Abstract
X-linked protoporphyria (XLP), a rare erythropoietic porphyria, results from terminal exon gain-of-function mutations in the ALAS2 gene causing increased ALAS2 activity and markedly increased erythrocyte protoporphyrin levels. Patients present with severe cutaneous photosensitivity and may develop liver dysfunction. XLP was originally reported as X-linked dominant with 100% penetrance in males and females. We characterized 11 heterozygous females from six unrelated XLP families and show markedly varying phenotypic and biochemical heterogeneity, reflecting the degree of X-chromsomal inactivation of the mutant gene. ALAS2 sequencing identified the specific mutation and confirmed heterozygosity among the females. Clinical history, plasma and erythrocyte protoporphyrin levels were determined. Methylation assays of the androgen receptor and zinc-finger MYM type 3 short tandem repeat polymorphisms estimated each heterozygotes X-chromosomal inactivation pattern. Heterozygotes with equal or increased skewing, favoring expression of the wild-type allele had no clinical symptoms and only slightly increased erythrocyte protoporphyrin concentrations and/or frequency of protoporphyrin-containing peripheral blood fluorocytes. When the wild-type allele was preferentially inactivated, heterozygous females manifested the disease phenotype and had both higher erythrocyte protoporphyrin levels and circulating fluorocytes. These findings confirm that the previous dominant classification of XLP is inappropriate and genetically misleading, as the disorder is more appropriately designated XLP.
Keywords: ALAS2, genotype-phenotype, X-chromosomal inactivation, X-linked protoporphyria
X-linked protoporphyria (XLP) (MIM 300752) is a recognized erythropoietic porphyria due to an increased enzymatic activity of erythroid-specific 5-aminolevulinate synthase 2 (ALAS2), the first enzyme in erythroid heme biosynthesis. The enzyme condenses glycine and succinyl-CoA to form 5-aminolevulinic acid (ALA) in the presence of its cofactor, vitamin B6 (pyridoxal 5′-phosphate) (1). Two exon 11 gain-of-function small deletions (c.1699_1700delAT and c.1706_1709delAGTG) of the X-linked erythroid-specific ALAS2 gene were originally identified as causing XLP (2). Recently, two additional ALAS2 mutations (c.1642C>T and c.1737delG) were described (3, 4). These mutations result in stop or frameshift lesions that prematurely truncate or abnormally elongate the wild-type enzyme, leading to increased ALAS2 activity (about 2–3 times fold normal in vitro) (4). Because most of ALA produced is metabolized to porphyrin and that the insertion of Fe2+ into protoporphyrin IX by ferrochelatase (FECH) is a rate-limiting step for erythroid heme synthesis, the continuously increased ALA formation leads to the erythroid accumulation of free and zinc-chelated protoporphyrin IX (PPIX) (5). The pathogenesis of XLP and its clinical manifestations characterized by acute, painful, cutaneous photosensitivity and liver disease are similar to those of autosomal recessive erythropoietic protoporphyria (EPP), which arises from loss-of-function mutations in the ferrochelatase (FECH) gene (3, 6–8). XLP has been described as an X-linked dominant trait with essentially 100% penetrance (2, 9). However, the presence of asymptomatic heterozygous females, in unrelated XLP families, makes the penetrance and the classification of its dominant inheritance incorrect and misleading for genetic and reproductive counseling. Generally, in female mammals, most genes on one X-chromosome are silenced as a result of the random X-chromosomal inactivation process (10). X-linked dominant inheritance typically refers to conditions in which the disorder is lethal in most males and is expressed in all heterozygous females such as Aicardi syndrome, Goltz-Gorlin syndrome and incontinentia pigmenti. Skewed X-chromosomal inactivation explains the phenotypic variability among females heterozygous for different X-linked conditions (11, 12). The inactivation process is synergistically established and maintained by constitutive heterochromatin-like histone hypoacetylation and methylation (13, 14). In somatic tissues, there is a strong correlation between global X-chromosomal inactivation and the methylation status of the deoxycytosine residues in CpG islands associated with promoters of X-linked genes (15, 16) allowing reduction in X-linked gene expression. Previous studies have specifically shown that methylation status of the human androgen receptor (AR or HUMARA) and zinc-finger MYM type 3 (ZMYM3 or ZNF261) genes is correlated with global X-inactivation in somatic tissues (17, 18). Thus, a method to analyze X-chromosomal inactivation in these genes is to use the methylation-sensitive restriction enzymes, HpaII and HhaI. The close proximity of these enzyme cleavage sites to a highly polymorphic short tandem repeats (STR) allows the use of polymerase chain reaction (PCR) assays to identify the methylation patterns of the maternally- and paternally derived X chromosomes when the phase of the disease and STR alleles are known from studies of family members.
