ABSTRACT
Trypanosoma cruzi is the etiologic agent of Chagas disease, and current methods for its genetic manipulation have been highly inefficient. We report here the use of the CRISPR (clustered regularly interspaced short palindromic repeats)/Cas9 (CRISPR-associated gene 9) system for disrupting genes in the parasite by three different strategies. The utility of the method was established by silencing genes encoding the GP72 protein, which is required for flagellar attachment, and paraflagellar rod proteins 1 and 2 (PFR1, PFR2), key components of the parasite flagellum. We used either vectors containing single guide RNA (sgRNA) and Cas9, separately or together, or one vector containing sgRNA and Cas9 plus donor DNA for homologous recombination to rapidly generate mutant cell lines in which the PFR1, PFR2, and GP72 genes have been disrupted. We demonstrate that genome editing of these endogenous genes in T. cruzi is successful without detectable toxicity of Cas9. Our results indicate that PFR1, PFR2, and GP72 contribute to flagellar attachment to the cell body and motility of the parasites. Therefore, CRISPR/Cas9 allows efficient gene disruption in an almost genetically intractable parasite and suggest that this method will improve the functional analyses of its genome.
IMPORTANCE
Trypanosoma cruzi is the agent of Chagas disease, which affects millions of people worldwide. Vaccines to prevent this disease are not available, and drug treatments are not completely effective. The study of the biology of this parasite through genetic approaches will make possible the development of new preventive or treatment options. Previous attempts to use the CRISPR/Cas9 in T. cruzi found a detectable but low frequency of Cas9-facilitated homologous recombination and fluorescent marker swap between exogenous genes, while Cas9 was toxic to the cells. In this report, we describe new approaches that generate complete disruption of an endogenous gene without toxicity to the parasites and establish the relevance of several proteins for flagellar attachment and motility.
INTRODUCTION
Trypanosoma cruzi is the etiologic agent of Chagas disease in humans. Millions of people are affected by this trypanosomiasis in North, Central, and South America, where the disease is endemic, as well as in countries where the disease is not endemic, where it occurs mostly in migrant workers or blood transfusion and organ transplant recipients. No vaccines are available to prevent this disease, and drug treatments have serious side effects or are not completely effective (1). The study of the biology of this eukaryotic organism will make possible the development of specific inhibitors as possible means of controlling the parasites without damaging the hosts.
Few cellular and genetic tools are available to work with T. cruzi (2, 3). The parasite is predominantly diploid, and thus, inactivation of most genes requires two rounds of gene replacement. However, attempts to generate null mutants by homologous recombination sometimes yield parasites bearing the two planned replacements but also bearing extra copies of the genes through aneuploidy or gene amplification (4, 5). The RNA interference (RNAi) technique for gene knockdowns has been well developed in Trypanosoma brucei, which belongs to the group of agents causing African trypanosomiasis, but the RNAi machinery is absent from T. cruzi (2, 6). Some recent genetic tools to work with T. cruzi include the development of stable tetracycline-regulated expression vectors (7), which are not always successful with membrane proteins, and new cloning systems for reverse genetics based on the gateway technology (8, 9), but the field is considerably behind what it is possible to do in comparison with T. brucei (2). Currently, the most efficient genetic tool available in T. cruzi is the overexpression of genes with the pTREX vector (10), a system that allows constitutive gene expression driven by the promoter of ribosomal genes, as well as sequence integration into the ribosomal DNA locus. This strategy has been used to upregulate gene expression in T. cruzi (11–13), but downregulation of genes in this organism still remains inefficient. In contrast to other trypanosomes, T. cruzi epimastigotes have a long generation time and amastigotes require a host cell to grow. Only a few trypomastigotes are enough to infect humans and laboratory accidents are also a concern (14, 15). The consequence of all of these problems is that these organisms are not very amenable to genetic or cellular analyses, while they possess a number of characteristics that cannot be studied in other models.
One of the peculiarities of kinetoplastida, shared with Euglenida, is the presence of the paraflagellar rod (PFR), a complex, trilaminar, lattice-like structure linked to the axonemal microtubules of the flagellum (16). Two core components of the PFR are PFR protein 1 (PFR1) and PFR2, which in T. cruzi correspond to what was known as PAR3 and PAR2 (17), respectively. The genes encoding PFR1 and PFR2 are distinct but related and are present in separate tandem arrays in T. brucei (16). Disruption of these genes has been used to test different RNAi approaches in both T. brucei (18) and Leishmania braziliensis (19). While disruption of these genes results in viable procyclic forms (PCF) of T. brucei (18, 20, 21) or Leishmania sp. promastigotes (18, 22, 23), disruption in bloodstream forms (BSF) of T. brucei is lethal (21). T. brucei PCF RNAi mutants (21) and Leishmania mexicana null mutants (22) for PFR2 show a dramatic decrease in flagellar motility and changes in morphology, while some authors were unable to obtain double-knockout PCF trypanosomes (24). A “blob” of accumulated PFR components is also detected at the tip of the flagellum of PCF RNAi mutants, but the flagellum remains attached to the cell body (21). Another peculiar protein of trypanosomes is an immunodominant glycoprotein localized to the junction between the flagellum and the cell body that is named GP72 in T. cruzi (25–28) or FLA1 in T. brucei (29) and is essential for flagellar attachment in both T. cruzi (28) and T. brucei (29).
The advent of the prokaryotic CRISPR (clustered regularly interspaced short palindromic repeats)/Cas9 (CRISPR-associated gene 9) system has been transformative in biology and has been used for genome editing in diverse organisms, ranging from yeast to mammals and including pathogenic protozoa (30–33). While our work was in preparation, an attempt to use it in T. cruzi was reported (34). The authors used in vitro-transcribed single guide RNA (sgRNA) to transfect cells and target a previously transfected exogenous gene (that for green fluorescent protein [GFP]) or single- and multiple-copy endogenous genes and found a detectable but low frequency of Cas9-facilitated homologous recombination and fluorescent marker swap between exogenous genes (GFP and tdTomato) (0.11 and 0.069% rates of fluorescent marker swap) (34). Cas9 expression was, in addition, toxic to these cells (34).
In this report, we demonstrate by three different strategies (using pTREX vectors containing sgRNA and Cas9, separately or together, or one pTREX vector containing sgRNA and Cas9 plus a donor DNA for homologous recombination) that genome editing of endogenous genes in T. cruzi is successful with no detectable toxicity of Cas9. Our results also indicate that PFR1, PFR2, and GP72 are all required for flagellar attachment to the cell body of the epimastigote stages of T. cruzi, demonstrating the utility of the CRISPR/Cas9 system in elucidating the role of specific genes in this organism.
RESULTS
To investigate whether the CRISPR/Cas9 system could be used to silence genes of T. cruzi, we used a genetically modified version of the type II CRISPR system from Streptococcus pyogenes, consisting of the endonuclease Cas9 and an engineered RNA chimera or sgRNA that conform a ribonucleoprotein complex able to recognize the target sequence and produce double-strand breaks (35). Double-strand breaks generated by Cas9 can be repaired by homologous recombination with donor DNA or by error-prone, nonhomologous end joining (NHEJ) (35). Trypanosomatids are deficient in canonical NHEJ, but they have a more recently characterized repair mechanism, named microhomology-mediated end joining (MMEJ) (36–38). MMEJ repairs DNA breaks via the use of substantial microhomology and always results in deletions (39, 40).
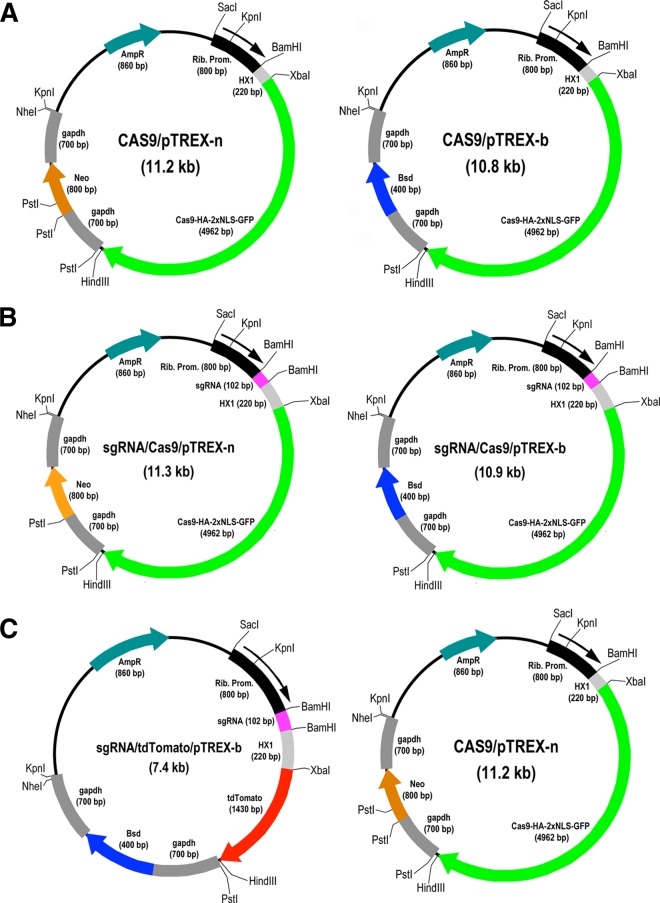
The constructs used in this study are shown in Fig. 1. We first investigated whether it was possible to target Cas9 to the nucleus of T. cruzi by using an exogenous nuclear localization signal. We transfected epimastigotes with either Cas9/pTREX-n or Cas9/pTREX-b (Fig. 1A) vector containing the S. pyogenes Cas9 sequence with a twice-repeated sequence of the simian virus 40 (SV40) nuclear localization signal (2×NLS) and GFP or with the empty vector (pTREX-b or pTREX-n). GFP expression was monitored to determine the transfection efficiency, revealing that ~20% of cells showed colocalization of GFP with 4′,6-diamidino-2-phenylindole (DAPI)-staining nuclei. Figure S1A in the supplemental material shows the results obtained with the Cas9/p-TREX-b vector after increasing the percentage of fluorescent cells by fluorescence-activated cell sorting (FACS). These cells exhibit green fluorescent nuclei, as observed by colocalization with DAPI, confirming the nuclear localization of the fusion endonuclease Cas9-GFP. Similar results were obtained with the Cas9/pTREX-n vector. Cells transfected with the empty vectors showed normal morphology and flagellar staining with antibodies against T. cruzi flagellar calcium binding protein (TcFCaBP) (see Fig. S1B in the supplemental material). Recombinant plasmids Cas9/pTREX-n and Cas9/pTREX-b were then used to develop two different approaches for gene silencing involving the transfection of epimastigotes with one (CRISPR-1V, Fig. 1B) or two (CRISPR-2V, Fig. 1C) vectors, respectively.
