10.1182/blood-2002-12-3809. [DOI] [PubMed] [Google Scholar]
- MOORE MAS, METCALF D. Ontogeny of the haemopoietic system: yolk sac origin of in vivo and in vitro colony forming cells in the developing mouse embryo. Br J Haematol. 1970;18:279–296. doi: 10.1111/j.1365-2141.1970.tb01443.x. [DOI] [PubMed] [Google Scholar]
- MUKOUYAMA Y, CHIBA N, HARA T, OKADA H, ITO Y, KANAMURU R, MIYAJIMA A, SATAKE M, WATANABE T. The AML1 transcription factor functions to develop and maintain hematogenic precursor cells in the embryonic aorta-gonad-mesonephros region. Dev Biol. 2000;220:27–36. doi: 10.1006/dbio.2000.9617. [DOI] [PubMed] [Google Scholar]
- MUKOUYAMA Y, CHIBA N, MUCENSKI ML, SATAKE M, MIYAJIMA A, HARA T, WATANABE T. Hematopoietic cells in cultures of the murine embryonic aorta-gonad-mesonephros region are induced by c-Myb. Curr Biol. 1999;9:833–836. doi: 10.1016/s0960-9822(99)80368-6. [DOI] [PubMed] [Google Scholar]
- MÜLLER AM, MEDVINSKY A, STROUBOULIS J, GROSVELD F, DZIERZAK E. Development of hematopoietic stem cell activity in the mouse embryo. Immunity. 1994;1:291–301. doi: 10.1016/1074-7613(94)90081-7. [DOI] [PubMed] [Google Scholar]
- NAKAGAWA M, ICHIKAWA M, KUMANO K, GOYAMA S, KAWAZU M, ASAI T, OGAWA S, KUROKAWA M, CHIBA S. AML1/Runx1 rescues Notch1-Null mutation-induced deficiency of para-aortic splanchnopleural hematopoiesis. Blood. 2006;108:3329–3334. doi: 10.1182/blood-2006-04-019570. [DOI] [PubMed] [Google Scholar]
- NISHIKAWA SI, NISHIKAWA S, KAWAMOTO H, YOSHIDA H, KIZUMOTO M, KATAOKA H, KATSURA Y. In vitro generation of lymphohematopoietic cells from endothelial cells purified from murine embryos. Immunity. 1998;8:761–769. doi: 10.1016/s1074-7613(00)80581-6. [DOI] [PubMed] [Google Scholar]
- NISHIMURA M, FUKUSHIMA-NAKASE Y, FUJITA Y, NAKAO M, TODA S, KITAMURA N, ABE T, OKUDA T. VWRPY motif-dependent and -independent roles of AML1/Runx1 transcription factor in murine hematopoietic development. Blood. 2004;103:562–570. doi: 10.1182/blood-2003-06-2109. [DOI] [PubMed] [Google Scholar]
- NORTH TE, DE BRUIJN MF, STACY T, TALEBIAN L, LIND E, ROBIN C, BINDER M, DZIERZAK E, SPECK NA. Runx1 expression marks long-term repopulating hematopoietic stem cells in the midgestation mouse embryo. Immunity. 2002;16:661–672. doi: 10.1016/s1074-7613(02)00296-0. [DOI] [PubMed] [Google Scholar]
- NORTH TE, GOESSLING W, PEETERS M, LI P, CEOL C, LORD AM, WEBER GJ, HARRIS J, CUTTING CC, HUANG P, et al. Hematopoietic stem cell development is dependent on blood flow. Cell. 2009;137:736–748. doi: 10.1016/j.cell.2009.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NORTH TE, GOESSLING W, WALKLEY CR, LENGERKE C, KOPANI KR, LORD AM, WEBER GJ, BOWMAN TV, JANG IH, GROSSER T, et al. Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature. 2007;447:1007–1011. doi: 10.1038/nature05883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NORTH TE, GU TL, STACY T, WANG Q, HOWARD L, BINDER M, MARÍN-PADILLA M, SPECK NA. Cbfa2 is required for the formation of intra-aortic hematopoietic clusters. Development. 1999;126:2563–2575. doi: 10.1242/dev.126.11.2563. [DOI] [PubMed] [Google Scholar]
