ABSTRACT
The prokaryotic CRISPR (clustered regularly interspaced short palindromic repeat)-Cas9, an RNA-guided endonuclease, has been shown to mediate efficient genome editing in a wide variety of organisms. In the present study, the CRISPR-Cas9 system has been adapted to Leishmania donovani, a protozoan parasite that causes fatal human visceral leishmaniasis. We introduced the Cas9 nuclease into L. donovani and generated guide RNA (gRNA) expression vectors by using the L. donovani rRNA promoter and the hepatitis delta virus (HDV) ribozyme. It is demonstrated within that L. donovani mainly used homology-directed repair (HDR) and microhomology-mediated end joining (MMEJ) to repair the Cas9 nuclease-created double-strand DNA break (DSB). The nonhomologous end-joining (NHEJ) pathway appears to be absent in L. donovani. With this CRISPR-Cas9 system, it was possible to generate knockouts without selection by insertion of an oligonucleotide donor with stop codons and 25-nucleotide homology arms into the Cas9 cleavage site. Likewise, we disrupted and precisely tagged endogenous genes by inserting a bleomycin drug selection marker and GFP gene into the Cas9 cleavage site. With the use of Hammerhead and HDV ribozymes, a double-gRNA expression vector that further improved gene-targeting efficiency was developed, and it was used to make precise deletion of the 3-kb miltefosine transporter gene (LdMT). In addition, this study identified a novel single point mutation caused by CRISPR-Cas9 in LdMT (M381T) that led to miltefosine resistance, a concern for the only available oral antileishmanial drug. Together, these results demonstrate that the CRISPR-Cas9 system represents an effective genome engineering tool for L. donovani.
IMPORTANCE
Leishmania donovani is the causative agent of fatal visceral leishmaniasis. To understand Leishmania infection and pathogenesis and identify new drug targets for control of leishmaniasis, more-efficient ways to manipulate this parasite genome are required. In this study, we have implemented CRISPR-Cas9 genome-editing technology in L. donovani. Both single- and dual-gRNA expression vectors were developed using a strong RNA polymerase I promoter and ribozymes. With this system, it was possible to generate loss-of-function insertion and deletion mutations and introduce drug selection markers and the GFP sequence precisely into the L. donovani genome. These methods greatly improved the ability to manipulate this parasite genome and will help pave the way for high-throughput functional analysis of Leishmania genes. This study further revealed that double-stranded DNA breaks created by CRISPR-Cas9 were repaired by the homology-directed repair (HDR) pathway and microhomology-mediated end joining (MMEJ) in Leishmania.
INTRODUCTION
The CRISPR (clustered regularly interspaced short palindromic repeat)-Cas9, a prokaryotic immune system derived from Streptococcus pyogenes is an RNA-guided endonuclease which has been shown to facilitate site-specific DNA cleavage in diverse organisms ranging from yeast to mammalian cells (1–3). The two key components of the CRISPR-Cas9 system are the Cas9 nuclease and a guide RNA (gRNA) that directs this nuclease to its specific DNA target. The gRNA consists of a 20-nucleotide (nt) guide (also known as protospacer) sequence, followed by an 82-nt chimeric sequence derived from the CRISPR RNA (crRNA) and the trans-activating RNAs (tracrRNAs) of the bacterial system. The 20-nt guide is complementary to the target DNA sequence which is followed by the triple nucleotides NGG known as the protospacer-adjacent motif (PAM). The 3′ portion of the gRNA (82-nt chimeric sequence) and the Cas9 nuclease form an RNA-protein complex that is required for Cas9 nuclease activities (4–6). The gRNA guides the Cas9 nuclease to the specific DNA target site, and subsequently Cas9 generates double-stranded breaks in the target DNA 3 bp upstream of the PAM site. In mammalian cells, double-stranded breaks introduced by Cas9 are repaired by the cellular machinery either through homologous recombination or the nonhomologous end-joining (NHEJ) pathway. DNA sequences homologous to the Cas9-targeted locus can be concurrently introduced to generate desired mutations and to insert epitope tags (5, 7–9). NHEJ in the targeted organism often leads to small deletions and insertions, resulting in a frameshift of the targeted gene. Given its simplicity and high efficiency, CRISPR-Cas9 has now been used for genome engineering in a wide variety of organisms, including human protozoan parasites Toxoplasma gondii, Plasmodium falciparum, Trypanosoma cruzi, and Leishmania major (1–3, 10–14).
Leishmania protozoans are the causative agent of leishmaniasis, one of the major human parasitic diseases (15, 16). The Leishmania genome contains about 8,000 genes, and the functions of most genes (hypothetical proteins) are still unknown (17). The genomes of different Leishmania species and new clinical isolates have been sequenced (18–20). However, due to the absence of an RNA interference (RNAi) pathway in most of the Leishmania species (21, 22), gene targeting in Leishmania mainly relies on homologous recombination with antibiotic selection markers, which is labor intensive and time consuming, and it is difficult to introduce specific mutations or epitope tags (23). A more efficient genome-editing technology is therefore required to better understand mechanisms involved with pathogenesis and to identify new drug targets for the control of leishmaniasis.
In this study, the CRISPR-Cas9 system has been adapted for use in Leishmania donovani. The RNA-guided Cas9 nuclease generated knockouts (deletions or insertions) with or without selection, and it was capable of introducing mutations, selection markers, and a green fluorescent protein (GFP) tag precisely into the L. donovani genome. These methods will streamline the functional analysis of Leishmania genes and pave the way for high-throughput engineering of the Leishmania genome (24, 25).
RESULTS
Expression of CRISPR-Cas9 in L. donovani and generation of gRNA expression vector constructs.
Two-vector systems were adopted to introduce CRISPR-Cas9 into L. donovani. For Cas9 expression, the Streptococcus pyogenes Cas9 nuclease with nuclear localization signals at both termini from plasmid pX330 (Addgene) was cloned into the Leishmania expression vector pLPhyg (LP stands for Leishmania promastigotes, and hyg stands for hygromycin) (5, 26) (Fig. 1A). L. donovani promastigotes transfected with pLPhygCas9 were selected and cultured in hygromycin-containing medium to generate a cell line that constitutively expresses Cas9 nuclease. The Cas9-expressing L. donovani cells proliferate as well as control plasmid pLPhyg-transfected cells do (data not shown). So far, RNA polymerase III promoters such as the human U6 promoter have been used in mammalian cells and other organisms to drive gRNA expression due to its precise initiation and termination of transcription. However, the L. donovani RNA polymerase III U6 promoter has not been characterized (27). We therefore explored whether the L. donovani ribosome RNA promoter (rRNAP, an RNA polymerase I promoter) could be used to drive gRNA expression in Leishmania (Fig. 1A; see Fig. S1 in the supplemental material). L. donovani rRNAP initiates transcription at the defined T-residue site. However, the sequence signals required for rRNAP transcription termination are not well defined (28, 29). As ribozymes are self-processing at specific cleavage sites and have been used to produce the correct size of gRNA in yeast (30, 31), we placed the 68-bp hepatitis delta virus (HDV) ribozyme sequence immediately after the gRNA coding sequence to ensure the precise termination of gRNA in Leishmania (Fig. 1, pSPneogRNAH [neo stands for neomycin]; see Fig. S1 in the supplemental material). For comparison, we also created a gRNA expression vector (pSPneogRNA [Fig. 1A and Fig. S2]) without the HDV ribozyme sequence that contained a 262-bp nonspecific DNA fragment (derived from the pX330 plasmid together with gRNA coding sequence) inserted between the gRNA coding sequence and the downstream pyrimidine track which provides the trans-splicing signal for the neomycin resistance gene (32). Since polyadenylation usually occurs 200 to 500 bp upstream of the trans-splicing site (33), the gRNA expressed in the pSPneogRNA vector can be longer or shorter than 102 nt and may contain a poly(A) tail.
FIG 1 .

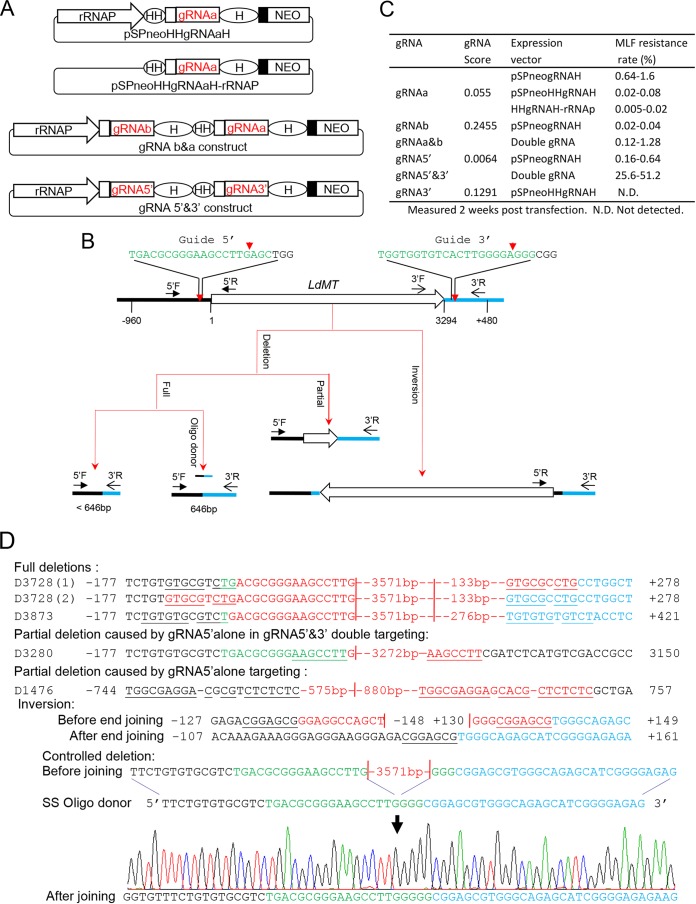
Introducing CRISPR-Cas9 into L. donovani. (A) Vectors used to express Cas9 and gRNA in L. donovani. The A2 intergenic sequence (A2-IGS), L. donovani rRNA promoter (rRNAP), and HDV ribozyme (H) are indicated. The small solid black box in the pSPneogRNAH and pSPneogRNA vectors represents the 92-bp pyrimidine track. The schematic is not drawn to scale. (B) Schematic of gRNAa and its guide sequence for targeting the LdMT gene. The first nucleotide U of gRNAa is highlighted (red) as L. donovani rRNAP initiates transcription at the T-residue site. (C) Coexpression of Cas9 and gRNAa in L. donovani resulted in MLF resistance. The MLF resistance rate was determined by limiting dilution as detailed in Materials and Methods. The values are averages from three independent experiments.
