Abstract
We discuss recent approaches for structure-based protein function annotation. We focus on template-based methods where the function of a query protein is deduced from that of a template for which both the structure and function are known. We describe the different ways of identifying a template. These are typically based on sequence analysis but new methods based on purely structural similarity are also being developed that allow function annotation based on structural relationships that can’t be recognized by sequence. The growing number of available structures of known function, improved homology modeling techniques and new developments in the use of structure allow template-based methods to be applied on a proteome-wide scale and in many different biological contexts. This progress significantly expands the range of applicability of structural information in function annotation to a level that previously was only achievable by sequence comparison.
Keywords: protein function prediction, template-based, machine learning
Introduction
It has been estimated that less than 1% of sequences in current sequence databases have an experimentally verified function [1] and, realistically, this situation is unlikely to change. Computational approaches offer the only viable solution to this problem. Numerous methods continue to be developed to infer protein function, most commonly based on sequence similarity, the presence of certain small sequence motifs, evolutionary history, and genomic location. Many of these methods are automatic and the best of them outperform simple orthology transfer, i.e., annotation transfer based on the best PSIBLAST hit [2]. Three-dimensional structure information generally plays only a minor role in automated methods but of course is invaluable in the manual annotation of the function of individual proteins. The overall limited use of protein structural information is due in large part to the small number of protein structures available relative to the numbers of sequences. However, this situation is changing and homology modeling is currently making structural information available for large numbers of proteins [3]. Moreover, it has been shown that modeled proteins can be effectively used to annotate function [4,5,6,7].
Structure-based methods for function annotation can be based on the properties of the structure of a given protein itself, such as the presence of surface cavities, surface patches containing evolutionarily conserved or covarying sets of residues, or biophysical features such as electrostatic potentials [8]. Here we focus exclusively on so-called “template”-based approaches, in which the function of a protein is assigned based primarily on its similarity to other proteins whose function is known. The wide applicability of such approaches is highlighted by the observation that, in general, there will be at least one, and usually several, proteins in structural databases that carries out a similar function using a mechanism similar to a query protein of interest [9,10,11,12]. This suggests that there are many new directions where protein structural information can be applied and, most significantly, used on a genome-wide scale [13*].
Templates are used in several ways in function annotation. Given a “query” protein with unknown function, a database of templates is searched for structurally similar proteins based on different metrics such as global sequence or structural similarity or local similarity of protein substructures. Whether the query has a function similar to the template is then evaluated by looking for similarities and differences sequence, geometric or biophysical features after superposing the query and template structures. By similar function we typically mean similar interaction properties (e.g., “these two proteins interact” or “this protein binds molecules of a certain type at this location”), but methods are also being developed to predict more specific functions such as enzyme class. Below we discuss recent progress in template-based function annotation. Although many of the methods are not new, their combination, especially in the context of machine learning approaches, is a recent development that has significantly expanded the role of structure in protein function annotation.
Exploiting Global Structural Similarity
The general strategy involved in using templates to identify binding properties of a given query is to search a database of protein complexes to identify those where one member of the complex (the template) shares some global similarity (both close and remote) with the query (Figure 1). The query and interacting partner of the template are placed in the same coordinate system using the transformation that structurally aligns the query and template structures, at which point it is necessary to determine if an interaction is likely to occur. The interaction partner can correspond to another protein, a peptide, a nucleic acid or a small molecule.
Figure 1. Function annotation using a template library.

The structure of a query protein (A) is used to scan a library of templates with known function (B). Templates can be proteins with various binding partners including other proteins (green), peptides (teal), RNA/DNA (brown) or small molecules (red star). For each complex in the library, the query, template and binding partner are placed in the same coordinate system by superposing the template and query based on global or local similarity (C, dotted line). An interaction model is then created which defines the parameters used to determine whether the query has functional properties similar to the template. These can range from an estimate of the physical interaction energy derived from residues interactions (D, yellow lines) in a 3-dimensional model of the interface, properties such as sequence conservation and covariation in the interface, or other features used as input to machine learning approaches.
