Abstract
A thorough review of the literature is the basis of all research and evidence-based practice. A gold-standard efficient and exhaustive search strategy is needed to ensure all relevant citations have been captured and that the search performed is reproducible. The PubMed database comprises both the MEDLINE and non-MEDLINE databases. MEDLINE-based search strategies are robust but capture only 89% of the total available citations in PubMed. The remaining 11% include the most recent and possibly relevant citations but are only searchable through less efficient techniques. An effective search strategy must employ both the MEDLINE and the non-MEDLINE portion of PubMed to ensure all studies have been identified. The robust MEDLINE search strategies are used for the MEDLINE portion of the search. Usage of the less robust strategies is then efficiently confined to search only the remaining 11% of PubMed citations that have not been indexed for MEDLINE. The current article offers step-by-step instructions for building such a search exploring methods for the discovery of medical subject heading (MeSH) terms to search MEDLINE, text-based methods for exploring the non-MEDLINE database, information on the limitations of convenience algorithms such as the “related citations feature,” the strengths and pitfalls associated with commonly used filters, the proper usage of Boolean operators to organize a master search strategy, and instructions for automating that search through “MyNCBI” to receive search query updates by email as new citations become available.
Keywords: nutrition, enteral nutrition, parenteral nutrition, nutrition support practice, outcomes research/quality, research and diseases
The ability to perform an effective search strategy comprises the foundation of any evidence-based discipline. Whether one is functioning in a clinical or research capacity, the assumption is that the expert suggestions offered arise from a complete review of the relevant literature. In systematic reviews and meta-analyses, this is taken a step further, necessitating that the methods of these exhaustive literature reviews be reproducible. The conveniences afforded by the basic PubMed search engine and algorithms, such as the “Related Citations” feature, provide powerful search tools but take much of the control out of the hands of the user. Blind usage of these hands-off methods weakens the user’s ability to ensure a thorough, systematic, and reproducible search. A common solution to this problem is to limit the search strategy to include only the MEDLINE portion of the PubMed database, allowing for the usage of more robust, user-controlled search methods. Unfortunately, depending on the journal indexed, MEDLINE is often as much as a year behind the current research,1 leaving many of the most recent and relevant research citations out of the search results. The strategies presented in this article offer a solution to this problem.
The general landscape of PubMed has been previously described in a recent article by Lindsey and Olin.2 The objective of the current article is to augment that landscape by proposing a detailed method through which its many components may be employed and integrated into one highly efficient, reproducible, and exhaustive search strategy. True comprehension of this system provides the ability to create a master search for any topic. Furthermore, the many free tools provided by the National Center for Biotechnology Information (NCBI) and the National Library of Medicine (NLM) through PubMed make it possible to receive timely notifications of citations, directly relevant to any area of one’s field as they become available. Part I of this article discusses theories, methods, and key concepts for using medical subject headings (MeSH) to search PubMed’s MEDLINE database, the less robust strategies necessary for searching the remaining non-MEDLINE database, and methods for restricting the less robust search tools to search only the non-MEDLINE database. Part II provides a step-by-step tutorial for implementing part I as well as methods for automating a search strategy so users may receive daily, weekly, or monthly ongoing updated results to their search query by email.
Part I: Theory, Methods, and Key Concepts for Performing a Master Search
PubMed, MEDLINE, and MeSH: The Filing System for Evidence
PubMed is a free citation database from the NLM. It currently houses >23 million citations dating back to 1809 with new citations added daily. The largest portion of PubMed citations comes from the MEDLINE database, which is an NLM database specific to the fields of biomedicine. The database covers citations from 1946 to present3 and currently presents 88.9% of all citations in PubMed. Of the remaining 11.1%, 23.3% are in the process of becoming MEDLINE indexed, 15.3% have been submitted by the publisher and may become MEDLINE indexed in the future, 51.7% are available on PubMed but will not be indexed by MEDLINE, and 9.7% belong to an older version of MEDLINE that was indexed under less rigorous standards.4 The distinctive feature of a MEDLINE-indexed citation is the manual rigor under which it is cataloged and categorized. Highly skilled information technicians read each article to determine the key points as well as its smaller discussion points. These points are then manually catalogued under a series of searchable Medical Subject Headings (MeSH terms) and subheadings.5
The current staff that manages citations and MeSH terms at the NLM generally have a master’s degree or higher in some form of biomedical field.6 Their collective expertise embraces a diverse range of biomedical topics. Each attends rigorous training in document analysis and cataloguing for MeSH (visible on the NLM website).7 They then complete an internship/apprenticeship with an indexing mentor who checks every indexed article for quality and consistency with MEDLINE cataloguing methodology.
The MeSH system is essentially a digital filing cabinet with 16 biomedical topic drawers that match to the broadest MeSH terms. These drawers are filled with a 12-tier hierarchy of folders within folders, which systematically narrow into increasingly refined topic areas; each topic area is accessible through its MeSH term. Each MeSH term comes with a selection of up to 83 possible subheadings to further refine the topic area to a specific focus of interest, such as the “adverse effects,” “economics,” “ethics,” or “methods,” associated with a specific term.7 This system is intricate, but all the tools necessary to simplify its navigation are available for use through PubMed.
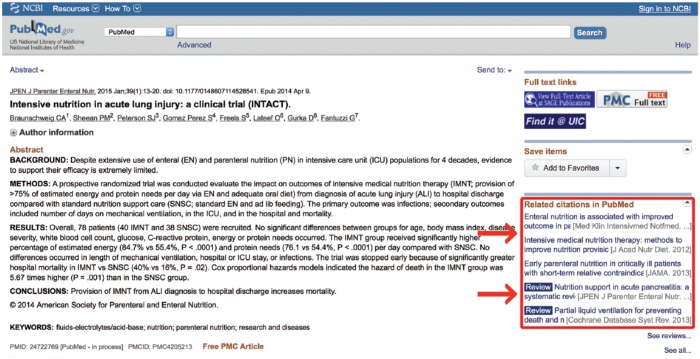
A general, unqualified PubMed search will retrieve citations through several different pathways. Wherever possible, PubMed will translate the user’s search terms into MeSH terminology, a process known as Automatic Term Mapping. Frequently, however, this is not possible, and PubMed must rely on the less sophisticated indexing system of word searching.8 When a search word does not map to a MeSH term, PubMed looks for the search phrase in other fields of the document. It first attempts to match the phrase to a journal name, then to an author name, then to a name of a research investigator, and finally it searches for the phrase in [all fields].5 A search in [all fields] includes areas such as the title field, the author field, the publication date field, and an abstract field when available.9 It then breaks the search phrase into its component words and repeats the above process for each individual word. While this may retrieve many relevant articles, these articles will be interspersed with an abundance of irrelevant articles. Many users circumnavigate this inconvenience by employing the “PubMed Related Citations” feature available on the right side of any PubMed search results page (Figure 1).
