Abstract
The rhesus macaque (Macaca mulatta) is the most widely used nonhuman primate for modeling the structure and function of the brain. Brain atlases, and particularly those based on magnetic resonance imaging (MRI), have become important tools for understanding normal brain structure, and for identifying structural abnormalities resulting from disease states, exposures, and/or aging. Diffusion tensor imaging (DTI) -based MRI brain atlases are widely used in both human and macaque brain imaging studies because of the unique contrasts, quantitative diffusion metrics, and diffusion tractography that they can provide. Previous MRI and DTI atlases of the rhesus brain have been limited by low contrast and/or low spatial resolution imaging. Here we present a microscopic resolution MRI/DTI atlas of the rhesus brain based on 10 postmortem brain specimens. The atlas includes both structural MRI and DTI image data, a detailed three-dimensional segmentation of 241 anatomic structures, diffusion tractography, cortical thickness estimates, and maps of anatomic variability amongst atlas specimens. This atlas incorporates many useful features from previous work, including anatomic label nomenclature and ontology, data orientation, and stereotaxic reference frame, and further extends prior analyses with the inclusion of high-resolution multi-contrast image data.
Keywords: brain atlas, diffusion tensor imaging, tractography, rhesus macaque, magnetic resonance imaging, Macaca mulatta
Graphical abstract

2. Introduction
The rhesus macaque (Macaca mulatta) is the primary nonhuman primate used to test theories about human brain structure and function. Brain atlases have become important tools for studying normal neuroanatomy and for localizing neuropathology that is the basis of disease states. Many different brain atlases have been generated for the rhesus macaque including those based on conventional histology (Paxinos et al., 2009; Saleem and Logothetis, 2012), anterograde/retrograde neuronal tracer studies (Schmahmann and Pandya, 2009), and magnetic resonance imaging (MRI) (Adluru et al., 2012; Dubach and Bowden, 2009; Frey et al., 2011; McLaren et al., 2009; Rohlfing et al., 2012; Saleem and Logothetis, 2012; Zakszewski et al., 2014). Although it is generally of lower resolution than other brain atlasing methods, MRI has several important advantages: 1) it is three-dimensional (3D) and volumetrically accurate; 2) it can provide multiple different image contrasts from the same tissue; and, 3) it allows probabilistic atlases to be generated from multiple subjects through non-linear image registration techniques. Previous MRI-based atlases of the rhesus brain have been limited by low resolution, a lack of multiple image contrasts, and/or absent or insufficient anatomic delineations.
In humans, diffusion tensor imaging (DTI) has emerged as an important imaging strategy for MRI-based brain atlases (Oishi et al., 2010; 2008). DTI provides a variety of unique MR image contrasts including fractional anisotropy (FA), axial diffusivity (AD), radial diffusivity (RD), and mean diffusivity (MD) (Mukherjee et al., 2008). These contrasts provide highly sensitive, quantitative metrics of tissue microstructure (Basser and Pierpaoli, 1996) and can improve the ability to segment many brain structures from MR images (Calabrese et al., 2012). DTI also allows 3D tractography of diffusion pathways, which has become a popular method for structural connectivity mapping in the brain (Mori et al., 1999). An increasing number of studies have reported on DTI changes resulting from neurologic and psychiatric diseases, both in humans and animal models (Konrad and Eickhoff, 2010; Kubicki et al., 2007; Song et al., 2002; Sundaram et al., 2008). These studies highlight the need for high-quality DTI-based brain atlases, particularly for important model systems like the rhesus macaque.
Several MRI and even DTI-based atlases of the rhesus macaque brain exist, but they are often limited by low contrast and/or low spatial resolution (Adluru et al., 2012; Frey et al., 2011; McLaren et al., 2009; Saleem and Logothetis, 2012; Wisco et al., 2008b; Zakszewski et al., 2014). Image resolution is particularly important because the macaque brain is 10–15 fold smaller volumetrically than the human brain (Herculano-Houzel, 2009), so image voxels must be 10–15 fold smaller to have comparable structural resolution. Postmortem MRI with exogenous contrast agents allows considerably higher resolution and higher quality images than would be possible with in vivo studies (Johnson et al., 2002; Lerch et al., 2012). Postmortem MRI brain atlasing has been used to great effect in small animal models (Calabrese et al., 2013; Chuang et al., 2011; Jiang and Johnson, 2011; Johnson et al., 2012; Veraart et al., 2011), but has seen only limited use in nonhuman primates (Ghosh et al., 1994; Hikishima et al., 2012), partially due to the difficulty with high resolution imaging of large specimens (D’Arceuil et al., 2007). In this article we present a high-resolution DTI atlas of the postmortem rhesus macaque brain based on 10 specimens. The atlas consists of eight different MRI image contrasts at microscopic image resolution as well as a volumetric segmentation of 241 anatomic structures. The anatomic variability of atlas specimens has been characterized, and correlated with demographic information. We further demonstrate the use of this atlas for cortical thickness measurements, and white matter modeling with diffusion tractography.
3. Methods
3.1. Brain specimens
The atlas was constructed based on a population of 10 young adult rhesus macaque brains. The sample population included both males (n = 7) and females (n = 3) with ages ranging from 1.8 to 11 years (average = 5.3 ± 2.8 years) and perimortem body weights ranging from 2.2 to 10 kg (average = 5.7 kg ± 2.8). Detailed information on age, weight, and gender is provided in Table 1. The brains were acquired following approved protocols from the Pathology Services and Tissue Distribution unit of the Wisconsin National Primate Research Center. All specimens were obtained at necropsy immediately following euthanasia for non-study related reasons. Brain specimens were extracted from the skull and submerged in a 500 mL solution of 10% formalin with 1% (5 mM) gadoteridol (ProHance, Bracco Imaging, Princeton, NJ) within minutes of euthanasia. After a fixation period of at least 4 weeks, brain specimens were transferred to a 500 mL solution of phosphate buffered saline with 0.5% (2.5 mM) gadoteridol for one week. Immediately prior to imaging, specimens were transferred to MRI compatible tubes and immersed in liquid fluorocarbon (Galden PFPE, Solvay, Brussels, Belgium) for susceptibility matching.
Table 1.
Descriptive information, including age and gender, for the 10 rhesus macaque brain specimens included in the atlas.
| Specimen ID | Sex | Scan Date | Age (years) |
Weight (kg) |
Brain Volume (mL) |
|---|---|---|---|---|---|
| rhesus 01 | M | 16-Dec-2013 | 2.8 | 3.4 | 80.80 |
| rhesus 02 | M | 20-Dec-2013 | 4.9 | 10.0 | 79.55 |
| rhesus 03 | M | 22-Jan-2014 | 5.2 | 8.0 | 77.50 |
| rhesus 04 | F | 27-Jan-2014 | 9.4 | 6.5 | 86.63 |
| rhesus 05 | M | 5-Feb-2014 | 5.2 | 10.0 | 79.00 |
| rhesus 06 | M | 9-Feb-2014 | 3.8 | 3.6 | 84.91 |
| rhesus 07 | M | 20-Feb-2014 | 4.1 | 3.4 | 69.47 |
| rhesus 08 | F | 2-May-2014 | 11.0 | 4.5 | 79.51 |
| rhesus 09 | M | 4-May-2014 | 1.8 | 2.2 | 69.85 |
| rhesus 10 | F | 15-Jun-2014 | 4.8 | 5.0 | 78.30 |
3.2. MRI acquisition
Imaging was performed on a 7 Tesla small animal MRI system (Magnex Scientific, Yarnton, Oxford, UK) controlled with an Agilent console running VnmrJ 4.0 (Agilent Technologies, Santa Clara, CA). Radiofrequency (RF) transmission and reception were achieved with a 65 mm inner-diameter quadrature RF coil (m2m Imaging, Cleveland, OH). Each brain specimen was imaged using two protocols, a high-resolution gradient echo anatomic scan and a DTI series.