Global similarity can be sequence or structure based. In sequence-based approaches if two query proteins are homologous to two other proteins that form a complex of known structure, the query proteins are first superimposed on their respective homologs in the complex. The likelihood of the query proteins forming a complex can be assessed using scoring schemes based on different factors such as overall sequence similarity, sequence similarity limited to the predicted interface [14] or sequence and structural similarity combined with biophysical properties of the predicted interface [15,16,17]. Interfacial residues in the query proteins are defined as those that align to interfacial residues in their respective templates. We use the term interaction model (Figure 1) to define the method used to score the putative interaction. This can range from an energetic analysis of the full three-dimensional structure to just a sequence analysis of interfacial residues.
In other structure-based approaches, templates are identified based solely on geometric similarity to the queries rather than on sequence similarity. Geometric similarity in principle enables a much broader coverage since there are many cases where structural and functional relationships are not detectable by sequence. The limited number of protein structures that have been determined experimentally limits the scope of this approach but homology modeling significantly expands the ability to exploit structural relationships. As an example, an experimental structure is available for at least one domain in about a quarter of the human genome but this number increases to about two thirds if homology models are used [13*]. The uncertain accuracy of many homology models may have limited their use in geometric alignments but we believe that they have in fact been underused. This is because, at the very least, they contain important information about a protein’s fold which can in turn be used to identify proteins with similar structures that may be functionally related.
We recently developed a structure-based approach to predict whether two proteins interact which relies heavily on homology models to provide extensive structural coverage of genomes [13*]. If the two putative interaction partners have one or more domains whose structure is found in the PDB or homology model databases, these structures are used to identify geometrically similar proteins. If any pair of these forms a complex of known structure, this complex is used to create an interaction model of the two query proteins. The confidence score for a modeled interaction is based on overall structural similarity, the quality of the alignment in the interfacial region and the nature of the predicted interfacial residues. These scores are combined using Bayesian statistics. Although the use of homology models generates less confident predictions for a PPI than obtained from crystal structures, they still generate many high confidence predictions. Moreover, when the structure-based score is combined with non-structural evidence, again using a Bayesian approach, a large number of high confidence predictions are generated with an accuracy similar to that obtained from high-throughput experimental methods. This study showed that structural information can be exploited on a genome-wide scale and suggests that there are many other ways that structural information can be exploited.
Template-based methods have also been used to identify protein-small molecule interactions. Here, templates that are structurally similar to a query protein and that bind small molecules are typically identified by global sequence similarity, as with protein-protein interactions. Interaction models are then used to determine whether a query is likely to bind the same or similar ligands. For example, a model of the interaction can be constructed by placing the ligand from the template complex in the coordinate system of the query using the transformation relating the template and query. The strength of the interaction is then estimated based on the contacts the ligand makes in that model [18]. In other recent implementations, the similarity of putative ligand-contacting residues determined from the alignment to the template is examined to determine whether the query will bind similar ligands. [19,20]. Such an approach has been applied on a proteome-wide scale and drugs targeting specific query proteins have been validated [21*]. As is the case for protein-protein interactions, templates are generally not identified based on structural similarity. However such an approach is likely to be quite fruitful since we have recently shown that even proteins that only share structural similarity frequently bind ligands at geometrically equivalent positions on their surfaces, making them suitable for ligand binding site prediction [9].
Template-based methods also been applied to the problem of identifying proteins that bind DNA and RNA [22,23*]. Here, the confidence in the inference of DNA/RNA binding was determined using an estimate of the binding affinity using a statistical potential specifically designed for protein-nucleic acid interactions. In one study [23*] previously unknown RNA-binding proteins were identified and validated.
Exploiting Local Structural Similarity
Proteins can have local structural similarity without an obvious global sequence or structural relationship, a phenomenon that suggests that protein fold space is continuous, at least in part [24] [25]. Local similarities are often indicative of functional similarity, a fact that is exploited by a number of template-based methods for function prediction. For example, structures of interacting proteins have been analyzed to identify common substructures consisting of small sets of secondary structure elements that mediate a protein-protein interaction [26]. These substructures are used as templates instead of the full protein, creating an interaction model by superposing the substructures if they are present in two potentially interacting query proteins [26]. A similar approach has been developed to predict interactions mediated by globular domains and peptides [27].
Local structural similarity is widely used in the identification of enzyme active sites. In these approaches, proteins with known enzymatic function are analyzed to extract a library of active sites which can be represented in several ways [28]. One commonly used representation is simply the three-dimensional coordinates the subset of residues in the protein present in the active site. A query protein of unknown function is then compared to each of these subsets using various techniques, such as graph theory [29,30] or dynamic programming [31] to determine whether the query has a similar active site and hence similar function.
