Summary
The Otx2 homeodomain transcription factor is essential for gastrulation and early neural development. We generated Otx2 conditional knockout (cKO) mice to investigate its roles in telencephalon development after neurulation (~E9.0). We conducted transcriptional profiling and in situ hybridization to identify genes de-regulated in Otx2 cKO ventral forebrain. In parallel, we used ChIP-seq to identify enhancer elements, the OTX2 binding motif, and de-regulated genes that are likely direct targets of OTX2 transcriptional regulation. We found that Otx2 was essential in: septum specification, regulation of Fgf signaling in the rostral telencephalon, and medial ganglionic eminence (MGE) patterning, neurogenesis, and oligodendrogenesis. Within the MGE, Otx2 was required for ventral but not dorsal identity, thus controlling the production of specific MGE derivatives.
Keywords: Otx2, Fgf, forebrain, telencephalon, patterning, MGE, POA, cholinergic, globus pallidus, microarray, ChIP-seq, enhancer, OTX2 motif
Graphical abstract
INTRODUCTION
Otx2 is one of two mammalian orthologs of the Drosophila homeodomain transcription factor (TF), Orthodenticle. Otx2 is essential in the visceral endoderm during gastrulation for specification of anterior neuroectoderm, where it induces early forebrain-specific genes (Acampora et al., 1995; Rhinn et al., 1998; Tian et al., 2002). Telencephalic expression of Otx2 continues in the rostral patterning center (RPC; septal anlage), ventral telencephalon (subpallium; ganglionic eminences), and caudodorsal telencephalon (including choroid plexus; Figure 1; (Simeone et al., 1992)). Abrogation of Otx2 function after early neurulation through enhancer deletion showed that Otx2 maintains forebrain identity after its specification, and is required later in the caudodorsal telencephalon for medial pallial morphogenesis (Kurokawa et al., 2004a; Kurokawa et al., 2004b; Sakurai et al., 2010).
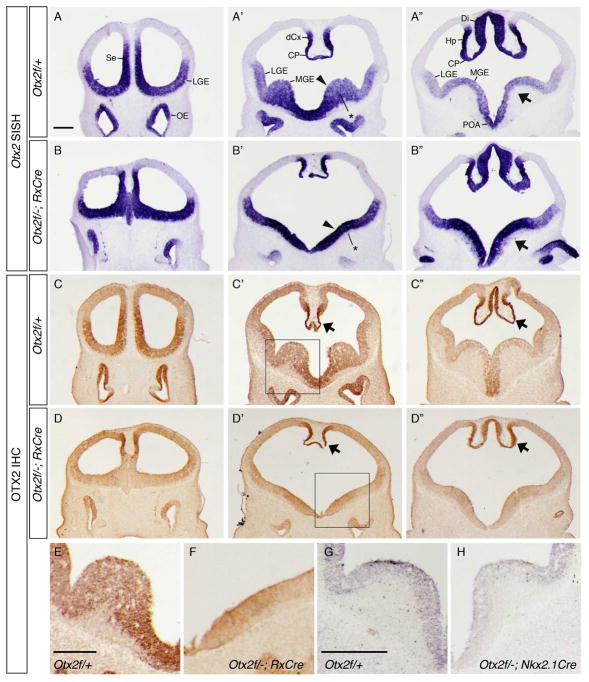
Figure 1. Otx2 expression in conditional knock outs (cKOs).
(A-B”) ISH on E11.5 coronal sections from (A–A”) Otx2f/+ and (B–B”) Otx2f/−; RxCre embryos using a full length Otx2 riboprobe. Otx2 transcription appears upregulated in the MGE of RxCre cKOs (arrowheads and arrows in A’, A”, B’, B”), and that the MGE SVZ and MZ are hypoplastic (asterisks in A’, B’). (C–H) Anti-OTX2 IHC: in RxCre cKOs (C–F), OTX2 protein expression is absent in cKO forebrains except in the dorsomedial caudal cortex, hippocampal anlage, and choroid plexus (arrows, C’-C”, D’–D”). E and F show higher magnification views of the boxed regions in C’, D’. (G–H) In Nkx2.1Cre cKOs, OTX2 expression was reduced in the MGE. A–D”: rostrocaudal series of coronal sections. Abbreviations: Se, septum; MGE, medial ganglionic eminence; LGE, lateral ganglionic eminence; dCx, dorsomedial cortex; Hp, hippocampal anlage; POA, preoptic area; Di, diencephalon; CP, choroid plexus; OE, olfactory epithelium. Scale bars: A, E = 0.25mm, G = 0.4mm.
Otx2 roles in the RPC and ventral telencephalon have not been reported. The RPC is a source of Fgfs (Fgf8, 17, 18) with essential roles in forebrain growth and patterning (Martinez et al., 1999; Crossley et al., 2001; Fukuchi-Shimogori and Grove, 2001; Chi et al., 2003; Storm et al., 2006; Cholfin and Rubenstein, 2007; Grove and Monucki, 2013). Notably, Otx2 expression also abuts the midbrain/hindbrain boundary (MHB) patterning center, which secretes the same array of Fgfs to help instruct development of midbrain/hindbrain structures (Simeone et al., 1992; Martinez et al., 1999; Chi et al., 2003).
Otx2 may be involved in regulating and responding to FGFs from the RPC and MHB. FGF8 bead implantation in prosencephalon, mesencephalon, and optic vesicle tissue indicated that FGF8 represses Otx2 expression (Martinez et al., 1999; Crossley et al., 2001). Conversely, there is evidence for Otx2 regulation of Fgf8 expression: reduced Otx2 expression results in anterior expansion of Fgf8 expression in the MHB (Acampora et al., 1997; Puelles et al., 2003).
Otx2 is expressed in the septal primordium (Se), the ganglionic eminences (GEs: MGE, LGE, CGE), and the preoptic area (POA; Figure 1). The MGE gives rise to the globus pallidus (GP) and to striatal and cortical interneurons (Flandin et al., 2010; Rubenstein and Campbell, 2013). The LGE gives rise to striatal neurons and olfactory bulb interneurons (Rubenstein and Campbell, 2013). The CGE gives rise to cortical interneurons (Batista-Britto and Fishell, 2013). The POA gives rise to preoptic nuclei and some of the amygdala and cortical interneurons (Hirata et al., 2009; Gelman et al., 2011). Fate mapping demonstrated that Fgf8+ ventral MGE and ventral Se progenitors generate cholinergic neurons of the basal ganglia (Hoch et al., 2015,). In addition to their neuronal derivatives, the embryonic MGE, LGE, and POA also generate oligodendrocytes that populate the basal ganglia and cortex (Kessaris et al., 2006).
The GEs are subdivided into molecularly and functionally distinct progenitor subdomains along the dorsoventral (D-V) axis that give rise to different subclasses of neurons (Flames et al., 2007; Flandin et al., 2010; Waclaw et al., 2010). Factors that establish these progenitor subdomains are unknown.
We used conditional mutagenesis to elucidate roles of Otx2 in the ventral forebrain and septum. These analyses revealed novel telencephalic roles of Otx2. In the RPC, Otx2 restricted Fgf8 and 17 expression along the rostral-caudal and D-V axes, and controlled Fgf-signaling through feedback regulation of Sprouty expression in the RPC, MGE and POA. In the GEs, Otx2 promoted MGE neurogenesis and oligodendrogenesis, and controlled MGE sub-regional identity. Based on mRNA expression changes and OTX2 ChIP-seq data, we propose molecular mechanisms how Otx2 patterns MGE regional subdivisions.
RESULTS
Conditional mutagenesis of Otx2 in the telencephalon
In the E11.5 telencephalon, Otx2 is expressed in the ventricular zone (VZ) of the Se, GEs, POA, choroid plexus, and hippocampal anlage (Figure 1A–A”). To investigate Otx2 functions in the telencephalon, we conducted conditional mutagenesis experiments using Otx2f mice (Tian et al., 2002). We used two Cre deleter alleles to abrogate Otx2 expression after gastrulation: RxCre, in which Cre is expressed in the telencephalic neural plate anlage (~E8.5; (Swindell et al., 2006)), and Nkx2.1Cre, in which Cre is expressed in the MGE, POA, and ventral Se (vSe) beginning around E9.5 (Xu et al., 2008).
We used immunohistochemistry (IHC) and in situ hybridization (ISH) to evaluate levels of protein and mRNA in Otx2 cKOs. Anti-OTX2 IHC showed that OTX2 protein expression was lost throughout the telencephalon in RxCre cKOs by E11.5, except in caudal dorsomedial structures (Figure 1C–F; arrows in C’-C” and D’-D” indicate maintained protein expression). Anti-OTX2 IHC confirmed that Nkx2.1Cre cKOs lacked OTX2 expression in the E11.5 MGE (Figure 1G–H).
ISH using a full-length Otx2 probe detected increased levels of the mutant transcript in RxCre cKOs, suggesting that conditional deletion of Otx2 leads to increased state-steady levels of Otx2 transcripts (Figure 1A–A”, B–B”), particularly in the MGE VZ and in the caudal MGE subventricular zone (SVZ; arrowheads, Figure A’, B’, and arrows, Figure 1A”, B”).
We performed OTX2 ChIP-seq three times from E12.5 wild type subpallium (described below in more detail). A ChIP-seq peak is presumptive evidence for OTX2 in vivo binding and possible function at this locus. We observed multiple ChIP-seq peaks near the Otx2 locus (Figure 2K). These data suggest that Otx2 negatively autoregulates its expression.
