Abstract
The structural integrity and conformational stability of various IgG1-Fc proteins produced from the yeast Pichia pastoris with different glycosylation site occupancy (di-, mono-, and non- glycosylated) was determined. In addition, the physical stability profiles of three different forms of non-glycosylated Fc molecules (varying amino acid residues at site 297 in the CH2 domain due to point mutations and enzymatic digestion of the Fc glycoforms) were also examined. The physical stability of these IgG1-Fc glycoproteins was examined as a function of pH and temperature by high throughput biophysical analysis using multiple techniques combined with data visualization tools (three index empirical phase diagrams and radar charts). Across the pH range of 4.0 to 6.0, the di- and mono- glycosylated forms of the IgG1-Fc showed the highest and lowest levels of physical stability respectively, with the non-glycosylated forms showing intermediate stability depending on solution pH. In the aglycosylated Fc proteins, the introduction of Asp (D) residues at site 297 (QQ vs. DN vs. DD forms) resulted in more subtle changes in structural integrity and physical stability depending on solution pH. The utility of evaluating the conformational stability profile differences between the various IgG1-Fc glycoproteins is discussed in the context of analytical comparability studies.
Keywords: conformation, stability, glycosylation, IgG, monoclonal antibody, Fc, formulation, mass spectrometry, spectroscopy
Introduction
Monoclonal antibodies (mAbs) are well established as the leading class of protein-based drugs due to their high target specificity and long half lifes.1, 2 The majority of mAbs developed to date are IgG1 proteins, consisting of four polypeptide chains (two heavy and two light chains) that arrange into 12 Ig domains that form into a Y-shaped molecule with two antigen binding (Fab) regions and one crystallizable (Fc) region. The homodimeric, horseshoe shaped, Fc region contains two interacting CH3 domains at the C-terminal ends and two CH2 domains at the N-terminal ends of the molecule. The two CH2 domains interact with each other through two buried N-linked glycosylation sites located at Asn 297. 3, 4
Glycosylation of the Asn 297 residue is one of the most common post-translational modifications found within mAbs. In the past few years, we have seen rapid growth in our understanding of the role of glycosylation with regard to both biological activity and pharmaceutical properties. Conformational changes of the CH2 domain, as a result of fully or partially removing the glycan residues, have been found responsible for altering the functionality3, physicochemical stability 5-8 and pharmacokinetic profile 9 of various mAbs. Additionally, protease resistance (using papain) has been shown to be significantly lowered in deglycosylated mAbs.10-11 These observations have been attributed to conformational differences due to the loss of both glycan-glycan and glycan-protein backbone non-covalent interactions upon deglycosylation. This results in the deglycosylated mAb to adopt a more open conformational state.
Mass spectrometric analyses of glycopeptides from mAbs have revealed significant heterogeneity in terms of glycosylation patterns of both currently marketed mAbs and those under development, depending on a variety of factors including the antibody type, expression systems and cell culture conditions.12-16 Among these glycoforms are the high mannose (HM) glycans consisting of 3 to 12 mannose units connected to two core GlcNAc units (N-acetyl glucosamine). In one study, an analysis of the glycan heterogeneity in Rituximab (a currently marketed drug for the treatment of non-Hodgkin's lymphoma) revealed that 1.7-5.4 % of the glycans present were of a HM nature. 17 Antibodies containing HM glycans are known to have faster clearance times compared to glycans having either GlcNAc, galactose or sialic acid units at the non-reducing termini of the oligosaccharide.18, 19 The effect of having enriched or depleted levels of HM IgG1 within a heterogeneous mixture of IgG1 glycoforms was shown to not affect the physical stability of the mAb preparation.20 Asymmetric mAb glycosylation (single arm glycosylation) has been reported for an IgG1 containing a single glycosylation in the Fab region.21 This results in the IgG1 losing its divalent binding capacity to its antigen. An asymmetrically glycosylated IgG1 in the CH2 domain was isolated and characterized by Ha et. al. (2011). 22 The authors purified the monoglycosylated form of the IgG1 to ~80-85%. Although minimal stability differences were noted by DSC at one solution pH (1°C lower for Tm1 and no difference for Tm2), Fc gamma receptor binding activity differences between the asymmetrically and the fully glycosylated IgG1 of 2-3 fold were reported. Comparisons of the physical stability profiles of a variety of different proteins have been performed in our laboratories recently including ten mutants of acidic fibroblast growth factor,23 three glycoforms of an IgG1 mAb generated by deglycosylation,24 and fifteen different formulations of GCSF protein.25 In this work, IgG1-Fc glycoforms containing well defined, homogeneous glycosylation patterns, produced using a Pichia pastoris yeast expression system followed by purification and specific enzymatic digestions, were utilized to more directly address the effect of glycosylation site occupancy and amino acid substitution (at Asn 297, the N-linked glycosylation site in CH2 domain) on the structural integrity and conformational stability of a human IgG1-Fc. The physical stability of this series of Fc glycoproteins was examined by high throughput biophysical analysis using multiple analytical techniques combined with data visualization tools (three-index empirical phase diagrams and radar charts). By using larger physical stability data sets acquired from multiple high throughput low-resolution biophysical techniques as a function of environmental stresses (pH and temperature), differences in the structural integrity and conformational stability in this series of Fc glycoforms were detected. These stability trends, as a function of site occupancy and amino acid substitution in the Fc glycoforms, were not necessarily observed using the same biophysical techniques under non stressed conditions. As a result, evaluating the conformational stability differences between the different IgG1-Fc glycoproteins may serve as a surrogate to monitor differences in higher-order structure between IgG1-Fc samples, an approach that could potentially be useful for analytical comparability studies.
Materials and Methods
Materials
Both the human IgG1-Fc sequence (comprising 446 amino acids with a theoretical molecular weight of 50132.92 Da) and a point mutant of the IgG1-Fc protein (with 446 amino acids and a theoretical molecular weight of 50160.96 Da) were prepared and expressed using a glycosylation deficient strain of Pichia pastoris as described by Xiao et. al. (2009).26 The nonglycosylated variant of the IgG1-Fc was made by mutating the N-linked glycosylation site at Asn 297 (EU numbering) to Gln 297, thus eliminating the Asn-X-Thr glycosylation site within the CH2 domain. This was achieved through PCR site-directed mutagenesis using Quikchange II site-directed mutagenesis kit (Agilent Technologies), followed by transfecting the yeast with the mutated plasmid after sequencing it for verification as described by Xiao et. al. (2009).26 After expression, purification and concentration of the different IgG1-Fc glycoproteins (as described below), samples were dialyzed into the storage buffer (10 % sucrose, 20 mM histidine, pH 6.0) and frozen at -80 °C in aliquots of 0.5 mL. For the initial characterization of the IgG1-Fc proteins (SDS-PAGE, mass spectroscopy and SE-HPLC), samples were analyzed without further dialysis. For biophysical characterization (far-UV circular dichroism, intrinsic/extrinsic fluorescence spectroscopy and turbidity measurements), samples were dialyzed against 20 mM citrate phosphate buffer (pH 4.0-6.0, 0.5 increments) and adjusted to an ionic strength of 0.15 with NaCl. Other chemicals and reagents not described below were obtained from Sigma–Aldrich (St. Louis, MO), Fisher Scientific (Pittsburg, PA), Invitrogen (Carlsbad, California) or Becton Dickinson and Company (Franklin Lakes, NJ).
Methods
Expression and purification of the IgG1-Fc proteins
IgG1-Fc proteins were cloned and then expressed using an OCH1 deleted strain of Pichia pastoris yeast expression system, followed by Protein G affinity purification, as described previously.26 To separate the differentially glycosylated forms of the IgG1-Fc, two different purification methods were used as described below. Since the purity of the IgG1-Fc variants was essentially the same from methods (data not shown), the purified fractions from each approach were combined (for each variant separately) to ensure the same material was being examined during biophysical studies.
First, a cation exchange chromatography (CEX) method using ProPac WCX-10 semi-preparative (9 × 250 mm) column, (Dionex, Sunnyvale, CA) was utilized.27 The column was equilibrated with Buffer A (20 mM sodium acetate pH 4.8) for 5 column volumes (CV). The protein G purified IgG1-Fc solution was then loaded onto the cation exchange column. Chromatographic separation was then performed in a linear gradient from 0 to 1M NaCl (10 CV) using Buffer B (20 mM sodium acetate pH 4.8, 1 M NaCl). Two mL fractions were collected throughout the gradient. Peaks were collected corresponding to the diglycosylated and monoglycosylated IgG1-Fc proteins (which are primarily used in this study and represented ~80% and ~15% of the material, respectively) in addition to an aglycosylated IgG1-Fc form (~5% of the material). After analyzing column fractions with SDS-PAGE to confirm glycoprotein identity, the two collected IgG1-Fc glycoforms were concentrated, dialyzed against 10% sucrose, 20 mM histidine pH 6.0 and frozen at -80 °C until further use.
For the second, scaled-up purification process, a hydrophobic interaction chromatography (HIC) method using a HiScreen fast flow phenyl sepharose column (4.7 mL bed volume and 7.7×100 mm bed dimensions, GE Healthcare) was adopted.26 The HIC column was equilibrated with Buffer A (20 mM sodium phosphate, 1M ammonium sulfate pH 7.0) for at least 5 CV. The protein G purified IgG1-Fc solution was then loaded onto the HIC column. Chromatographic separation was then conducted in a linear gradient from 1.0 to 0 M ammonium sulfate (10 CV) using Buffer B (20 mM sodium phosphate, pH 7.0). Ten mL fractions were collected throughout the gradient. Two peaks were observed and collected corresponding to the diglycosylated and monoglycosylated glycoforms of the IgG1-Fc proteins. After analyzing column fractions with SDS-PAGE, the collected glycoforms were concentrated, dialyzed against storage buffer (10% sucrose, 20 mM histidine pH 6.0) and frozen at -80 °C until further use.