- NORTH TE, STACY T, MATHENY CJ, SPECK NA, DE BRUIJN MF. Runx1 is expressed in adult mouse hematopoietic stem cells and differentiating myeloid and lymphoid cells, but not in maturing erythroid cells. Stem Cells. 2004;22:158–168. doi: 10.1634/stemcells.22-2-158. [DOI] [PubMed] [Google Scholar]
- NOTTINGHAM WT, JARRATT A, BURGESS M, SPECK CL, CHENG JF, PRABHAKAR S, RUBIN EM, LI PS, SLOANE-STANLEY J, KONG ASJ, et al. Runx1-mediated hematopoietic stem-cell emergence is controlled by a Gata/Ets/SCL-regulated enhancer. Blood. 2007;110:4188–4197. doi: 10.1182/blood-2007-07-100883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ODY C, VAIGOT P, QUERE P, IMHOF BA, CORBEL C. Glyco-protein IIb-IIIa is expressed on avian multilineage hematopoietic progenitor cells. Blood. 1999;93:2898–2906. [PubMed] [Google Scholar]
- OKADA H, WATANABE T, NIKI M, TAKANO H, CHIBA N, YANAI N, TANI K, HIBINO H, ASANO S, MUCENSKI ML, et al. AML1(−/−) embryos do not express certain hematopoiesis-related gene transcripts including those of the PU.1 gene. Oncogene. 1998;17:2287–2293. doi: 10.1038/sj.onc.1202151. [DOI] [PubMed] [Google Scholar]
- OKUDA T, TAKEDA K, FUJITA Y, NISHIMURA M, YAGYU S, YOSHIDA M, AKIRA S, DOWNING JR, ABE T. Biological characteristics of the leukemia-associated transcriptional factor AML1 disclosed by hematopoietic rescue of AML1-deficient embryonic stem cells by using a knock-in strategy. Mol Cell Biol. 2000;20:319–328. doi: 10.1128/mcb.20.1.319-328.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OKUDA T, VAN DEURSEN J, HIEBERT SW, GROSVELD G, DOWNING JR. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84:321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- OTTERSBACH K, DZIERZAK E. The murine placenta contains hematopoietic stem cells within the vascular labyrinth region. Dev Cell. 2005;8:377–387. doi: 10.1016/j.devcel.2005.02.001. [DOI] [PubMed] [Google Scholar]
- OTTO F, LUBBERT M, STOCK M. Upstream and downstream targets of RUNX proteins. J Cell Biochem. 2003;89:9–18. doi: 10.1002/jcb.10491. [DOI] [PubMed] [Google Scholar]
- PALIS J, ROBERTSON S, KENNEDY M, WALL C, KELLER G. Development of erythroid and myeloid progenitors in the yolk sac and embryo proper of the mouse. Development. 1999;126:5073–5084. doi: 10.1242/dev.126.22.5073. [DOI] [PubMed] [Google Scholar]
- PARDANAUD L, LUTON D, PRIGENT M, BOURCHEIX LM, CATALA M, DIETERLEN-LIÈVRE F. Two distinct endothelial lineages in ontogeny, one of them related to hemopoiesis. Development. 1996;122:1363–1371. doi: 10.1242/dev.122.5.1363. [DOI] [PubMed] [Google Scholar]
- PATTERSON LJ, GERING M, ECKFELDT CE, GREEN AR, VERFAILLIE CM, EKKER SC, PATIENT R. The transcription factors Scl and Lmo2 act together during development of the hemangioblast in zebrafish. Blood. 2007;109:2389–2398. doi: 10.1182/blood-2006-02-003087. [DOI] [PubMed] [Google Scholar]
- PETIT-COCAULT L, VOLLE-CHALLIER C, FLEURY M, PEAULT B, SOUYRI M. Dual role of Mpl receptor during the establishment of definitive hematopoiesis. Development. 2007;134:3031–40. doi: 10.1242/dev.001818. [DOI] [PubMed] [Google Scholar]
