Background: Hypusine modification of the eukaryotic initiation factor 5A (eIF-5A) represents a conserved post-translational modification that regulates translation.
Results: Deletion of hypusine modification enzymes exerts strong phenotypes. eIF-5A2-deleted animals are viable and fertile.
Conclusion: Both enzymatic steps of hypusine modification are essential for mammalian homeostasis, whereas the cancer-related isoform eIF-5A2 is dispensable.
Significance: eIF-5A2 might represent a safe therapeutic target.
Keywords: cancer therapy, eukaryotic initiation factor 5A (eIF5A), mouse genetics, post-translational modification (PTM), translation elongation factor
Abstract
Hypusine modification of the eukaryotic initiation factor 5A (eIF-5A) is emerging as a crucial regulator in cancer, infections, and inflammation. Although its contribution in translational regulation of proline repeat-rich proteins has been sufficiently demonstrated, its biological role in higher eukaryotes remains poorly understood. To establish the hypusine modification system as a novel platform for therapeutic strategies, we aimed to investigate its functional relevance in mammals by generating and using a range of new knock-out mouse models for the hypusine-modifying enzymes deoxyhypusine synthase and deoxyhypusine hydroxylase as well as for the cancer-related isoform eIF-5A2. We discovered that homozygous depletion of deoxyhypusine synthase and/or deoxyhypusine hydroxylase causes lethality in adult mice with different penetrance compared with haploinsufficiency. Network-based bioinformatic analysis of proline repeat-rich proteins, which are putative eIF-5A targets, revealed that these proteins are organized in highly connected protein-protein interaction networks. Hypusine-dependent translational control of essential proteins (hubs) and protein complexes inside these networks might explain the lethal phenotype observed after deletion of hypusine-modifying enzymes. Remarkably, our results also demonstrate that the cancer-associated isoform eIF-5A2 is dispensable for normal development and viability. Together, our results provide the first genetic evidence that the hypusine modification in eIF-5A is crucial for homeostasis in mammals. Moreover, these findings highlight functional diversity of the hypusine system compared with lower eukaryotes and indicate eIF-5A2 as a valuable and safe target for therapeutic intervention in cancer.
Introduction
Control of translational processes is essential to maintain cellular function and organismal integrity (1). In addition to the important role in normal physiology, there is accumulating evidence that alterations in the translation machinery (e.g. deregulated translation factors) lead to changes in protein biosynthesis and the development or progression of various diseases like cancer and viral infections (2, 3). Given that translation factors are frequently regulated by post-translational modifications, which are mediated by enzymes, those modifications can be harnessed therapeutically (4).
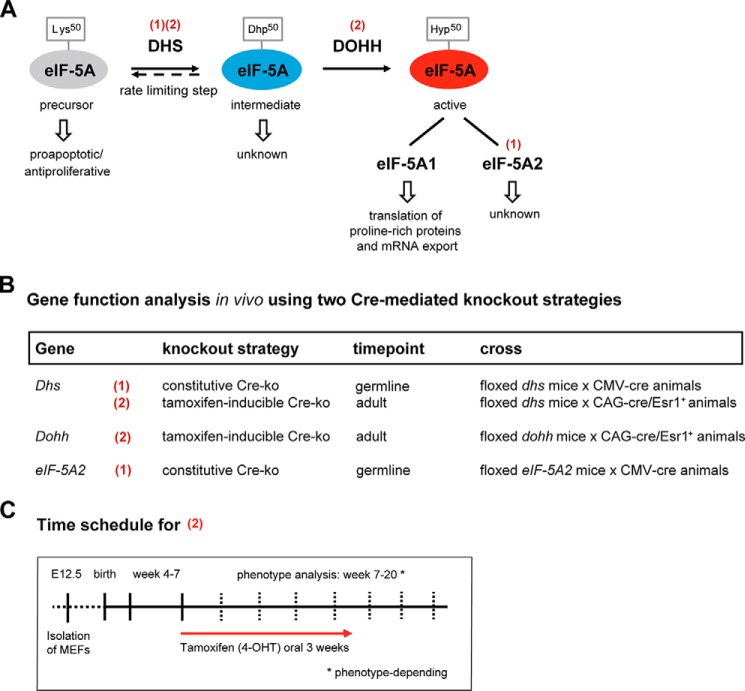
In this context, the eukaryotic initiation factor 5A (eIF-5A) represents a particularly interesting target for therapeutic intervention because it carries a highly specific post-translational modification, the unusual amino acid hypusine, which is unique in this protein (5, 6). The biosynthesis of hypusine is catalyzed from lysine in a two-step enzymatic reaction. First, the deoxyhypusine synthase (DHS)3 transfers a 4-aminobutyl moiety of spermidine to the ϵ-amino group of Lys50 to form a deoxyhypusine-containing intermediate. Second, the deoxyhypusine hydroxylase (DOHH) catalyzes the hydroxylation of the deoxyhypusine residue to generate hypusine-containing eIF-5A (Fig. 1A). Hypusine is mandatory for eIF-5A activity and function (7, 8), and only hypusinated eIF-5A was shown to control translation elongation as well as the nuclear export of specific mRNAs (9, 10). Moreover, recent studies linked eIF-5A to the translational control of proteins containing consecutive proline residues (11).
FIGURE 1.
Overview of the hypusine modification and experimental design to study its role in mammals. A, schematic representation of hypusine synthesis in eIF-5A catalyzed by the DHS and DOHH enzymes. Red numbers in parentheses indicate the knock-out strategy for the respective gene as outlined in B. Hyp, hypusine; Dhp, deoxyhypusine. B, conditional knock-out strategy using two Cre-deleter mouse strains for either an early constitutive (1) and/or a 4-OHT-inducible (2) knock-out of the respective gene. Cre mouse strains are as follows: (1) = B6.C-Tg(CMV-cre)1Cgn/J; (2) = B6.Cg-Tg(CAG-cre/Esr1*)5Amc/J113. C, time schedule for the 4-OHT-inducible knock-out (2) of the genes outlined in B.
Like other translation factors, eIF-5A and its modifying enzymes, DHS and DOHH, are highly conserved in eukaryotes as well as archaea. Prokaryotes express an orthologous factor, the elongation factor P (12). Although elongation factor P is not hypusine-modified, it is activated by a similar modification (lysinylation) at a comparable amino acid position (Lys34) and promotes translation of polyproline motifs in the same way as eIF-5A (13–15). Based on work in yeast, it seems that eIF-5A and its hypusine modification are essential in lower eukaryotes. However, a fundamental unanswered question is whether the hypusine axis is crucial for viability of mammals.
The observation that eIF-5A is also involved in diseases like cancer, infections, and diabetes highlights eIF-5A and particularly its hypusine modification as an attractive target for the development of new therapeutic strategies. Up-regulation of the eIF-5A1 isoform and the hypusine-forming enzymes can be found in many types of tumors, and inhibition of hypusination by siRNA or small molecules showed anti-proliferative effects in numerous tumor entities (16, 17). However, the hypusine system may have pleiotropic functions, as seen for the tumor suppressor activity of eIF-5A1 and DHS in lymphoma development (18). Moreover, perturbation of hypusination also preserves islet function in diabetes and blocks the replication cycle of HIV by affecting the export of specific mRNAs (19). Intriguingly, many cancers and tumor cell lines display elevated levels of eIF-5A2, an isoform of eIF-5A that shows no expression in normal tissue except brain and testis (20). In addition to its oncogenic activity (21, 22), eIF-5A2 was demonstrated to promote invasion and metastasis, and overexpression correlates with worse prognosis in various cancers (23, 24).
Together, a growing body of research now indicates that the enzymes of the hypusine system fulfill certain criteria for being a potential therapeutic target (25, 26), and consequently, lead substances have been developed (27, 28). However, a key question that still needs to be addressed before the hypusine system can be considered a novel target for therapies is to understand its function in mammalian homeostasis. In this regard, development of mouse models for manipulation of the hypusine system could provide the most valuable tool for addressing this question (29). Based on that consideration, we generated and characterized conditional knock-out mouse models for Dhs, Dohh, and eIF-5A2 that allowed either the general inhibition of both steps of hypusine modification or the selective depletion of the cancer-associated eIF-5A2 isoform in a particular temporal setting.
Experimental Procedures
Animal Studies
All animal experimental procedures were approved by the responsible Hamburg state authority according to German animal protection law. Mice were maintained in specific pathogen-free conditions at the University Medical Center Hamburg-Eppendorf animal facilities.
Generation of Conditional Knock-out Mice
Embryonic stem cell clones derived from C57BL/6N mice for the conditional knock-out of the Dhs (clones EPD0628_1_B06, EPD0628_1_C06, and EPD0628_1_F05) or eIF-5A2 (clone HEPD0734_3_A07) gene were obtained from the International Knock-out Mouse Consortium (30) and were verified by Southern blot and/or long range PCR. Clones were thawed, injected into E3.5 blastocysts of C57BL/6J mice, and transferred into the uterine horns of foster mothers. Male chimeric offspring were mated to C57BL/6J females and the resulting offspring analyzed for transmission of the targeted allele. Transgene-positive male offspring was mated to Flp-deleter (B6;SJL-Tg(ACTFLPe)9205Dym/J (31)) to remove the selection cassette. Resulting offspring were selected for the floxed allele, hereafter referred to with a superscript “p.” Wild type alleles are indicated with a superscript “+.” To generate a complete knock-out of Dhs or eIF-5A2, CMV-Cre-deleter (32) were mated to mice of the Dhs+/p or eIF-5A2+/p genotypes. After Cre-mediated deletion, the knock-out alleles are indicated with a superscript “−.” Mice of the resulting Dhs+/− or eIF-5A2+/− genotypes were further mated to individuals of the same genotype. To enable a 4-OHT-inducible knock-out of Dhs or Dohh, mice of the B6.Cg-Tg(CAG-cre/Esr1+)5Amc/J113 strain (The Jackson Laboratory, Bar Harbor, ME) were mated to mice of the Dhs+/p or Dohh+/p genotype. Mice of the resulting Dhs+/p;CAG-cre/Esr1+ or Dohh+/p;CAG-cre/Esr1+ genotypes were further mated to individuals of the same genotype (Fig. 1B). Tamoxifen (4-OHT)-inducible knock-out was induced by administering 4-OHT-containing feed (0.4 g/kg, LASvendi, Soest, Germany) for up to 3 weeks. Mice were monitored regularly and weighed at least every week (Fig. 1C).
Southern Blot Analysis
Southern blot was carried out as described previously (33).
Long Range PCR
Genomic DNA was isolated using QIAamp DNA blood mini kit (Qiagen, Venlo, NL) according to manufacturer's instructions. Long range PCR was performed using oligonucleotides listed in Table 1 and Long PCR Enzyme Mix (Thermo Fisher Scientific, Waltham, MA). PCRs were performed at 94 °C for 90 s, followed by 10 cycles of 95 °C for 15 s, 54 °C for 30 s, and 68 °C for 7 min and 25 cycles of 95 °C for 15 s, 54 °C for 30 s, and 68 °C for 7 min + 5 s/cycle and final elongation at 68 °C for 10 min.
TABLE 1.
Oligonucleotides used in this study
| Application | Oligonucleotide | Sequence/Qiagen QuantiTect number |
|---|---|---|
| Long range PCR | 3′ universal | CAC ACC TCC CCC TGA ACC TGA AAC |
| 3′ Dhs gene-specific | CAG GTT CTA TCG ATT CCA GTG TCC G | |
| 3′ Dhs gene-specific | GTG GCC ACG GCT ACG AAG TGC TAG | |
| 3′ eIF-5A2 gene-specific | GAT GAC TGC TGT GTG GAA TAG TAT CAT CTG | |
| 3′ eIF-5A2 gene-specific | GAG GAG GAC CAT GAG ATG GTG AGG ACA TG | |
| Genotyping | Dhs-forward | CCT CTG CCC TCT CAC AGA CCT GCG |
| Dhs-5′-arm | AGA GCG CCC AGG TCC AAG GCT ACG | |
| Dhs-3′-arm | AGC GTT AAG CTC CTC CTA CAA AGC | |
| Dhs-rev | ACC ATC CGC AGG AGA CCA CAC CTA | |
| eIF-5A2-5′-arm | AGC CGA GAT GCT TGG GAA CTG GAG G | |
| eIF-5A2-3′-arm | TGG AGT ATA CTT TGC CAT TCA GGC C | |
| eIF-5A2-reverse | AAG GCC AGC CTG AGA CCT AT | |
| Quantitative PCR | 18S-rRNA | QT01036875 |
| Dhs | QT02529093 | |
| Dohh | QT00163268 | |
| eIF-5A1 | QT01757861 | |
| eIF-5A2 | QT00148225 | |
| Dhs del forward | TTC AAC CGC GGC GTA GAT TA | |
| Dhs del reverse | TCT GCT CCA TTC CTC ATG GC |
Genotyping Using Genomic DNA from Tail Clippings, Organs, Embryos, or Culture Cells
Samples were digested overnight at 55 °C using proteinase K (Thermo Fisher Scientific, Waltham, MA) according to the manufacturer's instructions. PCR analyses of genomic DNA were performed using DreamTaq Green PCR MasterMix (Thermo Fisher Scientific, Waltham, MA) and 2 μl of lysate in a total volume of 20 μl. The oligonucleotide sequences are listed in Table 1.
