Background: There are multiple interactions between complement, coagulation, and fibrinolytic systems.
Results: C4b-binding protein (C4BP) and plasminogen form a complex and C4BP augments plasminogen activation.
Conclusion: C4BP affects the function of plasminogen.
Significance: The interaction may regulate fibrinolysis and complement at the site of injury and/or during acute inflammation to maintain homeostasis.
Keywords: complement, plasmin, plasminogen, protein-protein interaction, surface plasmon resonance (SPR), C4b-binding protein
Abstract
The complement, coagulation, and fibrinolytic systems are crucial for the maintenance of tissue homeostasis. To date numerous interactions and cross-talks have been identified between these cascades. In line with this, here we propose a novel, hitherto unknown interaction between the complement inhibitor C4b-binding protein (C4BP) and plasminogen of the fibrinolytic pathway. Binding of C4BP to Streptococcus pneumoniae is a known virulence mechanism of this pathogen and it was increased in the presence of plasminogen. Interestingly, the acute phase variant of C4BP lacking the β-chain and protein S binds plasminogen much stronger than the main isoform containing the β-chain and protein S. Indeed, the complement control protein (CCP) 8 domain of C4BP, which would otherwise be sterically hindered by the β-chain, primarily mediates this interaction. Moreover, the lysine-binding sites in plasminogen kringle domains facilitate the C4BP-plasminogen interaction. Furthermore, C4BP readily forms complexes with plasminogen in fluid phase and such complexes are present in human serum and plasma. Importantly, whereas the presence of plasminogen did not affect the factor I cofactor activity of C4BP, the activation of plasminogen by urokinase-type plasminogen activator to active plasmin was significantly augmented in the presence of C4BP. Taken together, our data demonstrate a novel interaction between two proteins of the complement and fibrinolytic system. Most complexes might be formed during the acute phase of inflammation and have an effect on the homeostasis at the site of injury or acute inflammation.
Introduction
The complement system is a major arm of the innate immune system and functions as a first line of defense against invading pathogens. In addition it also plays a critical role in the removal of apoptotic and necrotic cells, and in regulation of the adaptive immune system responses. It consists of about 40 proteins and depending on the initiating stimulus it may be activated via three different pathways: the classical (antibody dependent), the lectin (mannose binding lectin or ficolin dependent), and the alternative (spontaneous activation) pathways (1, 2). All pathways converge at the level of complement factor C3 resulting in the formation of the C3 and C5 convertases with subsequent deposition of the opsonin C3b, release of proinflammatory anaphylatoxin C3a and C5a, and finally formation of a lytic membrane attack complex, which disrupts membrane integrity of the target cell.
To restrict complement activation on foreign cells and avoid overconsumption as well as complement-mediated tissue injury, the process of complement activation is tightly regulated via both soluble and membrane-bound complement inhibitors (3, 4). C4b-binding protein (C4BP),3 a 570-kDa plasma glycoprotein, is the major soluble inhibitor of the classical and lectin complement pathways. C4BP not only inhibits the formation and accelerates the decay of the classical and lectin pathway C3 convertases (C4b2a), but it also serves as a cofactor to the serine protease factor I in the proteolytic degradation of C4b and C3b (5–7). C4BP consists of seven identical α-chains (70 kDa) and one β-chain (45 kDa), which are composed of repeating domains of ∼60 amino acids known as complement control protein (CCP) domains (8–10). Each α-chain contains eight CCP domains, and the β-chain contains three. Additionally, C4BP is also linked to the coagulation system because the β-chain binds with high affinity to the vitamin K-dependent anticoagulant protein S (C4BP-PS) (9).
Just like the complement system, the coagulation system is also important for the maintenance of tissue homeostasis, in addition to its essential role in the prevention of excessive blood loss upon vessel damage or injury. Two routes primarily initiate the coagulation cascade: the contact (intrinsic) pathway and the tissue factor (extrinsic) pathway, which eventually lead to thrombus formation by platelet aggregation and the conversion of soluble fibrinogen into an insoluble fibrin clot by thrombin (11, 12). Interestingly, similar to complement, the process of coagulation is also controlled and restricted to the site of vascular injury by natural anticoagulants (13).
When the clot is no longer needed, and to prevent its extension beyond the site of injury, the clot is gradually dissolved. The process of clot dissolution, known as fibrinolysis, is regulated by the plasminogen-plasmin system (14). Plasminogen is a 92-kDa single chain glycoprotein zymogen of the broad-spectrum protease plasmin. It is primarily synthesized in the liver and circulates at a concentration of ∼2 μm in blood and is composed of a pre-activation peptide (∼8 kDa), five homologous disulfide-bonded kringle domains (K1–5, 65 kDa), and a serine-protease domain (15). The first four kringle domains (K1–K4) of plasminogen are termed angiostatin because of its antiangiogenic role (16), the last kringle domain together with the enzymatic domain is referred to as mini-plasminogen (17). Plasminogen is converted into active serine protease plasmin by plasminogen activators like urokinase-type plasminogen activator (uPA) and tissue-type plasminogen activator (18). Upon activation, plasmin hydrolyzes polymerized fibrin clots into soluble fibrin degradation products. In addition, plasmin also degrades a variety of extracellular matrix and basal membrane components and thus contributes to maintaining homeostasis.
The complement, coagulation, and fibrinolytic systems play a central role in maintenance of tissue homeostasis. Considering the fact that these pathways have a common ancestry, multiple interactions and cross-talk between these cascades have been proposed (19–21). In line with these, we demonstrate a new interaction between the complement inhibitor C4BP and plasminogen, the main component in fibrinolysis.
Experimental Procedures
Proteins and Abs
C4BP-PS complexes (570 kDa) and C4BP without β-chain and protein S (−β/−PS) were purified from human plasma using differential precipitation with BaCl2 as described (22), whereas the total pool of C4BP present in plasma (C4BP total) was purified with affinity chromatography using monoclonal mouse anti-human C4BP mk104 (24). Recombinant factor I was expressed and purified from eukaryotic cells (23). Recombinant wild-type C4BP (rC4BP) and C4BP mutants lacking single CCP domains in the α-chains were expressed in eukaryotic cells and purified by affinity chromatography (24). BSA was purchased from AppliChem. The polyclonal rabbit anti-human C4BP 9008 Abs were a kind gift of Prof. B. Dahlbäck (Lund University, Sweden). This antibody recognizes all forms of C4BP (plasma purified C4BP-PS, rC4BP, and mutants) equally well (25). Human Glu-plasminogen and polyclonal sheep anti-human plasminogen Abs were purchased from Hematologic Technologies. A matched pair antibody set for ELISA of human plasminogen was obtained from Affinity Biologicals. Human angiostatin (Kringle 1–3) and uPA were obtained from Sigma, whereas the kringle 1–4 was purchased from Alpha Diagnostics. Monoclonal antibody (mAb) anti-C4BP mk104 was purified using a protein A-Sepharose column, whereas the mAb anti-plasminogen 4PG and tissue-type plasminogen activator were purchased from Hemochrom Diagnostica. Isotype control mouse IgG1 was from BD Biosciences, whereas the peroxidase-conjugated swine anti-rabbit, rabbit anti-sheep, and rabbit anti-mouse IgG were from Dako. The rabbit anti-sheep 650 antibody was obtained from eBioscience. Normal human serum was prepared from freshly drawn blood obtained from healthy volunteers with informed consent and according to the recommendations of the local ethical committee in Lund (permit 418/2008). The pooled blood was allowed to clot for 30 min at room temperature and then incubated for 1 h on ice. After two centrifugations, the serum fraction was frozen in aliquots and stored at −80 °C or used for preparing IgG-depleted serum. For IgG depletion, fresh serum was passed through a Protein G column (GE Healthcare). The flow-through was analyzed for IgG depletion, and the fractions lacking IgG were pooled and frozen in −80 °C. The Superose 12 HR10/30 column was from GE Healthcare.