To test whether these Cas9 and gRNA expression plasmids can function as predicted in Leishmania, we chose to target the L. donovani miltefosine transporter gene (LdMT) since a single point mutation in LdMT resulted in drug resistance to miltefosine (MLF) (34). As shown in Fig. 1B, a gRNA-targeting site was chosen near the N-terminal third of the coding sequence to ensure efficient inactivation of LdMT. The 19-bp guide sequence (guide a) was cloned into the pSPneogRNAH and pSPneogRNA vectors, respectively. The resulting gRNAa constructs were electroporated into the pLPhygCas9-containing L. donovani promastigotes, and the dual-drug-resistant cells were selected with hygromycin and G418. If the Cas9 nuclease and gRNAa were properly expressed in these transfectants, and the Cas9 nuclease was able to form a complex with the gRNAa to target the LdMT site resulting in mutations, these Leishmania cells would display resistance to MLF. Indeed, MLF-resistant cells were obtained (sequencing confirmed that the LdMT-targeting site was mutated [see below]) from both gRNA expression vectors, though the MLF resistance rate (the percentage of Leishmania cells surviving after MLF treatment) was higher for the gRNA construct containing the HDV ribozyme (Fig. 1C). Since both alleles of the LdMT gene need to be mutated to display MLF resistance, MLF resistance rates in this study are equivalent to the double allelic mutation frequencies of the LdMT gene. The inactivation mutations caused by CRISPR-Cas9 accumulated with time, the MLF resistance rate increased from 0.5 to 12% for the pSPneogRNAH vector containing the HDV ribozyme compared with 0.1 to 6% for the pSPneogRNA vector 6 weeks posttransfection. No MLF-resistant cells were observed in nontransfected wild-type or control gRNA vector-transfected L. donovani cells.
The double-strand DNA breaks generated by Cas9 are partially repaired by microhomology-mediated end joining; the nonhomologous end-joining pathway appears to be absent in L. donovani.
Doench et al. have identified sequence features that may improve gRNA activity after evaluations of thousands of gRNAs in mammalian cells (human and mouse) and developed a web-based tool to predict gRNA activity with a score ranging from 0 to 1 (35). Interestingly, according to this gRNA designer tool, our very first gRNA (gRNAa) described above had a score of only 0.055 and was among the least active of all gRNAs available from the sense and antisense sequences of the complete LdMT open reading frame. We therefore designed two additional gRNAs near the first gRNAa-targeting site with a score of 0.245 for gRNAb and a score of 0.717 for gRNAc (Fig. 2A), and they were cloned into the pSPneogRNAH vector. In contrast to gRNAa, gRNAc was ranked by the tool to be one of the most active gRNAs. However, although the MLF-resistant cells were observed in L. donovani cells expressing gRNAb or gRNAc, the resistance rates were not improved compared with the resistance rate for cells expressing gRNAa (Fig. 2B). This suggests that either this gRNA designer tool is not appropriate for Leishmania or an alternative and more sensitive method is required to measure gRNA activity in Leishmania.
FIG 2 .
The Cas9-created DSBs in the MLF-resistant cells were partially repaired by MMEJ. (A) Schematic of the LdMT locus showing the locations of three guide sequences for gRNAa, gRNAb, and gRNAc. The primers used to check the mutations caused by CRISPR-Cas9 are indicated above the schematic. (B) Comparison of MLF resistance rates of L. donovani cells targeted by gRNAa, gRNAb, and gRNAc as determined by the limiting dilution assay. The MLF resistance rates are shown as ranges from three independent experiments. (C) Sequences showing deletions caused by Cas9 and MMEJ. The red arrowhead and vertical line indicate putative Cas9 cleavage sites; the microhomologous sequences are underlined. Except for M1 (the red base C represents the mutation from T to C), all red bases and dashes represent deletions; the sizes of deletions flanking the cleavage sites are also indicated. (D) PCR analysis showing deletions caused by Cas9 cleavage and MMEJ. (Left) Lane 1, 1-kb DNA ladder; lanes 2 and 3 showing the L3R3 primer pair PCR products of wild-type (WT) L. donovani cells (lane 2) and L. donovani cells transfected with pSPneogRNAa (pooled) (lane 3). (Right) lane 1, 1-kb DNA ladder; lanes 2, the L3R3 primer pair PCR products of L. donovani cells transfected with pSPneogRNAaH (pooled). Note that several smaller PCR products resulting from deletion were seen in these gRNAa-targeted cells. (E) Chromatogram showing D10 deletion caused by gRNAa targeting.
Double-strand DNA breaks (DSBs) created by Cas9 in mammalian DNA are mainly repaired by the indel-forming nonhomologous end-joining (NHEJ) pathway, resulting in inactivation of the targeted gene. To examine the types of mutations created by CRISPR-Cas9 in the LdMT-targeting sites, we extracted genomic DNA from these MLF-resistant cells (pooled or individually cloned). PCRs were performed with primers of various distances flanking these targeting sites (Fig. 2A; see Fig. S3 in the supplemental material), followed by DNA sequencing analysis. If the NHEJ pathway is present in L. donovani, we would expect to see small (1- to 30-bp) insertions and deletions near the Cas9 cleavage sites. Surprisingly, as shown in Fig. 2C, we observed only one point mutation (M1 [Fig. 2C]) and deletions, but no insertion in the LdMT genes from these gRNA-targeted MLF-resistant cells. For gRNAa-targeted cells, nearly a third of the clones sequenced had a T-to-C point mutation (M1) at the expected Cas9 cleavage site, which resulted in a single amino acid change from methionine to threonine (M381T) for LdMT. Alignments show that methionine in this position is highly conserved in all phospholipid-transporting ATPase 1-like proteins aligned (Fig. S4). Like M, the only other substitute amino acid found in this position (isoleucine [I]) is hydrophobic, in contrast to the nucleophilic threonine. This provides an additional example that a single amino acid mutation can impair LdMT transport function (34). Another third of the clones sequenced all had the same 10-bp deletion surrounding the expected Cas9 cleavage site which caused a frameshift (Fig. 2C, D10). Half of the remaining clones from the gRNAa-targeted cells had a 330-bp deletion surrounding the gRNAa/Cas9 cleavage site (−275 to +55 bp). We also detected four large deletions ranging from 1,319 bp to 1,881 bp from gRNAa-transfected cells. An example of agarose gel electrophoresis showing deletion PCR products from a pooled Leishmania population caused by gRNAa targeting is shown in Fig. 2D. We identified two sizes of deletions from gRNAc-targeted cells: the 330-bp deletion (−119 to +211) and 1,438-bp deletion (−495 to +943) which are similar to deletions detected in gRNAa-transfected cells and one large 1,581-bp deletion (−22 to +1559) from gRNAb-targeted cells (Fig. 2C). No point mutations from gRNAb- or gRNAc-targeted cells were detected. From the limited number of individual clones examined, only one type of mutation was detected from each single clone, suggesting that most MLF-resistant cells had the same mutation in both alleles. This also demonstrates that the LdMT gene is not essential for Leishmania.
When inspecting the deletions more closely (Fig. 2C; also see Fig. 7D), we noticed that all sequences deleted (large or small) were flanked by two 5- to 21-bp microhomology sequences with one microhomology sequence remained after deletion. This indicated that the DSBs generated by CRISPR-Cas9 were at least partially repaired by microhomology-mediated end joining (MMEJ). Interestingly, unlike gRNAb and gRNAc, there are two microhomology sequences (GTGGA [Fig. 2C, D10]) adjacent to the gRNAa/Cas9 cleavage site. MMEJ efficiency is affected by the proximity of microhomology sequences to the DSB site (36, 37), which may explain why gRNAa was more efficient at inactivating the LdMT gene than gRNAb and gRNAc. As no small irregular insertions and deletions were detected, this suggests that the NHEJ pathway is absent in L. donovani.
FIG 7 .
Complete gene deletion with two gRNAs simultaneously expressed in L. donovani. (A) Single- and double-gRNA expression vectors containing Hammerhead (HH) and HDV (H) ribozymes. (B) Schematic of gene deletion strategy showing the gRNA5′- and gRNA3′-targeting sites in the LdMT gene locus and the gene deletion and inversion events detected after the double-gRNA targeting. The primers used to detect these genetic events are indicated below the schematic. Note that after the oligonucleotide donor was provided to the cells, the exact 646-bp 5′F and 3′R PCR band was obtained, indicating that the LdMT gene was precisely deleted as planned. (C) MLF resistance rates of L. donovani cells transfected with various single- and double-gRNA expression vectors. The MLF resistance rates are shown as ranges from three independent experiments. (D) Sequences showing deletions and inversion caused by gRNA5′&3′ targeting. Note that the deletion sequence caused by targeting gRNA5′ alone is also shown. The red vertical lines indicate putative Cas9 cleavage sites. The microhomologous sequences are underlined. The red bases and dashes indicate deletions. The sizes of deletions flanking the cleavage sites are also indicated. The sequence of oligonucleotide donor used for controlled gene deletion and a chromatogram showing controlled deletion are included. SS Oligo donor, single-strand oligonucleotide donor.
Insertion of a single-strand oligonucleotide donor into the CRISPR-Cas9 site by homology-directed repair.