Quantitative PCR
RNA was isolated from cell or tissues using TriFast (Peqlab, Erlangen, Germany) according to the manufacturer's instructions. cDNA was prepared by reverse transcription of 1 μg of total RNA using random hexamer primer and Moloney murine leukemia virus reverse transcriptase (Thermo Fisher Scientific, Waltham, MA). Quantitative real time PCR for Dhs, Dohh, eIF-5A1, and eIF-5A2 was performed in a 7500 Fast Real Time PCR System (Life Technologies, Inc.) using the oligonucleotides or QuantiTect primer mix (Qiagen, Venlo, Netherlands) listed in Table 1 and Platinum® SYBR® Green qPCR SuperMix-UDG (Life Technologies, Inc.). PCRs were performed at 95 °C for 3 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 30 s. PCRs were conducted in triplicate and normalized against 18S rRNA or Gapdh as reference gene using the 2−ΔΔCT method (34).
Expression of eIF-5A2 in normal mouse tissue was examined using the Tissue Scan qPCR Array MDRT101 (OriGene Technologies, Rockville, MD). Values were normalized to the expression of the housekeeping gene Gapdh and depicted as fold change based on the bone marrow.
One- and Two-dimensional Western Blot Analysis
One- and two-dimensional Western blot analyses were carried out as described before (16, 33) using anti-eIF-5A1 (Novus Biologicals, Littleton, CO) and anti-eIF-5A2 (Abcam, Cambridge, UK).
Generation and Culture of Mouse Embryonic Fibroblasts (MEFs)
MEFs were isolated from Dhs+/+ × Dhs+/p;CAG-cre/Esr1+ and Dhsp/p × Dhs+/p;CAG-cre/Esr1+ matings as described previously (33). The cells were cultured in DMEM (all cell culture media and additives were from Invitrogen) supplemented with 10% fetal bovine serum, 50 units/ml penicillin, 50 μg/ml streptomycin, 25 μm β-mercaptoethanol, and 4 mm l-glutamine (37 °C, 5% CO2, humidified atmosphere). For inducible knock-out experiments, 106 cells were seeded on a 10-cm culture dish, maintained at subconfluence, and treated with 100 nm 4-OHT for up to 10 days (Sigma) or 50 μm GC7 for 2 days (Biosearch Tech., Novato, CA).
Morphological and Histopathological Analysis of Mouse Tissue
Mice were transcardially perfused with a mixture of 4% paraformaldehyde (and 1% glutaraldehyde) in 0.1 m PB buffer at pH 7.4 for tissue preparation, and bone marrow was morphologically analyzed as described before (33). Briefly, after postfixation overnight at 4 °C, tibiae were decalcified for 4 days in 10% (w/v) EDTA in PBS and cut in 150-μm sagittal sections with a vibratome. The sections were then postfixed in 1% (v/v) OsO4, dehydrated, and embedded in Epon. Semi-thin sections (0.5 μm) were labeled with methylene blue and examined under the light microscope (Zeiss). Other tissues were processed to paraffin blocks using an ASP300S dehydration machine (Leica, Wetzlar, Germany) and an EG1160 tissue-embedding system (Leica, Wetzlar, Germany). Paraffin blocks were cut into 4-μm sections, which were stained with hematoxylin and eosin or with Turnbull's blue reagents following standard laboratory procedures.
Immunohistochemistry
For immunohistochemical staining the Ventana Benchmark XT machine (Ventana, Tucson, AZ) was used. Briefly, deparaffinated sections were boiled for 30–60 min in 10 mm citrate buffer, pH 6.0, for antigen retrieval. Primary antibodies were diluted in 5% goat serum (Dianova Immunodiagnostics, Hamburg, Germany), 45% Tris-buffered saline, pH 7.6 (TBS), and 0.1% Triton X-100 in antibody diluent solution (Zytomed, Berlin, Germany). Sections were then incubated with primary antibody against anti-Ki67 for proliferating cells (1:250; Abcam, Cambridge, UK) and anti-caspase-3 for apoptotic cells (1:1000; R&D Systems, Wiesbaden, Germany) for 1 h. Anti-rabbit histofine Simple Stain MAX PO universal immunoperoxidase polymers (Nichirei Biosciences, Wedel, Germany) were used as secondary antibodies. Detection of secondary antibodies and counterstaining was performed with an ultraview universal DAB detection kit from Ventana according to the standard settings of the machine. All sections were coverslipped using TissueTek® glove mounting media (Sakura Finetek, Staufen, Germany) and dried in an incubator at 60 °C. Pictures were taken using a light microscope (Axioskop 40, Zeiss, Jena, Germany or Olympus BH-2, Hamburg, Germany) equipped with a digital camera (AxioCam ICc3 Zeiss, Jena Germany).
eIF-5A1 Immunofluorescence of Immortalized 3T3 Dohh Knock-out Cells or Primary Dhs Knock-out MEFs
Knock-out of Dohh or Dhs was induced with 100 nm 4-OHT for 7 days in 3T3 Dohhp/p;CAG-cre/Esr1+ (33) or in MEFs Dhsp/p;CAG-cre/Esr1+, respectively. The cells were then seeded onto poly-l-lysine (Sigma)-coated 10-mm diameter round glass coverslips in 24-well plates and left to adhere for 1 more day in the presence or absence of 4-OHT. After washing with PBS, the cells were fixed with 3% (w/v) paraformaldehyde in PBS for 20 min at room temperature. After washing, residual paraformaldehyde was quenched with 50 mm NH4Cl in PBS for 10 min. For permeabilization, the cells were treated for 5 min with 0.1% (v/v) Triton X-100 in PBS, and then the samples were blocked with 0.5% (w/v) BSA in PBS for 30 min. Immunostaining was performed with mouse anti-eIF-5A1 (clone 26/eIF-5a; BD Biosciences) and Alexa Fluor 488-labeled goat anti-mouse antibody (Life Technologies, Inc.). Cell nuclei were stained with Hoechst 33342 (Life Technologies, Inc.). Images were taken as single confocal sections of 1 μm using a Nikon C2+ confocal laser scanning microscope (Nikon Instruments, Tokyo, Japan). For image acquisition and preparation, Nikon NIS Elements software and Adobe Photoshop CS4 (Adobe Systems Inc., San Jose, CA) were used.
Construction and Analysis of Protein-Protein Interaction Networks
For the construction of protein-protein interaction (PPI) networks for PPP (Pro-Pro-Pro)- and PPG (Pro-Pro-Gly)-rich proteins, genes with >1 PPP unit and >1 PPG unit were extracted as described by Mandal et al. (35). The number of PPP and PPG units were defined by the whole number of consecutive prolines divided by 3, as described recently (35). That means that 3–5 consecutive prolines were all counted as 1 PPP unit and 6–8 consecutive prolines as 2 PPP units and so on. STRING database version 10.0 was used as resource for contraction of PPI networks (36). PPI networks were visualized and analyzed using the software platform Cytoscape 3.2.1 with the plugins MCODE for cluster analysis and BinGO for gene ontology analysis of protein clusters (37–39). Global network topological analysis, including clustering coefficients, average number of neighbors (degree), and network heterogeneity as well as local network analysis using betweenness centrality value calculation were carried out with NetworkAnalyzer and the Cytoscape plugin CytoNCA (40, 41). The edges in all PPI networks were treated as undirected. Clustering coefficient for a network is the average of the clustering coefficient over all nodes and quantifies the local interconnectivity of a network. Node degree is the number of edges linked to a node. Network heterogeneity reflects tendency of a network to contain highly connected nodes (hubs). Betweenness centrality is a measure of the fraction of shortest paths passing through a node in the network, and nodes with higher betweenness are more globally central in the network (42).
Statistics
For statistical analyses GraphPad software (GraphPad Software, La Jolla, CA) was used. The statistical tests used in each analysis are stated in the corresponding figure legends.
Results
Dhs Is Crucial for Embryonic Development as Well as for Viability in Adult Mice
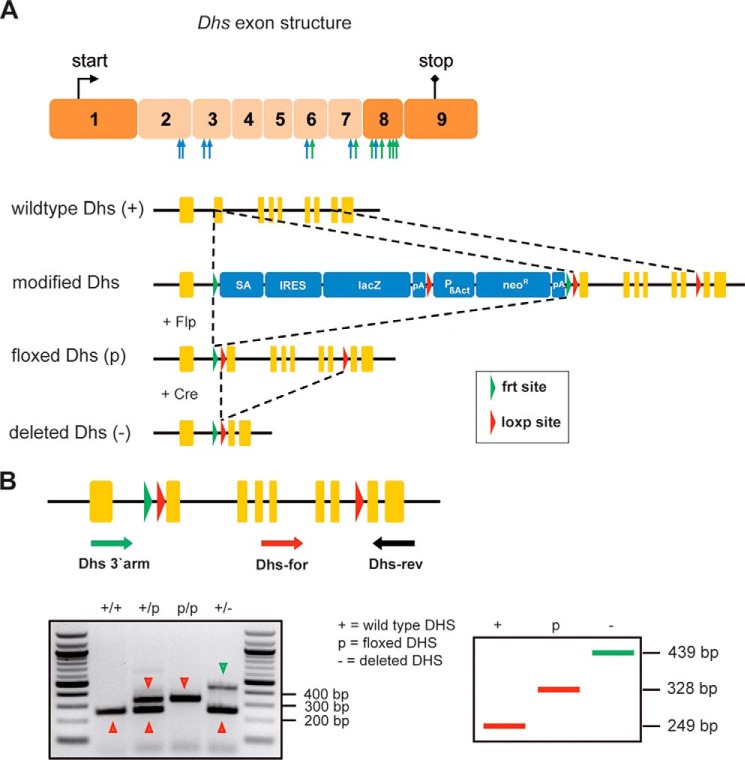
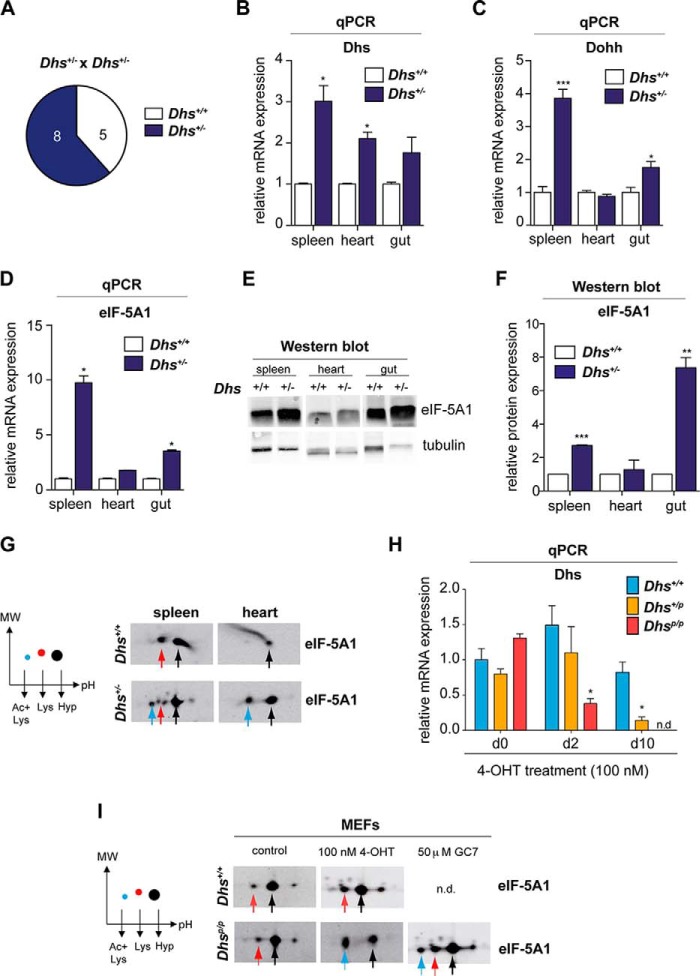
To study the biological relevance and functional requirement of the hypusine modification in embryonic development (43, 44) and for viability, we used a recently described conditional knock-out strain for Dohh (33) and established a new conditional knock-out mouse model for Dhs (B6.Dhpstm2a(EUCOMM)Wtsi). For the generation of the latter strain, embryonic stem cell clones were obtained from the International Knock-out Mouse Consortium (30). Detailed explanation for the Dhs gene targeting strategy that leads to deletion of exons two to seven resulting in a nonfunctional truncated Dhs is depicted in Fig. 2A. Information about the genotyping procedure is provided in Fig. 2B. For Cre-mediated knock-outs, we intercrossed B6.Dhpstm2a(EUCOMM)Wtsi to two different Cre-deleter strains allowing us to analyze how Dhs impairs either embryonic development or affects the viability of an adult organism after 4-OHT-mediated Cre induction (Fig. 1, B and C). After removal of the selection cassette, we induced a general knock-out using the CMV-Cre mouse model (Fig. 1B) (32). The loss of Dhs leads to embryonic lethality in a pure C57BL/6 background as we could not detect any homozygous Dhs−/− offspring in our crossings (Fig. 3A). This observation is well in line with data obtained in a mixed background published by others (43, 44). Heterozygous Dhs+/− animals were viable and did not show any obvious phenotype. Intriguingly, when we investigated mRNA levels in several organs of Dhs+/− mice, we detected a significant up-regulation of Dhs as well as Dohh and eIF-5A1 compared with wild type Dhs+/+ animals (Fig. 3, B–D). Moreover, eIF-5A1 protein levels were also elevated (Fig. 3, E and F), suggesting a compensatory feedback mechanism and up-regulation of the hypusine system in a Dhs-compromised setting protecting against the loss of DHS activity. We next asked whether heterozygous deletion of Dhs results in a reduced hypusine modification level in eIF-5A1. Two-dimensional Western blots revealed an accumulation of two additional eIF-5A1 spots in tissue from Dhs+/− mice (Fig. 3G). According to previous data (33, 45), these spots represent the unhypusinated (Fig. 3G, red arrow) and the more acidic unhypusinated and acetylated form of eIF-5A1 (Fig. 3G, blue arrow). Nevertheless, high amounts of hypusinated and therefore functional eIF-5A1 remained detectable (Fig. 3G, black arrow), explaining the normal phenotype of Dhs heterozygous animals. These observations indicate that under physiological conditions a reduced pool of fully modified eIF-5A1 is sufficient to support normal cellular function. As previous studies proposed a pro-apoptotic function for unhypusinated eIF-5A1 (46), our results further suggest that accumulation of low levels of unhypusinated eIF-5A1 does not affect embryonic development and viability.