Gradient echo anatomic images were acquired with a standard 3D gradient echo pulse sequence (TR/TE = 50/6.2 ms, α = 60°, bandwidth = 62.5 kHz). The acquisition matrix was 1060 × 800 × 680 over a 79.5 mm × 60 mm × 51 mm field of view (FOV) for a voxel size of 75 × 75 × 75 µm3
DTI data were acquired using a 3D diffusion weighted spin echo pulse sequence (TR/TE = 100/21.5 ms, bandwidth = 62.5 kHz). Diffusion preparation was accomplished using a pair of unipolar, half-sine diffusion gradient waveforms (Δ = 14 ms, δ = 4 ms, amplitude = 50 G/cm, b = 1500 s/mm2) on either side of a 2 ms hyperbolic-secant adiabatic inversion (180°) RF pulse. Twelve diffusion weighted images and a single non-diffusion weighted (b=0) were collected for each specimen. Diffusion sensitization vectors were generated using an electrostatic repulsion model and are provided as Supplementary Table 1. The acquisition matrix was 530 × 400 × 340 over a 79.5 mm × 60 mm × 51 mm field of view (FOV) for a voxel size of 150 × 150 × 150 µm3. Total acquisition time was approximately 46 hours per specimen.
3.3. Image processing and registration
All data processing was done on a high-performance computing cluster with 96 physical cores and 1.5 TB of RAM. After initial image reconstruction, the gradient echo image and all 12 diffusion-weighted images were registered to the b=0 image using the Advanced Normalization Tools (ANTs, http://picsl.upenn.edu/software/ants/) 12-parameter affine transform model to correct for the linear component of eddy current distortions. The rotational component of the affine transform for each diffusion-weighted image was applied to the corresponding gradient vector prior to tensor estimation. Diffusion tensors were estimated with Diffusion Toolkit (http://trackvis.org). Isotropic diffusion weighted images (DWI) were generated by taking the average of all 12 diffusion weighted images.
A laterally symmetric average image template was generated using an unbiased minimum deformation template strategy (Kochunov et al., 2001). In short, images were divided along the midline and the left side mirrored across the midline to match the right, resulting in 20 hemisphere images. Each unique pair of hemisphere images (190 pairs in total) was registered using a rigid-body transform followed by a non-linear (diffeomorphic) transform. All transforms from a given image to the rest of the group were averaged to generate a final transform between the individual and the final template. The DWI images were used to drive registration. Non-linear registration employed the ANTs greedy symmetric normalization model with cross-correlation as the image similarity metric (Avants et al., 2008). An iterative, multi-resolution scale-space approach was used with six down-sampling levels ranging from 8× to 1 × and Gaussian smoothing ranging from σ = 1.2 mm to 0 mm. Individual image volumes were transformed into the template space, averaged, and mirrored across the midline to generate the final atlas images. Diffusion tensors were transformed and averaged using Log-Euclidean math operations (Arsigny et al., 2006). Atlas images were rigidly aligned to an orientation consistent with previous work, and image origins were set to the center of the midline crossing of the anterior commissure (Frey et al., 2011; Paxinos et al., 2009). Average atlas templates for all eight image contrasts are available for download at http://www.civm.duhs.duke.edu/rhesusatlas/.
3.4. Anatomic segmentation
Anatomic segmentation was initialized through automated label registration using labels from the Paxinos et al. histology atlas of the rhesus macaque brain (Paxinos et al., 2009). This Nissl-stained histology atlas primarily focuses on forebrain gray matter structures, and features detailed delineations of brain nuclei and neocortical regions. First, digitized label diagrams were interpolated to 3D label volumes with each label represented by a unique integer value. Next, a binary version of the label volume was generated by setting all gray matter labels to one and all non-gray mater labels to zero. A corresponding binary mask of the average brain template was generated by simple thresholding of the gradient echo anatomic image to rapidly segment gray matter from non-gray matter. Finally, the binary label volume was non-linearly registered to the binary template mask, and the resultant transform was used to warp the integer labels into the MRI template space. Labels were then manually corrected with the 3D label editor module of Avizo 8 (FEI Visualization Sciences Group, Burlington, MA) using the Paxinos et al. histology atlas as a reference. The final segmentation consists of 241 structures that comprise the entire 3D volume of the brain (Supplementary Table 2). Labels were organized into a hierarchical ontology based on the Puelles et al. ontology of the mammalian brain (Puelles et al., 2013). The final label volume is available for download at http://www.civm.duhs.duke.edu/rhesusatlas/.
3.5. Voxel-based morphometry
To assess the morphologic variability of the 10 atlas brains, we generated a mean positional difference (MPD) map by taking the average magnitude of the diffeomorphic warp vector fields relating each specimen to the atlas template (Kovacevic et al., 2005). In addition, voxel-based morphometry (VBM) was used to test whether morphometric differences correlated with animal demographic information. VBM was performed in SPM8 (http://www.fil.ion.ucl.ac.uk/spm/software/spm8/) using the log-Jacobian determinants of the diffeomorphic warps after smoothing with a Gaussian kernel of σ = 1 mm. Data were fit to a multiple regression model that included hemisphere (left vs. right), sex, age, and bodyweight as covariates. We identified all voxel clusters with uncorrected p < 0.001 and extent of 10,000 pixels or greater and then corrected for multiple comparisons with the false discovery rate (FDR) method at the cluster level. Statistical significance was determined as FDR corrected p-value (q-value) < 0.05.
3.6. Cortical thickness mapping
Cortical thickness estimates were generated from the average atlas template. A 3D cortex segmentation was generated from the label set described previously by merging all neocortical labels. Cortical thickness maps were calculated from the cortex segmentation using the DiReCT (registration-based) method implemented in ANTs (Tustison et al., 2014). Relevant parameters included a convergence threshold of 10−4, a gradient step size of 0.01, an update field smoothing kernel of σ = 1 mm, and a thickness prior of 5 mm.
3.7. Diffusion tractography
Diffusion tractography was performed on the average atlas tensor volume using DSI Studio (http://dsi-studio.labsolver.org/). We used the streamline (Euler) tracking algorithm with a step size of 75 µm, an angle threshold of 30°, an FA threshold of 0.2, and a minimum length threshold of 5 mm. Seed and target regions were manually drawn for each fiber pathway of interest. In general, a single spherical seed region was placed near the larger end of a pathway, and a single hand-drawn target region was placed near the opposite end. Tractography was then seeded at random from within the seed region until 10,000 tracks were generated. Tracks were manually edited to remove obvious stray fibers.
4. Results
4.1. Atlas templates
We have constructed an MRI/DTI atlas template for the rhesus macaque based on an unbiased average of 10 different postmortem brain image volumes. Total brain volume as measured by MRI ranged from 69.47 to 86.63 mL (average = 78.55 ± 5.49 mL). This atlas covers a wide range of adolescent to adult rhesus macaques. Although the rhesus macaque brain continues to mature until approximately 3 years of age, volumetric changes are generally small after the first 2 years (Malkova et al., 2006). In addition, both standard anatomic (e.g. T1, T2) and DTI contrasts have been shown to stabilize after 3 years of age (Shi et al., 2013).
In total, eight different image contrasts are included in the average atlas (Fig. 1). Each image contrast is 3D, isotropic, and precisely aligned to the others, and each highlights different anatomic features. Table 2 provides the description, abbreviation, and resolution of each different image contrast. At 75 µm and 150 µm isotropic resolution respectively, the gradient echo and DTI templates are the highest spatial resolution rhesus brain MRI templates ever reported in the scientific literature. The combination of high-resolution, multiple image contrasts, and population averaging allows identification of many brain regions that would not be possible otherwise (Calabrese et al., 2012). In particular, population averaging can provide increased signal-to-noise ratio (SNR) and increased contrast-to-noise ratio (CNR) relative to single-subject images (Fig. 2), thus improving the ability to identify subtle anatomic features (Johnson et al., 2012). Importantly, both the GRE and b=0 contrasts from this atlas may differ substantially from analogous in vivo contrasts because of differential distribution of gadolinium in fixed gray/white matter and differences in pulse sequence timing.