Since the nonglycosylated (NN at site 297) form of the IgG1-Fc was difficult to purify and could only be produced in low, insufficient amounts, a nonglycosylated (QQ at sites 297) IgG1-Fc point mutant protein was expressed, using an OCH1 deleted Pichia pastoris strain, and purified in a similar fashion to that described above including the HIC purification method.26
Deglycosylation of IgG1-Fc glycoproteins
The purified diglycosylated and monoglycosylated forms of the IgG1-Fc were deglycosylated using the enzyme peptide-N-Glycosidase F (PNGase F) under non-denaturing conditions (made in-house as described elsewhere).28 Samples in storage buffer were thawed, treated with PNGase F in a protein to enzyme ratio of 500:1 (w/w) and incubated for 6 hrs. at 25 °C. Enzymatic reaction completion was confirmed by MS and SDS-PAGE analysis (see Table 1 and supplementary Figures S1 and S2).
Table 1.
Mass spectrometry analysis of reduced monomeric forms of the purified IgG1-Fc proteins. The molecular weight column shows the masses of the reduced non-glycosylated IgG1-Fc obtained from MS analysis along with the masses measured for the most abundant glycoforms (Man8GlcNAc2) of the glycosylated Fc. The values for experimentally observed change in mass, and predicted change in mass are also shown. Based on these results, the modification expected due to enzymatic treatment and/or glycan variation of the IgG1-Fc proteins is then presented.
| IgG1-Fc proteins | Observed Molecular Weight (Da) | Observed Δ Mass (Da) | Reduced Fc fragment Modificationsa | ||
|---|---|---|---|---|---|
| Arm 1 | Arm 2 | ||||
| Non-glycosylated IgG1-Fc | Mutant (QQ) | 25,077 | +11 | N → Q (+14) | N → Q (+14) |
| Mono-glycosylated + PNGase F (DN) | 25,063b | −3 | Deglycosylated N → D (+1) | N (0) | |
| Di-glycosylated + PNGase F (DD) | 25,063b | −3 | Deglycosylated N → D (+1) | Deglycosylated N → D (+1) | |
| Glycosylated IgG1-Fc | Mono-glycosylated c | 26,766d / 25,062b | +1,700/−4 | +Man8GlcNAc2 (+1,704) | N (0) |
| Di-glycosylated c | 26,766d | +1,700 | +Man8GlcNAc2 (+1,704) | +Man8GlcNAc2 (+1,704) | |
D, aspartic acid; N, asparagine; Q, glutamine; GlcNAc, N-acetylglucosamine; Man, Mannose; Numbers in ( ) indicates predicted Δ mass change in Da.
The theoretical mass of the reduced, non-glycosylated IgG1-Fc protein is 25,066 Da.
Man8-12GlcNAc2 glycoforms were observed during MS analysis for the diglycosylated IgG1-Fc. Man8-9GlcNAc2 glycoforms were observed for the mono-glycosylated IgG1-Fc.
theoretical mass of reduced monomelic form of the purified IgG1-Fc protein with Man8GlcNAc2 glycan is 26,770 Da.
Electrospray Ionization-Mass Spectrometry
IgG1-Fc proteins were analyzed by electrospray ionization mass spectrometry (ESI-MS). ESI spectra of reduced protein samples (15 mM dithiothreitol, pH 7.0, room temperature) were acquired on a SYNAPT G2 hybrid quadrupole / ion mobility / time of flight mass spectrometer (Waters Corp., Milford, MA). The instrument was operated in a sensitivity mode with all lenses optimized on the MH+ ion obtained from Enkephalin. The sample cone voltage was 30eV. Argon was admitted to the trap cell that was operated at 4eV for maximum transmission. Spectra were acquired at 9091 Hz pusher frequency covering the mass range from 100 to 3000 u and accumulating data for 1.5 seconds per cycle. Time to mass calibration was made with Nal cluster ions acquired under the same conditions. Samples were desalted on a reverse phase C4 column, 1 cm, 1 mm I.D. (Vydac, Midland, Canada, 300 A pore size. The 5 μm particles were packed by Micro-Tech Scientific) using a NanoAcquity chromatographic system (Waters Corp., Milford, MA). The solvents used were A (99.9% H2O, 0.1% formic acid) and B (99.9% acetonitrile, 0.1% formic acid). A short gradient was developed from 1 to 70% B in 4 min with a flow rate of 20 μl/min. Masslynx 4.1 software was used to collect the data. The MaxEnt 1routine was used for processing data to convert peaks of multiply charged protein ions into uncharged deconvoluted protein spectra.
Binding of IgG1-Fc glycoforms to Fc γ receptor IIIA
The interactions of the different IgG1-Fc proteins with Fc γ receptor IIIA (FcγRIIIA) were studied with bio-layer interferometry using a BLITZ instrument (Fortebio, Menlo Park, CA) with protein G biosensor tips. The soluble domain of the high affinity V158 polymorph of FcγRIIIA29 was expressed and purified using Pichia pastoris (manuscript in preparation) and binding studies using this receptor were conducted as follows. Briefly, an initial baseline was established with PBS buffer (50 mM sodium phosphate, 150 mM NaCl, pH = 7.4) and then the protein G biosensor tips were loaded with the various IgG1-Fc glycoforms at a concentration of 0.88 μM in PBS buffer. A new baseline was then established and then the association and dissociation of FcγRIIIA was measured in PBS buffer. To determine the dissociation constant (Kd) for the IgG1-Fc glycoforms, a range of FcγRIIIA concentrations were tested (50 nM, 100 nM, 200 nM, and 400 nM). Data generated from the binding of the receptor to IgG1-Fc glycoforms were generated in triplicate and fitted to a 1:1 binding model and analyzed using BLITZ Pro software.
Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis (SDS-PAGE) and Western Blot Analysis
Samples containing 20 μg protein, 10μl of a SDS solution, and for reduced samples, 5 μl of dithiothreitol (50 mM) were incubated at 80 °C for 2 min and loaded onto a NuPAGE 4-12 % Bis-tris gel (Invitrogen, Carlsbad, California) using 20x MES as a running buffer. The running time was 65 min at 160V. Gels were then stained with Coomassie blue and then destained using 10 % methanol, 10 % glacial acetic acid solution. Protein molecular weight standards (BIORAD, Hercules, CA) were loaded for molecular weight estimation. Band densitometry measurements were performed using the imageJ program (NIH) as described in detail elsewhere.30, 31 For Western blot analysis, proteins in SDS-PAGE gels were transferred onto a polyvinylidene fluoride (PVDF) membrane (Bio-RAD, Hercules, CA) using 25 mM Tris, 192 mM glycine, 20 % (v/v) methanol buffer (pH 8.3). Non-specific binding was blocked (5 % nonfat milk dissolved in Tris-buffered saline at pH 7.5, 3 hrs. at 4 °C). The PVDF membranes were washed with Tris-buffered saline containing 0.05 % Tween 20 followed by overnight incubation at 4 °C with alkaline phosphatase labeled anti-IgG-Fc specific secondary antibody (Thermo scientific, Waltham, MA). Protein bands were visualized by NBT/BCIP substrates (Thermo scientific, Waltham, MA).
Size-Exclusion Chromatography (SE-HPLC)
Experiments were performed using a Shimadzu high-performance liquid chromatography system equipped with a photodiode array detector and a temperature controlled autosampler. A Tosoh TSK-Gel Bioassist G3SWxL column (7.8 mm ID × 30.0 cm L) and a corresponding guard column (TOSOH Biosciences, King of Prussia, Pennsylvania) were used as described in detail elsewhere.32
Biophysical methods
Far-UV CD spectra ranging from 260 to 205 nm were collected in triplicate using a Chirascan spectropolarimeter (Applied Photophysics, Surrey, United Kingdom) with 200 μl of a 0.2 mg/ml protein solution in 0.1 cm path length quartz cuvette as described in detail elsewhere.24 Intrinsic (Trp) fluorescence was measured in triplicate using a Photon Technology International (PTI) Quantum master fluorometer (New Brunswick, New Jersey) with 200 μl of a 0.2 mg/ml protein solution in 0.2 cm path length quartz cuvette. For thermal stability measurements, the temperature was raised in increments of 1.25 °C from 10 to 90°C with a heating rate of 15°/hr. Data were collected and analyzed as described in detail elsewhere.24 The maximum peak position (λmax) values were calculated using a “spectral center of mass” method (MSM) that causes a red shift in the peak position by 10-14 nm compared to the actual value but produces a more reproducible measure of spectral shift and a higher signal to noise ratio. 24
Differential Scanning Fluorimetry (DSF) measurements were performed in triplicate using MX3005P QPCR system (Agilent Technologies). Samples contained 0.2 mg/ml protein and 1X SYPRO™ orange dye (Invitrogen, Carlsbad, CA), and data were collected from 25 to 90 °C using 60 °C/hr a heating rate and then analyzed as described in detail elsewhere.24
Turbidity measurements were performed in triplicate using a Cary 100 UV-Visible spectrophotometer (Varian Medical Systems, Inc. Palo Alto, CA) equipped with a 12 cell holder with peltier type temperature controller. Samples contained 0.2 mg/ml protein with a total volume of 200 μl in 1 cm path length quartz cells. Optical density at 350 nm was monitored as the temperature was raised in increments of 1.25 °C from 10 to 90 °C with a heating rate of 60°/hr. Corresponding buffer blanks were run and subtracted from each sample.
Data visualization techniques
The three-index Empirical Phase Diagram and Radar Chart data visualization methods, as described in detail elsewhere,33 were generated using in-house software (Middaugh Suite) and then utilized to describe biophysical stability characteristics of the various Fc proteins. In this work, three biophysical data sets as a function of pH and temperature were used for data visualization: Sypro Orange fluorescence, intrinsic Trp peak position fluorescence, and optical density at 350 nm. These three techniques were chosen for data visualization since data displayed clear thermal transitions; in contrast, circular dichroism thermal stability vs. pH data did not show clear thermal transitions (data not shown). For the three index EPDs, the Sypro Orange fluorescence stability data were mapped to the color red, Trp fluorescence peak position stability data to the color green, and optical density stability data to the color blue. The combination of these RGB values produced a single color at each point in pH and temperature coordinates. For each technique and protein, the minimum value was mapped to the loss of the color component, and the maximum value was mapped to the full color intensity.