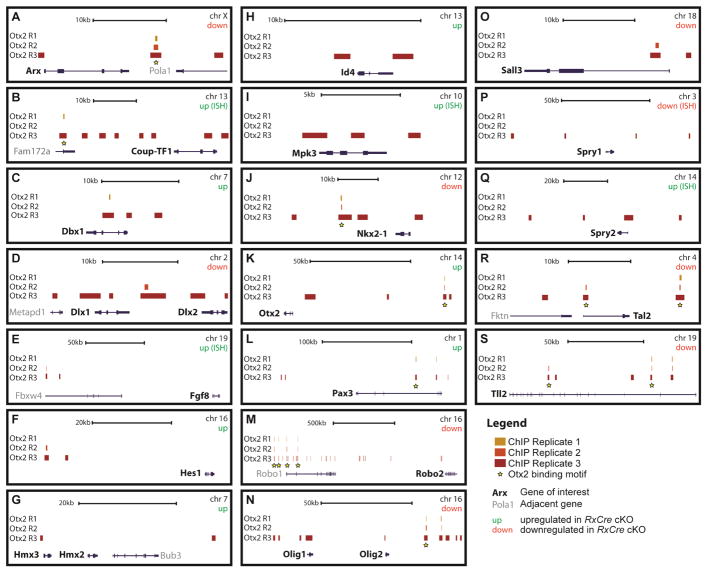
Figure 2. Anti-OTX2 ChIP-seq results.
“Called” peak locations relative to genomic loci are shown for genes (alphabetically organized) that are deregulated in Otx2 cKOs, and for which ChIP-seq peaks were identified within ~1 megabase (MB) of the gene body. Note the different scale bars for individual panels. Black arrows and text identify the Otx2-regulated gene of interest, grey arrows and text designate nearby genes. For each panel, italicized “up” or “down” indicates whether the gene was upregulated or downregulated in RxCre cKO forebrains. The yellow stars indicate that the OTX2-ChIP-seq peak had an OTX2 binding motif. Abbreviations: chr: chromosome; kb: kilobase
Otx1 is also expressed in the developing forebrain (Simeone et al., 1992). Otx1 and Otx2 had complementary expression patterns in the E11.5 telencephalon: Otx2 is expressed strongly in the subpallial VZ, whereas Otx1 is expressed predominantly in the pallial VZ and dorsal LGE, but is expressed at lower levels in the GEs (Figures 1A–A” and Figure S1A–C). Notably, Otx1 and Otx2 are expressed in the SE, LGE, caudal MGE, and dorsomedial cortical structures (Figures 1A–A”, asterisks in Figure S1A–C). Otx1 mRNA expression was not demonstrably altered in Otx2f/−; RxCre embryos at E11.5, (Figure S1B).
RxCre E11.5 cKOs had hypoplastic MGEs, with reductions in the SVZ and marginal zone (MZ; asterisks, Figure 1A’, B’). We conducted an RNA expression microarray experiment using RNA from control and RxCre cKO Se, MGE, and LGE (GEO accession number: GSE69727). This analysis identified 139 significantly deregulated genes (including those discussed in the results: Table S1). In parallel, we used microdissected subpallium (septum, MGE, and LGE) from wild type E12.5 embryos in three independent anti-OTX2 ChIP-seq experiments (Figure 2, data not shown)(GEO accession number: GSE69727). By comparing microarray and ChIP-seq datasets, we developed hypotheses as to which genes are direct targets of OTX2 that mediate its functions in the RPC and GEs.
Otx2 impacts Fgf signaling in the RPC and is required for septum specification
RxCre cKO telencephalons also are hypoplastic along the rostrocaudal axis at E11.5: the rostral pole>caudal septum distance was 528 +/− 32 μm in Otx2f/+ embryos (n=4) and 294 +/− 97 μm in Otx2f/−; RxCre embryos (n=5). Fgf8 and Fgf17 mRNA expression domains expanded rostrally and ventrolaterally into the MGE at E11.5 (arrowheads Figure 3A–E). Thus, Otx2 restricts RPC Fgf signaling. Fgf8 hypomorphs had reduced Otx2 expression in the rostral telencephalon (Figure S2A–B, (Storm et al., 2006)). Thus, Otx2 and Fgf8 function and expression are tightly coordinated.
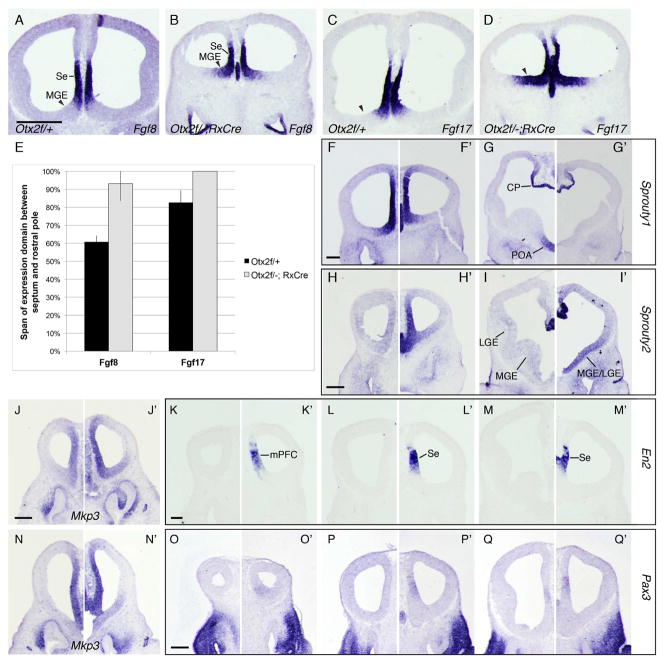
Figure 3. Otx2 restricts the domain of Fgf expression and controls regional specification of the RPC.
(A–D, F–Q’) ISH comparing gene expression in (A, C, F–Q) Otx2f/+ and (B, D, F’–Q’) Otx2f/-; RxCre embryos at E11.5. (A–B) Fgf8, (C–D) Fgf17, (F–G’) Sprouty1, (H–I’) Sprouty2, (J–J’, N-N’) Mkp3, (K–M’) En2, (O–Q’) Pax3. (E) Quantification of the rostral expansion of Fgf expression in cKO and control embryos at E11.5 (mean +/− st. dev.). For each embryo, we calculated the distance from the caudal septum (where Fgf8 and Fgf17 are expressed) to the rostral limit of Fgf expression, and expressed this as a percentage of the total (rostral telencephalic pole > caudal septum) distance. Rostral expansion was statistically significant for Fgf8 (p < 0.001) but not Fgf17 (p = 0.17; 2-tailed t tests, unequal variance). Abbreviation: mPFC, medial prefrontal cortex. Scale bars: A–D = 0.5mm, F–G’ = 0.2mm, H–Q’ = 0.25mm.
FGF signaling induces the expression of negative feedback inhibitors, including Sprouty1, Sprouty2, and MAP Kinase Phosphatase 3 (Mkp3, also called Dusp6; (Minowada et al., 1999; Eswarakumar et al., 2005; Thisse and Thisse, 2005; Faedo et al., 2010). These genes were deregulated in RxCre cKOs. Sprouty1 expression was diminished in the POA (Figure 3F–G’). Sprouty2 was upregulated in the RPC, MGE, and LGE (Figure 3H–I’), and Mkp3 was upregulated in the MGE (Figure 3J–J’, N-N’). Anti-OTX2 ChIP-seq data provided evidence that Fgf8, Sprouty1, Sprouty2, and Mkp3 were direct targets of OTX2 (Figure 2E, P, Q). We did not observe reproducible ChIP-seq peaks for Fgf17. Thus, Otx2 regulates FGF signaling in the Se and GEs in several ways.
Fgf8 and Fgf17 establish gradients of gene expression that pattern the cortical primordium (Fukuchi-Shimogori and Grove, 2001; Garel et al., 2003; Cholfin and Rubenstein, 2007). RxCre cKOs at E13.5 had altered dorsoventral gradients of COUP-TF1 and Sp8. COUP-TF1 expression was downregulated in the dorsal cortex (Figure S2C–D). Conversely, SP8 was upregulated in the dorsal and medial cortex and its graded expression extended further ventrally (Figure S2E–F). These changes are predictable consequences of increased Fgf8 signaling (Garel et al., 2003; Storm et al., 2006).
The RNA expression microarray identified two midbrain/hindbrain genes, En2 and Pax3, which were upregulated in RxCre cKO telencephalons (Figure 3K–M’, O-Q’; see also Allen Brain Atlas). Normally, En2 is highly expressed around the MHB (Liu and Joyner, 2001). Pax3 is expressed in the midbrain and hindbrain at E11.5 ((Goulding et al., 1991); Allen Brain Atlas). Both En2 and Pax3 were ectopically expressed in the RPC/Se and medial PFC (Figure 3K–M’, O–Q’). Notably, we detected low levels of Pax3 in a small subdomain of the caudal RPC in control forebrains (Figure 3Q). We did not detect ChIP-seq peaks near the En2 locus, but Pax3 had two strong intragenic peaks (Figure 2L), suggesting that this is a direct OTX2 target. Ectopic/elevated expression of En2 and Pax3 suggests that Se progenitors are mis-specified.