Direct Binding Assays
Microtiter plates were coated with plasminogen (10 μg/ml) or rC4BP (10 μg/ml) in PBS overnight at 4 °C. Wells coated with BSA were used as control. The plates were washed with 50 mm Tris-HCl (pH 8.0), 150 mm NaCl, and 0.1% Tween 20 between each step. The plates were then blocked with 250 μl of blocking solution (50 mm Tris HCl (pH 8.0), 150 mm NaCl, 0.1% Tween 20, 3% fish gelatin (Norland)) for 2 h at room temperature. After blocking, the plate was incubated with increasing concentrations of plasma-purified C4BP-PS or rC4BP for binding to plasminogen or with plasma-purified plasminogen for binding to rC4BP in binding buffer (50 mm HEPES (pH 7.4), 150 mm NaCl, 2 mm CaCl2, and 50 μg/ml of BSA) and incubated for 1 h 30 min at room temperature. Fixed concentrations (25 μg/ml) of C4BP-PS, C4BP–β/–PS, total C4BP or rC4BP (wild-type and mutants lacking individual domains) in TBS were added to the plate for binding to coated plasminogen and incubated for 1 h 30 min at room temperature or overnight at 4 °C. To determine the effect of EDTA or calcium on C4BP-PS binding to plasminogen, the assay was performed in binding buffer with or without 10 mm EDTA or 2 mm CaCl2. For investigation of the effect of ionic strength and the presence of lysine analog ϵ-ACA (ϵ-aminocaproic acid) (Sigma) on C4BP-plasminogen interactions, the binding buffer was supplemented with additional NaCl or NaBr (0 to 800 mm) or with ϵ-ACA to a final concentration ranging from 0 to 10 mm. Bound C4BP or plasminogen were detected using specific polyclonal Abs, followed by a swine anti-rabbit or rabbit anti-sheep peroxidase-conjugated Abs, respectively (Dako). The plates were developed with o-phenylenediamine (Dako) substrate and H2O2, and the absorbance at 490 nm was measured using a Varian Cary 50 MPR Microplate Reader.
Biacore Analysis
To measure the kinetics of C4BP-PS or rC4BP binding to plasminogen, surface plasmon resonance analysis was performed using Biacore 2000. Plasma-purified plasminogen was diluted to 50 μg/ml in 10 mm sodium acetate (pH 4.0) and immobilized on the surface of a CM5 sensor chip to reach 2953 response units. All experiments were performed at a continuous flow rate of 30 μl/min using Biacore buffer (150 mm NaCl, 10 mm HEPES, 2.5 mm CaCl2, 0.002% Tween 20, pH 7.4). The analyte, C4BP-PS or rC4BP, was injected in a concentration gradient from 5.5 to 175 nm followed by two consecutive injections of 2 m NaCl and 100 mm HCl for regeneration. The obtained sensorgrams were analyzed using Bio-evaluation software 3.0 using a 1:1 Langmuir binding model of interaction with a drifting baseline.
Capture ELISA
Microtiter plates coated with polyclonal sheep anti-human plasminogen Abs (10 μg/ml) were blocked and a mixture of C4BP and plasminogen diluted in binding buffer was added at concentrations indicated in figures. After incubation for 1 h 30 min at room temperature, bound C4BP was detected using polyclonal anti-C4BP. The plates were developed with o-phenylenediamine substrate and H2O2 and the absorbance at 490 nm was measured.
Gel Filtration
Pooled human plasma, physiological concentrations of rC4BP, plasminogen, or a mixture of rC4BP and plasminogen (0.2 mg/ml each) after overnight incubation at 37 °C were run on a Superose 12 gel filtration column in TBS. Collected fractions were tested for complexes of C4BP and plasminogen using ELISA.
Plasminogen-C4BP Sandwich ELISA
Microtiter plates were coated with anti-plasminogen capture antibody overnight at 4 °C. After blocking samples were incubated for 1 h at room temperature. C4BP was detected using monoclonal mk104 anti-C4BP antibody followed by goat anti-mouse antibody conjugated with HRP. The plates were developed with o-phenylenediamine substrate and H2O2 and the absorbance at 490 nm was measured.
Binding from Serum or Plasma
To investigate if C4BP-plasminogen complexes are present in human blood, microtiter plates were coated with polyclonal anti-C4BP (10 μg/ml). After blocking, human serum or human plasma in binding buffer was added at various concentrations for 1 h 30 min at room temperature. Bound C4BP-plasminogen complexes were detected using polyclonal sheep anti-human plasminogen Abs and rabbit anti-sheep peroxidase-conjugated antibodies (Dako).
Immunoprecipitation of C4BP-Plasminogen Complex from Serum
To demonstrate the presence of the C4BP-plasminogen complex, protein G beads pre-cleared 5% IgG-depleted serum was mixed with 2 μg of monoclonal anti-plasminogen Abs and protein G-Sepharose beads. Mouse IgG1 isotype control was used as a negative control. After incubation for 2 h at room temperature, samples were thoroughly washed with PBS containing 0.05% Tween 20. After washing, the samples were mixed with Laemmli buffer and boiled for 4 min at 96 °C. Finally the samples were subjected to SDS-PAGE, transferred to a polyvinylidene difluoride (PVDF) membrane, and submitted to Western blotting using a rabbit polyclonal anti-C4BP Abs and a secondary peroxidase-conjugated Abs. As positive control rC4BP was run on the same gel. The membrane was developed using ImmobilonTM Western chemiluminescence HRP substrate (Merck Millipore) and ChemiDoc Imaging system (Bio-Rad).
Plasminogen Activation Assay
To assess the effect of C4BP on the activation of plasminogen by uPA, plasminogen (0.5 μg/well) was incubated with or without C4BP-PS (5 μg/ml) in fluid phase. After incubation for 30 min at 37 °C, uPA (Sigma) (0.25 units/μl) was added and the activity of the newly generated plasmin was assayed with chromogenic substrate S-2251 (H-d-Valyl-l-leucyl-l-lysine-p-nitroaniline dihydrochloride) (Chromogenix). The plate was incubated at 37 °C, and cleavage of the chromogenic substrate was followed for the time periods indicated in the figure by measuring the absorbance at 405 nm. Plasminogen without uPA and uPA alone were used as controls.
Flow Cytometric Analysis of C4BP Binding to Pneumococci
Binding of plasma-purified C4BP and rC4BP in the presence or absence of plasminogen, and plasminogen in the presence or absence of rC4BP, to Streptococcus pneumoniae NCTC10319 was measured using flow cytometry. Pneumococci were cultured overnight on blood agar plates, washed in PBS, and adjusted to 109 cfu/ml. Bacteria (1 × 108 cfu) were incubated first with purified plasminogen or C4BP in 100 μl of PBS, followed by C4BP-PS and rC4BP or plasminogen for 30 min at 37 °C, respectively. Bound C4BP was detected using monoclonal anti-C4BP Abs (mk104) and Alexa Fluor 488-conjugated anti-mouse IgG for 45 min each on ice in the dark. Bound plasminogen was detected using sheep anti-plasminogen followed by Alexa Fluor 650-conjugated donkey anti-sheep IgG for 30 min each. The samples were washed between every step. Finally, bacteria were fixed using 1% paraformaldehyde (Sigma) and flow cytometry analysis was performed using CyFlow space (Partec) to detect the binding of C4BP. Bacteria were detected using log-forward and log-side scatter dot plots, and a gating region was set to exclude debris and larger aggregates of bacteria; 15,000 bacteria/events were analyzed for fluorescence using log-scale amplifications. The geometric mean fluorescence intensity was used as a measure for binding activity.