Figure 1.
Related citations.
Pros and Cons of “Related Citations”
When a user clicks on a citation, the “PubMed Related Citations” feature finds the 5 citations that contain the most verbiage in common with the selected citation. This algorithm-driven search utility is essentially an intricate word counter, which tracks the words in a citation and weighs them according to their frequency and location.10 It counts the number of times a word is repeated in the title, abstract, and list of MeSH terms in a document. It then applies different weights to the words according to where the word is found in the citation (ie, title, abstract, etc), the number of times it occurs in the document, and the frequency with which the word is found throughout all titles and abstracts in the database. A small mathematical adjustment is made for the length of the document, and a word score is assigned to the citation, which is used to determine similarity with other citations. This feature requires very little skill on the part of the user and is exceptionally good at quickly providing relevant articles. For more scientific pursuits, however, this feature may be inappropriate. While it is a useful feature, it is not a systematic search approach. Most research is built on the idea that an exhaustive literature review has been performed. With the related citations feature, an investigator does not know for sure which articles they have missed. Furthermore, science is built on the principle that someone’s methods of discovery must be reproducible. Every time a promising citation is selected, the “Related Citations” algorithm resets to that new citation and produces 5 more citations based on that new input, making the pathway of the search impossible to track. While this automated approach is a valuable feature, the ability to formulate an effective manual search strategy will enable investigators to know exactly which areas they are including and which they are ignoring, a vital component for any formal literature review.
Key Concepts of the Master Search Strategy for PubMed
As previously stated, 88.9% of the PubMed database comes from the MeSH-indexed MEDLINE database. While MEDLINE is a more robust search system, it can take weeks to months to years after release for an article to become MEDLINE indexed.1 The other 11.1% of non-MEDLINE citations are available in PubMed but without MeSH indexing. While this non-MEDLINE search system is less robust, its contents contain the most recent and temporally relevant research often many months before it would be found in a MEDLINE search. For this reason, a full review of the literature must have both a MEDLINE component and a non-MEDLINE component. The MEDLINE component may employ both MeSH and text searching (non-MeSH) strategies when necessary, but its results will always be limited to the MEDLINE database. By design, the non-MEDLINE component will use less robust non-MeSH search strategies, but its results will be limited to only include the 11.1% of citations that are not indexed in MEDLINE (Figure 2).
Figure 2.
Overview of the Master Search Strategy. Capital letters A, B, C, and D represent different or synonymous search terms in combination or separate. An “*” represents a truncated search term. MeSH, medical subject headings.
MeSH Strategies
The discovery of MeSH terms relevant to the chosen topic area is the foundation of an effective search strategy, as this prequalified collection of citations will comprise approximately 89% of the final search results. There are 2 main tools to determine which MeSH terms are most appropriate for a given search topic: (1) the MeSH database and (2) the “MeSH Terms” listed underneath the cataloged abstract of a relevant MEDLINE-indexed article.
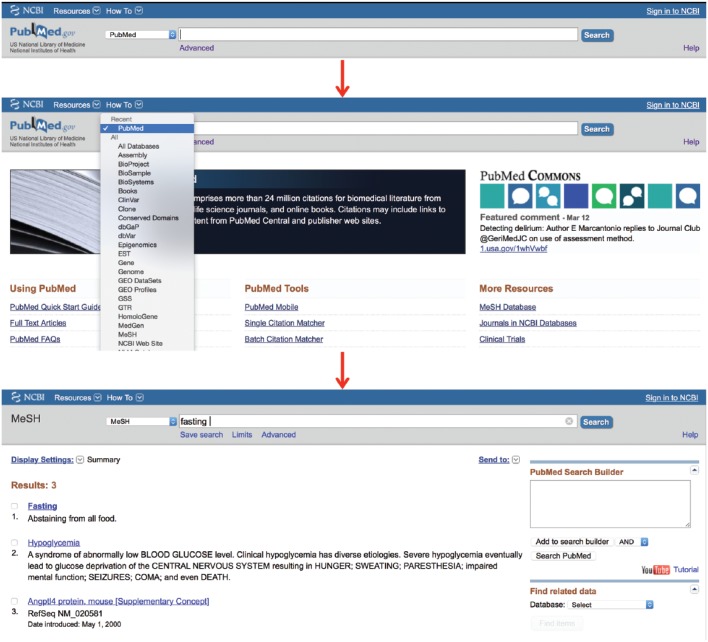
Key concept 1: The MeSH database
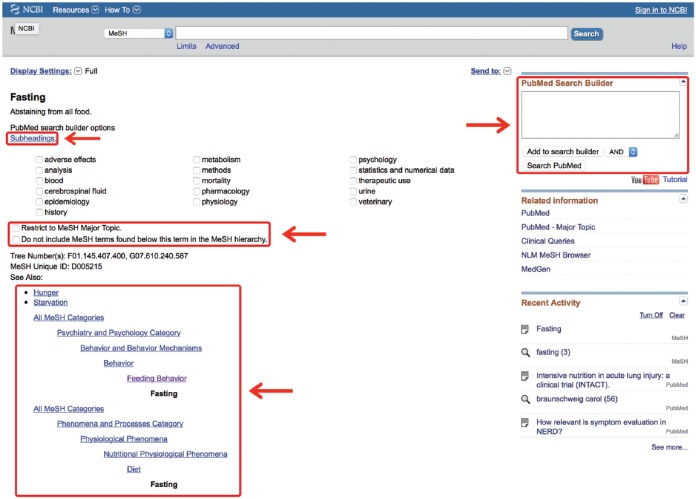
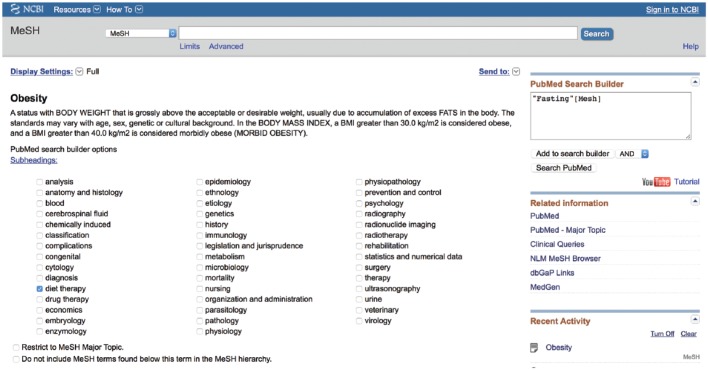
The MeSH database uses a Unified Medical Language System to automatically map a search term to its relevant MeSH terms and may be accessed from any PubMed page. A drop-down box to the left of the main PubMed search box may be activated and the “MeSH” option selected (Figure 3). Then, any term typed in the search box, providing it translates to MeSH terminology, will lead to a page of possible MeSH terms. Figure 3 provides an example using the search word “fasting.” This word yields 3 MeSH results (fasting, hypoglycemia and Angptl4 protein, mouse). The user may then click on any MeSH term to discover its definition, its possible subheadings, and its place in the MeSH hierarchy, allowing the user to view which broader term the MeSH term came from and which subterms that term includes (Figure 4; the MeSH term “fasting” was selected). In this way, the user may be very clear on exactly which article topics the term will cull if used in a search. This allows the user to restrict the search to one or more smaller MeSH terms or to conflate terms by choosing a single broader MeSH term to encompass them all. For example, “feeding behavior” is the next broadest term after “fasting.” This MeSH term will include the articles from the “fasting” MeSH folder, as well as other behavioral topics surrounding feeding. The user may also restrict a search to yield only citations wherein the MeSH term is the key topic of the article or restrict the search so that it does not contain any of the subterms within the main MeSH term.