FIG 1 .
Restriction map of molecular constructs generated for the CRISPR/Cas9 system in T. cruzi. (A) Cas9/pTREX-n and Cas9/pTREX-b are two plasmids derived from vectors pTREX-n and pTREX-b (10), respectively, by insertion of the Cas9-HA-2×NLS-GFP fusion gene through XbaI and HindIII restriction sites. (B) CRISPR-1V strategy: plasmids sgRNA/Cas9/pTREX-n and sgRNA/Cas9/pTREX-b were derived from the plasmids in panel A by insertion of a specific sgRNA fragment through the BamHI site. These plasmids differ only in the resistance gene (neomycin or blasticidin), and they allow the expression of a specific sgRNA together with Cas9 through a single transfection event of wild-type epimastigotes. (C) CRISPR-2V strategy. A specific sgRNA and the fusion gene Cas9-HA-2×NLS-GFP were individually cloned into two different plasmids to generate sgRNA/tdTomato/pTREX-b and Cas9/pTREX-n (the same as in panel A) for sequential transfection and coexpression of Cas9 and sgRNA in T. cruzi epimastigotes. In this strategy, sgRNA/tdTomato/pTREX-b (conferring cytoplasmic red fluorescence and blasticidin resistance) was used to transfect T. cruzi epimastigotes expressing Cas9 (green fluorescent and neomycin resistant) to generate CRISPR-ablated, red/green fluorescent parasites. Rib. Prom, ribosomal promoter; AmpR, ampicillin resistance gene; Neo, neomycin resistance gene; Bsd, blasticidin-S deaminase gene. HX1 and gapdh are trans-splicing regions described in reference 10. Cas9-HA-2×NLS-GFP is a fusion gene described in Materials and Methods. tdTomato encodes a red fluorescent tag protein. sgRNA is a fragment containing the sequence of a chimera sgRNA that, together with Cas9, targets a specific gene.
We initially chose TcPFR1 and TcPFR2 for gene silencing because their orthologs have been used before as knockdown targets in T. brucei and L. braziliensis to evaluate the efficiency of the RNAi system in these organisms, producing a nonlethal but morphologically easy-to-characterize phenotype (20, 41, 42). We also chose TcGP72 because knockout of this gene results in flagellar detachment without loss of viability (28). The sgRNA sequences used to target these genes contained a 20 (TcGP72)- or 25 (TcPFR1 and TcPFR2)-nucleotide (nt) region that targeted a complementary sequence (the protospacer) immediately upstream of a protospacer-adjacent motif (PAM, an NGG sequence required for S. pyogenes Cas9 recognition) (35). To ensure target specificity, we compared the protospacers to the T. cruzi genome to identify homologous sequences that contained both the protospacers and a PAM, and we were not able to identify any sequence with fewer than six mismatches with the ProtoMatch v1.0 script (31). We also transfected cells with a vector containing a scrambled sgRNA (pTREX-s) where the protospacer was replaced with a sequence of 20 scrambled nucleotides that do not target any gene on the T. cruzi genome.
For the CRISPR-1V strategy (one single vector for expression of sgRNA and Cas9), we took advantage of the fact that transcription in trypanosomatids is polycistronic. Therefore, we arranged the sequences of a specific sgRNA (or scrambled sgRNA) and Cas9 upstream and downstream of the HX1 trans-splicing signal in the pTREX vector (10), respectively (Fig. 1B). In this way, we obtained a construct to generate the CRISPR/Cas9 ribonucleoprotein complex in T. cruzi by using a one-step transfection protocol. The construct (sgRNA/Cas9/pTREX) allowed the transcription of both sequences (the sgRNA and the Cas9 gene) and the translation and nuclear import of Cas9 alone (Cas9-HA-2×NLS-GFP). The TcPFR1 and TcPFR2 sgRNAs were cloned into the Cas9/pTREX-b vector to generate the TcPFR1sgRNA/Cas9/pTREX-b and TcPFR2sgRNA/Cas9/pTREX-b constructs (blasticidin resistant), while the TcGP72 sgRNA was cloned into the Cas9/pTREX-n vector to generate the TcGP72sgRNA/Cas9/pTREX-n plasmid (neomycin resistant). These plasmids were then used to transfect epimastigotes and generate green fluorescent mutant cell lines. After selection in the presence of blasticidin or neomycin, respectively, cell sorting was done to enrich the populations of GFP-expressing parasites (CRISPR-1V strategy). Control growth curves of parasites transfected with the scrambled sgRNA and Cas9 were normal after a short lag period, suggesting lack of toxic effects of Cas9 or the scrambled sgRNA (see Fig. S1C in the supplemental material).
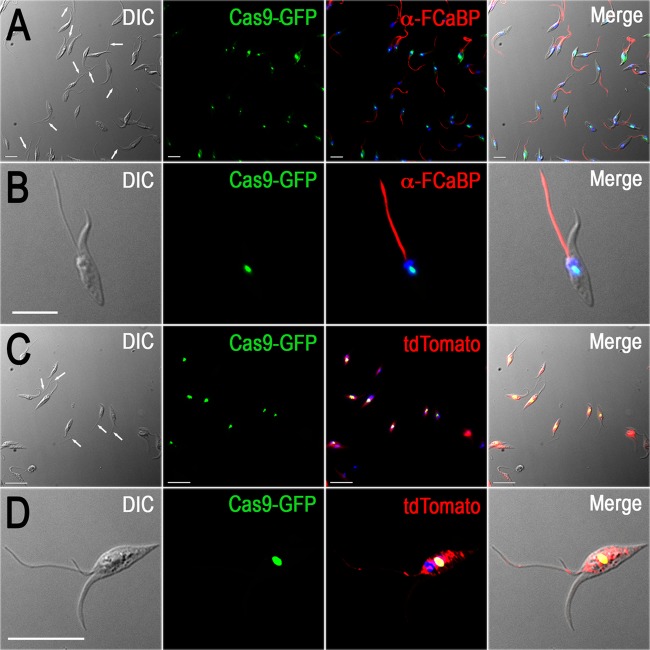
Figure 2A shows the presence of parasites with different degrees of flagellar detachment in the cells with PFR2 disrupted (Fig. 2A, white arrows). Figure 2B shows the complete detachment of the flagellum from the cell body in a PFR2-disrupted epimastigote. Alternatively, the TcPFR1 and TcPFR2 genes were disrupted by CRISPR by using a two-vector strategy (CRISPR-2V) to generate double-resistant (neomycin/blasticidin) red/green fluorescent cell lines. Phenotypic changes were similar to those obtained by the CRISPR-1V approach, thus confirming that TcPFR1 and TcPFR2 disruption results in flagellar detachment to the epimastigote cell body. Figure 2C shows different degrees of flagellar detachment in a population of PFR1-disrupted epimastigotes by the CRISPR-2V strategy (Fig. 2C, white arrows). Note the presence of a green nucleus and red cytoplasm in these cells (Fig. 2C and D). Not all epimastigotes showed flagellar detachment at any given time, and growing numbers of isolated flagella were observed in the culture medium (see Fig. S2A to C in the supplemental material). Epimastigote-like cells devoid of flagella (see Fig. S2D to F in the supplemental material) were also observed in these populations. The gradual development of the phenotypic changes was also revealed by the presence of blobs at the tip or close to the tip of the flagella, after staining with anti-FCaBP antibodies (see Fig. S3 in the supplemental material).
FIG 2 .
Fluorescence microscopy of TcPFR1 and TcPFR2 CRISPR-ablated epimastigotes. (A and B) The TcPRF2 gene in T. cruzi epimastigotes was disrupted by the CRISPR-1V strategy. Detached flagella are indicated by white arrows in the differential interference contrast (DIC) image in panel A, and the detail of a single cell is shown in panel B. Cas9-GFP fusion protein (green) colocalizes with DAPI-stained nuclei (blue). Antibodies against flagellar calcium-binding protein (FCaBP) were used to label flagella (red). (C and D) The TcPRF1 gene in T. cruzi epimastigotes was disrupted by the CRISPR-2V strategy. Detached flagella are indicated by white arrows in the DIC image in panel C, and a single-cell detail is shown in panel D. Cas9-GFP fusion protein (green) colocalizes with DAPI-stained nuclei (yellow). TdTomato expression (red) is cytosolic. Merged images are shown on the right side of each panel. Bars = 10 µm.
Examination of TcPFR1 and TcPFR2 CRISPR-disrupted epimastigotes demonstrated the lack of the paraflagellar rod in these mutants, as visualized by electron microscopy (EM) in longitudinal and cross sections (Fig. 3A). Western blot analysis confirmed the decreased labeling of both PFR1 (7 to 39% of the expression in wild-type cells) and PFR2 (0 to 56% of the expression in wild-type cells) when any one of the genes was disrupted by either the CRISPR-1V or the CRISPR-2V strategy (Fig. 3B), suggesting that lack of either protein prevents the complete assembly of the paraflagellar rod. Detailed examination by scanning EM also revealed flagellar detachment from the cell body (see Fig. S4B and C in the supplemental material).
FIG 3 .