Otx2 is required for early oligodendrogenesis in the basal ganglia
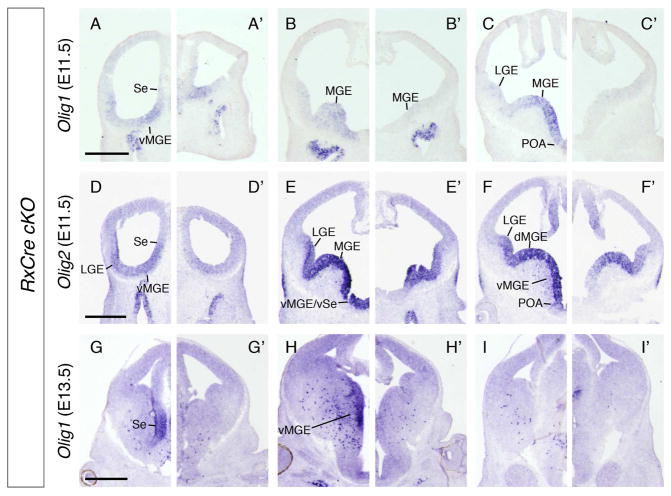
The bHLH TFs Olig1 and Olig2 were downregulated in the E12.5 RNA microarray experiment (Table S1). ISH at E11.5 confirmed that Olig1 was reduced throughout the caudal MGE and POA VZ, and Olig2 was selectively downregulated in the ventral subdomains of these structures (Figure 4B–C’, E–F’). At E13.5, RxCre cKOs had reduced Olig1 expression in the Se and MGE VZ, and fewer scattered Olig1+ immature oligodendrocytes in the MZ (Figure 4G–I’). E12.5 ChIP-seq data revealed numerous OTX2 binding sites in the vicinity of the Olig1/Olig2 locus (Figure 2N), suggesting these two genes are direct targets of OTX2 regulation.
Figure 4. Otx2 cKOs exhibit deficits in molecular markers of oligodendrogenesis.
ISH on coronal hemisections from (A–I) Otx2f/+, (A’–I’) Otx2f/−; RxCre, embryos: (A–C’) Olig1 at E11.5, (D–F’) Olig2 at E11.5, (G–I’) Olig1 at E13.5. Three planes of section are shown along the rostral-caudal axis. Abbreviations: vMGE, ventral MGE; dMGE, dorsal MGE; vSe, ventral septum. Scale bars: A–F’ = 0.5mm, G–I’ = 0.4mm.
Olig1 expression in Nkx2.1Cre cKOs was slightly reduced in the vMGE and POA at E11.5 (not shown). At E13.5, Olig1 expression in the Se VZ appeared normal, but was strongly reduced in the caudal MGE VZ, and there were fewer scattered Olig1+ cells in the MZ (not shown). Thus, Otx2 is required in the early MGE VZ for oligodendrogenesis.
Otx2 is required in the MGE for progenitor specification and neurogenesis
The GEs of RxCre cKOs were hypoplastic at E11.5 (Figure 4B–B’, Figure 5). We examined E11.5 expression of Dlx1, a homeobox gene expressed mosaically in the MGE and LGE VZ, homogeneously in their SVZs, and in differentiating neurons.. Dlx1 expression was reduced in the VZ, SVZ and MZ (Figure 5A’, B’). OTX2 binds to two enhancers in the Dlx1/2 locus (I12b and URE2) that are active in the embryonic subpallium (Figure 2D; S5G–J)(Ghanem et al., 2007). This phenotype was also observed in Nkx2.1Cre cKOs (Figure 5B, B’), suggesting that Otx2 participates in MGE neurogenesis.
Figure 5. Reduced neurogenesis and proliferation in the basal ganglia of E11.5 Otx2 cKOs.
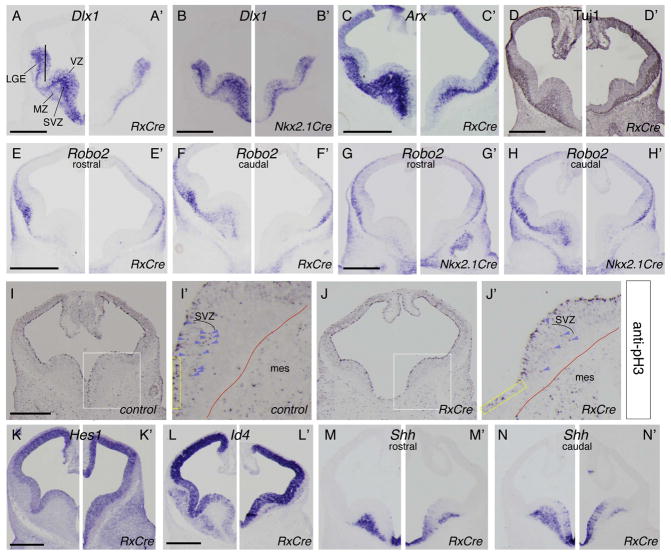
ISH and IHC on coronal hemisections (control: left, mutant: right). (A–H’) ISH: (A–H) Otx2f/+, (A’, C’, D’–F’) Otx2f/−; RxCre, and (B’, G’–H’) Otx2f/−; Nkx2.1Cre embryos using probes to (A–B’) Dlx1, (C–C’) Arx, (E–H’) Robo2. Anti-Tuj1 IHC (D) Otx2f/+ and (D’) Otx2f/−; RxCre. (I–J’) Anti- pH3 IHC: (I’, I) Otx2f/+ and (J, J’) Otx2f/−; RxCre embryos. I’–J’: high magnification of I, J. Red lines: neural/mesenchymal boundary; purple arrowheads: pH3+ SVZ cells; yellow rectangles highlight similar VZ regions of the vMGE in K–L’ showing upregulation of Hes1 and Id4, respectively. (K, L) Otx2f/+ and (K’, L’) Otx2f/-; RxCre embryos. (M–N’) Shh reduction in MZ and increase in VZ. Abbreviations: mes: mesenchyme; SVZ: subventricular zone; VZ ventricular zone. Scale bars: 0.5mm.
In support of this model, several markers of differentiating MGE neurons were downregulated (Table S1). For example Arx RNA was reduced ~1.7-fold in the microarray. OTX2 ChIP-seq peaks were identified near Arx (Figure 2A), including at two enhancers with subpallial activity (Figure S5A–D)(Ahituv et al., 2007; Colasante et al., 2008; Visel et al., 2013). At E11.5, Arx was expressed in MGE VZ, SVZ and MZ (Figure 5C), but was reduced in the RxCre cKOs (Figure 5C’). Similarly, Shh (MGE MZ), PlxnA4 (MGE and LGE SVZ/MZ) and Gbx2 (MGE MZ) expression was reduced (Figure 5M–N’, and data not shown). All three OTX2 ChIP-seq experiments detected OTX2 peaks on a Shh enhancer that promotes MGE expression (SBE4) (Jeong et al., 2006).
Markers of immature MGE-derived interneurons, including Lhx6, c-maf, Somatostatin, NPY, and Gad1, were also downregulated (Table S1). Furthermore, neurogenesis in the E11.5 MGE was reduced, as indicated by IHC to the pan-neuronal marker, β-III-tubulin (Tuj1 antibody)(Figure 5D–D’).
Unlike Arx, β-III-tubulin and Dlx1, which were moderately reduced in the cKOs, expression of Robo2 was barely detectable (Figure 5E–F’). Nkx2.1Cre cKOs exhibited a similar phenotype (Figure 5G–H’). Several ChIP-seq peaks were identified ~1MB from the Robo2 locus, within the Robo1 locus (Figure 2M).
We next examined proliferation in the E11.5 MGE of the RxCre cKO using IHC to phosphohistone H3 (pH3). pH3+ cells were strongly reduced in the SVZ (Figure 5I–J arrowheads), while the VZ did not show a clear phenotype. In considering the mutant’s neurogenesis deficit, and the reduction of mitotic SVZ (pH3+) cells, we were intrigued that anti-neurogenic factors Hes1 and Id4 were both upregulated in the E12.5 RxCre microarray (Table S1). Hes1 and Id4 are expressed in dorsal>ventral gradients in the MGE VZ at E11.5; these gradients were lost, as these genes were upregulated throughout the MGE (Figure 5K–L’). ChIP-seq data indicated that Otx2 binds genomic DNA near the Hes1 locus and may also bind near the Id4 locus (Figure 2F, H).
Together, these data support a model in which Otx2 directly regulates genes that control the generation and differentiation of MGE-derived neurons. Otx2 represses inhibitors of MGE differentiation (Hes1 and Id4), and thus, loss of Otx2 function results in reduced production of SVZ progenitors and neurons. Furthermore, Otx2 promotes neuronal maturation by positively regulating Arx and Dlx1 expression, genes that support the differentiation of MGE-derived neurons.
