ABSTRACT
A major challenge for the production of ethanol from biomass-derived feedstocks is to develop yeasts that can sustain growth under the variety of inhibitory conditions present in the production process, e.g., high osmolality, high ethanol titers, and/or elevated temperatures (≥40°C). Using adaptive laboratory evolution, we previously isolated seven Saccharomyces cerevisiae strains with improved growth at 40°C. Here, we show that genetic adaptations to high temperature caused a growth trade-off at ancestral temperatures, reduced cellular functions, and improved tolerance of other stresses. Thermotolerant yeast strains showed horizontal displacement of their thermal reaction norms to higher temperatures. Hence, their optimal and maximum growth temperatures increased by about 3°C, whereas they showed a growth trade-off at temperatures below 34°C. Computational analysis of the physical properties of proteins showed that the lethal temperature for yeast is around 49°C, as a large fraction of the yeast proteins denature above this temperature. Our analysis also indicated that the number of functions involved in controlling the growth rate decreased in the thermotolerant strains compared with the number in the ancestral strain. The latter is an advantageous attribute for acquiring thermotolerance and correlates with the reduction of yeast functions associated with loss of respiration capacity. This trait caused glycerol overproduction that was associated with the growth trade-off at ancestral temperatures. In combination with altered sterol composition of cellular membranes, glycerol overproduction was also associated with yeast osmotolerance and improved tolerance of high concentrations of glucose and ethanol. Our study shows that thermal adaptation of yeast is suitable for improving yeast resistance to inhibitory conditions found in industrial ethanol production processes.
IMPORTANCE
Yeast thermotolerance can significantly reduce the production costs of biomass conversion to ethanol. However, little information is available about the underlying genetic changes and physiological functions required for yeast thermotolerance. We recently revealed the genetic changes of thermotolerance in thermotolerant yeast strains (TTSs) generated through adaptive laboratory evolution. Here, we examined these TTSs’ physiology and computed their proteome stability over the entire thermal niche, as well as their preadaptation to other stresses. Using this approach, we showed that TTSs exhibited evolutionary trade-offs in the ancestral thermal niche, as well as reduced numbers of growth functions and preadaptation to other stresses found in ethanol production processes. This information will be useful for rational engineering of yeast thermotolerance for the production of biofuels and chemicals.
INTRODUCTION
In the production of ethanol from cellulosic-biomass feedstock, the most significant economic obstacles for the overall process are the enzyme costs and loss of productivity due to inactivation of yeast cells caused by a variety of inhibitory conditions associated with production processes (1, 2). Most of the large-scale production of fuel ethanol is typically accomplished in large-scale yeast fermentations operated at high glucose concentrations, elevated ethanol titers, and high temperatures, all conditions that cause stress for the yeast cells (3–5). Operation at high temperatures reduces cooling costs and contamination. Furthermore, for conversion of cellulosic biomass to ethanol, thermophilic enzymes and yeasts are often added at the same time to ensure simultaneous saccharification and fermentation (SFF). To reduce enzyme costs, it is therefore important to operate at high temperatures (5–7). Although temperatures between 38°C and 42°C allow significant reductions of production costs (3, 5), temperatures over 50°C would be more desirable (8). However, temperatures of 34°C or more greatly affect yeast metabolism and impair its growth.
Innate thermotolerance of yeast to heat, 40°C to 50°C for instance, can be stimulated by preadaptation with a variety of stressors, including high osmolality, high ethanol concentrations, and elevated nonlethal temperatures (e.g., 37°C) (9, 10). This capability, however, involves many cellular responses, including the overexpression of heat shock genes and low cyclic AMP-protein kinase (PK) activity, that are associated with reduced rates of growth and glucose consumption (9–11), and therefore, it does not represent a suitable option for biofuel production. So far, the thermotolerant yeast strains used in large-scale ethanol production have been isolated from industrial processes where they were exposed to high temperatures for long periods of time (3, 12). These strains grow and consume glucose faster than yeast cells adapted by induction of stress responses. There is, however, little basic knowledge of the genomic, biochemical, and physiological responses of these strains to the higher temperatures. For example, little information is available about the genetic changes and how these affect yeast adaptation across the ancestral range of temperatures (13). In addition, adaptation to heat can condition yeast responses to other stressors (14, 15).
Recently, we used adaptive laboratory evolution to select seven thermotolerant yeast strains (TTSs). These were able to grow, consume glucose, and produce ethanol about 1.92, 1.50, and 1.60 times faster, respectively, than the parental strain that underwent stress-based adaption at 40°C (16). Extensive characterization of the TTSs showed that, despite the accumulation of many structural changes in their genomes, nonsense mutations in the ERG3 gene appeared in all of the strains. These mutations disabled the capacity of yeast strains to synthesize ergosterol, and reverse engineering of the parental strain to insert these mutations resulted in 80% recuperation of the thermotolerant phenotype that involved replacement of ergosterol with fecosterol in the cellular membranes. Nonsense and other deleterious mutations also appeared in genes coding for proteins involved in the electron transport chain. Hence, the TTSs showed trade-offs in respiration and the metabolism of nonfermentative carbon sources.
Here, we evaluated whether the TTSs would show a growth trade-off in the ancestral thermal niche (13, 17) by studying them in the whole range of temperatures at which yeast can replicate. Hence, we cultivated the TTSs and the wild-type (WT) strain at temperatures between 15°C and 50°C to identify the temperatures at which the different strains can grow. With this experimental information and results from computational studies of the thermal stability of the yeast proteome at different temperatures, we inferred fitness (growth rate) and thermodynamic properties of growth for both the WT and the TTSs (18, 19). Additionally, we also evaluated whether changes in membrane composition and structure induced by the altered sterol composition affected cellular tolerance of stressful agents acting on the membrane (20–22), such as high osmolarity and elevated ethanol concentrations.
RESULTS
We earlier grew three clonal populations of the yeast strain CEN-PK113-7D at 39.5 ± 0.3°C for 90 days, obtaining over 300 generations (16). Three TTSs were randomly isolated from each population (hence, there were nine strains in total). Seven of these nine strains (TTS11, TTS12, TTS13, TTS21, TTS22, TTS31, and TTS33) grew at twice the rate of the WT at 40 ± 0.1°C and, thus, were sent for whole-genome sequencing. The genome sequences of TTS11, TTS12, and TTS13 were identical, so we selected TTS11, TTS21, TTS22, TTS31, and TTS33 for the evaluation of growth trade-off and preadaptation to other stresses.
Trade-offs and thermal niche.