Results
Synergistic Binding of C4BP and Plasminogen to S. pneumoniae
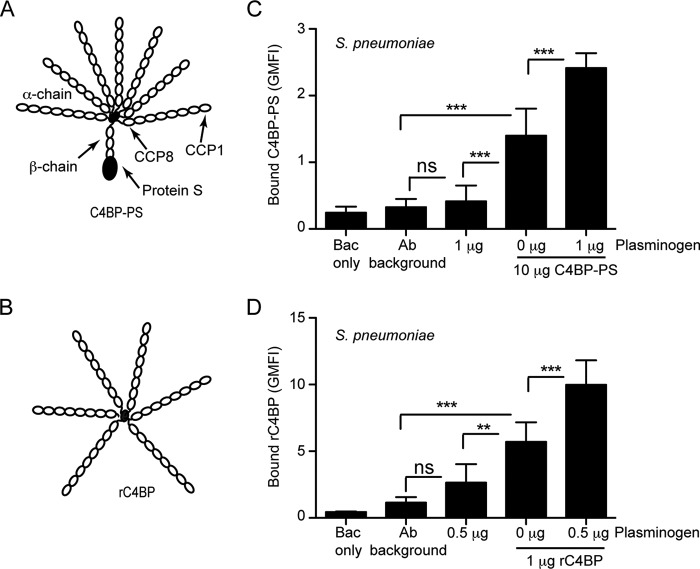
Pathogenic bacteria often recruit both complement inhibitor C4BP and plasminogen to their surface to facilitate complement evasion and invasion into tissues (26–32). Interestingly these pathogens often use the same receptor for binding of these two serum proteins (26–28, 31). One such pathogen that binds both C4BP and plasminogen is S. pneumoniae (26–28). Therefore we wondered whether the simultaneous presence of these proteins would have any synergistic effect on the binding efficiency of the other. To test this, pneumococci were incubated with plasminogen followed by addition of plasma-purified C4BP-PS or recombinant rC4BP (Fig. 1, A and B). C4BP readily bound pneumococci, however, in the presence of plasminogen the binding was significantly increased compared with pneumococci that had not been pre-treated with plasminogen (Fig. 1, C and D). The data suggest that the presence of plasminogen promotes C4BP binding over its basal level of binding to the pneumococcal surface. The same trend, although not statistically significant, was observed for increased plasminogen binding to the pneumococcal surface in the presence of C4BP (data not shown).
FIGURE 1.
Additive binding of C4BP and plasminogen to S. pneumoniae. A and B, schematic representation of the different variants of C4BP used: C4BP-PS (A) and rC4BP (B). The main isoform of C4BP contains seven identical α-chains and one unique β-chain. The β-chain-containing C4BP in circulation is bound to vitamin K-dependent anticoagulant Protein S (PS), forming a C4BP-PS complex. The rC4BP contains six α-chains and lacks the β-chain and the associated PS. C and D, increased binding of C4BP to pneumococci in the presence of plasminogen. Binding of C4BP-PS (C) and rC4BP (D) to pneumococci was analyzed in the presence or absence of plasminogen, by flow cytometry. The geometric mean fluorescence intensity (GMFI) and S.D. calculated from three independent experiments are shown. Bac only represents bacteria that were neither incubated with protein nor Abs, whereas the Ab background represents bacteria incubated with Abs only. A one-way analysis of variance test was used to calculate statistical significance. ns, not significant; **, p < 0.01; ***, p < 0.001.
C4BP Binds Plasminogen
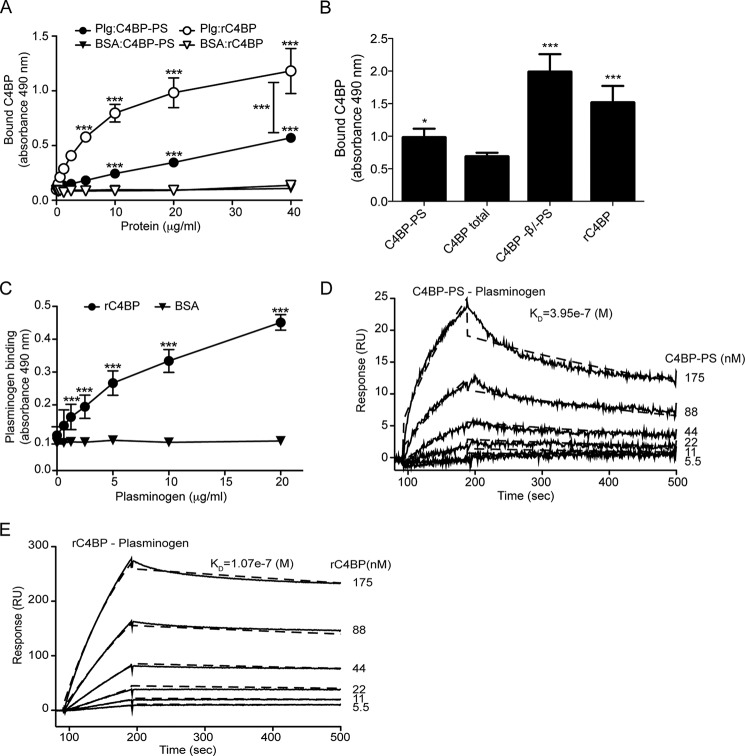
The above results showed that the presence of plasminogen enhances C4BP binding to the bacterial surface. However, it is not clear whether the enhanced C4BP binding observed is due to direct plasminogen-C4BP interaction or due to some indirect effect on the pneumococcal surface proteome. To test this, microtiter plates were coated with plasminogen and incubated with increasing concentrations of plasma-purified C4BP-PS or rC4BP (Fig. 1, A and B) followed by detection using specific antibodies. A dose-dependent binding of C4BP to plasminogen was observed (Fig. 2A). Interestingly, the binding of rC4BP lacking the β-chain was significantly stronger compared with plasma-purified C4BP-PS. This was confirmed by testing the ability of immobilized plasminogen to bind the acute phase C4BP variant, without the β-chain and protein S, which bound significantly better to plasminogen than C4BP-PS complex. Similarly strong binding was detected for rC4BP composed exclusively of 6 α-chains. The binding of total C4BP, which mainly contains the C4BP-PS variant, was similar to the binding of purified C4BP-PS complex (Fig. 2B). In addition, the binding of plasminogen to immobilized rC4BP was also dose-dependent (Fig. 2C). The affinity of the interaction between C4BP (C4BP-PS and rC4BP) and plasminogen was estimated using Biacore (Fig. 2, D and E). A concentration-dependent, high-affinity binding of C4BP to immobilized plasminogen was detected. The obtained sensorgrams yielded a KD of 39.5 μm for the interaction of plasma-purified C4BP-PS to plasminogen (Fig. 2B) and a KD of 10.7 μm for rC4BP binding to immobilized plasminogen (Fig. 2C). Representative sensorgrams show much lower response units for C4BP-PS binding to plasminogen compared with rC4BP, indicating that only a fraction could bind to the immobilized plasminogen. These data confirm that C4BP binds plasminogen and that the binding site for plasminogen is localized to the α-chains of C4BP, and moreover that plasminogen binds preferably to the acute phase C4BP form lacking the β-chain and protein S.
