Background: An understanding of the role of Nr4a2 in inflammation is needed.
Results: Nr4a2 is a transcription factor that induces expression of M2 characteristic genes, and adoptive transfer of macrophages overexpressing Nr4a2 gives protection against septic mortality.
Conclusion: Our data impart a new role for Nr4a2 in skewing macrophage plasticity to M2 type.
Significance: Therapeutic intervention of Nr4a2 may provide a cure for inflammatory diseases.
Keywords: gene transcription, inflammation, macrophage, nuclear receptor, sepsis, Arginase 1, M2 macrophages, Nr4a2
Abstract
The orphan nuclear receptor Nr4a2 is known to modulate both inflammatory and metabolic processes, but the mechanism by which it regulates innate inflammatory homeostasis has not been adequately addressed. This study shows that exposure to ligands for Toll-like receptors (TLRs) robustly induces Nr4a2 and that this induction is tightly regulated by the PI3K-Akt signaling axis. Interestingly, exogenous expression of Nr4a2 in macrophages leads to their alternative phenotype with induction of genes that are prototypical M2 markers. Moreover, Nr4a2 transcriptionally activates arginase 1 expression by directly binding to its promoter. Adoptive transfer experiments revealed that increased survival of animals in endotoxin-induced sepsis is Nr4a2-dependent. Thus our data identify a previously unknown role for Nr4a2 in the regulation of macrophage polarization.
Introduction
Acute inflammation provides the first line of defense against invading pathogens and is followed by events that remove damaged cells to restore tissue homeostasis. However, when inflammation goes awry, it leads to the development of a number of chronic inflammatory and autoimmune disorders including atherosclerosis, obesity, Parkinson disease, Alzheimer disease, and rheumatoid arthritis (1, 2). The early events that initiate inflammatory processes involve the ligation of pattern recognition molecules, such as Toll-like receptors (TLRs),2 with microbial motifs called pathogen-associated molecular patterns that have conserved molecular structures. This initial ligation results in the activation of multiple signaling events, which interdigitate to rapidly induce the expression of numerous genes that escalate the inflammatory process (3, 4). However, the same signaling processes also amplify genes that impede inflammation and ensure homeostasis. Major examples of this are the elevation in the levels of suppressor of cytokine signaling (SOCS) proteins that negatively regulate TLR signaling and induction of transcription factor aryl hydrocarbon that prevents exaggerated inflammation in response to TLR activation (5, 6). However, currently our knowledge of the genomic regulators that modulate and fine-tune inflammation, thereby contributing to homeostasis, is not adequately understood.
Macrophages are central to inflammatory processes and are involved in the initiation, perpetuation, and resolution of inflammation (7). They show considerable plasticity that permits them to adjust their phenotype and physiology in response to environmental cues. These adjustments lead to the emergence of different macrophage populations having the ability to perform different homeostatic functions (8). This functional heterogeneity is vital to inflammation and its resolution to ensure host survival. The two populations of macrophages that are at either end of this functional continuum have been established and are designated as classically activated macrophages, M1, and alternatively activated macrophages, M2. The M1 macrophages are mainly involved in initiating and perpetuating inflammation and are associated with secretion of high levels of pro-inflammatory cytokines and mediators, maintenance of Th1 response, and strong microbicidal activity. On the other hand, M2 macrophages are active in the resolution phase of inflammation, where their function is to secrete high levels of anti-inflammatory cytokines, support Th2 response, and initiate the arginase pathway for the synthesis of collagen and polyamines to promote tissue repair and wound healing (8, 9). Multiple signaling pathways and associated transcription factors contribute to these two activation states of macrophages. The signaling molecules IRF5 and STAT1 direct the macrophage phenotype toward M1 and are involved in the up-regulation of NF-κB- and AP-1-targeted gene expression (10, 11). Alternatively, signaling molecules IRF4 and STAT6 skew macrophages to the M2 phenotype that involves KLF4 and nuclear receptors PPARγ and PPARδ to control the expression of M2-associated genes (10, 12, 13).
Macrophages also express Nr4a members of the nuclear receptor family (14). The Nr4a subfamily of orphan nuclear receptors comprises three closely related members, Nr4a1 (Nur77), Nr4a2 (Nurr1), and Nr4a3 (NOR-1). Despite being a part of the nuclear receptor superfamily, these three receptors have no known endogenous ligands and are known as receptors that function in a ligand-independent manner. Structural and functional studies have revealed the presence of side chains of bulky hydrophobic amino acid residues in the ligand-binding pocket of these receptors, which is a plausible explanation as to why these receptors fail to bind ligand and lack any ligand-mediated regulation (15). However, this subfamily has evolved to be modulated by a bipartite regulation: first at the transcriptional level and second by specific post-translational modifications. Among Nr4a family members, Nr4a2 is rapidly and strongly induced by oxidized lipids, cytokines, and pathogen-associated molecular patterns, such as LPS (14, 16, 17). Abnormal expression of Nr4a2 has been correlated with many inflammatory disorders including multiple sclerosis, Parkinson disease, atherosclerosis, psoriasis, inflamed joints, and osteoarthritis (18–20). Silencing Nr4a2 in the substantia nigra of mice magnifies inflammatory responses to LPS that cause neurotoxicity, whereas mutations in the human Nr4a2 gene that result in loss of functional protein are correlated with late onset of Parkinson disease (17, 21). Nr4a2 also induces Foxp3 expression in naive CD4+ T cells and sustains the suppressive function of T regs while repressing IFNγ secretion from Th1 cells (22). This suggests an anti-inflammatory function for Nr4a2; however, many pro-inflammatory roles for this receptor have also been reported (23–25).
As a transcription factor, Na4a2 recognizes the short DNA sequences known as response elements as monomer, homodimer, or heterodimer, and its binding to these cis-elements in the promoter of the target genes subsequently activates their expression. The consensus sequence utilized by Nr4a2 to bind DNA as a monomer involves a core motif (AGGTCA) and a preceding AT-rich region, although it can also bind to DNA as a homodimer (26, 27). Nr4a2 heterodimerization with retinoid X receptor helps it mediate response to retinoids (28–30). However, recently a repressive function has been proposed in which Nr4a2-dependent transrepression of NF-κB represses inflammatory gene expression (17), but whether Nr4a2 transcriptional regulation has any bearing on macrophage polarization is unknown.
In this study, we report that Nr4a2 expression is induced in macrophages through the PI3K signaling pathway that attenuates innate inflammatory processes. Nr4a2 skews polarization of macrophages in favor of the alternative type and protects mice from endotoxin-induced sepsis. Thus, Nr4a2 appears to restrain inflammatory processes by polarizing macrophages to the M2 type.
Experimental Procedures
Mice and Ethics Statement
C57BL/6 mice were maintained and bred under specific pathogen-free conditions in the institute's animal house facility. Experiments with mice were approved by the Institutional Animal Ethics Committee of the Institute of Microbial Technology and performed according to the National Regulatory Guidelines issued by the Committee for the Purpose of Supervision of Experiments on Animals (Number 55/1999/Committee for the Purpose of Control and Supervision of Experiments on Animals (CPCSEA)), Ministry of Environment and Forest, Government of India.