The ability to insert an oligonucleotide sequence into a specific genome site would greatly improve genome editing such as creating point mutations, adding epitope tags, and introducing stop codons or frameshifts. To explore whether CRISPR-Cas9 would facilitate insertion of a single-strand oligonucleotide sequence into the Leishmania genome, we designed a 61-nt oligonucleotide which contained two homology sequence arms flanking the Cas9/gRNAa cleavage site (25 nt each side) and an 11-nt stop codon cassette (Fig. 3A). The oligonucleotide was electroporated into the Cas9/gRNAa-expressing Leishmania cells, and the cells were allowed to recover for 3 days before subjected to MLF selection. After the Cas9/gRNAa-expressing cells were provided with this oligonucleotide stop codon cassette donor, the MLF resistance rates were increased 2- to 6-fold compared with the rate for cells receiving no oligonucleotide donor (an example shown in Fig. 3B). Sequencing analysis of these MLF-resistant cells which had received the oligonucleotide donor transfection confirmed that more than 90% of the L-R primer pair (Fig. 2A; see Fig. S3 in the supplemental material) PCR products had the stop codon cassette insert as planned (Fig. 3). The fact that the L-R primer pair could miss larger deletions resulting from MMEJ could explain why the rate of the stop cassette insertion in the L-R PCR fragments was higher than the actual increase in the MLF resistance rate. We also provided the Cas9/gRNAc-expressing cells with the oligonucleotide stop cassette donor specific for the gRNAc-targeting site. Similarly, we observed that the MLF resistance rate for Cas9/gRNAc-expressing cells increased 4-fold after receiving the oligonucleotide donor. However, no MLF-resistant cells were observed in the Cas9-expressing L. donovani cells lacking the specific gRNA after being transfected with the oligonucleotide stop cassette donor, indicating that the DSB created by the Cas9/gRNA complex is essential for the specific integration of an oligonucleotide donor (data not shown). Homology-directed repair (HDR) relies on the homology sequence provided by another allele, repeated sequence, or the single-strand oligonucleotide transfected (9, 38). The high insertion efficiency of these oligonucleotides into the specific Cas9 cleavage sites strongly indicates that DSBs are mainly repaired by HDR in L. donovani and demonstrates the feasibility of this system for generating knockouts and point mutations as well as adding epitope tags.
FIG 3 .
Efficient and precise insertion of a single-strand oligonucleotide donor into the Cas9 cleavage site of the LdMT gene by HDR. (A) Insertion strategy by Cas9 cleavage and HDR showing the LdMT locus containing the gRNAa-targeting site, the 61-nt single-strand oligonucleotide donor with stop codons, and a chromatogram showing precise insertion of the oligonucleotide donor into the Cas9 cleavage site by HDR. The number of times a sequence was obtained after analyzing the donor-transfected cells is shown in parentheses after each sequence. (B) Oligonucleotide donor with stop codons and culture at 37°C significantly increased the MLF resistance rates of gRNAa-targeted L. donovani cells. The data shown are representative of the data from three independent experiments. The MLF resistance rates after oligonucleotide donor transfection and/or 4 days of 37°C treatment were determined in 96-well plates by limiting dilution culture at 27°C for 2 to 4 weeks (see Materials and Methods for details).
Unlike mammalian cells, Leishmania promastigotes grow faster and better at 27°C than 37°C. We reasoned that shifting the Cas9 transfectant cultures from 27°C to 37°C might improve this CRISPR-Cas9 system efficiency by increasing the Cas9 endonuclease activity and decreasing the cell duplication rate, which will allow more time for the Cas9/gRNA complex to scan and identify the targeting sequence in the genome. Indeed, after the CRISPR-Cas9 transfectant culture was shifted from 27°C to 37°C for 4 days, the MLF resistance rate was increased 2- to 4-fold (an example shown in Fig. 3B). Besides the temperature shift, the MLF resistance rates also increased with prolonged culture time posttransfection, which was likely due to accumulation of CRISPR-Cas9-mediated mutation. Taken together, the combination of a donor sequence, temperature shift, and extended culture time can further increase the efficiency of this CRISPR-Cas9 system, making it feasible to generate targeted gene disruption or other mutants without the need for drug selection.
Precise insertion of a drug selection marker and GFP tag into endogenous genes.
With 10 to 50% efficiency, specific mutants generated by this CRISPR-Cas9 system can be isolated in the absence of drug selection by cloning without significant difficulty. Nevertheless, it would be advantageous if the knockouts created for other genes can be isolated by rapid selection with a drug marker as was done above for the LdMT gene. Toward this end, we attempted to generate knockouts for the nonessential Ldbpk_241510.1 gene (a multidrug resistance protein-like gene) (39) by inserting a bleomycin resistance gene expression cassette into the Cas9 cleavage site. The gRNA-targeting site in the Ldbpk_241510.1 gene is as shown in Fig. 4A (see Fig. S5 in the supplemental material). The ble expression cassette (534 bp [Fig. S6]) with a pyrimidine track upstream of the ble gene was PCR amplified with a primer pair which had at its 5′ end a 25-nt 5′ or 3′ homology flanking sequence of the expected Cas9 cleavage site, respectively, to allow for HDR. Three days after transfection of the PCR product (584 bp) into the gRNALd241510-expressing cells, the cells were subjected to bleomycin selection. About 0.0025 to 0.005% gRNALd241510-expressing cells became bleomycin resistant after receiving the ble expression cassette donor fragment. PCR and sequencing analysis of these bleomycin-resistant cells confirmed that the ble expression cassette was inserted into the gRNALd241510-targeting site as planned (Fig. 4A). PCR analysis of two bleomycin-resistant cell clones revealed that both alleles of the Ldbpk_241510.1 gene were disrupted by the ble expression cassette, since only the 1,315-bp (781 plus 534) Ble inserting band but no 781-bp band from the wild-type allele was detected (Fig. 4B). This also confirmed our previous observation that the Ldbpk_241510.1 gene is not essential for Leishmania (39).
FIG 4 .

Precise insertion of the Ble expression cassette into the Cas9 cleavage site of an endogenous gene. (A) Insertion strategy by Cas9 cleavage and HDR showing the Ldbpk_241510.1 gene locus containing the gRNALd241510-targeting site, the double-strand DNA donor (PCR product) containing the 534-bp Ble gene expression cassette flanked by 25-bp homologous arms, and the chromatogram showing precise insertion of the Ble expression cassette into the Cas9 cleavage site by HDR. (B) PCR analysis showing that the 1,315-bp PCR product containing the Ble expression cassette, but not the 781-bp wild-type allele PCR product, was detected from two L. donovani bleomycin resistance clones. Lane 1, wild-type L. donovani cells; lanes 2 and 3, bleomycin-resistant clones 1 and 2.
To investigate whether this CRISPR-Cas9 system was able to inactivate essential Leishmania genes, we also used the above approach to target the essential L. donovani TOR1 gene Ldbpk_366580.1 (40) (Fig. 5A; see Fig. S7 in the supplemental material). Interestingly, while we were able to obtain bleomycin-resistant cells (resistance rate, 0.02 to 0.04%) from gRNALd366580- and Ble donor-transfected L. donovani cells, these cells grew 40 to 50% slower than gRNALd241510- and Ble donor-transfected cells from the previous experiment. This was likely because gRNALd366580/Cas9 was continually attacking the remaining TOR1 gene allele (Fig. 5B). Furthermore, half of these 10 gRNALd366580-transfected bleomycin-resistant cell clones that were initially isolated died out after continuous culture for 2 weeks. PCR analysis of the remaining five clones that survived revealed that although the ble expression cassette had disrupted one allele of the LdTOR1 gene, the other allele was still intact (Fig. 5C). This verified that this CRISPR-Cas9 system was able to partially inactivate essential Leishmania genes and further confirmed the ability to specifically target genes with a drug selection marker.
FIG 5 .
Insertion of the Ble expression cassette into an essential gene using this Cas9/gRNA system. (A) Insertion strategy by Cas9 cleavage and HDR, showing the Ldbpk_366580.1 gene locus containing the gRNALd366580-targeting site, the double-strand DNA donor (PCR product) containing the 534-bp Ble gene expression cassette flanked by 25-bp homologous arms, and the expected insertion of the Ble expression cassette into the Cas9 cleavage site by HDR. (B) Growth curves of L. donovani cells targeted by the gRNALd366580/ble or gRNALd241510/ble donor in culture medium containing hygromycin, G418, and phleomycin. The data shown represent averages from three independent experiments. (C) PCR analysis showing both the PCR products (1,051 bp or slightly larger) containing the Ble expression cassette and the 517-bp wild-type allele PCR product were detected from pooled L. donovani bleomycin-resistant cells (before cloning) and five L. donovani bleomycin-resistant clones, confirming that the Ldbpk_366580.1 gene is essential for Leishmania. From left to right, the lanes contain 1-kb DNA ladder, wild-type (WT) L. donovani, pooled bleomycin-resistant cells, and bleomycin-resistant clones 1 to 5.
The ability to tag an endogenous gene with GFP can be very useful to help determine the expression level of a gene and cellular localization of its protein product, especially for genes difficult to isolate due to their large size or repeated sequences. The LdMT gene is a single-copy gene encoding a membrane protein with a low level of expression (34). As a proof of principle, we attempted to tag the N-terminal portion (381 amino acids) of the LdMT protein with GFP using the same approach as inserting the ble expression cassette (Fig. 6; see Fig. S8 in the supplemental material). The gRNAa/Cas9-expressing cells were electroporated with a GFP donor PCR product and subjected to MLF selection. As shown in Fig. 6B, nearly 1% of the MLF-resistant cells showed some low level of green fluorescence by fluorescence-activated cell sorting (FACS) analysis. Confocal microscopy revealed that while the majority of the sorted MLF-resistant cells showed only faint green fluorescence, about a quarter of the cells displayed fairly bright green fluorescence, which indicated that the truncated LdMT-GFP fusion protein had relocated from the plasma membrane to inside the cell somewhere around the nucleus (Fig. 6C). PCR and sequencing analysis of these sorted cells confirmed that the N-terminal portion of the LdMT protein was correctly fused with GFP as planned (Fig. 6A). This demonstrates that this CRISPR-Cas9 system can be used to tag GFP precisely to an endogenous gene.