The WT and each of the five TTSs selected for further analysis were cultivated for at least 15 generations at the targeted temperature to confirm that the strains could maintain replicative growth at the target temperature. Following this, the specific growth rate at the target temperature was calculated. Figure 1A shows the average specific growth rates from triplicate cultivations of the five TTSs and a triplicate cultivation of the WT. A characteristic asymmetric growth rate was observed for both strains over the range of temperatures assayed. Compared with the thermal niche of the WT, the thermal niche of the TTSs was slightly displaced to higher temperatures. Thus, the optimal growth temperature was displaced from the ancestral value of ~31°C to the new value of ~34°C. In TTSs, the growth rate at temperatures higher than the ancestral optimal increased, whereas significant trade-off in the growth rate occurred at the ancestral temperatures below 34°C (Fig. 1B). At 42°C, the TTSs were able to replicate, while the WT was not (Fig. 1C). At 50°C, only the TTSs could consume glucose and produce ethanol, but neither the TTSs nor the WT were able to grow (Fig. 1D). Remarkably, the viability of the TTSs decayed faster than that of the WT at this temperature. If a trade-off exists between maximal performance at high temperatures and extent of performance at ancestral temperatures, such a trade-off may reflect structural constraints resulting from the compromise between the flexibility and stability of proteins.
FIG 1 .
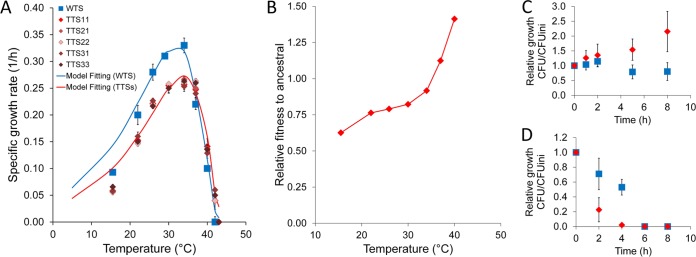
Effect of temperature on yeast growth. (A) Specific growth rates of thermotolerant and wild-type yeast strains at a temperature range between 15°C and 50°C. The line-fitting specific growth rate is the model using equation 1. (B) Fitness of thermotolerant yeast strains relative to that of the ancestral strain at a range of temperatures from 15°C to 42°C. (C and D) Relative growth for each strain type measured as the ratio of the CFU count at each time point and the initial CFU count at 42°C (C) and at 50°C (D). Blue symbols represent values for wild-type strains, and red symbols for thermotolerant strains. Error bars show standard deviations.
Yeast protein stability.
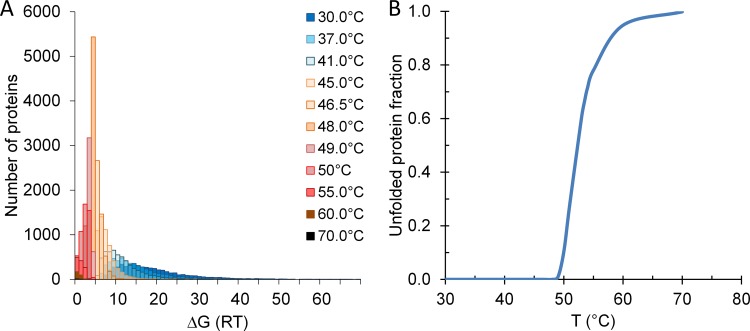
To gain insight into the effects of changes in genomic structure on TTS fitness in the ancestral thermal niche, we computed the thermal stability of proteins from the entire yeast genome. To carry out their functions, cellular proteins require a native state which is marginally more stable than the vast number of unfolded states called denatured states (typically 2 to 10 kcal/mol under physiological conditions) (23). The equilibrium between native and denatured states, defined as conformational stability, can be expressed as the change in Gibbs free energy (G) of native and denatured states (ΔG = GN − GD), and hence, a protein loses stability when ΔG = 0 (19). ΔG can be calculated from information on the length of a protein and its thermodynamic properties, which can be determined by the amino acid sequence (24–26). Assuming that loss of stability of any essential protein confers a lethal phenotype, a model for calculating growth rate based on the number of folded proteins required for replication was established (19, 27), as follows:
| (1) |
where r0 is an intrinsic rate, ΔH+ is the Arrhenius activation barrier for growth, the product term represents the stabilities of rate-determining proteins (i = 1, 2, 3, … and Γ) for growth at temperature T (26), with Γ being a fitted parameter, R is the ideal gas constant, and ΔG is the folding free energy. The distribution of ΔG values of the entire yeast proteome at temperatures between 30°C and 70°C is shown in Fig. 2A. The TTSs did not accumulate mutations that resulted in significant changes in the average proteome sequence and, hence, the ΔG distribution shown in Fig. 2A represents the proteome stability for both the TTSs and the WT strain. At the optimum growth temperature (30°C), the stability of the proteins is marginal since the average folding free energy of the yeast proteome is 8.5 to 9.1 kcal/mol from thermal denaturation. Also, it can be seen that the distribution of the proteome’s stability is very broad at 30°C, 37°C, and 41°C. Beyond this range, this is continuously narrowed down and moving left toward inactivation. At 48°C, most of the proteins are at the limit of instability, with an average ΔG of 2.55 kcal/mol. The model predicts that protein denaturation starts at 49°C, when 0.86% of proteins may get unfolded (including 21 essential proteins) (Fig. 2B; see also Table S1 in the supplemental material). Above this temperature, protein denaturation is very fast, as 10.4% of proteins may lose stability at 50°C, 27.9% at 51°C, and 60.5% at 53°C. Hence, the lethal temperature for TT and WT Saccharomyces cerevisiae strains is around 49°C, which is in agreement with experimental results from this and previous studies (28).
FIG 2 .
Thermal stability of yeast proteome. (A) Distribution of folding free energies (ΔG) of yeast proteome at different temperatures. The ΔG bin size unit is one RT [ΔG/(0.0083145 * T°K)]. (B) The fractions of unfolded protein at temperatures from 30°C and 70°C are represented.
With the data described above, we estimated that the ΔH+ values for the WT and TT strains are similar (52.6 and 53.1 kJ/mol), whereas the number of rate-determining proteins for growth (Γ) decreased from 215 to 144. Using these numbers in equation 1, we were able to fit the specific growth rate profiles for the two strains to the experimental data (lines in Fig. 1A). Because the fitness acquired during adaptive laboratory evolution for growth at higher temperatures did not involve any major changes in the primary structure of the proteome, the exponential changes of specific growth rates at temperatures below the optimum follow similar Arrhenius curves for the TTSs and the WT, which are seen as comparable values of ΔH+. Higher growth rates at temperatures above the optimum for the TTSs are due to lower Γ values, which suggests that these strains reduced the number of growth rate-determining gene functions to diminish complexity. An organism with a small number of growth rate-determining gene functions is less vulnerable to protein stability requirements, and hence, organisms living at high temperatures should have lower Γ values (19, 27).
The horizontal shift of the thermal niche in the TTSs was associated with the change in the optimal growth temperature, which was predicted to be ~34.9°C, compared with 30.8°C for the WT. The results from simulations using equation 1 also predicted that the maximum growth temperatures are 42°C for the parental strain and 45°C for the TTSs (Fig. 1A). These values are close to the experimental results (39.5°C and 42°C, respectively). Altogether, these results suggest that accumulations of nonsense mutations in TTSs lead to less complexity in terms of functions. For instance, mutations in genes associated with respiration resulted in a reduction of mitochondrial functions, including the electron transport chain, the tricarboxylic acid (TCA) cycle, and redox balance networks (16).