Macrophages and DC Culture
Macrophages were collected from the peritoneal cavity as described (31). The viability and absolute number of cells were determined by the trypan blue exclusion method. For the generation of bone marrow-derived macrophages (BMDMs), bone marrow precursors were cultured in RPMI 1640 medium (Gibco) supplemented with 10% new-born calf serum (Gibco), 1% penicillin/streptomycin (Gibco), and 50 ng/ml GM-CSF (eBioscience). The non-adherent cells were replated on day 3 and cultured for another 4 days. Adherent cells were harvested on day 7 with ∼98% purity as analyzed by flow cytometry based on the expression of CD11b and F4/80. Bone marrow-derived dendritic cells (BMDCs) were obtained by culturing bone marrow precursors in RPMI 1640 medium supplemented with 10% new born calf serum, 1% penicillin/streptomycin, 10 ng/ml GM-CSF, and 10 ng/ml IL-4 (eBioscience) for 7 days. Non-adherent cells were harvested on day 7 with ∼90% purity as analyzed by flow cytometry based on the expression of CD11c.
Adenovirus Production and Transduction
Recombinant adenovirus expressing LacZ (Ad-lacZ) and mouse Nr4a2 (Ad-Nr4a2) were produced using the Adeno-XTM Expression System 1 from Clontech according to the manufacturer's instructions. For transduction, BMDMs were incubated on the 7th day of culture with Ad-lacZ or Ad-Nr4a2 in RPMI 1640 medium supplemented with 10% new born calf serum and 1% penicillin/streptomycin. The cells were incubated in a humidified, CO2 (5%) incubator at 37 °C for 24 h followed by aspiration of supernatant and replacement of medium. Cells were further incubated for an additional 24 h before experimental assays. For in vivo transduction, macrophages were elicited in the peritoneal cavity by thioglycollate injection. Four days later, mice were given either Ad-lacZ or Ad-Nr4a2 intraperitoneally, and the cells were harvested from the peritoneal cavity after 48 h of adenovirus delivery.
For silencing, adenovirus expressing shRNA for LacZ or shRNA specific for murine Nr4a2 was generated by transfecting the adenoviral construct, pAd/BLOCK-iT-DEST expression clone, into the 293A cell line. The viral particles were then collected according to the manufacturer's guidelines (Invitrogen).
Endotoxin-induced Model of Sepsis
Sepsis was induced in C57BL/6 male mice by intraperitoneal injection of 60 mg/kg of body weight of Escherichia coli LPS (Sigma), and survival was monitored. In some experiments, animals received an intraperitoneal injection of 30 mg/kg of body weight of 6-mercaptopurine (Sigma) or vehicle 30 min prior to LPS challenge. For adoptive transfer studies, 2 × 106 BMDMs transduced with Ad-lacZ or Ad-Nr4a2 were injected intraperitoneally 24 h prior to LPS challenge. Serum was collected 6 h after LPS challenge for cytokine analysis, and liver was harvested after 24 h for histopathological evaluation.
RNA Isolation and Quantitative Real-time PCR
Total RNA was extracted from cells by the TRIzol method. cDNA synthesized from 1 μg of total RNA with the Verso cDNA kit (Thermo Scientific) was subjected to RT-qPCR by the SYBR Green method (DyNAmo ColorFlash SYBR Green qPCR kit, Thermo Scientific). The normalized expression of the target gene (Nt) for each sample was calculated by the equation Nt = 2−(Ct of target − Ct of β-act) (where β-act is β-actin). Relative gene expression was obtained by setting Nt in the control as 1.
Immunoblot Analysis
To prepare whole cell lysate, cells were incubated on ice for 30 min in lysis buffer and then centrifuged at 13,000 rpm at 4 °C. The supernatant was collected and the protein concentration was estimated using the Bio-Rad protein assay reagent (Bio-Rad). The extracts were run on SDS-PAGE and transferred to PVDF membranes (Immobilon-P; Millipore). Membranes were blocked for 1 h at room temperature with either 5% skim milk or 5% BSA in Tris-buffered saline (pH 7.4). Membranes were incubated overnight with antibody to Nr4a2, anti-β-actin (Santa Cruz Biotechnology), anti-Akt, or anti-phospho-Akt (Cell Signaling Technology) followed by incubation with HRP-conjugated secondary antibodies and visualization with Luminata Forte Western HRP substrate (Millipore).
Measurement of Cytokine Levels and Evolution of Urea
Cytokines in cell culture supernatants or serum were quantified by ELISA kits specific for mouse IL-10, IL-12p70, and TNFα (BD Biosciences) following the manufacturer's protocol. Challenged mice were cardiac-punctured at the stated time points. Blood was left to clot at room temperature for 10 min and then centrifuged at 1200 rpm for 10 min following which serum was collected and cytokines were assayed with ELISA kits. To determine arginase activity, production of urea from arginine was determined colorimetrically as described previously (32).
Immunofluorescence
For confocal imaging, BMDMs were treated with LPS, poly(I:C) (Sigma), and zymosan (Sigma) for 24 h. Cells were then fixed for 10 min in 4% paraformaldehyde in PBS, permeabilized for 30 min with 0.1% (v/v) Triton-X-100 in PBS, and incubated for 30 min with 5% (w/v) BSA. This was followed by overnight incubation at 4 °C with anti-Nr4a2. After three washes, cells were further incubated for 1 h at room temperature with Texas Red-conjugated secondary antibody. For nuclei staining, the cells were stained with DAPI (Sigma) and mounted with antifade (Invitrogen, Molecular Probes). Fluorescent images were acquired with an A1R Nikon confocal microscope.
EMSA
EMSA was performed with oligonucleotides, which were end-labeled using T4-polynucleotide kinase (New England Biolabs) and [γ-32P]ATP. The wild-type DNA probe containing the Nr4a2-binding site from arginase 1 promoter is described as End-labeled Arg1 and has sequence: 5′-GAAGTAAATGTAAGGTCAAGCGATTTTG-3′. The mutant in which the core site for Nr4a2 binding was mutated is described as Mutant 1 with the sequence: 5′-GAAGTAAATGTAATTTCAAGCGATTTTG-3′ (mutated bases are underlined), whereas the mutant in which the AT-rich region adjacent to the core site on either side was mutated is described as Mutant 2 and has the sequence: 5′-GAAGTCCCGGTAAGGTCAGGCGATTTTG-3′ (mutated bases are underlined). Recombinant Nr4a2 protein was prepared in vitro using the TnT Quick Coupled Transcription/Translation System (Promega) according to the manufacturer's instructions, and nuclear extract was prepared from BMDMs. For competition experiments, molar excess of unlabeled oligonucleotide was added to compete for the DNA binding. Supershift EMSAs were performed with anti-Nr4a2 antibody. Samples were run on a native polyacrylamide gel, and bands were visualized using a phosphorimaging device (Bio-Rad Molecular Imager FX).
ChIP
For isolation of chromatin, cells were fixed for 30 min in 1% (w/v) formaldehyde. Cross-linking was terminated by the addition of 150 mm glycine. The cells were then washed and lysed by sonication in SDS lysis buffer. Debris were removed by centrifugation, and the cleared lysates were used for immunoprecipitation with ChIP-grade antibody to Nr4a2 (M-196 X). Finally, the immunoprecipitated DNA was amplified.