FIGURE 2.
Binding of C4BP to plasminogen. Microtiter plates were coated with plasminogen (10 μg/ml) and (A) increasing amounts of plasma-purified C4BP-PS, or rC4BP, were added. Binding was detected using specific polyclonal Abs. BSA was used as negative control. B, binding of C4BP-PS complex, C4BP total (all forms found in blood, predominantly C4BP-PS complex), C4BP without β-chain and protein S (−β/−PS), and rC4BP (exclusively α-chains), all at 25 μg/ml, was measured. C, microplates were coated with rC4BP, and increasing concentrations of plasminogen were added. BSA was used as control. Mean ± S.D. of three independent experiments performed in duplicates are presented. Statistical significance was calculated using two-way analysis of variance test; ***, p < 0.001; *, p < 0.01. Binding of C4BP-PS (D) and rC4BP (E) to immobilized plasminogen as analyzed by surface plasmon resonance. Increasing concentrations of C4BP (5.5–175 nm) were injected onto a plasminogen-coated CM5 sensor chip. The amount of C4BP associating with plasminogen was measured in response units (RU). Representative sensorgrams are presented.
C4BP and Plasminogen Readily Form Complexes in Fluid Phase, Which Are Present in Normal Human Serum and Plasma
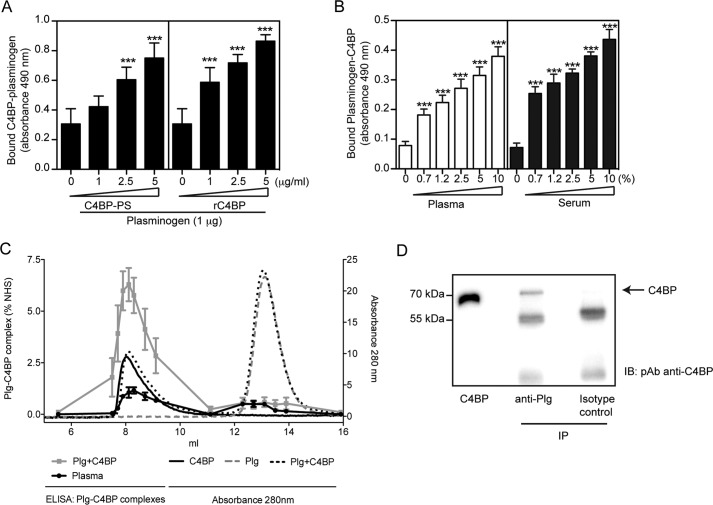
The above data suggest that both proteins interact when either of them is immobilized. However, we asked whether C4BP and plasminogen can readily form complexes in fluid phase and if so, whether this complex exists in serum or plasma. To demonstrate this, a capture ELISA was performed, whereby mixtures of various concentrations of either plasma-purified C4BP-PS or rC4BP (0 to 5 μg) and plasminogen (1 μg) were added to microtiter plates coated with polyclonal anti-plasminogen (10 μg/ml) Ab, and captured plasminogen in complex with C4BP was detected using specific polyclonal anti-C4BP Abs. A significant dose-dependent binding of C4BP-plasminogen complex to anti-plasminogen Ab was detected (Fig. 3A). To confirm these results and to verify that the complex exists in serum or plasma, microtiter plates coated with polyclonal anti-C4BP were incubated with various dilutions of human serum or plasma, and the deposited plasminogen-C4BP complexes were detected using polyclonal anti-plasminogen Abs. Similar to purified proteins, a dose-dependent binding of plasminogen-C4BP complex was detected from both human serum and plasma (Fig. 3B). Complexes were also formed when purified rC4BP and plasminogen were incubated at physiological concentrations (0.2 mg/ml for each) at 37 °C. Using gel filtration, plasminogen in complex with C4BP was separated from free plasminogen and detected using a plasminogen-C4BP sandwich ELISA (Fig. 3C). Small amounts of complexes could also be detected in the same fractions when human plasma was separated by gel filtration (Fig. 3C). This was confirmed by immunoprecipitation and Western blotting (Fig. 3D). Taken together, our data confirm that C4BP-plasminogen readily forms complexes that are present in human plasma.
FIGURE 3.
C4BP forms complexes with plasminogen. A and B, interaction between C4BP and plasminogen. Polyclonal sheep anti-plasminogen (10 μg/ml, A) or polyclonal anti-C4BP Ab (10 μg/ml, B) were coated on 96-well microtiter plates. A mixture of plasminogen with plasma-purified C4BP-PS or rC4BP (A), or normal plasma or serum (B) were added. Bound C4BP-plasminogen complexes were detected with a polyclonal rabbit anti-C4BP Ab (A) and sheep anti-plasminogen Ab (B) and peroxidase-conjugated respective secondary Abs and substrate. A one-way analysis of variance test was used to calculate statistical significance. ns, not significant; ***, p < 0.001. C, rC4BP (0.2 mg/ml) and plasminogen (0.2 mg/ml) were incubated separately or together overnight at 37 °C and analyzed on a Superose 12 gel filtration column. The A280 nm for rC4BP (black solid line), plasminogen (gray dotted line), and rC4BP with plasminogen (black dotted line) are shown. Complexes of rC4BP and plasminogen were measured using a sandwich ELISA and plotted as percentage of complex present in normal human serum (gray squares). Furthermore, ELISA was used to detect C4BP-plasminogen complexes in pooled human plasma (black dots). ELISA data represent mean ± S.D. of three independent experiments performed in duplicates. D, protein G-coated-Sepharose beads where incubated with a monoclonal anti-plasminogen Ab, and then incubated with IgG-depleted normal serum. C4BP was detected by Western blotting under reduced conditions using a polyclonal anti-C4BP Ab. Isotype control Ab was used as negative control.
Role of Lysine Residues on C4BP Binding to Plasminogen
The lysine-binding kringle domains of plasminogen mediate binding to fibrin, components of the ECM, or other lysine-containing host proteins (33, 34). To determine whether lysine residues in C4BP are involved with binding of plasminogen, increasing concentrations of the lysine analog ϵ-ACA were used. ϵ-ACA, when added at 0.15 mm, decreased the C4BP-PS-plasminogen interaction by 75% and the rC4BP-plasminogen interaction by more than 70%, whereas the addition of 10 mm ϵ-ACA reduced the signal to background level (Fig. 4A).
FIGURE 4.