FIG 6 .
Tagging GFP into LdMT using this Cas9/gRNA system. (A) Tagging strategy by Cas9 cleavage and HDR, showing the LdMT gene locus containing the gRNAa-targeting site, the double-strand DNA donor (PCR product) containing the 720-bp eGFP gene tag (in frame) flanked by 25-bp homologous arms, and chromatogram showing the precise tagging of the eGFP gene into the LdMT gene by HDR. (B) FACS analysis showing nearly 1% of the MLF-resistant cells had some low level of green fluorescence. FSC, forward scatter. (C) Live images of MLF-resistant promastigotes with green fluorescence by confocal microscopy (merged with differential interference contrast [DIC] image).
Whole-gene deletion with two gRNA sequences expressed from a single vector.
In mammalian cells, it has been shown that two or more gRNAs simultaneously expressed in the cell can be used to inactivate several target genes, to completely block a pathway, to delete a large gene cluster, and to improve the gene inactivation efficiency (5, 41–45). Likewise, it would require two gRNAs expressed simultaneously in Leishmania if complete deletion of an entire gene is preferred such as for generating a live attenuated vaccine strain. However, unlike mammalian cells, it is difficult to cotransfect two plasmids together into a Leishmania cell because of the low transfection efficiency (21). Therefore, we generated a vector that can express two gRNAs simultaneously in Leishmania by further exploiting the utility of ribozymes (Fig. 7A). We first developed a gRNA expression vector termed pSPneoHHgRNAH (HH stands for Hammerhead) in which a 43-bp Hammerhead ribozyme sequence was inserted between the rRNAP and the gRNA coding sequence in the pSPneogRNAH vector (Fig. 7A; see Fig. S9 in the supplemental material). The primary gRNA transcripts transcribed in this vector will be flanked at its 5′ end by the Hammerhead ribozyme and at its 3′ end by the HDV ribozyme. Similar to the HDV ribozyme which cleaves off from the 3′ end of gRNA, the Hammerhead ribozyme is expected to self-cleave off from the 5′ end of gRNA posttranscriptionally (30). We cloned the LdMT gene gRNAa sequence into this double-ribozyme vector to determine whether an active gRNA can be produced by this vector (pSPneoHHgRNAaH). Since this double-ribozyme design needs no defined transcription initiation, we also generated a vector (pSPneoHHgRNAaH-rRNAP) in which the rRNAP was removed from pSPneoHHgRNAaH to further test the functionality of the Hammerhead ribozyme (Fig. 7A). The MLF resistance assay and sequencing analysis confirmed that both pSPneoHHgRNAaH- and pSPneoHHgRNAaH-rRNAP-transfected Leishmania cells produced functional gRNA, indicating that the Hammerhead and HDV ribozymes were self-cleaved posttranscriptionally from the gRNA as expected. The additional time needed to process the Hammerhead ribozyme posttranscriptionally could have lowered the MLF resistance rate in cells transfected with pSPneoHHgRNAaH compared with cells transfected with pSPneogRNAaH (Fig. 7C). Meanwhile, cells transfected with pSPneoHHgRNAaH-rRNAP had an even lower MLF resistance rate likely because this vector relies on transcription by RNA polymerase II after removing the rRNA promoter (32). Knowing that the Hammerhead ribozyme was self-processed from the gRNAa, we inserted the fragment containing the rRNAP and gRNAbH from pSPneogRNAbH into the pSPneoHHgRNAaH-rRNAP vector in front of the HHgRNAaH coding sequence to generate a double-gRNA expression vector (pSPneogRNAbH-HHgRNAaH) (Fig. 7A). As shown in Fig. 7C, the percentage of MLF-resistant cells was increased more than 4-fold in Leishmania cells transfected with this double-gRNA expression vector compared with cells transfected with pSPneogRNAbH or pSPneoHHgRNAaH alone, suggesting that gRNAa and gRNAb were coexpressed and properly processed from this double-gRNA expression vector.
We then tested whether the dual-gRNA expression vector could be used to delete the complete LdMT gene from the L. donovani genome. Two new gRNAs were designed from the sequences flanking the 5′ and 3′ LdMT gene, respectively, and cloned into the double-gRNA expression vector (Fig. 7A and B). For comparison, we also created single-gRNA expression vectors pSPneogRNA5′H and pSPneoHHgRNA3′H. Leishmania cells were then transfected with these vectors and subjected to MLF selection. Not unexpectedly, a high percentage of Leishmania cells transfected with the dual 5′ and 3′ gRNA construct (gRNA5′&3′) developed resistance to MLF (25 to 50%) (Fig. 7C). PCR and sequencing analysis identified at least two types of gene deletion from the cells transfected with the dual-gRNA construct. A type 1 deletion was caused by gRNA5′&3′ as planned and had the whole LdMT gene together with the 5′ and 3′ flanking sequences deleted, and the two break sites were then joined by MMEJ (Fig. 7D). A type 2 deletion was caused by the gRNA5′ alone and had 149-bp 5′ flanking sequence and a large part of the LdMT coding sequence (3,131 bp) deleted. The observation that gRNA5′ alone was able to cause MLF resistance was further confirmed by cells transfected with plasmid pSPneogRNA5′H alone but with a much lower MLF resistance rate (Fig. 7C). Again, sequencing of a PCR fragment derived from these MLF-resistant cells expressing gRNA5′ alone showed that a large deletion had taken place, and the deletion included 723-bp 5′ flanking sequence and 752-bp LdMT coding sequence (Fig. 7D). Interestingly, we also detected a gene inversion with PCR primers 5′R and 3′R (Fig. 7B and D; see Fig. S3 in the supplemental material). Detection of this gene inversion event suggests that a DNA fragment as large as 3.5 kb can be inserted into the Cas9 cleavage site. However, no MLF-resistant cells were observed from cells transfected with the gRNA3′ construct alone, although large deletions resulting from MMEJ which affect the upstream LdMT coding sequence could occur.
As described above, the size of the deletion caused by MMEJ is often hard to predict and could affect the neighboring gene untranslated region (UTR) and coding sequence. We therefore attempted to have controlled gene deletion by providing the gRNA5′&3′-expressing cells with a 50-nt single-strand oligonucleotide donor containing 25-base homologous 5′ flanking sequence of the gRNA5′ cleavage site and 25-base homologous 3′ flanking sequence of the gRNA3′ cleavage site. After transfection of the oligonucleotide donor into the gRNA5′&3′-coexpressing cells, we were able to generate the nearly exact 646-bp PCR product with primers 5′F and 3′R, and sequencing confirmed that the two break sites were joined directly by the oligonucleotide donor as planned (Fig. 7B and D). This demonstrates the ability to delete an entire gene precisely in Leishmania.
DISCUSSION
In contrast to mammalian cells that use the nonhomologous end-joining (NHEJ) pathway, double-strand DNA breaks (DSBs) created by CRISPR-Cas9 in L. donovani were mainly repaired by HDR and MMEJ. This is in agreement with the genome analysis of trypanosomatids Trypanosoma cruzi, Trypanosoma brucei, and Leishmania major which had suggested that the NHEJ pathway is absent in trypanosomatids, since several key factors such as DNA ligase IV and XRCC4/Lif1 were not identified in these organisms (46). Our data are similar to what was observed in T. brucei where nearly 85% of DSBs generated by I-SceI meganuclease were repaired by interchromosomal homologous recombination and the remaining 15% were repaired by MMEJ and no NHEJ was detected (36, 37). Since NHEJ appears to be absent in trypanosomatids (13, 36, 37, 46), Leishmania must rely on HDR and MMEJ to repair the DSB generated by Cas9. As Leishmania is a diploid organism and some of its chromosomes are triploid or quadraploid (18–20), HDR of allelic recombination can be much more efficient than MMEJ. HDR domination and the absence of NHEJ for DSB repair in Leishmania may account for the relatively low initial gene inactivation frequency (caused by MMEJ alone) by this CRISPR-Cas9 system. In other words, although the Cas9/gRNA complex was continuously scanning and generating DNA breaks in the specific genome sites, most of these DNA breaks were faithfully repaired by interallelic homologous recombination, and only a small portion of these DNA breaks were repaired by MMEJ, resulting in deletion mutations. However, HDR domination for DSB repair will facilitate targeted modification of genomes. As demonstrated above, when the single-strand homology oligonucleotide donors with stop codons were provided to the cell, the MLF resistance rate or the LdMT gene inactivation frequency was increased by 2- to 6-fold. The capability of integrating an oligonucleotide donor into a specific genome site will also facilitate such genome-editing tasks as creating point mutations and adding epitope tags. HDR domination for DSB repair could also explain why only one type of mutation was detected in these MLF-resistant clones, as theoretically, different alleles could be repaired by MMEJ with different sizes of deletions. Based on a similar concept, a CRISPR-Cas9-mediated mutagenic chain reaction method for converting heterozygous mutations to homozygous mutations has been recently reported (47).