In another application of local similarity, conserved and covarying residues calculated from a multiple sequence alignment of query orthologs are used to identify subsets of residues that are clustered near each other in the 3-dimensional query structure. This 3-dimensional cluster is then taken to be a putative active site and is used to scan a library of known template functional sites [32,33].
Template Families
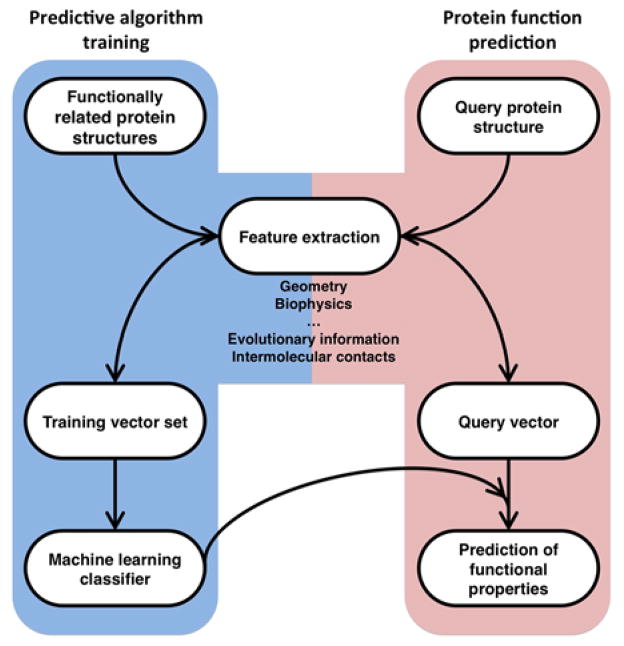
The existence of families of proteins with similar functions is enabling the development of a range of approaches to protein function annotation. Such template families can be used to train machine learning classifiers, using the strategy outlined in Figure 2. Classifiers discriminate, based on specific features, between sets of proteins that have a given function and sets of proteins that do not. Features can include, for example, geometric and biophysical properties or sequence conservation/covariation of amino acids in a binding pocket. The classifier is then used to derive a confidence score. This approach has been used recently to predict protein-protein interactions [34], protein-protein interfaces [35] small-molecule binding sites [36], and DNA and RNA binding sites [37,38,39].
Figure 2. Using a machine learning classifier for protein function annotation.
Blue panel: Proteins which share a functional relationship are collected, where the relationship can be specific (e.g., two proteins carrying out the same enzymatic reaction) or general (e.g., involved in protein-protein interaction). A vector of features (x,y…) is calculated for each structure in the collection, where x and y are the numerical quantification of some property of the structure (e.g., x may be the number of residues in the largest hydrophobic patch, and y might be the average degree of evolutionary conservation of those residues). A machine learning classifier takes this training set of feature vectors and attempts to identify patterns, i.e., the numerical values that are more likely to be associated with the function. This is typically done by comparing the feature vectors to those calculated for a collection of proteins known not to carry it out and quantifying any difference using statistical measures. Red panel: In annotation, the same set of features is calculated for a protein whose function is unknown and a confidence score for whether the protein has the given function is calculated based on its similarity to patterns found for the training set.
Template families can alternatively be used to train various potentials, i.e. identifying residues that occur more frequently in certain types of interfaces, as compared to a non-interfacial background. This approach has recently been used to identify novel proteins that interact with DNA or RNA [22,23*] and novel interactors of Bcl2 proteins [40*]. In another approach, a large set of protein-peptide structures was examined to construct, for each of the twenty amino acids, a three-dimensional grid of boxes containing the set of chemical groups that more frequently interact with that amino acid in protein-peptide interfaces [41]. These grids were used to predict protein-peptide interaction sites by placing them near a protein surface to identify positions where interaction with a given amino acid would be favorable.
Conclusion
The use of protein structure to annotate protein function is of course not new, but the use of structural alignments is a more recent development. As summarized above, these are generally used in two ways. First, once a structurally similar template has been identified, for example based on sequence based methods, structural alignment offers an approach to construct interaction models that enable the evaluation of the likelihood that a given complex will form. Second, structural alignment makes it possible to identify templates that cannot be identified from sequence. Improved homology modeling techniques, and technologies to efficiently search structural databases are ushering in an era where “structural BLAST” [9] can be used to annotate protein function on an unprecedented scale, comparable to that of sequence-based methods. Moreover, structural alignments make it possible to annotate function based on sets of related proteins, determined from sequence, alignments, annotated structural databases such as CATH [42] and SCOP [43], or interactively as in the MarkUs server [44].