Yeast physiology in the thermal niche.
The TTSs consumed glucose and produced glycerol and ethanol faster than the WT throughout the entire thermal niche (see Fig. S1A, B, and C in the supplemental material), regardless of the fact that the TTSs grew more slowly than the WT at temperatures between 15°C and 34°C (Fig. 1A). In both the TTSs and the WT, the yields of biomass and glycerol from glucose were nearly constant at cold temperatures from 15°C to 30°C and changed abruptly at warm temperatures between 34°C and 42°C (see Fig. S1D and E), whereas the ethanol yields remained nearly constant throughout the entire thermal niche (see Fig. S1F). The TTSs, however, produced around three times more glycerol than the WT in the range of cold temperatures, which provoked a 35% reduction in the biomass yield. At 42°C, both the TTSs and the WT consumed glucose, but only the TTSs were able to grow (Fig. 1C; see Fig. S1). Remarkably, the TTSs did not grow at 50°C, but they consumed glucose nevertheless.
In all strains, the specific rates of glucose consumption (rGlc), ethanol production (rEtOH) and glycerol production (rGly) clearly showed different trends at cold temperatures between 15°C and 30°C and at warm temperatures between 37°C and 42°C (see Fig. S2 in the supplemental material). At similar specific growth rates, TTSs growing in the range of cold temperatures consumed glucose at a higher rate (1.6 mmol/g cellular dry weight [CDW]/h) and produced ethanol at a higher rate (3.73 mmol/g CDW/h) than the wild-type strain (see Fig. S2A and B). In the range of warm temperatures, both the TTSs and the WT consumed glucose faster and produced ethanol faster than the wild-type strain growing at cold temperatures. The thermotolerant strains, however, sustained higher specific growth rates at high temperatures and could therefore consume glucose and produce ethanol faster than the wild-type strain at these temperatures. Compared with yeast growing under anaerobic conditions at 30°C in chemostats (29), both the TTSs and the WT growing at high temperatures consumed glucose and produced ethanol faster (3.9 and 2.78 mmol/g CDW/h more, respectively).
Even though high temperatures increased ethanol production, they also provoked high rates of glycerol production in both the WT and TT strains (see Fig. S2C in the supplemental material). Glycerol production increased by 1.08 mmol/g CDW/h in the TTSs growing at high temperatures compared with its production during growth at low temperatures. The specific glycerol production rates of TTSs at high temperatures were higher than for the WT growing at low temperatures (2.91 mmol/g CDW/h) or for the WT growing under anaerobic conditions at 30°C (2.6 mmol/g CDW/h). The glycerol production in the WT cultivated at high temperatures was also higher than the production of the same strain growing under anaerobic conditions at 30°C, suggesting that other mechanisms besides anaerobiosis are implicated in glycerol overproduction. For instance, cell wall integrity stress responses can also activate the hyperosmotic stress response (21).
Respiration deficiency and glycerol overproduction in TTSs.
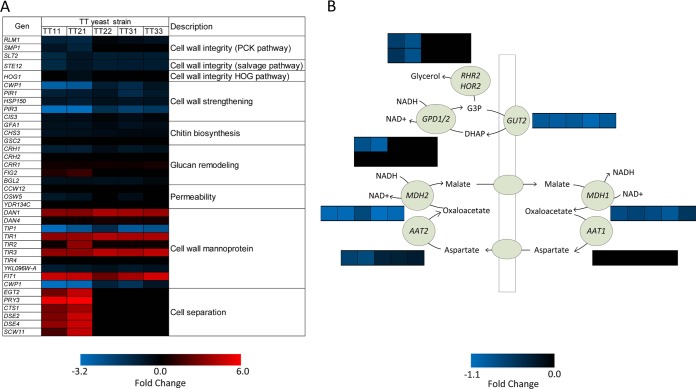
Increased glycerol production can be triggered by any of the pathways for cell wall integrity stress responses, namely, the protein kinase C (PKC)-mediated mitogen-activated protein (MAP) kinase pathway, the sterile vegetative growth (SVG) pathway, and the high-osmolarity glycerol response (HOG) pathway (21, 30). Based on genome-wide transcriptome analysis using Affymetrix microarrays, which have shown good consistency with quantitative reverse transcription-PCR (31), these pathways were found to be slightly down-regulated in the TTSs, which have a membrane structure that is more tolerant of high temperatures, due to the replacement of ergosterol with fecosterol (16). SMP1 and RLM1, encoding transcription factors (TFs) that regulate the PKC-MAP pathway, and their activator kinase, encoded by SLT2, were slightly down-regulated in the TTSs compared with their transcription levels in the wild type when both strain types were cultured at 40°C, as was the STE12-encoded TF that is involved in the SVG pathway. In addition, the expression of the protein kinase gene HOG1 that is involved in the signal transduction cascade of the HOG pathway did not change (Fig. 3A). These TFs and kinases can activate glycerol accumulation in different ways (32, 33), for example, via HAP1 (34), which is a possible target of HOG1 (35). Furthermore, the down-regulation of genes encoding proteins associated with cell wall strengthening/permeability regulated by the cell wall integrity pathway (CWP1, HSP150, PIR1, and PIR3) (36), chitin biosynthesis induced by the PKC-MAP and SVG pathways (GFA1 and CHS3) (35, 37), and glucan remodeling (CRH1 and BGL2) induced by RLM1 and STE12 (35) was also observed. Some of the TTSs (TT11 and TT21) carried a duplication of a segment in chromosome III that resulted in gene duplication of the MAP kinase kinase SSK22 of the HOG pathway, and these strains showed lower expression of some of these genes than the TTSs without such a duplication. However, genes associated with cell separation processes were only up-regulated in the TTSs carrying this duplication. Transcription of the cell wall mannoprotein genes DAN1, TIR1, TIR3, and FIT1, however, increased more than 4 times in all the TTSs, and increased expression of the first 3 of these mannoproteins has been observed previously under anaerobic growth (38). Overall, these results suggest that the increased glycerol production in the TTSs was not stimulated by a cell wall integrity response. This is consistent with findings that the genes associated with glycerol production are down-regulated (Fig. 3B).
FIG 3 .
Transcriptional responses of thermotolerant and wild-type yeast strains growing at 40°C. (A) Changes in transcription of genes associated with stress responses related to cell wall integrity. (B) Changes in gene expression associated with glycerol synthesis and metabolism.