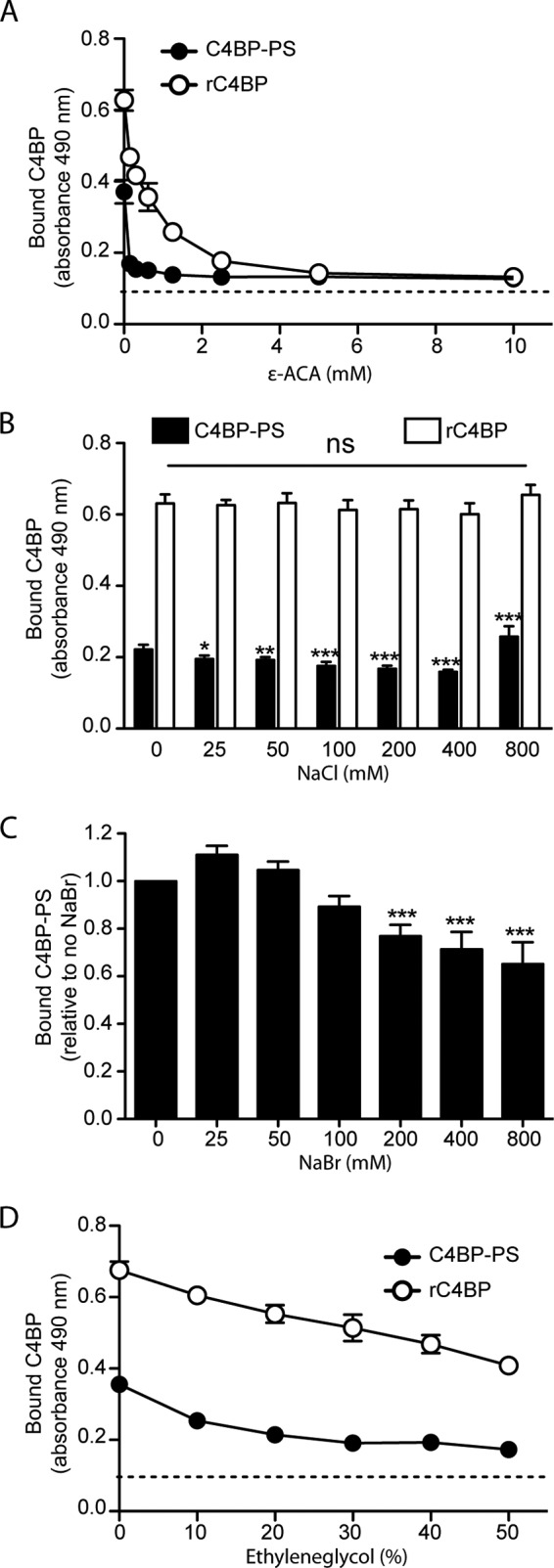
The C4BP-plasminogen interaction is mediated by lysine residues and is partially ionic and hydrophobic in nature. A, microtiter plates were coated with plasminogen (10 μg/ml) and a constant amount of C4BP-PS or rC4BP was added in the presence of increasing concentrations of the lysine analog ϵ-ACA. Binding of C4BP-PS (B–D) or rC4BP (B and D) was analyzed under increasing amounts of NaCl (B), NaBr (C), or ethylene glycol (D). Bound C4BP was detected using a specific polyclonal Abs. Dotted line represents the Ab background. Data represent mean ± S.D. of three independent experiments conducted in duplicates. Statistical significance was calculated using one-way analysis of variance and Dunnett's post-test to compare the binding in the absence of inhibitor; ns, not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001.
The Binding of C4BP to Plasminogen Is Partially of Ionic Nature
The effect of ionic strength on the C4BP-plasminogen interaction was also investigated. Binding of C4BP-PS and rC4BP to plasminogen was measured in the presence of increasing concentrations of NaCl. At a NaCl concentration of 400 mm higher than the physiological concentration, a 30% reduction in the binding of plasma purified C4BP-PS was observed, whereas no such effect was observed for rC4BP (Fig. 4B). Interestingly, at 800 mm NaCl an increase in binding of C4BP-PS to plasminogen was observed, which could be due to the effect on the hydrophobic part of the interaction that tends to increase with high salt. Moreover, even at 6 times the physiological concentration of NaCl, the affinity of rC4BP for plasminogen remained unaffected (Fig. 4B). Because plasminogen is known to bind chloride anions, it is likely that changing the ionic strength with NaCl was not sufficient. We therefore studied the effect of NaBr on C4BP-PS binding to plasminogen (Fig. 4C). Starting at a concentration of 200 mm a statistically significant reduction in C4BP-PS plasminogen binding was detected. A reduction of almost 25% was observed in binding in the presence of 200 mm NaBr compared with the absence of salt. These data suggest that ionic strength does have an impact on the C4BP-plasminogen interaction. Interestingly, the absence of Ca2+ or the presence of EDTA had no effect on binding efficiency (data not shown).
Additionally, to estimate the influence of hydrophobic interactions between C4BP and plasminogen, we incubated C4BP-PS or rC4BP with immobilized plasminogen in the presence of increasing amounts of ethylene glycol. We found that a concentration of 20% ethylene glycol already reduced by ∼55 and ∼20% the binding of C4BP-PS and rC4BP to plasminogen, respectively (Fig. 4D). Interestingly, increasing the concentration of ethylene glycol to 50% further reduced the respective binding by ∼70 and ∼45% compared with binding in the absence of ethylene glycol. Taken together, the interaction between C4BP and plasminogen seems to depend on both electrostatic as well as hydrophobic interactions.
Localization of Plasminogen Binding Site on C4BP
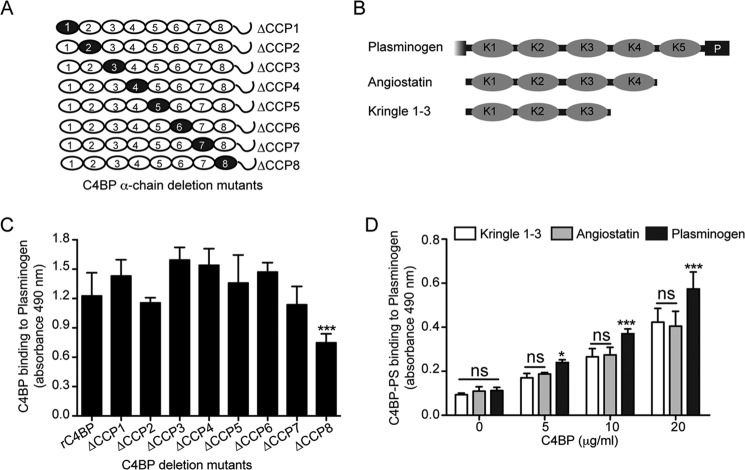
We further characterized the interaction between C4BP and plasminogen using different forms of C4BP, which are represented schematically in Figs. 1, A and B, and 5A. Strong binding of rC4BP to plasminogen indicated that the binding site is localized to the α-chains of C4BP (Fig. 1C). Binding of rC4BP and C4BP mutants lacking one CCP domain at a time showed that mutant ΔCCP8 had significantly decreased binding capacities for plasminogen (Fig. 5C). This result suggests that C4BP contacts plasminogen via binding sites, which are primarily localized within CCP8 of the α-chain. This further explains the weaker binding observed using plasma-purified C4BP-PS compared with the rC4BP, where the β-chain sterically hinders the interaction meditated via CCP8.
FIGURE 5.
Localization of binding domains within C4BP and plasminogen. A and B, schematic representation of different C4BP α-chain deletion mutants (A) and various plasminogen derivatives (B) used. Black circles within each α-chain represent the C4BP α-chain deletion mutants lacking single CCP domains. Although for plasminogen, the kringle domains are depicted by K1 to K5 and the serine protease domain is depicted by P. C, the C4BP variants were allowed to bind to plasminogen immobilized on a plate. Bound C4BP was detected with polyclonal Abs. The graph represents data from three independent experiments done in duplicates ± S.D. Statistical significance was calculated using a one-way analysis of variance test. **, p = 0.01; ***, p = 0.001. D, microtiter plates were coated with variants of plasminogen (10 μg/ml) and incubated with plasma-purified C4BP-PS. Bound C4BP was detected using specific Abs. The data represent mean ± S.D. of three independent experiments performed in duplicate. Statistical significance was calculated using a two-way analysis of variance test. ns, not significant; *, p < 0.05; ***, p < 0.001.