The use of structural information and homology modeling in template-based methods has several advantages. For proteins that are closely related in sequence, identifying any functional differences between them generally requires examination of the biophysical properties of the structure such as electrostatic potential [45]. Machine learning using features derived from structure lowers the computational cost of using structure compared to other techniques, allowing them to be applied in a genome-wide scale. Moreover, template-based methods combined with machine learning are not as sensitive to modeling errors as detailed structural models. Steric clashes due, for example, to inaccurate modeling or conformational changes associated with binding are typically not a problem for machine-learning approaches but can limit the applicability of docking [46]. Other errors in modeling that can be accommodated by machine learning include the absence, in a modeled structure, of a post-translational modification. This can confound the experimental identification of protein-protein interactions which may not sample a cellular state in which the modification is present [47]. All these issues make template-based approaches an important complement to purely sequence-based approaches, to those based on high-resolution, computationally intensive modeling, and to high-throughput experimental approaches to determine function.
Most notably, template-based methods are having considerable biological impact, and have been applied to identify, almost always with experimental validation, previously unknown protein-protein interactions [13*,40*], RNA-binding proteins [23*], membrane binding-proteins [48*], enzyme families [49*], drug targets [21*], to analyze biological pathways [50,51] and to understand the relationship between genetic variation and disease [52*]. Protein structure is, of course, a key component of these approaches. In particular, the ability to examine and compare structural features and to construct approximate models of proteins with their interacting partners is enabling the development of techniques to add confidence to computational function annotation transfer that are not possible with purely sequence-based approaches. Although they still for the most part limited to the community of biologists familiar with three-dimensional structure, structure-based function annotation methods are becoming more accessible as new web servers are created to facilitate their use (see [18,36,41,45,53,54,55] for just a few recent examples). As new structures become available and new techniques are developed, the applicability and utility of these methods will almost certainly grow.
Acknowledgments
This work is supported by NIH grants GM030518, GM094597, and CA121852.
Contributor Information
T. Scott Chen, Email: tc2640@columbia.edu.
Lei Deng, Email: leideng@csu.edu.en.
Jose Ignacio Garzon, Email: jig2114@columbia.edu.
Howook Hwang, Email: hh2533@columbia.edu.
Gorka Lasso, Email: gl2411@columbia.edu.
Antonina Silkov, Email: as2137@columbia.edu.
Barry Honig, Email: bh6@columbia.edu.
References
- 1.The UniProt C. Reorganizing the protein space at the Universal Protein Resource (UniProt) Nucleic Acids Research. 2012;40:D71–D75. doi: 10.1093/nar/gkr981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Radivojac P, Clark WT, Oron TR, Schnoes AM, Wittkop T, Sokolov A, Graim K, Funk C, Verspoor K, Ben-Hur A, et al. A large-scale evaluation of computational protein function prediction. Nat Meth. 2013;10:221–227. doi: 10.1038/nmeth.2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwede T. Protein Modeling: What Happened to the “Protein Structure Gap”? Structure. 2013;21:1531–1540. doi: 10.1016/j.str.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gallo Cassarino T, Bordoli L, Schwede T. Assessment of ligand binding site predictions in CASP10. Proteins: Structure, Function, and Bioinformatics. 2014;82:154–163. doi: 10.1002/prot.24495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Skolnick J, Zhou H, Gao M. Are predicted protein structures of any value for binding site prediction and virtual ligand screening? Current Opinion in Structural Biology. 2013;23:191–197. doi: 10.1016/j.sbi.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodrigues JPGLM, Melquiond ASJ, Karaca E, Trellet M, van Dijk M, van Zundert GCP, Schmitz C, de Vries SJ, Bordogna A, Bonati L, et al. Defining the limits of homology modeling in information-driven protein docking. Proteins: Structure, Function, and Bioinformatics. 2013;81:2119–2128. doi: 10.1002/prot.24382. [DOI] [PubMed] [Google Scholar]