Figure 1.
The eight different image contrasts included in the average atlas. For each contrast, a single axial slice through the basal ganglia is shown. GRE, gradient recalled echo; b=0, non-diffusion weighted image; DWI, isotropic diffusion weighted image; MD, mean diffusivity; RD, radial diffusivity; AD, axial diffusivity; FA, fractional anisotropy; FAC, directionally colored fractional anisotropy.
Table 2.
Abbreviations, descriptions, and isotropic resolution of the eight different image contrasts included in the atlas.
| Abbreviation | Description | Isotropic resolution |
|---|---|---|
| GRE | T2*-weighted gradient echo | 75 µm |
| b=0 | T2-weighted non-diffusion spin echo | 150 µm |
| DWI | Isotropic (average) diffusion weighted image | 150 µm |
| AD | Axial diffusivity | 150 µm |
| RD | Radial diffusivity | 150 µm |
| MD | Mean diffusivity | 150 µm |
| FA | Fractional anisotropy | 150 µm |
| FAC | Directionally colored fractional anisotropy | 150 µm |
Figure 2.
Comparison of single-subject vs. group average gradient echo anatomic images. A coronal slice through the hippocampus shows differences between a single-subject image (left half) and the group average image (right). Several anatomic structures are more apparent in the group average image (yellow arrowheads). In addition, some image artifacts are reduced in the group image due to averaging (red arrowheads). SLF II, superior longitudinal fasciculus II; Cl, claustrum; MD, mediodorsal thalamic nucleus; LGN, lateral geniculate nucleus; SNVL, substantia nigra compact part, ventrolateral division.
4.2. Anatomic labels
The final atlas segmentation contains 241 anatomic labels, including the entire complement of neocortical labels from the Paxinos et al. histology atlas of the rhesus brain (Fig. 3). The label nomenclature follows that of the Paxinos et al. atlas, and all structures have been organized into a consistent ontology based on embryologic origins (Puelles et al., 2013). Neocortical labels have been further organized into seven major cortical regions: cingulate cortex, insular cortex, occipital cortex, parietal cortex, temporal cortex, frontal cortex, and orbitofrontal cortex. While these large cortical classifications are somewhat arbitrary, they are still widely used to describe gross brain structure, and have been included here to aid in organization. We have also annotated the atlas template with more traditional gyri and sulci nomenclature, which may be more familiar to some researchers (Fig. 4, A).
Figure 3.
Demonstration of the atlas anatomic labels in two dimensions in the coronal plane. Five different coronal slices are presented as indicated by the slice diagram (top left). Adjacent slices are 7.5 mm apart. Each slice shows anatomic labels as red outlines (left half of images) and as semi-transparent color overlays (right half of images).
Figure 4.
Demonstration of the atlas anatomic labels in three dimensions. (A) Lateral (top) and dorsal (bottom) views of the atlas brain surface. Major gyri and sulci are labeled for reference. (B) Lateral (top) and dorsal (bottom) views of atlas label surface renderings. Each different anatomic region is displayed with a different color surface. (C) Lateral (top) and dorsal (bottom) views of a surface rendering of the cerebral white matter tree (red). A semi-transparent rendering of the brain surface is included for reference. (D) Lateral (top) and posterior (bottom) views of surface renderings of five major subcortical structures: putamen (blue), caudate (yellow), hippocampus (green), external globus pallidus (teal), internal globus pallidus (orange). AG, angular gyrus; aspd, anterior supraprincipal dimple; ASPG, anterior superior parietal gyrus; cs, central sulcus; ecal, external calcarine sulcus; iar, inferior arcuate sulcus; IFG, inferior frontal gyrus; ios, inferior occipital sulcus; ipd, infraprincipal dimple; ips, intraparietal sulcus; lf, lateral fissure; lu, lunate sulcus; MFG, middle frontal gyrus; MG, marginal gyrus; MTG, middle temporal gyrus; OG, occipital gyrus; OTG, occipito-temporal gyrus; pos, parietooccipital sulcus; PrCG, precentral gyrus; PSPG, posterior superior parietal gyrus; sar, superior arcuate sulcus; SFG, superior frontal gyrus; spcd, superior precentral dimple; STG, superior temporal gyrus; sts, superior temporal sulcus; su, superior postcentral dimple; TPG, temporal polar gyrus.
The complete list of atlas structures, as well as their abbreviations and ontological classification are provided as Supplementary Table 2. These labels cover the entire 3D volume of the brain without any unlabeled brain regions (Fig. 4, B). As a result, a small number of “catch-all” labels were created such as “cerebral white matter,” which contains all major white matter structures in the cerebrum (Fig. 4, C). The focus of this segmentation was primarily on forebrain structures, so the midbrain and hindbrain have considerably less detailed anatomic delineations. Anatomic labels have been manually corrected in three orthogonal planes to ensure volumetric accuracy, and to allow accurate 3D rendering of brain structures (Fig. 4, D). Manual correction resulted in considerably smoother, and more anatomically accurate 3D labels (Supplementary Fig. 1, A–C). In some cases, small subdivisions from the Paxinos atlas were combined into their parent structures. For example, the superior and inferior portions of the ventral cortical amygdaloid nucleus were combined into a single structure. Six atlas structures have two distinct, disconnected regions on each side of the brain: area 11, area 13, visual area 3 dorsal, visual area 3 ventral, secondary somatosensory cortex, and juxtastriate area. This is consistent with the Paxinos atlas where these structures also have two distinct, disconnected parts.
The 3D, isotropic nature of these labels makes them suitable for automated atlas-based segmentation of 2D (multi-slice) MR images of the rhesus brain acquired in any arbitrary plane, assuming that morphometric and image contrast differences can be adequately accounted for with automated image registration. Additionally, because of the hierarchical organization of the labels, smaller structures can be collapsed into their larger parent structures to accommodate different resolution scales. The complete 3D label volume, as well as atlas templates for all eight image contrasts are available for download and online visualization at http://www.civm.duhs.duke.edu/rhesusatlas/.