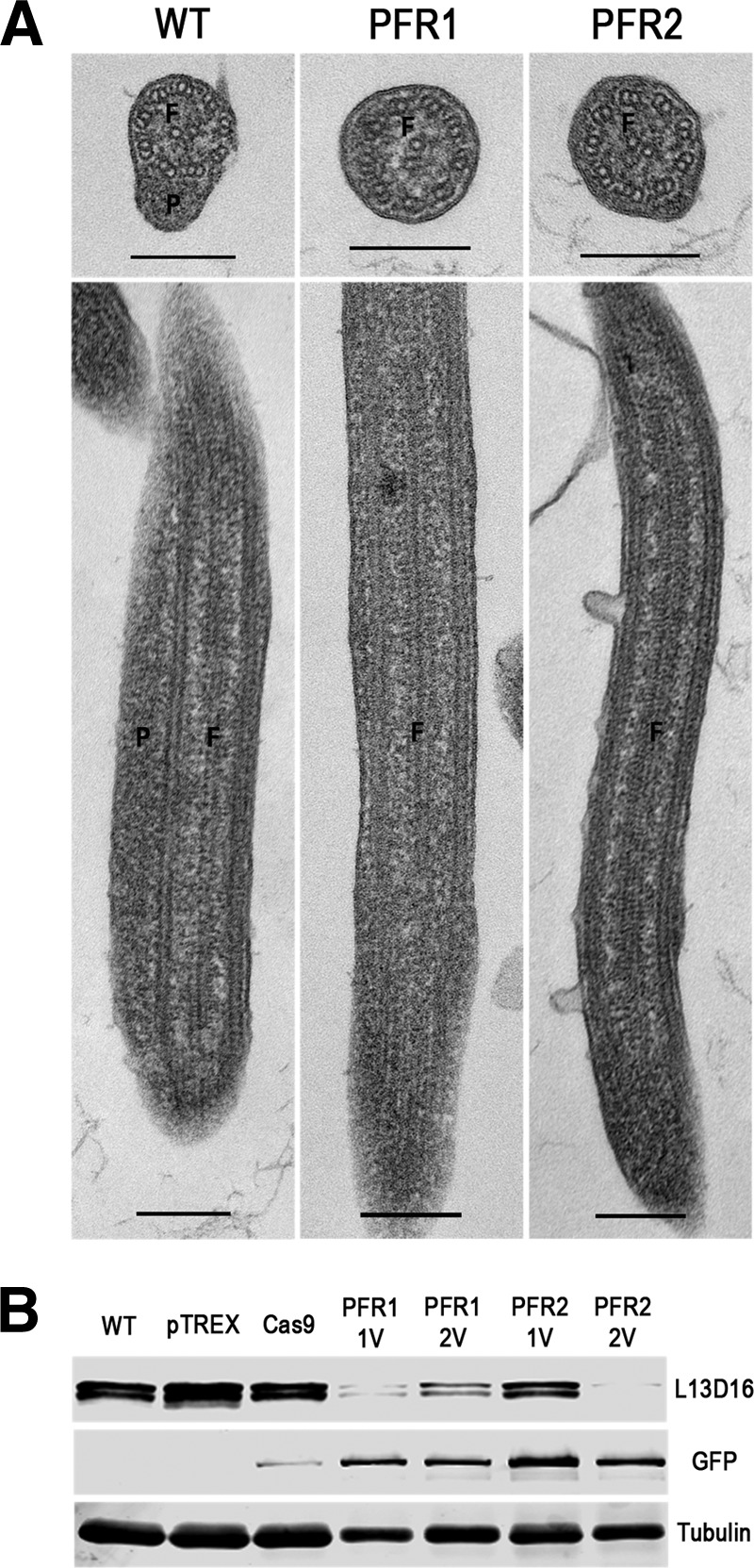
TEM and Western blot analysis of TcPFR1 and TcPFR2 CRISPR-ablated epimastigotes. (A) T. cruzi epimastigotes with mutant PFR1 (CRISPR-1V) and PFR2 (CRISPR-2V) were fixed, stained, and subjected to TEM. The localization of the paraflagellar rod (P) adjacent to the flagellar axonemes (F) was observed in wild-type (WT) cells, while it was absent from mutant parasites (PFR1 and PFR2), as shown in cross sections (top images) and longitudinal sections of the flagellum (bottom images). Scale bars: 200 nm. (B) Western blot analysis of control and CRISPR-ablated epimastigotes. Total lysates (20 µg/lane) of wild-type (WT) epimastigotes, control parasites transfected with plasmids pTREX-b (pTREX) and Cas9/pTREX-n (Cas9), and mutant cell lines producing PFR1 and PFR2 (CRISPR-1V and -2V) were analyzed by Western blot analysis with MAb L13D16 (44), which recognizes the PFR1 and PFR2 proteins (73 and 69 kDa, respectively). The same blot was incubated with anti-GFP and anti-β-tubulin antibodies (loading control). Cell lines are indicated above the blot, and antibodies are shown on the right.
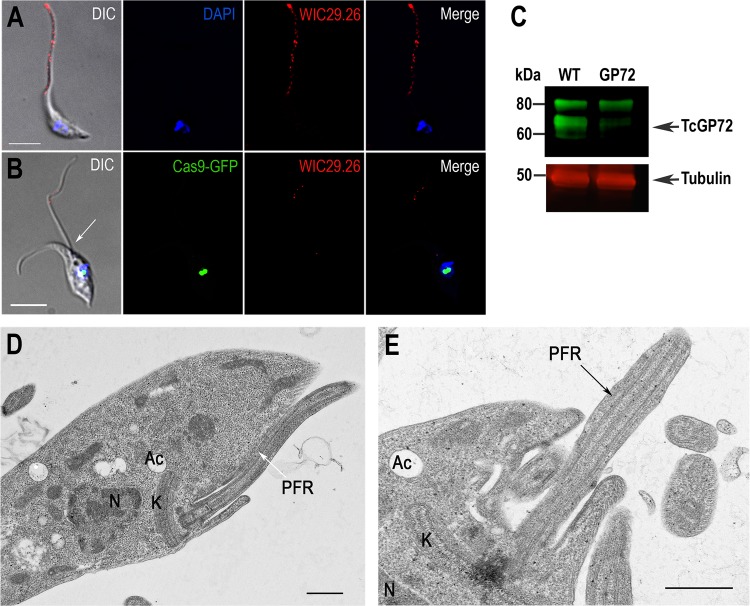
As reported before in a knockout mutant (27), disruption of TcGP72 by CRISPR/Cas9 also resulted in flagellar detachment (compare Fig. 4A and B). While the glycan epitope recognized by monoclonal antibody (MAb) WIC29.26 (43) was evenly distributed over the entire surface of the epimastigote, including the flagellum (Fig. 4A), very little staining remained in the detached flagella of mutant epimastigotes (Fig. 4B). The abnormal flagellar attachment affected the motility of these epimastigotes, which tended to sink faster than the wild-type epimastigotes. Western blot analysis of TcGP72 mutants with MAb WIC29.26 showed a significant decrease in the labeling of the 72-kDa band compared to that in wild-type epimastigotes (Fig. 4C). As reported before (28), this antibody recognizes the glycan epitope in additional lower- and higher-molecular-weight proteins (Fig. 4C). Transmission EM (TEM) examination of the wild type (Fig. 4D) and the TcGP72 CRISPR mutant (Fig. 4E) revealed the flagellar detachment with preservation of the paraflagellar rod and no other morphological changes, as reported for the GP72 knockout mutants (28). Flagellar detachment was also evident on examination by scanning EM (see Fig. S4D in the supplemental material).
FIG 4 .
Phenotype of TcGP72 CRISPR-ablated epimastigotes. Fluorescence microscopy of wild-type epimastigotes (A) and epimastigotes with TcGP72 ablated by the CRISPR-1V strategy (B). Cas9-GFP fusion protein (green) colocalizes with DAPI-stained nuclei (blue). MAb WIC29.26 was used to label TcGP72 in the cells (red). A detached flagellum is indicated by a white arrow in the DIC image. Bars = 10 µm. (C) Western blot analysis of control and CRISPR-ablated epimastigotes. Total lysates (20 µg/lane) of wild-type (WT) epimastigotes and epimastigotes with TcGP72 ablated were analyzed by Western blot with MAb WIC29.26, which recognizes GP72 and other glycoproteins. There is a significant decrease in labeling of TcGP72 (arrowhead) but not of other proteins. The same blot was incubated with anti-β-tubulin antibodies (loading control). Molecular masses of markers are indicated at the left. Transmission electron micrographs of wild-type epimastigotes (D) and epimastigotes with TcGP72 ablated by CRISPR (E) showing the preservation of the paraflagellar rod (PFR) and detached flagellum in the mutant cell. Ac, acidocalcisomes; K, kinetoplast; N, nucleus. Bars: 0.5 µm.
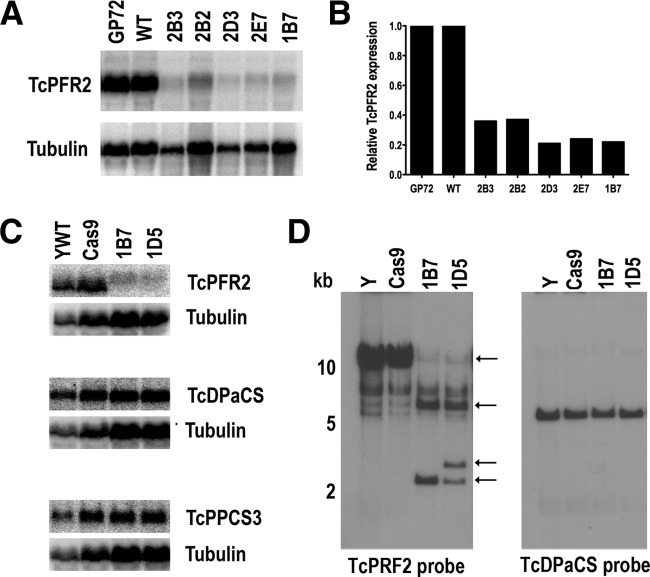
After obtaining mutant-enriched mixed populations by FACS, clonal populations of epimastigotes with TcPFR2 disrupted were obtained by serial dilutions in 96-well plates as described in Materials and Methods. To investigate the mutations caused by CRISPR/Cas9, we amplified a portion of the TcPFR2 genomic locus with primers flanking the Cas9-targeted sequence. The amplified loci were cloned, and individual clones were sequenced. Initially, we used primers to amplify 200- and 500-bp fragments, but after several attempts to find mutations, we designed primers to amplify longer regions (1 and 1.5 kb) where the protospacer of TcPFR2 was approximately in the middle of the sequence. However, after analyzing more than 100 clones, we were unable to find deletions. Northern blot analyses showed a marked decrease in mRNA expression in several of the clones analyzed (Fig. 5A and B). To rule out the simultaneous downregulation of genes located in the same polycistron, we also performed Northern blot analyses of genes situated upstream (trafficking protein particle complex subunit 3, TcPPCS3) and downstream (DNA polymerase catalytic subunit, TcDPaCS) of the TcPFR2 gene, but there was no downregulation of the expression of these genes (Fig. 5C). However, Southern blot analysis of some of these clones revealed dramatic changes suggesting large deletions of TcPFR2 (Fig. 5D). The use of EcoRI digestion of genomic DNA (gDNA) was based on the T. cruzi Esmeraldo-like genomic sequence that would generate a unique band of 4,658 bp in wild-type parasites. However, the pattern obtained was different from that expected. As the Y strain genome sequence is not yet available, we are unable to predict the labeling pattern generated with any restriction enzyme/probe used. However, this result is strong evidence of genome editing in PFR2 CRISPR mutants.