Redox balancing in the cytosol of the TTSs was compromised due to the inactivation of respiration. Thus, genes involved in glycerol and aspartate-malate shuttles, responsible for operating proton transport across the mitochondrial membrane, were down-regulated (Fig. 3B). Therefore, the conversion of dihydroxyacetone phosphate (DHAP) to glycerol-3-phosphate (G3P) by glycerol-3-phosphate dehydrogenases 1 and 2 (GPD1/2) became the sole route for preventing NADH accumulation in the cytosol. Remarkably, glycerol accumulation in TTS11 and TTS21 occurred despite down-regulation of the enzymes involved in its production, which suggests that the increased flux to glycerol is solely kinetically determined by the accumulation of NADH and G3P.
Ethanol production under different stress conditions.
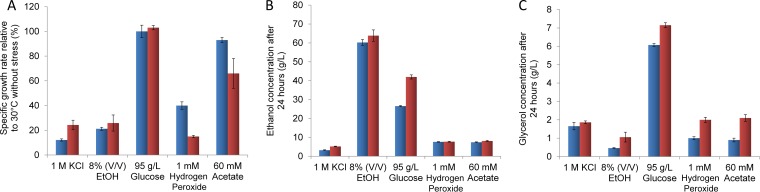
We also evaluated growth and the production of glycerol and ethanol in fermentations containing one of several stressors found in ethanol production from cellulose hydrolysates, i.e., 95 g/liter glucose, 8% (vol/vol) ethanol, 1 M KCl, and 60 mM acetic acid, as well as in a fermentation containing 1 mM hydrogen peroxide to explore the TTSs’ tolerance of reactive oxygen species (ROS) (Fig. 4). All of the stress-causing conditions except the high glucose concentration negatively impacted the specific growth rates of both the TTSs and the WT (Fig. 4A). However, TTSs grew 2.0 and 1.2 times faster than the WT under conditions of osmotic and ethanol stress. When exposed to acetic acid and oxygen peroxide, TTSs showed less tolerance than the ancestral strain, with the greater decrease in the growth rate at 1 mM of H2O2. Ethanol was accumulated to similar levels in both strains under each condition except for cultivations with 1 mM KCl and 95 g/liter glucose, where it was accumulated to a level 1.6 times higher than in the WT (Fig. 4B). Glycerol accumulated at higher levels in TTSs cultivated under every condition except for fermentations with 1 M KCl, where both types of strains produced similar amounts (Fig. 4C). After 24 h of fermentation, TTSs growing at 95 g/liter glucose completely consumed the glucose and accumulated 42 g/liter ethanol, while in cultivations of the WT, 21 g/liter glucose remained after 24 h and 26.6 g/liter ethanol was produced. Hence, the ethanol productivity of the TTSs was 1.75 g/liter/h, and that of the WT was 1.1 g/liter/h. Using these values, we used Kumar’s flowchart (7) to compare the costs of ethanol production with an industrial process using 2,000 tons (dry basis) of lignocellulose per day and a 24-h fermentation. Under these conditions, ethanol production costs may decrease by $0.50 per gallon by using the thermotolerant yeast strains.
FIG 4 .
Fitness relative to that under optimal conditions (30°C without stress) (A) and production of ethanol (B) and glycerol (C) in TT (red bars) and WT (blue bars) yeast strains cultivated with 1 M KCl, 8% (vol/vol) ethanol, 95 g/liter glucose, 60 mM acetic acid, or 1 mM hydrogen peroxide. Error bars show standard deviations.
DISCUSSION
Implications for thermal norms.
Adaptation to heat by evolution imposes changes in shape and in the limits of thermal reaction norms (39). We earlier used adaptive laboratory evolution (ALE) to isolate seven thermotolerant yeast strains (TTSs) with substantial improvements in fitness at up to 39.5°C, measured as improvement in the specific growth rate (16). Here, we show that these TTSs had a higher optimal growth temperature than the ancestral wild-type (WT) strain and displaced the upper limit of the ancestral thermal norm from 40°C to ~42°C (Fig. 1A). The fitness of the TTSs relative to that of the ancestral strain was considerably increased at temperatures higher than 37°C, whereas it was decreased at temperatures below ~34°C (Fig. 1B). This trade-off in relative fitness at ancestral temperatures was not observed in populations of Escherichia coli evolved at 42°C (40), but it was observed in E. coli populations evolved at 45°C and 48°C (41). Changes in thermal norms have also been observed during evolution of wasps, copepods, and flies at high nonlethal temperatures (13, 39, 41–45). A remarkable early study on the evolution of thermotolerance was reported by William Dallinger (1839 to 1909), who found that gradually increasing the temperature of flagellates (infusoria) in cultivation resulted in populations able to survive at 70°C (41). Dallinger’s report included a perceptive quote from Darwin that stated “the fact which you mention about their being adapted to certain temperatures, but becoming gradually accustomed to much higher ones, is very remarkable. It explains the existence of algae in hot springs.”
Implications for proteome stability and cellular complexity.
The shapes of thermal reaction norms and growth rates (fitness) can be inferred from the folding free energy (ΔG) of proteins, with the assumption that loss of stability by any essential protein confers a lethal phenotype on the organism (18, 24, 26, 27). Computational analysis showed that at 30°C, most yeast proteins are marginally stable, since the average ΔG of the proteome is about 9 kcal/mol from denaturation (Fig. 2A). The average proteome ΔG decreased rapidly with increasing temperatures, which was associated with a sharp decrease of the specific growth rate (Fig. 1A and 2A). Thermal instability of the proteome starts at 49°C, indicating that this is the lethal temperature for yeast, which was confirmed by experimental data. The influence of temperature on the specific growth rate can also be calculated using equation 1 (27), and with this approach, we found that the optimal growth rate of TTSs was about 3°C higher in the TTSs than for the WT and that the estimated limit for TTSs to sustain growth was 44°C, which is close to the observed experimental value of 42°C. Remarkably, these changes were associated with a decrease in the number of rate-determining proteins for growth (parameter Γ in equation 1), since the ΔH+ values were similar for the TTSs and the WT. Reduction of the cellular complexity by decreasing the number of growth-related cellular functions has been pointed out as a critical feature for organisms to acquire thermotolerance (19). Organisms with fewer functions may have less pressure for keeping a large number of enzymes functional, and hence, the number of growth-related functions encoded in the genome decreases (19, 25, 27). In the TTSs, loss of respiration capacity decreased the cellular functions associated with this trait, including the stress response associated with ROS. These losses are associated with lower fitness at ancestral temperatures and inadequate responses upon exposure to ROS.