Figure 3.
The medical subject headings (MeSH) database.
Figure 4.
Inside the medical subject headings (MeSH) term.
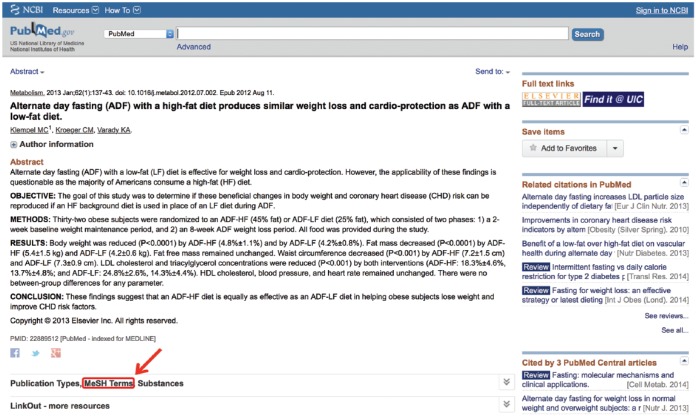
Key concept 2: Articles of known relevance
Often, it is helpful to search documents of known relevance to the search topic to assess which MeSH terms have been ascribed to that article. This may be done simply to make sure the users did not miss any terms, which may be helpful to their search, or because the non-MeSH term they searched did not successfully translate into a MeSH term. This is accomplished by clicking on the link located below the article’s abstract. This link typically contains a string of names, which vary from article to article. Provided the article has been MEDLINE indexed, one of those names will be “MeSH Terms” (Figure 5). Clicking on this link provides the user with all the possible MeSH terms under which this article may be found. This list may then be searched for other synonymous or relevant MeSH terms that match the concepts being queried. It should be noted that some terms have an asterisk (*) after their names. This is used to signify a term that represents a main topic of the article. All other MeSH terms listed will have been discussion points in the article but were not the main thrust of the article.
Figure 5.
Search medical subject headings (MeSH) terms of relevant articles.
Key concept 3: Working with Boolean operators
Three highly useful commands may be used in both PubMed and MEDLINE searching systems. They are AND, OR, and NOT. These are known as Boolean operators. They must be written in all caps, and they provide the search engine information regarding the relationship between different terms in a search query. Using AND between terms will yield only citation results that contain both terms. In this way, AND tends to contract and limit a search. Using OR between terms yields any citation that contains one or both terms. In this way, OR is an additive term. Using NOT between terms will tell the search engine to exclude any results with the term or terms it precedes. NOT is a risky operator, as it will remove any study that has been categorized according to the term being excluded regardless of its many other MeSH terms. If any of these terms are relevant, relevant citations will be lost.1 A good rule of thumb is to exclude with NOT only when the presence of the search term invalidates the purpose of the search. For example, if the query of interest were dietary patterns of normal healthy eaters in a particular population, it may be appropriate to exclude any studies that have been catalogued under “eating disorders.” Adding an expression such as “NOT “eating disorders” [MeSH]” would effectively remove any citations that have been cross-referenced in the “eating disorders” MeSH folder.
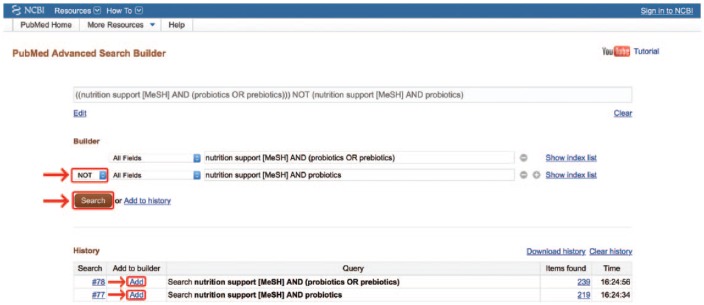
Perhaps the most appropriate use of NOT is when an entire search strategy must be excluded. For example, when researchers perform systematic reviews, they often must scan hundreds to thousands of articles for relevance under a carefully chosen search strategy. After lengthy scanning through citation titles, they sometimes realize they have left an important search term out of their strategy. They notice that adding the term into their strategy only increases their citation results by 30–40 citations, but these citations are interspersed within and indistinguishable from the hundreds of citations they have already scanned. To begin the scanning process over will result in hours of lost effort. A quick solution for this is to click on the “Advanced” link directly under the search box (Figure 6). This link leads to a page where recent searches have been saved in a search history. The “Advanced” page comes with a Search Builder that allows entire searches to be added and manipulated with Boolean operators. The new, expanded search strategy may be added to the first box by clicking “Add.” The original, more limited search strategy may be similarly added to the second box (Figure 7). From here, selecting the Boolean operator NOT from the drop-down menu and clicking “Search” will yield a search results page where only the 30–40 citations missed by the original search may be scanned for relevance without having to start the entire search over.
Figure 6.

Click the “Advanced” link.
Figure 7.
Boolean manipulation of entire searches. MeSH, medical subject headings.