Flow Cytometry
Cells were washed, blocked, and then incubated for 30 min with allophycocyanin-conjugated anti-mouse F4/80, allophycocyanin-conjugated anti-mouse CD11c, and phycoerythrin-conjugated anti-mouse CD11b using the appropriate isotype controls. For indirect staining, cells were washed, blocked, and then incubated with anti-CD36 for 1 h followed by washing and incubation with fluorochrome-conjugated secondary antibody for 30 min. The stained cells were acquired on a BD Accuri flow cytometer (BD Biosciences) and analyzed with FlowJo software (Tree Star, Inc.).
Statistical Analysis
The statistical analysis was performed using SigmaPlot and GraphPad Prism software. The results are expressed as mean and standard deviation unless otherwise mentioned.
Results
TLR Ligands Induce Nr4a2 Expression in Macrophages and Dendritic Cells
The orphan receptors of the Nr4a family have been shown to be induced in macrophages by pathogenic components (14, 16, 17), which raised the possibility that the recognition of molecular pathogen-associated molecular patterns by pattern recognition receptors might be responsible for their expression. As anticipated, we observed elevated levels of Nr4a1, Nr4a2, and Nr4a3 mRNA with the maximum induction seen for Nr4a2 in LPS-stimulated BMDMs as compared with that in control cells (Fig. 1A). When protein levels were examined, elevation in Nr4a2 expression was seen as early as 2 h after LPS treatment, with levels remaining elevated even at 24 h (Fig. 1B). To evaluate whether ligation of other TLRs can also induce Nr4a2 expression, BMDMs were treated with different TLR ligands: zymosan, a ligand for TLR2; poly(I:C), a ligand for TLR3; and LPS, a ligand for TLR4. The engagement of each of these TLRs in response to their respective ligand did induce Nr4a2 expression (Fig. 1C). Furthermore, both peritoneal macrophages elicited by thioglycollate treatment and BMDCs also showed increased levels of Nr4a2 expression when stimulated by TLR ligands (Fig. 1, D and E), which suggests that these innate cells with myeloid lineage from different physiological niches might show increased levels of Nr4a2 upon activation, although the levels of induction varied with different stimuli. Additionally, confocal imaging further showed the induction of Nr4a2, which interestingly was localized in cytosol with a significant amount also present in the nucleus (Fig. 1F). Together, these results indicate that Nr4a2 expression is controlled by TLR signaling.
FIGURE 1.
Ligation of TLRs with their respective ligands induces Nr4a2 expression. A, expression of Nr4a family members (Nr4a1, Nr4a2, and Nr4a3) was measured by RT-qPCR in BMDMs left either unstimulated (control) or stimulated with LPS (500 ng/ml). B, immunoblot analysis of Nr4a2 in BMDMs stimulated with LPS (500 ng/ml) for 0–24 h. C–E, immunoblot analysis of Nr4a2 in BMDMs (C), peritoneal macrophages (D), and BMDCs (E) stimulated with LPS (500 ng/ml), poly(I:C) (5 μg/ml), and zymosan (25 μg/ml) for 24 h. F, confocal microscopy of BMDMs unstimulated (control) or stimulated with LPS (500 ng/ml), poly(I:C) (5 μg/ml), and zymosan (25 μg/ml) for 24 h and then stained with anti-Nr4a2 and DAPI. DIC is the differential interference contrast. Scale bars, 10 μm. Asterisks denote significant differences (* indicates p < 0.05). Data in B–F are representative of three independent experiments, whereas in A, data are averaged from three independent experiments (mean and S.D.).
Nr4a2 Expression Is Induced by the PI3K-Akt-mTOR Pathway
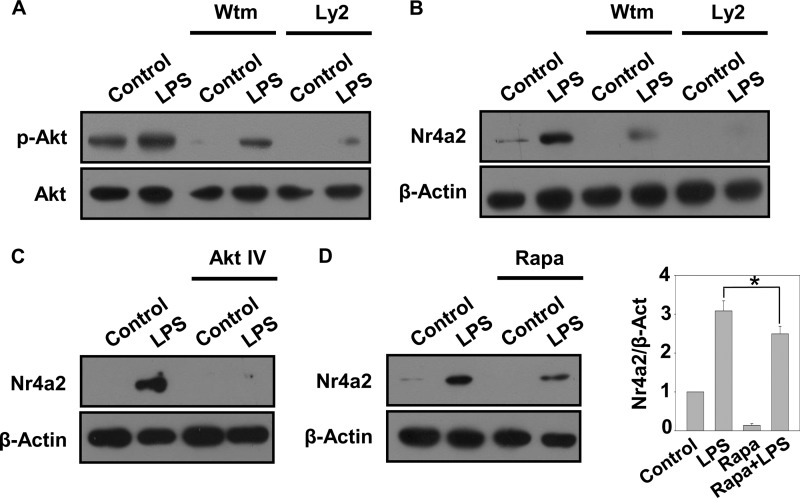
The various signaling pathways downstream of TLRs exert distinct biological effects (33). One such pathway that is elicited upon LPS recognition by TLR4 is the PI3K-Akt pathway, and several studies have shown the involvement of this pathway in LPS-induced gene expression (34–36). To determine whether the PI3K-Akt pathway also regulates expression of Nr4a2, we studied the effects of inhibitors for various mediators of this pathway. As reported, we found LPS-induced Akt phosphorylation, which was inhibited when cells were pretreated with the PI3K inhibitors wortmannin and Ly294002 (Fig. 2A). Interestingly, such inhibition of Akt phosphorylation resulted in abrogation of LPS-induced expression of Nr4a2 (Fig. 2B). Similar loss of LPS-induced Nr4a2 expression was also observed when cells were pretreated with an Akt-specific pharmacological inhibitor, Akt inhibitor IV (Fig. 2C). As mTOR is a well studied downstream effector target for activated Akt (37), we determined the expression of Nr4a2 in the presence of the mTOR inhibitor rapamycin. Pretreatment of cells with rapamycin significantly reduced LPS-dependent Nr4a2 expression (Fig. 2D). These results suggest that the PI3K-Akt-mTOR pathway mediates LPS-elicited Nr4a2 expression.
FIGURE 2.
P13K/Akt pathway modulates the expression of Nr4a2. A–D, immunoblot analyses were performed on BMDMs either left unstimulated (control) or stimulated with LPS (500 ng/ml) with or without preincubation for 1 h with PI3K inhibitors wortmannin (Wtm, 200 nm) or Ly294002 (Ly2, 50 μm) (A and B), Akt inhibitor IV (Akt IV, 10 μm) (C), or mTOR inhibitor rapamycin (Rapa, 100 nm) (D). A, the levels in whole cell lysates of Akt and its phosphorylated form were determined by immunoblot analysis. B–D, expression of Nr4a2 and β-actin (loading control) in whole cell lysates was determined by immunoblot analysis. Densitometry was performed and represented as the ratio of intensity of Nr4a2 to β-actin. Asterisks denote significant differences (* indicates p < 0.05). Data are representative of three independent experiments.