All but Kringle-4 Mediates Plasminogen Binding to C4BP
Binding of plasminogen to host proteins is mediated by the five kringle domains of plasminogen (33, 35). To narrow down the domain(s) of plasminogen required for the interaction with C4BP, plasminogen fragments comprising of various lysine-binding kringle domains were assayed for their abilities to bind C4BP-PS. As can be observed in Fig. 5, B and D, the constructs consisting of kringle 1–3 and kringle 1–4 showed no difference in binding, however, this binding was relatively weaker compared with the full-length plasminogen containing all five domains and the protease domain. Thus, based on the available constructs our data suggest kringle domains 1–3 and 5, but not kringle domain 4, contain the probable binding sites for C4BP (Fig. 5D).
Presence of C4BP Enhances the Activation of Plasminogen to Active Plasmin
During fibrinolysis plasminogen is converted to the serine protease plasmin by plasminogen activators, such as uPA or tissue-type plasminogen activator (36). Moreover, because C4BP readily forms a complex with plasminogen, we asked whether the presence of plasma-purified C4BP-PS would affect the extent of plasminogen activation by its activator uPA, and subsequent conversion to the active protease plasmin. Plasminogen was therefore preincubated with or without plasma-purified C4BP-PS, and then incubated with uPA, and the generated plasmin activity was evaluated by incubating with the chromogenic substrate S-2251. A time-dependent cleavage of the substrate by the generated plasmin was observed (Fig. 6A and B). Interestingly, the presence of C4BP-PS significantly enhanced the conversion of plasminogen to active plasmin, as compared with the condition when C4BP-PS was not present (Fig. 6, A and B). Moreover, no cleavage of the substrate was observed when plasminogen was incubated with only C4BP-PS without addition of uPA, or in the presence of only uPA, demonstrating the specific protease activity of the generated plasmin. A similar enhanced activation of plasminogen to plasmin was observed in the presence of rC4BP, whereas the incubation of C4BP-PS or rC4BP with uPA alone did not initiate the cleavage of the substrate (data not shown). Interestingly, the presence of plasminogen did not affect the cofactor activity of C4BP as determined by FI-mediated C4b degradation (data not shown). Taken together, the interaction between C4BP and plasminogen enhances the activation of plasminogen to active serine protease plasmin and therefore suggest an important unidentified role in maintaining homeostasis.
FIGURE 6.
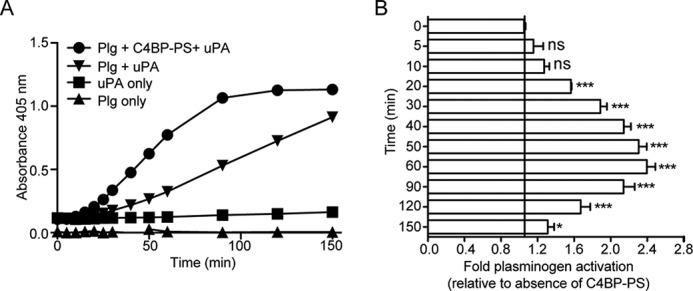
C4BP enhances plasminogen activation in fluid phase. A and B, activation of plasminogen (Plg) to active serine protease plasmin was determined using the chromogenic substrate S-2251. Plasminogen incubated with or without plasma-purified C4BP-PS was mixed together with uPA and S-2251 and the plate was read at 405 nm in 10-min intervals up to 150 min. uPA or plasminogen alone incubated with S-2251 were used as negative control. A representative graph of three independent experiments is shown (A) and the fold-activation of plasminogen in the presence of C4BP-PS compared with the absence is presented as mean ± S.D. of three independent experiments conducted in duplicate (B). Statistical significance was calculated using a one-way analysis of variance and Dunnett's post test to compare the binding in the absence of inhibitor; ns, not significant; *, p < 0.05; ***, p < 0.001.
Discussion
Tissue injury, trauma, or systemic inflammation are considered major triggering factors for the activation of complement as well as the coagulation and fibrinolysis pathways, which are crucial for the maintenance of tissue homeostasis (19–21). Although complement as a part of the innate immune system majorly helps in protection from infections as well as clearance of unwanted debris such as dying cells, the coagulation and fibrinolytic systems work in tandem to repair the damaged blood vessels. Complement, in addition to its primary role in neutralization of invading pathogens, also orchestrates various immunological and inflammatory processes (1, 2). During the recent decade, studies have demonstrated multiple interactions and cross-talks between the complement and coagulation pathways. Complement has been shown to potentiate the coagulation cascade by inhibiting fibrinolysis, through C5a, which induces the expression of tissue factor and plasminogen-activator inhibitor 1 (37), whereas proteases of the coagulation and fibrinolytic system were shown to cleave and activate C3 and generate C5a (21, 38). Additionally, during infections, localized complement-mediated coagulation activation enhances clotting, which not only provides a mechanical barrier against the spread of invading bacteria, but also helps in generating antimicrobial peptides and support the inflammatory response (21, 39).
However, to restrict the action of the complement, coagulation, and fibrinolytic systems to the site of injury, the cascades are tightly regulated by numerous soluble as well as cell surface inhibitors (2–4, 14). C4BP is a major soluble inhibitor of the classical and lectin complement pathways, whereas plasminogen is a circulating zymogen for broad-spectrum serine protease plasmin, which mediates fibrin clot lysis. Additionally, plasmin(ogen) has also been characterized as a complement inhibitor (40). Plasminogen not only binds complement proteins such as C3 and C5, but has also been shown to cleave C3b and C5 upon activation to active plasmin (40). Interestingly, many pathogenic microorganisms acquire these inhibitors (including but not limited to C4BP and plasminogen) to establish an infection and evade the constant attack of complement (27, 29, 41–44).
Here in this study, we describe for the first time a novel interaction between C4BP and plasminogen. A dose-dependent binding to plasminogen was observed for plasma-purified full-length C4BP, which contains seven identical α-chains and one unique β-chain with attached vitamin K-dependent protein S (C4BP-PS), as well as for the recombinant C4BP (rC4BP), consisting of six α-chains. The specificity of the interaction between C4BP and plasminogen was demonstrated by Biacore studies, capture ELISA, and gel filtration, which demonstrated that the proteins readily form a complex in fluid phase and that the complexes are also present in both plasma and serum. Moreover, the co-immunoprecipitation studies using anti-plasminogen Ab further confirmed the C4BP-plasminogen interaction.
The interaction with both variants of C4BP indicated that the binding site for plasminogen is localized to the α-chains of C4BP. Interestingly, the binding of rC4BP was stronger compared with that of C4BP-PS, as observed in the direct binding assay and the Biacore analysis, suggesting that the presence of β-chain and protein S has an inhibitory effect on the affinity of C4BP to plasminogen. Similar differential binding was obtained using different physiological isoforms of C4BP (−β/−PS and total), where the strongest binding was observed for the isoform lacking the β-chain, similar to that of rC4BP. Indeed, our binding experiment with rC4BP mutants, each deficient in one CCP domain, indicated the presence of binding sites in domain CCP8, which would otherwise be masked by the β-chain.