- 7.Petrey D, Honig B. Protein structure prediction: inroads to biology. Mol Cell. 2005;20:811–819. doi: 10.1016/j.molcel.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 8.Dukka BK. STRUCTURE-BASED METHODS FOR COMPUTATIONAL PROTEIN FUNCTIONAL SITE PREDICTION. Computational and Structural Biotechnology Journal. 2013;8:1–8. doi: 10.5936/csbj.201308005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dey F, Cliff Zhang Q, Petrey D, Honig B. Toward a “Structural BLAST”: Using structural relationships to infer function. Protein Science. 2013;22:359–366. doi: 10.1002/pro.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vakser IA. Low-resolution structural modeling of protein interactome. Current Opinion in Structural Biology. 2013 doi: 10.1016/j.sbi.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kundrotas PJ, Vakser IA, Janin J. Structural templates for modeling homodimers. Protein Science. 2013;22:1655–1663. doi: 10.1002/pro.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kundrotas PJ, Zhu Z, Janin J, Vakser IA. Templates are available to model nearly all complexes of structurally characterized proteins. Proceedings of the National Academy of Sciences. 2012 doi: 10.1073/pnas.1200678109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13*.Zhang QC, Petrey D, Deng L, Qiang L, Shi Y, Thu CA, Bisikirska B, Lefebvre C, Accili D, Hunter T, et al. Structure-based prediction of protein-protein interactions on a genome-wide scale. NATURE. 2012;490:556–560. doi: 10.1038/nature11503. An example of how homology modeling and machine learning were used to apply template-based method on a genome-wide scale to predict protein-protein interactions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee H, Park H, Ko J, Seok C. GalaxyGemini: a web server for protein homo-oligomer structure prediction based on similarity. Bioinformatics. 2013;29:1078–1080. doi: 10.1093/bioinformatics/btt079. [DOI] [PubMed] [Google Scholar]
- 15.Tyagi M, Hashimoto K, Shoemaker BA, Wuchty S, Panchenko AR. Large-scale mapping of human protein interactome using structural complexes. EMBO Reports. 2012;13:266–271. doi: 10.1038/embor.2011.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guerler A, Govindarajoo B, Zhang Y. Mapping Monomeric Threading to Protein–Protein Structure Prediction. Journal of Chemical Information and Modeling. 2013;53:717–725. doi: 10.1021/ci300579r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu L, Arakaki AK, Lu H, Skolnick J. Multimeric threading-based prediction of protein-protein interactions on a genomic scale: application to the Saccharomyces cerevisiae proteome. Genome Research. 2003;13:1146–1154. doi: 10.1101/gr.1145203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Beer TAP, Laskowski RA, Duban M-E, Chan AWE, Anderson WF, Thornton JM. LigSearch: a knowledge-based web server to identify likely ligands for a protein target. Acta Crystallographica Section D. 2013;69:2395–2402. doi: 10.1107/S0907444913022294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou H, Skolnick J. FINDSITEcomb: A Threading/Structure-Based, Proteomic-Scale Virtual Ligand Screening Approach. Journal of Chemical Information and Modeling. 2012;53:230–240. doi: 10.1021/ci300510n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roy A, Yang J, Zhang Y. COFACTOR: an accurate comparative algorithm for structure-based protein function annotation. Nucleic Acids Research. 2012;40:W471–W477. doi: 10.1093/nar/gks372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21*.Srinivasan B, Zhou H, Kubanek J, Skolnick J. Experimental validation of FINDSITEcomb virtual ligand screening results for eight proteins yields novel nanomolar and micromolar binders. Journal of Cheminformatics. 2014;6:16. doi: 10.1186/1758-2946-6-16. An example of how template-based methods were used it identify novel protein-small molecule interactions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao H, Wang J, Zhou Y, Yang Y. Predicting DNA-Binding Proteins and Binding Residues by Complex Structure Prediction and Application to Human Proteome. PLoS ONE. 2014;9:e96694. doi: 10.1371/journal.pone.0096694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23*.Zhao H, Yang Y, Janga SC, Kao CC, Zhou Y. Prediction and validation of the unexplored RNA-binding protein atlas of the human proteome. Proteins: Structure, Function, and Bioinformatics. 