4.3. Morphometric variations in the atlas
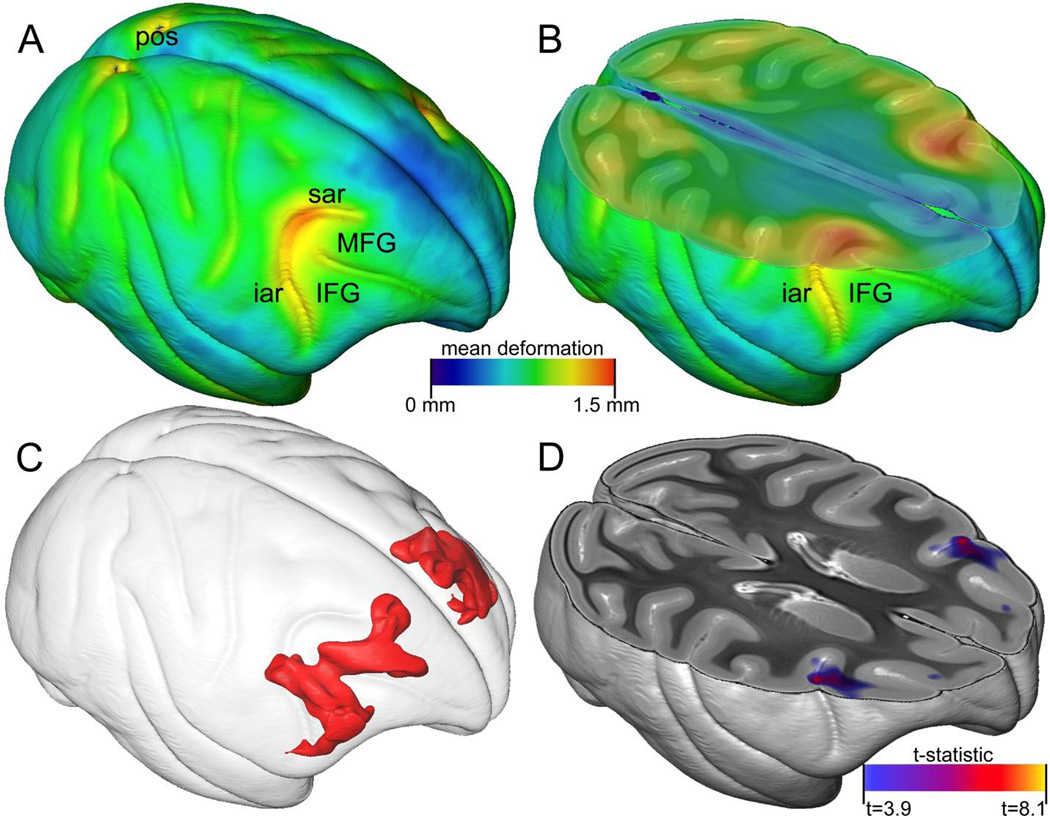
To assess morphometric variability in the atlas specimens, we generated mean positional difference (MPD) maps, which show the average voxelwise deformation in millimeters required to transform individual images to the atlas template (Fig. 5, A). Overall morphologic variability was quite low, with an average MPD across all brain voxels of only 0.74 mm and a maximum MPD of 1.43 mm. The maximum positional difference of any individual image voxel was 3.61 mm. Moderate MPD values (~1 mm) were detected in the parietooccipital sulcus and the occipital gyri, but the highest values were observed near the inferior and middle frontal gyri and the arcuate sulcus (Fig. 5, A–B). This area of the frontal cortex is known to be a highly variable in rhesus macaques, and a number have studies have demonstrated morphologic changes in this region that correlate with animal age (Koo et al., 2012; Luebke et al., 2010; Peters et al., 1994; Wisco et al., 2008a). We used voxel-based morphometry with multiple regression analysis to test whether the observed morphologic variability correlated with demographic features of atlas animals. Of the covariates analyzed (age, weight, sex, and hemisphere), only age showed a significant correlation that survived multiple comparison correction. The largest significant voxel cluster for the age correlation co-localized with the highest observed MPD values in the middle and inferior frontal gyri (Fig. 5, C–D). These results indicate that the volume of the middle and inferior frontal gyri, relative to total brain volume, decreases with age across the range studied in the atlas (1.8 to 11 years). These results are consistent with previous observations in rhesus macaques and humans where reduction in prefrontal cortex volume is a commonly identified characteristic of brain maturation and aging, and is sometimes associated with cognitive decline in older adults (Luebke et al., 2010; Tisserand et al., 2002; Wellington et al., 2013; Wisco et al., 2008a). In this case, due to the relatively young age of the animals included in this study, it seems likely that the observed changes are related to brain maturation and development rather than cognitive decline. We found no differences related to brain hemisphere (left vs. right) in this area, even as an interaction with age, suggesting that this effect is bilateral. DTI parameters (FA, AD, RD, MD) were also investigated, but we found no significant correlation with animal age, weight, sex, or hemisphere.
Figure 5.
Morphologic variation in the atlas. (A) A volume rendering of the mean positional difference (MPD) map generated from atlas diffeomorphic transforms. The color scale for the MPD map is shown in the center of the Figure. Relevant gyri and sulci are labeled for reference. (B) A volume rendered MPD map similar to (A), sliced axially through the arcuate sulcus. (C) Surface rendering of statistically significant voxel clusters for the voxel-based morphometry age correlation (false discovery rate adjusted, q<0.05). (D) t-statistic map for the voxel-based morphometry age correlation. The t-statistic color scale is shown in bottom right of Figure. Only regions that survived multiple comparison correction are colored. IFG, inferior frontal gyrus; MFG, middle frontal gyrus; iar, inferior arcuate sulcus; sar, superior arcuate sulcus.
4.4. Cortical thickness measurements
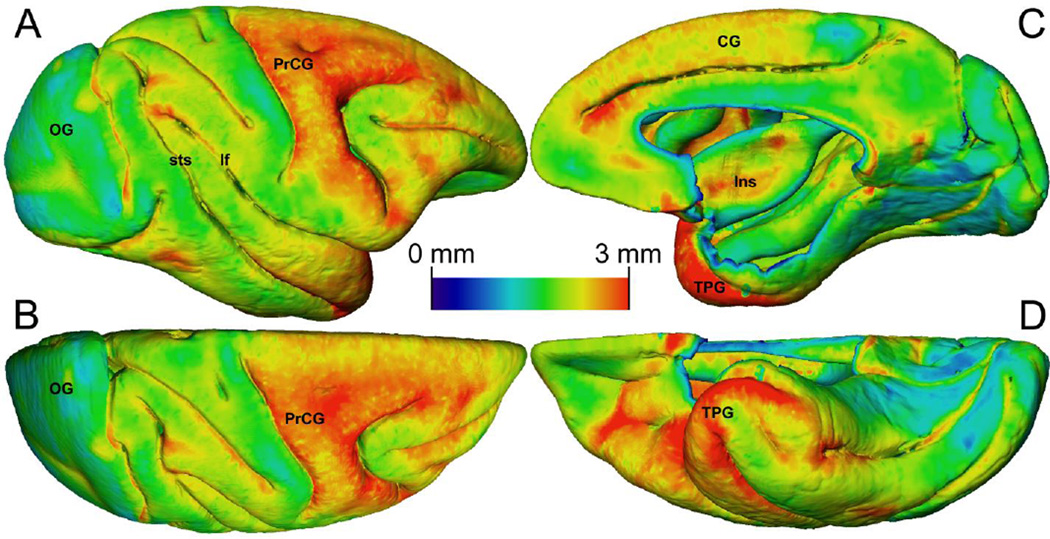
Automated cortical thickness mapping software has been shown to produce robust and reproducible estimates of cortical thickness even in the presence of complex gyral structure. In particular, the DiReCT method, implemented in ANTs has proven to be quite reliable, and requires nothing more than a simple gray/white segmentation of the brain volume (Tustison et al., 2014). This volume-based cortical thickness estimation technique was applied directly to the average atlas template to generate cortex-wide thickness estimates (Fig. 6). These data show a general decreasing thickness gradient from rostral to caudal, consistent with previously published results (Koo et al., 2012). The precentral gyrus (Fig. 6, A–B) and temporal polar gyrus (Fig. 6, C–D) gave the highest thickness estimates with values near 3 mm, and the occipital gyrus (Fig. 6, A–B) was the thinnest with some portions being only 1 mm thick. These data also allow visualization of the cingulate gyrus and insular cortex (Fig. 6, C), which are normally obscured by the contralateral side of the brain. Some small regions appear artificially thicker than expected due to limitations of the DiReCT method (Das et al., 2009). For example, a small area at the intersection of the lateral fissure and superior temporal sulcus appears thicker than surrounding areas (Fig. 6, A), but this is likely due to registration error during DiReCT processing rather than a true thickness difference. Such errors could be avoided by using a two-surface Laplacian-based thickness estimate (S. E. Jones et al., 2000), which will be an area of future work for these data.
Figure 6.