C4BP is an acute-phase protein. Normally in the plasma of healthy individuals, the α7β1 oligomer is the most abundant isoform of C4BP (46). However, during the acute inflammatory phase, the α- and β-chains of C4BP undergo differential regulation resulting in expression of the C4BP protein without β-chain (i.e. C4BP with α7β0). This in turn results in a molar excess of free anticoagulant PS over PS bound to C4BP, despite high levels of C4BP in circulation. Our data demonstrate that plasminogen has preferential high affinity for the C4BP isoform lacking the β-chain, and in addition, the presence C4BP substantially enhances the activation of zymogen plasminogen to the active serine protease plasmin. Therefore, we hypothesize that this interaction might potentially play a role in regulating the fibrinolysis pathway and complement activation at the site of injury or during acute inflammation.
Plasminogen primarily interacts with the components of the fibrinolytic system and other serum proteins, including bacterial ligands, via lysine binding sites located within the kringle domains (45). Interestingly, although lysine residues play a role in the C4BP-plasminogen interaction, alteration of ionic strength with NaCl had a negligible effect on C4BP binding. However, in contrast to NaCl, the presence of NaBr had a significant effect on C4BP binding to plasminogen, suggesting that the interaction is partially dependent on ionic strength. Interestingly, the alteration of the hydrophobicity by ethylene glycol had a drastic effect on the binding of C4BP to plasminogen. Taken together, the C4BP-plasminogen interaction seems to depend on a combined effect of hydrophobic and ionic interactions.
S. pneumoniae recruits both C4BP and plasminogen through multiple surface-exposed proteins such as enolase, PepO, PspC, as well as multiple other plasminogen-binding proteins. We have previously shown that C4BP and plasminogen bind to different sites on enolase (26, 27). However, it is not clear if it is also true for other C4BP and plasminogen-binding pneumococcal proteins. Moreover in this study, we demonstrate that C4BP and plasminogen form complexes both in serum/plasma as well as using purified proteins. We observed an enhanced binding of C4BP to the pneumococcal surface in the presence of plasminogen, which is primarily due to the ability of these two proteins to interact directly. However, the bacterial protein to which plasminogen is binding could be any of the multiple plasminogen-binding pneumococcal proteins (and not just enolase alone). Additionally, whereas the pathogens could benefit from the surge in the relative amount of complement inhibitors on their surfaces, further investigations are required to completely understand the implication of this interaction. Taken together, this novel interaction opens a new chapter in the cross-talk between the complement and coagulation and fibrinolytic systems. Therefore, more studies and in-depth analysis are required to completely understand the implications of this interaction both on the process of homeostasis as well as on the pathogenesis of microorganisms.
Author Contributions
V. A. and A. M. B. conceived and coordinated the study and wrote the paper. V. A. and A. M. G. performed the majority of presented experiments. S. T. performed experiments requested during revisions. All authors reviewed the results and approved the final version of the manuscript.
This work was supported in part by Swedish Research Council Grant K2012-66X-14928-09-5, Foundations of Torsten Söderberg, Österlund, Greta and Johan Kock, King Gustav V's 80th Anniversary, Knut and Alice Wallenberg, Inga-Britt and Arne Lundberg, Åke Wiberg, Royal Physiographic Society in Lund, and the Skåne University Hospital. The authors declare that they have no conflicts of interest with the contents of this article.
- C4BP
- C4b-binding protein
- CCP
- complement control protein
- ϵ-ACA
- ϵ-aminocaproic acid
- uPA
- urokinase-type plasminogen activator.
References
- 1. Holers V. M. (2014) Complement and its receptors: new insights into human disease. Annu. Rev. Immunol. 32, 433–459 [DOI] [PubMed] [Google Scholar]
- 2. Ricklin D., Hajishengallis G., Yang K., Lambris J. D. (2010) Complement: a key system for immune surveillance and homeostasis. Nat. Immunol. 11, 785–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sjöberg A. P., Trouw L. A., Blom A. M. (2009) Complement activation and inhibition: a delicate balance. Trends Immunol. 30, 83–90 [DOI] [PubMed] [Google Scholar]
- 4. Zipfel P. F., Skerka C. (2009) Complement regulators and inhibitory proteins. Nat. Rev. Immunol. 9, 729–740 [DOI] [PubMed] [Google Scholar]
- 5. Fujita T., Gigli I., Nussenzweig V. (1978) Human C4-binding protein: II. role in proteolysis of C4b by C3b-inactivator. J. Exp. Med. 148, 1044–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fujita T., Nussenzweig V. (1979) The role of C4-binding protein and β1H in proteolysis of C4b and C3b. J. Exp. Med. 150, 267–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gigli I., Fujita T., Nussenzweig V. (1979) Modulation of the classical pathway C3 convertase by plasma proteins C4 binding protein and C3b inactivator. Proc. Natl. Acad. Sci. U.S.A. 76, 6596–6600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dahlbäck B. (1991) Protein S and C4b-binding protein: components involved in the regulation of the protein C anticoagulant system. Thromb. Haemost. 66, 49–61 [PubMed] [Google Scholar]
- 9. Dahlbäck B., Stenflo J. (1981) High molecular weight complex in human plasma between vitamin K-dependent protein S and complement component C4b-binding protein. Proc. Natl. Acad. Sci. U.S.A. 78, 2512–2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Norman D. G., Barlow P. N., Baron M., Day A. J., Sim R. B., Campbell I. D. (1991) Three-dimensional structure of a complement control protein module in solution. J. Mol. Biol. 219, 717–725 [DOI] [PubMed] [Google Scholar]
- 11. Wolberg A. S. (2007) Thrombin generation and fibrin clot structure. Blood Rev. 21, 131–142 [DOI] [PubMed] [Google Scholar]
- 12. Wolberg A. S., Campbell R. A. (2008) Thrombin generation, fibrin clot formation and hemostasis. Transfus. Apher. Sci. 38, 15–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rosenberg R. D., Rosenberg J. S. (1984) Natural anticoagulant mechanisms. J. Clin. Investig. 74, 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cesarman-Maus G., Hajjar K. A. (2005) Molecular mechanisms of fibrinolysis. Br. J. Haematol. 129, 307–321 [DOI] [PubMed] [Google Scholar]
- 15. Miyashita C., Wenzel E., Heiden M. (1988) Plasminogen: a brief introduction into its biochemistry and function. Haemostasis 18, 7–13 [DOI] [PubMed] [Google Scholar]
- 16. Chavakis T., Athanasopoulos A., Rhee J. S., Orlova V., Schmidt-Wöll T., Bierhaus A., May A. E., Celik I., Nawroth P. P., Preissner K. T. (2005) Angiostatin is a novel anti-inflammatory factor by inhibiting leukocyte recruitment. Blood 105, 1036–1043 [DOI] [PubMed] [Google Scholar]