MMEJ is distinguished from the other repair mechanisms by its use of 5- to 25-bp microhomologous sequences to align the broken strands before joining. Since MMEJ is a more error-prone method of repair and results in deletion mutations, it is used only when the NHEJ pathway is not available or disrupted (48–50). As reported for T. brucei, the break proximity, microhomology length, and GC content may affect MMEJ efficiency (36, 37); likewise, we observed that the small deletions (Fig. 2, D10 and D330) accounted for more than half of all deletions detected in gRNAa-expressing cells. It is not clear how the single point mutation arose in the LdMT gene from gRNAa-expressing cells (Fig. 2, M1). The point mutation and short deletions together accounted for more than 80% of all deletions detected in gRNAa-expressing cells, indicating that close proximity of microhomologous sequences to the DSB favors MMEJ in Leishmania. Unlike for gRNAa, there were no microhomologous sequences immediately adjacent to the gRNAb or gRNAc Cas9 cleavage sites, which could explain why gRNAa was more efficient than gRNAb and -c to inactivate the LdMT gene, although the latter two gRNAs had higher activity scores by the designer tool (35). Therefore, it is important to consider the accessibility of microhomologous sequences surrounding the Cas9 cleavage site when designing a gRNA if a drug selection marker or an oligonucleotide donor is not going to be used for gene inactivation in Leishmania (13). In addition, RAD51, a DNA recombinase, has been shown to play a central role in homologous recombination during DSB repair in mammalian cells, T. brucei, and Leishmania but is not involved in MMEJ (36, 37, 50, 51). As HDR dominates DSB repair in Leishmania, it will be of considerable interest to determine whether inhibition of the HDR pathway by disrupting L. donovani RAD51 would increase the frequency of MMEJ-mediated DSB repair, so as to improve gene inactivation efficiency. Generating an L. donovani RAD51 null mutant with this CRISPR/Cas9 system is under way.
To our knowledge, this is the first time that an RNA polymerase I promoter has been used to drive gRNA expression in any organism. The L. donovani rRNA promoter is a strong RNA polymerase I promoter which has been shown to induce gene expression by more than 60-fold (28). Indeed, compared with the gRNA vector pSPneoHHgRNAaH-rRNAP, which relies on the RNA polymerase II promoter for transcription, many more cells transfected with pSPneoHHgRNAaH (containing the rRNA promoter) became G418 and MLF resistant (Fig. 7C). It will be of interest in the future to compare the efficacies of RNA polymerase I and III promoters to drive gRNA expression once the L. donovani RNA polymerase III U6 promoter is identified. It would also be of interest to investigate using the RNA polymerase I promoter for improving gRNA expression in other organisms, including human cells. Although some sequence differences are observed between rRNA promoters of different Leishmania species, we predict that this CRISPR-Cas9 system would also work in Leishmania infantum, L. major, and several other Leishmania species since transient-transfection experiments suggested that the L. donovani rRNA promoter could be active in these species (28, 29).
To develop a safe stable live attenuated Leishmania vaccine strain, complete deletion of one or several virulence genes would be required. With the use of Hammerhead and HDV ribozymes and the rRNA promoter, a double-gRNA expression vector was developed to express two gRNAs simultaneously. This double-gRNA vector was used successfully to delete the whole LdMT gene, and it significantly improved gene inactivation efficiency. If required, for example, to completely block a pathway or a multigene family, a plasmid able to express several more gRNAs flanked by Hammerhead and HDV ribozymes in tandem array can be constructed using this vector system (52). Moreover, one gRNA can be used to target the LdMT gene and the other gRNA can be used to target a different gene of interest, and following selection for MLF resistance, the gene of interest could be mutated with high efficiency (53, 54).
Although manipulating the T. cruzi genome is usually more difficult than in Leishmania with the traditional gene-targeting approach, the CRISPR-Cas9 approach works efficiently in T. cruzi with 60% double allelic mutation frequency (13). T. cruzi proliferates much more slowly than Leishmania in culture (doubling time of 36 h for T. cruzi versus 8 h for L. donovani), and therefore, the high efficiency could be due in part to the long doubling time allowing the Cas9/gRNA complex more time to scan the genome and cause mutations before DNA replication. The work with T. cruzi involved transfection of in vitro-synthesized gRNA directly into the parasite in contrast to this study of Leishmania which developed plasmid vectors for stable expression of gRNA. Direct transfection of in vitro-synthesized gRNA can bypass the cloning and selection steps, and several gRNA species could be delivered simultaneously (13). However, if the gene-targeting efficiency is low because of repair by HDR such as in Leishmania, sequential transfection of the gRNA would be required. The gRNA expression vector approach described in this study can also be used to construct a gRNA-targeting library.
CRISPR-Cas9-mediated gene deletion in L. major was recently reported where the U6 promoter was used to drive gRNA expression, and the flanking donor sequences were also included in the gRNA expression plasmid to promote HDR (14). Since the study of L. major tested only a single gRNA and a plasmid donor to disrupt one gene locus (the nonessential paraflagellar rod-2 locus), it is difficult to fully compare these two systems. However, in a similar experiment set up to target a gene locus with a drug selection marker donor, in this study, all bleomycin-resistant clones examined contained the desired insertion in the targeted L. donovani genome sites (Fig. 4, 5). In contrast, only a quarter of the drug-resistant L. major cells had the correct PFR2 replacement with the puromycin selection marker donor. Those authors suspected that the L. major U6 promoter could be weak and the presence of circular donor plasmid contributed to the low CRISPR-Cas9 gene-targeting efficiency in L. major. In comparison, it was simpler in the present study to prepare the drug selection marker donor as a PCR product with 25-bp homology arms than to construct a donor plasmid with two 1-kb-long homology sequences flanking a drug resistance gene similar to the traditional gene-targeting approach in L. major (14).
With carefully designed gene-specific gRNAs, CRISPR-Cas9-induced off-target mutations are rare in mammalian cells (55). Likewise, no off-target mutations were detected in T. cruzi and L. major (13, 14). To address this concern, one can design several specific gRNAs for a gene of interest such as the gRNAa, -b, and -c used to target the LdMT gene in this study and determine whether each results in a similar phenotype. A phenotype can be further verified by gene complementation, as with traditional gene-targeting approaches.
Successful genome-scale CRISPR-Cas9 knockout screenings have been carried out in human cells (24, 25). However, before a similar genome-scale screening can be performed in Leishmania, the direct gene-targeting efficiency (without a donor) by this CRISPR-Cas9 system has to be improved. To achieve this goal, one approach as described above would be to generate a RAD51 null mutant to increase MMEJ. The rRNA promoter could be used to try to increase Cas9 nuclease expression. Additional gRNAs targeting various LdMT sites can be tested to help ascertain sequence features which enhance gRNA activity and promote MMEJ (35–37). Likewise, a series of mutated gRNAs with some unmatched nucleotides in the guide sequence against an LdMT site can be designed to identify sequence elements which could cause off-target mutations (5). The use of a bleomycin selection marker donor in this study has significantly eased isolation of gene disruption mutants. However, to further assess the efficacy of this CRISPR-Cas9 system, it will be helpful to examine additional bleomycin-resistant clones to determine whether most of these clones have both alleles disrupted for the nonessential Ldbpk_241510.1 gene (Fig. 4). Likewise, it will be useful to determine the frequency of each type of the LdMT gene deletion mutations mediated by the double gRNA (gRNA5′ and gRNA3′) targeting with or without the oligonucleotide donor (Fig. 7) in a follow-up study.
In summary, this study describes the implementation of CRISPR-Cas9-mediated genome editing in L. donovani. With this simple and efficient system, it was possible to introduce mutations, antibiotic selection markers, and a protein tag sequence (GFP) precisely into the L. donovani genome and to delete an entire gene. Together, these methods significantly improve the ability to manipulate the L. donovani genome and will help in the identification of virulence genes, new drug targets, and drug resistance mechanisms and enable an overall better understanding of biological processes in Leishmania.
MATERIALS AND METHODS
Leishmania donovani strain and culture medium.
The L. donovani 1S/Cl2D strain used in this study was routinely cultured at 27°C in M199 medium (pH 7.4) supplemented with 10% heat-inactivated fetal bovine serum, 40 mM HEPES (pH 7.4), 0.1 mM adenine, 5 mg liter−1 hemin, 1 mg liter−1 biotin, 1 mg liter−1 biopterine, 50 U ml−1 penicillin, and 50 µg ml−1 streptomycin. Cultures were passaged to fresh medium in a 20-fold dilution once a week. Unless indicated, all Leishmania cultures were carried out at 27°C in this study.
Plasmid construction.
All primer sequences used in this study are listed in Fig. S10 in the supplemental material.
The pLPhygCas9 plasmid was generated by cloning the humanized Streptococcus pyogenes Cas9 nuclease with nuclear localization signals (4.4 kb) from plasmid pX330 (Addgene) (5) into the HindIII and BamHI sites of Leishmania expression vector pLPhyg (26) with primers pX330Cas9F (F stands for forward) and pX330Cas9R (R stands for reverse).
The pSPneogRNA plasmid was generated as follows. (i) The 180-bp L. donovani rRNA promoter was derived from L. donovani genomic DNA by PCR with primers LdrRNApF and LdrRNApR. (ii) A 370-bp DNA fragment that contains the guide sequence insertion site and the 82-bp Cas9 binding RNA coding sequence was obtained from the pX330 plasmid with primer pX330gRNAF and primer pX330gRNAR. (iii) The 180-bp LdrRNA promoter sequence from step 1 and the 370-bp guide RNA coding sequence from step 2 (there are 26-bp overlap sequence between these two sequences) were joined together by PCR with primer pair LdrRNApF and pX330gRNAR. (iv) The joined sequence (550 bp) from step 3 was digested with HindIII and BamHI and cloned into the corresponding sites of pSPneo (32) to generate gRNA expression plasmid pSPneogRNA.
The pSPneogRNAH plasmid was generated as follows. (i) The 313-bp DNA sequence which contains the L. donovani rRNA promoter and the complete gRNA coding sequence from the pSPneogRNA plasmid was amplified by PCR with primer LdrRNApF and primer HDVRiboR1 which also contains at its 5′ end 28 nucleotides that overlap the HDV ribozyme coding sequence. (ii) The 313-bp PCR fragment of the LdrRNA promoter and gRNA sequence from step 1 was mixed with the following two HDV ribozyme coding sequence oligonucleotides, HDVRiboF1 and HDVRiboR2. Note that HDVRiboF1 and HDVRiboR2 have 19-nucleotide complementary sequences at their 3′ ends. (iii) The PCR fragment and the HDV ribozyme coding sequence oligonucleotide mixture from step 2 were joined together by PCR with primer pair LdrRNApF and HDVRiboR3. (iv) The joined sequence (359 bp) from step 3 was digested with HindIII and BamHI and cloned into the corresponding sites of pSPneo (32) to generate gRNA expression plasmid pSPneogRNAH.