Automated cortical thickness estimates for the average atlas generated using the DiReCT method. Thickness estimates are displayed as a colored surface rendering of the cortex. The color scale for cortical thickness estimates is shown in the center of the Figure. Relevant gyri and sulci are labeled for reference. (A) Lateral view. (B) Dorsal view. (C) Medial (midline) view. (D) Ventral view. CG, cingulate gyrus; Ins, insular cortex; lf, lateral fissure; OG, occipital gyrus; PrCG, precentral gyrus; sts, superior temporal sulcus; TPG, temporal polar gyrus.
4.5. Tractography
Diffusion tractography was used for 3D white matter pathway modeling and segmentation. In total, we segmented 42 major white matter tracks including 24 midline pathways and 18 lateralized pathways on either side of the brain. The complete list of tracks and their abbreviations are included as Supplementary Table 3. Midline pathways include the corpus callosum, which has been divided into seven segments based on the Witelson classification (Witelson, 1989) (Fig. 7, A), as well as the inferior, middle, and superior cerebellar peduncles (Fig. 7, B). Lateralized pathways include several cortical association pathways like the cingulum, middle longitudinal fasciculus, inferior longitudinal fasciculus, and uncinate fasciculus (Fig. 7, C). Some white matter pathways were difficult to reconstruct due to the limitations of the diffusion tensor model, such as the arcuate fasciculus and the three superior longitudinal fasciculi, so these pathways were omitted from the atlas. Despite these shortcomings, the high spatial resolution of the data allowed tracking of several smaller pathways that are difficult to identify in lower resolution data, such as the cranial nerves, the fasciculus retroflexus, and the stria medullaris (Supplementary Fig. 2, A–F). All tractography data are available for download in TrackVis and Visualization Toolkit formats at http://www.civm.duhs.duke.edu/rhesusatlas/.
Figure 7.
Atlas tractography results. (A) Corpus callosum tractography divided into seven segments following the Witelson classification: genu (red), segment I (orange), segment II (yellow), segment III (green), segment IV (light blue), segment V (dark blue), splenium (purple). (B) Inferior (blue), middle (red), and superior cerebellar peduncles (green). (C) Cortical association pathways including: cingulum (red), middle longitudinal fasciculus (green), uncinate fasciculus (blue), and inferior longitudinal fasciculus (yellow).
5. Discussion
5.1. Postmortem brain atlasing
Compared to previously published atlases of the rhesus macaque brain, this work has two major differences. First, these data were generated from considerably higher resolution images, and are therefore likely to be more anatomically precise than previous atlases. For example, the DTI data presented here are volumetrically over 800 times higher resolution than a recently published in vivo DTI atlas of the rhesus brain (Adluru et al., 2012). This increased precision comes at the cost of the second important difference, which is a potential loss of accuracy from imaging of fixed, postmortem specimens. Both brain extraction and formalin fixation are known to affect total brain volume. However, these effects can be counteractive, and one study of human brain specimens found the net brain volume reduction from a 70-day fixation in 10% formalin to be only 3.3% ± 0.5% (Schulz et al., 2011). It has also been suggested that brain shrinkage from formalin fixation is both non-uniform and anisotropic, which could further influence these results (Quester and Schröder, 1997). Nonetheless, the total brain volume measurements from our atlas are quite similar to established values for the rhesus macaque. For example, total brain volume measurements from two previously published in vivo MRI atlases of the rhesus brain are both within one standard deviation of our measurements (Adluru et al., 2012; Frey et al., 2011). Our cortical thickness measurements are also very comparable to in vivo results using similar methods (Koo et al., 2012). While volume changes from fixation should certainly be considered when using these data, we do not believe it will significantly reduce the utility of the atlas, particularly for atlas-based segmentation where global volume differences are easily accounted for by affine registration.
The potential effects of tissue fixation are perhaps more important when considering diffusion tensor data. Formalin fixation produces an approximately 4-fold reduction in the diffusivity of water in brain tissues (Alexander et al., 2010; Dyrby et al., 2011). As a result, DTI derived scalars, such as MD, AD, and RD are typically 4-fold lower than in in vivo DTI data. Although reports vary to some degree, FA appears to be preserved in postmortem DTI, suggesting that diffusivity reductions are uniform and proportional (D’Arceuil and de Crespigny, 2007; Sun et al., 2005; Widjaja et al., 2009). Another consequence of the diffusivity reduction from fixation is that 4-fold larger b-values are required to achieve in vivo equivalent diffusion weighting. Reduced diffusion weighting can negatively impact tensor estimation accuracy, particularly with regard to diffusion tractography. Fortunately, postmortem diffusion imaging offers several image quality improvements that improve the ability to estimate diffusion tensors (Lerch et al., 2012). Postmortem studies typically have higher SNR, are not complicated by physiologic/subject motion, and avoid EPI-induced eddy current distortions, each of which affects tensor estimation (D. K. Jones, 2004; Le Bihan et al., 2006; Skare and Andersson, 2001). Further, combining diffusion tensors from multiple subjects, as was done for this atlas, can reduce tensor errors through averaging. Despite these considerations, atlas tractography results would likely be further improved by using a higher b-value. Unfortunately, this was not possible for the current study due to the high hardware requirements of postmortem diffusion imaging.
5.2. The average atlas
In constructing this atlas, we attempted to combine the large body of existing neuroanatomical data for the rhesus brain with modern high-resolution brain image techniques. By using the anatomic labels from the Paxinos et al. histology atlas of the rhesus brain as a starting point, we hope to facilitate multi-modality anatomic comparisons and the use of a standardized nomenclature. We have also taken care to organize the 241 atlas structures into a developmentally defined ontology, which allows direct comparison of homologous brain regions in virtually any mammalian species, including mice and humans. All data are provided in the NIfTI standard data format, and are oriented in accordance with previously published MRI atlases of the rhesus brain (Frey et al., 2011). The stereotaxic coordinate system and image origin of the atlas have also been borrowed from previous work to improve interoperability, and allow direct comparisons between atlases (Frey et al., 2011; Paxinos et al., 2009).
This atlas builds on previous work with the addition of high-resolution, multi-contrast, 3D image data, cortical thickness estimates, and tractography. The imaging protocols and labeling scheme were developed to be above and beyond what is common in the rhesus brain in order to be both comprehensive and flexible enough to accommodate future advances in brain imaging technology. The 3D isotropic nature of the image data and the hierarchical organization of the anatomic labels also allow the atlas to be sub-sampled to accommodate lower resolution datasets. Regional cortical thickness measurements, like those presented here, are a commonly used biomarker that can be extracted from MR images (Fischl and Dale, 2000). Several studies in both macaques and humans have demonstrated correlations between cortical thickness measurements and aging, behaviors, or the presence of underlying neurologic disease (Hardan et al., 2006; Koo et al., 2012; Lerch et al., 2008). DTI tractography has been widely used in both humans and macaques to explore white matter structure and organization (Adluru et al., 2012; Catani and Thiebaut de Schotten, 2008; Oishi et al., 2010). Computer generated tracks can also be used as a framework for statistical comparisons of local DTI parameters in disease models (Corouge et al., 2006; Goodlett et al., 2008). Unfortunately, diffusion tractography is inherently and practically limited in its ability to generate anatomically accurate tracks, particularly within the confines of the diffusion tensor model (Calabrese et al., 2014; Thomas et al., 2014). The tractography data presented here should be interpreted within the context of these limitations, but are nonetheless representative of the diffusion tractography data that are commonly derived from DTI studies of the rhesus macaque brain. Both the high spatial resolution and the population averaging of tensor data in our atlas may serve to improve the accuracy of tractography results relative to previously published data, but neither addresses the fundamental limitations of tractography.