Otx2 specifies vMGE fate and repress POA fate
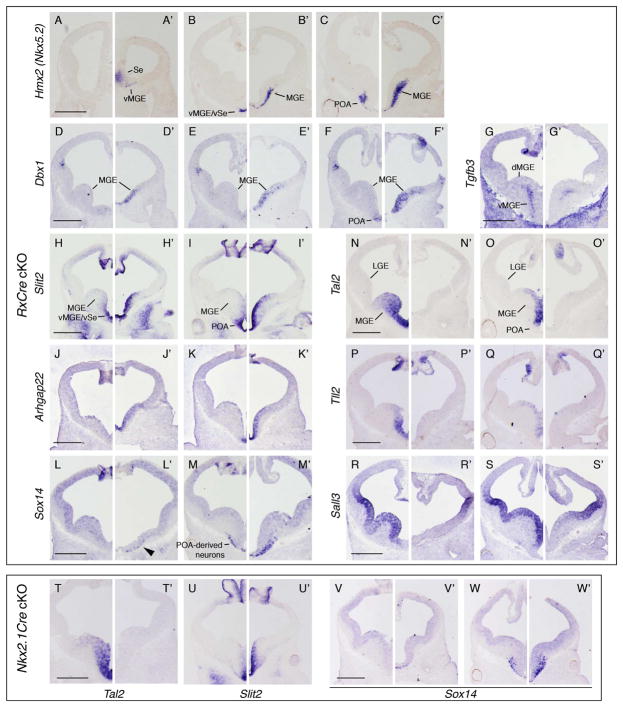
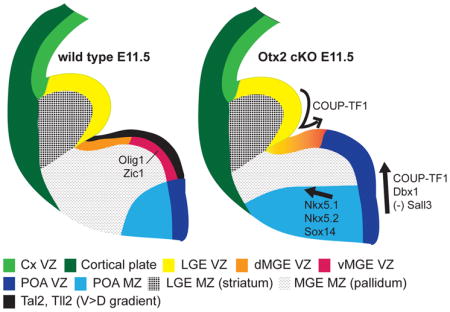
Olig2, Fgf, and Sprouty1 expression changes in RxCre cKOs indicated that the vMGE and POA were particularly affected by the loss of Otx2. Thus, we hypothesized that Otx2 may play a role in regional specification of the basal ganglia. To investigate this, we examined microarray data (Table S1) for expression changes relevant to MGE and POA patterning, and performed ISHs at E11.5. Multiple genes that had restricted expression in the POA (progenitors and/or neurons) were upregulated (Table S1); ISH revealed that their expression expanded rostrally and/or dorsally into the ventral MGE (vMGE). These “POA genes” included Nkx5.2, Dbx1, Slit2, Arhgap22, Sox3, Sox14, and mShisa (Figure 6A–C, D–F, H–M, and data not shown). Conversely, several MGE markers were identified as downregulated on the microarray (Table S1). ISHs validated these results and demonstrated that the RxCre cKO MGE VZ failed to express Tal2 and Tll2, which are novel markers of the vMGE (Figure 6N–Q). Furthermore, Sall3 was downregulated within the vMGE VZ (Figure 6R–S), and Tgfb3 was downregulated in the vMGE SVZ (Figure 6G). COUP-TF2, which is strongly expressed in the CGE and dMGE of wild type E11.5 embryos, was overexpressed in the vMGE VZ and SVZ of Otx2 cKOs (data not shown); this could reflect a rostral and/or ventral shift in this gene’s expression domain. Together, these findings (summarized in Figure S7) provide evidence that Otx2 patterns MGE regional identity by specifying vMGE properties and by repressing POA identity.
Figure 6. Re-specification of the vMGE towards the POA fate in Otx2 RxCre cKOs.
ISH on E11.5 coronal hemisections from (A–S) Otx2f/+, (A’–O’) Otx2f/−; RxCre, and (P’S’) Otx2f/−; Nkx2.1Cre embryos showing expanded expression of POA genes (A–M’, U–W’) and diminished expression of vMGE genes (N–S’, T–T’) in cKOs. For each experiment (except Tal2 and Slit2 in Nkx2.1Cre cKOs), two or three sections are shown to demonstrate effects at different rostral-caudal planes. (A–C’) Hmx2 (Nkx5.2), (D–F’) Dbx1, (G–G’) Tgfb3, (H–I’, U–U’) Slit2, (J–K’) Arhgap22, (L–M’, V–W’) Sox14, (N–O’, T–T’) Tal2, (P–Q’) Tll2, (R–S’) Sall3. Scale bars: 0.5mm.
Importantly, ChIP-seq data revealed OTX2 binding peaks in or near several downregulated MGE genes (Tal2, Tll2, Sall3)(Figures 2O,R,S; S5K,L). In contrast, most upregulated POA genes, including Nkx5.2, Slit2, Arhgap22, and Sox3, did not have nearby OTX2 binding peaks (Figure 2 and data not shown). One notable exception was Dbx1, a POA marker that had OTX2 ChIP-seq peaks (Figure 2C). Furthermore, OTX2 occupied enhancer elements, with subpallial activity at E11.5, near genes with reduced expression in the Otx2 RxCre cKOs (Arx, Dlx1&2 and Sall3; Figure S5). These data support a model in which Otx2 patterns the basal ganglia by direct, positive transcriptional regulation of MGE genes, and by repressive effects on POA gene expression that are predominantly indirect (Figure S7).
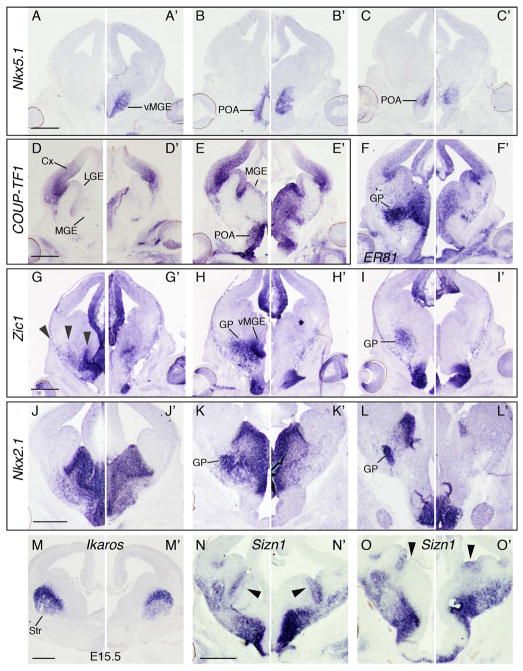
At E13.5, RxCre cKOs phenotypes continue to demonstrate expansion of POA identity into the MGE. Nkx5.1 expression, which is normally restricted to a POA SVZ subdomain and a subset of POA MZ cells, expanded dorsally and rostrally (Figure 7A–C and data not shown). COUP-TF1 is normally expressed in the LGE VZ, the POA VZ and MZ, and in a dorsal>ventral gradient in the caudal MGE VZ (Figure 7D,E). In cKOs COUP-TF1 had increased expression MGE MZ and was ectopically expressed in the caudal MGE MZ (Figure 7D’, E’). ChIP-seq data suggest that COUP-TF1 may be directly regulated by OTX2 (Figure 2). Zic1 is a marker of the vMGE VZ, the GP, and a subset of other MGE MZ cells at E13.5 (Figure 7G–I). In cKOs Zic1 was not detectable in the vMGE VZ or GP, and labeled fewer MGE MZ cells (Figure 7G’–I’). ChIP-seq data revealed multiple peaks near the Zic1 locus (data not shown) suggesting direct regulation by OTX2. GP hypoplasia in RxCre cKOs was confirmed by Nkx2.1 and ER81 ISH at E13.5, and with ER81 and NPAS1 ISH at P0 (Figure 7F, K, data not shown). Nkx2.1Cre cKOs at E13.5 exhibited similar, though less severe POA and MGE phenotypes (Figure S3).
Figure 7. E13.5-E15.5 MGE and POA development in Otx2 cKOs. ISH on (A–O).
Otx2f/+ and (A’–O’) Otx2f/−; RxCre coronal hemisections, using the following probes: (AC’) Nkx5.1, (D–E’) COUP-TF1, (F–F’) ER81, (G–I’) Zic1, (J–L’) Nkx2.1, (M–M’) Ikaros, (NO’) Sizn1. Arrowheads in G indicate three streams of neurons or progenitors that appear to emanate from the POA or vMGE and migrate toward the MGE, LGE, and ventral cortex; these streams are not apparent in cKOs. Arrowheads in N–O’ point to the dMGE: Sizn1 expression appears to shift ventrally from its vLGE domain into the dMGE in cKOs. All are E13.5 except for Ikaros, which is E15.5. Note that in L’, the dark region in the CGE (*) is a tissue fold, not ISH signal. Abbreviations: GP, globus pallidus; Str, striatum. Scale bars: 0.5mm.
While the ventral MGE had the clearest patterning defects in RxCre cKOs, we also observed subtle deficits in the dorsal MGE. At E13.5, transcriptional co-activator Sizn1 (MGI nomenclature: Zcchc12) is expressed in the VZ of the ventral LGE (Figure 7N, O). In cKOs, Sizn1 expression extended ventrally into dMGE (Figure 7N’, O’); we did not observe OTX2 ChIP-seq peaks near Sizn1, suggesting this was an indirect effect of Otx2 functions.
Although Otx2 is expressed in the VZ of the LGE and MGE, the LGE had only a mild phenotype in RxCre cKOs at E11.5, E13.5 and P0. For example, Ikaros, which labels neurons in the MZ of the LGE (striatum), was expressed in the normal domain, albeit at lower levels, at E15.5 (Figure 7M).