FIGURE 2.
Generation of a conditional Dhs knock-out mouse strain using ES-cell clones harboring a targeted mutation of the Dhs gene. A, schematic representation of the strategy for achieving a knock-out of the Dhs gene. The seven exons designed for deletion are depicted in a lighter color. Blue arrows indicate regions encoding amino acids that are essential for binding of NAD; green arrows indicate regions essential for binding of spermidine (86). The targeted mutation of the Dhs gene consists of a lacZ-reporter cassette (SA, splice acceptor; IRES, internal ribosome entry site; lacZ, β-galactosidase gene; pA, polyadenylation signal), a neomycin-selection cassette (PβAct, eukaryotic β-actin promotor; neoR, neomycin resistance gene), and three loxP and two FRT sites. B, genotyping strategy for Cre-mediated knock-out of the Dhs gene. Animals harboring the floxed Dhs genes were mated to Cre-deleter mice (B6.C-Tg(CMV-cre)1Cgn/J strain or B6.Cg-Tg(CAG-cre/Esr1+)5Amc/J113 strain plus oral 4-OHT administration. Usage of three oligonucleotides (Dhs-3′-arm, Dhs-for, and Dhs-rev) allows simultaneous detection of all possible alleles (+, p, and −), including the deleted allele (−), which is achieved by Cre-mediated recombination.
FIGURE 3.
Homozygous deletion of Dhs is embryonically lethal in mice, whereas heterozygous Dhs knock-out animals are viable and show compensatory up-regulation of the hypusine system. A, representative litter size of the indicated breeding. Note that no Dhs−/− pups were born after Dhs+/− × Dhs+/− breeding. B–D, mRNA levels of Dhs, Dohh, and eIF-5A1 in different organs of wild type or heterozygous mice as assessed by quantitative real time PCR. Expression values were plotted relative to the expression of the housekeeping gene 18S rRNA. E and F, expression of eIF-5A1 protein in different organs using Western blot. Protein expression was quantified relative to the loading control α-tubulin using the Odyssey Infrared Imaging System. G, two-dimensional Western blot detecting different eIF-5A1 forms in the outlined organs. H and I, analysis of MEFs isolated from Dhs-deficient Dhsp/+ and Dhsp/p-CAG-cre/Esr1+ mice as well as wild type Dhs+/+; CAG-cre/Esr1+ animals after 4-OHT treatment in vitro. H, mRNA expression status of Dhs after 100 nm 4-OHT treatment at the indicated time points relative to the housekeeping gene 18S as assessed by quantitative real time PCR. I, two-dimensional Western blot for eIF-5A1 after treatment with 4-OHT (100 nm; 8 days) or incubation with 50 μm GC7 (2 days). Significances were calculated using the unpaired t test and marked with an asterisk if significant (***, p < 0.001; **, p < 0.01; *, p < 0.05). +, wild type allele; −, deleted allele. Colored arrows in the representative blots correspond to the different eIF-5A1 forms as outlined in the schematic plot on the left: black = fully hypusinated Lys50, pH 5.2; red = unmodified Lys50, pH 5.1; blue = unmodified Lys50 plus acetylated Lys47, pH 5.0. qPCR, quantitative PCR. n.d., not detectable.
First Step of Hypusine Modification Is Essential for Viability
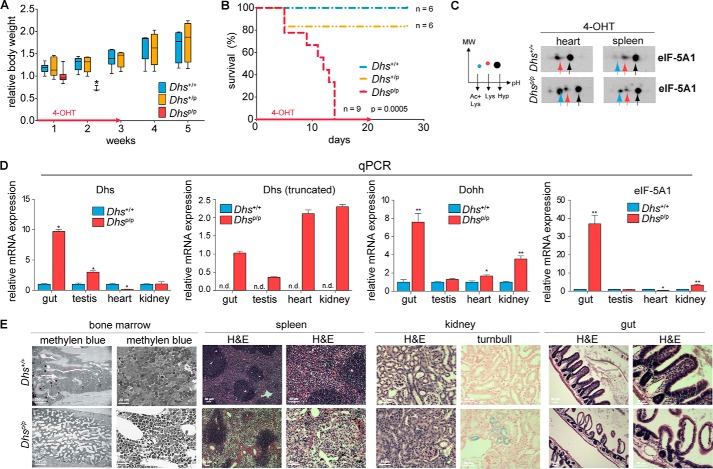
Given the crucial function of Dhs in embryonic development, we next investigated whether Dhs is also essential for viability of an adult organism. For this reason, we utilized a 4-OHT-inducible conditional CAG/Cre-Esr1+ mouse strain (Fig. 1, B and C), allowing us to induce a general knock-out of Dhs in most tissues (47). First, we employed Dhs+/+ and Dhsp/p mouse embryonic fibroblasts to investigate the functionality of the Cre-mediated Dhs knock-out in vitro. The analysis outlined in Fig. 3H revealed a clear reduction of wild type Dhs transcript after 4-OHT treatment and impaired hypusination of eIF-5A1 in those cells (Fig. 3I), showing loss of DHS activity. Next, we induced Dhs deletion in 5–7-week-old mice using 4-OHT-containing feed. Intriguingly, after 4-OHT administration Dhsp/+ animals behaved similarly to Dhs+/+ mice, whereas the body weight of Dhsp/p animals drastically decreased (Fig. 4A), and mice died or had to be euthanized due to a wasting syndrome (weight loss >20%) 5–14 days after induction of the knock-out (median 12 days; Fig. 4B). Notably, the Dhsp/p mice expressed a truncated form of Dhs suggesting a successful knock-out (Fig. 4D) that leads to the accumulation of the unhypusinated eIF-5A1 precursor protein as shown in heart and spleen by two-dimensional Western blot (Fig. 4C). However, those animals also displayed a significant up-regulation of the hypusine system in different organs at a terminal stage involving a remarkable increase in mRNA of Dhs, Dohh, and eIF-5A1 (Fig. 4D), compared with their normal Dhs+/+ counterparts, as well as a detectable level of hypusinated eIF-5A1 (Fig. 4C). This argues for a partial knock-out in organs of Dhsp/p animals due to an incomplete mosaic-like expression of the Cre recombinase in CAG/Cre-Esr1+ mice, as is known for this particular Cre-deleter strain (47). Furthermore, this implicates the activation of a compensatory positive feedback mechanism in the remaining wild type cells, similarly as observed in heterozygous Dhs+/− mice (Fig. 3, B–F), to keep the hypusine modification in a range that allows viability.
FIGURE 4.
Loss of Dhs confers lethality in adult mice and affects organ integrity. A–E, analysis of adult Dhs+/+, Dhsp/+, and Dhsp/p-CAG-cre/Esr1+-positive mice after 4-OHT administration resulting from a breeding according to the scheme in B and C of Fig. 1. A, bodyweight of animals after 4-OHT-mediated knock-out in vivo at the indicated time points. Red arrow indicates duration of 4-OHT treatment. B, Kaplan-Meier plot showing overall survival of mice after 4-OHT-mediated knock-out in vivo at the indicated time points. Red arrow indicates duration of 4-OHT treatment. C, two-dimensional Western blot for eIF-5A1 after 4-OHT treatment in vivo. Heart and spleen were isolated from mice showing more than 20% weight loss. Colored arrows in the representative blots correspond to the different eIF-5A1 forms as outlined in the schematic plot on the left: black = fully hypusinated Lys50, pH 5.2; red = unmodified Lys50, pH 5.1; blue = unmodified Lys50 plus acetylated Lys47, pH 5.0. D, quantitative real time PCR assessing the mRNA expression status of Dhs, Dohh, and eIF-5A1 in several tissues after 4-OHT treatment in vivo expressed relative to the housekeeping gene 18S rRNA. Organs were isolated from animals showing more than 20% weight loss. E, comprehensive histopathological analysis of various mouse organs after 4-OHT-mediated knock-out in vivo. Organs were isolated from animals showing more than 20% weight loss. Staining with methylene blue or H&E showed reduced bone marrow cellularity (4/4 animals) and an impaired cellularity of the red pulpa in the spleen (4/4 animals), respectively, in Dhs-deficient animals. Turnbull staining unveiled enrichment of ferrous iron in the kidney of Dhs-deficient mice (3/4 animals). Epithelium of the intestine was severely damaged in Dhs-deficient animals as demonstrated by H&E (1/4 animals). Significances were calculated using the unpaired t test and marked with an asterisk if significant (***, p < 0.001; **, p < 0.01; *, p < 0.05). +, wild type allele, p, floxed allele; n.d., not detectable. qPCR, quantitative PCR.
Deletion of Dhs Affects Bone Marrow and Spleen Cellularity
To identify putative morphological abnormalities accounting for the observed strong lethal phenotype associated with the loss of Dhs, we performed comprehensive histological analyses. Although these analyses did not point to a single recurrent tissue defect causing lethality, histopathological examination unveiled gross changes in various organs of Dhsp/p animals compared with their normal wild type Dhs+/+ counterparts. The hematopoietic system seemed to be severely affected because the bone marrow and the spleen showed reduced overall cellularity in Dhsp/p animals. As shown in Fig. 4E, this was mainly due to a reduction of all three cell lines in the bone marrow and in the red pulpa of the spleen. Moreover, in two out of four Dhs-deficient animals, the kidney showed enhanced incorporation of ferrous iron (Fig. 4E); however, histological examination revealed no obvious alteration in kidney tissue architecture (data not shown). In addition, the epithelial structures of the intestine were markedly destroyed in one out of four Dhsp/p animals. Thus, it can be assumed that loss of Dhs confers lethality by induction of multiple phenotypic changes particularly in highly proliferative tissues (bone marrow and intestine) in adult mice.