These data also demonstrate the ability to use MRI-based brain atlases to test anatomic theories related to growth, aging, sex, and brain lateralization. Although the population of animals use for this study was both relatively small and unevenly distributed in terms of demographics (e.g. age, sex), we were still able to detect statistically significant correlations between morphometric parameters and age. It is likely that the ability to detect such anatomic correlations would improve significantly with larger n and/or a more evenly distributed population of animals. For example, a wider range of ages would allow identification of potential non-linear dependencies between age and anatomic parameters. Similarly, a more balanced gender ratio would provide additional sensitivity for detecting sex differences. Incorporating additional datasets into the atlas, including multi-modal image data from other sources, could prove useful for testing other, more complex anatomic hypotheses.
5.3. Data sharing and future work
All atlas data, including average image data and derived datasets, are available for online visualization and download at http://www.civm.duhs.duke.edu/rhesusatlas/. The entire collection of atlas data is approximately 6 GB in size, but each component can be downloaded individually as needed. Future work pertaining to the atlas will be added as it becomes available. For example, two-surface Laplacian-based cortical thickness calculations are currently in development, and are expected to be added to the atlas by late-2015. There are several other areas for potential future work relating to the atlas. First, several of the specimens used for the atlas were also imaged with high angular resolution diffusion imaging (HARDI) and diffusion spectrum imaging (DSI) protocols, as part of a separate study (Calabrese et al., 2014). The improved tractography results from these specialized datasets could be transformed into the atlas space and incorporated into this work. Second, we have compiled a similar collection of brain specimens of the closely related cynomolgus or crab-eating macaque, Macaca fascicularis, which is now frequently being used in neuroscience and pharmaceutical research. Construction of a similar atlas of the cynomolgus brain could provide interesting insights into the behavioral and functional consequences of neuroanatomical differences between these two macaque species, as suggested by a previous MRI brain atlasing study (Frey et al., 2011). Finally, the addition of conventional histology and immunohistochemical studies is possible because of the non-destructive nature of postmortem MRI. As with any brain atlas, the value of our resource will become more meaningful if utilized by others in the scientific community. We hope that future use of these data by collaborators and other researchers will help drive refinements and improvements to the atlas.
Supplementary Material
Supplementary Figure 1. Manual correction and smoothing of anatomic labels in three dimensions. Surface renderings of five major subcortical structures are shown within a semi-transparent rendering of the whole brain: putamen (blue), caudate (yellow), hippocampus (green), external globus pallidus (teal), and internal globus pallidus (orange). Pre-correction labels are shown on the left side of the brain, and post-correction labels are shown on the right side of the brain. (A) Anterior view. (B) Posterior view. (C) Dorsal view.
Supplementary Figure 2. Tractography of small white matter pathways afforded by high spatial resolution DTI. (A) Cranial nerve III. (B) Cranial nerve IV. (C) Cranial nerve VII. (D) Cranial nerve VIII. (E) Fasciculus retroflexus. (F) Stria medullaris. All background images are gradient echo anatomic images in the coronal plane except for (F), which is in the axial plane.
Supplementary Table 1. The 12-direction diffusion gradient encoding scheme used for diffusion tensor MRI acquisition.
Supplementary Table 2. The complete list of atlas anatomic labels, including abbreviations and hierarchical organization.
Supplementary Table 3. The complete list of atlas tractography structures, including abbreviations and hierarchical organization.
Highlights.
We present a high-resolution DTI/MRI atlas of 10 postmortem rhesus macaque brains.
The atlas includes 3D segmentations of 241 brain regions, and 42 tracts.
We analyze morphometric variation and cortical thickness across the atlas group.
Acknowledgements
This work was supported by several National Institutes of Health grants: NIBIB grant number P41 EB015897, NA-MIC Roadmap for Medical Research grant number U54 EB005149-01, NIMH grant number R01 MH091645, NICHD grant number U54 HD079124, and NIA grant number K01 AG041211 to AB.
The invaluable assistance of the pathology unit of the Wisconsin National Primate Research Center in providing brain specimens is gratefully acknowledged (grant number P51 OD011106).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adluru N, Zhang H, Fox AS, Shelton SE, Ennis CM, Bartosic AM, Oler JA, Tromp DPM, Zakszewski E, Gee JC, Kalin NH, Alexander AL. A diffusion tensor brain template for rhesus macaques. Neuroimage. 2012;59:306–318. doi: 10.1016/j.neuroimage.2011.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander DC, Hubbard PL, Hall MG, Moore EA, Ptito M, Parker GJM, Dyrby TB. Orientationally invariant indices of axon diameter and density from diffusion MRI. Neuroimage. 2010;52:1374–1389. doi: 10.1016/j.neuroimage.2010.05.043. [DOI] [PubMed] [Google Scholar]
- Arsigny V, Fillard P, Pennec X, Ayache N. Log-Euclidean metrics for fast and simple calculus on diffusion tensors. Magn Reson Med. 2006;56:411–421. doi: 10.1002/mrm.20965. [DOI] [PubMed] [Google Scholar]
- Avants BB, Epstein CL, Grossman M, Gee JC. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med Image Anal. 2008;12:26–41. doi: 10.1016/j.media.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basser PJ, Pierpaoli C. Microstructural and physiological features of tissues elucidated by quantitative-diffusion-tensor MRI. J Magn Reson B. 1996;111:209–219. doi: 10.1006/jmrb.1996.0086. [DOI] [PubMed] [Google Scholar]
- Calabrese E, Badea A, Coe CL, Lubach GR, Styner MA, Johnson GA. Investigating the tradeoffs between spatial resolution and diffusion sampling for brain mapping with diffusion tractography: Time well spent? Hum Brain Mapp. 2014 doi: 10.1002/hbm.22578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese E, Badea A, Watson C, Johnson GA. A quantitative magnetic resonance histology atlas of postnatal rat brain development with regional estimates of growth and variability. Neuroimage. 2013;71C:196–206. doi: 10.1016/j.neuroimage.2013.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese E, Johnson GA, Watson C. An ontology-based segmentation scheme for tracking postnatal changes in the developing rodent brain with MRI. Neuroimage. 2012;67C doi: 10.1016/j.neuroimage.2012.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catani M, Thiebaut de Schotten M. A diffusion tensor imaging tractography atlas for virtual in vivo dissections. Cortex. 2008;44:1105–1132. doi: 10.1016/j.cortex.2008.05.004. [DOI] [PubMed] [Google Scholar]
- Chuang N, Mori S, Yamamoto A, Jiang H, Ye X, Xu X, Richards LJ, Nathans J, Miller MI, Toga AW, Sidman RL, Zhang J. An MRI-based atlas and database of the developing mouse brain. Neuroimage. 2011;54:80–89. doi: 10.1016/j.neuroimage.2010.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corouge I, Fletcher PT, Joshi S, Gouttard S, Gerig G. Fiber tract-oriented statistics for quantitative diffusion tensor MRI analysis. Med Image Anal. 2006;10:786–798. doi: 10.1016/j.media.2006.07.003. [DOI] [PubMed] [Google Scholar]