Otx2 regulates development of interneurons and cholinergic neurons derived from the MGE and POA
The MGE and POA give rise to cortical and striatal interneurons and basal ganglia cholinergic neurons, as such RxCre cKOs affected these neurons. Lhx6 and Lhx8 expression were reduced in these regions (Figure S4A’–C’). Lhx8+ striatal interneurons were reduced (Figure S4D–F’). Lhx6 and c-maf ISHs suggested that RxCre cKOs may have reduced numbers of cortical interneurons at P0 (Figure 8A–C’, data not shown). However, at P13-15 we did not observe significant changes in somatostatin and parvalbumin IHC-positive cortical interneuron numbers (data not shown).
~80% of cholinergic neurons in the basal ganglia originate in the vMGE and septum from RPC-derived progenitors (Hoch et al., 2015). As this domain was severely affected in RxCre cKOs, we examined cholinergic neurons numbers. Gbx1 and TrkA are expressed in basal ganglia cholinergic neurons at P0 (Asbreuk et al., 2002; Sanchez-Ortiz et al., 2012); both markers were reduced in the mutants (Figure S4G–I’), as were ChAT+ neurons at P13–P15 (Figure S4J–L’). Nkx2.1Cre cKOs exhibited a milder reduction of ChAT+ neurons (Figure S3J–L’).
Bioinformatics identifies OTX-binding motifs
OTX2 ChIP-seq was performed three times from E12.5 subpallium. Replica #1 had 995 peaks, replica #2 had 1416 peaks and replica #3 had 19881 peaks (Figure S5M). We focused on ChIP-seq peaks present in all three replicas, yielding 590 regions (Figure S5M), which were analyzed Regulatory Sequence Analysis Tools (RSAT) (Thomas-Chollier, et al., 2011). Four variations of a frequently detected motif were assigned to CRX [GGATTA (TAATCC)] by the JASPAR database. CRX and OTX2 both have bicoid homeodomains that bind to the same motif (Zhang et al., 2002). 269/590 of the OTX2 ChIP-seq peaks (45.6%) had the core binding sequence GGATTA. These regulatory domains also had motifs for other homeodomain proteins [HOXA5/PDX1 (36%) or NOBOX (12.5%)], and for HMG box proteins, (e.g. SOX2 motifs; 32%) (Figure S6A). 53% of OTX2 motif-containing enhancers had either the other homeodomain or HMG box motifs; 61% of OTX2 motif-negative enhancers did not have either of these motifs.
Several of the dysregulated genes in the Otx2 mutants had OTX2 ChIP-seq peaks (Figure 2); those with OTX2 motifs have yellow stars in Figure 2. In some cases, broad domains of OTX2 binding lacked the OTX2 motif (e.g. in the Dlx1/2, Dbx1, Mpk3 loci), suggesting that OTX2 binding in these cases may be through protein-protein interactions.
Gene ontologies (GO) were computed using the GREAT tool (McLean et al., 2010)(Figure S6B). The most frequent GO molecular function terms showed that OTX2 target genes were highly enriched for transcription regulators. The most frequent GO biological function terms showed OTX2 target genes were highly enriched for regulators of neural development. Thus, ChIP-seq analysis on E12.5 ganglionic eminences provided strong support for OTX2 binding in vivo to regulatory elements containing the OTX consensus sequence near genes that regulate transcription.
DISCUSSION
Using transcriptional profiling and conditional mutagenesis with two Cre alleles, we demonstrated that Otx2 regulates: 1) RPC identity and signaling, 2) specification of the vMGE, and 3) promotes MGE neurogenesis and oligodendrogenesis. OTX2 ChIP-seq provided evidence that a subset of genes deregulated in cKOs were direct transcriptional targets of OTX2. To our knowledge, this is the first report of a genome-wide TF ChIP-seq analysis from embryonic basal ganglia. It enabled us to deduce the in vivo binding site motifs for OTX2 (Figure S6), to provide evidence for the other TFs that bind in adjacent regions (Figure S6), and to make predictions about which domains have OTX2 binding that do not depend on its association with the OTX2 core motif.
Otx2 specifies rostral patterning center (RPC) identity
Otx2 plays pivotal roles in the early specification of the forebrain and midbrain (Ang et al., 1996; Suda et al., 1996; Acampora and Simeone, 1999), and, at later stages, in midbrain/hindbrain patterning and differentiation (Joyner et al., 2000; Simeone, 2000; Puelles et al., 2003; Vernay et al., 2005; Sakurai et al., 2010). Here we show using RxCre that Otx2 expression after E8.5 is required for specifying RPC function and identity based on: 1) Fgf expression domains were expanded, and 2) genes expressed in the MHB patterning center (Pax3, En2) were ectopically expressed in the RPC (Figure 3). In the developing midbrain, ectopic Pax3 can induce transcription of Fgf8, En2, and Pax3 (Matsunaga et al., 2001). OTX2 ChIP-seq identified two Pax3 intragenic peaks (Figure 2L); no peaks were found near En2. These data suggest that direct repression of Pax3 by OTX2 is required to inhibit En2 expression and restrict Fgf8 and Pax3 expression in the RPC. This may be a crucial step in defining the identity of the Se, and/or in distinguishing forebrain and midbrain/hindbrain fates. Misspecification of the Se likely contributes to Se hypoplasia (Figure S4J’) and reduction in Se cholinergic neurons (Figure S4G–L’).
FGF8-bead experiments demonstrated that FGF8 represses Otx2 expression (Martinez et al., 1999; Crossley et al., 2001). Furthermore, Fgf8 gain of function studies show that increased Fgf8 represses growth (Crossley et al., 2001; Assimacopoulos et al., 2012) that is consistent with the hypoplastic rostral telencephalon (Figure 3A–E). Furthermore, Otx2 cKO and Fgf8 hypomorph analyses revealed that Otx2 spatially restricts Fgf8 to the RPC (Figure 3A–D), whereas Fgf8 positively regulates Otx2 in the early rostral telencephalon (Figure S2 A, B; Storm et al., 2006). These data support a model in which Fgf8 and Otx2 regulation are interdependent. Indeed, Otx2 controls feedback regulators of Fgf signaling (Spry1, Spry2, Mkp3) (Figure 3F–J). ChIP-seq data suggest that Fgf8, Spry1, Spry2, and Mkp3 are direct OTX2 targets (Figure 2). Deregulated Fgf signaling in Otx2 mutants likely contributes to deficits in cell populations that arise from, or adjacent to, the RPC.
Otx2 promotes neurogenesis and oligodendrogenesis in the MGE
Otx2 expression in the E9.5–E12.5 subpallium is restricted to the VZ and SVZ (Figure 1A–A”), where in the MGE it is required to generate normal numbers of SVZ progenitors and MZ neurons (Figure 5). Otx2 cKOs overexpress anti-neurogenic TFs (Hes1, Id4; Table S1; Figure 5) that inhibit neurogenic TFs such as Ascl1 (Mash1) (Casarosa et al., 1999; Yun et al., 2002). Furthermore, Otx2 promotes oligodendrogenesis through positive regulation of Olig1 and Olig2 (Figure 4)(Petryniak et al., 2007; Silbereis et al., 2014). These mechanisms for neurogenesis and oligodendrogenesis appear to be mediated by direct binding of OTX2 at genomic loci of key regulatory TFs (Figure 2). Later, compensatory mechanisms may rescue these phenotypes as neurogenesis has improved by E13.5–E15.5 (Figure 7). This compensation may in part be mediated by Otx1 (Acampora et al., 1997; Suda et al., 1997), which is expressed at low levels in the MGE (Figure S1).
Otx2 regulates rostrocaudal and dorsoventral patterning of the MGE
There is evidence that vMGE generates most of the GP, whereas the dMGE may principally generate interneurons (Flandin et al., 2010). Whereas Nkx2.1 function is required throughout the MGE (Sussel et al., 1999; Flandin et al., 2010), Otx2 preferentially controls the identity of the vMGE. This is a surprising result, given that Otx2 mRNA and protein are expressed throughout the MGE.
Analysis of gene expression changes and OTX2 genomic binding sites provides three lines of evidence that Otx2 specifies vMGE identity through direct regulation of TF genes expressed in the vMGE and POA (Figure S7): 1) Otx2 autoregulates its transcription in the MGE and POA (Figure 1, 2); 2) Otx2 drives vMGE expression of Sall3, Tal2 and Tll2 (Figure 6; Table S1). OTX2 has binding sites near these genes (Figures 2, S5); 3) Otx2 represses POA identity in the MGE by blocking Dbx1, Slit2 and Sox3 expression (Figure 6 and not shown). There is OTX2 binding near Dbx1 (Figure 2). Note that Nkx2.1 expression persists in vMGE progenitors in Otx2 mutants (Figure 7J–L); thus Otx2 and Nkx2.1 may specify vMGE identity via parallel pathways.
vMGE respecification in Otx2 mutants has consequences for subpallial development, including GP agenesis (or, loss of molecular identity based on ER81, Lhx6, Lhx8, Nkx2.1, Zic1; Figures 7, S4). There is dorsal and rostral expansion of POA progenitor (Arhgap22, Dbx1, Slit2, Sox3), and neuronal (Nkx5.2 and Sox14), properties (Figures 6, 7). Other vMGE neuronal cell types are reduced, including Lhx6+ neurons in the ventral pallidum, and cholinergic neurons in the nucleus basalis, diagonal band and striatum (ChAT+, Lhx8+, and TrkA+; Figure S4).