- PIMANDA JE, CHAN WY, DONALDSON IJ, BOWEN M, GREEN AR, GOTTGENS B. Endoglin expression in the endothelium is regulated by Fli-1, Erg, and Elf-1 acting on the promoter and a −8-kb enhancer. Blood. 2006;107:4737–4745. doi: 10.1182/blood-2005-12-4929. [DOI] [PubMed] [Google Scholar]
- PIMANDA JE, DONALDSON IJ, DE BRUIJN MF, KINSTON S, KNEZEVIC K, HUCKLE L, PILTZ S, LANDRY JR, GREEN AR, TANNAHILL D, et al. The SCL transcriptional network and BMP signaling pathway interact to regulate RUNX1 activity. Proc Natl Acad Sci USA. 2007a;104:840–845. doi: 10.1073/pnas.0607196104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PIMANDA JE, OTTERSBACH K, KNEZEVIC K, KINSTON S, CHAN WY, WILSON NK, LANDRY JR, WOOD AD, KOLB-KOKOCINSKI A, GREEN AR, et al. Gata2, Fli1, and Scl form a recursively wired gene-regulatory circuit during early hematopoietic development. Proc Natl Acad Sci USA. 2007b;104:17692–17697. doi: 10.1073/pnas.0707045104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PORCHER C, SWAT W, ROCKWELL K, FUJIWARA Y, ALT FW, ORKIN SH. The T-cell leukemia oncoprotein SCL/tal-1 is essential for development of all hematopoietic lineages. Cell. 1996;86:47–57. doi: 10.1016/s0092-8674(00)80076-8. [DOI] [PubMed] [Google Scholar]
- POUGET C, GAUTIER R, TEILLET MA, JAFFREDO T. Somite-derived cells replace ventral aortic hemangioblasts and provide aortic smooth muscle cells of the trunk. Development. 2006;133:1013–1022. doi: 10.1242/dev.02269. [DOI] [PubMed] [Google Scholar]
- POZNER A, LOTEM J, XIAO C, GOLDENBERG D, BRENNER O, NEGREANU V, LEVANON D, GRONER Y. Developmentally regulated promoter-switch transcriptionally controls Runx1 function during embryonic hematopoiesis. BMC Dev Biol. 2007;7:84–93. doi: 10.1186/1471-213X-7-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PUTZ G, ROSNER A, NUESSLEIN I, SCHMITZ N, BUCHHOLZ F. AML1 deletion in adult mice causes splenomegaly and lymphomas. Oncogene. 2006;25:929–939. doi: 10.1038/sj.onc.1209136. [DOI] [PubMed] [Google Scholar]
- REBEL VI, KUNG AL, TANNER EA, YANG H, BRONSON RT, LIVINGSTON DM. Distinct roles for CREB-binding protein and p300 in hematopoietic stem cell self-renewal. Proc Natl Acad Sci USA. 2002;99:14789–14794. doi: 10.1073/pnas.232568499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RHODES KE, GEKAS C, WANG Y, LUX CT, FRANCIS CS, CHAN DN, CONWAY S, ORKIN SH, YODER MC, MIKKOLA HK. The emergence of hematopoietic stem cells is initiated in the placental vasculature in the absence of circulation. Cell Stem Cell. 2008;2:252–263. doi: 10.1016/j.stem.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROBERT-MORENO A, ESPINOSA L, DE LA POMPA JL, BIGAS A. RBPjkappa-dependent Notch function regulates Gata2 and is essential for the formation of intra-embryonic hematopoietic cells. Development. 2005;132:1117–1126. doi: 10.1242/dev.01660. [DOI] [PubMed] [Google Scholar]