The resulting color map was generated: The color black indicates minimum values of all three techniques, which can be interpreted as the structure closest to the native state (least amount of structural change). The green or red color appears within the three index EPD as the Trp fluorescence peak positions changes (displays red shift) or Sypro Orange binding increases, respectively. The blue appears when high turbidity is measured by optical density values at 350 nm. To determine common regions within the three index EPD, a k-Means clustering algorithm is first applied to each EPD to identify boundaries of similarly color regions. The resulting clusters can scatter as small blocks of colors, especially over the structurally altered regions. Then, using visual assessments and comparisons to the raw physical stability profiles for each instrument, only certain commonly observed clusters were selected. Radar charts were generated to display similar characteristics (changes in readouts from each of the three biophysical instruments) for each of these commonly observed clusters. The same three analytical techniques and min-max range were used for preparing radar charts. The minimum value was mapped to the center of the radar chart, while the maximum value was mapped onto the outer circumference. All values within the cluster were averaged from multiple runs and values were connected to yield a final polygon to represent a specific structural state.
Results
Initial characterization of the different IgG1-Fc glycoforms
A pictorial presentation of the five different types of IgG1-Fc proteins (diglycosylated, monoglycosylated and three different non-glycosylated forms) examined in this study is presented in Figure 1. High mannose (HM) IgG1-Fc glycoforms were expressed in yeast and purified using a combination of Protein G and cation exchange/hydrophobic interaction chromatography. Purified protein fractions of each variant from both cation exchange and hydrophobic interaction chromatography were combined to ensure the same material was being examined during biophysical studies (see Methods section). Expression of IgG1-Fc in Pichia pastoris produced IgG1-Fc with incomplete glycosylation at the Asn 297 site. Glycosylation site occupancy is ~ 90% at the Asn 297 site, and since IgG1-Fc is a disulfide-linked dimer, a mixture of diglycosylated dimer, monoglycosylated dimer and trace amount of nonglycosylated dimer is obtained. First, an IgG1-Fc bearing HM glycans at both CH2 domains of the dimer (~80 % of the amount purified) designated diglycosylated IgG1-Fc was identified. Second, an IgG1-Fc containing HM glycan at a single CH2 domain of the dimer (~15 % of the amount purified) labeled monoglycosylated IgG1-Fc was found. In addition, IgG1-Fc proteins with variations in amino acid residues at site 297 of the CH2 domain were prepared. IgG1-Fc with Asp/Asp (DD) and Asp/Asn (DN) residues were made by treating the diglycosylated and monoglycosylated IgG1-Fc proteins with the enzyme PNGase F. PNGase F cleaves the glycan residue(s) and converts Asn to Asp, adding one or two negative charges to the IgG1-Fc molecules (depending on the initial glycosylation site occupancy). Finally, a N297Q IgG1-Fc mutant, with Asn/Asn (NN) substituted with Gln/Gln (QQ) through site directed mutagenesis, was also expressed and purified (Figure 1).
Figure 1.
Summary of the five different IgG1-Fc proteins examined in this study. The left side shows the two glycosylated forms of the Fc protein (diglycosylated and monoglycosylated), while, the right side shows the three different nonglycosylated IgG1-Fc protein variants (two from PNGase treatment of glycoforms and one point mutant). Identity of amino acid residue at site 297 in the CH2 domain (Asn, Asp, Gln) and the nature of glycoforms (Man, mannose and GlcNAc, N-acetylglucosamine) are indicated in the figure.
To confirm the correct masses for each of the purified IgG1-Fc glycoproteins, mass spectrometric analysis of reduced samples was performed (Table 1 and supplementary Figure S1). The nonglycosylated (QQ) Fc mutant had a mass of 25,077 Da, in close agreement (- 3 Da) with the mass predicted from the sequence. MS analysis of diglycosylated IgG1-Fc protein revealed a mass of 26,766 Da reflecting a predominant Man8GlcNAc2 glycosylation form, in close agreement to the predicted mass (26,770 Da). Additionally, peaks corresponding to one, two, three and four additional mannoses were also present indicating glycoforms with 9, 10, 11 and 12 mannoses, respectively, but in lower abundance (see supplemental Figure S1). The monoglycosylated IgG1-Fc protein had two distinct peaks with masses of 25,062 and 26,766 Da in agreement with nonglycosylated and Man8GlcNAc2 glycosylated arms of the glycoprotein. A heavier peak reflecting the presence of Man9GlcNAc2 IgG1-Fc glycoform was detected as well (see supplemental Figure S1). Upon PNGase F enzymatic treatment of the diglycosylated and monoglycosylated IgG1-Fc samples, a mass of 25,063 Da was observed for each protein indicating fully deglycosylated Fc proteins.
The functionality of the proteins produced in this study was assessed using bio-layer interferometry. All IgG1-Fc proteins used in this study were functional in their ability to bind protein G as evidenced by the use of protein G affinity chromatography in their initial purification and the ability to immobilize all of the purified IgG1-Fc proteins on protein G biosensor tips. In addition, those IgG1-Fc glycoforms that are expected to bind to FcγRIIIA based on literature reports were indeed functional in that they bound FcγRIIIA. The high mannose di-glycosylated IgG1-Fc bound tightly to FcγRIIIA with a Kd of 32 ± 3 nM, as would be expected for a non-fucosylated IgG1-Fc glycoform.34, 35 The high mannose monoglycosylated IgG1-Fc glycoform bound less tightly with a Kd of 100 ± 8 nM, also as would be expected from previous studies.22 PNGase F treated di- and mono-glycosylated IgG1-Fc (DD and DN) and mutant IgG1-Fc (QQ) all showed no binding at the highest receptor concentration tested which is consistent with previous studies of the importance of Fc glycosylation in IgG1 Fc receptor interactions.34
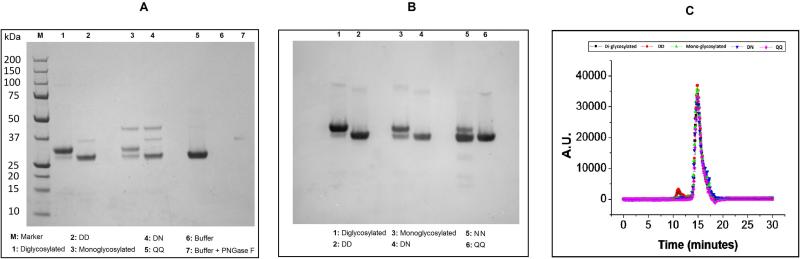
The purity of the various IgG1-Fc glycoproteins was determined under denaturing (SDS-PAGE and Western Blot) and non-denaturing conditions (SEC) as summarized in Figures 2A, 2B and 2C, respectively. The di-, mono- and non- glycosylated Fc proteins migrated to their expected positions (e.g., under reduced conditions, ~28 kDa, ~28/~26 kDa mixture and ~26 kDa, respectively) as shown in Figure 2A. Densitometry analysis of the reduced SDS-PAGE gels showed diglycosylated, nonglycosylated DD, and the nonglycosylated mutant (QQ) had ~97-99 % purity. The monoglycosylated Fc protein (and the corresponding nonglycosylated DN form derived from PNGase treatment of the monoglycosylated Fc) showed somewhat lower purity values of ~84% Visual inspection of the Western blot analysis showed high purify levels for all samples and confirmed the PNGase treatment led to the expected deglycosylation patterns (Figure 2B). The purity of the IgG1-Fc samples under non-denaturing conditions was also determined by SEC (Figure 2C). The diglycosylated Fc protein showed 91.8% vs. 91.1% monomer, and 7.8% vs. 8.2% aggregate before and after PNGase treatment. The monoglycosylated Fc protein showed 95.3% vs. 93.7% monomer, and 3.4% vs. 4.0% aggregate before and after PNGase treatment. The QQ aglycosylated mutant showed 96.6% monomer and 1.8% aggregate. The remaining area can be accounted for by the presence of small amounts of fragments at the level of ~0.2-2.0% (data not shown).
Figure 2.
Purity evaluation of the five Fc proteins under denaturing and non-denaturing conditions. SDS-PAGE (A) and Western Blot (B) analysis of reduced IgG1-Fc samples are shown along with size exclusion chromatograms (SEC) of the non-reduced IgG1-Fc proteins. See Figure 1 for nomenclature describing the different IgG1-Fc proteins and text for summary of results.
In summary, the monoglycosylated Fc protein, which was only ~15 % of the expressed protein, was more difficult to purify compared to the diglycosylated form with more notable differences in purity levels (97% vs. 84%) by SDS-PAGE and more minor differences by (~92 vs. ~95% monomer) by SEC. PNGase treatment of these samples had minor to no effects on the overall protein purity levels, but as expected, removed the oligosaccharides from the Fc glycoproteins. Although all of the Fc proteins bind protein G, the asymmetrical, monoglycosylated Fc protein showed ~3X decreased functional binding to FcγRIIIA receptor while the aglycosylated forms did not bind the same receptor. Similar purity levels and functional receptor binding results have been reported recently for an intact monoglycosylated mAb 22 (see discussion section). As shown in the next section, the asymmetrical monoglycosylated Fc protein displayed notable differences in structural integrity and thermal stability as a function of solution pH. .