Together, these data indicate that Dhs-mediated hypusination of eIF-5A1 is essential for embryonic development and is mandatory to ensure viability of an adult organism. Moreover, a potential positive feedback mechanism is activated protecting heterozygous Dhs mice from gene dosage effects, underlining the importance of the hypusine system for cellular integrity.
Dohh Is Equally Important for an Adult Organism but Consequences of a Dohh Knock-out Are Less Pronounced Compared with the Dhs Knock-out
The second step of hypusine modification in eIF-5A1 was shown to be not essential in lower eukaryotes like Saccharomyces cerevisiae (48). However, we have shown recently that deletion of Dohh and subsequent loss of hypusine modification induced an embryonic lethal phenotype in mice (33).
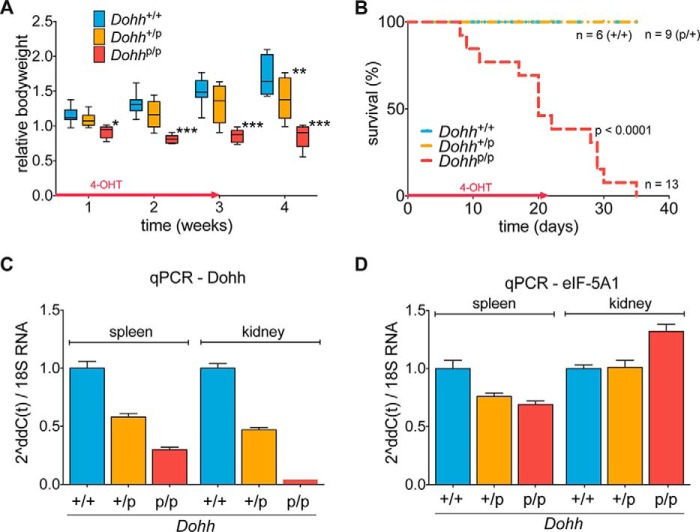
To investigate the role of Dohh in adult mice, we intercrossed our previously described B6.DOHHtm2a/bal animals (33) with the CAG/Cre-Esr1+ mouse strain and used 4-OHT to induce Dohh knock-out in 4–7-week-old mice as described for the Dhs knock-out mouse model (Fig. 1, B and C). Notably, heterozygous Dohhp/+ animals behaved like wild type controls, but the body weight of Dohhp/p mice stagnated, and animals finally died 1–5 weeks after knock-out induction due to a wasting condition (median 20 days; Fig. 5, A and B). Although we observed an expected decrease in Dohh mRNA in spleen and kidney from those animals at a terminal stage (Fig. 5C), we could not detect a consistent compensatory up-regulation of the hypusine system as in Dhs-deficient mice (Fig. 5D). As a consequence, loss of Dohh resulted in a shift toward the unhypusinated eIF-5A1 precursor protein as seen in the spleen of Dohhp/p animals (Fig. 6A, red arrow), even though hypusinated eIF-5A1 was still detectable (Fig. 6A, black arrow). In agreement with our recent in vitro results (33) and in contrast to the ablation of Dhs, unhypusinated and acetylated eIF-5A1 was not observed (Fig. 6A).
FIGURE 5.
Dohh-mediated hypusination of eIF-5A1 is essential in adult mice. A–D, analysis of adult Dohh+/+, Dohhp/+, and Dohhp/p-CAG-cre/Esr1+-positive mice after 4-OHT administration resulting from a breeding according to the scheme in Fig. 1, B and C. A, bodyweight of mice after 4-OHT-mediated knock-out in vivo at the indicated time points. Red arrow indicates duration of 4-OHT treatment. B, Kaplan-Meier plot showing overall survival of mice after 4-OHT-mediated knock-out in vivo at the indicated time points. Red arrow indicates duration of 4-OHT treatment. C and D, real time PCR assessing the mRNA expression status of Dohh and eIF-5A1 in spleen and kidney after 4-OHT treatment in vivo expressed relative to the housekeeping gene 18S rRNA. Organs were isolated from animals showing more than 20% weight loss. qPCR, quantitative PCR.
FIGURE 6.
Loss of Dohh reveals histological anomalies. A, two-dimensional Western blot for eIF-5A1 in spleen tissue after 4-OHT treatment in vivo. Spleen was isolated from mice showing more than 20% weight loss. Colored arrows in the representative blots correspond to the different eIF-5A1 forms as outlined in the schematic plot in Fig. 4C. Black = fully hypusinated Lys50, pH 5.2; red = unmodified Lys50, pH 5.1. B, H&E staining of kidney sections after 4-OHT induction in vivo unveiled tubular necrosis in one of the Dohh-deficient mice (1/4 animals; red arrow). Organs were isolated from animals showing more than 20% weight loss. C, methylene blue staining revealed no overt change in cellular composition of the bone marrow. D, histological analysis of the spleen using H&E staining did not show any major difference. Turnbull staining unveiled enrichment in ferrous iron in Dohh-deficient mice (2/2 animals). E and F, H&E, Ki67, and caspase-3 staining of liver tissue compared with control in 5–7 weeks (E) and 6–7-month-old mice (F). Red arrow indicates liver necrosis, and green arrow indicates focal inflammation.
Although homozygous deletion of Dohh was lethal in all animals, rigorous histopathological examinations of adult Dohh knock-out mice (homozygote deletion of Dohh was induced in seven mice at the age of 5–7 weeks and in two mice at the age of 6–7 months) revealed no relevant recurrent tissue alterations (Fig. 6, B–F) except histological signs for a tubular necrosis in the kidney of one animal (Fig. 6B) and increased incorporation of ferrous iron in the spleen of both analyzed adult (6–7 months old) animals (Fig. 6D). Furthermore, we have detected moderate necrosis in the liver of two knock-out mice (group of 5–7 weeks old mice) (Fig. 6E) and moderate focal liver inflammation in one knock-out animal (group of 6–7 month old mice) (Fig. 6F). Compared with the Dhs knock-out animals, bone marrow was apparently not affected in Dohh-deleted mice (Fig. 6C). All other animals died without obvious cause, and no consistent pathology could be identified.
Together with the observation that an early general knock-out of Dohh is embryonically lethal as published by our group recently (33), we conclude here that Dohh is equally as important for early embryonic development and adult organisms as Dhs, yet the lethal phenotype of Dohh-deficient animals seems to be less pronounced. However, it must be pointed out that the exact reason for the observed lethal phenotype has to be elucidated.
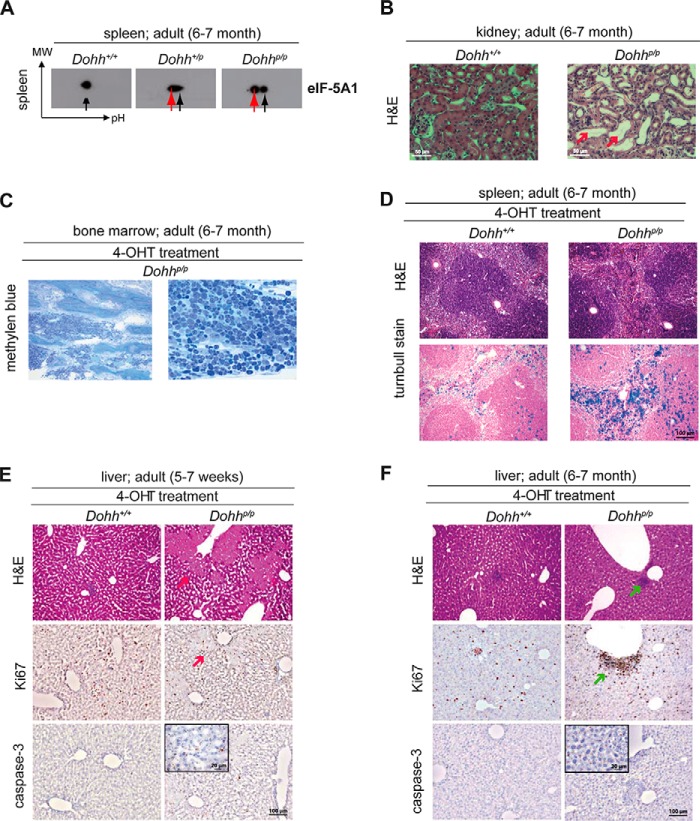
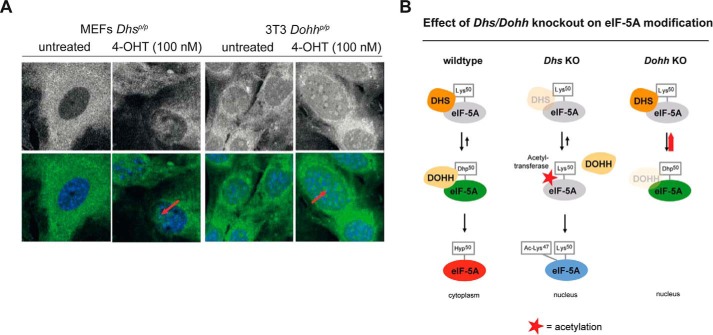
eIF-5A Is Also Localized in the Nucleus after Deletion of Dhs or Dohh
Because modifications of eIF-5A1 influence its cellular localization (49, 50), the accumulation of different eIF-5A1 forms may therefore account for the different phenotype of the Dhs and the Dohh knock-out. Although the knock-out of Dohh causes an accumulation of unhypusinated eIF-5A1 precursor (Fig. 6A, red arrow), Dhs deficiency provokes its additional acetylation (Fig. 4C, blue arrow). Investigating the localization of eIF-5A1 by immunofluorescence in a Dhs- and Dohh-dependent setting in vitro, we found that the 4-OH-induced knock-out of Dhs and Dohh leads to nuclear accumulation of eIF-5A1 (Fig. 7A). Because fully hypusinated eIF-5A1 was recently shown to be located in the cytoplasm (where it regulates translation of specific proteins (11)), the observed nuclear eIF-5A in the Dhs- and Dohh-deficient setting likely reflects the unhypusinated (acetylated and nonacetylated) eIF-5A. Fig. 7B summarizes an hypothesis explaining why eIF-5A1 is differently modified in these scenarios. In a Dohh-deficient background, DHS is constantly linked to eIF-5A1 due to the missing hydroxylation of deoxyhypusine (Dhp50) and subsequently prevents binding of acetyltransferases like HDAC6, SIRT2, p300, and CBP (49, 51). In contrast, Dhs depletion prevents DOHH binding and further allows acetylation of eIF-5A1.
FIGURE 7.
Knockdown of the hypusine modification enzymes affect localization and post-translational modification of eIF-5A1. A, immunofluorescence for endogenous eIF-5A1 in a Dhs or Dohh-deficient cellular background, respectively. Upper panels depict eIF-5A1 fluorescence alone; the lower panels show the overlay of the eIF-5A1 signal (green) and the Hoechst 33342 nuclear DNA stain (blue). Primary MEFs from Dhsp/p;CAG-cre/Esr1+ mice (left) and immortalized MEFs (3T3) from Dohhp/p;CAG-cre/Esr1+ animals (right) were treated with or without 4-OHT (100 nm; 7 days) in vitro to induce the respective knock-out. Red arrows indicate nuclear accumulation of eIF-5A1. B, predicted model explaining how loss of Dhs and Dohh could influence eIF-5A1's modification.