Often, a search strategy contains several terms connected by many Boolean operators. When this occurs, it becomes important that these terms are grouped properly. Similar to algebraic equations, parentheses are used to delineate whether the operator is meant to act upon a single term, a grouping of terms, or even upon 2 or more full searches.1 For example, (a OR b) NOT (a AND b) would retrieve all the studies that are catalogued under “a” and all the studies that are catalogued under “b,” excluding those studies that are catalogued under both “a” and “b.” Analyzing the arrangement of parentheses in any search is vital to obtaining the intended results.
Key concept 4: The MeSH Search Builder
The PubMed Search Builder box, located on any MeSH Term page, allows the user to directly add the page’s MeSH terms to a search strategy and modify them with Boolean operators.11 From the web page of the chosen MeSH term, one simply clicks “Add to Search Builder” and chooses the appropriate Boolean operator from the drop-down menu (Figure 4). The MeSH term will then remain in the box as other terms are searched, allowing the user to build on that term. The Search Builder tool will attempt to use parentheses to group the items properly, but often the user will need to alter the parenthetical grouping of the terms. Fortunately, the search box is directly editable, allowing users to perfect their MeSH search before submission. Once the search is built, clicking “Search PubMed” will yield a robust list of article citations.
Non-MeSH Search Strategies
Depending on the availability of MeSH terminology for a particular search topic, non-MeSH strategies for both the MEDLINE and non-MEDLINE portion of the search may be necessary. Unlike the MeSH database, which uses the intelligence of trained humans to catalog the meaning of an article, the non-MeSH system uses computer intelligence to catalog the words. In fact, the non-MeSH system of searching does not even include the text in the article but focuses only on the text available in the citation and abstract. The word-based nature of a non-MeSH search places the responsibility on the user to use many different varieties of the same word.
Key concept 5: Non-MeSH search tips
The following tips1,8,11 may be helpful when constructing a non-MeSH search:
Use “quotations” to search a specific phrase or exact spelling of a word.
Be sure to include alternate spellings (eg, (color OR colour)).
Use a thesaurus to look up common synonyms or related words.
Include plural versions of appropriate words.
Include any acronyms with and without periods.
Include different endings for the same word (eg, microbiology, microbiologist, microbiologia, etc).
Using an asterisk (*) to truncate a word (eg, microbiolog*) will automate the culling of plurals and alternate suffixes as well as include all subheadings associated with the word in MeSH.
Key concept 6: The Search Details box
In a nonqualified default search, PubMed attempts to interpret the search terms into MeSH terms or non-MeSH variations. Unfortunately, its efforts are often incomplete or incorrect. If a search phrase is entered into PubMed nonqualified, it will first attempt to find MeSH terms to match the words. Often, the MeSH terms chosen will match the words but not the meaning of the phrase. For example, a search for Medium Chain Triglycerides will match the word Medium to the MeSH term for Communications Media[MeSH] and try to find citations within that term that contain the words Chain[All Fields] and Triglycerides[All Fields].
This valuable information regarding how PubMed interpreted the search may be found in the “Search Details” box on the middle right side of any PubMed results screen (Figure 8).11 The Search Details box is an invaluable tool for diagnosing problems in a search strategy. It instantly tells the user whether MeSH terms were found, lists what liberties PubMed may have taken to expand the search, and provides a snapshot of the cause-and-effect relationship between a search strategy and its outcome.
Figure 8.
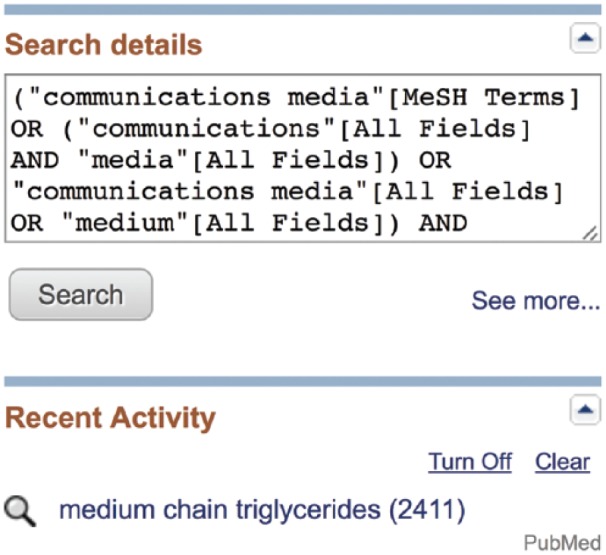
Search details box.
Key concept 7: Filters
As previously stated, any exhaustive search strategy will have a MEDLINE component and a non-MEDLINE component. The MEDLINE component should use MeSH-based strategies wherever possible and non-MeSH strategies only where necessary. The non-MEDLINE component must use only non-MeSH strategies, because only MEDLINE articles are cataloged with MeSH terms. It is then necessary to restrict the non-MEDLINE portion of the search to only the small fraction of citations that are not indexed in MEDLINE. Several filters, known as “subsets,” select for these non-MEDLINE citations. Subset filters may be recognized by their tag, [sb]. Examples of non-MEDLINE subset filters include publisher[sb], inprocess[sb], oldmedline[sb], and pubmednotmedline[sb].12 The simplest filter that will collect citations from all 4 of these categories is to add a Boolean NOT to the filter medline[sb] and connect this at the end of the non-MEDLINE portion of the search.13 This will exclude all MEDLINE citations from that portion of the search strategy.
It is important to acknowledge that any articles culled from the non-MEDLINE portion of the search may become MEDLINE articles by the time another researcher attempts to reproduce the search. If one is performing a systematic review, it is imperative for methods reproducibility that they provide a list of which articles were chosen from the non-MEDLINE search to account for their repositioning from the PubMed to MEDLINE indexing system.
Some Further Filter Issues
For the convenience of the user, PubMed makes several filters available on the left-hand side of any search results page with checkboxes.14 Many of these automatic filters on the left of the main search page are MeSH-based filters. They will only pull articles that have been indexed with MeSH terms. When these MeSH filters are selected, they are automatically attached to the entire current search with an AND Boolean operator. This means the entire search has now been limited to yield only MEDLINE citations. To use these filters without losing valuable non-MEDLINE citations, they must be added manually. The trick is to keep the MeSH side of the search separate from the non-MeSH side, connected only with an OR Boolean operator.
Filters that use MeSH include:
Article Types, Species, Sex, and Age filters.