Nr4a2 Favors Alternative Activation of Macrophages
Nr4a2 has been reported to keep a check on inflammation by regulating the expression of various genes in immune cells (17). To examine whether Nr4a2 modulates inflammation by supporting alternative activation of macrophages, we overexpressed Nr4a2 and looked at its effects on BMDMs. As determined by flow cytometry, no significant change in the expression of F4/80 or CD11b was observed in BMDMs transduced with adenovirus encoding either LacZ or Nr4a2 (Fig. 3A). On the other hand, Nr4a2 appears to enhance the surface expression of CD36 (Fig. 3B). Further, in BMDMs transduced with adenovirus encoding Nr4a2, we observed elevated expression of M2 prototype genes such as arginase 1, mannose receptor, and Ym1 and decreased chemokine (CXC motif) ligand 9 (CXCL9) mRNA levels (Fig. 3C). Consistent with these observations, the secretion of IL-10 was significantly enhanced, whereas that of TNFα and IL-12p70 was significantly reduced in BMDMs overexpressing Nr4a2 (Fig. 3D). We also observed an augmented expression of CD36 on macrophages derived from the peritoneal cavity of mice injected with adenovirus expressing Nr4a2 (Fig. 3E). Together, these results indicate that by enhancing the expression of M2 marker genes, Nr4a2 primes macrophages toward the alternative activation state.
FIGURE 3.
Nr4a2 influences the expression of alternatively activated macrophage markers. A–D, BMDMs were transduced with adenovirus expressing LacZ (Ad-lacZ) or Nr4a2 (Ad-Nr4a2) for 48 h. A, FACS analysis to determine CD11b and F4/80; the numbers indicate the percentage of cells in each of the four quadrants. B, FACS analysis of CD36. C, RT-qPCR analysis to determine the relative expression of arginase 1, mannose receptor (MR), CXCL9, and Ym1. D, ELISA of the secretion of IL-10, IL-12p70, and TNFα in supernatants. E, FACS analysis of CD36 on thioglycollate-elicited macrophages obtained from the peritoneal cavity of mice injected with Ad-lacZ or Ad-Nr4a2. Asterisks denote significant differences (* and ** indicates p < 0.05 and p < 0.01 respectively). Data in A, B, and E are representative of three independent experiments, whereas in C and D, data are averaged from three independent experiments (mean and S.D.).
Arginase 1 Is a Direct Target Gene for Nr4a2
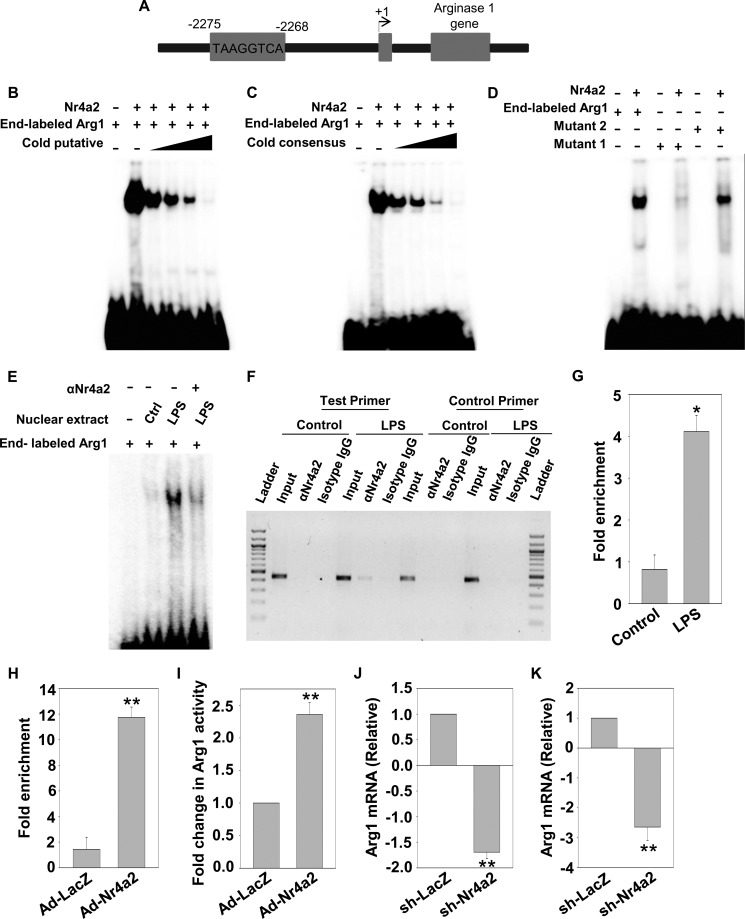
Nr4a2 regulates gene expression by binding to the conserved motifs on the promoters of its target genes. The promoters of M2 marker genes were extracted using the Cold Spring Harbor Laboratory mammalian promoter database (CSHLmpd) and examined for Nr4a2-binding sites. We found a putative Nr4a2-response element in the distal region of the arginase 1 promoter (Fig. 4A). The binding on this putative motif was confirmed by performing EMSA with a double-stranded radiolabeled oligonucleotide containing the sequence of Nr4a2-response element in the region of arginase 1 promoter. Nr4a2 bound to this putative response element (Fig. 4, B and C). The specificity of this binding was confirmed by performing a competition assay with unlabeled oligonucleotide containing either the putative sequence or the consensus sequence for the Nr4a2-binding site (Fig. 4, B and C). Additionally, we generated mutants to determine the bases in the putative region that are essential for binding. We observed that binding was completely abrogated when mutation was in the core motif, whereas it was significantly decreased when adjacent AT-rich region was mutated (Fig. 4D), suggesting that core motif is required for Nr4a2 binding. Next, we performed EMSAs in the presence of nuclear extracts obtained from unstimulated and LPS-stimulated BMDMs. Nuclear extracts from stimulated BMDMs bound to the putative binding site on arginase 1, but extracts from unstimulated BMDMs failed to do so (Fig. 4E). The interaction between radiolabeled oligonucleotide and nuclear extract from stimulated BMDMs was decreased after preincubation with antibody to Nr4a2, suggesting that Nr4a2 in nuclear extract of stimulated BMDMs interacted with the binding element present in arginase 1 promoter (Fig. 4E). In addition, ChIP assay showed that Nr4a2 interacted with its binding site in arginase 1 promoter in stimulated BMDMs but not in unstimulated cells as seen with amplification of DNA isolates with test primer (Fig. 4, F and G). No amplification of DNA isolates was observed with control primers (Fig. 4F). The recruitment of Nr4a2 on arginase 1 promoter was also observed in cells transduced with Ad-Nr4a2 (Fig. 4H). We also elucidated that overexpression of Nr4a2 induces arginase enzymatic activity (Fig. 4I). Earlier studies have highlighted that apart from IL-4, pro-inflammatory mediators such as LPS induce arginase 1 expression and its activity (38, 39). So we determined the effect of Nr4a2 silencing on LPS-induced arginase 1 in BMDMs and peritoneal macrophages and found that loss of Nr4a2 decreased arginase 1 expression (Fig. 4, J and K). Thus, it is clear that Nr4a2 interacts with arginase 1 promoter during inflammatory conditions.
FIGURE 4.