The pSPneoHHgRNAH plasmid was generated as follows. (i) The following five oligonucleotides, HH241510+1, HHBbsI+2, HHBbsI-1, HH241510-2, and HH241510-3, which contain a HindIII site and a BglII site, Hammerhead ribozyme sequence, and two BbsI sites were annealed in T4 DNA ligase buffer. (ii) The annealed DNA fragment from step 1 was ligated into BbsI-digested pSPneogRNAH plasmid after the unique BglII site was eliminated with DNA polymerase I Klenow fragment.
The pSPneogRNA5′H-HHgRNA3′H plasmid (gRNA5′&3′ construct) was generated by inserting the 360-bp HindIII and BamHI fragment, which contains LdrRNAP and Ld1315905′gRNA coding sequence from pSPneogRNA5′H, into the HindIII and BglII sites of pSPneoHHgRNA3′H after removing the 180-bp LdrRNAP sequence.
gRNA and primer design and synthesis.
The gRNA-targeting sequences were selected manually or with the gRNA designer tool (http://www.broadinstitute.org/rnai/public/analysis-tools/sgrna-design). The primers for PCR were picked manually or by Primer3 (http://gmdd.shgmo.org/primer3). All primers and oligonucleotides used in this study were synthesized and purchased from Alpha DNA (Montreal, Canada).
Cloning guide sequence into various gRNA expression vectors.
The complementary oligonucleotides of guide sequence were first phosphorylated in T4 DNA ligase buffer with T4 polynucleotide kinase and then annealed by heating at 95°C for 5 min in a 1-liter water glass beaker, which was allowed to cool to room temperature for about an hour. The annealed guide DNA linkers (4 µM) were then cloned into the various BbsI-digested gRNA expression vectors (see Fig. S1, S2, and S9 in the supplemental material).
Leishmania transfection and limiting dilution assay.
Leishmania transfections were performed as described previously (21, 56). Four to 10 µg of plasmid DNA, 10 µl of 100-µm single-strand oligonucleotide donor, and 2 to 4 µg of purified PCR products were used for each transfection. To generate a cell line that constantly expresses Cas9, L. donovani promastigotes transfected with pLPhygCas9 were selected and maintained at culture medium containing 100 µg/ml hygromycin. The Cas9-expressing Leishmania cells transfected with various gRNA expression plasmids were selected with G418 (100 µg/ml) and maintained in medium containing both hygromycin and G418 (100 µg/ml each) as a pooled population. Aliquots (<100 µl) of these cultures were taken out for MLF resistance assay by limiting dilution culture at various time points as indicated in the figures (for example, 4 and 6 weeks posttransfection). To determine whether transfection of an oligonucleotide donor would increase LdMT inactivation frequency, the gRNA transfectant culture was split into two flasks. One flask remained in a 27°C incubator. Cells from the other flask were harvested, transfected with the oligonucleotide donor, and then resuspended in culture medium containing both hygromycin and G418 in a 27°C incubator. After continuous culture for 3 days, the cells were taken out from the non-oligonucleotide donor-transfected or oligonucleotide donor-transfected culture flasks, counted, and then directly used for MLF resistance assay by limiting dilution culture. To determine whether culture at 37°C would improve the LdMT inactivation frequency, the gRNA transfectant cultures with or without the oligonucleotide donor were split into two flasks, one remained in a 27°C incubator, and the other flask was transferred to a 37°C incubator. After 4 days of culture, cells were taken out from the 27°C and 37°C culture flasks, counted, and then used for MLF resistance assay by limiting dilution culture.
Limiting dilution cultures used to clone the transfectants and to quantify MLF resistance and Ble donor insertion rates were performed at 27°C in 96-well plates that contained Leishmania culture medium (100 µl/well) with 40 µM miltefosine or 100 µg/ml phleomycin in addition to hygromycin and G418 (100 µg/ml each). The gRNA transfectants were inoculated into the wells in the first column at two thousand to one mi llion cells in 200 µl medium per well and then serially diluted by 2-fold from left to right until column 12. Each sample was serially diluted in triplicate or quadruplicate and cultured at 27°C for 2 to 4 weeks. The resistance rates were determined by counting back from the last well containing a single parasite to determine the number of resistant parasites, and this was compared to the number of parasites added in the first well before drug selection. Unlike phleomycin and hygromycin, which take days to kill wild-type L. donovani promastigote cells, miltefosine requires less than 20 h to completely kill wild-type L. donovani cells.
The primer pair 241510BleF and 241510BleR with pSPble plasmid as the PCR template were used to generate the bleomycin resistance gene expression cassette donor (see Fig. S6 in the supplemental material). Two to 4 µg of purified Ble donor PCR products was used to transfect the Cas9- and gRNALd241510-expressing Leishmania cells. Three days posttransfection, the cells were subjected to drug selection with 100 µg/ml phleomycin.
The primer pair 131590GFPF and 131590GFPR with pEGFP-N1 (EGFP stands for enhanced green fluorescent protein) plasmid as the PCR template were used to generate the GFP tag donor (see Fig. S8 in the supplemental material). Two to 4 µg of purified GFP donor PCR products was used to transfect the Cas9- and gRNAa-expressing Leishmania cells. Three days posttransfection, the cells were subjected to drug selection with 40 µM miltefosine.
Sequencing analysis.
All plasmid constructs generated in this study were verified by DNA sequencing.
Genomic DNAs from wild-type and gRNA construct-transfected Leishmania promastigotes were extracted with a minipreparation method as described previously (57). The sequences surrounding the gRNA-targeting sites were amplified by KAPA Taq DNA polymerase with various primer pairs listed in the supplemental material. The PCR products were purified from agarose gels and sent directly for sequencing or subcloned into the pSP72 vector before sequencing. All gRNA expression plasmids were sequenced by a modified protocol for DNA with secondary structure. Sequencing was performed at the McGill University and Genome Quebec Innovation Center.
Flow cytometry and fluorescence microscopy.
Flow cytometric analysis was performed using a FACSariaII cytometer from Becton Dickinson (BD Biosciences, USA), and data were collected by using FACSDiva version 6.1.3. Imaging of live Leishmania promastigotes by confocal microscopy (Olympus) was performed as described previously (58).
The pLPhygCas9, pSPneogRNAH, pSPneoHHgRNAH, and pSPble plasmids have been deposited in Addgene with identification (ID) no. 63555, 63556, 63557, and 63561, respectively. Partial sequences for some of these plasmids are provided in Fig. S1, S6, and S9 in the supplemental material.
SUPPLEMENTAL MATERIAL
L. donovani gRNA expression vector pSPneogRNAH and its partial sequence. (A) Schematic of pSPneogRNAH and its guide sequence insertion site. The L. donovani rRNA promoter (rRNAP) and HDV ribozyme (H) are indicated. The small solid black box represents the 92-bp pyrimidine track (see Fig. S6 in the supplemental material). The first nucleotide U of gRNA is indicated in black, as L. donovani rRNAP initiates transcription at the T-residue site. The schematic is not drawn to scale. (B) The partial sequence of pSPneogRNAH (HindIII and BamHI fragment). The 180-bp rRNAP sequence (black) and its transcription initiation site T (bold) are underlined. The 82-bp Cas9 binding RNA coding sequence (green), the 68-bp HDV ribozyme coding sequence (blue), and the HindIII, BbsI, and BamHI restriction enzyme sites (red) are indicated. Download
L. donovani gRNA expression vector pSPneogRNA partial sequence (HindIII and BamHI fragment). The 180-bp rRNAP sequence (black) and its transcription initiation site T (bold) are underlined. The 82-bp Cas9 binding RNA coding sequence (green), the 262-bp nonspecific DNA sequence derived from the pX330 plasmid (black), and the HindIII, BbsI, and BamHI restriction enzyme sites are highlighted in red. Download
L. donovani miltefosine transporter gene (LdMT; LdBPK_131590.1) sequence and the sequences of gRNA guides, oligonucleotide donors, and primers used to generate and detect LdMT mutants. The locations and directions of gRNA guides and primers in the LdMT gene are indicated and underlined with an arrow. Some of these primers had a restriction enzyme site added to its 5′ end to facilitate cloning of the PCR product. Download
Partial amino acid sequence alignments of phospholipid-transporting ATPase 1-like proteins (miltefosine transporters). The methionine (M) at position 381 is highly conserved in all phospholipid-transporting ATPase 1-like proteins aligned. Like M, the only other substitute amino acid found at this position (isoleucine [I]) is hydrophobic. LdMT, L. donovani miltefosine transporter (LdBPK_131590.1); XP 001681897, L. major miltefosine transporter (MT); AHM92900, Leishmania amazonensis MT; XP 003873354, Leishmania mexicana MT; XP 001563280, Leishmania braziliensis MT; XP 010697220, Leishmania panamensis MT. Download
LdBPK_241510.1 gene partial sequence and the sequences of gRNA guide and primers used to generate and detect LdBPK_241510.1 mutants. Download
Bleomycin resistance gene expression cassette sequence and primers used to generate the LdBPK_241510.1 gene disruption PCR product donor. The 92-bp pyrimidine track upstream of the bleomycin resistance gene is underlined. Download
LdBPK_366580.1 gene (target of rapamycin [TOR] kinase 1) partial sequence and the sequences of the gRNA guide and primers used to generate and detect LdBPK_366580.1 mutants. Download
EGFP gene sequence and primers used to generate the LdMT-GFP fusion PCR product donor. Download
L. donovani gRNA expression vector pSPneoHHgRNAH and its partial sequence. (A) Schematic of pSPneoHHgRNAH and its guide sequence insertion site. The L. donovani rRNA promoter (rRNAP), Hammerhead ribozyme (HH), and HDV ribozyme (H) are indicated. The small solid black box represents the 92-bp pyrimidine track (see Fig. S6 in the supplemental material). (B) The partial sequence of pSPneoHHgRNAH (HindIII and BamHI fragment). The 183-bp rRNAP sequence (black) and its transcription initiation site T (bold) are underlined. The 82-bp Cas9 binding RNA coding sequence (green), the 49-bp Hammerhead ribozyme (blue and underlined), the 68-bp HDV ribozyme sequence (blue), and the HindIII, BglII, BbsI, and BamHI restriction enzyme sites (red) are indicated. Download
Primers used to generate pLPHygCas9 and gRNA expression vectors. Download
ACKNOWLEDGMENTS
We thank Feng Zhang for plasmid pX330, Marc Ouellette and Barbara Papadopoulou for plasmids pSPneo and pSPhyg, and Brendan D. Snarr for help with confocal microscopy.