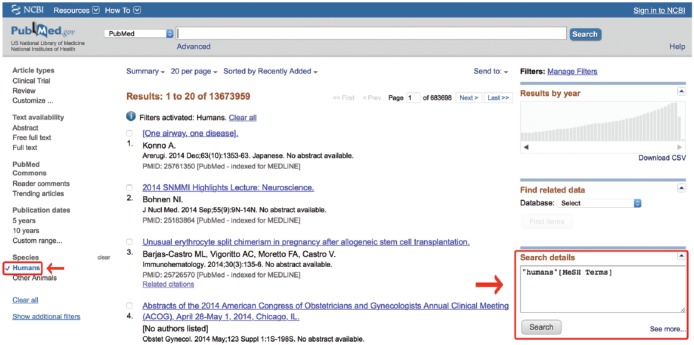
To enter these filters by hand, one must know how they are coded. Discovering this involves entering any term into the search box and clicking “Search.” Then, deleting that term and checking the box of the desired filter will cause the “Search Details” box to reveal how that particular MeSH term is catalogued (Figure 9). An alternative option is to find the term in the MeSH database and add it with the PubMed Search Builder discussed earlier. In either case, that exact term, including its [MeSH] tag, may be inserted into the MeSH section of the search with an AND Boolean operator to restrict the filter’s limiting effect to only the MEDLINE portion of the search strategy.
Figure 9.
Obtaining a filter tag from the Search Details box. MeSH, medical subject headings.
Key concept 8: The Master Search Strategy
With all the components of an exhaustive but efficient search in place, it is possible to assemble the search strategy. Below is a simplified way to state the basic Master Search Formula for PubMed from Figure 2:
Simple version:
This formula allows the users to use their knowledge of MeSH terms to focus their search on human-cataloged MEDLINE articles while limiting their default PubMed search to only those articles that have no MEDLINE equivalents. In this way, their search will be exhaustive but efficient.
Part II: A Step-by-Step Tutorial
The previous sections of this article have outlined the components of the Master Search Strategy. This next section demonstrates these components in action as one performs a Master Search. A screen-capture tutorial of this process may be found at www.ResearchRDN.com. Building a Master Search occurs in phases. Some phases are simple and linear, while others require more reflection and exploration of terms. Key concepts (KCs) from the previous sections of this article will be referred to throughout the search to provide clarity. While learning, it is helpful to open a document where one may copy and paste the components of the search strategy as they are created. These components may then be assembled within that document and pasted back into the PubMed search box once the search strategy is complete.
As an example, consider the nonqualified search terms Alternate Day Fasting entered into the PubMed search box. Currently, this search yields 121 results. Scrolling down to the “Search Details” box (KC-6) sheds light on how PubMed processed this search. It is as follows:
alternate[All Fields] AND day[All Fields] AND (“fasting”[MeSH Terms] OR “fasting”[All Fields])
Notice that the only term that mapped to a MeSH term was “Fasting”[MesH]. The current search is indeed 2 separate searches. One is searching within the “Fasting”[MesH] term for citations that also include the words “alternate” AND “day.” This is good in the sense that it will be MEDLINE articles but inadequate by itself, since it will exclude all non-MEDLINE articles. The other search is looking for any citations anywhere that contain the words “Alternate” AND “Day” AND “Fasting.” This is good in the sense that it will capture both MEDLINE and non-MEDLINE articles but bad in the sense that it will cull many articles that contain these 3 words in combinations irrelevant to the search query. To improve this search, the MEDLINE search and the non-MEDLINE search must be cleanly separated (Figure 2).
The MEDLINE Portion
We will begin by constructing the MEDLINE portion of the search.
Action steps:
1. From the PubMed.gov homepage, make sure the “MeSH” option is selected from the drop-down menu to the left of the PubMed search box.
2. Enter the word “Fasting” and click “Search.”
This will display a page with the term’s description, possible subheadings, and a visual of the term’s place within the hierarchy of MeSH terms (Figure 4) (KC-1). “Fasting”[MeSH] is the narrowest subterm for a long lineage of behavioral MeSH terms and nutrition physiology MeSH terms.
Action step:
3. Perform a similar search for “Alternate” and “Day.”
When “Alternate” and “Day” are searched in the MeSH database, the Search Detail box reveals they do not directly translate to MeSH terms. It may be useful to search an article in PubMed of known relevance to alternate-day fasting (KC-2) for its catalogued MeSH terms to discover how the MEDLINE staff indexes these concepts. One such article is by Klempel et al,15 entitled “Alternate day fasting (ADF) with a high-fat diet produces similar weight loss and cardio-protection as ADF with a low-fat diet.”
Action steps:
4. Click on the drop-down box to the left of the PubMed search box and change the selection from “MeSH” to “PubMed.”
5. Type the name of the above article into the PubMed search box and click “Search.”
Below the abstract of this article in PubMed is a string of links entitled “Publications Types, MeSH Terms, Substances” (Figure 5). Clicking on this will reveal how the document is MeSH indexed.
Action step:
6. Click on “Publications Types, MeSH Terms, Substances.”
It appears from this list that some additional relevant MeSH terms may include “obesity/diet therapy” and “weight loss/physiology” (Figure 10). The terms after the “/” signify subheadings. Record these terms. They will be added to the search shortly. At this point, it may be beneficial to further restrict the search to human subjects.
Figure 10.