Implications for changes in underlying protein functions
The stability and structural flexibility of growth-related proteins can be altered by changes in amino acid residues and by the accumulation of stabilizing solutes (e.g., trehalose and glycerol) (46, 47). The replacement of lysine/glycine with arginine and of serine with alanine or threonine, among a few other amino acid substitutions, has been shown to be useful for increasing the thermal stability of proteins (48, 49). In the TTSs, we only found one replacement of glycine by arginine, in the product of the MTM1 gene, and various replacements of serine by tyrosine, asparagine, or proline, as well as of proline by serine (16). Among the instances of replacement of serine by asparagine, one resulted in instability of the product of LCB3, which is involved in sphingolipid biosynthesis. The TTSs accumulated other deleterious mutations, including novel stop codons in the products of ERG3, ATP2, and ATP3, which accounted for 45% of the total number of mutations. Mutations in ERG3 substantially changed the sterol composition in the membrane and were responsible for most of the thermotolerance. On the other hand, loss of respiration caused by mutations in ATP2 or ATP3 predisposed cells to produce glycerol at rates three times higher than the WT. Since glycerol protects proteins from irreversible thermal denaturation (47), we hypothesize that the TTS yeast proteome is less prone to thermal denaturation. Loss of respiration, however, reduced the specific growth rates in the ancestral thermal niche. Mutations in genes encoding proteins with critical growth functions, for instance, cell replication, morphogenesis, host attachment, transcription, integrity of the cell wall membrane, and catabolism of carbon sources, have been reported for the evolution of thermal sensitivity (16, 45, 50). These functions depend on the organism: whereas bacteria and yeast accumulated mutations in genes related to catabolism of a carbon source (16, 50), phages accumulated mutations in genes for replication and host attachment (45). Gene mutations associated with the integrity of the cell wall and morphogenesis have been found in yeast and phages (16, 45). In E. coli, considerable increases in GroEL/GroES chaperone levels were shown to be essential for resistance at 48°C (41). In TT S. cerevisiae strains, neither the heat shock protein 60 (HSP60)/HSP10 complex nor the HSP104/HSP70/HSP40 complex, homologous to bacterial GroEL/GroES, were up-regulated compared with their transcription in the WT when grown at 40°C.
From our computational analysis of proteome stability, we found that proteins larger than the average protein size (in terms of number of amino acids) are more sensitive to heat. For instance, we predicted that essential proteins, including helicases (encoded by BRR2, SEN1, and SLH1), global catabolic regulators (CYR1, IRA1, and TOR2), components of fatty acid synthase (FAS1 and FAS2), and the catalytic subunits of 1,3-beta-d-glucan synthase (FKS1 and GSC2), are unstable at 49°C. IRA1 and TOR2, interestingly, were targets of the evolutionary process in TTSs, whereas helicases have been associated with thermal resistance in thermophiles (51). Our thermodynamic analysis indicated that all glycolytic enzymes are stable at temperatures below 53°C. However, only the TTSs and not the WT were able to consume glucose and produce ethanol and glycerol at 50°C. There is therefore the possibility that impaired glucose consumption is due to loss of glucose transport shortly after being exposed to 50°C.
The challenge of industrial biotechnology is to generate more robust cell factories that are able to survive inhibitory conditions while keeping the proper catalytic properties for conversion of raw materials to the target biochemicals or biofuels. A lack of basic information about the mechanisms underlying yeast adaptation is currently limiting the development of robust biocatalysts. Here, we show the genetic basis of thermal adaptation of TTSs and the related trade-offs, and we present insights on the preadaptation of TTSs to other stresses found in ethanol production processes. This information will be valuable for reverse engineering of robust yeast strains for the production of ethanol and other fuels and chemicals.
MATERIALS AND METHODS
Strains.
The thermotolerant yeast strains (TTSs) were previously isolated from evolutionary laboratory experiments (16). These experiments were carried out by serial dilution of three independent clonal populations of the parental yeast strain CEN-PK113-7D (WT) cultivated at 39.5 ± 0.3°C in minimal medium. This procedure was repeated until a significant change in specific growth rate was detected. This was performed for more than 320 generations, when we randomly selected three strains from each of the three populations. From these nine strains (TTS11, TTS12, TTS13, TTS21, TTS22, TTS23, TTS31, TTS32, and TTS33), seven were sent for genome sequencing (TTS11, TTS12, TTS13, TTS21, TTS22, TTS31, and TTS33). The strains from population one (TTS11, TTS12, and TTS13) showed identical genome sequences, and therefore, only one of them, TTS11, was chosen for this study, along with TTS21, TTS22, TTS31, and TTS33. The genotypes of these strains were recently reported (16).
Culture media.
We used minimal medium for cultivation of TTSs and the WT under all conditions. This contained 2% glucose and 5 g (NH3)2SO4, 3 g (NH4)2PO4, and 0.5 g MgSO4 per liter, in addition to 1 ml of trace element solution and 1 ml of vitamin solution. The trace element solution contained, per liter (pH 4), 15.0 g EDTA (sodium salt), 4.5 g ZnSO4·7H2O, 0.84 g MnCl2·2H2O, 0.3 g CoCl2·6H2O, 0.3 g CuSO4·5H2O, 0.4 g Na2MoO4·2H2O, 4.5 g CaCl2·2H2O, 3.0 g FeSO4·7H2O, 1.0 g H3BO3, and 0.10 g KI. The vitamin solution contained, per liter (pH 6.5), 0.05 g biotin, 0.2 g p-amino benzoic acid, 1 g nicotinic acid, 1 g Ca-pantothenate, 1 g pyridoxine-HCl, 1 g thiamine-HCl, and 25 g myo-inositol. The initial pH of the medium was 5.2.
Physiology in the thermal niche.
Cultivation of the TTSs and the WT was performed at temperatures from 15°C to 50°C to detect the range of temperatures at which both strains were able to replicate. The strains were first propagated at 30°C for around 15 generations. Then, they were cultivated at the target temperature for another 16 to 20 generations to eliminate the effects of innate adaptation and population dynamics of yeast adaptation to the target temperature. After that, aliquots of the cultures were transferred to fresh medium at an initial optical density of 0.15 at 600 nm (OD600). Samples were taken every one or two hours to measure the OD600, glucose, and fermentation metabolites. The yield from glucose and the specific rates of glucose consumption and metabolite production were calculated for at least 6 time points.
Tolerance of other stresses.
Both the TTSs and the WT were propagated at 30°C. Aliquots from cultivations in the mid-log phase were transferred to fresh minimal medium to adjust the OD600 to 0.1. We then let the cultivations grow until they reached an OD600 of 0.4 before applying the stressor. As stressors, we used 1 M KCl, 8% (vol/vol) ethanol, 95 g/liter glucose, or 60 mM acetic acid, which are representative of processing conditions for ethanol production. Furthermore, 1 mM of hydrogen peroxide was also used to check the ROS tolerance of TTSs. Samples were taken every hour to determinate the growth rate, glucose consumption, and ethanol and glycerol production. The results from these experiments were compared with the results from experiments without stressors at 30°C.
Biomass production, glucose uptake, and fermentation metabolites.