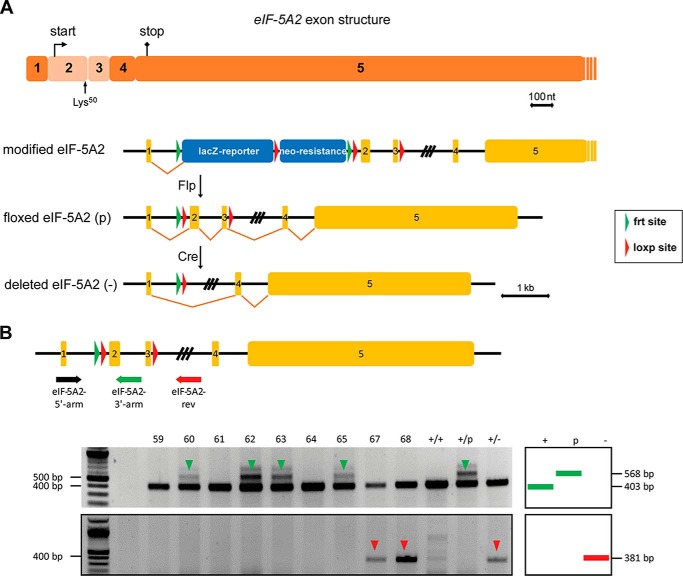
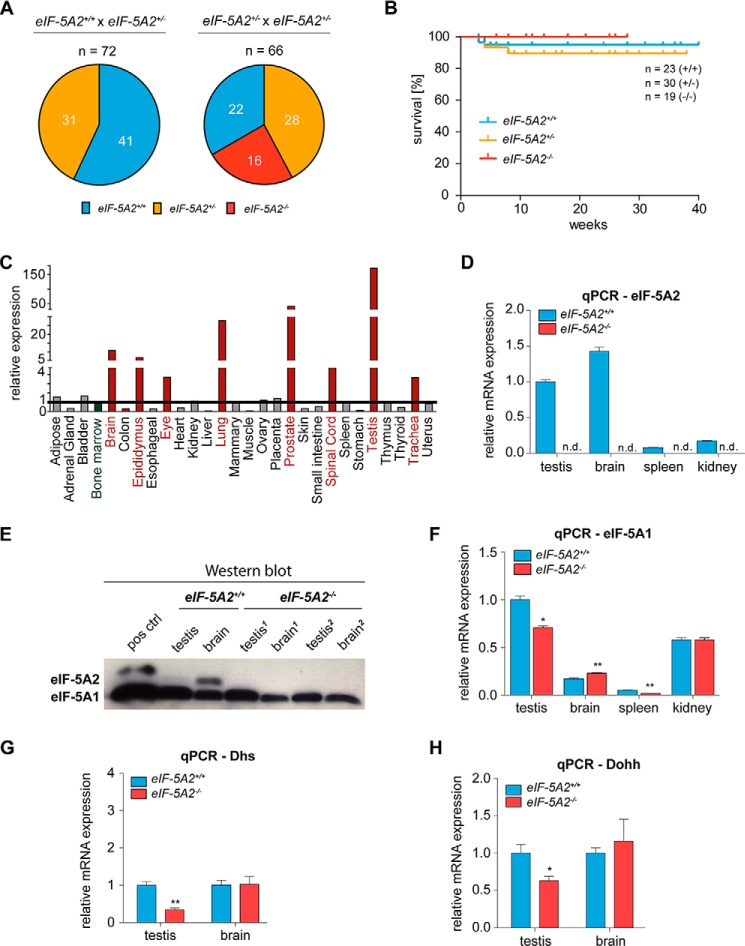
eIF-5A2 Is Not Essential for Normal Development and Steady State Viability
As the general use of DHS and DOHH inhibitors seems to require more refined analyses due to their essential role in higher eukaryotes, we reasoned whether eIF-5A2, which is only expressed in few tissues of the body, but highly abundant in cells of various tumor types, might represent a better target for therapy. To address this question and to investigate the effects of eIF-5A2 deficiency in development and in adult mice, we generated eIF-5A2 conditional knock-out mice (B6.eIF5A2tm2a(EUCOMM)wtsi) (Fig. 8, A and B) and intercrossed them with the CMV-Cre-deleter strain to induce an early general knock-out (Fig. 1B). In contrast to Dhs- and Dohh-deficient animals, both heterozygous eIF-5A2+/− and homozygous eIF5-A2−/− mice were viable (Fig. 9, A and B) and did not show any abnormalities regarding body weight (data not shown) or survival over the indicated time period. Expression of eIF-5A2 is thought to be limited to a few human tissues like lung, brain, testis, or prostate (52). Quantitative PCR revealed a comparable expression pattern in mouse tissues (Fig. 9C). As expected, eIF-5A2 mRNA and protein are not detectable in testis and brain tissue of eIF5-A2−/− mice (Fig. 9, D and E), indicating the functional and successful knock-out. Moreover and in contrast to a loss of DHS, a consistent compensatory overexpression of the hypusine modification system was not detectable in eIF5-A2−/− mice (Fig. 9, F–H). Taken together, these data suggest that loss of the eIF-5A2 isoform is not essential for embryonic development or for steady state viability in an adult organism because eIF-5A2 knock-out mice are viable, fertile, and do not show an obvious phenotype. Fig. 10 summarizes phenotypes induced by deletion of the hypusine modification systems.
FIGURE 8.
Generation of a conditional eIF-5A2 knock-out mouse strain using an ES cell clone harboring a targeted mutation of the eIF-5A2 gene. A, schematic representation of the knock-out strategy for achieving a conditional knock-out of the eIF-5A2 gene. The two exons designed for deletion are depicted in a lighter color. Note that this region encodes the translational start point and the critical hypusine modification residue Lys50. The targeted mutation of the eIF-5A2 gene consists of a lacZ reporter cassette (SA, splice acceptor; IRES, internal ribosome entry site; lacZ, β-galactosidase gene; pA, polyadenylation signal), a neomycin-selection cassette (PβAct, eukaryotic β-actin promotor; neoR, neomycin resistance gene), three loxP and two FRT sites. B, genotyping strategy for Cre-mediated knock-out of the eIF-5A2 gene. Mice harboring the floxed eIF-5A2 gene were mated to Cre-deleter mice (B6.C-Tg(CMV-cre)1Cgn/J strain). Usage of three oligonucleotides (eIF-5A2-3′-arm, eIF-5A2-5′-arm, and eIF-5A2-rev) allows detection of all possible alleles (+, p, green arrow), including the deleted allele (−, red arrow), which is achieved by Cre-mediated recombination.
FIGURE 9.
eIF-5A2 knock-out mice are viable and fertile. A, offspring analysis of the indicated breedings. B, Kaplan-Meier plot showing overall survival of mice at the indicated time points after birth. C, mRNA levels of eIF-5A2 in different tissues of wild type mice as assessed by quantitative real time PCR using the Tissue Scan qPCR Array MDRT101 (OriGene Technologies, Rockville, MD). Values were normalized to the expression of the housekeeping gene Gapdh and depicted as fold changes with bone marrow as the reference. D, mRNA status of eIF-5A2 in different organs of mice as assessed by quantitative real time PCR. Expression values were plotted relative to the expression of the housekeeping gene 18S. E, protein expression of eIF-5A2 in testis and brain of eIF-5A2−/− animals (n = 2) and eIF-5A2+/+ mice (n = 1) assessed by Western blot. eIF-5A1 expression served as a loading control. Organs were isolated from 7- to 10-month-old animals. F–H, mRNA status of eIF-5A1 as well as Dhs and Dohh in different organs of mice as assessed by quantitative real time PCR. Expression values were plotted relative to the expression of the housekeeping gene 18S. Significances were calculated using the unpaired t test and marked with an asterisk if significant (**, p < 0.001; *, p < 0.05). n.d., not detectable.
FIGURE 10.
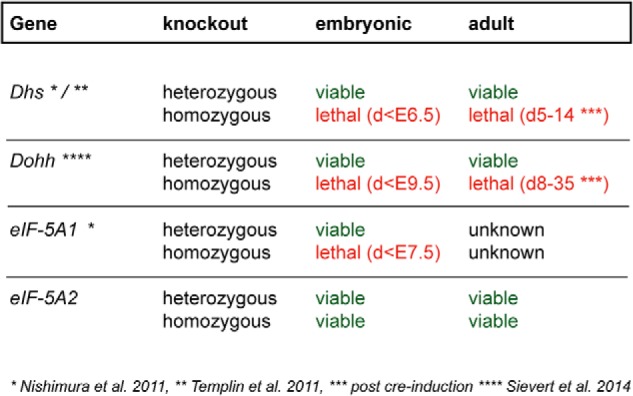
Summary of observed phenotypes after genetic manipulation of the hypusine modification system. Whereas a heterozygous knock-out of any of the genes of the hypusine modification system does not affect the viability of mice, a homozygous depletion of Dhs, Dohh, or eIF-5A1 causes lethality in embryonic and adult mice with different penetrance. In contrast, the cancer-associated isoform eIF-5A2 is dispensable for normal development and viability.
eIF-5A Targets Are Organized in Highly Connected Protein-Protein Interaction Networks
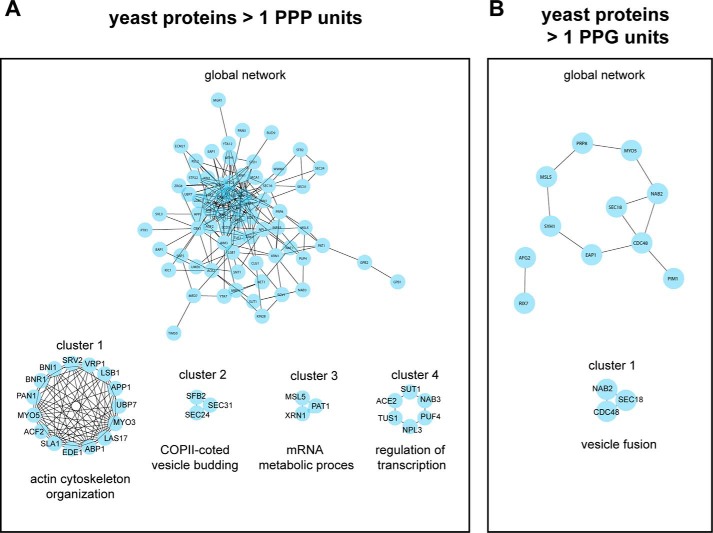
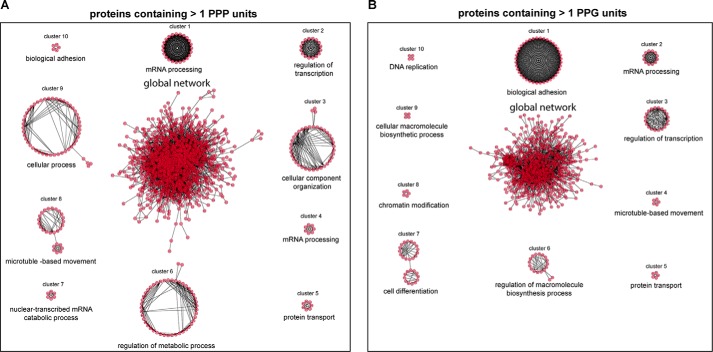
It was recently shown that fully hypusinated eIF-5A1 is a sequence-specific elongation factor, regulating the translation of proteins containing consecutive prolines in the form of repetitive PPP (Pro-Pro-Pro) and/or PPG (Pro-Pro-Gly) sequence units (11). Translation of proteins working together in complexes is regulated in a “proportional synthesis strategy” to provide the cell with the correct stoichiometric amounts for each member of the protein complex (53). Based on these observations, we hypothesize that eIF-5A in being a sequence-specific elongation factor might regulate the optimal translation of PPP and PPG repeat-rich protein complexes and that the observed phenotype after homozygous deletion of Dhs or Dohh is based on disruption of those complexes. Bioinformatic network analysis using STRING database and the Cytoscape software platform with the aim to find network clusters and essential proteins (hubs) was performed for yeast and mouse genes expressing PPP or PPG units. To reduce the complexity of the networks and to focus on proteins with long consecutive proline sequences, we analyzed genes with more than 1 PPP or PPG unit. The exact number of proteins that have been used for network construction and number of nodes in the networks are summarized in Table 2. As depicted in Figs. 11 and 12 PPP and PPG repeat-rich proteins are organized in networks with a considerable variance in the structure of the networks between both species. We exploited three fundamental network parameters (clustering coefficient, average degree, and network heterogeneity) to determine the network interconnectivity (Table 2). Together, the global networks for murine genes show a higher degree of interconnectivity. The average number of neighbors (average degree) and the network heterogeneity have shown higher values for the murine interactome. Interestingly, yeast PPP-rich proteins form a highly connected network with higher clustering coefficients than murine PPP-rich proteins. Furthermore, detailed information for each node inside the networks is available in supplemental Tables 1–4.
TABLE 2.