Thus, the methylation differences near the STRs in the AR and ZMYM3 genes have been used to investigate the hypothesis that non-random X-chromosomal inactivation is responsible for the variable phenotypic expression in heterozygous females in XLP families.
Material and methods
Patients
Peripheral blood and/or saliva samples were collected from 33 subjects of six unrelated families (four of Italian, one of Irish/English and one of German ancestry). For most subjects, the percentage of fluorescent erythrocytes (i.e. fluorocytes), the plasma peak wavelength (nm) and the level of erythrocyte PPIX were measured. In all subjects, ALAS2 molecular analysis was performed, while only in women was the percent inactivation for both X-chromosomes determined by methylation analyses. The genotyping and pedigree studies were conducted in accordance with the ethical principles of the World Medical Association Declaration of Helsinki for medical research involving human subjects, with its subsequent amendments. All subjects provided informed consent.
Biochemical analyses
Erythrocytes fluorescence
Tenmicroliters of ethylenediaminetetraacetic acid whole blood were diluted in 10 μl of phosphate-buffered solution, pH 7.2, and directly processed in a flow cytometer (Becton Dickinson FACScan, BD, Franklin Lake, NJ, USA) without staining. Fluorescent erythrocytes emit red fluorescence at λ>620 nm when excited at 488 nm by an argon laser (19, 20). Results are expressed as percentage of cells beyond the marker of autofluorescence of control subjects.
Fluorometric scanning of plasma porphyrins
The plasma fluorescence emission spectrum was determined using 2 μl of undiluted plasma in a ND 3300 NanoDrop Fluorometer (NanoDrop Technologies, Wilmington, DE) as previously described by Di Pierro (21) or by the spectrofluorometrical method of Poh-Fitzpatrick (22). Total plasma porphyrins were measured as reported by Egger et al. (23). Erythrocyte PPIX Erythrocyte free protoporphyrin (PPIX) and zinc protoporphyrin (ZPP) were measured after extraction with acetone and 4% aqueous formic acid as previously described (24). Filtered samples (0.45 μm cellulose filters, 30mm diameter, National Scientific, Rockwood, TN) were analyzed by liquid chromatography (quaternary pump Agilent Technologies Series 1200, Agilent Technologies, Santa Clara, CA, USA) interfaced with a fluorescence spectrophotometer (G1321A Model, Agilent Technologies series 1200) equipped with a 8 μl cell. Separation was achieved using a C18 reversed-phase Chromsytems 44100 column, Chromsystems Instrumens and Chemicals GmbH, Gräfelfing, Germany (15 cm× 4.6mm ID, 3 μm) with a Chromolith guard column and a gradient of methanol and 1% aqueous acetic acid as eluent. ZPP was detected at 30.5min, setting the fluorescence spectrophotometer at 400 nm as the excitation wave length (λex) and 620 nm as the emission wave length (λem); PPIX was detected at 33.7min with λex 387 nm and λem 633. The photomultiplier gain was set at 17. The quantification was performed vs a calibration curve prepared by dissolving the pure chemicals (from Frontier Scientific Porphyrin Product, Logan, UT) in dimethylformamide and then in a mixture of acetone, water, and formic acid. The concentration of each erythrocyte porphyrin was adjusted by hemoglobin (Hb). The limit of quantification of the assay was 0.5 μg/g Hb for both PPIX and ZPP.
DNA analysis
DNA extraction, PCR and sequencing of exon 11 ALAS2
Genomic DNA was extracted from buffy coats of peripheral blood using the Maxwell 16 DNA Blood Kit with an automatic extractor Maxwell®16 (Promega Corp., Madison City, WI) or from saliva using the Oragene DNA OG500 kit, DNA Genotek Inc., Ottawa, Canada. Saliva DNA was extracted by manual purification and the QIAamp DNA mini kit Qiagen, Valencia, California, CA, USA (25).
For Italian families, exon 11 of ALAS2 was amplified using forward and reverse primers listed in Table 1 (10 pmol each) in a reaction containing 0.2mM dNTPs, 1.5mM MgCl2, 67mM Tris–HCl at pH8.8, 16mM (NH4)2SO2, 0.01% Tween-20 and 2.5 U of BioTaq DNA polymerase (Bioline, London, UK) in a final volume of 50 μl. The PCR was carried out with an initial denaturation step at 94°C for 5min, followed by 30 s at 94°C, 30 s at 60°C and 30 s at 72°C for 30 cycles. The PCR products were subsequently submitted for direct Sanger sequencing of both strands (AbiPrism 310 Genetic Analyzer, Appliedbiosystems by Lifetechnologies, Carlsbad, CA, USA). For the North American families, the entire ALAS2 exon 11 coding region and ~30–40 bp intronic and 3′untranslated sequence were amplified using the forward and reverse primers F-GGGGGATCAATATCTTGGCTC and R-CCAACAAGTGACCTATGGTTACCT as previously described (3). Sequences were analyzed by using Sequencer version 4.8 software (Gene Codes) (reference sequences: ALAS2 transcript GenBank NM_000032.4).