2014;82:640–647. doi: 10.1002/prot.24441. An example of how template-based methods were used to identify novel protein-RNA interactions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petrey D, Honig B. Is protein classification necessary? Toward alternative approaches to function annotation. Current Opinion in Structural Biology. 2009;19:363–368. doi: 10.1016/j.sbi.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nepomnyachiy S, Ben-Tal N, Kolodny R. Global view of the protein universe. Proceedings of the National Academy of Sciences. 2014;111:11691–11696. doi: 10.1073/pnas.1403395111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baspinar A, Cukuroglu E, Nussinov R, Keskin O, Gursoy A. PRISM: a web server and repository for prediction of protein–protein interactions and modeling their 3D complexes. Nucleic Acids Research. 2014;42:W285–W289. doi: 10.1093/nar/gku397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verschueren E, Vanhee P, Rousseau F, Schymkowitz J, Serrano L. Protein-Peptide Complex Prediction through Fragment Interaction Patterns. Structure. 2013;21:789–797. doi: 10.1016/j.str.2013.02.023. [DOI] [PubMed] [Google Scholar]
- 28.Laskowski RA, Watson JD, Thornton JM. Protein function prediction using local 3D templates. Journal of Molecular Biology. 2005;351:614–626. doi: 10.1016/j.jmb.2005.05.067. [DOI] [PubMed] [Google Scholar]
- 29.Sanjaka BVMV, Changhui Y. Prediction of enzyme catalytic sites on protein using a graph kernel method. Systems Biology (ISB), 2013 7th International Conference; 23–25 Aug. 2013; 2013. pp. 31–33. [Google Scholar]
- 30.Nilmeier JP, Kirshner DA, Wong SE, Lightstone FC. Rapid Catalytic Template Searching as an Enzyme Function Prediction Procedure. PLoS ONE. 2013;8:e62535. doi: 10.1371/journal.pone.0062535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao M, Skolnick J. APoc: large-scale identification of similar protein pockets. Bioinformatics. 2013;29:597–604. doi: 10.1093/bioinformatics/btt024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amin SR, Erdin S, Ward RM, Lua RC, Lichtarge O. Prediction and experimental validation of enzyme substrate specificity in protein structures. Proceedings of the National Academy of Sciences. 2013;110:E4195–E4202. doi: 10.1073/pnas.1305162110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Erdin S, Venner E, Lisewski A, Lichtarge O. Function prediction from networks of local evolutionary similarity in protein structure. BMC Bioinformatics. 2013;14:S6. doi: 10.1186/1471-2105-14-S3-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maleki M, Hall M, Rueda L. Using structural domains to predict obligate and non-obligate protein-protein interactions. Computational Intelligence in Bioinformatics and Computational Biology (CIBCB), 2012 IEEE Symposium; 9–12 May 2012; 2012. pp. 9–15. [Google Scholar]
- 35.Bendell C, Liu S, Aumentado-Armstrong T, Istrate B, Cernek P, Khan S, Picioreanu S, Zhao M, Murgita R. Transient protein-protein interface prediction: datasets, features, algorithms, and the RAD-T predictor. BMC Bioinformatics. 2014;15:82. doi: 10.1186/1471-2105-15-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brylinski M, Feinstein W. eFindSite: Improved prediction of ligand binding sites in protein models using meta-threading, machine learning and auxiliary ligands. Journal of Computer-Aided Molecular Design. 2013;27:551–567. doi: 10.1007/s10822-013-9663-5. [DOI] [PubMed] [Google Scholar]
- 37.Liu R, Hu J. DNABind: A hybrid algorithm for structure-based prediction of DNA-binding residues by combining machine learning- and template-based approaches. Proteins: Structure, Function, and Bioinformatics. 2013;81:1885–1899. doi: 10.1002/prot.24330. [DOI] [PubMed] [Google Scholar]
- 38.Yang X-X, Deng Z-L, Liu R. RBRDetector: Improved prediction of binding residues on RNA-binding protein structures using complementary feature- and template-based strategies. Proteins: Structure, Function, and Bioinformatics. 2014;82:2455–2471. doi: 10.1002/prot.24610. [DOI] [PubMed] [Google Scholar]