Structural integrity at ambient temperature conditions
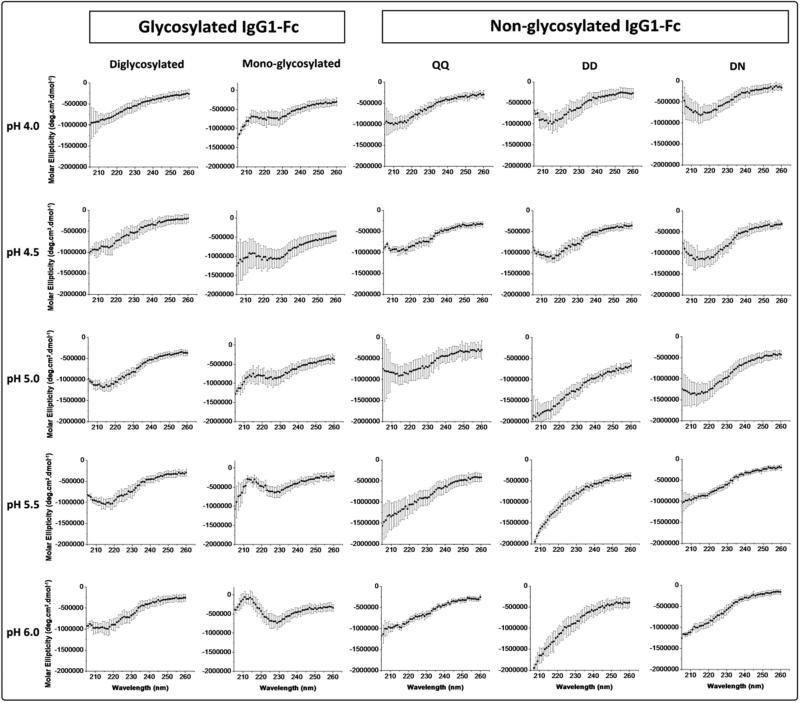
The overall secondary structure of the five IgG1-Fc proteins was evaluated at 10 °C by far-UV circular dichroism (CD) analysis from 260 to 205 nm under various pH conditions (Figure 3). The CD spectra of the diglycosylated IgG1-Fc show a clear minimum at 217 nm from pH 5.0-6.0, and to a lesser extent at pH 4.0-5.0, indicating a β-sheet rich structure as expected from immunoglobulin folded domains. The same overall β-sheet rich secondary structure is seen for the nonglycosylated DD (from PNGase treatment of the diglycosylated form) at pH 4.0-4.5, whereas, at pH 5.0, 5.5 and 6.0 a constant decrease in CD signal is observed indicating some alteration in the protein secondary structure, under these pH conditions, compared to the fully glycosylated form of the protein. Interestingly, for the nonglycosylated mutant (QQ) IgG1-Fc, CD spectra show a clear minimum at 217 nm from pH 4.0-5.0, and to a lesser extent at pH 5.0-6.0.
Figure 3.
Far-UV CD spectra from 205-260 nm at 10 °C for the two different Fc glycoforms (di- and mono glycosylated) and three different nonglycosylated IgG1-Fc samples (QQ, DD, DN) at pH values of 4.0, 4.5, 5.0, 5.5, 6.0. See Figure 1 for a pictorial summary of the five Fc proteins examined.
For the monoglycosylated IgG1-Fc protein, a different CD spectrum is observed compared to the other Fc protein samples with a positive hump present at 212 nm that increases in intensity as the solution pH increases. This could potentially be attributed to an increasing influence from type I β-turns. When the monoglycosylated IgG1-Fc protein was deglycosylated using PNGase F (nonglycosylated DN), more native-like structural features in the CD spectra were detected in the form of a minimum at 217 nm in the pH range of 4.0-5.0, and to lesser extent at pH 5.5 and 6.0. These results indicate the monoglycosylated Fc protein is in a structurally perturbed state, and that upon deglycosylation, the effect is at least partially reversible with the regaining of native-like secondary structure seen by CD (monoglycosylated vs. DN nonglycosylated forms of the Fc protein in Figure 3). The CD spectra intensity signals as a function of increasing temperature were monitored. Due to noisy data, however, as well as the lack of intensity change as a function of temperature for many of the Fc proteins (e.g., monoglycosylated as well its deglycosylated form), the CD thermal melting curves were not further employed in this study (data not shown).
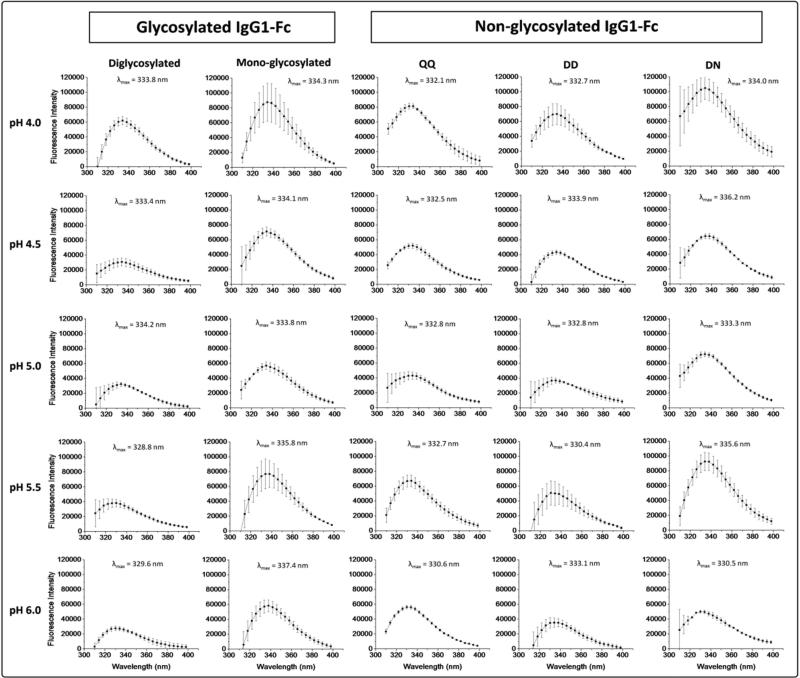
The structural integrity of the overall tertiary structure of the five IgG1-Fc proteins was then evaluated using intrinsic (Trp) fluorescence spectroscopy (Figure 4). Diglycosylated and nonglycosylated mutant (QQ) IgG1-Fc proteins manifest fluorescence spectra at pH 4.0, with relatively high peak intensities, and λmax values of 333.8 and 332.1 nm, respectively. As the pH is raised, the λmax starts shifting to lower wavelengths (with λmax values of 329.6 and 330.6 nm at pH 6.0 accompanied by relatively lower peak intensity), indicating the average Trp residues in a more apolar environment, and presumably more folded state. For the nonglycosylated DD IgG1-Fc protein, the λmax does not shift to lower wavelength as the solution pH is raised, remaining at ~333 nm, suggesting that deglycosylated protein remains in the same structure in this pH range, presumably an effect due to deglycosylation and amino acid substitution (Asn to Asp at site 297). Monoglycosylated IgG1-Fc protein, however, shows a small red shift in λmax as a function of increasing solution pH, indicating potentially less folded structure at pH 6.0 (vs. pH 4.0). In addition, the λmax value at pH 6.0 (337.4 nm) for the monoglycosylated IgG1-Fc is significantly red-shifted by 4-7 nm compared to the other samples. Upon PNGase treatment of the monoglycosylated protein, the nonglycosylated DN protein is generated and at pH 6.0 shows a more native like λmax value of (330.5 nm). It also behaves more like the diglycosylated and QQ nonglycosylated mutant with a noticeable blue shift in λmax as the pH is raised from pH 4.0 to 6.0 (~334 to ~330 nm). These intrinsic fluorescence spectroscopy results, similar to the CD results above, indicate that the monoglycosylated Fc protein is in a structurally perturbed state, and that upon deglycosylation, the effect is at least partially reversible.
Figure 4.
Intrinsic (Trp) fluorescence spectra at 10 °C for the two different glycosylated (di- and mono glycosylated) and three different nonglycosylated forms (QQ, DD, NN) of the IgG1-Fc across the pH range of 4.0-6.0. The λmax values are also shown for each protein at each pH condition. Each of the samples were run in triplicate and the standard deviation of λmax values the ranged from 0.2-0.6 nm. See Figure 1 for pictorial summary of the five Fc proteins examined.
Physical stability of IgG1-Fc proteins as a function of pH and temperature
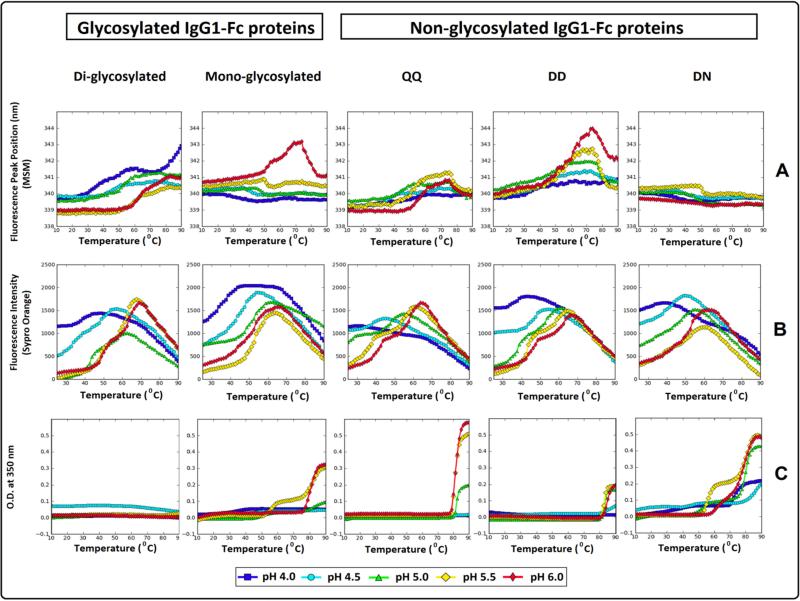
The conformational stability of the tertiary structure of the five IgG1-Fc proteins was evaluated using a combination of intrinsic (Trp) fluorescence and extrinsic fluorescence (i.e., differential scanning fluorimetry, DSF) spectroscopy (see Figures 5A, 5B, and 5C with the same physical stability data shown in Supplemental Figure S2 with corresponding standard deviation error bars from triplicate measurements). The conformational stability of the five Fc proteins was first evaluated by following changes in λmax as a function of increasing temperature (Figure 5A). It should be noted that the λmax values in Figure 4A are mathematically shifted compared to the actual values (see Figure 4), due to the use of a spectral central of mass (MSM) method as described in the methods section. Diglycosylated and nonglycosylated mutant (QQ) both had relatively low λmax peak positions at the starting temperature across the pH conditions tested (~339-340 nm using the MSM method) with higher thermal stability being observed for the diglycosylated IgG1-Fc protein. The nonglycosylated DD IgG1-Fc protein, with λmax peak positions of ~340 nm, showed a subtle, steady increase in peak position as the temperature was raised followed by a more dramatic increase in peak position. The monoglycosylated and nonglycosylated DN forms (derived from PNGase treatment) had λmax peak positions of ~340-341 nm with little to no detectable shift in peak position as the temperature increases, except for the monoglycosylated Fc protein at pH 6.0 where a transition curve is detected (Figure 5A).