PPP and PPG repeat-rich proteins showing different network topology in yeast and mouse
Overview of network parameters for protein-protein interaction networks of murine and yeast proteins containing >1 PPP or >1 PPG units.
| Network parameters |
S. cerevisiae |
M. musculus |
||
|---|---|---|---|---|
| >1 PPP unit | >1 PPG unit | >1 PPP unit | >1 PPG unit | |
| No. of proteinsa used for network construction in STRING | 76 | 14 | 1523 | 913 |
| No. of nodes | 70 | 11 | 1256 | 710 |
| Clustering coefficient | 0.392 | 0.136 | 0.238 | 0.251 |
| Average no. of neighbors (average degree) | 7.0 | 2 | 11.1 | 9.5 |
| Network heterogeneity | 0.87 | 0.43 | 1.28 | 1.32 |
a Data were extracted from Ref. 35.
FIGURE 11.
PPP- and PPG-rich proteins form functional networks in yeast. STRING-network analysis for yeast (S. cerevisiae) genes encoding >1 PPP units (A) or >1 PPG units (B). The complete protein-protein interaction networks are depicted in the center of A and B, respectively. Closely connected clusters were directly extracted from each network using the MCODE algorithm. The highest scoring clusters are represented around the whole networks for genes encoding >1 PPP units (A) or >1 PPG units (B). For each cluster, the highest significant GO biological process based on BinGO analysis is indicated. Additional information for complete networks and for all clusters is available in supplemental Tables 1 and 2.
FIGURE 12.
Murine PPP- and PPG-rich proteins are organized in highly interconnected networks. STRING-network analysis for murine (M. musculus) genes encoding >1 PPP units (A) or >1 PPG units (B). The complete protein-protein interaction networks are depicted in the center of A and B, respectively. Closely connected clusters were directly extracted from each network using the MCODE algorithm. The 10 highest scoring clusters are represented around the whole networks for genes encoding >1 PPP units (A) or >1 PPG units (B). For each cluster the highest significant GO biological process based on BinGO analysis is indicated. Additional information for complete networks and for all clusters is available in supplemental Tables 3 and 4.
Identification of Protein Complexes and Biological Processes in Proline Repeat-rich Proteins
To detect densely connected subnetworks (clusters) potentially representing biological modules or protein complexes, we performed MCODE cluster analysis for yeast and murine PPP- or PPG-rich networks. This resulted in a higher number of protein clusters for Mus musculus as compared with S. cerevisiae (Figs. 11 and 12), further supporting the higher complexity of proline repeat-rich proteins in multicellular eukaryotes compared with yeast. We identified 28 and 21 clusters for PPP- and PPG-rich for murine proteins, respectively. In S. cerevisiae, MCODE analysis revealed four and one clusters for PPP-rich and PPG-rich proteins, respectively. Detailed information about the proteins forming these clusters is available in supplemental Tables 1–4. To predict the biological functionality of the clusters, we used BinGO to determine enriched biological processes. Highest scoring gene ontology (GO) terms are depicted in Figs. 11 and 12 for each cluster. Strikingly, the functional activity of these clusters partially overlapped between S. cerevisiae and M. musculus. These include biological processes involved in cytoskeletal organization and regulation, mRNA metabolism and processing, as well as regulation of transcription. However, GO terms associated with cellular differentiation, chromatin modification, and DNA replication appeared exclusively in protein clusters extracted from M. musculus (Fig. 12), pointing further to specialized function of proline repeat-rich proteins in multicellular eukaryotes.
Identification of Highly Connected Network Hubs
Given that the most connected proteins in the network are the most important for cellular survival (54), we used betweenness centrality analysis to identify those network hubs for murine PPP- or PPG-rich proteins, respectively. Betweenness centrality describes the level of control that one node exerts over the interaction of other nodes in a network (55). Information about the betweenness centrality score, for each protein in the global networks for murine PPP- or PPG-rich proteins is available in supplemental Tables 3 and 4. The 10 highest scoring proteins are listed in Table 3. Furthermore, we performed a literature search for known phenotypes associated with a knockdown of these proteins in M. musculus (Table 3). These data suggested that the observed phenotypes in Dhs and Dohh knock-out animals might be based on dysregulated translation of these proteins after inhibition of the hypusine modification.
TABLE 3.
Summary of the 10 highest connected proteins (hubs) in networks from murine proteins containing >1 PPP or >1 PPG units and known knockout phenotype of the genes found in the literature
| Gene name | Knock-out phenotype | |
|---|---|---|
| M. musculus >1 PPP unit | Abl1 | Neonatal lethality (∼75%); thymic and splenic atrophy, T and B cell lymphopenia (65) |
| Crebbp | Embryonic lethality (E9–10.5); defects in hemopoiesis, blood vessel formation, and neural tube closure (66) | |
| Notch1 | Embryonic lethality (before E11.5) (87), neonatally induced KO: growth retardation, defect in thymocyte development (67) | |
| Ep300 | Embryonic lethality (E9–11.5); defects in neurulation, cell proliferation, and heart development (88) | |
| Abl2 | Viable; no hematopoietic abnormalities, reduced litter frequency, behavioral phenotypes (89) | |
| Erbb2 | Embryonic lethality (E10.5); defects in cardiac and neural development (90) | |
| Ptpn23 | Embryonic lethality (E9.5) (91) | |
| Erbb4 | Embryonic lethality (E11); cardiac defects, alterations in hindbrain development (92) | |
| Jund | Viable; reduced postnatal growth, defects in male reproduction (93) | |
| Smarca4 | Embryonic lethality (before E6.5) (94) | |
| M. musculus >1 PPG unit | Abl1 | Neonatal lethality (∼75%); thymic and splenic atrophy, T and B cell lymphopenia (65) |
| Notch1 | Embryonic lethality (before E11.5) (87), neonatally induced KO: growth retardation, defect in thymocyte development (67) | |
| Jund | Viable; reduced postnatal growth, defects in male reproduction (93) | |
| Mapk7 | Embryonic lethality (E9.5–10.5); abnormal embryonic cardiac and vascular development (95) | |
| Ep300 | Embryonic lethality (E9–11.5); defects in neurulation, cell proliferation, and heart development (88) | |
| Dhx8 | No information for knock-out mice available | |
| Crebbp | Embryonic lethality (E9–10.5); defects in hemopoiesis, blood vessel formation, and neural tube closure (66) | |
| Cad | No information for knock-out mice available | |
| Smarca4 | Embryonic lethality (before E6.5) (94) | |
| Ptpn23 | Embryonic lethality (E9.5) (91) |
Discussion
The highly specific and conserved post-translational hypusine modification system has a crucial function in translation regulation (9, 11), and aberrant activation or inhibition is implicated in disparate disorders like cancer and infectious diseases. Notably, hypusine modification of eIF-5A displays an attractive platform for therapeutic intervention. First proof-of-principle studies have already shown promising results in that direction (17, 56–58). However, this modification occurs in all eukaryotic cells and has been shown to be essential for proliferation of lower eukaryotes and mammalian cell lines (59). As these data have been mostly generated in yeast and in vitro, they do not entirely reflect the in vivo situation in mammals. Not surprisingly, there are concerns about whether such a crucial modification can be safely targeted by drugs. From a clinical point of view, further preclinical studies analyzing the biological relevance of hypusinated eIF-5A are needed to evaluate the hypusine axis as a drug target and to predict possible side effects. The well known observation that ∼20–30% of all new therapeutic strategies fail in early clinical trials due to unexpected safety concerns clearly emphasizes that need (60).
Given that mouse models represent an appropriate tool to predict possible side effects (61), we generated several conditional knock-out mouse models and here present the first comprehensive analysis of the biological consequences of the inhibition of the hypusine system in adult mammals (summarized in Fig. 10). Based on our results, we propose that particularly the isoform eIF-5A2 represents a promising target for the treatment of malignant tumors. In contrast, inhibition of the hypusine-mediating enzymes (i.e. DHS and DOHH) seems to have a smaller therapeutic index but nevertheless is tolerated up to the level of haploinsufficiency. Therefore, they might be targeted without relevant side effects in a pathological condition where cells depend on an increased rate of hypusine modification, as compared with normal cells.
Of note, we observed drastic effects upon a complete deletion of either Dhs or Dohh. This is in line with a number of previous observations, indicating that proliferation of eukaryotic cells depends on accurate modification of eIF-5A1. One possible explanation for the strong embryonic and adult phenotype observed after deletion of Dhs and Dohh is that insufficient hypusine modification of eIF-5A1 affects the correct translation of a huge number of proline repeat-rich proteins on the level of single proteins and in terms of protein complexes. We have shown here that murine proline repeat-rich proteins are organized in networks with an increasing connectivity and complexity in multicellular eukaryotes compared with yeast. In fact, recent data suggest that control of phenotypes is partially regulated by multiprotein complexes rather than by single genes (62, 63) and that protein complexes can be used to predict phenotypic effects (64). We believe that disruption of these complexes causes at least some of the knock-out phenotypes described in this study. Differences in biological processes enriched in protein complexes from M. musculus compared with S. cerevisiae may reflect the fact that in higher eukaryotes the hypusine modification is involved in more complex cellular functions and that both hypusine-modifying enzymes are essential. In addition, centrality analysis of global murine networks revealed highly connected hubs (single highly connected proteins) in the networks with crucial biological function for development and viability. Literature analysis uncovers that a single knock-out of most of the 10 highest scoring network hubs leads to embryonic lethality in mice caused by diverse defects (Table 3). Strikingly, dysregulation of each of the top three proteins (Abl1, Crebbp, and Notch1) is associated with defects in hematopoiesis (65–67). This meets our observation of reduced cellularity in bone marrow and spleen after Dhs depletion, indicating a severe defect in hematopoiesis as a consequence of reduced hypusine modification. Together, the observed phenotypes of Dhs and Dohh knock-out animals can be interpreted as a synergistic effect of functional loss of single essential proteins as well as multiprotein complexes due to inhibition of the hypusine modification of eIF-5A. Further in vitro and in vivo studies are needed to prove the functional relevance of these bioinformatic-driven results. Furthermore, because DHS and DOHH might have other cellular targets than eIF-5A, we cannot entirely exclude hypusine-independent effects after deletion of Dhs and Dohh. Studies using mutated hypusine-deficient eIF-5A (eIF-5A-K50 mutant) will be needed to further address this question.
Interestingly, the knock-out of Dohh, although lethal, exerted a lower phenotype penetrance compared with the depletion of Dhs, most likely because of the different modification states of the resulting nonhypusinated eIF-5A1. In contrast to fully hypusinated eIF-5A1, which is localized in the cytoplasm where it regulates translation of specific proteins (11), the observed unhypusinated acetylated and nonacetylated eIF-5A forms in the Dhs- and Dohh-deficient setting, respectively, seem to be located in the nucleus. Therefore, we assume that the phenotypical differences in Dhs and Dohh knock-out animals are caused by yet unknown distinct nuclear functions of the acetylated and the nonacetylated unhypusinated eIF-5A1 precursor. This observation points to modification-dependent and translation-independent functions of the nonhypusinated eIF-5A as it has been demonstrated for other translational factors or ribosomal proteins (68, 69). However, the molecular functions of the differently modified nonhypusinated eIF-5A1 forms have to be elucidated. As current studies highlight that eIF-5A2 has additional transcriptional activity (24), it can be assumed that eIF-5A1 might have similar nuclear activity, which is responsible for its strong phenotype.
It is common knowledge that tumor cells accelerate their translational activity to adapt to the increasing cellular demands (2). Accordingly, previous work unveiled that the hypusine modification system is frequently overexpressed in cancer tissue (16, 17, 70), supporting the hypothesis that eIF-5A facilitates translation of genes with tumor promoting activity as described for other translation factors (71). Indeed, certain oncogenes like c-abl contain polyproline stretches and should therefore be translationally regulated by eIF-5A1. Furthermore, our studies on glioblastoma cell lines have shown a higher sensitivity of tumor cells against the inhibition of hypusine synthesis compared with normal human astrocytes (16), suggesting that certain malignant cells depend on an activated hypusine-dependent translation that is above the activation level in normal cells. An intriguing observation made by using another tumor model is that haploinsufficiency of ribosomal proteins attenuates Myc-dependent malignant transformation without affecting normal cells (72). This clearly highlights the dependence of tumor cells on elevated translational activity. Therefore, we propose that a pharmacological inhibition of the hypusine-modifying enzymes below a level that is required for tumor cell proliferation and disease progression might represent a reliable therapeutic intervention. Interestingly, the concept of submaximal inhibition of hypusine synthesis has been suggested recently for inflammatory diseases (44). However, further studies to define the correct therapeutic index are needed to prevent devastating effects on normal homeostasis. Given the less pronounced phenotype of DOHH depletion, an inhibition of the second step of hypusine modification might represent the preferential target. In that regard, recent studies have demonstrated antiproliferative effects of DOHH inhibition in cancer cells (17, 70, 73).