Otx2 also impacts patterning of the telencephalon along the rostrocaudal axis. RPC Fgf expression domains are expanded rostrally by E11.5 in Otx2 mutants (Figure 3). COUP-TF1, a TF expressed in the caudal MGE, is repressed by Otx2, as COUP-TF1 is ectopically expressed rostrally by E13.5 in the mutants (Figure 6). In addition, several POA markers expand rostrally as well as dorsally into the MGE (Nkx5.2, Dbx1; Figure 6). Thus, Otx2 controls both rostrocaudal and dorsoventral MGE patterning.
In summary, Otx2 is essential in the E8.5–E13.5 telencephalon for regional specification of the RPC and vMGE, and for MGE neurogenesis and oligodendrogenesis. In the absence of Otx2, the RPC takes on MHB properties and the vMGE takes on POA properties, leading to Se, GP and cholinergic deficits. OTX2 ChIP-seq provided evidence for direct mechanisms through which Otx2 controls regional and cell type identity in the subpallium.
EXPERIMENTAL PROCEDURES
Mouse lines
We used the following published mouse lines: Fgf8neo (G. Martin, UCSF; (Meyers et al., 1998), non-hypomorphic Otx2f (S. Aizawa, CDB, RIKEN; Acc. No. CDB0013K, http://www.cdb.riken.jp/arg/mutant%20mice%20list.html; (Tian et al., 2002)), RxCre (Y. Zhao, NIH; (Swindell et al., 2006)), Nkx2.1Cre (Stewart Anderson, U. Penn. (Xu et al., 2008)), Fgf8CreER (Hoch et al, manuscript in preparation). Otx2f/+ mice were crossed to βactin::Cre (Lewandoski et al., 1997) to generate Otx2+/− mice. Unless otherwise specified, conditional knockouts were of the genotype Otx2f/−; Cre+, generated by crossing Otx2f/f mice to Cre lines maintained on an Otx2+/− background. Mice were maintained in social cages in a specific pathogen-free barrier facility at UCSF on a 12 hour light/dark cycle with free access to food and water.
For embryonic (timed mating) experiments, day 0.5 was designated as noon on the day a vaginal plug was observed. At the time of experiment, mice were euthanized by CO2 inhalation followed by cervical dislocation. Embryonic heads (E10.5–E12.5) or isolated brains (E13.5 and older) were fixed overnight in 4% PFA (made in 1x PBS), transferred to 30% sucrose for cryoprotection, and then embedded and frozen in OCT for cryosectioning. Section thickness ranged from 10–20 μm depending on stage.
For postnatal experiments (>P7), animals were anesthetized with i.p. avertin (0.015 ml/g of 2.5% solution), perfused transcardially with 1X PBS and with 4% PFA, followed by brain isolation, fixation, cryoprotection, and freezing/embedding.
Immunohistochemistry (IHC)
We used the following antibodies: ChAT (Chemicon AB144P), Otx2 (R&D systems goat, catalog # AF1979), Tuj1 (Covance MMS-435P), pH3 (Millipore 06-570), parvalbumin (Millipore MAB1572), somatostatin (Santa Cruz sc-7819).
Cryosections were rinsed in PBS, blocked in 10% normal serum/PBST (1x PBS, 0.1% Triton X-100), incubated in primary antibody overnight (4° C), washed in PBST, incubated in secondary antibody 1–3 hours (room temperature), and washed in PBS. For fluorescent detection, we used Alexa 488- and Alexa 594-conjugated secondary antibodies (Invitrogen). For colorimetric detection, biotinylated secondary antibodies (Vector) were used with the ABC (Vector)/DAB detection method.
For ChAT IHC, antigen retrieval was achieved by incubating slides in 2.94g/L trisodium citrate dehydrate, 0.05% Tween-20, pH 6.0 for 15 minutes at 90° C. Blocking and antibody incubations were done in 1% BSA in PBST. Sections were incubated two days at 4° C with primary antibody, and signal was amplified with biotinylated anti-goat (Vector) prior to fluorescent detection with streptavidin-594 (Invitrogen).
For OTX2 IHC, we modified the IHC protocol according to the recommendations of Yuki Muranishi in the Furakawa lab (Osaka, Japan). Briefly, antigen retrieval was achieved as for ChAT IHC, and samples were blocked in 4% donkey serum in PBST.
In situ hybridization (ISH)
We performed ISH on a minimum of n=2 and n=3 biological replicates for controls and mutants, respectively. In each case, a rostrocaudal series of at least 10 sections was examined. Reduced expression was interpreted as reduced RNA per cell, unless otherwise stated. Section ISHs were performed using digoxigenin-labeled riboprobes as described (Schaeren-Wiemers and Gerfin-Moser, 1993) with the following modifications. Prior to acetylation, sections were incubated with proteinase K (1μg/ml) and post-fixed in 4% PFA. Slides were equilibrated in NTT prior to antibody incubation (overnight, 4° C, AP-antidigoxigenin (Roche)), and then washed in NTT 3 x 30 minutes at room temperature. They were then washed 3 times in NTTML (NTT + 50 mM MgCl2, 2 mM Levamisole) and transferred to BM Purple (Roche) for colorimetric detection (dark, 37° C). Slides were rinsed in water, then postfixed (4% PFA overnight), dehydrated, incubated briefly in xylene, and coverslipped using Permount. Acetylation buffer: 1.33% (v/v) triethanolamine, 0.065% HCl, 0.375% (v/v) acetic anhydride. Riboprobe block/hybridization buffer: 50% formamide, 5x SSC pH 4.5, 1% SDS, 50 μg/ml yeast tRNA, 50 μg/ml heparin. Antibody blocking buffer (NTT): 0.15M NaCl, 0.1M Tris pH 8.0, 0.1% Tween-20.
E12.5 RNA isolation and microarray
Subpallial (GE and Se) tissue was microdissected from E12.5 female brains (n=4 control, n=3 CKO), snap frozen, and stored at −80°C. Total RNA was isolated using the Qiagen RNAeasy kit. RNA was amplified (labeled with Cy3-CTP) with Agilent low RNA input fluorescent linear amplification kits, and cRNA was assessed using the Nandrop ND-100 (Nanodrop Technologies, Inc., Wilmington DE). Equal amounts of Cy3 labeled target were hybridized to Agilent whole mouse genome 4x44K Ink-jet arrays, by the UCSF Genomics Core, who then performed the differential gene expression analysis (http://www.arrays.ucsf.edu; http://www.agilent.com) (Holm, 1979) Bolstad et al., 2003; Gentleman et al., 2004; Smyth, 2004). Significant changes in gene expression were defined as B value greater than zero. B = log 10 posterior odds ratio, ratio between the probability that a given gene is differentially expressed (DE) over the probability that a given gene is not differentially expressed. B≥ 0 means equal or greater probability that a gene is DE than non-DE (Lonnstedt and Speed, 2002).
Chromatin immunoprecipitation-sequencing experiment (ChIP-seq) and informatics
ChIP was performed using anti-OTX2 (R&D #AF1979) (McKenna et al., 2011). E12.5 CD1 GEs were fixed in 1.5% formaldehyde for 20 min. and neutralized with glycine. Fixed chromatin was lysed and sheared into 200–1000 bp fragments using a bioruptor (Diagenode). Immunoprecipitation (IP) reactions were performed in duplicates using Goat IgG as negative controls. Precipitated fractions were purified using Dynabeads (Invitrogen). Libraries were prepared using an Ovation Ultralow DR Multiplex System (Nugen), size selected in the range of 200–300 bp on a LabChip (Lifesciences), quality control tested on a Bioanalyzer (Agilent) and sequenced on a HiSeq (Illumina). Reads from ChIP, input, and negative control (IgG) libraries were mapped to the mouse genome (mm9) using BWA and peaks were called using MACS considering both input and IgG as the control sample with filtering to remove peaks in repeat regions.
For downstream analysis of ChIP-seq data, only peaks that overlapped in each of the 3 OTX2 ChIP-seq replicates were selected (590 regions). Nucleotide motifs were identified using the Regulatory Sequence Analysis Tools (RSAT) peak-motifs tool (Thomas-Chollier et al., 2011). Gene Ontology for biological process and molecular function was computed using the Genomic Regions Enrichment of Annotations Tool (GREAT) (McLean et al., 2010).
Supplementary Material
Highlights.
OTX2 directs enhancer regulation.
Otx2 regulates Fgf8 expression and Fgf signaling responses.
Otx2 promotes MGE neurogenesis and oligodendrogenesis.
Otx2 is required for specification of vMGE versus POA fate.
Acknowledgments
This work was supported by NIH Grant #F32MH081431 to R.V.H., NIH T32 Predoctoral Training in Developmental Biology, Grant #: T32 HD 007470 to J.D.P., and by funds from Nina Ireland, Weston Havens Foundation, NINDS (NS34661), and NIMH (MH049428 and MH081880) to J.L.R.R. We thank Shin Aizawa for sharing the Otx2f mouse line for this project, Alex Nord and Axel Visel for aiding in the analysis of ChIP-Seq data, Luis Puelles for assistance with neuroanatomical nomenclature, and colleagues in the Rubenstein lab for their helpful suggestions over the course of the project. John Rubenstein is a founder of Neurona is on their scientific board. The work in this paper was not supported by a company.
Footnotes
ACCESSION NUMBERS
The GEO accession number for both the RNA array and ChIP-Seq data sets is GSE69727, which gives access to the two different data-sets: GSE69547: RNA array data and GSE69724: ChIP-Seq data.