Samples from shake flasks were taken every 1 to 2 h and processed at 4°C for biomass, glucose, and fermentation metabolite analyses. Biomass was first measured indirectly by determining the optical density of the sample at 600 nm. Samples were also vacuum filtered, and the biomass pellets obtained were washed twice with isotonic solution, dried for 15 min in a microwave oven at medium power, and kept in a desiccator until they reached a constant weight. The supernatants were stored at −20°C for further analysis of fermentation metabolites by high-performance liquid chromatography. An Aminex HPX-87H column (Bio-Rad, CA) coupled to refractive index and photodiode array detectors was used to separate and quantify glucose and fermentation metabolites. The mobile phase was an 8 mM solution of H2SO4 at 0.5 ml/min, and the assay was run at 50°C. Pure glucose, organic acids, glycerol, and ethanol from Sigma Aldrich were used to construct the calibration curves used to quantify these compounds in the culture. Calculation of the yield coefficients, stoichiometries, and degrees of redox balance and consistency testing of experimental data were performed according to Villadsen et al. (52).
Thermal stability of the yeast proteome.
Thermodynamic analysis of proteins was carried out based on the entire yeast proteome. Extensive data on the reversible folding stabilities have shown that the dominant factor affecting the thermal properties of proteins is the chain length. These data were recently captured in a model that calculates the free energy of protein folding, ΔG, as a function of temperature (T) and chain length (L) (18, 19, 25), as follows:
| (2) |
Differences in entropy (ΔS), enthalpy (ΔH), and specific heat (ΔCp) between native and denatured states were calculated using average correlations obtained from data on the reversible folding stabilities of 63 ideal mesophilic proteins (25), which showed a good correlation between thermodynamic parameters and chain length.
| (3) |
| (4) |
| (5) |
The reference temperatures Th and Ts are the temperatures at which enthalpy and entropy from sequence and hydrophobic effects are near zero (18, 25).
From equations 2 to 5, we calculated the protein stability of the entire proteome at different temperatures from 30°C to 70°C. We also extended our analysis to calculate the stability distribution of the entire proteome, which followed a gamma distribution, as follows:
| (6) |
Using the proteome chain length distribution and equation 2, we calculated the free energy distribution P(ΔG) of the entire proteome to calculate the specific growth rate using equation 1.
Analysis of gene expression.
Analysis of gene expression was performed with experimental data generated in a previous work (16). Briefly, samples for transcriptome analysis were taken at mid-exponential phase from the WT and TTSs cultivated at 40°C in bioreactors under fully aerobic conditions. They were quickly put on ice and centrifuged at 3,000 × g for 5 min at 4°C. The supernatants were discarded, and biomass pellets were frozen in liquid nitrogen and then stored at −80°C. Total RNA was extracted from pellets using the RiboPure-yeast kit (Ambion, TX) with the help of RNase-free solvents (Ambion, TX). For transcriptome analysis, we used the Affymetrix GeneChip yeast genome 2.0 array. Bioconductor (53) was used to process the CEL files using R version 3.1.2. The Affymetrix chip description file (CDF file) was obtained from the microarray developers and imported to R using the Bioconductor package makecdfenv. Data normalization was performed with the probe logarithmic intensity error (PLIER) normalization method (54) using only perfectly matched probes. The moderated t statistic was applied to identify pairwise differences in gene expression between each of the evolved strains. For multiple-testing corrections of the P values, we used the Benjamini-Hochberg method (55). A cutoff of <0.001 for adjusted P values was used to determine differential expression of genes between each of the two conditions. The PIANO package was used for reporter gene ontology (GO) term and gene set enrichment analysis (56).
SUPPLEMENTAL MATERIAL
Physiological responses of thermotolerant (red diamonds) and wild-type S. cerevisiae (blue squares) strains cultivated at temperatures between 15°C and 50°C. Specific rates of glucose consumption (A), glycerol production (B), and ethanol production (C) and yields of biomass (D), glycerol (E), and ethanol (F) per unit of glucose consumed were calculated for exponential-phase cells growing at the targeted temperature. Download
Linear correlations of specific rates of glucose consumption (A), ethanol production (B), and glycerol production (C) with temperatures and specific growth rates at temperature ranges from 15°C to 30°C and from 37°C to 42°C. Equations for linear correlations and their R-squared values are shown below the corresponding plots. Blue squares, red diamonds, and gray circles denote results from experiments with the WT and the TTSs at different temperatures and with the WT at 30°C, respectively. Data for the WT cultivated anaerobically at 30°C were obtained from Verduyn et al. (29). Download
Protein unfolding at lethal temperatures between 49°C and 60°C.
ACKNOWLEDGMENTS
We thank Payam Ghiaci for assistance with the experimental work and Carmen Gutierrez for fruitful discussion about the thermodynamics of protein folding.
This work was financed by the Novo Nordisk Foundation, the Knut and Alice Wallenberg Foundation, and the the Europe Research Council (grant no. 247013).
Footnotes
Citation Caspeta L, Nielsen J. 2015. Thermotolerant yeast strains adapted by laboratory evolution show trade-off at ancestral temperatures and preadaptation to other stresses. mBio 6(4):e00431-15. doi:10.1128/mBio.00431-15.
REFERENCES
- 1.Caspeta L, Buijs NAA, Nielsen J. 2013. The role of biofuels in the future energy supply. Energy Environ Sci 6:1077–1082. doi: 10.1039/c3ee24403b. [DOI] [Google Scholar]
- 2.Lynd LR, Cushman JH, Nichols RJ, Wyman CE. 1991. Fuel ethanol from cellulosic biomass. Science 251:1318–1323. doi: 10.1126/science.251.4999.1318. [DOI] [PubMed] [Google Scholar]
- 3.Abreu-Cavalheiro A, Monteiro G. 2013. Solving ethanol production problems with genetically modified yeast strains. Braz J Microbiol 44:665–671. doi: 10.1590/S1517-83822013000300001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mussatto SI, Dragone G, Guimarães PMR, Silva JPA, Carneiro LM, Roberto IC, Vicente A, Domingues L, Teixeira JA. 2010. Technological trends, global market, and challenges of bio-ethanol production. Biotechnol Adv 28:817–830. doi: 10.1016/j.biotechadv.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 5.Abdel-Banat BM, Hoshida H, Ano A, Nonklang S, Akada R. 2010. High-temperature fermentation: how can processes for ethanol production at high temperatures become superior to the traditional process using mesophilic yeast? Appl Microbiol Biotechnol 85:861–867. doi: 10.1007/s00253-009-2248-5. [DOI] [PubMed] [Google Scholar]
- 6.Caspeta L, Caro-Bermúdez MA, Ponce-Noyola T, Martinez A. 2014. Enzymatic hydrolysis at high-solids loadings for the conversion of agave bagasse to fuel ethanol. Appl Energy 113:277–286. doi: 10.1016/j.apenergy.2013.07.036. [DOI] [Google Scholar]
- 7.Kumar D, Murthy GS. 2011. Impact of pretreatment and downstream processing technologies on economics and energy in cellulosic ethanol production. Biotechnol Biofuels 4:27. doi: 10.1186/1754-6834-4-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kate Y. 2014. Modified yeast tolerate alcohol, heat. The Scientist. http://www.the-Scientist.com/?articles.view/articleNo/41142/title/Modified-Yeast-Tolerate-Alcohol--Heat/.