FIG 5 .
Northern and Southern blot analyses of TcPRF2 mutant parasites. Northern blot analysis (A) and densitometric quantitation (B) indicate that TcPFR2 mRNA is significantly downregulated in the subclones indicated while the two nearby genes (C) are not affected. The membranes were first hybridized with the indicated TcPFR2, TcPPCS3, and TcDPaCS probes and then washed and hybridized to the alpha-tubulin gene used as a loading control. Northern blot analyses with the TcPFR2 probe were performed with three different RNA preparations. Densitometric analysis was performed with ImageJ v1.48 software. Values were normalized against the wild type (YWT). The GP72 CRISPR mutant was used as a control. (D) Southern blot analysis indicates that the TcPFR2 gene has been rearranged. The genomic DNAs were extracted from the cell lines indicated and digested with EcoRI. After separated on 1% agarose gels and transfer to nylon membranes, a TcPFR2 probe was used for hybridization (left). Arrows indicate changes in the pattern of bands observed in PFR2 CRISPR clones. A duplicate membrane was hybridized to TcDPaCS (right).
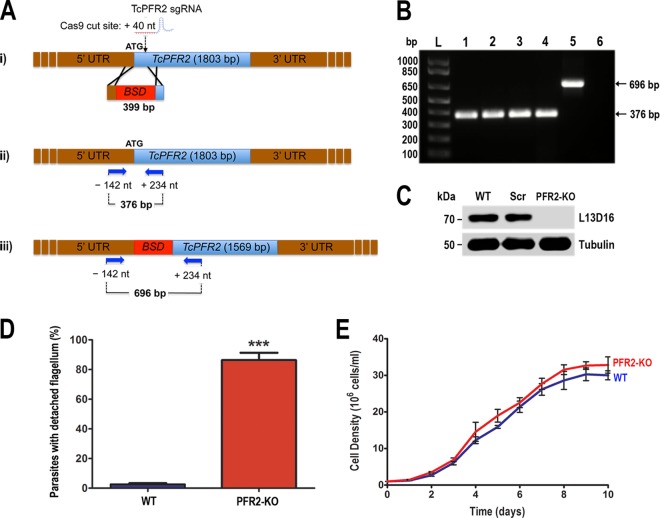
Since these experiments failed to demonstrate mutations in the targeted endogenous genes, we investigated whether the CRISPR/Cas9 system could be used to facilitate gene knockout by integration of a selectable marker (CRISPR/HR strategy). We cotransfected the TcPFR2sgRNA/Cas9/pTREX-n plasmid with a donor DNA fragment consisting of a linear cassette with the blasticidin resistance gene (Bsd) flanked by homology arms corresponding to 100-bp regions upstream of the start codon and downstream of the Cas9 target site in the TcPFR2 gene. Following cotransfection, epimastigotes were selected with neomycin and blasticidin and the stable double-resistant cells were subjected to PCR and Western blot analyses. Disruption of the TcPFR2 gene was verified by PCR with primers annealing outside the homology regions on the blasticidin cassette (primers 13 and 14; see Table S1 in the supplemental material), to amplify a fragment from nt −142 to nt +234 of the TcPFR2 locus (Fig. 6A). A fragment of 696 bp was amplified from gDNA isolated from a TcPFR2 knockout (TcPFR2-KO) mutant obtained by CRISPR/Cas9 by the donor DNA strategy (Fig. 6B, lane 5), indicating disruption of the PFR2 gene, which generates a fragment of 376 bp in the original locus (Fig. 6B, lanes 1 to 4). Western blot analysis with MAb L13D16 (44), which recognizes the TcPFR1 and TcPFR2 proteins (73 and 69 kDa, respectively), showed the absence of TcPFR2-encoded protein in this mutant cell line (Fig. 6C). This result confirmed our previous observation of a significant decrease in the level of TcPFR1 protein accompanying the disappearance of TcPFR2, which we attribute to the role of TcPFR2 in the assembly of the paraflagellar rod structure. Finally, flagellar detachment was observed in 86% of the cells by light microscopy (Fig. 6D) without significant changes in cell growth (Fig. 6E). Cells that did not exhibit flagellar detachment showed abnormally thin flagella, while a loss of forward cell motility was observed in the entire population. This motility defect was compatible with the cell sinking phenotype observed for this mutant in culture flasks. Taken together, our results indicate that the CRISPR/Cas9 system allowed efficient genome editing in T. cruzi, and the cotransfection of epimastigotes with a linear cassette for homology recombination improved the editing efficiency by generating a mutant cell line where the entire population exhibits gene disruption and ablation of the TcPFR2 protein involved in paraflagellar rod assembly, flagellar attachment, and cell motility.
FIG 6 .
Phenotype analysis of TcPFR2 knockout cell line. (A) Schematic representation of the strategy used to generate a TcPFR2-KO mutant by CRISPR/Cas9-induced homologous recombination. A double-stranded gDNA break was produced by Cas9 at nt +40 of the TcPFR2 open reading frame. DNA was repaired with a linear blasticidin S deaminase (BSD) cassette containing 100-bp homologous regions from the TcPFR2 locus (i). Arrows represent primers 13 and 14 (see Table S1 in the supplemental material), which were used to verify gene disruption by PCR. The intact locus generates a PCR product of 376 bp (ii), while the disrupted locus generates a fragment of 696 bp (iii). (B) PCR products obtained with gDNA from different cell lines were analyzed by electrophoresis on 1% agarose gel. Lanes: L, 1-kb plus ladder; 1, wild type; 2, pTREX-n control cells; 3, Cas9/pTREX-n control cells; 4, scrambled sgRNA/Cas9/pTREX-n control cells; 5, TcPFR2-KO mutant cell line; 6, PCR negative control (H2O). (C) Western blot analysis of control and TcPFR2-KO epimastigotes. Total lysates (20 µg/lane) of wild-type (WT), scrambled control (Scr), and TcPFR2-KO epimastigotes were analyzed by Western blotting with MAb L13D16. Anti-β-tubulin antibody was used as a loading control. (D) Percentages of wild-type (WT) and TcPFR2-KO parasites exhibiting a detached flagellum, as visualized by light microscopy. Observation of fixed parasites was performed in 20 fields with a 100× objective lens. (E) Growth curve of wild-type (blue) and TcPFR2-KO epimastigotes (red). Results in panels D and E are expressed as mean values of three independent experiments ± the standard deviations. Comparison was performed by Student’s t test with a significance level of P < 0.05 (indicated by asterisks).
DISCUSSION
T. cruzi has been especially refractory to genetic manipulation, and the implementation of the CRISPR/Cas9 technology in this organism will greatly facilitate the functional analysis of its genome. Here we report the use of separate or single plasmids containing sgRNA and Cas9 to promote rapid and efficient editing of T. cruzi genomic loci. In addition, CRISPR/Cas9 provided a very efficient system for site-specific insertion of a selectable marker through homologous recombination and resulted in 100% disruption of an endogenous gene. In previous work (34), Cas9-facilitated homologous recombination in T. cruzi was only tested by swapping one exogenous gene (that for GFP) with another (that for tdTomato). However, this method yielded only 0.11 and 0.069% rates of fluorescent marker swapping. CRISPR/Cas9-mediated homologous recombination was also used recently in Leishmania major to replace PFR2 genes with a puromycin resistance cassette (23). However, in that work, mixed cell populations were obtained, suggesting that genome editing was not 100% efficient. In contrast, our homologous recombination method resulted in a clean site-specific disruption of TcPFR2, as detected by PCR and Western blot analyses. Moreover, this mutant cell line was obtained from a unique transfection event without the need to select clonal populations, which is usually time consuming. While previous work with kinetoplastids required cotransfection of cells with in vitro-transcribed sgRNA and two plasmids (34) or cotransfection with three plasmids (23) to induce CRISPR/Cas9-facilitated homologous recombination, we achieved highly efficient genome editing by using a single plasmid (sgRNA/Cas9/pTREX-n), cotransfecting it with a PCR-amplified blasticidin cassette for homologous recombination. We were able to generate this mutant cell line after exactly 5 weeks of selection, a goal that was not previously achieved with T. cruzi. The high efficiency observed with this system is possibly a consequence of the constitutive expression of Cas9 and the sgRNA cloned together into the pTREX-n vector, which permanently produces a site-directed double-strand DNA break that is further repaired by homologous recombination with the blasticidin homology template. When the selectable marker is inserted into at least one allele of the gene, this allele works as the template to repair any other Cas9-induced break occurring in the second allele, or in other gene copies possibly present, by a mechanism similar to the mutagenic chain reaction recently described (45). A schematic representation of the strategies developed for CRISPR/Cas9-mediated genome editing in T. cruzi is shown in Fig. S5 in the supplemental material.