This work was supported by Canadian Institute of Health Research grant MOP125996 to G.M.
Footnotes
Citation Zhang W-W, Matlashewski G. 2015. CRISPR-Cas9-mediated genome editing in Leishmania donovani. mBio 6(4):e00861-15. doi:10.1128/mBio.00861-15.
REFERENCES
- 1.Doudna JA, Charpentier E. 2014. The new frontier of genome engineering with CRISPR-Cas9. Science 346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 2.Harrison MM, Jenkins BV, O’Connor-Giles KM, Wildonger J. 2014. A CRISPR view of development. Genes Dev 28:1859–1872. doi: 10.1101/gad.248252.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hsu PD, Lander ES, Zhang F. 2014. Development and applications of CRISPR-Cas9 for genome engineering. Cell 157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, Ishitani R, Zhang F, Nureki O. 2014. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 156:935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Böttcher R, Hollmann M, Merk K, Nitschko V, Obermaier C, Philippou-Massier J, Wieland I, Gaul U, Förstemann K. 2014. Efficient chromosomal gene modification with CRISPR/cas9 and PCR-based homologous recombination donors in cultured Drosophila cells. Nucleic Acids Res 42:e89. doi: 10.1093/nar/gku289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paix A, Wang Y, Smith HE, Lee CY, Calidas D, Lu T, Smith J, Schmidt H, Krause MW, Seydoux G. 2014. Scalable and versatile genome editing using linear DNAs with microhomology to Cas9 sites in Caenorhabditis elegans. Genetics 198:1347–1356. doi: 10.1534/genetics.114.170423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin S, Staahl B, Alla RK, Doudna JA. 2014. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. Elife 3:e04766. doi: 10.7554/eLife.04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen B, Brown KM, Lee TD, Sibley LD. 2014. Efficient gene disruption in diverse strains of Toxoplasma gondii using CRISPR/CAS9. mBio 5(3):e01114-14. doi: 10.1128/mBio.01114-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghorbal M, Gorman M, Macpherson CR, Martins RM, Scherf A, Lopez-Rubio JJ. 2014. Genome editing in the human malaria parasite Plasmodium falciparum using the CRISPR-Cas9 system. Nat Biotechnol 32:819–821. doi: 10.1038/nbt.2925. [DOI] [PubMed] [Google Scholar]
- 12.Wagner JC, Platt RJ, Goldfless SJ, Zhang F, Niles JC. 2014. Efficient CRISPR-Cas9-mediated genome editing in Plasmodium falciparum. Nat Methods 11:915–918. doi: 10.1038/nmeth.3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peng D, Kurup SP, Yao PY, Minning TA, Tarleton RL. 2015. CRISPR-Cas9-mediated single-gene and gene family disruption in Trypanosoma cruzi. mBio 6(1):e02097-14. doi: 10.1128/mBio.02097-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sollelis L, Ghorbal M, MacPherson CR, Martins RM, Kuk N, Crobu L, Bastien P, Scherf A, Lopez-Rubio JJ, Sterkers Y. 19 June 2015. First efficient CRISPR-Cas9-mediated genome editing in Leishmania parasites. Cell Microbiol doi: 10.1111/cmi.12456. [DOI] [PubMed] [Google Scholar]
- 15.Alvar J, Vélez ID, Bern C, Herrero M, Desjeux P, Cano J, Jannin J, den Boer M, WHO Leishmaniasis Control Team . 2012. Leishmaniasis worldwide and global estimates of its incidence. PLoS One 7:e35671. doi: 10.1371/journal.pone.0035671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ready PD. 2014. Epidemiology of visceral leishmaniasis. Clin Epidemiol 6:147–154. doi: 10.2147/CLEP.S44267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peacock CS, Seeger K, Harris D, Murphy L, Ruiz JC, Quail MA, Peters N, Adlem E, Tivey A, Aslett M, Kerhornou A, Ivens A, Fraser A, Rajandream MA, Carver T, Norbertczak H, Chillingworth T, Hance Z, Jagels K, Moule S. 2007. Comparative genomic analysis of three Leishmania species that cause diverse human disease. Nat Genet 39:839–847. doi: 10.1038/ng2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Downing T, Imamura H, Decuypere S, Clark TG, Coombs GH, Cotton JA, Hilley JD, de Doncker S, Maes I, Mottram JC, Quail MA, Rijal S, Sanders M, Schönian G, Stark O, Sundar S, Vanaerschot M, Hertz-Fowler C, Dujardin JC, Berriman M. 2011. Whole genome sequencing of multiple Leishmania donovani clinical isolates provides insight into population structure and mechanisms of resistance. Genome Res 21:2143–2156. doi: 10.1101/gr.123430.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rogers MB, Hilley JD, Dickens NJ, Wilkes J, Bates PA, Depledge DP, Harris D, Her Y, Herzyk P, Imamura H, Otto TD, Sanders M, Seeger K, Dujardin JC, Berriman M, Smith DF, Hertz-Fowler C, Mottram JC. 2011. Chromosome and gene copy number variations allow major structural change between species and strains of Leishmania. Genome Res 21:2129–2142. doi: 10.1101/gr.122945.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang WW, Ramasamy G, McCall LI, Haydock A, Ranasinghe S, Abeygunasekara P, Sirimanna G, Wickremasinghe R, Myler P, Matlashewski G. 2014. Genetic analysis of Leishmania donovani tropism using a naturally attenuated cutaneous strain. PLoS Pathog 10:e1004244. doi: 10.1371/journal.ppat.1004244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robinson KA, Beverley SM. 2003. Improvements in transfection efficiency and tests of RNA interference (RNAi) approaches in the protozoan parasite Leishmania. Mol Biochem Parasitol 128:217–228. doi: 10.1016/S0166-6851(03)00079-3. [DOI] [PubMed] [Google Scholar]
- 22.Lye LF, Owens K, Shi H, Murta SM, Vieira AC, Turco SJ, Tschudi C, Ullu E, Beverley SM. 2010. Retention and loss of RNA interference pathways in trypanosomatid protozoans. PLoS Pathog 6:e1001161. doi: 10.1371/journal.ppat.1001161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dean S, Sunter J, Wheeler RJ, Hodkinson I, Gluenz E, Gull K. 2015. A toolkit enabling efficient, scalable and reproducible gene tagging in trypanosomatids. Open Biol 5:140197. doi: 10.1098/rsob.140197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang T, Wei JJ, Sabatini DM, Lander ES. 2014. Genetic screens in human cells using the CRISPR-Cas9 system. Science 343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F. 2014. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang WW, Charest H, Matlashewski G. 1995. The expression of biologically active human p53 in Leishmania cells: a novel eukaryotic system to produce recombinant proteins. Nucleic Acids Res 23:4073–4080. doi: 10.1093/nar/23.20.4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Padilla-Mejía NE, Florencio-Martínez LE, Figueroa-Angulo EE, Manning-Cela RG, Hernández-Rivas R, Myler PJ, Martínez-Calvillo S. 2009. Gene organization and sequence analyses of transfer RNA genes in trypanosomatid parasites. BMC Genomics 10:232. doi: 10.1186/1471-2164-10-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan S, Lodes MJ, Fox M, Myler PJ, Stuart K. 1999. Characterization of the Leishmania donovani ribosomal RNA promoter. Mol Biochem Parasitol 103:197–210. doi: 10.1016/S0166-6851(99)00126-7. [DOI] [PubMed] [Google Scholar]
- 29.Martínez-Calvillo S, Sunkin SM, Yan S, Fox M, Stuart K, Myler PJ. 2001. Genomic organization and functional characterization of the Leishmania major Friedlin ribosomal RNA gene locus. Mol Biochem Parasitol 116:147–157. doi: 10.1016/S0166-6851(01)00310-3. [DOI] [PubMed] [Google Scholar]
- 30.Gao Y, Zhao Y. 2014. Self-processing of ribozyme-flanked RNAs into guide RNAs in vitro and in vivo for CRISPR-mediated genome editing. J Integr Plant Biol 56:343–349. doi: 10.1111/jipb.12152. [DOI] [PubMed] [Google Scholar]
- 31.Ryan OW, Cate JH. 2014. Multiplex engineering of industrial yeast genomes using CRISPRm. Methods Enzymol 546:473–489. doi: 10.1016/B978-0-12-801185-0.00023-4. [DOI] [PubMed] [Google Scholar]
- 32.Papadopoulou B, Roy G, Ouellette M. 1994. Autonomous replication of bacterial DNA plasmid oligomers in Leishmania. Mol Biochem Parasitol 65:39–49. doi: 10.1016/0166-6851(94)90113-9. [DOI] [PubMed] [Google Scholar]
- 33.LeBowitz JH, Smith HQ, Rusche L, Beverley SM. 1993. Coupling of poly(A) site selection and trans-splicing in Leishmania. Genes Dev 7:996–1007. doi: 10.1101/gad.7.6.996. [DOI] [PubMed] [Google Scholar]
- 34.Pérez-Victoria FJ, Gamarro F, Ouellette M, Castanys S. 2003. Functional cloning of the miltefosine transporter. A novel P-type phospholipid translocase from Leishmania involved in drug resistance. J Biol Chem 278:49965–49971. doi: 10.1074/jbc.M308352200. [DOI] [PubMed] [Google Scholar]