Table 1.
Primers used for DNA analysis
| ALAS2-11-F | 5′-AGATTTGGAAGATCTAGTCTAACCCA-3′ |
| ALAS2-11-R | 5′-TCTGAGGGAGTCAGAATGCAC-3′ |
| AR-FW | 5′-FAM-TCCAGAATCTGTTCCAGAGCGTGC-3′ |
| AR-RW | 5′-GCTGTGAAGGTTGCTGTTCCTCAT-3′ |
| ZMYM-FW | 5′-HEX-ATGCTAAGGACCATCCAGGA-3′ |
| ZMYM-RW | 5′-GGAGTTTTCCTCCCTCACCA-3′ |
PCR of the STR in AR and ZMYM3 genes
To correlate the pattern of X-chromosomal inactivation with the clinical phenotype, we used the methylation-based analysis of the human AR and zinc-finger MYM type 3 (ZMYM3) genes, which contain a CAG-repeat sequence and GC highly polymorphic dinucleotides, respectively (17, 18, 26). To determine the size of the microsatellites in the AR and ZMYM3 genes, 50 ng of DNA from each individual was amplified in different tubes using FAM and HEX-labeled forward primers and reverse primers (10 pmol each) as listed in Table 1, in a reaction containing 0.2mM dNTPs, 1.5mM MgCl2, 67mM Tris–HCl at pH8.8, 16mM (NH4)2SO2, 0.01% Tween-20 and 2.5 U of BioTaq DNA polymerase (Bioline, London, UK). PCR condition was as follows: 95°C for 5min, then 35 cycles of 96°C for 30 s, 65°C for 30 s, and 72°C for 30 s. One microliter of the PCR products were added to 0,5 μl of ROX-500 Size Standard, 13,5 μl of HiDi Formammide (Appliedbiosystems by Lifetechnologies, Carlsbad, CA, USA) and dH2O to a final volume of 15 μl, denaturated for 3 min at 96°C and electrophoresed on an automatic sequence analyzer (AbiPrism 310 Genetic Analyzer, Appliedbiosystems by Lifetechnologies, Carlsbad, CA, USA) for fragment analysis. To determine the extent of inactivation, the genes in each heterozygous female, DNA sample (500 ng) were digested with 20U HpaII and 20U HhaI (Promega Corp., Madison City, WI) in a 20-μl total volume reaction at 37°C overnight and with an inactivation step at 65°C for 20 min. Two microliters (50 ng DNA) of digested DNA was PCR amplified as above, and the sample was run on an automated sequencer after the undigested PCR-amplified DNA of the same patient.
Quantification of X-chromosomal inactivation
Quantification of the relative X-chromosomal inactivation of the AR and ZMYM3 alleles for each patient was calculated as previously reported by Lau et al. with the following modifications (27). The degree of skewing was defined as the peak height of each allele, relative to the sum of the heights of both alleles. The peak heights corresponding to each allele from the digested and undigested samples were determined using the Gene Mapper program (Applied Biosystems). Peak heights values for the digested samples were normalized with those for the undigested samples, to account for preferential allele amplification. Thus, the degree of skewing of each gene was calculated as follows: (AlleleA_ d/AlleleA_u)/(AlleleA_d/AlleleA_u) +(AlleleB_d/AlleleB_u), where allele A_d and allele B_d represent the peaks heights of the digested sample, while allele A_u and allele B_u the corresponded to the peaks heights from the undigested sample.
Results
Biochemical analyses
Nine patients (seven males and two females) from six unrelated XLP families had history of photosensitivity from childhood and all had the typical PPIX plasma fluorescence peak at 632–635 nm, markedly increased (~30- to 50-fold higher than normal) fluorescent erythrocytes, and accumulation of both free and zinc-chelated PPIX. In contrast, there was a remarkable heterogeneity of biochemical markers and clinical manifestations between the nine heterozygous carrier females. In particular, seven females showed only slight increases (from twofold to sixfold higher than normal) in fluorescent erythrocytes with slight increases in both free and zinc-chelated PPIX, absence of the plasma fluorescent peak at 632–635 nm, and lack of clinical photosensitivity from childhood. Of particular relevance, the other two heterozygous females were completely asymptomatic at both biochemical and clinical levels. The clinical and biochemical findings for these individuals are summarized in Tables 2 and 3.