- ROBERT-MORENO A, GUIU J, RUIZ-HERGUIDO C, LOPEZ ME, INGLES-ESTEVE J, RIERA L, TIPPING A, ENVER T, DZIERZAK E, GRIDLEY T, et al. Impaired embryonic haematopoiesis yet normal arterial development in the absence of the Notch ligand Jagged1. EMBO J. 2008;27:1886–1895. doi: 10.1038/emboj.2008.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RODRIGUES NP, JANZEN V, FORKERT R, DOMBKOWSKI DM, BOYD AS, ORKIN SH, ENVER T, VYAS P, SCADDEN DT. Haploinsufficiency of GATA-2 perturbs adult hematopoietic stem-cell homeostasis. Blood. 2005;106:477–484. doi: 10.1182/blood-2004-08-2989. [DOI] [PubMed] [Google Scholar]
- SABIN FR. Studies on the origin of blood vessels and of red corpuscles as seen in the living blastoderm of the chick during the second day of incubation. Contributions to Embryology. 1920;9:213–262. [Google Scholar]
- SAKAI E, KITAJIMA K, SATO A, NAKANO T. Increase of hematopoietic progenitor and suppression of endothelial gene expression by Runx1 expression during in vitro ES differentiation. Exp Hematol. 2009;37:334–345. doi: 10.1016/j.exphem.2008.11.007. [DOI] [PubMed] [Google Scholar]
- SANCHEZ M, GOTTGENS B, SINCLAIR AM, STANLEY M, BEGLEY CG, HUNTER S, GREEN AR. An SCL 3′ enhancer targets developing endothelium together with embryonic and adult haematopoietic progenitors. Development. 1999;126:3891–3904. doi: 10.1242/dev.126.17.3891. [DOI] [PubMed] [Google Scholar]
- SANDBERG ML, SUTTON SE, PLETCHER MT, WILTSHIRE T, TARANTINO LM, HOGENESCH JB, COOKE MP. c-Myb and p300 regulate hematopoietic stem cell proliferation and differentiation. Dev Cell. 2005;8:153–166. doi: 10.1016/j.devcel.2004.12.015. [DOI] [PubMed] [Google Scholar]
- SASAKI K, YAGI H, BRONSON RT, TOMINAGA K, MATSUNASHI T, DEGUCHI K, TANI Y, KISHIMOTO T, KOMORI T. Absence of fetal liver hematopoiesis in mice deficient in transcriptional coactivator core binding factor beta. Proc Natl Acad Sci USA. 1996;93:12359–12363. doi: 10.1073/pnas.93.22.12359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHINDLER JW, VAN BUREN D, FOUDI A, KREJCI O, QIN J, ORKIN SH, HOCK H. TEL-AML1 corrupts hematopoietic stem cells to persist in the bone marrow and initiate leukemia. Cell Stem Cell. 2009;5:43–53. doi: 10.1016/j.stem.2009.04.019. [DOI] [PubMed] [Google Scholar]
- SCHLAEGER TM, MIKKOLA HK, GEKAS C, HELGADOTTIR HB, ORKIN SH. Tie2Cre mediated gene ablation defines the Stem Cell Leukemia gene (SCL/tal1)-dependent window during hematopoietic stem cell development. Blood. 2005;105:3871–3874. doi: 10.1182/blood-2004-11-4467. [DOI] [PubMed] [Google Scholar]
- SINCLAIR AM, GOTTGENS B, BARTON LM, STANLEY ML, PARDANAUD L, KLAINE M, GERING M, BAHN S, SANCHEZ M, BENCH AJ, et al. Distinct 5′ SCL enhancers direct transcription to developing brain, spinal cord, and endothelium: neural expression is mediated by GATA factor binding sites. Dev Biol. 1999;209:128–142. doi: 10.1006/dbio.1999.9236. [DOI] [PubMed] [Google Scholar]
- SMITH RA, GLOMSKI CA. «Hemogenic endothelium» of the embryonic aorta: does it exist? Dev Comp Immunol. 1982;6:359–368. doi: 10.1016/s0145-305x(82)80019-0. [DOI] [PubMed] [Google Scholar]