- 9.Piper PW. 1993. Molecular events associated with acquisition of heat tolerance by the yeast Saccharomyces cerevisiae. FEMS Microbiol Rev 11:339–355. doi: 10.1111/j.1574-6976.1993.tb00005.x. [DOI] [PubMed] [Google Scholar]
- 10.Lindquist S. 1986. The heat-shock response. Annu Rev Biochem 55:1151–1191. doi: 10.1146/annurev.bi.55.070186.005443. [DOI] [PubMed] [Google Scholar]
- 11.Lu C, Brauer MJ, Botstein D. 2009. Slow growth induces heat-shock resistance in normal and respiratory-deficient yeast. Mol Biol Cell 20:891–903. doi: 10.1091/mbc.E08-08-0852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edgardo A, Carolina P, Manuel R, Juanita F, Baeza J. 2008. Selection of thermotolerant yeast strains Saccharomyces cerevisiae for bioethanol production. Enzyme Microb Technol 43:120–123. doi: 10.1016/j.enzmictec.2008.02.007. [DOI] [Google Scholar]
- 13.Bennett AF, Lenski RE. 1993. Evolutionary adaptation to temperature. II. Thermal niches of experimental lines of Escherichia coli. Evolution 47:1–12. doi: 10.2307/2410113. [DOI] [PubMed] [Google Scholar]
- 14.Mitchell A, Romano GH, Groisman B, Yona A, Dekel E, Kupiec M, Dahan O, Pilpel Y. 2009. Adaptive prediction of environmental changes by microorganisms. Nature 460:220–224. doi: 10.1038/nature08112. [DOI] [PubMed] [Google Scholar]
- 15.Cullum AJ, Bennett AF, Lenski RE. 2001. Evolutionary adaptation to temperature. IX. Preadaptation to novel stressful environments of Escherichia coli adapted to high temperature. Evolution 55:2194–2202. doi: 10.1111/j.0014-3820.2001.tb00735.x. [DOI] [PubMed] [Google Scholar]
- 16.Caspeta L, Chen Y, Ghiaci P, Feizi A, Buskov S, Hallström BM, Petranovic D, Nielsen J. 2014. Altered sterol composition renders yeast thermotolerant. Science 346:75–78. doi: 10.1126/science.1258137. [DOI] [PubMed] [Google Scholar]
- 17.Bennett AF, Lenski RE. 2007. An experimental test of evolutionary trade-offs during temperature adaptation. Proc Natl Acad Sci U S A 104(Suppl 1):8649–8654. doi: 10.1073/pnas.0702117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghosh K, Dill K. 2010. Cellular proteomes have broad distributions of protein stability. Biophys J 99:3996–4002. doi: 10.1016/j.bpj.2010.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeldovich KB, Chen P, Shakhnovich EI. 2007. Protein stability imposes limits on organism complexity and speed of molecular evolution. Proc Natl Acad Sci U S A 104:16152–16157. doi: 10.1073/pnas.0705366104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wojda I, Alonso-Monge R, Bebelman J-P, Mager WH, Siderius M. 2003. Response to high osmotic conditions and elevated temperature in Saccharomyces cerevisiae is controlled by intracellular glycerol and involves coordinate activity of MAP kinase pathways. Microbiology 149:1193–1204. doi: 10.1099/mic.0.26110-0. [DOI] [PubMed] [Google Scholar]
- 21.Alonso-Monge R, Real E, Wojda I, Bebelman JP, Mager WH, Siderius M. 2001. Hyperosmotic stress response and regulation of cell wall integrity in Saccharomyces cerevisiae share common functional aspects. Mol Microbiol 41:717–730. doi: 10.1046/j.1365-2958.2001.02549.x. [DOI] [PubMed] [Google Scholar]
- 22.Lam FH, Ghaderi A, Fink GR, Stephanopoulos G. 2014. Engineering alcohol tolerance in yeast. Science 346:71–75. doi: 10.1126/science.1257859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pace CN, Treviño S, Prabhakaran E, Scholtz JM. 2004. Protein structure, stability and solubility in water and other solvents. Philos Trans R Soc Lond B Biol Sci 359:1225–1235. doi: 10.1098/rstb.2004.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghosh K, Dill KA. 2009. Computing protein stabilities from their chain lengths. Proc Natl Acad Sci U S A 106:10649–10654. doi: 10.1073/pnas.0903995106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sawle L, Ghosh K. 2011. How do thermophilic proteins and proteomes withstand high temperature? Biophys J 101:217–227. doi: 10.1016/j.bpj.2011.05.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zeldovich KB, Berezovsky IN, Shakhnovich EI. 2007. Protein and DNA sequence determinants of thermophilic adaptation. PLoS Comput Biol 3:e5. doi: 10.1371/journal.pcbi.0030005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen P, Shakhnovich EI. 2010. Thermal adaptation of viruses and bacteria. Biophys J 98:1109–1118. doi: 10.1016/j.bpj.2009.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hottiger T, Schmutz P, Wiemken A. 1987. Heat-induced accumulation and futile cycling of trehalose in Saccharomyces cerevisiae. J Bacteriol 169:5518–5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Verduyn C, Postma E, Scheffers WA, van Dijken JP. 1990. Physiology of Saccharomyces cerevisiae in anaerobic glucose-limited chemostat cultures. J Gen Microbiol 136:395–403. doi: 10.1099/00221287-136-3-395. [DOI] [PubMed] [Google Scholar]
- 30.Siderius M, Rots E, Mager WH. 1997. High-osmolarity signalling in Saccharomyces cerevisiae is modulated in a carbon-source-dependent fashion. Microbiology 143:3241–3250. doi: 10.1099/00221287-143-10-3241. [DOI] [PubMed] [Google Scholar]
- 31.André B, Canelas AB, Harrison N, Fazio A, Zhang J, Pitkänen JP, van den Brink J, Bakker BM, Bogner L, Bouwman J, Castrillo JI, Cankorur A, Chumnanpuen P, Lapujade PD, Dikicioglu D, van Eunen K, Ewald JC, Heijnen JJ, Kirdar B, Mattila I, Mensonides FIC, Niebel A, Penttilä M, Pronk JT, Reuss M, Salusjärvi S, Sauer U, Sherman D, Siemann-Herzberg M, Westerhoff H, de Winde J, Petranovic D, Oliver SG, Workman ChT, Zamboni N, Nielsen J. 2010. Integrated multilaboratory systems biology reveals differences in protein metabolism between two reference yeast strains. Nat Commun 1:145. doi: 10.1038/ncomms1150. [DOI] [PubMed] [Google Scholar]
- 32.De Nadal E, Casadomé L, Posas F. 2003. Targeting the MEF2-like transcription factor Smp1 by the stress-activated Hog1 mitogen-activated protein kinase. Mol Cell Biol 23:229–237. doi: 10.1128/MCB.23.1.229-237.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hahn JS, Thiele DJ. 2002. Regulation of the Saccharomyces cerevisiae Slt2 kinase pathway by the stress-inducible Sdp1 dual specificity phosphatase. J Biol Chem 277:21278–21284. doi: 10.1074/jbc.M202557200. [DOI] [PubMed] [Google Scholar]