Table 2.
Clinical and biochemical findings in Italian XLP families
| Family/patient | Sex | Peak (nm) | Fluorocytes (%) n.v.: 0–1% |
Erythrocyte protoporphyrins (mcg/gHgB) n.v.: <3 mcg/gHB (%) |
Photosensitivity from childhood | Genotype |
|---|---|---|---|---|---|---|
| A | ||||||
| Pt 1 (III.1) | F | 632 | 30.9 | 92.5 (Zn62PPIX38) | Yes | c.1706-1709 delAGTG |
| Pt 2 (III.2) | M | 630 | 38.9 | 25.4 (Zn54PPIX46) | Yes | c.1706-1709 delAGTG |
| Mother (II.2) | F | No peak | 5.84 | 4.8 (Zn53PPIX47) | No | c.1706-1709 delAGTG |
| Pt 3 (III.3) | M | 627 | 59.85 | 136.4 (Zn46PPIX54) | Yes | c.1706-1709 delAGTG |
| Mother (II.3) | F | No peak | 4.54 | 6.3 (Zn51PPIX49) | No | c.1706-1709 delAGTG |
| Grandmother (I.2) | F | No peak | 2.90 | 5.2 (Zn58PPIX42) | No | c.1706-1709 delAGTG |
| B | ||||||
| Pt 4 (II.1) | M | 635 nm | 19.25 | 29.2 (Zn84PPIX16) | Yes | c.1706-1709 delAGTG |
| Mother (I.2) | F | No peak | 0.24 | 1.7 (Zn92PPIX8) | No | c.1706-1709 delAGTG |
| C | ||||||
| Pt 5 (II.1) | M | 632 nm | 36.90 | 69 (Zn43PPIX57) | Yes | c.1706-1709 delAGTG |
| Mother (I.2) | F | no peak | 1.06 | 1.1 (Zn93PIX7) | No | wt-wt |
| D | ||||||
| Pt 6 (I.2) | F | 632 nm | 35.48 | 82 (Zn45PPIX55) | Yes | c.1706-1709 delAGTG |
Table 3.
Clinical and biochemical findings in North American XLP families
| Family | Sex | Peak (nm) | Plasma total porphyrins (mcg/dl) n.v.: 0–0.9 mcg/dl |
Erythrocyte protoporphyrins (mcg/dl) n.v.: 20–80 mcg/dl |
Photosensitivity from childhood | Genotype |
|---|---|---|---|---|---|---|
| E | ||||||
| Pt 7 (III.3) | M | 634 | 2.6 | 1600 (Zn50PPIX50) | Yes | c.1734delG |
| Mother (II.2) | F | no peak | 0.4 | 201 (Zn56PPIX44) | No | c.1734delG |
| Aunt (II.3) | F | no peak | 0.2 | 379 (Zn47PPIX53) | No | c.1734delG |
| F | ||||||
| Pt 8 (II.1) | M | 634 | 23.4 | 2948 (Zn34PPIX66) | Yes | c.1706-1709 delAGTG |
| Pt 9 (II.4) | M | n.d. | n.d. | n.d. | Yes | c.1706-1709 delAGTG |
| Sister (II.2) | F | n.d. | n.d. | n.d. | No | c.1706-1709 delAGTG |
| Mother (I.2) | F | 630 | 1.8 | 1112 (Zn26PPIX74) | No | c.1706-1709 delAGTG |
Genetic analysis
Three known mutations in exon 11 of ALAS2 gene were identified by sequencing of PCR-amplified genomic DNA, confirming the diagnosis of XLP in the members of the six families. The small four base deletion c.1706_1709delAGTG was present in Familes A-D and F, while Family E had the single base deletion c. 1734delG. None of the family members had a mutation in their FECH genes. Pedigree analyses revealed that all mothers had the family mutation with the only exception being Family C, suggesting that the mutation was de novo.