- 17. Duboscq C., Genoud V., Parborell M. F., Kordich L. C. (1997) Impaired clot lysis by rt-PA catalyzed mini-plasminogen activation. Thromb. Res. 86, 505–513 [DOI] [PubMed] [Google Scholar]
- 18. Castellino F. J., Powell J. R. (1981) Human plasminogen. Methods Enzymol. 80, 365–378 [DOI] [PubMed] [Google Scholar]
- 19. Amara U., Flierl M. A., Rittirsch D., Klos A., Chen H., Acker B., Brückner U. B., Nilsson B., Gebhard F., Lambris J. D., Huber-Lang M. (2010) Molecular intercommunication between the complement and coagulation systems. J. Immunol. 185, 5628–5636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Amara U., Rittirsch D., Flierl M., Bruckner U., Klos A., Gebhard F., Lambris J. D., Huber-Lang M. (2008) Interaction between the coagulation and complement system. Adv. Exp. Med. Biol. 632, 71–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Markiewski M. M., Nilsson B., Ekdahl K. N., Mollnes T. E., Lambris J. D. (2007) Complement and coagulation: strangers or partners in crime? Trends Immunol. 28, 184–192 [DOI] [PubMed] [Google Scholar]
- 22. Dahlbäck B. (1983) Purification of human C4b-binding protein and formation of its complex with vitamin K-dependent protein S. Biochem. J. 209, 847–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nilsson S. C., Trouw L. A., Renault N., Miteva M. A., Genel F., Zelazko M., Marquart H., Muller K., Sjöholm A. G., Truedsson L., Villoutreix B. O., Blom A. M. (2009) Genetic, molecular and functional analyses of complement factor I deficiency. Eur. J. Immunol. 39, 310–323 [DOI] [PubMed] [Google Scholar]
- 24. Blom A. M., Kask L., Dahlbäck B. (2001) Structural requirements for the complement regulatory activities of C4BP. J. Biol. Chem. 276, 27136–27144 [DOI] [PubMed] [Google Scholar]
- 25. Avirutnan P., Hauhart R. E., Somnuke P., Blom A. M., Diamond M. S., Atkinson J. P. (2011) Binding of flavivirus nonstructural protein NS1 to C4b binding protein modulates complement activation. J. Immunol. 187, 424–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Agarwal V., Hammerschmidt S., Malm S., Bergmann S., Riesbeck K., Blom A. M. (2012) Enolase of Streptococcus pneumoniae binds human complement inhibitor C4b-binding protein and contributes to complement evasion. J. Immunol. 189, 3575–3584 [DOI] [PubMed] [Google Scholar]
- 27. Agarwal V., Kuchipudi A., Fulde M., Riesbeck K., Bergmann S., Blom A. M. (2013) Streptococcus pneumoniae endopeptidase O (PepO) is a multifunctional plasminogen- and fibronectin-binding protein, facilitating evasion of innate immunity and invasion of host cells. J. Biol. Chem. 288, 6849–6863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Agarwal V., Sroka M., Fulde M., Bergmann S., Riesbeck K., Blom A. M. (2014) Binding of Streptococcus pneumoniae endopeptidase O (PepO) to complement component C1q modulates the complement attack and promotes host cell adherence. J. Biol. Chem. 289, 15833–15844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lambris J. D., Ricklin D., Geisbrecht B. V. (2008) Complement evasion by human pathogens. Nat. Rev. Microbiol. 6, 132–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Smeesters P. R., McMillan D. J., Sriprakash K. S. (2010) The streptococcal M protein: a highly versatile molecule. Trends Microbiol. 18, 275–282 [DOI] [PubMed] [Google Scholar]
- 31. Souza N. M., Vieira M. L., Alves I. J., de Morais Z. M., Vasconcellos S. A., Nascimento A. L. (2012) Lsa30, a novel adhesin of Leptospira interrogans binds human plasminogen and the complement regulator C4bp. Microbial Pathogenesis 53, 125–134 [DOI] [PubMed] [Google Scholar]
- 32. Bergmann S., Schoenen H., Hammerschmidt S. (2013) The interaction between bacterial enolase and plasminogen promotes adherence of Streptococcus pneumoniae to epithelial and endothelial cells. Int. J. Med. Microbiol. 303, 452–462 [DOI] [PubMed] [Google Scholar]
- 33. Angles-Cano E. (1994) Overview on fibrinolysis: plasminogen activation pathways on fibrin and cell surfaces. Chem. Phys. Lipids 67–68, 353–362 [DOI] [PubMed] [Google Scholar]
- 34. Ponting C. P., Marshall J. M., Cederholm-Williams S. A. (1992) Plasminogen: a structural review. Blood Coagul. Fibrinolysis 3, 605–614 [PubMed] [Google Scholar]
- 35. Plow E. F., Herren T., Redlitz A., Miles L. A., Hoover-Plow J. L. (1995) The cell biology of the plasminogen system. FASEB J. 9, 939–945 [DOI] [PubMed] [Google Scholar]
- 36. Castellino F. J., McCance S. G. (1997) The kringle domains of human plasminogen. Ciba Found. Symp. 212, 46–60; discussion 60–45 [DOI] [PubMed] [Google Scholar]
- 37. Ritis K., Doumas M., Mastellos D., Micheli A., Giaglis S., Magotti P., Rafail S., Kartalis G., Sideras P., Lambris J. D. (2006) A novel C5a receptor-tissue factor cross-talk in neutrophils links innate immunity to coagulation pathways. J. Immunol. 177, 4794–4802 [DOI] [PubMed] [Google Scholar]
- 38. Huber-Lang M., Sarma J. V., Zetoune F. S., Rittirsch D., Neff T. A., McGuire S. R., Lambris J. D., Warner R. L., Flierl M. A., Hoesel L. M., Gebhard F., Younger J. G., Drouin S. M., Wetsel R. A., Ward P. A. (2006) Generation of C5a in the absence of C3: a new complement activation pathway. Nat. Med. 12, 682–687 [DOI] [PubMed] [Google Scholar]
- 39. Frick I. M., Akesson P., Herwald H., Mörgelin M., Malmsten M., Nägler D. K., Björck L. (2006) The contact system: a novel branch of innate immunity generating antibacterial peptides. EMBO J. 25, 5569–5578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Barthel D., Schindler S., Zipfel P. F. (2012) Plasminogen is a complement inhibitor. J. Biol. Chem. 287, 18831–18842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Barthel D., Singh B., Riesbeck K., Zipfel P. F. (2012) Haemophilus influenzae uses the surface protein E to acquire human plasminogen and to evade innate immunity. J. Immunol. 188, 379–385 [DOI] [PubMed] [Google Scholar]
- 42. Blom A. M., Hallström T., Riesbeck K. (2009) Complement evasion strategies of pathogens-acquisition of inhibitors and beyond. Mol. Immunol. 46, 2808–2817 [DOI] [PubMed] [Google Scholar]
- 43. Koch T. K., Reuter M., Barthel D., Böhm S., van den Elsen J., Kraiczy P., Zipfel P. F., Skerka C. (2012) Staphylococcus aureus proteins Sbi and Efb recruit human plasmin to degrade complement C3 and C3b. PloS One 7, e47638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Koenigs A., Hammerschmidt C., Jutras B. L., Pogoryelov D., Barthel D., Skerka C., Kugelstadt D., Wallich R., Stevenson B., Zipfel P. F., Kraiczy P. (2013) BBA70 of Borrelia burgdorferi is a novel plasminogen-binding protein. J. Biol. Chem. 288, 25229–25243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wiman B., Lijnen H. R., Collen D. (1979) On the specific interaction between the lysine-binding sites in plasmin and complementary sites in α2-antiplasmin and in fibrinogen. Biochim. Biophys. Acta 579, 142–154 [DOI] [PubMed] [Google Scholar]
- 46. Hillarp A., Hessing M., Dahlbäck B. (1989) Protein S binding in relation to the subunit composition of human C4b-binding protein. FEBS Lett. 259, 53–56 [DOI] [PubMed] [Google Scholar]