- 34.Ter Linde JJ, Steensma HY. 2002. A microarray-assisted screen for potential Hap1 and Rox1 target genes in Saccharomyces cerevisiae. Yeast 19:825–840. doi: 10.1002/yea.879. [DOI] [PubMed] [Google Scholar]
- 35.MacIsaac KD, Wang T, Gordon DB, Gifford DK, Stormo GD, Fraenkel E. 2006. An improved map of conserved regulatory sites for Saccharomyces cerevisiae. BMC Bioinformatics 7:113. doi: 10.1186/1471-2105-7-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jung US, Levin DE. 1999. Genome-wide analysis of gene expression regulated by the yeast cell wall integrity signalling pathway. Mol Microbiol 34:1049–1057. doi: 10.1046/j.1365-2958.1999.01667.x. [DOI] [PubMed] [Google Scholar]
- 37.Watzele G, Tanner W. 1989. Cloning of the glutamine:fructose-6-phosphate amidotransferase gene from yeast. Pheromonal regulation of its transcription. J Biol Chem 264:8753–8758. [PubMed] [Google Scholar]
- 38.Klis FM, Mol P, Hellingwerf K, Brul S. 2002. Dynamics of cell wall structure in Saccharomyces cerevisiae. FEMS Microbiol Rev 26:239–256. doi: 10.1111/j.1574-6976.2002.tb00613.x. [DOI] [PubMed] [Google Scholar]
- 39.Huey RB, Kingsolver JG. 1989. Evolution of thermal sensitivity of ectotherm performance. Trends Ecol Evol 4:131–135. doi: 10.1016/0169-5347(89)90211-5. [DOI] [PubMed] [Google Scholar]
- 40.Bennett AF, Lenski RE. 1993. Evolutionary adaptation to temperature. II. Thermal niches of experimental lines of Escherichia coli. Evolution 47:1–12. doi: 10.2307/2410113. [DOI] [PubMed] [Google Scholar]
- 41.Rudolph B, Gebendorfer KM, Buchner J, Winter J. 2010. Evolution of Escherichia coli for growth at high temperatures. J Biol Chem 285:19029–19034. doi: 10.1074/jbc.M110.103374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dallinger WH. 1887. The President’s address. J R Microsc Soc 7:185–199. doi: 10.1111/j.1365-2818.1887.tb01566.x. [DOI] [Google Scholar]
- 43.White EB, Debach P, Garber MJ. 1970. Artificial selection for genetic adaptation to temperature extremes in Aphytis lingnanensis Compere (Hymenoptera: Aphelinidae). Hilgardia 40:161–192. doi: 10.3733/hilg.v40n06p161. [DOI] [Google Scholar]
- 44.Bradley B. 1982. Models for physiological and genetic adaptation to variable environments, p 33–50. In Dingle H, Hegmann JP (ed), Evolution and genetics of life histories. Springer-Verlag, New York, NY. [Google Scholar]
- 45.Knies JL, Izem R, Supler KL, Kingsolver JG, Burch CL. 2006. The genetic basis of thermal reaction norm evolution in lab and natural phage populations. PLoS Biol 4:e201. doi: 10.1371/journal.pbio.0040201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fields PA. 2001. Review: protein function at thermal extremes: balancing stability and flexibility. Comp Biochem Physiol A Mol Integr Physiol 129:417–431. doi: 10.1016/S1095-6433(00)00359-7. [DOI] [PubMed] [Google Scholar]
- 47.Meng FG, Hong YK, He HW, Lyubarev AE, Kurganov BI, Yan YB, Zhou HM. 2004. Osmophobic effect of glycerol on irreversible thermal denaturation of rabbit creatine kinase. Biophys J 87:2247–2254. doi: 10.1529/biophysj.104.044784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vogt G, Woell S, Argos P. 1997. Protein thermal stability, hydrogen bonds, and ion pairs. J Mol Biol 269:631–643. doi: 10.1006/jmbi.1997.1042. [DOI] [PubMed] [Google Scholar]
- 49.Nishio Y, Nakamura Y, Kawarabayasi Y, Usuda Y, Kimura E, Sugimoto S, Matsui K, Yamagishi A, Kikuchi H, Ikeo K, Gojobori T. 2003. Comparative complete genome sequence analysis of the amino acid replacements responsible for the thermostability of Corynebacterium efficiens. Genome Res 13:1572–1579. doi: 10.1101/gr.1285603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chang RL, Andrews K, Kim D, Li Z, Godzik A, Palsson BO. 2013. Structural systems biology evaluation of metabolic thermotolerance in Escherichia coli. Science 340:1220–1223. doi: 10.1126/science.1234012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.López-García P. 1999. DNA supercoiling and temperature adaptation: a clue to early diversification of life? J Mol Evol 49:439–452. doi: 10.1007/PL00006567. [DOI] [PubMed] [Google Scholar]
- 52.Villadsen J, Nielsen J, Lidén G. 2011. Biochemical engineering principles, 3rd ed. Springer, New York, NY. [Google Scholar]
- 53.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Affymetrix 2005. Guide to probe logarithmic intensity error (plier) estimation. Technical report. Affymetrix, Inc., Santa Clara, CA. [Google Scholar]
- 55.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B Stat Methodol 57:289–300. [Google Scholar]
- 56.Väremo L, Nielsen J, Nookaew I. 2013. Enriching the gene set analysis of genome-wide data by incorporating directionality of gene expression and combining statistical hypotheses and methods. Nucleic Acids Res 41:4378–4391. doi: 10.1093/nar/gkt111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Physiological responses of thermotolerant (red diamonds) and wild-type S. cerevisiae (blue squares) strains cultivated at temperatures between 15°C and 50°C. Specific rates of glucose consumption (A), glycerol production (B), and ethanol production (C) and yields of biomass (D), glycerol (E), and ethanol (F) per unit of glucose consumed were calculated for exponential-phase cells growing at the targeted temperature. Download
Linear correlations of specific rates of glucose consumption (A), ethanol production (B), and glycerol production (C) with temperatures and specific growth rates at temperature ranges from 15°C to 30°C and from 37°C to 42°C. Equations for linear correlations and their R-squared values are shown below the corresponding plots. Blue squares, red diamonds, and gray circles denote results from experiments with the WT and the TTSs at different temperatures and with the WT at 30°C, respectively. Data for the WT cultivated anaerobically at 30°C were obtained from Verduyn et al. (29). Download
Protein unfolding at lethal temperatures between 49°C and 60°C.