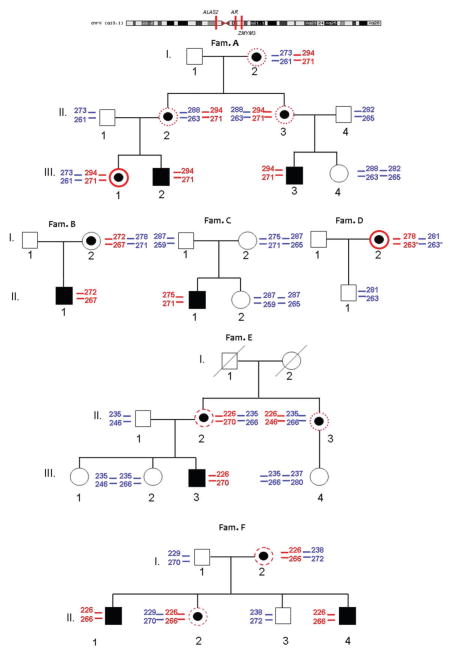
To test the hypothesis that X-chromosomal inactivation directly influenced the photosensitivity in XLP, we conducted methylation-based assays. For each family member, the informative regions of the AR and ZMYM3 genes were PCR amplified and the fragment sizes determined. Fragment size analysis was initially conducted on the PCR amplicon of a male subject to identify the size of microsatellites associated with the disease allele in each family, and to define for each female the parental origin of the two alleles (Fig. 1). PCR and fragment analyses were performed on digested and undigested DNA from heterozygous females to estimate the degree of inactivation of both their alleles. The ZMYM3 assay was uninformative only in Family D because the patient was homozygous for the polymorphic STR. The results of six independent experiments were averaged to determine the degree of X-chromosomal inactivation for both alleles in each heterozygote and are summarized in Table 4.
Fig. 1.
Family trees; X chromosome alleles segregation. The position of ALAS2, AR and ZMYM3 genes are reported in the upper panel with respect to the X-Chromosome. In the family trees in red is indicated the mutated allele and in blue the wild-type allele of the X-chromosome. For each allele the size of AR and ZMYM3 polymorphisms are reported: the upper one is referred to AR and the lower one to ZMYM3. Symptomatic males are in black. Black dots indicate the presence of mutation in females; red circles indicate symptomatic females, dashed red circles indicate late onset of symptoms and dotted red circles indicates females showing only slight increase of PPIX in erythrocytes.
Table 4.
X-Chromosomal inactivation studiesa
| AR | Relative size | Mean value (%) | Min (%) | Max (%) | ZMYM3 (%) | Relative size | Mean value (%) | Min (%) | Max (%) | Mean % X-Inac |
|---|---|---|---|---|---|---|---|---|---|---|
| Family A | ||||||||||
| I.2 | 273 | 21.31 | 16.95 | 24.68 | I.2 | 261 | 26.44 | 24.85 | 27.98 | 23.88 |
| 294 | 78.69 | 75.32 | 83.05 | 271 | 73.56 | 72.02 | 75.15 | 76.13 | ||
| II.2 | 288 | 24.09 | 21.33 | 25.53 | II.2 | 263 | 48.26 | 43.46 | 58.86 | 36.18 |
| 294 | 75.92 | 74.47 | 78.67 | 271 | 51.74 | 41.14 | 56.54 | 63.83 | ||
| II.3 | 288 | 35.90 | 23.42 | 40.16 | II.3 | 263 | 40.50 | 32.13 | 49.13 | 38.20 |
| 294 | 64.10 | 59.84 | 76.58 | 271 | 59.51 | 50.87 | 67.87 | 61.81 | ||
| III.1 | 273 | 85.27 | 80.01 | 94.99 | III.1 | 261 | 87.31 | 81.28 | 92.25 | 86.29 |
| 294 | 14.74 | 5.01 | 19.99 | 271 | 12.69 | 7.75 | 18.72 | 13.72 | ||
| Family B | ||||||||||
| I.2 | 272 | 92.38 | 85.89 | 95.75 | I.2 | 267 | 84.42 | 8.26 | 85.88 | 88.40 |
| 278 | 7.62 | 4.25 | 14.11 | 271 | 15.58 | 14.12 | 1.74 | 11.60 | ||
| Family D | ||||||||||
| I.2 | 278 | 26.70 | 22.32 | 32.34 | I.2 | 263 | Ni | Ni | Ni | 26.70 |
| 281 | 73.30 | 67.66 | 77.68 | 263 | Ni | Ni | Ni | 73.30 | ||
| Family E | ||||||||||
| II.2 | 226 | 75.43 | 69.95 | 79.57 | II.2 | 270 | 66.53 | 61.17 | 69.76 | 70.98 |
| 235 | 24.57 | 20.43 | 30.05 | 266 | 33.47 | 30.24 | 38.83 | 29.02 | ||
| II.3 | 226 | 52.46 | 49.57 | 55.47 | II.3 | 246 | 66.23 | 61.08 | 72.61 | 59.35 |
| 235 | 47.54 | 44.53 | 50.43 | 266 | 33.77 | 27.39 | 38.92 | 40.66 | ||
| Family F | ||||||||||
| I.2 | 226 | 83.85 | 81.28 | 85.99 | I.2 | 266 | 71.11 | 6.57 | 76.44 | 77.48 |
| 238 | 16.16 | 14.01 | 18.72 | 272 | 28.89 | 23.56 | 3.43 | 22.53 | ||
| II.2 | 226 | 57.88 | 5.61 | 59.57 | II.2 | 266 | 73.37 | 70.25 | 75.47 | 65.63 |
| 229 | 42.12 | 40.43 | 4.39 | 270 | 26.63 | 24.53 | 29.75 | 34.38 | ||
Ni, not informative for homozygous allele.