Figure 5.
Thermal melting curves for the two different glycosylated (di- and mono glycosylated) and three different nonglycosylated forms (QQ, DD, NN) of the IgG1-Fc across the pH range of 4.0-6.0. Biophysical measurements include (A) Intrinsic Trp fluorescence, (B) Extrinsic Sypro Orange fluorescence spectroscopy and (C) optical density at 350 nm. See Figure 1 for pictorial summary of the five Fc proteins examined.
In DSF analysis, SYPRO orange dye was used as a probe for detecting structurally disrupted proteins, seen in the form of increased fluorescence intensity upon exposure to apolar environments due to structural alternations and/or aggregate formation (See Figure 5B). Diglycosylated IgG1-Fc was the most thermally stable protein manifesting two thermal transitions (presumably sequential unfolding events of the CH2 and CH3 domains). This was seen across most of the pH conditions tested (except at pH 4.0) with an observed increase in thermal stability as a function of increasing pH. Additionally, fluorescence peak intensity at the starting temperature in the range of pH 5.0-6.0 is decreased compared to the values at the lower pH conditions. For the monoglycosylated Fc protein at pH between 4.0 and 5.5, and nonglycosylated DN protein at pH between 4.0 and 5.0, a single thermal transition is seen, consistent with the presence of a more structurally disrupted CH2 domain. Two thermal transitions were seen for the nonglycosylated mutant (QQ) and the nonglycosylated DD Fc proteins under most of the pH conditions tested, except at pH of 4.0 and 4.5, again consistent with the presence of a more structurally altered CH2 domain under lower pH conditions (compared to the higher pH range) for these two proteins.
Temperature induced aggregation was evaluated by monitoring optical density changes at 350 nm as a function of increasing temperature and pH (Figure 5C). Differences between the IgG1-Fc proteins were observed in terms of the onset temperature of aggregation as well as the profile of the optical density transitions. Diglycosylated IgG1-Fc protein did not show detectable thermal transitions in optical density across all pH conditions tested. The monoglycosylated Fc protein, on the other hand, displayed a single transition in optical density at pH 4.0, 4.5 and 6.0 starting at 21°, 33°, and 76 °C, respectively, whereas, at pH 5.0 and 5.5, two transitions were detected with the first transitions starting at 45° and 79 °C and the second at 52° and 75 °C, respectively. For the nonglycosylated mutant (QQ), a single thermal event of increasing optical density was detected at ~79 °C at pH 5.0-6.0, with no thermal transitions noted at lower pH values of pH 4.0 and 4.5. Similarly, the nonglycosylated DD Fc protein showed no detectable thermal transition at pH 4.0, a single thermal transition in optical density starting at 78 °C at pH 4.5, and at 82 °C for pH 5.0, 5.5 and 6.0. In contrast, the nonglycosylated DN Fc protein (derived from PNGase treatment of the monoglycosylated form) manifested two optical density thermal transitions under all pH conditions examined. At pH 4.0, the nonglycosylated DN protein showed an increase in optical density starting from 10 °C followed by a second major increase at 69 °C. At pH 4.5, 5.0 and 5.5, an initial increase in optical density was detected at 40°, 46° and 52 °C, followed by a second increase at 75°, 78° and 74 °C, respectively. Two adjacent transitions in optical density were seen at pH 6.0, with the first transition initiating at 57 °C followed by a second transition at 74 °C.
Physical stability evaluations using various data visualization techniques
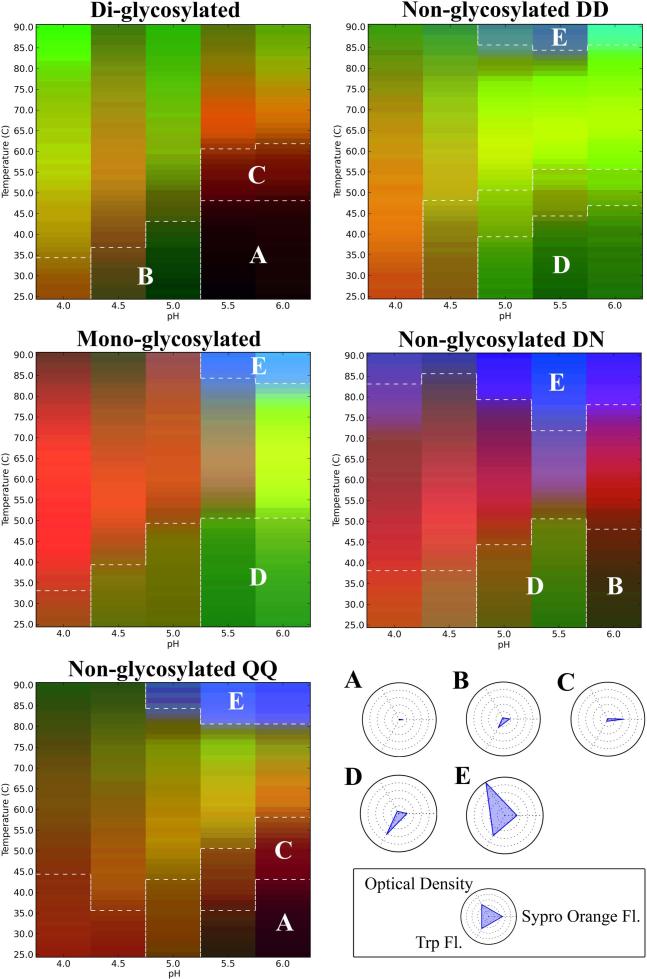
Using the physical stability data acquired from differential scanning fluorimetry, intrinsic fluorescence spectroscopy (peak position), and turbidity measurements, three index EPDs and radar charts were constructed for each of the individual IgG1-Fc proteins. A summary figure was constructed showing the overall results, from the three-index EPD analysis from each of the five individual IgG1-Fc proteins, merged into one display (Figure 6). A summary of the radar plot analysis as a function of temperature for each of the five individual IgG1-Fc proteins is provided in supplemental Figure S3. The five color plots in Figure 6 highlight five regions with similar structural characteristics designated regions A-E, as observed in the three-index EPDs for the five different IgG1-Fc proteins with various glycosylation occupancy (diglycosylated, monoglycosylated) as well as the nonglycosylated forms of the Fc protein containing amino acid variations (QQ, DD and DN) in the CH2 domain. In addition, as shown in the legend box in Figure 5, radar plots were prepared to better define the five different conformational regions (Regions A through E) that can be observed in each of the five Fc proteins under the various pH and temperature conditions.
Figure 6.
Three index EPDs for the two different glycosylated (di- and mono glycosylated) and three different nonglycosylated forms (QQ, DD, NN) forms of the IgG1-Fc. The legend box contains radar plots defining the five different conformational stability regions observed in the five different Fc proteins. Region A (Black) and B (Dark green) represent regions where the protein exists in a native-like and slightly altered structures, respectively. Regions C (Dark red) and D (Green) represent regions where the Fc proteins exist at an altered structural state as seen from an increase in SYPRO orange and Trp fluorescence signals, respectively. Region E (Blue) represents a region where the Fc proteins are in an aggregated state as seen by an increased optical density signal.
Region A represents a state in which the IgG1-Fc protein exists in its most native-like conformation. As shown in the radar chart legend in Figure 6, there are minimal changes in signals for each of the three biophysical techniques (intrinsic Trp and extrinsic SYPRO orange fluorescence spectroscopy as well as turbidity at OD 350nm), which results in a black color for Region A as shown in the EPDs for the five Fc proteins in Figure 6. For Region B, there are small changes in the signals from the biophysical instruments, which represents protein with a slightly altered conformation (minor increases in SYPRO orange fluorescence intensity and Trp fluorescence peak position) which manifest as a dark green color in the EPDs shown in Figure 6. Regions C and D represents more altered conformational states with either a substantial increase in SYPRO orange fluorescence intensity (Region C as a dark red color) or notable changes in Trp fluorescence peak position (Region D as a green color). Finally, Region E represents an aggregated state of the Fc protein with increased signal in optical density at 350 nm (see also the radar chart) as shown as a blue color in Figure 6.
A comparison of the IgG1-Fc proteins with variations in glycosylation occupancy (see the vertical direction in the three color EPD plots on left side of Figure 6) shows different degrees of structural integrity and conformational stability. At low temperatures, the diglycosylated Fc protein at pH 5.5 and 6.0 is present in a native-like structural state (black color, region A). A structural transition to a structurally altered state (dark red, region C) starts as Fc diglycosylated Fc protein reaches temperatures around 48 °C in this pH range. At lower temperatures in the pH range of 4.5 and 5.0, the diglycosylated Fc protein is present in a slightly altered conformation (dark green, region B), where it transitions to a more extensive altered structures at temperatures of 38° and 44 °C, respectively. At pH 4.0 at all temperatures, the diglycosylated protein is present in this more structurally altered structural state. Diglycosylated protein did not show any detectable aggregation by optical density measurements, thus no Region E was observed in the EPD for the fully glycosylated Fc protein.