AUTHOR CONTRIBUTIONS
R.V.H. designed, conducted, and analyzed data for all experiments described in this manuscript, except the ChIP-seq study. S.L. performed and helped to analyze ChIP-seq experiments, and provided comments on the manuscript. J.D.P. performed informatic analyses of the ChIP-Seq data. J.L.R.R. provided funding and laboratory resources for this study, and helped guide the project and analyze results. R.V.H. and J.L.R.R. prepared the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Acampora D, Avantaggiato V, Tuorto F, Simeone A. Genetic control of brain morphogenesis through Otx gene dosage requirement. Development. 1997;124(18):3639–50. doi: 10.1242/dev.124.18.3639. [DOI] [PubMed] [Google Scholar]
- Acampora D, Mazan S, Lallemand Y, Avantaggiato V, Maury M, Simeone A, Brulet P. Forebrain and midbrain regions are deleted in Otx2−/ − mutants due to a defective anterior neuroectoderm specification during gastrulation. Development. 1995;121(10):3279–90. doi: 10.1242/dev.121.10.3279. [DOI] [PubMed] [Google Scholar]
- Acampora D, Simeone A. The TINS Lecture. Understanding the roles of Otx1 and Otx2 in the control of brain morphogenesis. Trends Neurosci. 1999;22(3):116–22. doi: 10.1016/s0166-2236(98)01387-3. [DOI] [PubMed] [Google Scholar]
- Ahituv N, Zhu Y, Visel A, Holt A, Afzal V, Pennacchio LA, Rubin EM. Deletion of ultraconserved elements yields viable mice. PLoS Biol. 2007;5(9):e234. doi: 10.1371/journal.pbio.0050234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang SL, Jin O, Rhinn M, Daigle N, Stevenson L, Rossant J. A targeted mouse Otx2 mutation leads to severe defects in gastrulation and formation of axial mesoderm and to deletion of rostral brain. Development. 1996;122(1):243–52. doi: 10.1242/dev.122.1.243. [DOI] [PubMed] [Google Scholar]
- Asbreuk CH, van Schaick HS, Cox JJ, Kromkamp M, Smidt MP, Burbach JP. The homeobox genes Lhx7 and Gbx1 are expressed in the basal forebrain cholinergic system. Neuroscience. 2002;109(2):287–98. doi: 10.1016/s0306-4522(01)00466-3. [DOI] [PubMed] [Google Scholar]
- Assimacopoulos S, Kao T, Issa NP, Grove EA. Fibroblast growth factor 8 organizes the neocortical area map and regulates sensory map topography. J Neurosci. 2012;232(21):7191–7201. doi: 10.1523/JNEUROSCI.0071-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista-Britto R, Fishell G. The generation of cortical interneurons. In: Rubenstein JLR, Rakic P, editors. Patterning and Cell Type Specification in the Developing CNS and PNS. Vol. 1. Academic Press; San Diego, CA: 2013. [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19(2):185–93. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Casarosa S, Fode C, Guillemot F. Mash1 regulates neurogenesis in the ventral telencephalon. Development. 1999;126(3):525–34. doi: 10.1242/dev.126.3.525. [DOI] [PubMed] [Google Scholar]
- Chi CL, Martinez S, Wurst W, Martin GR. The isthmic organizer signal FGF8 is required for cell survival in the prospective midbrain and cerebellum. Development. 2003;130(12):2633–44. doi: 10.1242/dev.00487. [DOI] [PubMed] [Google Scholar]
- Cholfin JA, Rubenstein JLR. Patterning of frontal cortex subdivisions by Fgf17. Proc Natl Acad Sci U S A. 2007;104:7652–7657. doi: 10.1073/pnas.0702225104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colasante G, Collombat P, Raimondi V, Bonanomi D, Ferrai C, Maira M, Yoshikawa K, Mansouri A, Valtorta F, Rubenstein JL, Broccoli V. Arx is a direct target of Dlx2 and thereby contributes to the tangential migration of GABAergic interneurons. J Neurosci. 2008;28(42):10674–86. doi: 10.1523/JNEUROSCI.1283-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossley PH, Martinez S, Ohkubo Y, Rubenstein JL. Coordinate expression of Fgf8, Otx2, Bmp4, and Shh in the rostral prosencephalon during development of the telencephalic and optic vesicles. Neuroscience. 2001;108(2):183–206. doi: 10.1016/s0306-4522(01)00411-0. [DOI] [PubMed] [Google Scholar]
- Eswarakumar VP, Lax I, Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine & growth factor reviews. 2005;16(2):139–49. doi: 10.1016/j.cytogfr.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Faedo A, Borello U, Rubenstein JL. Repression of Fgf signaling by sprouty1-2 regulates cortical patterning in two distinct regions and times. J Neurosci. 2010;30(11):4015–23. doi: 10.1523/JNEUROSCI.0307-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flames N, Pla R, Gelman DM, Rubenstein JL, Puelles L, Marin O. Delineation of multiple subpallial progenitor domains by the combinatorial expression of transcriptional codes. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2007;27(36):9682–95. doi: 10.1523/JNEUROSCI.2750-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flandin P, Kimura S, Rubenstein JL. The progenitor zone of the ventral medial ganglionic eminence requires Nkx2-1 to generate most of the globus pallidus but few neocortical interneurons. J Neurosci. 2010;30(8):2812–23. doi: 10.1523/JNEUROSCI.4228-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuchi-Shimogori T, Grove EA. Neocortex patterning by the secreted signaling molecule FGF8. Science. 2001;294(5544):1071–4. doi: 10.1126/science.1064252. [DOI] [PubMed] [Google Scholar]
- Ghanem N, Yu M, Long J, Hatch G, Rubenstein JL, Ekker M. Distinct cis-regulatory elements from the Dlx1/Dlx2 locus mark different progenitor cell populations in the ganglionic eminences and different subtypes of adult cortical interneurons. J Neurosci. 2007;27(19):5012–22. doi: 10.1523/JNEUROSCI.4725-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garel S, Huffman KJ, Rubenstein JL. Molecular regionalization of the neocortex is disrupted in Fgf8 hypomorphic mutants. Development. 2003;130(9):1903–14. doi: 10.1242/dev.00416. [DOI] [PubMed] [Google Scholar]
- Gelman D, Griveau A, Dehorter N, Teissier A, Varela C, Pla R, Pierani A, Marin O. A wide diversity of cortical GABAergic interneurons derives from the embryonic preoptic area. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2011;31(46):16570–80. doi: 10.1523/JNEUROSCI.4068-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome biology. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulding MD, Chalepakis G, Deutsch U, Erselius JR, Gruss P. Pax-3, a novel murine DNA binding protein expressed during early neurogenesis. The EMBO journal. 1991;10(5):1135–47. doi: 10.1002/j.1460-2075.1991.tb08054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove EA, Monucki ES. Morphogens, patterning centers, and their mechanisms of action. In: Rubenstein JLR, Rakic P, editors. Patterning and Cell Type Specification in the Developing CNS and PNS. Vol. 1. Academic Press; San Diego, CA: 2013. [Google Scholar]
- Hirata T, Li P, Lanuza GM, Cocas LA, Huntsman MM, Corbin JG. Identification of distinct telencephalic progenitor pools for neuronal diversity in the amygdala. Nature neuroscience. 2009;12(2):141–9. doi: 10.1038/nn.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch RV, Clarke JA, Rubenstein JL. Fgf Signaling Controls the Telencephalic Distribution of Fgf-Expressing Progenitors Generated in the Rostral Patterning Center. Neural Development. 2015;10:8. doi: 10.1186/s13064-015-0037-7. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm S. A simple sequentially rejective multiple test procedure. Scandinavian Journal of Statistics. 1979;6:65–70. [Google Scholar]
- Jeong Y, El-Jaick K, Roessler E, Muenke M, Epstein DJ. A functional screen for sonic hedgehog regulatory elements across a 1 Mb interval identifies long-range ventral forebrain enhancers. Development. 2006;133(4):761–772. doi: 10.1242/dev.02239. [DOI] [PubMed] [Google Scholar]
- Joyner AL, Liu A, Millet S. Otx2, Gbx2 and Fgf8 interact to position and maintain a mid-hindbrain organizer. Current opinion in cell biology. 2000;12(6):736–41. doi: 10.1016/s0955-0674(00)00161-7. [DOI] [PubMed] [Google Scholar]
- Kessaris N, Fogarty M, Iannarelli P, Grist M, Wegner M, Richardson WD. Competing waves of oligodendrocytes in the forebrain and postnatal elimination of an embryonic lineage. Nature neuroscience. 2006;9(2):173–9. doi: 10.1038/nn1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa D, Kiyonari H, Nakayama R, Kimura-Yoshida C, Matsuo I, Aizawa S. Regulation of Otx2 expression and its functions in mouse forebrain and midbrain. Development. 2004a;131(14):3319–31. doi: 10.1242/dev.01220. [DOI] [PubMed] [Google Scholar]