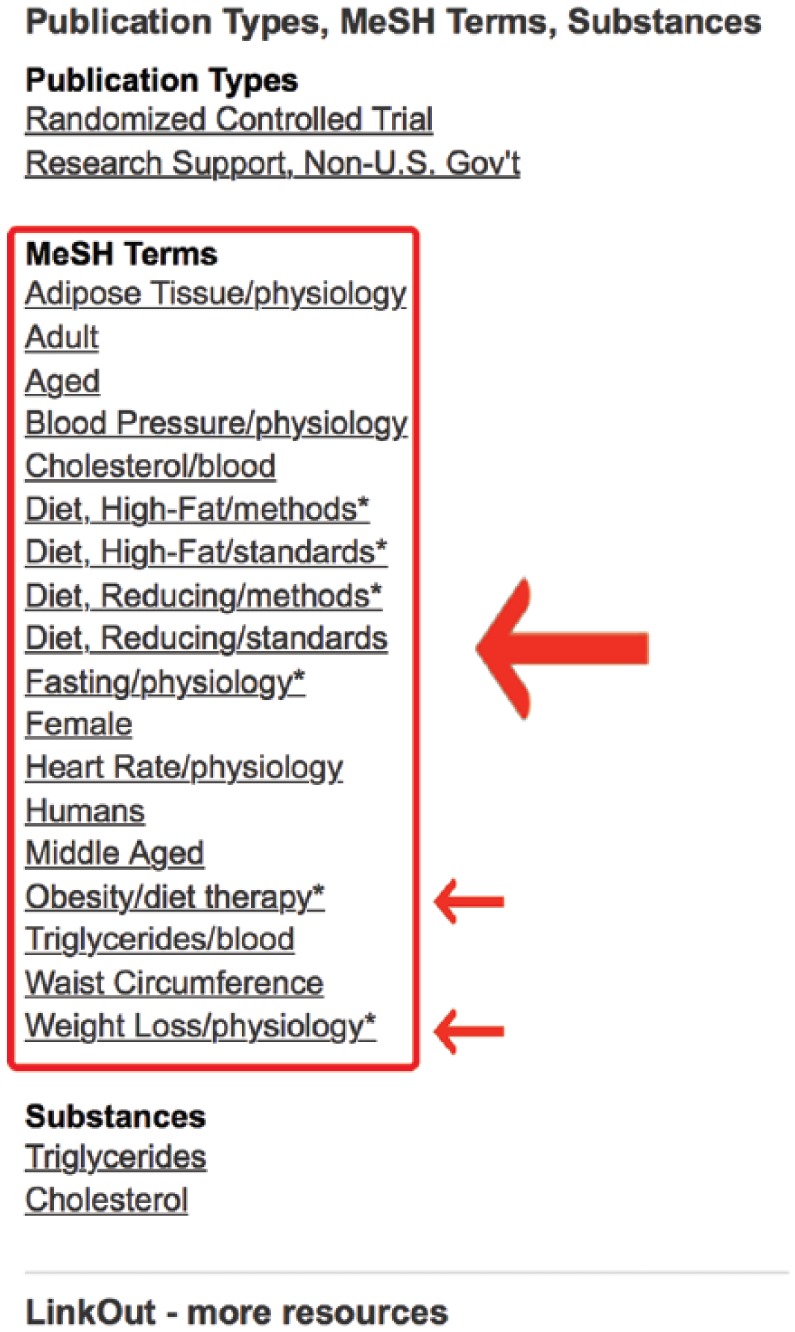
Medical subject headings (MeSH) terms for article of known relevance.
Action step:
7. Check the humans box on the left-hand side of any PubMed search results page and look in the Search Details box to discover its code (Figure 9) (KC-7).
In this case, “Humans” is a MeSH term (“Humans”[MeSH]), so it would go on the MEDLINE side of the search strategy, connected with a Boolean AND (KC-3). Record this term as well.
Now that all the relevant MeSH terms are known, it is time to build the MeSH portion of the search. Begin by systematically building the search in the Search Builder utility in the MeSH database (KC-4).
Action steps:
8. Click the drop-down menu next to the search box and select “MeSH.”
9. Type in the MeSH term “Fasting” and click “Search.”
10. Select the “Fasting” MeSH term from the resulting search output page. (This will load the dedicated “Fasting” MeSH term page.)
11. Click “Add to Search Builder.”
12. Return to the main “MeSH” search box at the middle of the screen and type “Obesity” and click “Search.”
13. Select “Obesity” from the result MeSH Search results page.
14. From the list of checkboxes, check the subterm “diet therapy.” (Notice the term “Fasting” has been retained in your Search Builder box so that you may simply add to it.)
15. Set the drop-down Boolean operator in the Search Builder box to “OR” and click “Add to Search Builder” (Figure 11).
16. Return to the MeSH search box at the top middle of the screen; type in the known MeSH term, “weight loss”; and click “Search.”
17. Select “Weight Loss” from the output screen and check the subheading “physiology” on the resulting MeSH term page.
18. Set the Boolean operator to OR in the PubMed search box and click “Add to Search Builder” to continue building on the prior two terms.
19. Return to the MeSH search box in the top middle of the screen, type “Humans,” and click “Search.”
20. Click on “Humans” in the resulting output.
21. Set the Boolean operator to AND in the PubMed search box and click “Add to Search Builder” to continue building on the prior 3 terms.
Figure 11.
Building the medical subject headings (MeSH) portion of the search (example: obesity/diet therapy [MeSH]).
This will create a grouping of properly worded MeSH terms all connected with an OR except for “Humans,” which will be connected with an AND. Make sure to adjust the parentheses so that the words connected with OR are grouped together with “AND Humans [MeSH]” on the outside (see below). Then, group the entire MeSH section in parentheses (KC-3). Copy and paste this into a separate document.
((“Fasting”[MeSH] OR “Obesity/diet therapy”[MeSH] OR “Weight Loss/physiology”[Mesh]) AND “Humans”[MeSH])
Recall that the words “Alternate” and “Day” did not map to MeSH terms. These may still be added as text terms to the MEDLINE search. The rules associated with non-MeSH strategies may be used, such as creating suitable synonyms and related words (KC-5). If this grouping of terms is connected to the current MeSH search with a Boolean AND, the search is effectively limited to include only those citations contained within the stated MeSH terms. The non-MeSH terms should be connected among themselves with a Boolean OR (KC-3).
Action steps:
22. Create a string of synonyms for any words that did not map into MeSH terms.
23. Connect them to each other with Boolean ORs and enclose them in parentheses.
24. Connect this grouping of synonyms to the rest of the MEDLINE search with a Boolean AND.
25. Contain the entire MEDLINE strategy in its own set of parentheses
This completes the MEDLINE portion of the search strategy (see below).
The MEDLINE portion of the search strategy is assembled as follows:
(((“Fasting”[MeSH] OR “Obesity/diet therapy”[MeSH] OR “Weight Loss/physiology”[Mesh]) AND “Humans”[MeSH]) AND (“alternate day fasting” OR “fasting on alternating days” OR “alternate-day fasting”))
This alone yields 17 relevant citations but may be missing newer articles that have not yet been indexed in MEDLINE.
The Non-MEDLINE Portion
The non-MEDLINE portion requires the creation of synonyms, alternate suffixes, and any other combination of necessary terms (KC-5). It is then followed by NOT medline[sb] (KC-3) to restrict the search to the 11% of citations that are not MEDLINE indexed.
Action steps:
26. Create a string of synonyms for the expression “alternate day fasting” that you might expect to find in the title or abstract of a paper relevant to your topic.
27. Connect them to each other with Boolean ORs, and enclose them in parentheses.
28. Outside of these parentheses, connect the MEDLINE filter with the Boolean operator NOT by typing “NOT medline[sb].”
The non-MEDLINE portion of the search strategy could be assembled as follows:
((“Alternate Day Fasting” OR “fasting on alternating days” OR “alternate-day fasting”) NOT medline[sb])
This yields 12 results.