In blue the wild-type allele; in red the mutated allele; in green the symptomatic patients (photosensitivity from childhood); in pink the full asymptomatic patients (no photosensitivity and no storage of IX); in black all mild females (late-onset photosensitivity or only mild storage of PPIX).
In general, the results were comparable for both AR and ZMYM3 assays. Of note, the two photosensitive heterozygous females (Family A: III-1 and Family D I-2) had a skewed X-chromosomal inactivation pattern of the wild-type allele, 86.3% and 73.3% inactivated, respectively. Of the two completely asymptomatic females, one had a skewed X-chromosomal inactivation of mutant allele (88.4%) while the other one was not heterozygous for the mutant ALAS2 allele. Importantly, the other seven heterozygous females who had no or late-onset photosensitivity had relatively balanced X-chromosomal inactivation patterns with preferential expression of the wild-type ALAS2 allele.
Discussion
The X-chromosomal inactivation process affects most regions of the X chromosome and is synergistically established and maintained by constitutive heterochromatin-like histone hypoacetylation and methylation (28). In total, only ~15% of X-linked genes escape inactivation to some degree depending on their position on the X chromosome. An additional 10% of X-linked genes show variable profiles of inactivation and are expressed to different extents from some inactive X chromosomes. Carrel and Willard (10) established that the ALAS2 gene is subject to complete inactivation. Thus, the methylation status of the AR and ZMYM3 promoters correlate with the expression of the ALAS2 gene.
The X-chromosomal inactivation (Xi) process occurs early in female development to silence one of the two X-chromosomes allowing dosage compensation between human males and females. The choice of chromosome to be inactivated is generally random and it is then clonally propagated during subsequent cell divisions, resulting in females being mosaics for cells with either the maternal or paternal X-chromosome inactivated (29). The relative ratio of these two cell populations in a given female is known as the Xi pattern. Most normal females have a ratio close to 50:50. An abnormal Xi pattern has most often been defined as greater than 80:20, although a stricter value of 90:10 has also been suggested (30). Non-random X-inactivation can occur due to a selective advantage of one cell population or by chance (31, 32). For X-linked diseases, favorable skewing occurs when the normal allele is preferentially active, while unfavorable skewing occurs when the mutant allele is preferentially active. Both favorable and unfavorable Xi skewing have been found in heterozygous females with various X-linked mutations (11, 30).
In this study, female heterozygotes for XLP that had balanced Xi ratios were clinically asymptomatic. They had only very slightly increased percentages of fluorescent erythrocytes and only slightly increased levels of free and zinc-chelated PPIX which was not sufficient to give rise to photosensitivity from childhood. Only in the presence of unfavorable skewing and inactivation of the wild-type allele did the heterozygous females have clinical symptoms. Notably, the heterozygous females with favorable skewing were asymptomatic clinically and biochemically with normal levels of erythrocyte PPIX. Thus, for asymptomatic heterozygotes in XLP families, it is necessary to perform ALAS2 molecular analyses for accurate genetic and reproductive counseling.
In this study, we demonstrated that the Xi pattern directly influences the penetrance and the severity of the phenotype in heterozygous females for XLP. As suggested by Dobyns et al. (33) in revisiting 32 X-linked diseases, we recommend that use of the term X-linked dominant be discontinued and that this disorder be appropriately recognized as having X-linked inheritance with its clinical manifestations determined primarily by random X-inactivation. Thus, the change in disease nomenclature is appropriate and the disease should be designated XLP.
Acknowledgments
We thank the patients and their families for participation in this study; Alessandra Cattaneo and Cecilia Frugoni for their excellent flow cytometer assistance and Dario Tavazzi for biochemical analysis. This research was supported in part by grants from the Italian Ministry of Health (RF-2009-1519531 and RF-2010-2303934) and from Fondazione IRCCS Ca’ Granda Ospedale Maggiore Policlinico. Thiswork alsowas supported in part by the National Institutes of Health (NIH) grant for the Porphyrias Consortium (U54 DK083909) which is a part Rare Disease Clinical Research Network (RDCRN), supported through collaboration between the NIH Office of Rare Diseases Research (ORDR) at the National Center for Advancing Translational Science (NCATS), and the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Finally, M. B. is the recipient of a grant from the National Institute of Health (K23 DK095946-02).