The nonglycosylated Fc mutant (QQ) at lower temperatures and at pH 5.5 and 6.0 also shows a native like region (black color, Region A). Thermal transition temperatures to the structurally altered region C (dark red color), however, initiate at lower temperatures (35 and 44 °C, respectively) compared to the diglycosylated protein (Figure 6). Additionally, the nonglycosylated mutant (QQ) at pH 4.5 and 5.0 exists at a more extensively structurally altered conformational region, unlike the diglycosylated protein which shows a more native-like conformation in this pH range (Region B, dark green). In addition, Region E (blue color), representing the Fc protein at an aggregated state is detected for the nonglycosylated mutant (QQ) at elevated temperatures in the pH range of 5.0 to 6.0.
In comparison to the di- and non- glycosylated forms, the monoglycosylated Fc protein at low temperatures from pH 4.5 to 6.0, exists in a structurally altered Region D (green color) as shown in Figure 6. Thus, no native like states (Region A, black color) can be detected. The region representing aggregated protein (Region E, blue color) is also seen at high temperature for the monoglycosylated Fc. The monoglycosylated Fc protein was the least stable compared to the diglycosylated and nonglycosylated (QQ) Fc proteins with no stable Region A being detected and with lower temperature thermal transitions noted depending on the pH and biophysical readout.
A comparison of the three different nonglycosylated IgG1-Fc proteins with variations in the amino acid residue in site 297 (also see Figure 6) also shows different degrees of structural integrity and conformational stability. The nonglycosylated DD and nonglycosylated DN Fc proteins, containing two and one Asp 297, respectively, were created by PNGase F enzymatic treatment of the corresponding di- and mono- glycosylated Fc molecules (See Figure 1). The nonglycosylated mutant (QQ) with two Gln residues was also prepared and evaluated as a nonglycosylated Fc protein containing an uncharged amino acid residue at the 297 site. As shown in Figure 6, the nonglycosylated DD IgG1-Fc protein at a pH between 5.0 and 6.0 exists in an altered state (region D, green color). A small region of aggregated protein (region E, blue color) is detected at higher temperature. The aglycosylated DN Fc protein shows an enhanced structural integrity at pH 6.0, compared to the nonglycosylated DD form where a more native like conformation (region B, dark green) is detected. A structurally altered state (region D, green) is observed for the nonglycosylated DN protein at pH 5.0 and 5.5 (similar to that seen for the nonglycosylated DD form). A relatively large region of aggregated protein (region E, blue color) is present at high temperature for the nonglycosylated DN protein, indicating that this form is most prone to aggregation upon heating. Finally, the nonglycosylated mutant (QQ), as already discussed above, defines a region where the proteins exists in a native-like conformation at pH 5.5 and 6.0 that is not seen for the nonglycosylated DD or DN species.
In summary, the presence of a charged amino acid residue at site 297 in the CH2 domain (through the conversion of Asn to Asp as a result of the PNGase F enzymatic treatment) had a measurable influence on the IgG-1 Fc conformational stability. Comparing the nonglycosylated DD to the nonglycosylated mutant (QQ), a stability advantage was detected for the nonglycosylated mutant (QQ) with a stable, native like Region A being detected, along with higher temperature thermal transitions noted, depending on the pH and the method of detection employed. As discussed below, these differences in physical stability profiles may not only be due to charge differences of the amino acid residues at site 297, but also due to differences in the overall folded state and conformational integrity of the different forms.
Discussion
Comparability exercises are routinely performed in the biopharmaceutical industry as a result of manufacturing process changes (e.g., cell culture or purification steps) or alterations in final product presentation (e.g., formulation composition or different packaging material) for protein based drugs under development or currently marketed.36, 37 In these studies, the critical quality attributes of the pre and post-change protein-drug are evaluated in a head-to-head fashion to better understand the effect (if any) of the process or product changes on key comparability elements such as physiochemical integrity, storage stability, functionality, immunogenicity and pharmacokinetics. With regard to physiochemical characterization, numerous advances have been made in terms of characterization of a protein's primary structure and post translational modifications through multiple chromatographic (size exclusion, reverse phase and ion exchange HPLC) and electrophoretic (capillary isoelectric focusing and capillary sodium dodecyl sulfate) methods typically applied with mass spectrometric detection (intact molecular weight, peptide maps, and oligosaccharides maps). Challenges regarding the development of analytical techniques for the structural determination of higher order structures still remain, however, resulting in the requirement for functional potency assays to ensure biological activity as a surrogate marker to demonstrate the protein is properly folded. Although multiple high-resolution analytical techniques have been used for the characterization of the higher order structures of proteins, ranging from nuclear magnetic resonance (NMR)38, X-ray crystallography39, small angle x-ray scattering (SAXS) 4 and hydrogen-deuterium exchange mass spectrometry (HDX-MS),40-43 multiple practical drawbacks such as the requirement of isotope labeling, generation of protein crystals, interference of excipients, and/or the complicated and time consuming nature of these approaches have limited their use in biocomparability studies.
Alternatively, comparing the physical stability profiles of proteins using lower resolution techniques may be a useful complement to these higher sensitivity analytical methods to examine subtle differences in protein structure in formulated drug product dosage forms. The effect of variations introduced into the glycan structure of an IgG1mAb, through enzymatic glycan truncation, protein structural integrity was recently evaluated in our laboratory by this alternative approach using more readily available biophysical instruments (e.g., fluorescence spectroscopy, light scattering and differential scanning calorimetry).24 By using a combination of lower resolution biophysical techniques to study the physical stability of mAb glycoforms over a specific pH and temperature range (at 0.5 pH intervals from 4-6 and 1.25 °C temperature increments from 10-90°C), combined with recently developed data visualization tools, structural and conformational stability differences between the different IgG1glycoforms were demonstrated. In the present study, a comprehensive evaluation of the overall structural integrity and conformational stability of five different IgG1-Fc glycoproteins using multiple biophysical methods across a wide range of pH and temperature conditions is undertaken and comparisons are made between their corresponding global physical stability profiles (three index EPDs and radar charts).
First, initial characterization of the five well-defined glycoforms of IgG1-Fc (Figures 1 and 2) was carried out using a combination of mass spectrometry (MS), sodium dodecyl sulfate polyacrylamide gel (SDS-PAGE), size exclusion chromatography (SEC) and spectral characterization (CD and fluorescence spectroscopy) to evaluate mass (degree of glycosylation), purity and overall structural integrity prior to initiating physical stability studies. The five proteins had measured masses in close agreement to their expected masses due to their glycosylation status (Table 1). The five proteins were shown to have relatively high purity (~91-97%) in primarily monomeric forms as seen from SEC with some relatively low levels of aggregation and fragmentation, under non-denaturing conditions. Purity under denaturing conditions was analyzed by SDS-PAGE under reduced conditions and showed values ranging from ~97-99% for the diglycosylated Fc derived proteins and ~84% for the monoglycosylated Fc derived proteins. The impurities were further analyzed by non-reduced SDS-PAGE and Western Blot analysis (data not shown). One of the minor impurity bands was accounted for by the presence of residual PNGase remaining from the enzymatically treated Fc samples. The migration of the second minor protein band (~39 kDa) found in the monoglycosylated derived proteins was not affected by PNGase enzymatic treatment or by reduction with DTT (and did not react with the anti-human IgG-Fc antibody by western blot analysis). This impurity could not be further removed despite Protein G affinity purifications combined with cation exchange or HIC columns. These differences in the purification behavior between the di- and mono- glycosylated IgG1-Fc proteins, along with identification of the minor band at ~39 kDa, are the subject of ongoing work in our laboratories. Interestingly, similar to slightly lower purity levels (~80-85%) were recently reported for a monoglycosylated IgG1 mAb.22 The lower purity of the monoglycosylated forms of IgG-Fc (this work) and IgG1 mAb22 are likely related to the fact that the monoglycosylated forms are a minor fraction of the total protein (IgG1-Fc and/or IgG1) produced from yeast expression. In this study, the monoglycosylated form is only ~15% of the total IgG1-Fc produced by yeast expression and displayed structural perturbations as well as lower conformational stability; taken together, these attributes resulted in significantly more difficulty in purifying this asymmetrical, monoglycosylated Fc species compared to the diglycosylated IgG1-Fc. It would be of interest in future work to isolate, characterize and compare the soluble aggregates, observed at relatively low levels of 3.4-7.8% by SEC, from both the diglycosylated and monoglycosylated Fc preparations.
The secondary and tertiary structure of the five proteins using circular dichroism (CD) and intrinsic Trp fluorescence spectroscopy was then evaluated (Figures 3 and 4, respectively). The diglycosylated and nonglycosylated mutant (QQ) were both present in a more native-like structure, especially at pH 5-6 (as determined by λmax values and CD minima near 217 nm). Monoglycosylated Fc at all pH conditions tested was structurally disturbed as indicated by the altered CD spectra shape and the red shifted λmax values. Interestingly, upon deglycosylation of the monoglycosylated Fc (creating the nonglycosylated DN form), the Fc protein showed some recovery of native like secondary structure, more similar to the CD spectra of the diglycosylated Fc form with increasing solution pH. This observation further supports the hypothesis that somewhat lower purity level of the monoglycosylated Fc does not affect the interpretability of the measurements of structure.