Action step:
29. Place the entire non-MEDLINE search in its own set of parentheses and connect it to the MEDLINE search with a Boolean OR.
30. Switch the search drop-down box to PubMed if needed.
31. Copy and paste the completed search strategy into the main PubMed search box.
32. Click Search.
The combined master search is
(((“Fasting”[MeSH] OR “Obesity/diet therapy”[MeSH] OR “Weight Loss/physiology”[Mesh]) AND “Humans”[MeSH]) AND (“alternate day fasting” OR “fasting on alternating days” OR “alternate-day fasting”)) OR ((“Alternate Day Fasting” OR “fasting on alternating days” OR “alternate-day fasting”) NOT medline[sb])
This yields a manageable 29 exhaustive yet highly efficient results covering 13 years of research on the topic of alternate-day fasting in humans.
Automated Citation Updates: Saved Searches in MyNCBI
Once a search strategy has been created, PubMed has a powerful tool to help any researcher or clinician stay as current in his or her field as possible. Once the past literature has been reviewed through an exhaustive search strategy, that strategy may be saved into a MyNCBI account. MyNCBI is a free service offered by the National Center for Biotechnology Information. By clicking on the MyNCBI link in the upper right corner of the PubMed search page, the user is led through a series of steps to set up a free account (Figure 12). Once completed, MyNCBI may be used to save a current search strategy by clicking “Create Alert” under the search box. Here, the user is given the option to set up a plan whereby he or she may receive an email monthly, weekly, or daily with any new citations indexed in PubMed that meet his or her search criteria (Figure 13). In this way, a well-created search strategy allows the user to be the first to read any new articles related to his or her search.
Figure 12.

Signing into MyNCBI.
Figure 13.
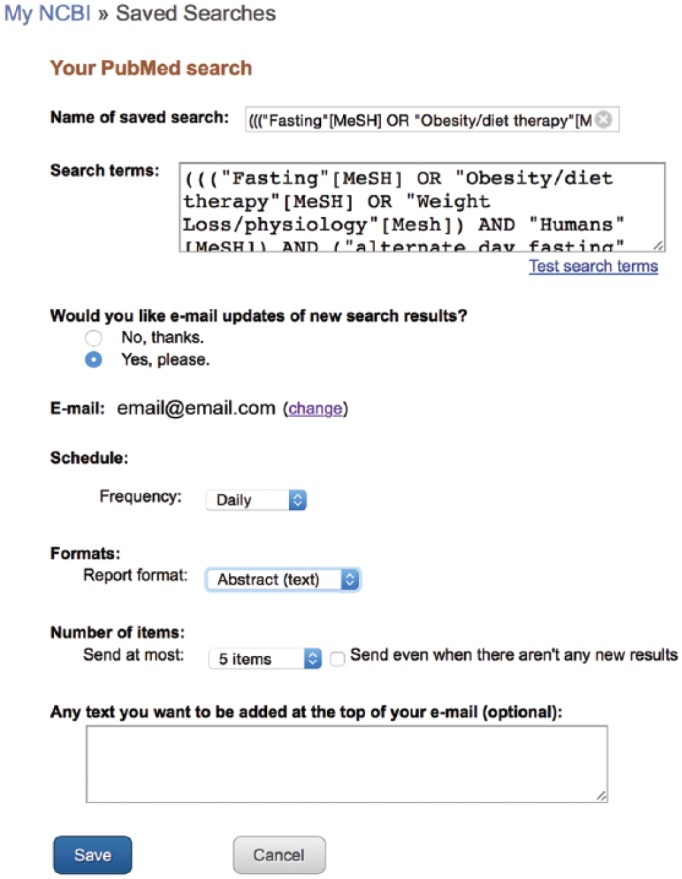
Setting up automated citation delivery.
Steps to Assembling a Master Search Strategy (Summary)
The steps to assembling a master search strategy may be summarized as follows:
Determine which terms best describe the search concept.
Create the MEDLINE portion of the search using the MeSH database to see if the search terms map to acceptable MeSH terms and articles of known relevance to capture any terms you may have missed.
For terms that map well, use the PubMed Search Builder to construct a MeSH search strategy.
If a portion of the terms does not map well to MeSH terms, create a grouping of words using the non-MeSH default search techniques. Connect the words with OR, place them all in one set of parentheses, and connect the non-MeSH group to the MeSH group with the Boolean operator AND. This is also the location to place MeSH filters such as Article Types, Species, Sex, and Age filters.
When the entire MEDLINE portion of the search is complete, click “Search.”
To construct the non-MEDLINE portion of the search, use non-MeSH techniques to generate appropriate search terms, connecting the synonyms with OR. Unique terms may be connected with AND if you wish to restrict your search to yield only citations containing every unique term. Place this whole non-MEDLINE portion in parentheses. On the non-MeSH side, an “*” is acceptable to truncate terms as long as the term is not in quotations.
Next, filter for only non-MEDLINE citations by connecting the filter “medline[sb]” to the non-MEDLINE search with a Boolean NOT.
Connect the MEDLINE search to the non-MEDLINE search with a Boolean OR.
Log into MyNCBI and click on “Create Alert,” selecting the option of receiving daily updates for any new citations that meet the search parameters.
A tip for those who do not have access to a medical library: There is a simple filter that will allow users to search only articles that offer free access to their publications. These results may be filtered by connecting loattrfree full text[sb] to the entire search strategy.
Example:
(The Entire Master Search Strategy) AND (“loattrfree full text”[sb])
For articles that are not automatically free in PubMed, it may be worthwhile to visit the journal’s homepage. Often, simply registering with the journal will be enough to allow access to free articles. If no free method is available, the main service for ordering articles is Loansome Doc. Loansome Doc connects PubMed users with medical libraries for the delivery of paid articles. To learn more and set up a Loansome Doc account, visit their webpage.16
Conclusion
The ability to competently search a citation database is the foundation of all evidence-based practice and research. Timely access to the right information is the currency of an expert in any field. The MEDLINE system continues to be a powerful resource for prequalified information but only if intelligently used and combined with select subsets of non-MEDLINE articles. Armed with these skills, researchers and clinicians may be assured that their recommendations are well informed and that they have performed a truly exhaustive review of the literature.
Footnotes
Statement of Authorship: L. McKeever, V. Nguyen, S. J. Peterson, S. Gomez-Perez, and C. Braunschweig contributed to the conception or design of the research; L. McKeever contributed to the acquisition, analysis, and interpretation of the data; L. McKeever drafted the manuscript; L. McKeever, V. Nguyen, S. J. Peterson, S. Gomez-Perez, and C. Braunschweig critically revised the manuscript; and L. McKeever, V. Nguyen, S. J. Peterson, S. Gomez-Perez, and C. Braunschweig agree to be fully accountable for ensuring the integrity and accuracy of the work. All authors read and approved the final manuscript.