Arginase 1 is a direct target gene of Nr4a2. A, pictorial representation of arginase 1 promoter having a response element for Nr4a2 binding. B and C, EMSA analysis using end-labeled oligonucleotide containing the sequence for the conserved Nr4a2-binding site on the arginase 1 regulatory region in the presence of in vitro translated Nr4a2, competed either in the absence or in the presence of 5-, 10-, 50-, and 100-fold excess of unlabeled oligonucleotide containing sequence for putative arginase 1 Nr4a2-response element (B) or unlabeled oligonucleotide containing sequence for the consensus Nr4a2-response element (C). Lane 1 contains free radiolabeled oligonucleotide. D, EMSA was performed using end-labeled oligonucleotide containing sequence for the wild-type arginase 1 regulatory region, Mutant 1 (in which the core motif was mutated) or Mutant 2 (in which AT-rich regions adjacent to core site were mutated) in the presence of in vitro translated Nr4a2. E, EMSA to determine binding of Nr4a2 on the arginase 1 regulatory region using nuclear extracts obtained from unstimulated (Ctrl) or LPS-stimulated BMDMs (500 ng/ml). The DNA-protein binding specificity was determined using anti-Nr4a2 antibody. Lane 1 contains free radiolabeled oligonucleotide. F–H, ChIP analysis of Nr4a2 binding on arginase 1 promoter was performed using chromatin isolated from unstimulated (control) and LPS-stimulated (500 ng/ml) BMDMs (F and G) or BMDMs transduced with Ad-lacZ or Ad-Nr4a2 (H). Cross-linked lysates were immunoprecipitated with either isotype control antibody or ChIP-grade Nr4a2 antibody, and DNA isolates were subjected to amplification. I, BMDMs were transduced with adenovirus expressing LacZ (Ad-lacZ) or Nr4a2 (Ad-Nr4a2) for 48 h, and an arginase assay was performed to determine the enzymatic activity of arginase 1. J and K, BMDMs (J) and peritoneal macrophages (K) were transduced with adenovirus particles expressing sh-LacZ or sh-Nr4a2. 48 h after transduction, cells were stimulated with LPS, and 24 h after LPS stimulation, the samples were harvested and RT-qPCR was performed to determine the relative expression of arginase 1. Asterisks denote significant differences (* and ** indicate p < 0.05 and p < 0.01, respectively).
Nr4a2 Imparts Protection in Septic Animals
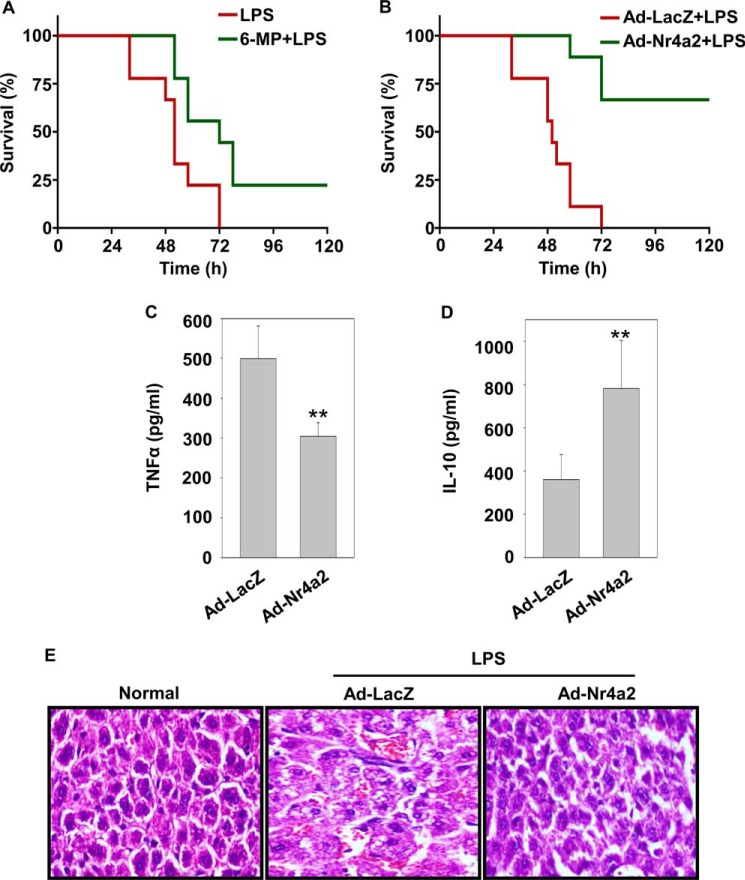
To ascertain the pathological relevance of Nr4a2 in vivo, we evaluated systemic inflammation induced by an intraperitoneal injection of a lethal dose of LPS with or without 6-mercaptopurine. 6-Mercaptopurine was used because it induces the transcriptional activity of Nr4a2 (40). Within 60 h of LPS administration, 90% of the control mice died, whereas mice that received prior treatment of 6-mercaptopurine had better survival during the same period (Fig. 5A). To further confirm the effect of Nr4a2 against endotoxemia mortality, we adoptively transferred 2 × 106 BMDMs transduced with adenovirus expressing LacZ or Nr4a2, intraperitoneally. One day later, mice were challenged with a lethal dose of LPS, and survival was monitored. In this experiment, we noted improved survival in mice that received BMDMs transduced with adenovirus expressing Nr4a2 (Fig. 5B). The higher survival rates in these mice correlated with lower serum concentration of pro-inflammatory cytokine TNFα (Fig. 5C), which is known to add to the pathological manifestation of endotoxin shock. In contrast, mice that had received BMDMs transduced with adenovirus expressing Nr4a2 had a higher expression of anti-inflammatory cytokine IL-10 than mice that were treated with BMDMs transduced with adenovirus expressing LacZ (Fig. 5D). Analysis of liver samples showed substantial decrease in LPS-induced damage in mice that received BMDMs transduced with adenovirus expressing Nr4a2 in comparison with those that received BMDMs transduced with adenovirus expressing LacZ (Fig. 5E). Together, these data suggest that Nr4a2 rescued mice from endotoxin-induced mortality by keeping a check on uncontrolled systemic inflammation.
FIGURE 5.
Nr4A2 provides protection against endotoxic shock in mice. A, Kaplan-Meier survival analysis of mice (n = 9 per group) injected intraperitoneally with a lethal dose of LPS (60 mg/kg of body weight) with or without prior treatment with 6-mecrcaptopurine (6-MP, 30 mg/kg of body weight) given intraperitoneally 30 min before LPS challenge. B, Kaplan-Meier survival analysis of mice (n = 9 per group) injected intraperitoneally with 2 × 106 BMDMs transduced with adenovirus expressing LacZ (Ad-lacZ) or Nr4a2 (Ad-Nr4a2). 24 h after adoptive transfer, mice were challenged intraperitoneally with a lethal dose of LPS (60 mg/kg of body weight). C and D, animals were treated as in B and assayed by ELISA after 6 h of LPS challenge for cytokines TNFα (C) and IL-10 (D). E, animals were treated as in B, and hematoxylin and eosin staining of liver sections from untreated control and treated groups was obtained. Magnification: ×200. Asterisks denote significant differences (** indicates p < 0.01). Data in C and D are averaged of five individual mice (mean and S.D.).