Expression of sgRNA and Cas9 alone (one- or two-vector strategy) resulted in genome editing and visible phenotypic changes in nonclonal populations, but it was not possible to detect gene deletions at the sgRNA target sites. Gene mutations at the sgRNA target sites were also undetected in clonal cell lines obtained by limiting dilution of these TcPFR2 mutants, but Southern blot analysis of these clones revealed dramatic changes suggesting large deletions of TcPFR2. Deletions attributed to the function of a microhomology-mediated end-joining double-strand break repair system were previously detected only in an exogenous transfected gene (that for GFP) (34). It is unclear why there is still a wild-type sequence with the target site intact while the chromosomal TcPFR2 locus is clearly rearranged after expressing the sgRNA that targets that locus and Cas9 alone. One possible explanation is that the parasites can maintain this wild-type sequence in a location or structure that Cas9 cannot access. This copy of the gene was not transcribed either, as observed by the decrease in mRNA levels shown by Northern blot analysis.
The results obtained by transfection of sgRNA and Cas9 alone are comparable to the downregulation of gene expression that can be observed in T. brucei upon RNAi. In our case, deletion of the TcPFR1, TcPFR2, and TcGP72 genes also allowed the detection of progressive loss of the proteins and the gradual appearance of the final phenotypes, probably reflecting the slow turnover of the proteins. As demonstrated by the silencing of PFR2 genes in L. major (23), which are present in separate tandem arrays, this method allows the silencing of families of genes containing homologous sequences, an achievement that will allow the discovery of the function of large gene families, such as those of trans-sialidases (>1,400 genes) and mucins (>800 genes) (46). An important advantage of this technique is that upon initial antibiotic selection of epimastigotes, not all cells became deficient in the disrupted gene. If the target gene in question is required for cell survival, this partial selection results in only some epimastigotes showing a phenotypic change, i.e., flagellar detachment in the case of TcPFR1, TcPFR2, and TcGP72, but they could also exhibit a growth defect at the population level, suggesting an essential role of the gene.
PRF1 and PFR2 gene disruptions have been done before by using T. brucei procyclic forms (18, 20, 21) and L. braziliensis promastigotes (18, 22) to test different RNAi approaches and investigate their potential roles. In both cases, disruption mutants were viable and loss of the paraflagellar rod was observed, suggesting that the assembly of this structure depends on the presence of both PFR1 and PFR2. In the case of the procyclic form trypanosomes, a blob of accumulated PFR components was present at the tip of the flagellum of RNAi mutants but the flagellum remained attached to the cell body (21). Double-knockout mutants of procyclic forms could not be obtained (24), while mutants obtained by transfection of antisense PFR2 were not viable in case of the bloodstream forms (21). We report here that partial disruption of either PFR1 or PFR2 in T. cruzi epimastigotes with the CRISPR/Cas9 system results in accumulation of PFR components at the tip of the flagella and subsequent loss of the paraflagellar rod structure with flagellar detachment from the cell body. The cells appear to lose the flagellum in the medium, and aflagellate epimastigote-like cells appeared. The mutant cells showing flagellar detachment have reduced cell motility and sink to the bottom of the culture flasks. It has been reported previously that knockout of TcGP72 (28) (and RNAi ablation of the ortholog FLA1 in T. brucei procyclic forms [29]) also results in flagellar detachment from the cell body, and we confirmed these results with the CRISPR/Cas9 system.
In both T. cruzi and T. brucei, flagellar detachment affects the motility of the parasites but does not affect the viability of the cells, and this was confirmed here after the disruption of TcPFR2 by an antibiotic-resistance marker. In addition, T. cruzi GP72 mutant epimastigotes are able to differentiate into metacyclic stages and infect tissue culture cells but produce no tissue culture-derived trypomastigotes but only round forms with short flagella (microamastigotes) or elongated forms with flagella emerging from the middle of the cell (mesoamastigotes) that are deficient in infecting mice and had lower viability in the insect vector (27, 47). It is also interesting that disruption of the PFR assembly in T. brucei by RNAi ablation of PFR-located calmodulin (48) results in loss of the PFR structure and flagellar detachment. Our results support the hypothesis that the assembly of the PFR is essential to maintain flagellar attachment to the cell body of trypanosomes.
In summary, the highly efficient gene disruption achieved by CRISPR/Cas9-induced homologous recombination will greatly improve our ability to perform genome editing in T. cruzi and, as a consequence, facilitate the study of gene function in this parasite.
MATERIALS AND METHODS
Culture methods.
T. cruzi epimastigotes (Y strain) were grown at 28°C in liver infusion tryptose (LIT) medium (49) supplemented with 10% heat-inactivated fetal bovine serum (FBS). CRISPR mutant cell lines were maintained in medium containing 250 µg/ml G418 and/or 100 µg/ml blasticidin. We determined the growth of epimastigotes by measuring the change in optical density at 600 nm in a Gilford spectrophotometer with a starting culture of 4.5 × 106 epimastigotes or by counting cells in a Neubauer chamber.
Chemicals and reagents.
Taq polymerase, BenchMark Prestained Protein Ladder, MagicMark XP Western Protein Standard, Alexa-conjugated secondary antibodies, blasticidin S HCl, GENEART Site-Directed Mutagenesis System and Topo TA Cloning kit were purchased from Life Technologies (Grand Island, NY). G418 was from Calbiochem (Darmstadt, Germany). Pierce BCA Protein Assay Reagent was from Thermo Fisher Scientific Inc. (Rockford, IL). T4 DNA ligase was from Promega (Madison, WI). Antitubulin MAb, anti-GFP polyclonal antibody, phleomycin and protease inhibitor cocktail (catalog number p8840) were from Sigma (St. Louis, MO). IRDye-conjugated secondary antibodies were from LI-COR Biosciences (Lincoln, NE). QIAprep Spin Miniprep and Midiprep kits, QIAquick gel extraction kit, and MinElute PCR purification kit were from Qiagen (Valencia, CA). The primers were purchased from Integrated DNA Technologies (Coralville, IA). Antibiotics and all other reagents of analytical grade were from Sigma (St. Louis, MO). MAb L13D16 (44) was kindly provided by Philippe Bastin (Institute Pasteur, Paris), MAb WIC29.26 (43) was a gift from George Cross (Rockefeller Institute, NY), and anti-T. cruzi flagellar calcium-binding protein MAb (MAb FCaBP) was a gift from David Engman (Northwestern University, Evanston, IL). The pTREX-b and pMJ920 vectors were kindly provided by Rick Tarleton and Vasant Muralidharan (University of Georgia, Athens, GA), respectively. Plasmid pUC_sgRNA was custom made by Blue Heron Biotech, LLC (Bothell, WA).
Molecular constructs.
The CAS9-HA-2×NLS-GFP fusion gene, containing the S. pyogenes codon-optimized synthetic Cas9 sequence, the human influenza virus hemagglutinin (HA) tag, a twice-repeated sequence of the SV40 nuclear localization signal (2×NLS), and GFP, was amplified from plasmid pMJ920 (Addgene) with primers containing XbaI and HindIII restriction sites (primers 1 and 2; see Table S1 in the supplemental material) to perform in-frame subcloning of the PCR product into the pTREX-n (neomycin resistance) and pTREX-b (blasticidin resistance) vectors (10). After confirming the sequence of the recombinant plasmid, the CAS9-HA-2×NLS-GFP fusion gene was modified to remove a BamHI restriction site without changing the predicted amino acid sequence, with the GENEART site-directed mutagenesis system and primers 3 and 4 (see Table S1 in the supplemental material). The final constructs were named Cas9/pTREX-n and Cas9/pTREX-b, respectively (Fig. 1A). These recombinant plasmids were used to develop three different strategies for gene silencing involving transfection of epimastigotes with one vector (CRISPR-1V, Fig. 1B), two vectors (CRISPR-2V, Fig. 1C), and one vector plus donor DNA for homology recombination (CRISPR-HR), respectively (see Fig. S5 in the supplemental material). Chimeric sgRNA sequences to target genes TcPFR1 (TryTripDB accession no. TcCLB.509617.20), TcPFR2 (TryTripDB accession no. TcCLB.511215.119), and TcGP72 (GenBank accession no. AF507906) were PCR amplified from plasmid pUC_sgRNA, containing the sgRNA backbone sequence (82 bp) (50), with specific forward primers where protospacer regions (20- to 25-nt base-pairing region) were present (primers 5, 6, and 7; see Table S1 in the supplemental material) and the same reverse primer (primer 8; see Table S1 in the supplemental material) for all of the sgRNAs amplified. The presence of the protospacer-adjacent motif NGG (PAM, a motif required for Cas9 recognition of the target sequence) was verified in all chosen protospacer regions. ProtoMatch v1.0, a script kindly provided by Sebastián Lourido (31), was used to verify that the chosen protospacers do not generate Cas9 off targeting on the T. cruzi genome. Primers 5 to 8, designed to amplify sgRNAs, also contained a BamHI restriction site for the individual cloning of these sequences into the Cas9/pTREX-n and Cas/pTREX-b vectors, upstream of the HX1 trans-splicing signal (10). The TcPFR1 and TcPFR2 sgRNAs were cloned by BamHI into the Cas9/pTREX-b vector to generate the TcPFR1sgRNA/Cas9/pTREX-b and TcPFR2sgRNA/Cas9/pTREX-b constructs (blasticidin resistant). TcGP72 sgRNA was cloned by BamHI into the Cas9/pTREX-n vector (neomycin resistant) to generate the TcGP72sgRNA/Cas9/pTREX-n plasmid (all-in-one vector strategy, CRISPR-1V; Fig. 1B), which was used to transfect T. cruzi epimastigotes and generate green fluorescent mutant cell lines. Alternatively, TcPFR1 and TcPFR2 sgRNA sequences were cloned by BamHI into the tdTomato/pTREX-b vector to generate the TcPFR1sgRNA/tdTomato/pTREX-b and TcPFR2sgRNA/tdTomato/pTREX-b constructs. The tdTomato/pTREX-b plasmid was generated by amplifying the tdTomato tag sequence with primers 9 and 10 (see Table S1 in the supplemental material) and subcloning it into the pTREX-b vector by using XbaI and HindIII restriction sites. For the two-vector strategy (CRISPR-2V, Fig. 1C), the TcPFR3sgRNA/tdTomato/pTREX-b and TcPFR2sgRNA/tdTomato/pTREX-b constructs were used together with the Cas9/pTREX-n plasmid to transfect T. cruzi epimastigotes and generate double-resistant (neomycin and blasticidin) red and green fluorescent cell lines. All of the molecular constructs generated in this study were verified by sequencing.