In contrast to eIF-5A1, the expression of eIF-5A2 is limited to tissue such as testis and few parts of the adult brain (52), but it is highly abundant in cancers such as ovarian, lung, and melanoma (74–76). The eIF-5A2 gene is located on 3q26, a chromosomal region that is frequently amplified in cancer. Moreover, the expression of eIF-5A2 in those entities correlates with survival (77, 78), disease stage, as well as metastasis (79), suggesting that eIF-5A2 function is crucial for tumor development and maintenance but not for normal tissue homeostasis. As eIF-5A2 shares almost 84% homology with eIF-5A1, it can be speculated that eIF-5A2 is also implicated in translation elongation and is responsible for the selective transport and translation of specific mRNA subsets. This is supported by the observation that eIF-5A2 is detectable in the riboproteome of prostate cancer cells compared with normal prostate cells (80).
However, although eIF-5A1 was shown to preferentially regulate the translation of proline-rich proteins (11), the spectrum may be different for eIF-5A2. Furthermore, there are data that propose that eIF-5A2 might also control transcriptional processes in the nucleus, yet more studies are necessary to address this question (24). In this regard, Zender and et al. (21) have shown that nuclear eIF-5A2 exerts oncogenic activity. Therefore, it is conceivable that the two isoforms differ in their function. This hypothesis is supported by our previous observation that the protein interaction networks of both isoforms show isoform-specific protein-protein interaction patterns (81). Although eIF-5A1 seems to control basic cellular processes, eIF-5A2 is rather involved in disease regulation and is essential during tumorigenesis, making it more attractive as a therapeutic target. Encouraging reports in the direction of isoform-specific drug targeting have been described for other essential genes. Muller et al. (82) introduced the concept of collateral vulnerability for the inhibition of enolase 2 in glioblastoma cells. Although targeting enolase 2 in tumor cells inhibited cell growth, normal astrocytes were protected by the expression of enolase 1. Other promising examples for isoform-specific tumor targeting have been demonstrated for pyruvate kinase M (83) and phosphoinositide 3-kinase (84).
Indeed, recent data from in vitro studies support the hypothesis that eIF-5A2 has oncogenic potential (21, 22). In particular, inhibition of eIF-5A2 by shRNA blocked invasion in a melanoma cell line and inhibited growth of ovarian cancer (22, 76). Moreover, foci formation capacity was markedly reduced in liver cancer cells transfected with eIF-5A2 shRNA (21). Intriguingly, in terms of oncogenic activity, the presence of eIF-5A1 in these cancer cells could not compensate for the loss of the second isoform. This leads to the assumption that certain tumor cells are dependent on functional eIF-5A2, and the specific interference with eIF-5A2 activity displays an effective strategy to block proliferation and invasion in eIF-5A2-overexpressing cancers. Remarkably, we found that knock-out of eIF-5A2 is not lethal in adult mice and that animals do not show any conspicuous phenotype, suggesting that the expected toxic effects may be quite low in normal tissues after eIF-5A2 interference. This finding differs from the data from Caenorhabditis elegans studies where phenotypical alterations after deletion of both isoforms were shown (85). Nevertheless, the exact role of eIF-5A2 still needs to be elucidated. This research will be of great interest to address eIF-5A2's detailed molecular function and may enable therapeutic targeting of downstream molecules or pathways in eIF-5A2-dependent tumor entities.
Together, our knock-out mouse models have revealed important aspects of the biology of the hypusine modification system in mammals and constitute a useful resource for further investigation into the molecular function of the hypusine modification under normal and pathological conditions. Furthermore, our data suggest that the hypusine biosynthesis can be harnessed for therapeutic intervention. Even more intriguing is the observation that the cancer-related isoform eIF-5A2 is not crucial for normal homeostasis in mammals. Therefore, we propose that specific inhibitors of eIF-5A2 will exhibit selective toxicity toward malignant cells, making them promising therapeutic targets in the treatment of eIF-5A2-dependent tumors.
Supplementary Material
This work was supported by Deutsche Forschungsgemeinschaft Grants BA 3506/1-1 (to S. B.) and HA 2580/4-1 (to J. H.), Eppendorfer Krebs and Leukämiehilfe e.V., and Swiss National Science Foundation Grant 31003A_150066 (to S. B.). The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Tables S1–S4.
- DHS
- deoxyhypusine synthase
- DOHH
- deoxyhypusine hydroxylase
- EF-P
- elongation factor P
- 4-OHT
- 4-hydroxytamoxifen
- MEF
- mouse embryonic fibroblast
- Esr1
- estrogen receptor 1
- CAG
- CMV early enhancer/chicken β-actin promoter
- PPI
- protein-protein interaction.
References
- 1. Vázquez-Laslop N., Mankin A. S. (2014) Protein accounting in the cellular economy. Cell 157, 529–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ruggero D. (2013) Translational control in cancer etiology. Cold Spring Harb. Perspect. Biol. 5, a012336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Walsh D., Mohr I. (2011) Viral subversion of the host protein synthesis machinery. Nat. Rev. Microbiol. 9, 860–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Grzmil M., Hemmings B. A. (2012) Translation regulation as a therapeutic target in cancer. Cancer Res. 72, 3891–3900 [DOI] [PubMed] [Google Scholar]
- 5. Cooper H. L., Park M. H., Folk J. E. (1982) Post-translational formation of hypusine in a single major protein occurs generally in growing cells and is associated with activation of lymphocyte growth. Cell 29, 791–797 [DOI] [PubMed] [Google Scholar]
- 6. Cooper H. L., Park M. H., Folk J. E., Safer B., Braverman R. (1983) Identification of the hypusine-containing protein hy+ as translation initiation factor eIF-4D. Proc. Natl. Acad. Sci. U.S.A. 80, 1854–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Park M. H., Lee Y. B., Joe Y. A. (1997) Hypusine is essential for eukaryotic cell proliferation. Biol. Signals 6, 115–123 [DOI] [PubMed] [Google Scholar]
- 8. Landau G., Bercovich Z., Park M. H., Kahana C. (2010) The role of polyamines in supporting growth of mammalian cells is mediated through their requirement for translation initiation and elongation. J. Biol. Chem. 285, 12474–12481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Saini P., Eyler D. E., Green R., Dever T. E. (2009) Hypusine-containing protein eIF5A promotes translation elongation. Nature 459, 118–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hauber J. (2010) Revisiting an old acquaintance: role for eIF5A in diabetes. J. Clin. Invest. 120, 1806–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gutierrez E., Shin B.-S., Woolstenhulme C. J., Kim J.-R., Saini P., Buskirk A. R., Dever T. E. (2013) eIF5A promotes translation of polyproline motifs. Mol. Cell 51, 35–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bailly M., de Crécy-Lagard V. (2010) Predicting the pathway involved in post-translational modification of elongation factor P in a subset of bacterial species. Biol. Direct. 5, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Peil L., Starosta A. L., Virumäe K., Atkinson G. C., Tenson T., Remme J., Wilson D. N. (2012) Lys34 of translation elongation factor EF-P is hydroxylated by YfcM. Nat. Chem. Biol. 8, 695–697 [DOI] [PubMed] [Google Scholar]
- 14. Doerfel L. K., Wohlgemuth I., Kothe C., Peske F., Urlaub H., Rodnina M. V. (2013) EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science 339, 85–88 [DOI] [PubMed] [Google Scholar]
- 15. Ude S., Lassak J., Starosta A. L., Kraxenberger T., Wilson D. N., Jung K. (2013) Translation elongation factor EF-P alleviates ribosome stalling at polyproline stretches. Science 339, 82–85 [DOI] [PubMed] [Google Scholar]
- 16. Preukschas M., Hagel C., Schulte A., Weber K., Lamszus K., Sievert H., Pällmann N., Bokemeyer C., Hauber J., Braig M., Balabanov S. (2012) Expression of eukaryotic initiation factor 5A and hypusine forming enzymes in glioblastoma patient samples: implications for new targeted therapies. PLoS One 7, e43468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Balabanov S., Gontarewicz A., Ziegler P., Hartmann U., Kammer W., Copland M., Brassat U., Priemer M., Hauber I., Wilhelm T., Schwarz G., Kanz L., Bokemeyer C., Hauber J., Holyoake T. L., et al. (2007) Hypusination of eukaryotic initiation factor 5A (eIF5A): a novel therapeutic target in BCR-ABL-positive leukemias identified by a proteomics approach. Blood 109, 1701–1711 [DOI] [PubMed] [Google Scholar]
- 18. Scuoppo C., Miething C., Lindqvist L., Reyes J., Ruse C., Appelmann I., Yoon S., Krasnitz A., Teruya-Feldstein J., Pappin D., Pelletier J., Lowe S. W. (2012) A tumour suppressor network relying on the polyamine-hypusine axis. Nature 487, 244–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Maier B., Ogihara T., Trace A. P., Tersey S. A., Robbins R. D., Chakrabarti S. K., Nunemaker C. S., Stull N. D., Taylor C. A., Thompson J. E., Dondero R. S., Lewis E. C., Dinarello C. A., Nadler J. L., Mirmira R. G. (2010) The unique hypusine modification of eIF5A promotes islet beta cell inflammation and dysfunction in mice. J. Clin. Invest. 120, 2156–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Clement P. M., Henderson C. A., Jenkins Z. A., Smit-McBride Z., Wolff E. C., Hershey J. W., Park M. H., Johansson H. E. (2003) Identification and characterization of eukaryotic initiation factor 5A-2. Eur. J. Biochem. 270, 4254–4263 [DOI] [PubMed] [Google Scholar]
- 21. Zender L., Xue W., Zuber J., Semighini C. P., Krasnitz A., Ma B., Zender P., Kubicka S., Luk J. M., Schirmacher P., McCombie W. R., Wigler M., Hicks J., Hannon G. J., Powers S., Lowe S. W. (2008) An oncogenomics-based in vivo RNAi screen identifies tumor suppressors in liver cancer. Cell 135, 852–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Guan X.-Y., Fung J. M., Ma N.-F., Lau S.-H., Tai L.-S., Xie D., Zhang Y., Hu L., Wu Q.-L., Fang Y., Sham J. S. (2004) Oncogenic role of eIF-5A2 in the development of ovarian cancer. Cancer Res. 64, 4197–4200 [DOI] [PubMed] [Google Scholar]
- 23. Zhu W., Cai M. Y., Tong Z. T., Dong S. S., Mai S. J., Liao Y. J., Bian X. W., Lin M. C., Kung H. F., Zeng Y. X., Guan X. Y., Xie D. (2012) Overexpression of EIF5A2 promotes colorectal carcinoma cell aggressiveness by upregulating MTA1 through C-myc to induce epithelial-mesenchymal transition. Gut 61, 562–575 [DOI] [PubMed] [Google Scholar]
- 24. Li Y., Fu L., Li J. B., Qin Y., Zeng T. T., Zhou J., Zeng Z. L., Chen J., Cao T. T., Ban X., Qian C., Cai Z., Xie D., Huang P., Guan X. Y. (2014) Increased expression of EIF5A2, via hypoxia or gene amplification, contributes to metastasis and angiogenesis of esophageal squamous cell carcinoma. Gastroenterology 146, 1701–1713 [DOI] [PubMed] [Google Scholar]
- 25. Overington J. P., Al-Lazikani B., Hopkins A. L. (2006) How many drug targets are there? Nat. Rev. Drug Discov. 5, 993–996 [DOI] [PubMed] [Google Scholar]
- 26. Hopkins A. L., Groom C. R. (2002) The druggable genome. Nat. Rev. Drug Discov. 1, 727–730 [DOI] [PubMed] [Google Scholar]
- 27. Schroeder M., Kolodzik A., Pfaff K., Priyadarshini P., Krepstakies M., Hauber J., Rarey M., Meier C. (2014) In silico design, synthesis, and screening of novel deoxyhypusine synthase inhibitors targeting HIV-1 replication. ChemMedChem 9, 940–952 [DOI] [PubMed] [Google Scholar]
- 28. von Koschitzky I., Kaiser A. (2013) Chemical profiling of deoxyhypusine hydroxylase inhibitors for antimalarial therapy. Amino Acids 45, 1047–1053 [DOI] [PubMed] [Google Scholar]