- D’Arceuil H, de Crespigny A. The effects of brain tissue decomposition on diffusion tensor imaging and tractography. Neuroimage. 2007;36:64–68. doi: 10.1016/j.neuroimage.2007.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Arceuil HE, Westmoreland S, de Crespigny AJ. An approach to high resolution diffusion tensor imaging in fixed primate brain. Neuroimage. 2007;35:553–565. doi: 10.1016/j.neuroimage.2006.12.028. [DOI] [PubMed] [Google Scholar]
- Das SR, Avants BB, Grossman M, Gee JC. Registration based cortical thickness measurement. Neuroimage. 2009;45:867–879. doi: 10.1016/j.neuroimage.2008.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubach MF, Bowden DM. BrainInfo online 3D macaque brain atlas: a database in the shape of a brain. Presented at the Society for Neuroscience Annual Meeting; Chicago, IL. 2009. [Google Scholar]
- Dyrby TB, Baaré WFC, Alexander DC, Jelsing J, Garde E, Søgaard LV. An ex vivo imaging pipeline for producing high-quality and high-resolution diffusion-weighted imaging datasets. Hum Brain Mapp. 2011;32:544–563. doi: 10.1002/hbm.21043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Dale AM. Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proc Natl Acad Sci USA. 2000;97:11050–11055. doi: 10.1073/pnas.200033797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey S, Pandya DN, Chakravarty MM, Bailey L, Petrides M, Collins DL. An MRI based average macaque monkey stereotaxic atlas and space (MNI monkey space) Neuroimage. 2011;55:1435–1442. doi: 10.1016/j.neuroimage.2011.01.040. [DOI] [PubMed] [Google Scholar]
- Ghosh P, O’Dell M, Narasimhan PT, Fraser SE, Jacobs RE. Mouse lemur microscopic MRI brain atlas. Neuroimage. 1994;1:345–349. doi: 10.1006/nimg.1994.1019. [DOI] [PubMed] [Google Scholar]
- Goodlett CB, Fletcher PT, Gilmore JH, Gerig G. Group statistics of DTI fiber bundles using spatial functions of tensor measures. Med Image Comput Comput Assist Interv. 2008;11:1068–1075. doi: 10.1007/978-3-540-85988-8_127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardan AY, Muddasani S, Vemulapalli M, Keshavan MS, Minshew NJ. An MRI study of increased cortical thickness in autism. Am J Psychiatry. 2006;163:1290–1292. doi: 10.1176/appi.ajp.163.7.1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herculano-Houzel S. The human brain in numbers: a linearly scaled-up primate brain. Front Hum Neurosci. 2009;3:31. doi: 10.3389/neuro.09.031.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikishima K, Sawada K, Murayama AY, Komaki Y, Kawai K, Sato N, Inoue T, Itoh T, Momoshima S, Iriki A, Okano HJ, Sasaki E, Okano H. Atlas of the developing brain of the marmoset monkey constructed using magnetic resonance histology. Neuroscience. 2012 doi: 10.1016/j.neuroscience.2012.09.053. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Johnson GA. Microscopic diffusion tensor atlas of the mouse brain. Neuroimage. 2011;56:1235–1243. doi: 10.1016/j.neuroimage.2011.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson GA, Calabrese E, Badea A, Paxinos G, Watson C. A multidimensional magnetic resonance histology atlas of the Wistar rat brain. Neuroimage. 2012;62:1848–1856. doi: 10.1016/j.neuroimage.2012.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson GA, Cofer GP, Fubara B, Gewalt SL, Hedlund LW, Maronpot RR. Magnetic resonance histology for morphologic phenotyping. J Magn Reson Imaging. 2002;16:423–429. doi: 10.1002/jmri.10175. [DOI] [PubMed] [Google Scholar]
- Jones DK. The effect of gradient sampling schemes on measures derived from diffusion tensor MRI: a Monte Carlo study. Magn Reson Med. 2004;51:807–815. doi: 10.1002/mrm.20033. [DOI] [PubMed] [Google Scholar]
- Jones SE, Buchbinder BR, Aharon I. Three-dimensional mapping of cortical thickness using Laplace’s equation. Hum Brain Mapp. 2000;11:12–32. doi: 10.1002/1097-0193(200009)11:1<12::AID-HBM20>3.0.CO;2-K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochunov P, Lancaster JL, Thompson P, Woods R, Mazziotta J, Hardies J, Fox P. Regional spatial normalization: toward an optimal target. J Comput Assist Tomogr. 2001;25:805–816. doi: 10.1097/00004728-200109000-00023. [DOI] [PubMed] [Google Scholar]
- Konrad K, Eickhoff SB. Is the ADHD brain wired differently? A review on structural and functional connectivity in attention deficit hyperactivity disorder. Hum Brain Mapp. 2010;31:904–916. doi: 10.1002/hbm.21058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo B-B, Schettler SP, Murray DE, Lee J-M, Killiany RJ, Rosene DL, Kim D-S, Ronen I. Age-related effects on cortical thickness patterns of the Rhesus monkey brain. Neurobiol Aging. 2012;33:200.e23–200.e31. doi: 10.1016/j.neurobiolaging.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacevic N, Henderson JT, Chan E, Lifshitz N, Bishop J, Evans AC, Henkelman RM, Chen XJ. A three-dimensional MRI atlas of the mouse brain with estimates of the average and variability. Cereb Cortex. 2005;15:639–645. doi: 10.1093/cercor/bhh165. [DOI] [PubMed] [Google Scholar]
- Kubicki M, McCarley R, Westin C-F, Park H-J, Maier S, Kikinis R, Jolesz FA, Shenton ME. A review of diffusion tensor imaging studies in schizophrenia. J Psychiatr Res. 2007;41:15–30. doi: 10.1016/j.jpsychires.2005.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bihan D, Poupon C, Amadon A, Lethimonnier F. Artifacts and pitfalls in diffusion MRI. J Magn Reson Imaging. 2006;24:478–488. doi: 10.1002/jmri.20683. [DOI] [PubMed] [Google Scholar]
- Lerch JP, Gazdzinski L, Germann J, Sled JG, Henkelman RM, Nieman BJ. Wanted dead or alive? The tradeoff between in-vivo versus ex-vivo MR brain imaging in the mouse. Frontiers in Neuroinformatics. 2012;6:6. doi: 10.3389/fninf.2012.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerch JP, Pruessner J, Zijdenbos AP, Collins DL, Teipel SJ, Hampel H, Evans AC. Automated cortical thickness measurements from MRI can accurately separate Alzheimer’s patients from normal elderly controls. Neurobiol Aging. 2008;29:23–30. doi: 10.1016/j.neurobiolaging.2006.09.013. [DOI] [PubMed] [Google Scholar]
- Luebke J, Barbas H, Peters A. Effects of normal aging on prefrontal area 46 in the rhesus monkey. Brain Res Rev. 2010;62:212–232. doi: 10.1016/j.brainresrev.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkova L, Heuer E, Saunders RC. Longitudinal magnetic resonance imaging study of rhesus monkey brain development. Eur J Neurosci. 2006;24:3204–3212. doi: 10.1111/j.1460-9568.2006.05175.x. [DOI] [PubMed] [Google Scholar]
- McLaren DG, Kosmatka KJ, Oakes TR, Kroenke CD, Kohama SG, Matochik JA, Ingram DK, Johnson SC. A population-average MRI-based atlas collection of the rhesus macaque. Neuroimage. 2009;45:52–59. doi: 10.1016/j.neuroimage.2008.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori S, Crain BJ, Chacko VP, van Zijl PC. Three-dimensional tracking of axonal projections in the brain by magnetic resonance imaging. Ann Neurol. 1999;45:265–269. doi: 10.1002/1531-8249(199902)45:2<265::aid-ana21>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Mukherjee P, Berman JI, Chung SW, Hess CP, Henry RG. Diffusion tensor MR imaging and fiber tractography: theoretic underpinnings. AJNR Am J Neuroradiol. 2008;29:632–641. doi: 10.3174/ajnr.A1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi K, Faria AV, van Zijl PCM, Mori S. MRI Atlas of Human White Matter. 2nd ed. San Diego: Academic Press; 2010. [Google Scholar]