Footnotes
Conflict of interest
All authors declare no conflict of interest.
References
- 1.Anderson KE, Sassa S, Bishop DF, Desnick RJ. Disorders of heme biosynthesis: X-linked sideroblastic anemia and the porphyrias. In: Scriver CS, Beaudet AL, Sly WS, Valle D, editors. The molecular and metabolic bases of inherited disease. New York, NY: McGraw-Hill; 2001. pp. 2961–3062. [Google Scholar]
- 2.Whatley SD, Ducamp S, Gouya L, et al. C-terminal deletions in the ALAS2 gene lead to gain of function and cause X-linked dominant protoporphyria without anemia or iron overload. Am J Hum Genet. 2008;83 (3):408–414. doi: 10.1016/j.ajhg.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balwani M, Doheny D, Bishop DF, et al. Loss-of-function ferrochelatase and gain-of-function erythroid 5-aminolevulinate synthase mutations causing erythropoietic protoporphyria and X-linked protoporphyria in North American patients reveal novel mutations and a high prevalence of X-linked protoporphyria. Mol Med. 2013;19:26–35. doi: 10.2119/molmed.2012.00340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bishop DF, Tchaikovskii V, Nazarenko I, Desnick RJ. Molecular expression and characterization of erythroid-specific 5-aminolevulinate synthase gain-of-function mutations causing X-linked protoporphyria. Mol Med. 2013;19:18–25. doi: 10.2119/molmed.2013.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Puy H, Gouya L, Deybach JC. Porphyrias. Lancet. 2010;375 (971):924–937. doi: 10.1016/S0140-6736(09)61925-5. [DOI] [PubMed] [Google Scholar]
- 6.Thunell S, Harper P, Brun A. Porphyrins, porphyrin metabolism and porphyrias. IV. Pathophysiology of erythyropoietic protoporphyria-- diagnosis, care and monitoring of the patient. Scand J Clin Lab Invest. 2000;60 (7):581–604. [PubMed] [Google Scholar]
- 7.Di Pierro E, Brancaleoni V, Besana V, Ausenda S, Drury S, Cappellini MD. A 10376 bp deletion of FECH gene responsible for erythropoietic protoporphyria. Blood Cells Mol Dis. 2008;40 (2):233–236. doi: 10.1016/j.bcmd.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 8.Di Pierro E, Brancaleoni V, Moriondo V, Besana V, Cappellini MD. Co-existence of two functionalmutations on the same allele of the human ferrochelatase gene in erythropoietic protoporphyria. Clin Genet. 2007;71 (1):84–88. doi: 10.1111/j.1399-0004.2007.00733.x. [DOI] [PubMed] [Google Scholar]
- 9.Seager MJ, Whatley SD, Anstey AV, Millard TP. X-linked dominant protoporphyria: a new porphyria. Clin Exp Dermatol. 2014;39 (1):35–37. doi: 10.1111/ced.12202. [DOI] [PubMed] [Google Scholar]
- 10.Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434 (7031):400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- 11.Plenge RM, Stevenson RA, Lubs HA, Schwartz CE, Willard HF. Skewed X-chromosome inactivation is a common feature of X-linked mental retardation disorders. Am J Hum Genet. 2002;71 (1):168–173. doi: 10.1086/341123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Orstavik KH. X-chromosome inactivation in clinical practice. HumGenet. 2009;126 (3):363–373. doi: 10.1007/s00439-009-0670-5. [DOI] [PubMed] [Google Scholar]
- 13.Heard E. Recent advances in X-chromosome inactivation. Curr Opin Cell Biol. 2004;16 (3):247–255. doi: 10.1016/j.ceb.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 14.Jeppesen P, Turner BM. The inactive X chromosome in female mammals is distinguished by a lack of histone H4 acetylation, a cytogenetic marker for gene expression. Cell. 1993;74 (2):281–289. doi: 10.1016/0092-8674(93)90419-q. [DOI] [PubMed] [Google Scholar]
- 15.Norris DP, Brockdorff N, Rastan S. Methylation status of CpG-rich islands on active and inactive mouse X chromosomes. Mamm Genome. 1991;1 (2):78–83. doi: 10.1007/BF02443782. [DOI] [PubMed] [Google Scholar]