IgG1-Fc proteins with varying glycosylation site occupancy showed differences in their conformational and colloidal stability with the diglycosylated IgG1-Fc being the most stable protein followed by the nonglycosylated mutant (QQ). Perhaps surprisingly the monoglycosylated IgG1-Fc manifested the lowest relative physical stability (Figures 5 and 6). These observed differences in conformational stability as are consistent with the differences in overall secondary and tertiary structures noted at lower temperatures. To the best of our knowledge, the effect of Fc glycosylation site occupancy on physical stability has only been reported in one limited study by Ha et. al. (2011), which evaluated the thermal stability of an asymmetrically glycosylated (single arm glycosylated) IgG1 using one method (differential scanning calorimetry) at one pH value (pH 6.0).22 The authors reported the CH2 domain of the fully glycosylated IgG1 was thermally more stable than the CH2 domain of the asymmetrically glycosylated IgG1 by ~1 °C, with no detectable change in transition temperatures involving the other domains within the IgG1 protein.
The three index EPDs (Figure 6) and radar charts (supplemental Figure S3) were also able to detect similar structural states among the three IgG1-Fc glycoforms with different conformational stability profiles. Region A (Black color), where the proteins exist in a more native-like conformation as determined by these three biophysical methods, was only detected for the diglycosylated and nonglycosylated QQ at pH 5.5 and 6.0. In addition, a higher transition temperature to an altered conformational form (Region C) was observed for the diglycosylated protein in this pH range. Diglycosylated protein at lower temperatures and pH conditions (4.5 and 5.0) was present in a slightly altered conformational state (Region B), a conformational state that the nonglycosylated QQ protein doesn't seem to access, reflecting the important role glycosylation may play in stabilization of the CH2 domain in acidic environments.8 Due to fluorescence λmax peak position values shifting to higher wavelengths relative to the diglycosylated and nonglycosylated QQ, the monoglycosylated Fc protein at pH 5.5 and 6.0 was determined to be present in an altered conformational state (Region D). The monoglycosylated Fc protein was then deglycosylated, and showed recovery of native like structure under these conditions, further indicating the monoglycosylated form has partially altered higher-order structures, the extent of which depends on the solution pH examined.
In terms of the effect of amino acid substitution and negative charge introduction at site 297 in the CH2 domain, the nonglycosylated QQ, DN and DD forms of the IgG1-Fc were evaluated using the same biophysical techniques. Compared to the nonglycosylated DD and DN forms (with two and one additional Asp residues, respectively), the nonglycosylated mutant (QQ) demonstrated increased conformational stability (Figures 5 and 6). However, since some differences were noted in the soluble aggregate levels between these aglycosylated Fc samples (see Figure 2), future work will need to further evaluate the inter-relationships between aggregate formation and conformational stability. Additional work also is required to further determine if there are any additional differences in the overall charge heterogeneity profiles of the five IgG1-Fc samples; initial attempts at peptide mapping and capillary isoelectric focusing analysis were unsuccessful due to instability during sample preparation (precipitation) and/or lack of reproducibility (data not shown). Structural differences under ambient temperature conditions between the nonglycosylated IgG1-Fc proteins with variations in amino acid substitutions at site 297 (QQ, DN and DD) were detected as well. Nonglycosylated QQ was the only Fc protein that existed at a native-like conformation (region A) at low temperatures at pH 5.5 and 6.0, whereas, nonglycosylated DN and DD both existed in a slightly and more extensively altered forms (Regions B and D) suggesting the important role charge may play in the conformational stability of the CH2 domain of these Fc proteins and potentially in mAbs in general.44-46
In summary, by assessing large conformational stability data sets as a function of environmental stress (pH, temperature), subtle differences in IgG1-Fc glycoform structural integrity were detected. These differences were not readily apparent under non-stressed conditions (i.e., lower temperatures at more neutral pH). Thus, the use of conformational stability data combined with advanced data visualization methods may be an effective surrogate to monitor subtle differences in higher order structure due to post-translational modifications such as glycosylation and amino acid substitution/mutation. During analytical comparability studies of protein drugs within their pharmaceutical dosage forms, it is essential to gather higher order structural information without altering formulation conditions (such as protein concentration, pH, ionic strength, excipients, etc.). Biophysical analysis of conformational stability using lower resolution techniques in a high throughput setup, combined with EPD/radar chart analysis for data visualization, may be a useful complement to higher sensitivity analytical methods such as NMR and hydrogen-deuterium exchange mass spectrometry to examine subtle differences in protein higher order structure in formulated drug product dosage forms.
Supplementary Material
Acknowledgements
The authors wish to acknowledge the NIH for the financial support of T.J.T. via NIH grant NIGMS RO1 GM090080, and S.Z.O via NIH biotechnology training grant 5-T32-GM008359. The authors also acknowledge King Saud University for the financial support of M.A. Alsenaidy. We also wish to acknowledge and thank Dr. Nadezhda Galeva of the KU Mass Spectrometry/Analytical Proteomics Laboratory for her efforts in acquiring the ESI-LC/MS spectra. The Waters Synapt G2 and NanoAcquity were purchased with support from federal grants HRSAC76HF16266 and NIH P20RRl7708.
References
- 1.Aggarwal SR. What's fueling the biotech engine—2011 to 2012. Nature Biotechnology. 2012;30:1191–1197. doi: 10.1038/nbt.2437. [DOI] [PubMed] [Google Scholar]
- 2.Reichert JM. Antibodies to watch in 2013: Mid-year update. mAbs. 2013;5(4):0–1. doi: 10.4161/mabs.24990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnold JN, Wormald MR, Sim RB, Rudd PM, Dwek RA. The Impact of Glycosylation on the Biological Function and Structure of Human Immunoglobulins. Annual Review of Immunology. 2007;25(1):21–50. doi: 10.1146/annurev.immunol.25.022106.141702. [DOI] [PubMed] [Google Scholar]
- 4.Borrok MJ, Jung ST, Kang TH, Monzingo AF, Georgiou G. Revisiting the Role of Glycosylation in the Structure of Human IgG Fc. ACS Chemical Biology. 2012;7(9):1596–1602. doi: 10.1021/cb300130k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li CH, Narhi LO, Wen J, Dimitrova M, Wen Z-Q, Li J, Pollastrini J, Nguyen XC, Tsuruda T, Jiang Y. The Effect of pH, temperature and salt on the stability of E. coli and CHO derived IgG1 Fc. Biochemistry. 2012 doi: 10.1021/bi300702e. [DOI] [PubMed] [Google Scholar]
- 6.Kayser V, Chennamsetty N, Voynov V, Forrer K, Helk B, Trout BL. Glycosylation influences on the aggregation propensity of therapeutic monoclonal antibodies. Biotechnology Journal. 2011;6(1):38–44. doi: 10.1002/biot.201000091. [DOI] [PubMed] [Google Scholar]
- 7.Wang W, Antonsen K, Wang YJ, Wang DQ. pH dependent effect of glycosylation on protein stability. European Journal of Pharmaceutical Sciences. 2008;33(2):120–127. doi: 10.1016/j.ejps.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 8.Latypov RF, Hogan S, Lau H, Gadgil H, Liu D. Elucidation of Acid-induced Unfolding and Aggregation of Human Immunoglobulin IgG1 and IgG2 Fc. Journal of Biological Chemistry. 2012;287(2):1381–1396. doi: 10.1074/jbc.M111.297697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu L, Stadheim A, Hamuro L, Pittman T, Wang W, Zha D, Hochman J, Prueksaritanont T. Pharmacokinetics of IgG1 monoclonal antibodies produced in humanized Pichia pastoris with specific glycoforms: A comparative study with CHO produced materials. Biologicals. 2011;39(4):205–210. doi: 10.1016/j.biologicals.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 10.Raju TS, Scallon BJ. Glycosylation in the Fc domain of IgG increases resistance to proteolytic cleavage by papain. Biochemical and Biophysical Research Communications. 2006;341(3):797–803. doi: 10.1016/j.bbrc.2006.01.030. [DOI] [PubMed] [Google Scholar]
- 11.Raju TS, Briggs JB, Chamow SM, Winkler ME, Jones AJS. Glycoengineering of Therapeutic Glycoproteins: In Vitro Galactosylation and Sialylation of Glycoproteins with Terminal N-Acetylglucosamine and Galactose Residues. Biochemistry. 2001;40(30):8868–8876. doi: 10.1021/bi010475i. [DOI] [PubMed] [Google Scholar]