Gene disruption by CRISPR-Cas9 with donor DNA.
The TcPFR2 sgRNA sequence was cloned into the Cas9/pTREX-n vector by using the BamHI site as described above and then used to cotransfect T. cruzi epimastigotes with a linear donor DNA template to induce DNA repair by homologous recombination (CRISPR-HR). This cassette was generated by PCR with 120-bp ultramers (primers 11 and 12; see Table S1 in the supplemental material) from which 100 bp correspond to regions located upstream of the start codon (forward primer) and downstream of the Cas9 target site (reverse primer) of the TcPFR2 gene and 20 bp for annealing on the blasticidin-S deaminase gene (Bsd) on the pTREX-b vector, which was used as a PCR template. Epimastigotes cotransfected with TcPFR2sgRNA/Cas9/pTREX-n and a blasticidin resistance cassette were cultured with G418 and blasticidin for selection of double-resistant parasites. Primers 13 and 14 (see Table S1 in the supplemental material) were used in a PCR assay to verify disruption of the TcPFR2 gene by the blasticidin resistance cassette. For a detailed protocol for gene disruption in T. cruzi by CRISPR/Cas9-mediated homologous recombination, see Supplemental Methods S1 in the supplemental material.
Cell transfections.
T. cruzi Y strain epimastigotes (4 × 107 cells, at room temperature [RT], suspended in phosphate-buffered saline [PBS], pH 7.4) were transfected in ice-cold CytoMix (25 mM HEPES, 120 mM KCl, 0.15 mM CaCl2, 10 mM K2HPO4, 2 mM EGTA, 5 mM MgCl2, 0.5% glucose, 100 µg/ml bovine serum albumin [BSA], 1 mM hypoxanthine [pH 7.6]) containing 25 µg of each plasmid construct or donor DNA in 4-mm electroporation cuvettes with three pulses (1,500 V, 25 µF) delivered by a Gene Pulser II (Bio-Rad). Stable cell lines were established and maintained under drug selection (250 µg/ml G418 and/or 100 µg/ml blasticidin). Until stable cell lines were obtained, LIT medium was supplemented with 20% heat-inactivated FBS. Transfectant cell lines were sorted by either green or green/red fluorescence before clonal populations were obtained by serial dilutions in 96-well plates with conditioned medium (40% fresh LIT, 40% conditioned LIT, 20% heat-inactivated FBS) supplemented with selective drugs.
FACS.
After selection of transfectant cell lines, mixed populations of epimastigotes were sorted at the CTEGD Flow Cytometry Core Facility (University of Georgia, Athens, GA) with an S3 sorter (Bio-Rad, Hercules, CA). Green fluorescence was excited with a 488-nm laser and read through a 525/30-nm band-pass filter in log. Red fluorescence was excited with a 561-nm laser and read through a 586/25-nm laser in log. Acquisition was triggered on forward scatter (FSC), and light scatter was collected on log (log FSC versus log side scatter).
Sequence analysis.
Genomic DNA was extracted from parental and mutant cell lines and used as the template to amplify fragments of 200 to 1,500 bp containing the protospacer regions chosen for Cas9 targeting of the TcPFR1, TcPFR2, and TcGP72 genes. PCR products were cloned into the pCR2.1-TOPO vector, and several clones were sequenced with universal primers M13 reverse and T7 promoter. Sequences were analyzed to verify the presence of mutations with DNAMAN software (version 7.212; Lynnon Corp., Quebec, Canada).
Western blot analysis.
Parental and mutant cell lines were harvested separately. Parasites were washed twice in PBS and resuspended in radioimmunoprecipitation assay buffer (150 mM NaCl, 20 mM Tris-HCl [pH 7.5], 1 mM EDTA, 1% SDS, 0.1% Triton X-100) plus a mammalian cell protease inhibitor mixture (Sigma P8340, diluted 1:250), 1 mM EDTA, 1-mM phenylmethylsulfonyl fluoride, 2.5 mM tosyl phenylalanyl chloromethyl ketone (TPCK), 100 µM N-(trans-epoxysuccinyl)-l-leucine 4-guanidinobutylamide (E64), and Benzonase nuclease (25 U/ml of culture). The cells were then incubated for 1 h on ice. Cell lysis was verified under a light microscope, and protein concentration was determined by a bicinchoninic acid protein assay (Pierce). Twenty micrograms of protein from each cell lysate was mixed with 4× Laemmli sample buffer and analyzed by SDS-PAGE in 10% gels. Separated proteins were transferred onto nitrocellulose membranes (Bio-Rad) with a Bio-Rad Transblot apparatus. Membranes were blocked with 5% nonfat dried skim milk in PBST (PBS containing 0.1% [vol/vol] Tween 20) overnight at 4°C. Next, blots were incubated for 1 h at RT with a primary antibody, i.e., MAb L13D16 (1:50 dilution) (44), MAb WIC29.26 (1:1,000) (43), an antitubulin MAb (1:50,000), or an anti-GFP polyclonal antibody (1:1,000). After three washes with PBST, blots were incubated with the appropriate secondary antibody for 1 h at RT in the dark, i.e., IRDye 680RD-conjugated goat anti-rabbit IgG (1:15,000) or IRDye 800CW-conjugated goat anti-mouse IgG (1:15,000). Membranes were washed three times with PBST, and Western blot images were obtained and processed with the Odyssey infrared imaging system (LI-COR Biosciences).
Northern and Southern blot analyses.
For Northern blot analysis, total RNA was isolated from epimastigotes with TRI reagent. RNA was subjected to electrophoreses in 1% agarose gels containing 2.2 M formaldehyde, 20 mM morpholinepropanesulfonic acid (MOPS; pH 7.0), 1 mM EDTA, and 8 mM sodium acetate; transferred to nylon membranes; and hybridized with radiolabeled probes containing 500 bp of the genes obtained by PCR. The alpha-tubulin gene was used as a loading control. The results were quantified by PhosphorImager analyses. DNA probes were labeled with [α-32P]dCTP (Perkin-Elmer) with random hexanucleotide primers and the Klenow fragment of DNA polymerase (Prim-a-Gene Labeling System). For Southern blot analysis, total genomic DNA was isolated from epimastigotes by phenol-chloroform extraction, digested with EcoRI, separated on a 1% agarose gel, and transferred to nylon membranes. The blot was probed with [α-32P]dCTP-labeled TcPFR2, TcCLB.508837.180 (DNA polymerase catalytic subunit, putative), or TcCLB.508839.30 (trafficking protein particle complex subunit 3, putative).
Immunofluorescence microscopy.
Epimastigotes were washed with PBS and fixed with 4% paraformaldehyde in PBS for 1 h on ice. Cells were allowed to adhere to poly-l-lysine-coated coverslips and then permeabilized for 5 min with 0.1% Triton X-100. Permeabilized cells were incubated for 1 h at RT with 100 mM NH4Cl in PBS and blocked overnight at 4°C with 3% BSA in PBS (pH 8.0). Cells were incubated with a primary antibody (MAb FCaBP [1:10] or MAb WIC29.26 [1:100]) for 1 h at RT. Excess primary antibody was removed from the cells with three washes with 1% BSA in PBS (pH 8.0), and then cells were incubated for 1 h at RT in the dark with an Alexa Fluor 546-conjugated goat anti-mouse secondary antibody (1:1,500). Following incubation with the secondary antibody, cells were washed and mounted on slides. DAPI (5 µg/ml) was included in the mounting medium to stain DNA. Secondary-antibody controls were performed as described above but in the absence of a primary antibody. Differential interference contrast and fluorescence optical images were captured under nonsaturating conditions with an Olympus IX-71 inverted fluorescence microscope with a Photometrix CoolSnapHQ charge-coupled device camera driven by DeltaVision software (Applied Precision, Issaquah, WA). Deconvolution was done as described before (13).
Scanning EM and TEM.
Epimastigotes were processed for TEM at the Electron Microscopy Facility of the Department of Pathology, College of Veterinary Medicine, University of Georgia, Athens, GA. TEM images were obtained with a JEM-1210 transmission electron microscope (JEOL United States Inc., Peabody, MA) equipped with an XR41C bottom-mount CCD camera from Advanced Microscopy Techniques, Corp., Woburn, MA. Parasites for scanning EM were processed and viewed with a Zeiss 1450EP scanning electron microscope at the Center for Advanced Ultrastructural Research, University of Georgia.