- Oishi K, Zilles K, Amunts K, Faria A, Jiang H, Li X, Akhter K, Hua K, Woods R, Toga AW, Pike GB, Rosa-Neto P, Evans A, Zhang J, Huang H, Miller MI, van Zijl PCM, Mazziotta J, Mori S. Human brain white matter atlas: identification and assignment of common anatomical structures in superficial white matter. Neuroimage. 2008;43:447–457. doi: 10.1016/j.neuroimage.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos G, Huang X-F, Toga AW. The Rhesus Monkey Brain in Stereotaxic Coordinates. 2nd ed. San Diego: Academic Press; 2009. [Google Scholar]
- Peters A, Leahu D, Moss MB, McNally KJ. The effects of aging on area 46 of the frontal cortex of the rhesus monkey. Cereb Cortex. 1994;4:621–635. doi: 10.1093/cercor/4.6.621. [DOI] [PubMed] [Google Scholar]
- Puelles L, Harrison M, Paxinos G, Watson C. A developmental ontology for the mammalian brain based on the prosomeric model. Trends Neurosci. 2013;36:570–578. doi: 10.1016/j.tins.2013.06.004. [DOI] [PubMed] [Google Scholar]
- Quester R, Schröder R. The shrinkage of the human brain stem during formalin fixation and embedding in paraffin. J Neurosci Methods. 1997;75:81–89. doi: 10.1016/s0165-0270(97)00050-2. [DOI] [PubMed] [Google Scholar]
- Rohlfing T, Kroenke CD, Sullivan EV, Dubach MF, Bowden DM, Grant KA, Pfefferbaum A. The INIA19 Template and NeuroMaps Atlas for Primate Brain Image Parcellation and Spatial Normalization. Frontiers in Neuroinformatics. 2012;6:27. doi: 10.3389/fninf.2012.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleem KS, Logothetis NK. A Combined MRI and Histology Atlas of the Rhesus Monkey Brain in Stereotaxic Coordinates. San Diego: Academic Press; 2012. [Google Scholar]
- Schmahmann JD, Pandya D. Fiber Pathways of the Brain. New York: Oxford University Press; 2009. [Google Scholar]
- Schulz G, Crooijmans HJA, Germann M, Scheffler K, Müller-Gerbl M, Müller B. Three-dimensional strain fields in human brain resulting from formalin fixation. J Neurosci Methods. 2011;202:17–27. doi: 10.1016/j.jneumeth.2011.08.031. [DOI] [PubMed] [Google Scholar]
- Shi Y, Short SJ, Knickmeyer RC, Wang J, Coe CL, Niethammer M, Gilmore JH, Zhu H, Styner MA. Diffusion tensor imaging-based characterization of brain neurodevelopment in primates. Cereb Cortex. 2013;23:36–48. doi: 10.1093/cercor/bhr372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skare S, Andersson JL. On the effects of gating in diffusion imaging of the brain using single shot EPI. Magn Reson Imaging. 2001;19:1125–1128. doi: 10.1016/s0730-725x(01)00415-5. [DOI] [PubMed] [Google Scholar]
- Song SK, Sun SW, Ramsbottom MJ, Chang C, Russell J, Cross AH. Dysmyelination revealed through MRI as increased radial (but unchanged axial) diffusion of water. Neuroimage. 2002;17:1429–1436. doi: 10.1006/nimg.2002.1267. [DOI] [PubMed] [Google Scholar]
- Sun S-W, Neil JJ, Liang H-F, He YY, Schmidt RE, Hsu CY, Song S-K. Formalin fixation alters water diffusion coefficient magnitude but not anisotropy in infarcted brain. Magn Reson Med. 2005;53:1447–1451. doi: 10.1002/mrm.20488. [DOI] [PubMed] [Google Scholar]
- Sundaram SK, Kumar A, Makki MI, Behen ME, Chugani HT, Chugani DC. Diffusion tensor imaging of frontal lobe in autism spectrum disorder. Cereb Cortex. 2008;18:2659–2665. doi: 10.1093/cercor/bhn031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C, Ye FQ, Irfanoglu MO, Modi P, Saleem KS, Leopold DA, Pierpaoli C. Anatomical accuracy of brain connections derived from diffusion MRI tractography is inherently limited. Proc Natl Acad Sci USA. 2014;111:16574–16579. doi: 10.1073/pnas.1405672111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisserand DJ, Pruessner JC, Sanz Arigita EJ, van Boxtel MPJ, Evans AC, Jolles J, Uylings HBM. Regional frontal cortical volumes decrease differentially in aging: an MRI study to compare volumetric approaches and voxel-based morphometry. Neuroimage. 2002;17:657–669. [PubMed] [Google Scholar]
- Tustison NJ, Cook PA, Klein A, Song G, Das SR, Duda JT, Kandel BM, van Strien N, Stone JR, Gee JC, Avants BB. Large-scale evaluation of ANTs and FreeSurfer cortical thickness measurements. Neuroimage. 2014;99:166–179. doi: 10.1016/j.neuroimage.2014.05.044. [DOI] [PubMed] [Google Scholar]
- Veraart J, Leergaard TB, Antonsen BT, Van Hecke W, Blockx I, Jeurissen B, Jiang Y, Van der Linden A, Johnson GA, Verhoye M, Sijbers J. Population-averaged diffusion tensor imaging atlas of the Sprague Dawley rat brain. Neuroimage. 2011;58:975–983. doi: 10.1016/j.neuroimage.2011.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellington RL, Bilder RM, Napolitano B, Szeszko PR. Effects of age on prefrontal subregions and hippocampal volumes in young and middle-aged healthy humans. Hum Brain Mapp. 2013;34:2129–2140. doi: 10.1002/hbm.22054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widjaja E, Wei X, Vidarsson L, Moineddin R, Macgowan CK, Nilsson D. Alteration of diffusion tensor parameters in postmortem brain. Magn Reson Imaging. 2009;27:865–870. doi: 10.1016/j.mri.2008.11.009. [DOI] [PubMed] [Google Scholar]
- Wisco JJ, Killiany RJ, Guttmann CRG, Warfield SK, Moss MB, Rosene DL. An MRI study of age-related white and gray matter volume changes in the rhesus monkey. Neurobiol Aging. 2008a;29:1563–1575. doi: 10.1016/j.neurobiolaging.2007.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisco JJ, Rosene DL, Killiany RJ, Moss MB, Warfield SK, Egorova S, Wu Y, Liptak Z, Warner J, Guttmann CRG. A rhesus monkey reference label atlas for template driven segmentation. J. Med. Primatol. 2008b;37:250–260. doi: 10.1111/j.1600-0684.2008.00288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witelson SF. Hand and sex differences in the isthmus and genu of the human corpus callosum. A postmortem morphological study. Brain. 1989;112(Pt 3):799–835. doi: 10.1093/brain/112.3.799. [DOI] [PubMed] [Google Scholar]
- Zakszewski E, Adluru N, Tromp DPM, Kalin N, Alexander AL. A diffusion-tensor-based white matter atlas for rhesus macaques. PLoS ONE. 2014;9:e107398. doi: 10.1371/journal.pone.0107398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Manual correction and smoothing of anatomic labels in three dimensions. Surface renderings of five major subcortical structures are shown within a semi-transparent rendering of the whole brain: putamen (blue), caudate (yellow), hippocampus (green), external globus pallidus (teal), and internal globus pallidus (orange). Pre-correction labels are shown on the left side of the brain, and post-correction labels are shown on the right side of the brain. (A) Anterior view. (B) Posterior view. (C) Dorsal view.
Supplementary Figure 2. Tractography of small white matter pathways afforded by high spatial resolution DTI. (A) Cranial nerve III. (B) Cranial nerve IV. (C) Cranial nerve VII. (D) Cranial nerve VIII. (E) Fasciculus retroflexus. (F) Stria medullaris. All background images are gradient echo anatomic images in the coronal plane except for (F), which is in the axial plane.
Supplementary Table 1. The 12-direction diffusion gradient encoding scheme used for diffusion tensor MRI acquisition.
Supplementary Table 2. The complete list of atlas anatomic labels, including abbreviations and hierarchical organization.
Supplementary Table 3. The complete list of atlas tractography structures, including abbreviations and hierarchical organization.