- 12.Leymarie N, Zaia J. Effective Use of Mass Spectrometry for Glycan and Glycopeptide Structural Analysis. Analytical Chemistry. 2012;84(7):3040–3048. doi: 10.1021/ac3000573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zauner G, Selman MHJ, Bondt A, Rombouts Y, Blank D, Deelder AM, Wuhrer M. Glycoproteomic Analysis of Antibodies. Molecular & Cellular Proteomics. 2013;12(4):856–865. doi: 10.1074/mcp.R112.026005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sinha S, Pipes G, Topp EM, Bondarenko PV, Treuheit MJ, Gadgil HS. Comparison of LC and LC/MS Methods for Quantifying N-Glycosylation in Recombinant IgGs. Journal of the American Society for Mass Spectrometry. 2008;19(11):1643–1654. doi: 10.1016/j.jasms.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 15.Wuhrer M. Glycomics using mass spectrometry. Glycoconjugate Journal. 2013;30(1):11–22. doi: 10.1007/s10719-012-9376-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baković MP, Selman MHJ, Hoffmann M, Rudan I, Campbell H, Deelder AM, Lauc G, Wuhrer M. High-Throughput IgG Fc N-Glycosylation Profiling by Mass Spectrometry of Glycopeptides. Journal of Proteome Research. 2013;12(2):821–831. doi: 10.1021/pr300887z. [DOI] [PubMed] [Google Scholar]
- 17.Visser J, Feuerstein I, Stangler T, Schmiederer T, Fritsch C, Schiestl M. Physicochemical and Functional Comparability Between the Proposed Biosimilar Rituximab GP2013 and Originator Rituximab. BioDrugs. 2013:1–13. doi: 10.1007/s40259-013-0036-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Putnam WS, Prabhu S, Zheng Y, Subramanyam M, Wang Y-MC. Pharmacokinetic, pharmacodynamic and immunogenicity comparability assessment strategies for monoclonal antibodies. Trends in Biotechnology. 2010;28(10):509–516. doi: 10.1016/j.tibtech.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 19.Goetze AM, Liu YD, Zhang Z, Shah B, Lee E, Bondarenko PV, Flynn GC. High-mannose glycans on the Fc region of therapeutic IgG antibodies increase serum clearance in humans. Glycobiology. 2011;21(7):949–959. doi: 10.1093/glycob/cwr027. [DOI] [PubMed] [Google Scholar]
- 20.Lu Y, Westland K, Ma Y.-h., Gadgil H. Evaluation of effects of Fc domain high-mannose glycan on antibody stability. Journal of Pharmaceutical Sciences. 2012;101(11):4107–4117. doi: 10.1002/jps.23284. [DOI] [PubMed] [Google Scholar]
- 21.Labeta MO, Margni RA, Leoni J, Binaghi RA. Structure of asymmetric non-precipitating antibody: Presence of a carbohydrate residue in only one Fab region of the molecule. Immunology. 1986;57:311–317. [PMC free article] [PubMed] [Google Scholar]
- 22.Ha S, Ou Y, Vlasak J, Li Y, Wang S, Vo K, Du Y, Mach A, Fang Y, Zhang N. Isolation and Characterization of IgG1 with Asymmetrical Fc Glycosylation. Glycobiology. 2011;21(8):1087–1096. doi: 10.1093/glycob/cwr047. [DOI] [PubMed] [Google Scholar]
- 23.Alsenaidy MA, Wang T, Kim JH, Joshi SB, Lee J, Blaber M, Volkin DB, Middaugh CR. An empirical phase diagram approach to investigate conformational stability of “second-generation” functional mutants of acidic fibroblast growth factor-1. Protein Science. 2012;21(3):418–432. doi: 10.1002/pro.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alsenaidy MA, Kim JH, Majumdar R, Weis DD, Joshi SB, Tolbert TJ, Middaugh CR, Volkin DB. High-Throughput Biophysical Analysis and Data Visualization of Conformational Stability of an IgG1 Monoclonal Antibody After Deglycosylation. Journal of Pharmaceutical Sciences. 2013;102(11):3942–3956. doi: 10.1002/jps.23730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iyer V, Maddux N, Hu L, Cheng W, Youssef AK, Winter G, Joshi SB, Volkin DB, Middaugh CR. Comparative signature diagrams to evaluate biophysical data for differences in protein structure across various formulations. Journal of Pharmaceutical Sciences. 2013;102(1):43–51. doi: 10.1002/jps.23367. [DOI] [PubMed] [Google Scholar]
- 26.Xiao J, Chen R, Pawlicki MA, Tolbert TJ. Targeting a Homogeneously Glycosylated Antibody Fc To Bind Cancer Cells Using a Synthetic Receptor Ligand. Journal of the American Chemical Society. 2009;131(38):13616–13618. doi: 10.1021/ja9045179. [DOI] [PubMed] [Google Scholar]
- 27.Wang S, Ionescu R, Peekhaus N, Leung J.-y., Ha S, Vlasak J. Separation of post-translational modifications in monoclonal antibodies by exploiting subtle conformational changes under mildly acidic conditions. Journal of Chromatography A. 2010;1217(42):6496–6502. doi: 10.1016/j.chroma.2010.08.044. [DOI] [PubMed] [Google Scholar]
- 28.Loo T, Patchett ML, Norris GE, Lott JS. Using Secretion to Solve a Solubility Problem: High-Yield Expression in Escherichia coli and Purification of the Bacterial Glycoamidase PNGase F. Protein Expression and Purification. 2002;24(1):90–98. doi: 10.1006/prep.2001.1555. [DOI] [PubMed] [Google Scholar]
- 29.Koene HR, Kleijer M, Algra J, Roos D, Kr EG, von dem Borne A, de Haas M. FcγRIIIa-158V/F Polymorphism Influences the Binding of IgG by Natural Killer Cell FcγRIIIa, Independently of the FcγRIIIa-48L/R/H Phenotype. Blood. 1997;90(3):1109–1114. [PubMed] [Google Scholar]
- 30.Rasband WS, ImageJ US. National Institutes of Health; Bethesda, Maryland, USA: pp. 1997–2012. http://imagej.nih.gov/ij/ [Google Scholar]
- 31.Caroline A, Schneider WSRKWE. NIH Image to ImageJ: 25 years of image analysis. Nature Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bond MD, Panek ME, Zhang Z, Wang D, Mehndiratta P, Zhao H, Gunton K, Ni A, Nedved ML, Burman S, Volkin DB. Evaluation of a dual-wavelength size exclusion HPLC method with improved sensitivity to detect protein aggregates and its use to better characterize degradation pathways of an IgG1 monoclonal antibody. Journal of Pharmaceutical Sciences. 2010;99(6):2582–2597. doi: 10.1002/jps.22034. [DOI] [PubMed] [Google Scholar]
- 33.Kim JH, Iyer V, Joshi SB, Volkin DB, Middaugh CR. Improved data visualization techniques for analyzing macromolecule structural changes. Protein Science. 2012;21(10):1540–1553. doi: 10.1002/pro.2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jefferis R. Glycosylation as a strategy to improve antibody-based therapeutics. Nat Rev Drug Discov. 2009;8(3):226–234. doi: 10.1038/nrd2804. [DOI] [PubMed] [Google Scholar]
- 35.Ferrara C, Grau S, Jager C, Sondermann P, Brunker P, Waldhauer I, Hennig M, Ruf A, Rufer AC, Stihle M, Umana P, Benz J. Unique carbohydrate-carbohydrate interactions are required for high affinity binding between FcγRIII and antibodies lacking core fucose. Proc Natl Acad Sci. 2011;108(31):12669–12674. doi: 10.1073/pnas.1108455108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Federici M, Lubiniecki A, Manikwar P, Volkin DB. Analytical lessons learned from selected therapeutic protein drug comparability studies. Biologicals. 2013;41(3):131–147. doi: 10.1016/j.biologicals.2012.10.001. [DOI] [PubMed] [Google Scholar]
- 37.Lubiniecki A, Volkin DB, Federici M, Bond MD, Nedved ML, Hendricks L, Mehndiratta P, Bruner M, Burman S, DalMonte P, Kline J, Ni A, Panek ME, Pikounis B, Powers G, Vafa O, Siegel R. Comparability assessments of process and product changes made during development of two different monoclonal antibodies. Biologicals. 2011;39(1):9–22. doi: 10.1016/j.biologicals.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 38.Amezcua CA, Szabo CM. Assessment of higher order structure comparability in therapeutic proteins using nuclear magnetic resonance spectroscopy. Journal of Pharmaceutical Sciences. 2013;102(6):1724–1733. doi: 10.1002/jps.23531. [DOI] [PubMed] [Google Scholar]
- 39.Berkowitz SA, Engen JR, Mazzeo JR, Jones GB. Analytical tools for characterizing biopharmaceuticals and the implications for biosimilars. Nat Rev Drug Discov. 2012;11(7):527–540. doi: 10.1038/nrd3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Houde D, Arndt J, Domeier W, Berkowitz S, Engen JR. Characterization of IgG1 Conformation and Conformational Dynamics by Hydrogen/Deuterium Exchange Mass Spectrometry. Analytical Chemistry. 2009;81(7):2644–2651. doi: 10.1021/ac802575y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Houde D, Peng Y, Berkowitz SA, Engen JR. Post-translational Modifications Differentially Affect IgG1 Conformation and Receptor Binding. Molecular & Cellular Proteomics. 2010;9(8):1716–1728. doi: 10.1074/mcp.M900540-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Manikwar P, Majumdar R, Hickey JM, Thakkar SV, Samra HS, Sathish HA, Bishop SM, Middaugh CR, Weis DD, Volkin DB. Correlating excipient effects on conformational and storage stability of an IgG1 monoclonal antibody with local dynamics as measured by hydrogen/deuterium-exchange mass spectrometry. Journal of Pharmaceutical Sciences. 2013;102(7):2136–2151. doi: 10.1002/jps.23543. [DOI] [PubMed] [Google Scholar]
- 43.Majumdar R, Manikwar P, Hickey JM, Samra HS, Sathish HA, Bishop SM, Middaugh CR, Volkin DB, Weis DD. Effects of Salts from the Hofmeister Series on the Conformational Stability, Aggregation Propensity, and Local Flexibility of an IgG1 Monoclonal Antibody. Biochemistry. 2013;52(19):3376–3389. doi: 10.1021/bi400232p. [DOI] [PubMed] [Google Scholar]
- 44.Khawli LA, Goswami S, Hutchinson R, Kwong ZW, Yang J, Wang X, Yao Z, Sreedhara A, Cano T, Tesar DB, Nijem I, Allison DE, Wong PY, Kao Y-H, Quan C, Joshi A, Harris RJ, Motchnik P. Charge variants in IgG1: Isolation, characterization, in vitro binding properties and pharmacokinetics in rats. mAbs. 2010;2(6):613–624. doi: 10.4161/mabs.2.6.13333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vlasak J, Bussat MC, Wang S, Wagner-Rousset E, Schaefer M, Klinguer-Hamour C, Kirchmeier M, Corvaïa N, Ionescu R, Beck A. Identification and characterization of asparagine deamidation in the light chain CDR1 of a humanized IgG1 antibody. Analytical Biochemistry. 2009;392(2):145–154. doi: 10.1016/j.ab.2009.05.043. [DOI] [PubMed] [Google Scholar]
- 46.Yadav S, Laue TM, Kalonia DS, Singh SN, Shire SJ. The Influence of Charge Distribution on Self-Association and Viscosity Behavior of Monoclonal Antibody Solutions. Molecular Pharmaceutics. 2012;9(4):791–802. doi: 10.1021/mp200566k. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.