Discussion
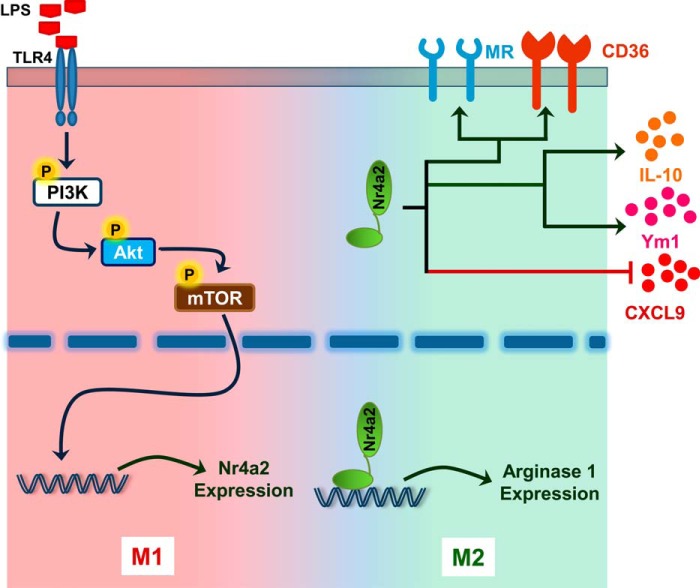
Members of the Nr4a subfamily of orphan nuclear receptors transcriptionally regulate expression of genes. In this study, we identify Nr4a2 as a gene induced in macrophages in response to TLR ligands. The presence of this receptor skews macrophage polarization to the M2 type (Fig. 6). Additionally, we showed that Nr4a2 binds to the promoter of arginase 1 and transcriptionally enhances its expression. Moreover, the adoptive transfer of macrophages expressing Nr4a2 protects mice from endotoxemia by curtailing pro-inflammatory cytokine production. Overall, this strongly supports an anti-inflammatory role for Nr4a2.
FIGURE 6.
Schematic representation highlighting the role of Nr4a2 in macrophages. Circled P indicates phosphorylation.
Recent studies have made clear that macrophage polarization is a complex process and is accompanied by a characteristic gene expression program. However, identification of the transcription factors that guide macrophage polarization remains exceptionally challenging as macrophages are constantly vigilant, sensing changes in the microenvironment and rapidly switching their phenotype to mount an appropriate immune response. Given the role of Nr4a2 in checking aberrant inflammatory processes, our goal was to unravel its relationship to macrophage participation in these processes. We observed elevated Nr4a2 expression in macrophages in response to ligands of various TLRs (Fig. 1, C, D, and F). Nr4a2 expression has been shown to be rapidly induced in cells after an encounter with inflammatory mediators, which has been the basis for categorizing Nr4a2 as an immediate-early response gene (14, 16, 17). Additionally, cAMP-PKA activators and agonists of PKC and calcium signaling are also known to up-regulate Nr4a2 expression (26). On the other hand, loss of Nr4a2 significantly increases the expression of TNFα, IL-1β, and inducible nitric-oxide synthase (iNOS) in activated microglia (17). Depending upon the stimuli, distinct signaling pathways may modulate different transcriptional programs to regulate expression. We showed that LPS-elicited Nr4a2 expression is mediated by the PI3K-Akt-mTOR pathway (Fig. 2). The activation of macrophages by LPS involves Akt phosphorylation in a PI3K-dependent manner (36, 41), and mTOR is a crucial downstream target for activated Akt (37, 42). The PI3K-Akt pathway has also been shown to relay its effect in association with other TLRs in different cells (43–45).
Only a few transcription factors crucial for macrophage polarization have been identified and functionally characterized. We examined whether Nr4a2 influences the phenomenon of macrophage polarization and observed that overexpression of Nr4a2 in macrophages induces prototypical mouse M2 marker genes (Fig. 3, B–E). We have also identified arginase 1 as a direct target gene of Nr4a2 (Fig. 4, B–E). Although the effect of Nr4a2 on arginase 1 was genomic, its non-genomic role leading to changes in the expression of macrophage marker genes cannot be ruled out. The role of arginase 1 in the immune system is already well appreciated. Arginase 1 is expressed in alternatively activated macrophages and competes for arginine as a common substrate with iNOS in classically activated macrophages. iNOS catalyzes the conversion of arginine to microbicidal nitric oxide and citrulline, whereas arginase 1 converts arginine to urea and ornithine. Ornithine is a precursor for polyamines and intermediates that give rise to collagen for wound healing and tissue repair (46). Moreover, BMDMs obtained from mice lacking arginase 1 are more susceptible to LPS-induced endotoxin as they show increased production of pro-inflammatory mediators as compared with BMDMs from wild-type mice (47). Various transcription factors have been shown to regulate arginase 1 expression in response to various stimuli. STAT6 and CCAAT/enhancer-binding protein β (C/EBPβ) are recruited at an enhancer 3 kb upstream of the transcription start site to regulate arginase 1 expression in response to IL-4 (48). STAT6 also acts in synergy with KLF4 to regulate arginase 1 expression; however, arginase 1 expression in response to Bacillus Calmette-Guérin (BCG) is STAT6-independent and depends only upon CCAAT/enhancer-binding protein β transcriptional activity (13, 49). PU.1 is also known to bind to the arginase 1 promoter at two sites, one in the enhancer region 3 kb upstream of transcription start site and another 700 bp upstream of the basal promoter (48, 50). Furthermore, arginase 1 is known to be a PPAR-responsive gene (51). Thus the expression of arginase 1 is tightly regulated, and any abnormality in its expression leads to inflammatory disorders.
Nr4a2 is a “true orphan receptor” as it lacks a disengaged ligand-binding pocket because the site remains indefinitely inhabited by side chains of colossal hydrophobic amino acids (15). In the absence of ligands, the N-terminal AF1 domain of Nr4a2, which is involved in ligand-independent transcriptional activity and is the target for various post-translational modifications, is crucial for the regulation of the receptor (52). Ligands including 6-mercaptopurine and methylene-substituted diindolylmethanes (C-DIM), such as 1, 1-bis (3′-indolyl)-1-(p-chlorophenyl) methane (DIM-C-pPHCI), transactivate Nr4a2, possibly by targeting the post-translational modifications in the AF1 domain (40, 53). In this study, we observed that 6-mercaptopurine prevented mortality in animals challenged with a lethal dose of LPS (Fig. 5A). Given the role of 6-mercaptopurine as an Nr4a2 transactivator, we believe that the protective effects of 6-mercaptopurine against endotoxin shock are at least partially dependent on Nr4a2. 6-Mercaptopurine does activate other members of Nr4a family, and hence ligand that can specifically activate Nr4a2 may prove to be more effective. Moreover, adoptive transfer of macrophages overexpressing Nr4a2 also proved protective in septic animals.
Overall this work highlights the role of Nr4a2 in guiding alternative activation of macrophages as a vital factor in providing a survival advantage in endotoxin shock. Given this, the role of Nr4a2 in imparting a protective mechanism must be investigated in other inflammatory and autoimmune disorders. Importantly, because DCs were also found to have up-regulated expression of Nr4a2, future studies will take into account the effects of Nr4a2 in both macrophages and DCs in an integrated host response to inflammatory disorders.
Acknowledgments
We thank Dr. B. N. Datta for histopathology; Dr. Girish Sahni for support; and the Council of Scientific and Industrial Research-Institute of Microbial Technology (IMTECH) for providing facilities.
This work was supported by the Department of Biotechnology-India project BT/01/IYBA/2009 and Council of Scientific and Industrial Research 12th Plan Network project Bugs to Drugs and Infectious Disease (Grants BSC0211 and BSC0210) (to P. G.). The authors declare that they have no conflicts of interest with the contents of this article.
This article was selected as a Paper of the Week.