- SPENDER LC, WHITEMAN HJ, KARSTEGL CE, FARRELL PJ. Transcriptional cross-regulation of RUNX1 by RUNX3 in human B cells. Oncogene. 2005;24:1873–1881. doi: 10.1038/sj.onc.1208404. [DOI] [PubMed] [Google Scholar]
- SPYROPOULOS DD, PHARR PN, LAVENBURG KR, JACKERS P, PAPAS TS, OGAWA M, WATSON DK. Hemorrhage, impaired hematopoiesis, and lethality in mouse embryos carrying a targeted disruption of the Fli1 transcription factor. Mol Cell Biol. 2000;20:5643–5652. doi: 10.1128/mcb.20.15.5643-5652.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SROCZYNSKA P, LANCRIN C, PEARSON S, KOUSKOFF V, LACAUD G. In vitro differentiation of mouse embryonic stem cells as a model of early hematopoietic development. Methods Mol Biol. 2009;538:317–334. doi: 10.1007/978-1-59745-418-6_16. [DOI] [PubMed] [Google Scholar]
- SUONPAA P, KOHONEN P, KOSKELA K, KOSKINIEMI H, SALMINEN-MANKONEN H, LASSILA O. Development of early PCLP1-expressing haematopoietic cells within the avian dorsal aorta. Scand J Immunol. 2005;62:218–223. doi: 10.1111/j.1365-3083.2005.01655.x. [DOI] [PubMed] [Google Scholar]
- TAOUDI S, MORRISON AM, INOUE H, GRIBI R, URE J, MEDVINSKY A. Progressive divergence of definitive haematopoietic stem cells from the endothelial compartment does not depend on contact with the foetal liver. Development. 2005;132:4179–4191. doi: 10.1242/dev.01974. [DOI] [PubMed] [Google Scholar]
- TAVIAN M, COULOMBEL L, LUTON D, SAN CLEMENTE H, DIETERLEN-LIÈVRE F, PEAULT B. Aorta-associated CD34+ hematopoietic cells in the early human embryo. Blood. 1996;87:67–72. [PubMed] [Google Scholar]
- TELFER JC, ROTHENBERG EV. Expression and function of a stem cell promoter for the murine CBFa2 gene: distinct roles and regulation in natural killer and T cell development. Dev Biol. 2001;229:363–382. doi: 10.1006/dbio.2000.9991. [DOI] [PubMed] [Google Scholar]
- THOMPSON MA, RANSOM DG, PRATT SJ, MACLENNAN H, KIERAN MW, DETRICH HW, 3RD, VAIL B, HUBER TL, PAW B, BROWNLIE AJ, et al. The cloche and spadetail genes differentially affect hematopoiesis and vasculogenesis. Dev Biol. 1998;197:248–269. doi: 10.1006/dbio.1998.8887. [DOI] [PubMed] [Google Scholar]
- TOBER J, KONISKI A, MCGRATH KE, VEMISHETTI R, EMERSON R, DE MESY-BENTLEY KK, WAUGH R, PALIS J. The megakaryocyte lineage originates from hemangioblast precursors and is an integral component both of primitive and of definitive hematopoiesis. Blood. 2007;109:1433–1441. doi: 10.1182/blood-2006-06-031898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TSAI JY, KELLER T, KUO FC, WEISS M, CHEN J, ROSENBLATT M, ALT FW, ORKIN SH. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature. 1994;371:221–226. doi: 10.1038/371221a0. [DOI] [PubMed] [Google Scholar]
- WANG Q, STACY T, BINDER M, MARÍN-PADILLA M, SHARPE AH, SPECK NA. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc Natl Acad Sci USA. 1996a;93:3444–3449. doi: 10.1073/pnas.93.8.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG Q, STACY T, MILLER JD, LEWIS AF, HUANG X, BORIES JC, BUSHWELLER JH, ALT FW, BINDER M, MARÍN-PADILLA M, et al. The CBFb subunit is essential for CBFa2 (AML1) function in vivo. Cell. 1996b;87:697–708. doi: 10.1016/s0092-8674(00)81389-6. [DOI] [PubMed] [Google Scholar]