- 39.Walia RR, Xue LC, Wilkins K, El-Manzalawy Y, Dobbs D, Honavar V. RNABindRPlus: A Predictor that Combines Machine Learning and Sequence Homology-Based Methods to Improve the Reliability of Predicted RNA-Binding Residues in Proteins. PLoS ONE. 2014;9:e97725. doi: 10.1371/journal.pone.0097725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40*.DeBartolo J, Taipale M, Keating AE. Genome-Wide Prediction and Validation of Peptides That Bind Human Prosurvival Bcl-2 Proteins. PLoS Comput Biol. 2014;10:e1003693. doi: 10.1371/journal.pcbi.1003693. An example of how template-based methods were used to identify novel-protein peptide interactions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trabuco LG, Lise S, Petsalaki E, Russell RB. PepSite: prediction of peptide-binding sites from protein surfaces. Nucleic Acids Research. 2012;40:W423–W427. doi: 10.1093/nar/gks398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sillitoe I, Cuff AL, Dessailly BH, Dawson NL, Furnham N, Lee D, Lees JG, Lewis TE, Studer RA, Rentzsch R, et al. New functional families (FunFams) in CATH to improve the mapping of conserved functional sites to 3D structures. Nucleic Acids Research. 2013;41:D490–D498. doi: 10.1093/nar/gks1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Andreeva A, Howorth D, Chothia C, Kulesha E, Murzin AG. SCOP2 prototype: a new approach to protein structure mining. Nucleic Acids Research. 2013 doi: 10.1093/nar/gkt1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fischer M, Zhang QC, Dey F, Chen BY, Honig B, Petrey D. MarkUs: a server to navigate sequence–structure–function space. Nucleic Acids Research. 2011;39:W357–W361. doi: 10.1093/nar/gkr468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bachman BJ, Venner E, Lua RC, Erdin S, Lichtarge O. ETAscape: analyzing protein networks to predict enzymatic function and substrates in Cytoscape. Bioinformatics. 2012;28:2186–2188. doi: 10.1093/bioinformatics/bts331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vreven T, Hwang H, Pierce BG, Weng Z. Evaluating template-based and template-free protein–protein complex structure prediction. Briefings in bioinformatics. 2013 doi: 10.1093/bib/bbt047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu BA, Engelmann BW, Nash PD. High-throughput analysis of peptide-binding modules. Proteomics. 2012;12:1527–1546. doi: 10.1002/pmic.201100599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48*.Chen Y, Sheng R, Källberg M, Silkov A, Tun Moe P, Bhardwaj N, Kurilova S, Hall Randy A, Honig B, Lu H, et al. Genome-wide Functional Annotation of Dual-Specificity Protein- and Lipid-Binding Modules that Regulate Protein Interactions. Molecular Cell. 2012;46:226–237. doi: 10.1016/j.molcel.2012.02.012. An example of how template-based methods were used to identify novel membrane-binding properties of protein families. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49*.Wang Z, Yin P, Lee J, Parasuram R, Somarowthu S, Ondrechen M. Protein function annotation with Structurally Aligned Local Sites of Activity (SALSAs) BMC Bioinformatics. 2013;14:S13. doi: 10.1186/1471-2105-14-S3-S13. An example of how template-based methods were used to identify novel members of enzyme families. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chang RL, Andrews K, Kim D, Li Z, Godzik A, Palsson BO. Structural Systems Biology Evaluation of Metabolic Thermotolerance in Escherichia coli. Science. 2013;340:1220–1223. doi: 10.1126/science.1234012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guven Maiorov E, Keskin O, Gursoy A, Nussinov R. The structural network of inflammation and cancer: Merits and challenges. Seminars in Cancer Biology. 2013;23:243–251. doi: 10.1016/j.semcancer.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 52.Das J, Fragoza R, Lee HR, Cordero NA, Guo Y, Meyer MJ, Vo TV, Wang X, Yu H. Exploring mechanisms of human disease through structurally resolved protein interactome networks. Molecular Biosystems. 2014;10:9–17. doi: 10.1039/c3mb70225a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tan KP, Varadarajan R, Madhusudhan MS. DEPTH: a web server to compute depth and predict small-molecule binding cavities in proteins. Nucleic Acids Research. 2011 doi: 10.1093/nar/gkr356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Simonetti FL, Teppa E, Chernomoretz A, Nielsen M, Marino Buslje C. MISTIC: mutual information server to infer coevolution. Nucleic Acids Research. 2013;41:W8–W14. doi: 10.1093/nar/gkt427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mosca R, Ceol A, Aloy P. Interactome3D: adding structural details to protein networks. Nat Meth. 2013;10:47–53. doi: 10.1038/nmeth.2289. [DOI] [PubMed] [Google Scholar]