Financial disclosure:None declared.
Glossary
Article of known relevance: an article that is known to be closely related to the query of interest, which has already been indexed for MEDLINE with visible MeSH terms.
Automatic Term Mapping: a process by which PubMed translates a user’s query into the MEDLINE system of medical subject headings.
Boolean operator: a command such as OR, AND, or NOT that respectively includes, restricts, or eliminates a search term or grouping of search terms in a larger search strategy.
Citation: the reference to a scholarly article obtained from the results of a search query.
Loansome Doc: a service through which articles may be purchased.
Master Search Formula: the subject of this article—an efficient and exhaustive search strategy that uses robust MeSH search strategies to search the MEDLINE database and limits less robust strategies to search only those citations in PubMed that have not yet been indexed for MEDLINE.
Medical subject headings (MeSH): a clearly named, defined, and searchable folder within the MEDLINE system.
MEDLINE: The National Library of Medicine’s database specific to the biomedical field.
MESH term: see Medical subject headings.
MyNCBI: a free service offered by the National Center for Biotechnology Information through which a search strategy may be saved for the purpose of automated citation updates through email.
National Center for Biotechnology Information: part of the United States National Library of Medicine (NLM), a branch of the National Institutes of Health responsible for providing access to biomedical and genomic information.
National Library of Medicine: the world’s largest library of biomedical information and the source for the PubMed/MEDLINE database.
PubMed: a free citation database from the National Library of Medicine, which includes both MEDLINE and non-MEDLINE citations.
Related Citations Feature: a convenience algorithm used by PubMed to match the current selected citation with 5 other citations of likely relevance.
Search Details box: a box of text in the lower right-hand corner of any search results page that details how PubMed interpreted your search query.
Text searching: a least robust form of searching that involves the typing of words relevant to one’s query into a search box and relying on PubMed’s automated processes to execute the search.
References
- 1. Katcher BS. MEDLINE: A Guide to Effective Searching in PubMed & Other Interfaces. San Francisco, CA: Ashbury Press; 2006. [Google Scholar]
- 2. Lindsey WT, Olin BR. PubMed searches: overview and strategies for clinicians. Nutr Clin Pract. 2013;28(2):165-176. [DOI] [PubMed] [Google Scholar]
- 3. U.S. National Library of Medicine. MEDLINE fact sheet. http://www.nlm.nih.gov/pubs/factsheets/medline.html. Accessed August 31, 2013.
- 4. U.S. National Library of Medicine. OLDMEDLINE data. http://www.nlm.nih.gov/databases/databases_oldmedline.html. Accessed August 31, 2013.
- 5. U.S. National Library of Medicine. Medical Subject Headings (MeSH) in MEDLINE/PubMed: a tutorial. http://www.nlm.nih.gov/bsd/disted/meshtutorial/introduction/index.html. Accessed August 28, 2013.
- 6. U.S. National Library of Medicine. Medical Subject Headings: MeSH staff. http://www.nlm.nih.gov/mesh/staff.html. Accessed August 29, 2013.
- 7. U.S. National Library of Medicine. MEDLINE indexing: online training course. http://www.nlm.nih.gov/bsd/indexing/training/USE_010.htm. Accessed August 28, 2013.
- 8. Richter RR, Austin TM. Using MeSH (medical subject headings) to enhance PubMed search strategies for evidence-based practice in physical therapy. Phys Ther. 2012;92(1):124-132. [DOI] [PubMed] [Google Scholar]
- 9. Mount Sinai School of Medicine. Search fields in PubMed. http://library.mssm.edu/reference/pdfs/fieldsearch.pdf. Accessed August 29, 2013.
- 10. U.S. National Library of Medicine. PubMed related citations algorithm. http://ii.nlm.nih.gov/MTI/related.shtml. Accessed August 30, 2013.
- 11. U.S. National Library of Medicine. PubMed tutorial. http://www.nlm.nih.gov/bsd/disted/pubmedtutorial. Accessed September 2, 2013.
- 12. U.S. National Library of Medicine. Advanced PubMed searching resource packet. http://nnlm.gov/training/resources/pubmedpacket.pdf. Accessed September 1, 2013.
- 13. Motschall E, Falck-Ytter Y. Searching the MEDLINE literature database through PubMed: a short guide. Onkologie. 2005;28(10):517-522. [DOI] [PubMed] [Google Scholar]
- 14. National Center for Biotechnology Information. PubMed help. http://www.ncbi.nlm.nih.gov/books/NBK3827. Accessed September 2, 2013.
- 15. Klempel MC, Kroeger CM, Varady KA. Alternate day fasting (ADF) with a high-fat diet produces similar weight loss and cardio-protection as ADF with a low-fat diet. Metabolism. 2013;62(1):137-143. [DOI] [PubMed] [Google Scholar]
- 16. U.S. National Library of Medicine. Fact sheet: Loansome Doc. http://www.nlm.nih.gov/pubs/factsheets/loansome_doc.html. Accessed September 1, 2013.
Suggested Reading
A video tutorial for the methods in this article is available free of charge at http://www.researchrdn.com.
- Katcher BS. MEDLINE: A Guide to Effective Searching in PubMed & Other Interfaces. San Francisco, CA: Ashbury Press; 2006. [Google Scholar]
- Lindsey WT, Olin BR. PubMed searches: Overview and strategies for clinicians. Nutr Clin Pract. 2013;28(2):165-176. [DOI] [PubMed] [Google Scholar]
- National Center for Biotechnology Information. PubMed help. http://www.ncbi.nlm.nih.gov/books/NBK3827
- U.S. National Library of Medicine. Advanced PubMed searching resource packet. http://nnlm.gov/training/resources/pubmedpacket.pdf
- U.S. National Library of Medicine. Medical Subject Headings (MeSH) in MEDLINE/PubMed: a Tutorial. http://www.nlm.nih.gov/bsd/disted/meshtutorial/introduction/index.html. Accessed August 28, 2013.
- U.S. National Library of Medicine. MEDLINE indexing: online training course. http://www.nlm.nih.gov/bsd/indexing/training/USE_010.htm
- U.S. National Library of Medicine. PubMed related citations algorithm. http://www.ncbi.nlm.nih.gov/books/NBK3827/#pubmedhelp.Computation_of_Related_Citati