- 16.Toniolo D, Martini G, Migeon BR, Dono R. Expression of the G6PD locus on the human X chromosome is associated with demethylation of three CpG islands within 100 kb of DNA. EMBO J. 1988;7 (2):401–406. doi: 10.1002/j.1460-2075.1988.tb02827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Allen RC, Zoghbi HY, Moseley AB, Rosenblatt HM, Belmont JW. Methylation of HpaII and HhaI sites near the polymorphic CAG repeat in the human androgen-receptor gene correlates with X chromosome inactivation. Am J Hum Genet. 1992;51 (6):1229–1239. [PMC free article] [PubMed] [Google Scholar]
- 18.Beever C, Lai BP, Baldry SE, et al. Methylation of ZNF261 as an assay for determining X chromosome inactivation patterns. Am J Med Genet A. 2003;120A (3):439–441. doi: 10.1002/ajmg.a.20045. [DOI] [PubMed] [Google Scholar]
- 19.Cordiali FP, Macri A, Trento E, et al. Flow cytometric analysis of fluorocytes in patients with erythropoietic porphyria. Eur J Histochem. 1997;41S2:9–10. [PubMed] [Google Scholar]
- 20.Mathews-Roth MM, Wise RJ, Miller BA. Burst-forming units-erythroid from erythropoietic protoporphyria patients fluoresce under 405 nm light. Blood. 1996;87 (10):4480–4481. [PubMed] [Google Scholar]
- 21.Di Pierro E, Ventura P, Brancaleoni V, et al. Clinical, biochemical and genetic characteristics of Variegate Porphyria in Italy. Cell Mol Biol. 2009;55 (2):79–88. [PubMed] [Google Scholar]
- 22.Poh-Fitzpatrick MB. A plasma porphyrin fluorescence marker for variegate porphyria. Arch Dermatol. 1980;116 (5):543–547. [PubMed] [Google Scholar]
- 23.Egger NG, Motamedi M, Pow-Sang M, Orihuela E, Anderson KE. Accumulation of porphyrins in plasma and tissues of dogs after delta-aminolevulinic acid administration: implications for photodynamic therapy. Pharmacology. 1996;52 (6):362–370. doi: 10.1159/000139403. [DOI] [PubMed] [Google Scholar]
- 24.Bailey GG, Needham LL. Simultaneous quantification of erythrocyte zinc protoporphyrin and protoporphyrin IX by liquid chromatography. Clin Chem. 1986;32 (12):2137–2142. [PubMed] [Google Scholar]
- 25.Looi ML, Zakaria H, Osman J, Jamal R. Quantity and quality assessment of DNA extracted from saliva and blood. Clin Lab. 2012;58 (3–4):307–312. [PubMed] [Google Scholar]
- 26.Pegoraro E, Schimke RN, Arahata K, et al. Detection of new paternal dystrophin gene mutations in isolated cases of dystrophinopathy in females. Am J Hum Genet. 1994;54 (6):989–1003. [PMC free article] [PubMed] [Google Scholar]
- 27.Lau AW, Brown CJ, Penaherrera M, Langlois S, Kalousek DK, Robinson WP. Skewed X-chromosome inactivation is common in fetuses or newborns associated with confined placental mosaicism. Am J Hum Genet. 1997;61 (6):1353–1361. doi: 10.1086/301651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brockdorff N. Chromosome silencing mechanisms in X-chromosome inactivation: unknown unknowns. Development. 2011;138 (23):5057–5065. doi: 10.1242/dev.065276. [DOI] [PubMed] [Google Scholar]
- 29.Lyon MF. Gene action in the X-chromosome of the mouse (Mus musculus L. ) Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- 30.Willard HF. The sex chromosomes and X chromosome inactivation. In: Scriver CR, Beaudet AL, Sly WS, Valle D, Chids B, Vogelstein B, editors. The metabolic and molecular bases of inherited disease. New York, NY: McGraw-Hill; 2000. pp. 1191–1221. [Google Scholar]
- 31.Plenge RM, Hendrich BD, Schwartz C, et al. A promoter mutation in the XIST gene in two unrelated families with skewed X-chromosome inactivation. Nat Genet. 1997;17 (3):353–356. doi: 10.1038/ng1197-353. [DOI] [PubMed] [Google Scholar]
- 32.Schmidt M, Du SD. Functional disomies of the X chromosome influence the cell selection and hence the X inactivation pattern in females with balanced X-autosome translocations: a review of 122 cases. Am J Med Genet. 1992;42 (2):161–169. doi: 10.1002/ajmg.1320420205. [DOI] [PubMed] [Google Scholar]
- 33.Dobyns WB, Filauro A, Tomson BN. Inheritance of most X-linked traits is not dominant or recessive, just X-linked. Am J Med Genet A. 2004;129A (2):136–143. doi: 10.1002/ajmg.a.30123. [DOI] [PubMed] [Google Scholar]