- TLR
- Toll-like receptors
- M2
- alternatively activated macrophages
- M1
- classically activated macrophages
- BMDM
- bone marrow-derived macrophage
- BMDC
- bone marrow-derived dendritic cell
- PAMP
- pathogen-associated molecular pattern
- mTOR
- mammalian target of rapamycin
- Ad
- adenovirus
- qPCR
- quantitative PCR
- iNOS
- inducible nitric-oxide synthase
- PPAR
- peroxisome proliferator-activated receptor.
References
- 1. Hotamisligil G. S. (2006) Inflammation and metabolic disorders. Nature 444, 860–867 [DOI] [PubMed] [Google Scholar]
- 2. Handschin C., Spiegelman B. M. (2008) The role of exercise and PGC1α in inflammation and chronic disease. Nature 454, 463–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. O'Neill L. A., Golenbock D., Bowie A. G. (2013) The history of Toll-like receptors: redefining innate immunity. Nat. Rev. Immunol. 13, 453–460 [DOI] [PubMed] [Google Scholar]
- 4. Akira S., Takeda K. (2004) Toll-like receptor signalling. Nat. Rev. Immunol. 4, 499–511 [DOI] [PubMed] [Google Scholar]
- 5. Murray P. J., Smale S. T. (2012) Restraint of inflammatory signaling by interdependent strata of negative regulatory pathways. Nat. Immunol. 13, 916–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kondo T., Kawai T., Akira S. (2012) Dissecting negative regulation of Toll-like receptor signaling. Trends Immunol. 33, 449–458 [DOI] [PubMed] [Google Scholar]
- 7. Fujiwara N., Kobayashi K. (2005) Macrophages in inflammation. Curr. Drug Targets Inflamm. Allergy 4, 281–286 [DOI] [PubMed] [Google Scholar]
- 8. Mosser D. M., Edwards J. P. (2008) Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 8, 958–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sica A., Mantovani A. (2012) Macrophage plasticity and polarization: in vivo veritas. J. Clin. Invest. 122, 787–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lawrence T., Natoli G. (2011) Transcriptional regulation of macrophage polarization: enabling diversity with identity. Nat. Rev. Immunol. 11, 750–761 [DOI] [PubMed] [Google Scholar]
- 11. Tugal D., Liao X., Jain M. K. (2013) Transcriptional control of macrophage polarization. Arterioscler. Thromb. Vasc. Biol. 33, 1135–1144 [DOI] [PubMed] [Google Scholar]
- 12. Chawla A. (2010) Control of macrophage activation and function by PPARs. Circ. Res. 106, 1559–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liao X., Sharma N., Kapadia F., Zhou G., Lu Y., Hong H., Paruchuri K., Mahabeleshwar G. H., Dalmas E., Venteclef N., Flask C. A., Kim J., Doreian B. W., Lu K. Q., Kaestner K. H., Hamik A., Clément K., Jain M. K. (2011) Kruppel-like factor 4 regulates macrophage polarization. J. Clin. Invest. 121, 2736–2749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pei L., Castrillo A., Chen M., Hoffmann A., Tontonoz P. (2005) Induction of NR4A orphan nuclear receptor expression in macrophages in response to inflammatory stimuli. J. Biol. Chem. 280, 29256–29262 [DOI] [PubMed] [Google Scholar]
- 15. Wang Z., Benoit G., Liu J., Prasad S., Aarnisalo P., Liu X., Xu H., Walker N. P., Perlmann T. (2003) Structure and function of Nurr1 identifies a class of ligand-independent nuclear receptors. Nature 423, 555–560 [DOI] [PubMed] [Google Scholar]
- 16. Barish G. D., Downes M., Alaynick W. A., Yu R. T., Ocampo C. B., Bookout A. L., Mangelsdorf D. J., Evans R. M. (2005) A Nuclear Receptor Atlas: macrophage activation. Mol. Endocrinol. 19, 2466–2477 [DOI] [PubMed] [Google Scholar]
- 17. Saijo K., Winner B., Carson C. T., Collier J. G., Boyer L., Rosenfeld M. G., Gage F. H., Glass C. K. (2009) A Nurr1/CoREST pathway in microglia and astrocytes protects dopaminergic neurons from inflammation-induced death. Cell 137, 47–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shi Y. (2007) Orphan nuclear receptors in drug discovery. Drug Discov. Today 12, 440–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Han Y. F., Cao G. W. (2012) Role of nuclear receptor NR4A2 in gastrointestinal inflammation and cancers. World J. Gastroenterol. 18, 6865–6873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. O'Kane M., Markham T., McEvoy A. N., Fearon U., Veale D. J., FitzGerald O., Kirby B., Murphy E. P. (2008) Increased expression of the orphan nuclear receptor NURR1 in psoriasis and modulation following TNF-α inhibition. J. Invest. Dermatol. 128, 300–310 [DOI] [PubMed] [Google Scholar]
- 21. Le W. D., Xu P., Jankovic J., Jiang H., Appel S. H., Smith R. G., Vassilatis D. K. (2003) Mutations in NR4A2 associated with familial Parkinson disease. Nat. Genet. 33, 85–89 [DOI] [PubMed] [Google Scholar]
- 22. Sekiya T., Kashiwagi I., Inoue N., Morita R., Hori S., Waldmann H., Rudensky A. Y., Ichinose H., Metzger D., Chambon P., Yoshimura A. (2011) The nuclear orphan receptor Nr4a2 induces Foxp3 and regulates differentiation of CD4+ T cells. Nat. Commun. 2, 269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Doi Y., Oki S., Ozawa T., Hohjoh H., Miyake S., Yamamura T. (2008) Orphan nuclear receptor NR4A2 expressed in T cells from multiple sclerosis mediates production of inflammatory cytokines. Proc. Natl. Acad. Sci. U.S.A. 105, 8381–8386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Aherne C. M., McMorrow J., Kane D., FitzGerald O., Mix K. S., Murphy E. P. (2009) Identification of NR4A2 as a transcriptional activator of IL-8 expression in human inflammatory arthritis. Mol. Immunol. 46, 3345–3357 [DOI] [PubMed] [Google Scholar]
- 25. Mix K. S., McMahon K., McMorrow J. P., Walkenhorst D. E., Smyth A. M., Petrella B. L., Gogarty M., Fearon U., Veale D., Attur M. G., Abramson S. B., Murphy E. P. (2012) Orphan nuclear receptor NR4A2 induces synoviocyte proliferation, invasion, and matrix metalloproteinase 13 transcription. Arthritis Rheum. 64, 2126–2136 [DOI] [PubMed] [Google Scholar]
- 26. Pirih F. Q., Tang A., Ozkurt I. C., Nervina J. M., Tetradis S. (2004) Nuclear orphan receptor Nurr1 directly transactivates the osteocalcin gene in osteoblasts. J. Biol. Chem. 279, 53167–53174 [DOI] [PubMed] [Google Scholar]
- 27. Maira M., Martens C., Batsché E., Gauthier Y., Drouin J. (2003) Dimer-specific potentiation of NGFI-B (Nur77) transcriptional activity by the protein kinase A pathway and AF-1-dependent coactivator recruitment. Mol. Cell. Biol. 23, 763–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Perlmann T., Jansson L. (1995) A novel pathway for vitamin A signaling mediated by RXR heterodimerization with NGFI-B and NURR1. Genes Dev. 9, 769–782 [DOI] [PubMed] [Google Scholar]