SUPPLEMENTAL MATERIAL
Supplemental methods. Download
Fluorescence microscopy of control cell lines. (A) Cas9-expressing epimastigotes generated by transfection of wild-type parasites with plasmid Cas9/pTREX-n. Cells exhibiting normal morphology can be observed by differential interference contrast (DIC). Cas9-GFP fusion protein (green) colocalizes with DAPI-stained nuclei (blue), as shown in the merged image. (B) Epimastigotes transfected with plasmid pTREX-b also exhibit a normal phenotype, as shown by DIC and DAPI-stained nuclei. Antibodies against flagellar calcium-binding protein (FCaBP) were used to label flagella (red). Scale bars = 10 µm. (C) Growth of scrambled sgRNA/Cas9-expressing parasites (green) and wild-type epimastigotes (orange). Download
Differential interference contrast microscopy of TcPFR2 mutant epimastigotes. Panels A to C show the presence of free flagella (arrowheads) in the medium, while panels D to F show epimastigote-like cells without flagella. Bars = 10 µm. Download
Fluorescence microscopy of TcPFR2 mutants. Panels A to C show TcPFR2 mutant epimastigotes stained with antibodies against FCaBP (red) showing accumulation of material in blobs (arrows) close to the tip (A and B) or at the tip (C) of the flagella. DNA is labeled with DAPI (blue), and GFP (nucleus) and tdTomato (cytosol) are green. Bars = 5 µm. Download
Scanning electron micrographs of normal (A) and TcPFR2 (B and C) and TcGP72 (D) mutant epimastigotes showing detached flagella. Arrowheads show the emergence of detached flagella. Bars: 10 µm. Download
Schematic representation of currently available strategies for CRISPR/Cas9-mediated genome editing in T. cruzi. (A) Strategy developed by Peng et al. (34), where T. cruzi epimastigotes are transfected with an in vitro-transcribed sgRNA molecule and plasmid pTREX-b-NLS-hSpCas9 for Cas9 expression, generating a heterogeneous population of cells with DNA repaired by MMEJ. (B) CRISPR-2V strategy developed in the present work, where T. cruzi epimastigotes are cotransfected with vectors sgRNA/tdTomato/pTREX-b and Cas9/pTREX-n for expression of sgRNA and Cas9, respectively. This strategy generates a heterogeneous population that can be further enriched in red/green fluorescent epimastigotes by cell sorting. DNA is probably repaired by MMEJ. (C) CRISPR-1V strategy developed in the present work, where T. cruzi epimastigotes are transfected with a single vector (sgRNA/Cas9/pTREX-n) for simultaneous expression of sgRNA and Cas9. This strategy generates a heterogeneous population that can be further enriched in green fluorescent epimastigotes by cell sorting. DNA is probably repaired by MMEJ. (D) CRISPR-HR strategy developed in the present work, where T. cruzi epimastigotes are cotransfected with vector sgRNA/Cas9/pTREX-n and a linear DNA cassette containing the selectable marker blasticidin-S deaminase (bsd) flanked by homology regions HR1 and HR2. This strategy generates a homogeneous population of green fluorescent epimastigotes where the target gene is disrupted by the bsd gene and the DNA break is repaired by homologous recombination. Download
Primers used to generate CRISPR/Cas9 constructs for T. cruzi.
ACKNOWLEDGMENTS
We thank Vasant Muralidharan (pMJ920 vector) and Rick Tarleton (pTREX-b vector) for vectors; Philippe Bastin, George A. M. Cross, and David Engman for antibodies; Sebastian Lourido for ProtoMatch v1.0 script; Julie Nelson for assistance in cell sorting; Mary Ard for sample processing and cell imaging by TEM; and Miguel Angel Chiurillo for drawing Fig. 6 and Fig. S5.
Funding for this work was provided by the U. S. National Institutes of Health (grant AI107663) and the São Paulo Research Foundation (FAPESP), Brazil (2013/50624-0). N.L. is a postdoctoral fellow of FAPESP (2014/08995-4).
Footnotes
Citation Lander N, Li Z-H, Niyogi S, Docampo R. 2015. CRISPR/Cas9-induced disruption of paraflagellar rod protein 1 and 2 genes in Trypanosoma cruzi reveals their role in flagellar attachment. mBio 6(4):e01012-15. doi:10.1128/mBio.01012-15.
REFERENCES
- 1.Urbina JA, Docampo R. 2003. Specific chemotherapy of Chagas disease: controversies and advances. Trends Parasitol 19:495–501. doi: 10.1016/j.pt.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 2.Docampo R. 2011. Molecular parasitology in the 21st century. Essays Biochem 51:1–13. doi: 10.1042/bse0510001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taylor MC, Huang H, Kelly JM. 2011. Genetic techniques in Trypanosoma cruzi. Adv Parasitol 75:231–250. doi: 10.1016/B978-0-12-385863-4.00011-3. [DOI] [PubMed] [Google Scholar]
- 4.Obado SO, Taylor MC, Wilkinson SR, Bromley EV, Kelly JM. 2005. Functional mapping of a trypanosome centromere by chromosome fragmentation identifies a 16-kb GC-rich transcriptional “strand-switch” domain as a major feature. Genome Res 15:36–43. doi: 10.1101/gr.2895105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Minning TA, Weatherly DB, Atwood J III, Orlando R, Tarleton RL. 2009. The steady-state transcriptome of the four major life-cycle stages of Trypanosoma cruzi. BMC Genomics 10:370. doi: 10.1186/1471-2164-10-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DaRocha WD, Otsu K, Teixeira SM, Donelson JE. 2004. Tests of cytoplasmic RNA interference (RNAi) and construction of a tetracycline-inducible T7 promoter system in Trypanosoma cruzi. Mol Biochem Parasitol 133:175–186. doi: 10.1016/j.molbiopara.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 7.Taylor MC, Kelly JM. 2006. pTcINDEX: a stable tetracycline-regulated expression vector for Trypanosoma cruzi. BMC Biotechnol 6:32. doi: 10.1186/1472-6750-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Batista M, Marchini FK, Celedon PA, Fragoso SP, Probst CM, Preti H, Ozaki LS, Buck GA, Goldenberg S, Krieger MA. 2010. A high-throughput cloning system for reverse genetics in Trypanosoma cruzi. BMC Microbiol 10:259. doi: 10.1186/1471-2180-10-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu D, Brandán CP, Basombrío MA, Tarleton RL. 2009. Evaluation of high efficiency gene knockout strategies for Trypanosoma cruzi. BMC Microbiol 9:90. doi: 10.1186/1471-2180-9-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vazquez MP, Levin MJ. 1999. Functional analysis of the intergenic regions of TcP2beta gene loci allowed the construction of an improved Trypanosoma cruzi expression vector. Gene 239:217–225. doi: 10.1016/S0378-1119(99)00386-8. [DOI] [PubMed] [Google Scholar]
- 11.Galizzi M, Bustamante JM, Fang J, Miranda K, Soares Medeiros LC, Tarleton RL, Docampo R. 2013. Evidence for the role of vacuolar soluble pyrophosphatase and inorganic polyphosphate in Trypanosoma cruzi persistence. Mol Microbiol 90:699–715. doi: 10.1111/mmi.12392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ulrich PN, Lander N, Kurup SP, Reiss L, Brewer J, Soares Medeiros LC, Miranda K, Docampo R. 2014. The acidocalcisome vacuolar transporter chaperone 4 catalyzes the synthesis of polyphosphate in insect-stages of Trypanosoma brucei and T. cruzi. J Eukaryot Microbiol 61:155–165. doi: 10.1111/jeu.12093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Niyogi S, Mucci J, Campetella O, Docampo R. 2014. Rab11 regulates trafficking of trans-sialidase to the plasma membrane through the contractile vacuole complex of Trypanosoma cruzi. PLoS Pathog 10:e1004224. doi: 10.1371/journal.ppat.1004224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hofflin JM, Sadler RH, Araujo FG, Page WE, Remington JS. 1987. Laboratory-acquired Chagas disease. Trans R Soc Trop Med Hyg 81:437–440. doi: 10.1016/0035-9203(87)90162-3. [DOI] [PubMed] [Google Scholar]
- 15.Brener Z. 1985. Host-parasite relations in Chagas disease: mechanisms of infection and disease. Ann Soc Belg Med Trop 65(Suppl 1):9–13. [PubMed] [Google Scholar]
- 16.Portman N, Gull K. 2010. The paraflagellar rod of kinetoplastid parasites: from structure to components and function. Int J Parasitol 40:135–148. doi: 10.1016/j.ijpara.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gadelha C, LeBowitz JH, Manning J, Seebeck T, Gull K. 2004. Relationships between the major kinetoplastid paraflagellar rod proteins: a consolidating nomenclature. Mol Biochem Parasitol 136:113–115. doi: 10.1016/j.molbiopara.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 18.LaCount DJ, Bruse S, Hill KL, Donelson JE. 2000. Double-stranded RNA interference in Trypanosoma brucei using head-to-head promoters. Mol Biochem Parasitol 111:67–76. doi: 10.1016/S0166-6851(00)00300-5. [DOI] [PubMed] [Google Scholar]
- 19.Lye LF, Owens K, Shi H, Murta SM, Vieira AC, Turco SJ, Tschudi C, Ullu E, Beverley SM. 2010. Retention and loss of RNA interference pathways in trypanosomatid protozoans. PLoS Pathog 6:e1001161. doi: 10.1371/journal.ppat.1001161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bastin P, Ellis K, Kohl L, Gull K. 2000. Flagellum ontogeny in trypanosomes studied via an inherited and regulated RNA interference system. J Cell Sci 113:3321–3328. [DOI] [PubMed] [Google Scholar]
- 21.Bastin P, Sherwin T, Gull K. 1998. Paraflagellar rod is vital for trypanosome motility. Nature 391:548. doi: 10.1038/35300. [DOI] [PubMed] [Google Scholar]
- 22.Santrich C, Moore L, Sherwin T, Bastin P, Brokaw C, Gull K, LeBowitz JH. 1997. A motility function for the paraflagellar rod of Leishmania parasites revealed by PFR-2 gene knockouts. Mol Biochem Parasitol 90:95–109. doi: 10.1016/S0166-6851(97)00149-7. [DOI] [PubMed] [Google Scholar]
- 23.Sollelis L, Ghorbal M, MacPherson CR, Martins RM, Kuk N, Crobu L, Bastien P, Scherf A, Lopez-Rubio J, Sterkers Y. 4 May 2015. First efficient CRISPR-Cas9-mediated genome editing in Leishmania parasites. Cell Microbiol doi: 10.1111/cmi.12456. [DOI] [PubMed] [Google Scholar]