- Kurokawa D, Takasaki N, Kiyonari H, Nakayama R, Kimura-Yoshida C, Matsuo I, Aizawa S. Regulation of Otx2 expression and its functions in mouse epiblast and anterior neuroectoderm. Development. 2004b;131(14):3307–17. doi: 10.1242/dev.01219. [DOI] [PubMed] [Google Scholar]
- Lewandoski M, Meyers EN, Martin GR. Analysis of Fgf8 gene function in vertebrate development. Cold Spring Harb Symp Quant Biol. 1997;62:159–68. [PubMed] [Google Scholar]
- Liu A, Joyner AL. EN and GBX2 play essential roles downstream of FGF8 in patterning the mouse mid/hindbrain region. Development. 2001;128(2):181–91. doi: 10.1242/dev.128.2.181. [DOI] [PubMed] [Google Scholar]
- Lonnstedt I, Speed TP. Replicated microarray data. Stat Sinica. 2002;2:31–46. [Google Scholar]
- Martinez S, Crossley PH, Cobos I, Rubenstein JL, Martin GR. FGF8 induces formation of an ectopic isthmic organizer and isthmocerebellar development via a repressive effect on Otx2 expression. Development. 1999;126(6):1189–200. doi: 10.1242/dev.126.6.1189. [DOI] [PubMed] [Google Scholar]
- Matsunaga E, Araki I, Nakamura H. Role of Pax3/7 in the tectum regionalization. Development. 2001;128(20):4069–77. doi: 10.1242/dev.128.20.4069. [DOI] [PubMed] [Google Scholar]
- McKenna WL, Betancourt J, Larkin KA, Abrams B, Guo C, Rubenstein JL, Chen B. Tbr1 and Fezf2 regulate alternate corticofugal neuronal identities during neocortical development. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2011;31(2):549–64. doi: 10.1523/JNEUROSCI.4131-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28(5):495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers EN, Lewandoski M, Martin GR. An Fgf8 mutant allelic series generated by Cre- and Flp-mediated recombination. Nat Genet. 1998;18(2):136–41. doi: 10.1038/ng0298-136. [DOI] [PubMed] [Google Scholar]
- Minowada G, Jarvis LA, Chi CL, Neubuser A, Sun X, Hacohen N, Krasnow MA, Martin GR. Vertebrate Sprouty genes are induced by FGF signaling and can cause chondrodysplasia when overexpressed. Development. 1999;126(20):4465–75. doi: 10.1242/dev.126.20.4465. [DOI] [PubMed] [Google Scholar]
- Petryniak MA, Potter GB, Rowitch DH, Rubenstein JL. Dlx1 and Dlx2 control neuronal versus oligodendroglial cell fate acquisition in the developing forebrain. Neuron. 2007;55(3):417–33. doi: 10.1016/j.neuron.2007.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puelles E, Acampora D, Lacroix E, Signore M, Annino A, Tuorto F, Filosa S, Corte G, Wurst W, Ang SL, et al. Otx dose-dependent integrated control of antero-posterior and dorso-ventral patterning of midbrain. Nature neuroscience. 2003;6(5):453–60. doi: 10.1038/nn1037. [DOI] [PubMed] [Google Scholar]
- Rhinn M, Dierich A, Shawlot W, Behringer RR, Le Meur M, Ang SL. Sequential roles for Otx2 in visceral endoderm and neuroectoderm for forebrain and midbrain induction and specification. Development. 1998;125(5):845–56. doi: 10.1242/dev.125.5.845. [DOI] [PubMed] [Google Scholar]
- Rubenstein JLR, Campbell K. Neurogenesis in the basal ganglia. In: Rubenstein JLR, Rakic P, editors. Patterning and Cell Type Specification in the Developing CNS and PNS. Vol. 1. Academic Press; San Diego, CA: 2013. [Google Scholar]
- Sakurai Y, Kurokawa D, Kiyonari H, Kajikawa E, Suda Y, Aizawa S. Otx2 and Otx1 protect diencephalon and mesencephalon from caudalization into metencephalon during early brain regionalization. Developmental biology. 2010;347(2):392–403. doi: 10.1016/j.ydbio.2010.08.028. [DOI] [PubMed] [Google Scholar]
- Sanchez-Ortiz E, Yui D, Song D, Li Y, Rubenstein JL, Reichardt LF, Parada LF. TrkA gene ablation in basal forebrain results in dysfunction of the cholinergic circuitry. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2012;32(12):4065–79. doi: 10.1523/JNEUROSCI.6314-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeren-Wiemers N, Gerfin-Moser A. A single protocol to detect transcripts of various types and expression levels in neural tissue and cultured cells: in situ hybridization using digoxigenin-labelled cRNA probes. Histochemistry. 1993;100(6):431–40. doi: 10.1007/BF00267823. [DOI] [PubMed] [Google Scholar]
- Silbereis JC, Nobuta H, Tsai HH, Heine VM, McKinsey GL, Meijer DH, Howard MA, Petryniak MA, Potter GB, Alberta JA, et al. Olig1 function is required to repress dlx1/2 and interneuron production in Mammalian brain. Neuron. 2014;81(3):574–87. doi: 10.1016/j.neuron.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeone A. Positioning the isthmic organizer where Otx2 and Gbx2meet. Trends in genetics : TIG. 2000;16(6):237–40. doi: 10.1016/s0168-9525(00)02000-x. [DOI] [PubMed] [Google Scholar]
- Simeone A, Acampora D, Gulisano M, Stornaiuolo A, Boncinelli E. Nested expression domains of four homeobox genes in developing rostral brain. Nature. 1992;358(6388):687–90. doi: 10.1038/358687a0. [DOI] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Statistical applications in genetics and molecular biology. 2004;3:Article 3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Storm EE, Garel S, Borello U, Hebert JM, Martinez S, McConnell SK, Martin GR, Rubenstein JL. Dose-dependent functions of Fgf8 in regulating telencephalic patterning centers. Development. 2006;133(9):1831–44. doi: 10.1242/dev.02324. [DOI] [PubMed] [Google Scholar]
- Suda Y, Matsuo I, Aizawa S. Cooperation between Otx1 and Otx2 genes in developmental patterning of rostral brain. Mechanisms of development. 1997;69(1–2):125–41. doi: 10.1016/s0925-4773(97)00161-5. [DOI] [PubMed] [Google Scholar]
- Suda Y, Matsuo I, Kuratani S, Aizawa S. Otx1 function overlaps with Otx2 in development of mouse forebrain and midbrain. Genes to cells : devoted to molecular & cellular mechanisms. 1996;1(11):1031–44. doi: 10.1046/j.1365-2443.1996.900288.x. [DOI] [PubMed] [Google Scholar]
- Sussel L, Marin O, Kimura S, Rubenstein JL. Loss of Nkx2.1 homeobox gene function results in a ventral to dorsal molecular respecification within the basal telencephalon: evidence for a transformation of the pallidum into the striatum. Development. 1999;126(15):3359–70. doi: 10.1242/dev.126.15.3359. [DOI] [PubMed] [Google Scholar]
- Swindell EC, Bailey TJ, Loosli F, Liu C, Amaya-Manzanares F, Mahon KA, Wittbrodt J, Jamrich M. Rx-Cre, a tool for inactivation of gene expression in the developing retina. Genesis. 2006;44(8):361–3. doi: 10.1002/dvg.20225. [DOI] [PubMed] [Google Scholar]
- Thisse B, Thisse C. Functions and regulations of fibroblast growth factor signaling during embryonic development. Developmental biology. 2005;287(2):390–402. doi: 10.1016/j.ydbio.2005.09.011. [DOI] [PubMed] [Google Scholar]
- Tian E, Kimura C, Takeda N, Aizawa S, Matsuo I. Otx2 is required to respond to signals from anterior neural ridge for forebrain specification. Dev Biol. 2002;242(2):204–23. doi: 10.1006/dbio.2001.0531. [DOI] [PubMed] [Google Scholar]
- Thomas-Chollier M, Herrmann C, Defrance M, Sand O, Thieffry D, van Helden J. RSAT peak-motifs: motif analysis in full-size ChIP-seq datasets. Nucleic Acids Research. 2011:9. doi: 10.1093/nar/gkr1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernay B, Koch M, Vaccarino F, Briscoe J, Simeone A, Kageyama R, Ang SL. Otx2 regulates subtype specification and neurogenesis in the midbrain. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2005;25(19):4856–67. doi: 10.1523/JNEUROSCI.5158-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waclaw RR, Ehrman LA, Pierani A, Campbell K. Developmental origin of the neuronal subtypes that comprise the amygdalar fear circuit in the mouse. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2010;30(20):6944–53. doi: 10.1523/JNEUROSCI.5772-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Tam M, Anderson SA. Fate mapping Nkx2.1-lineage cells in the mouse telencephalon. J Comp Neurol. 2008;506(1):16–29. doi: 10.1002/cne.21529. [DOI] [PubMed] [Google Scholar]
- Yun K, Fischman S, Johnson J, Hrabe de Angelis M, Weinmaster G, Rubenstein JL. Modulation of the notch signaling by Mash1 and Dlx1/2 regulates sequential specification and differentiation of progenitor cell types in the subcortical telencephalon. Development. 2002;129(21):5029–5040. doi: 10.1242/dev.129.21.5029. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Miki T, Iwanaga T, Koseki Y, Okuno M, Sunaga Y, Ozaki N, Yano H, Koseki H, Seino S. Identification, tissue expression, and functional characterization of Otx3, a novel member of the Otx family. J Biol Chem. 2002;277(31):28065–28069. doi: 10.1074/jbc.C100767200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.