- 29. Zetterström R. H., Solomin L., Mitsiadis T., Olson L., Perlmann T. (1996) Retinoid X receptor heterodimerization and developmental expression distinguish the orphan nuclear receptors NGFI-B, Nurr1, and Nor1. Mol. Endocrinol. 10, 1656–1666 [DOI] [PubMed] [Google Scholar]
- 30. Maira M., Martens C., Philips A., Drouin J. (1999) Heterodimerization between members of the Nur subfamily of orphan nuclear receptors as a novel mechanism for gene activation. Mol. Cell. Biol. 19, 7549–7557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mahajan S., Chandra V., Dave S., Nanduri R., Gupta P. (2012) Stem bromelain-induced macrophage apoptosis and activation curtail Mycobacterium tuberculosis persistence. J. Infect. Dis. 206, 366–376 [DOI] [PubMed] [Google Scholar]
- 32. Mahajan S., Dkhar H. K., Chandra V., Dave S., Nanduri R., Janmeja A. K., Agrewala J. N., Gupta P. (2012) Mycobacterium tuberculosis modulates macrophage lipid-sensing nuclear receptors PPARγ and TR4 for survival. J. Immunol. 188, 5593–5603 [DOI] [PubMed] [Google Scholar]
- 33. O'Neill L. A., Bowie A. G. (2007) The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat. Rev. Immunol. 7, 353–364 [DOI] [PubMed] [Google Scholar]
- 34. Ruse M., Knaus U. G. (2006) New players in TLR-mediated innate immunity: PI3K and small Rho GTPases. Immunol. Res. 34, 33–48 [DOI] [PubMed] [Google Scholar]
- 35. Park S. J., Lee S. C., Hong S. H., Kim H. M. (2002) Degradation of IκBα in activated RAW264.7 cells is blocked by the phosphatidylinositol 3-kinase inhibitor LY294002. Cell Biol. Toxicol. 18, 121–130 [DOI] [PubMed] [Google Scholar]
- 36. Ojaniemi M., Glumoff V., Harju K., Liljeroos M., Vuori K., Hallman M. (2003) Phosphatidylinositol 3-kinase is involved in Toll-like receptor 4-mediated cytokine expression in mouse macrophages. Eur. J. Immunol. 33, 597–605 [DOI] [PubMed] [Google Scholar]
- 37. LoPiccolo J., Blumenthal G. M., Bernstein W. B., Dennis P. A. (2008) Targeting the PI3K/Akt/mTOR pathway: effective combinations and clinical considerations. Drug Resist. Updat. 11, 32–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Menzies F. M., Henriquez F. L., Alexander J., Roberts C. W. (2010) Sequential expression of macrophage anti-microbial/inflammatory and wound healing markers following innate, alternative and classical activation. Clin. Exp. Immunol. 160, 369–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cho D. I., Kim M. R., Jeong H. Y., Jeong H. C., Jeong M. H., Yoon S. H., Kim Y. S., Ahn Y. (2014) Mesenchymal stem cells reciprocally regulate the M1/M2 balance in mouse bone marrow-derived macrophages. Exp. Mol. Med. 46, e70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ordentlich P., Yan Y., Zhou S., Heyman R. A. (2003) Identification of the antineoplastic agent 6-mercaptopurine as an activator of the orphan nuclear hormone receptor Nurr1. J. Biol. Chem. 278, 24791–24799 [DOI] [PubMed] [Google Scholar]
- 41. Monick M. M., Carter A. B., Robeff P. K., Flaherty D. M., Peterson M. W., Hunninghake G. W. (2001) Lipopolysaccharide activates Akt in human alveolar macrophages resulting in nuclear accumulation and transcriptional activity of β-catenin. J. Immunol. 166, 4713–4720 [DOI] [PubMed] [Google Scholar]
- 42. Hay N., Sonenberg N. (2004) Upstream and downstream of mTOR. Genes Dev. 18, 1926–1945 [DOI] [PubMed] [Google Scholar]
- 43. Arbibe L., Mira J. P., Teusch N., Kline L., Guha M., Mackman N., Godowski P. J., Ulevitch R. J., Knaus U. G. (2000) Toll-like receptor 2-mediated NF-κB activation requires a Rac1-dependent pathway. Nat. Immunol. 1, 533–540 [DOI] [PubMed] [Google Scholar]
- 44. Sarkar S. N., Peters K. L., Elco C. P., Sakamoto S., Pal S., Sen G. C. (2004) Novel roles of TLR3 tyrosine phosphorylation and PI3 kinase in double-stranded RNA signaling. Nat. Struct. Mol. Biol. 11, 1060–1067 [DOI] [PubMed] [Google Scholar]
- 45. Ishii K. J., Takeshita F., Gursel I., Gursel M., Conover J., Nussenzweig A., Klinman D. M. (2002) Potential role of phosphatidylinositol 3 kinase, rather than DNA-dependent protein kinase, in CpG DNA-induced immune activation. J. Exp. Med. 196, 269–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Munder M. (2009) Arginase: an emerging key player in the mammalian immune system. Br. J. Pharmacol. 158, 638–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wijnands K. A., Hoeksema M. A., Meesters D. M., van den Akker N. M., Molin D. G., Briedé J. J., Ghosh M., Köhler S. E., van Zandvoort M. A., de Winther M. P., Buurman W. A., Lamers W. H., Poeze M. (2014) Arginase-1 deficiency regulates arginine concentrations and NOS2-mediated NO production during endotoxemia. PLoS One 9, e86135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pauleau A. L., Rutschman R., Lang R., Pernis A., Watowich S. S., Murray P. J. (2004) Enhancer-mediated control of macrophage-specific arginase I expression. J. Immunol. 172, 7565–7573 [DOI] [PubMed] [Google Scholar]
- 49. El Kasmi K. C., Qualls J. E., Pesce J. T., Smith A. M., Thompson R. W., Henao-Tamayo M., Basaraba R. J., König T., Schleicher U., Koo M. S., Kaplan G., Fitzgerald K. A., Tuomanen E. I., Orme I. M., Kanneganti T. D., Bogdan C., Wynn T. A., Murray P. J. (2008) Toll-like receptor-induced arginase 1 in macrophages thwarts effective immunity against intracellular pathogens. Nat. Immunol. 9, 1399–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pourcet B., Feig J. E., Vengrenyuk Y., Hobbs A. J., Kepka-Lenhart D., Garabedian M. J., Morris S. M., Jr., Fisher E. A., Pineda-Torra I. (2011) LXRα regulates macrophage arginase 1 through PU.1 and interferon regulatory factor 8. Circ. Res. 109, 492–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gallardo-Soler A., Gómez-Nieto C., Campo M. L., Marathe C., Tontonoz P., Castrillo A., Corraliza I. (2008) Arginase I induction by modified lipoproteins in macrophages: a peroxisome proliferator-activated receptor-γ/δ-mediated effect that links lipid metabolism and immunity. Mol. Endocrinol. 22, 1394–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sacchetti P., Carpentier R., Ségard P., Olivé-Cren C., Lefebvre P. (2006) Multiple signaling pathways regulate the transcriptional activity of the orphan nuclear receptor NURR1. Nucleic Acids Res. 34, 5515–5527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Li X., Lee S. O., Safe S. (2012) Structure-dependent activation of NR4A2 (Nurr1) by 1,1-bis(3′-indolyl)-1-(aromatic)methane analogs in pancreatic cancer cells. Biochem. Pharmacol. 83, 1445–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]