- 24.Hunger-Glaser I, Seebeck T. 1997. Deletion of the genes for the paraflagellar rod protein PFR-A in Trypanosoma brucei is probably lethal. Mol Biochem Parasitol 90:347–351. doi: 10.1016/S0166-6851(97)00139-4. [DOI] [PubMed] [Google Scholar]
- 25.Cooper R, Inverso JA, Espinosa M, Nogueira N, Cross GA. 1991. Characterization of a candidate gene for GP72, an insect stage-specific antigen of Trypanosoma cruzi. Mol Biochem Parasitol 49:45–59. doi: 10.1016/0166-6851(91)90129-T. [DOI] [PubMed] [Google Scholar]
- 26.Haynes PA, Russell DG, Cross GA. 1996. Subcellular localization of Trypanosoma cruzi glycoprotein Gp72. J Cell Sci 109:2979–2988. [DOI] [PubMed] [Google Scholar]
- 27.de Jesus AR, Cooper R, Espinosa M, Gomes JE, Garcia ES, Paul S, Cross GA. 1993. Gene deletion suggests a role for Trypanosoma cruzi surface glycoprotein GP72 in the insect and mammalian stages of the life cycle. J Cell Sci 106:1023–1033. [DOI] [PubMed] [Google Scholar]
- 28.Cooper R, de Jesus AR, Cross GA. 1993. Deletion of an immunodominant Trypanosoma cruzi surface glycoprotein disrupts flagellum-cell adhesion. J Cell Biol 122:149–156. doi: 10.1083/jcb.122.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.LaCount DJ, Barrett B, Donelson JE. 2002. Trypanosoma brucei FLA1 is required for flagellum attachment and cytokinesis. J Biol Chem 277:17580–17588. doi: 10.1074/jbc.M200873200. [DOI] [PubMed] [Google Scholar]
- 30.Shen B, Brown KM, Lee TD, Sibley LD. 2014. Efficient gene disruption in diverse strains of Toxoplasma gondii using CRISPR/CAS9. mBio 5:e01114-01114. doi: 10.1128/mBio.01114-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sidik SM, Hackett CG, Tran F, Westwood NJ, Lourido S. 2014. Efficient genome engineering of Toxoplasma gondii using CRISPR/Cas9. PLoS One 9:e100450. doi: 10.1371/journal.pone.0100450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ghorbal M, Gorman M, Macpherson CR, Martins RM, Scherf A, Lopez-Rubio JJ. 2014. Genome editing in the human malaria parasite Plasmodium falciparum using the CRISPR-Cas9 system. Nat Biotechnol 32:819–821. doi: 10.1038/nbt.2925. [DOI] [PubMed] [Google Scholar]
- 33.Wagner JC, Platt RJ, Goldfless SJ, Zhang F, Niles JC. 2014. Efficient CRISPR-Cas9-mediated genome editing in Plasmodium falciparum. Nat Methods 11:915–918. doi: 10.1038/nmeth.3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peng D, Kurup SP, Yao PY, Minning TA, Tarleton RL. 2015. CRISPR-Cas9-mediated single-gene and gene family disruption in Trypanosoma cruzi. mBio 6:e02097-02014. doi: 10.1128/mBio.02097-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burton P, McBride DJ, Wilkes JM, Barry JD, McCulloch R. 2007. Ku heterodimer-independent end joining in Trypanosoma brucei cell extracts relies upon sequence microhomology. Eukaryot Cell 6:1773–1781. doi: 10.1128/EC.00212-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Glover L, Jun J, Horn D. 2011. Microhomology-mediated deletion and gene conversion in African trypanosomes. Nucleic Acids Res 39:1372–1380. doi: 10.1093/nar/gkq981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Glover L, McCulloch R, Horn D. 2008. Sequence homology and microhomology dominate chromosomal double-strand break repair in African trypanosomes. Nucleic Acids Res 36:2608–2618. doi: 10.1093/nar/gkn104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McVey M, Lee SE. 2008. MMEJ repair of double-strand breaks (director’s cut): deleted sequences and alternative endings. Trends Genet 24:529–538. doi: 10.1016/j.tig.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nussenzweig A, Nussenzweig MC. 2007. A backup DNA repair pathway moves to the forefront. Cell 131:223–225. doi: 10.1016/j.cell.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 41.Atayde VD, Ullu E, Kolev NG. 2012. A single-cloning-step procedure for the generation of RNAi plasmids producing long stem-loop RNA. Mol Biochem Parasitol 184:55–58. doi: 10.1016/j.molbiopara.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Durand-Dubief M, Kohl L, Bastin P. 2003. Efficiency and specificity of RNA interference generated by intra- and intermolecular double stranded RNA in Trypanosoma brucei. Mol Biochem Parasitol 129:11–21. doi: 10.1016/S0166-6851(03)00071-9. [DOI] [PubMed] [Google Scholar]
- 43.Ferguson MA, Allen AK, Snary D. 1983. Studies on the structure of a phosphoglycoprotein from the parasitic protozoan Trypanosoma cruzi. Biochem J 213:313–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kohl L, Sherwin T, Gull K. 1999. Assembly of the paraflagellar rod and the flagellum attachment zone complex during the Trypanosoma brucei cell cycle. J Eukaryot Microbiol 46:105–109. doi: 10.1111/j.1550-7408.1999.tb04592.x. [DOI] [PubMed] [Google Scholar]
- 45.Gantz VM, Bier E. 2015. Genome editing. The mutagenic chain reaction: a method for converting heterozygous to homozygous mutations. Science 348:442–444. doi: 10.1126/science.aaa5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.El-Sayed NM, Myler PJ, Bartholomeu DC, Nilsson D, Aggarwal G, Tran AN, Ghedin E, Worthey EA, Delcher AL, Blandin G, Westenberger SJ, Caler E, Cerqueira GC, Branche C, Haas B, Anupama A, Arner E, Aslund L, Attipoe P, Bontempi E, Bringaud F, Burton P, Cadag E, Campbell DA, Carrington M, Crabtree J, Darban H, da Silveira JF, de Jong P, Edwards K, Englund PT, Fazelina G, Feldblyum T, Ferella M, Frasch AC, Gull K, Horn D, Hou L, Huang Y, Kindlund E, Klingbeil M, Kluge S, Koo H, Lacerda D, Levin MJ, Lorenzi H, Louie T, Machado CR, McCulloch R, McKenna A. 2005. The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science 309:409–415. doi: 10.1126/science.1112631. [DOI] [PubMed] [Google Scholar]
- 47.Basombrío MA, Gómez L, Padilla AM, Ciaccio M, Nozaki T, Cross GA. 2002. Targeted deletion of the gp72 gene decreases the infectivity of Trypanosoma cruzi for mice and insect vectors. J Parasitol 88:489–493. doi: 10.1645/0022-3395(2002)088[0489:TDOTGG]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 48.Ginger ML, Collingridge PW, Brown RW, Sproat R, Shaw MK, Gull K. 2013. Calmodulin is required for paraflagellar rod assembly and flagellum-cell body attachment in trypanosomes. Protist 164:528–540. doi: 10.1016/j.protis.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 49.Bone GJ, Steinert M. 1956. Isotopes incorporated in the nucleic acids of Trypanosoma mega. Nature 178:308–309. doi: 10.1038/178308a0. [DOI] [PubMed] [Google Scholar]
- 50.Larson MH, Gilbert LA, Wang X, Lim WA, Weissman JS, Qi LS. 2013. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat Protoc 8:2180–2196. doi: 10.1038/nprot.2013.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental methods. Download
Fluorescence microscopy of control cell lines. (A) Cas9-expressing epimastigotes generated by transfection of wild-type parasites with plasmid Cas9/pTREX-n. Cells exhibiting normal morphology can be observed by differential interference contrast (DIC). Cas9-GFP fusion protein (green) colocalizes with DAPI-stained nuclei (blue), as shown in the merged image. (B) Epimastigotes transfected with plasmid pTREX-b also exhibit a normal phenotype, as shown by DIC and DAPI-stained nuclei. Antibodies against flagellar calcium-binding protein (FCaBP) were used to label flagella (red). Scale bars = 10 µm. (C) Growth of scrambled sgRNA/Cas9-expressing parasites (green) and wild-type epimastigotes (orange). Download
Differential interference contrast microscopy of TcPFR2 mutant epimastigotes. Panels A to C show the presence of free flagella (arrowheads) in the medium, while panels D to F show epimastigote-like cells without flagella. Bars = 10 µm. Download
Fluorescence microscopy of TcPFR2 mutants. Panels A to C show TcPFR2 mutant epimastigotes stained with antibodies against FCaBP (red) showing accumulation of material in blobs (arrows) close to the tip (A and B) or at the tip (C) of the flagella. DNA is labeled with DAPI (blue), and GFP (nucleus) and tdTomato (cytosol) are green. Bars = 5 µm. Download
Scanning electron micrographs of normal (A) and TcPFR2 (B and C) and TcGP72 (D) mutant epimastigotes showing detached flagella. Arrowheads show the emergence of detached flagella. Bars: 10 µm. Download
Schematic representation of currently available strategies for CRISPR/Cas9-mediated genome editing in T. cruzi. (A) Strategy developed by Peng et al. (34), where T. cruzi epimastigotes are transfected with an in vitro-transcribed sgRNA molecule and plasmid pTREX-b-NLS-hSpCas9 for Cas9 expression, generating a heterogeneous population of cells with DNA repaired by MMEJ. (B) CRISPR-2V strategy developed in the present work, where T. cruzi epimastigotes are cotransfected with vectors sgRNA/tdTomato/pTREX-b and Cas9/pTREX-n for expression of sgRNA and Cas9, respectively. This strategy generates a heterogeneous population that can be further enriched in red/green fluorescent epimastigotes by cell sorting. DNA is probably repaired by MMEJ. (C) CRISPR-1V strategy developed in the present work, where T. cruzi epimastigotes are transfected with a single vector (sgRNA/Cas9/pTREX-n) for simultaneous expression of sgRNA and Cas9. This strategy generates a heterogeneous population that can be further enriched in green fluorescent epimastigotes by cell sorting. DNA is probably repaired by MMEJ. (D) CRISPR-HR strategy developed in the present work, where T. cruzi epimastigotes are cotransfected with vector sgRNA/Cas9/pTREX-n and a linear DNA cassette containing the selectable marker blasticidin-S deaminase (bsd) flanked by homology regions HR1 and HR2. This strategy generates a homogeneous population of green fluorescent epimastigotes where the target gene is disrupted by the bsd gene and the DNA break is repaired by homologous recombination. Download
Primers used to generate CRISPR/Cas9 constructs for T. cruzi.