- WANG XP, ABERG T, JAMES MJ, LEVANON D, GRONER Y, THESLEFF I. Runx2 (Cbfa1) inhibits Shh signaling in the lower but not upper molars of mouse embryos and prevents the budding of putative successional teeth. J Dent Res. 2005;84:138–143. doi: 10.1177/154405910508400206. [DOI] [PubMed] [Google Scholar]
- WASTESON P, JOHANSSON BR, JUKKOLA T, BREUER S, AKYUREK LM, PARTANEN J, LINDAHL P. Developmental origin of smooth muscle cells in the descending aorta in mice. Development. 2008;135:1823–1832. doi: 10.1242/dev.020958. [DOI] [PubMed] [Google Scholar]
- WILKINSON RN, POUGET C, GERING M, RUSSELL AJ, DAVIES SG, KIMELMAN D, PATIENT R. Hedgehog and Bmp polarize hematopoietic stem cell emergence in the zebrafish dorsal aorta. Dev Cell. 2009;16:909–916. doi: 10.1016/j.devcel.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WONG PM, CHUNG SW, WHITE JS, REICHELD SM, PATTERSON M, CLARKE BJ, CHUI DH. Adult hemoglobins are synthesized in murine fetal hepatic erythropoietic cells. Blood. 1983;62:1280–1288. [PubMed] [Google Scholar]
- WOTTON S, TERRY A, KILBEY A, JENKINS A, HERZYK P, CAMERON E, NEIL JC. Gene array analysis reveals a common Runx transcriptional programme controlling cell adhesion and survival. Oncogene. 2008;27:5856–5866. doi: 10.1038/onc.2008.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YOKOMIZO T, HASEGAWA K, ISHITOBI H, OSATO M, EMA M, ITO Y, YAMAMOTO M, TAKAHASHI S. Runx1 is involved in primitive erythropoiesis in the mouse. Blood. 2008;111:4075–4080. doi: 10.1182/blood-2007-05-091637. [DOI] [PubMed] [Google Scholar]
- YOKOMIZO T, OGAWA M, OSATO M, KANNO T, YOSHIDA H, FUJIMOTO T, FRASER S, NISHIKAWA S, OKADA H, SATAKE M, et al. Requirement of Runx1/AML1/PEBP2aB for the generation of haematopoietic cells from endothelial cells. Genes Cells. 2001;6:13–23. doi: 10.1046/j.1365-2443.2001.00393.x. [DOI] [PubMed] [Google Scholar]
- YOKOMIZO T, TAKAHASHI S, MOCHIZUKI N, KUROHA T, EMA M, WAKAMATSU A, SHIMIZU R, OHNEDA O, OSATO M, OKADA H, et al. Characterization of GATA-1(+) hemangioblastic cells in the mouse embryo. EMBO J. 2007;26:184–196. doi: 10.1038/sj.emboj.7601480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YOON MJ, KOO BK, SONG R, JEONG HW, SHIN J, KIM YW, KONG YY, SUH PG. Mind bomb-1 is essential for intraembryonic hematopoiesis in the aortic endothelium and the subaortic patches. Mol Cell Biol. 2008;28:4794–4804. doi: 10.1128/MCB.00436-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZEIGLER BM, SUGIYAMA D, CHEN M, GUO Y, DOWNS KM, SPECK NA. The allantois and chorion, when isolated before circulation or chorio-allantoic fusion, have hematopoietic potential. Development. 2006;133:4183–4192. doi: 10.1242/dev.02596. [DOI] [PubMed] [Google Scholar]
- ZOVEIN AC, HOFMANN JJ, LYNCH M, FRENCH WJ, TURLO KA, YANG Y, BECKER MS, ZANETTA L, DEJANA E, GASSON JC, et al. Fate tracing reveals the endothelial origin of hematopoietic stem cells. Cell Stem Cell. 2008;3:625–636. doi: 10.1016/j.stem.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]