- 35.Doench JG, Hartenian E, Graham DB, Tothova Z, Hegde M, Smith I, Sullender M, Ebert BL, Xavier RJ, Root DE. 2014. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat Biotechnol 32:1262–1267. doi: 10.1038/nbt.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Glover L, McCulloch R, Horn D. 2008. Sequence homology and microhomology dominate chromosomal double-strand break repair in African trypanosomes. Nucleic Acids Res 36:2608–2618. doi: 10.1093/nar/gkn104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Glover L, Jun J, Horn D. 2011. Microhomology-mediated deletion and gene conversion in African trypanosomes. Nucleic Acids Res 39:1372–1380. doi: 10.1093/nar/gkq981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morrical SW. 2015. DNA-pairing and annealing processes in homologous recombination and homology-directed repair. Cold Spring Harb Perspect Biol 7:a016444. doi: 10.1101/cshperspect.a016444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang WW, Matlashewski G. 2012. Deletion of an ATP-binding cassette protein subfamily C transporter in Leishmania donovani results in increased virulence. Mol Biochem Parasitol 185:165–169. doi: 10.1016/j.molbiopara.2012.07.006. [DOI] [PubMed] [Google Scholar]
- 40.Madeira da Silva L, Beverley SM. 2010. Expansion of the target of rapamycin (TOR) kinase family and function in Leishmania shows that TOR3 is required for acidocalcisome biogenesis and animal infectivity. Proc Natl Acad Sci U S A 107:11965–11970. doi: 10.1073/pnas.1004599107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ota S, Hisano Y, Ikawa Y, Kawahara A. 2014. Multiple genome modifications by the CRISPR/Cas9 system in zebrafish. Genes Cells 19:555–564. doi: 10.1111/gtc.12154. [DOI] [PubMed] [Google Scholar]
- 42.Zhou J, Wang J, Shen B, Chen L, Su Y, Yang J, Zhang W, Tian X, Huang X. 2014. Dual sgRNAs facilitate CRISPR/Cas9-mediated mouse genome targeting. FEBS J 281:1717–1725. doi: 10.1111/febs.12735. [DOI] [PubMed] [Google Scholar]
- 43.Chen X, Xu F, Zhu C, Ji J, Zhou X, Feng X, Guang S. 2014. Dual sgRNA-directed gene knockout using CRISPR/Cas9 technology in Caenorhabditis elegans. Sci Rep 4:7581. doi: 10.1038/srep07581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li W, Teng F, Li T, Zhou Q. 2013. Simultaneous generation and germline transmission of multiple gene mutations in rat using CRISPR-Cas systems. Nat Biotechnol 31:684–686. doi: 10.1038/nbt.2652. [DOI] [PubMed] [Google Scholar]
- 45.Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. 2013. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Passos-Silva DG, Rajão MA, Nascimento de Aguiar PH, Vieira-da-Rocha JP, Machado CR, Furtado C. 2010. Overview of DNA repair in Trypanosoma cruzi, Trypanosoma brucei, and Leishmania major. J Nucleic Acids 2010:840768. doi: 10.4061/2010/840768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gantz VM, Bier E. 2015. The mutagenic chain reaction: a method for converting heterozygous to homozygous mutations. Science 348:442–444. doi: 10.1126/science.aaa5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chapman JR, Taylor MR, Boulton SJ. 2012. Playing the end game: DNA double-strand break repair pathway choice. Mol Cell 47:497–510. doi: 10.1016/j.molcel.2012.07.029. [DOI] [PubMed] [Google Scholar]
- 49.McVey M, Lee SE. 2008. MMEJ repair of double-strand breaks (director’s cut): deleted sequences and alternative endings. Trends Genet 24:529–538. doi: 10.1016/j.tig.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Decottignies A. 2013. Alternative end-joining mechanisms: a historical perspective. Front Genet 4:48. doi: 10.3389/fgene.2013.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ubeda JM, Raymond F, Mukherjee A, Plourde M, Gingras H, Roy G, Lapointe A, Leprohon P, Papadopoulou B, Corbeil J, Ouellette M. 2014. Genome-wide stochastic adaptive DNA amplification at direct and inverted DNA repeats in the parasite Leishmania. PLoS Biol 12:e1001868. doi: 10.1371/journal.pbio.1001868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xie K, Minkenberg B, Yang Y. 2015. Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc Natl Acad Sci U S A 112:3570–3575. doi: 10.1073/pnas.1420294112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arribere JA, Bell RT, Fu BX, Artiles KL, Hartman PS, Fire AZ. 2014. Efficient marker free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics 198:837–846. doi: 10.1534/genetics.114.169730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim H, Ishidate T, Ghanta KS, Seth M, Conte D Jr, Shirayama M, Mello CC. 2014. A co-CRISPR strategy for efficient genome editing in Caenorhabditis elegans. Genetics 197:1069–1080. doi: 10.1534/genetics.114.166389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu X, Kriz AJ, Sharp PA. 2014. Target specificity of the CRISPR-Cas9 system. Quant Biol 2:59–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang WW, Matlashewski G. 2015. Screening Leishmania donovani complex-specific genes required for visceral disease. Methods Mol Biol 1201:339–361. doi: 10.1007/978-1-4939-1438-8_20. [DOI] [PubMed] [Google Scholar]
- 57.Medina-Acosta E, Cross GA. 1993. Rapid isolation of DNA from trypanosomatid protozoa using a simple “mini-prep” procedure. Mol Biochem Parasitol 59:327–329. doi: 10.1016/0166-6851(93)90231-L. [DOI] [PubMed] [Google Scholar]
- 58.Zhang WW, Chan KF, Song Z, Matlashewski G. 2011. Expression of a Leishmania donovani nucleotide sugar transporter in Leishmania major enhances survival in visceral organs. Exp Parasitol 129:337–345. doi: 10.1016/j.exppara.2011.09.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
L. donovani gRNA expression vector pSPneogRNAH and its partial sequence. (A) Schematic of pSPneogRNAH and its guide sequence insertion site. The L. donovani rRNA promoter (rRNAP) and HDV ribozyme (H) are indicated. The small solid black box represents the 92-bp pyrimidine track (see Fig. S6 in the supplemental material). The first nucleotide U of gRNA is indicated in black, as L. donovani rRNAP initiates transcription at the T-residue site. The schematic is not drawn to scale. (B) The partial sequence of pSPneogRNAH (HindIII and BamHI fragment). The 180-bp rRNAP sequence (black) and its transcription initiation site T (bold) are underlined. The 82-bp Cas9 binding RNA coding sequence (green), the 68-bp HDV ribozyme coding sequence (blue), and the HindIII, BbsI, and BamHI restriction enzyme sites (red) are indicated. Download
L. donovani gRNA expression vector pSPneogRNA partial sequence (HindIII and BamHI fragment). The 180-bp rRNAP sequence (black) and its transcription initiation site T (bold) are underlined. The 82-bp Cas9 binding RNA coding sequence (green), the 262-bp nonspecific DNA sequence derived from the pX330 plasmid (black), and the HindIII, BbsI, and BamHI restriction enzyme sites are highlighted in red. Download
L. donovani miltefosine transporter gene (LdMT; LdBPK_131590.1) sequence and the sequences of gRNA guides, oligonucleotide donors, and primers used to generate and detect LdMT mutants. The locations and directions of gRNA guides and primers in the LdMT gene are indicated and underlined with an arrow. Some of these primers had a restriction enzyme site added to its 5′ end to facilitate cloning of the PCR product. Download
Partial amino acid sequence alignments of phospholipid-transporting ATPase 1-like proteins (miltefosine transporters). The methionine (M) at position 381 is highly conserved in all phospholipid-transporting ATPase 1-like proteins aligned. Like M, the only other substitute amino acid found at this position (isoleucine [I]) is hydrophobic. LdMT, L. donovani miltefosine transporter (LdBPK_131590.1); XP 001681897, L. major miltefosine transporter (MT); AHM92900, Leishmania amazonensis MT; XP 003873354, Leishmania mexicana MT; XP 001563280, Leishmania braziliensis MT; XP 010697220, Leishmania panamensis MT. Download
LdBPK_241510.1 gene partial sequence and the sequences of gRNA guide and primers used to generate and detect LdBPK_241510.1 mutants. Download
Bleomycin resistance gene expression cassette sequence and primers used to generate the LdBPK_241510.1 gene disruption PCR product donor. The 92-bp pyrimidine track upstream of the bleomycin resistance gene is underlined. Download
LdBPK_366580.1 gene (target of rapamycin [TOR] kinase 1) partial sequence and the sequences of the gRNA guide and primers used to generate and detect LdBPK_366580.1 mutants. Download
EGFP gene sequence and primers used to generate the LdMT-GFP fusion PCR product donor. Download
L. donovani gRNA expression vector pSPneoHHgRNAH and its partial sequence. (A) Schematic of pSPneoHHgRNAH and its guide sequence insertion site. The L. donovani rRNA promoter (rRNAP), Hammerhead ribozyme (HH), and HDV ribozyme (H) are indicated. The small solid black box represents the 92-bp pyrimidine track (see Fig. S6 in the supplemental material). (B) The partial sequence of pSPneoHHgRNAH (HindIII and BamHI fragment). The 183-bp rRNAP sequence (black) and its transcription initiation site T (bold) are underlined. The 82-bp Cas9 binding RNA coding sequence (green), the 49-bp Hammerhead ribozyme (blue and underlined), the 68-bp HDV ribozyme sequence (blue), and the HindIII, BglII, BbsI, and BamHI restriction enzyme sites (red) are indicated. Download
Primers used to generate pLPHygCas9 and gRNA expression vectors. Download