- 29. Sharpless N. E., Depinho R. A. (2006) The mighty mouse: genetically engineered mouse models in cancer drug development. Nat. Rev. Drug Discov. 5, 741–754 [DOI] [PubMed] [Google Scholar]
- 30. Brown S. D., Moore M. W. (2012) The international mouse phenotyping consortium: past and future perspectives on mouse phenotyping. Mamm. Genome 23, 632–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rodríguez C. I., Buchholz F., Galloway J., Sequerra R., Kasper J., Ayala R., Stewart A. F., Dymecki S. M. (2000) High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat. Genet. 25, 139–140 [DOI] [PubMed] [Google Scholar]
- 32. Schwenk F., Baron U., Rajewsky K. (1995) A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res. 23, 5080–5081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sievert H., Pällmann N., Miller K. K., Hermans-Borgmeyer I., Venz S., Sendoel A., Preukschas M., Schweizer M., Boettcher S., Janiesch P. C., Streichert T., Walther R., Hengartner M. O., Manz M. G., Brümmendorf T. H., et al. (2014) A novel mouse model for inhibition of DOHH mediated hypusine modification reveals crucial function for embryonic development, proliferation and oncogenic transformation. Dis. Model. Mech. 7, 963–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Livak K. J., Schmittgen T. D. (2001) Analysis of relative gene expression data using real time quantitative PCR and the 2(−ΔΔC(T)) method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 35. Mandal A., Mandal S., Park M. H. (2014) Genome-wide analyses and functional classification of proline repeat-rich proteins: potential role of eIF5A in eukaryotic evolution. PLoS One 9, e111800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K. P., Kuhn M., Bork P., Jensen L. J., von Mering C. (2015) STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43, D447–D452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Saito R., Smoot M. E., Ono K., Ruscheinski J., Wang P. L., Lotia S., Pico A. R., Bader G. D., Ideker T. (2012) A travel guide to cytoscape plugins. Nat. Methods 9, 1069–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bader G. D., Hogue C. W. (2003) An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 4, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Maere S., Heymans K., Kuiper M. (2005) BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21, 3448–3449 [DOI] [PubMed] [Google Scholar]
- 40. Doncheva N. T., Assenov Y., Domingues F. S., Albrecht M. (2012) Topological analysis and interactive visualization of biological networks and protein structures. Nat. Protoc. 7, 670–685 [DOI] [PubMed] [Google Scholar]
- 41. Tang Y., Li M., Wang J., Pan Y., Wu F. X. (2015) CytoNCA: a cytoscape plugin for centrality analysis and evaluation of protein interaction networks. Bio Systems 127, 67–72 [DOI] [PubMed] [Google Scholar]
- 42. Pritykin Y., Singh M. (2013) Simple topological features reflect dynamics and modularity in protein interaction networks. PLoS Comput. Biol. 9, e1003243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nishimura K., Lee S. B., Park J. H., Park M. H. (2012) Essential role of eIF5A-1 and deoxyhypusine synthase in mouse embryonic development. Amino Acids 42, 703–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Templin A. T., Maier B., Nishiki Y., Tersey S. A., Mirmira R. G. (2011) Deoxyhypusine synthase haploinsufficiency attenuates acute cytokine signaling. Cell Cycle 10, 1043–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bevec D., Jaksche H., Oft M., Wöhl T., Himmelspach M., Pacher A., Schebesta M., Koettnitz K., Dobrovnik M., Csonga R., Lottspeich F., Hauber J. (1996) Inhibition of HIV-1 replication in lymphocytes by mutants of the Rev cofactor eIF-5A. Science 271, 1858–1860 [DOI] [PubMed] [Google Scholar]
- 46. Sun Z., Cheng Z., Taylor C. A., McConkey B. J., Thompson J. E. (2010) Apoptosis induction by eIF5A1 involves activation of the intrinsic mitochondrial pathway. J. Cell. Physiol. 223, 798–809 [DOI] [PubMed] [Google Scholar]
- 47. Hayashi S., McMahon A. P. (2002) Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Dev. Biol. 244, 305–318 [DOI] [PubMed] [Google Scholar]
- 48. Park J.-H., Aravind L., Wolff E. C., Kaevel J., Kim Y. S., Park M. H. (2006) Molecular cloning, expression, and structural prediction of deoxyhypusine hydroxylase: a HEAT-repeat-containing metalloenzyme. Proc. Natl. Acad. Sci. U.S.A. 103, 51–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ishfaq M., Maeta K., Maeda S., Natsume T., Ito A., Yoshida M. (2012) Acetylation regulates subcellular localization of eukaryotic translation initiation factor 5A (eIF5A). FEBS Lett. 586, 3236–3241 [DOI] [PubMed] [Google Scholar]
- 50. Lee S. B., Park J. H., Kaevel J., Sramkova M., Weigert R., Park M. H. (2009) The effect of hypusine modification on the intracellular localization of eIF5A. Biochem. Biophys. Res. Commun. 383, 497–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lee S. B., Park J. H., Folk J. E., Deck J. A., Pegg A. E., Sokabe M., Fraser C. S., Park M. H. (2011) Inactivation of eukaryotic initiation factor 5A (eIF5A) by specific acetylation of its hypusine residue by spermidine/spermine acetyltransferase 1 (SSAT1). Biochem. J. 433, 205–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jenkins Z. A., Hååg P. G., Johansson H. E. (2001) Human eIF5A2 on chromosome 3q25-q27 is a phylogenetically conserved vertebrate variant of eukaryotic translation initiation factor 5A with tissue-specific expression. Genomics 71, 101–109 [DOI] [PubMed] [Google Scholar]
- 53. Li G. W., Burkhardt D., Gross C., Weissman J. S. (2014) Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell 157, 624–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jeong H., Mason S. P., Barabási A. L., Oltvai Z. N. (2001) Lethality and centrality in protein networks. Nature 411, 41–42 [DOI] [PubMed] [Google Scholar]
- 55. Yoon J., Blumer A., Lee K. (2006) An algorithm for modularity analysis of directed and weighted biological networks based on edge-betweenness centrality. Bioinformatics 22, 3106–3108 [DOI] [PubMed] [Google Scholar]
- 56. Taylor C. A., Liu Z., Tang T. C., Zheng Q., Francis S., Wang T. W., Ye B., Lust J. A., Dondero R., Thompson J. E. (2012) Modulation of eIF5A expression using SNS01 nanoparticles inhibits NF-κB activity and tumor growth in murine models of multiple myeloma. Mol. Ther. 20, 1305–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hauber I., Bevec D., Heukeshoven J., Krätzer F., Horn F., Choidas A., Harrer T., Hauber J. (2005) Identification of cellular deoxyhypusine synthase as a novel target for antiretroviral therapy. J. Clin. Invest. 115, 76–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Moore C. C., Martin E. N., Lee G., Taylor C., Dondero R., Reznikov L. L., Dinarello C., Thompson J., Scheld W. M. (2008) Eukaryotic translation initiation factor 5A small interference RNA-liposome complexes reduce inflammation and increase survival in murine models of severe sepsis and acute lung injury. J. Infect. Dis. 198, 1407–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Park M. H. (2006) The post-translational synthesis of a polyamine-derived amino acid, hypusine, in the eukaryotic translation initiation factor 5A (eIF5A). J. Biochem. 139, 161–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Arrowsmith J., Miller P. (2013) Trial watch: phase II and phase III attrition rates 2011–2012. Nat. Rev. Drug Discov. 12, 569. [DOI] [PubMed] [Google Scholar]
- 61. Hoehndorf R., Hiebert T., Hardy N. W., Schofield P. N., Gkoutos G. V., Dumontier M. (2014) Mouse model phenotypes provide information about human drug targets. Bioinformatics 30, 719–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Dudley A. M., Janse D. M., Tanay A., Shamir R., Church G. M. (2005) A global view of pleiotropy and phenotypically derived gene function in yeast. Mol. Syst. Biol. 1, 2005.0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Gavin A. C., Aloy P., Grandi P., Krause R., Boesche M., Marzioch M., Rau C., Jensen L. J., Bastuck S., Dümpelfeld B., Edelmann A., Heurtier M. A., Hoffman V., Hoefert C., Klein K., et al. (2006) Proteome survey reveals modularity of the yeast cell machinery. Nature 440, 631–636 [DOI] [PubMed] [Google Scholar]
- 64. Fraser H. B., Plotkin J. B. (2007) Using protein complexes to predict phenotypic effects of gene mutation. Genome Biol. 8, R252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tybulewicz V. L., Crawford C. E., Jackson P. K., Bronson R. T., Mulligan R. C. (1991) Neonatal lethality and lymphopenia in mice with a homozygous disruption of the c-abl proto-oncogene. Cell 65, 1153–1163 [DOI] [PubMed] [Google Scholar]
- 66. Oike Y., Takakura N., Hata A., Kaname T., Akizuki M., Yamaguchi Y., Yasue H., Araki K., Yamamura K., Suda T. (1999) Mice homozygous for a truncated form of CREB-binding protein exhibit defects in hematopoiesis and vasculo-angiogenesis. Blood 93, 2771–2779 [PubMed] [Google Scholar]
- 67. Radtke F., Wilson A., Stark G., Bauer M., van Meerwijk J., MacDonald H. R., Aguet M. (1999) Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity 10, 547–558 [DOI] [PubMed] [Google Scholar]
- 68. Pederson T., Tsai R. Y. (2009) In search of nonribosomal nucleolar protein function and regulation. J. Cell Biol. 184, 771–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Valouev I. A., Fominov G. V., Sokolova E. E., Smirnov V. N., Ter-Avanesyan M. D. (2009) Elongation factor eEF1B modulates functions of the release factors eRF1 and eRF3 and the efficiency of translation termination in yeast. BMC Mol. Biol. 10, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Mémin E., Hoque M., Jain M. R., Heller D. S., Li H., Cracchiolo B., Hanauske-Abel H. M., Pe'ery T., Mathews M. B. (2014) Blocking eIF5A modification in cervical cancer cells alters the expression of cancer-related genes and suppresses cell proliferation. Cancer Res. 74, 552–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ruggero D., Montanaro L., Ma L., Xu W., Londei P., Cordon-Cardo C., Pandolfi P. P. (2004) The translation factor eIF-4E promotes tumor formation and cooperates with c-Myc in lymphomagenesis. Nat. Med. 10, 484–486 [DOI] [PubMed] [Google Scholar]
- 72. Barna M., Pusic A., Zollo O., Costa M., Kondrashov N., Rego E., Rao P. H., Ruggero D. (2008) Suppression of Myc oncogenic activity by ribosomal protein haploinsufficiency. Nature 456, 971–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Epis M. R., Giles K. M., Kalinowski F. C., Barker A., Cohen R. J., Leedman P. J. (2012) Regulation of expression of deoxyhypusine hydroxylase (DOHH), the enzyme that catalyzes the activation of eIF5A, by miR-331–3p and miR-642–5p in prostate cancer cells. J. Biol. Chem. 287, 35251–35259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lee N. P., Tsang F. H., Shek F. H., Mao M., Dai H., Zhang C., Dong S., Guan X.-Y., Poon R. T., Luk J. M. (2010) Prognostic significance and therapeutic potential of eukaryotic translation initiation factor 5A (eIF5A) in hepatocellular carcinoma. Int. J. Cancer 127, 968–976 [DOI] [PubMed] [Google Scholar]
- 75. He L.-R., Zhao H.-Y., Li B.-K., Liu Y.-H., Liu M.-Z., Guan X.-Y., Bian X.-W., Zeng Y.-X., Xie D. (2011) Overexpression of eIF5A-2 is an adverse prognostic marker of survival in stage I non-small cell lung cancer patients. Int. J. Cancer 129, 143–150 [DOI] [PubMed] [Google Scholar]
- 76. Khosravi S., Wong R. P., Ardekani G. S., Zhang G., Martinka M., Ong C. J., Li G. (2014) Role of EIF5A2, a downstream target of Akt, in promoting melanoma cell invasion. Br. J. Cancer 110, 399–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Yang G.-F., Xie D., Liu J.-H., Luo J.-H., Li L.-J., Hua W.-F., Wu H.-M., Kung H.-F., Zeng Y.-X., Guan X.-Y. (2009) Expression and amplification of eIF-5A2 in human epithelial ovarian tumors and overexpression of EIF-5A2 is a new independent predictor of outcome in patients with ovarian carcinoma. Gynecol. Oncol. 112, 314–318 [DOI] [PubMed] [Google Scholar]
- 78. Chen W., Luo J.-H., Hua W.-F., Zhou F.-J., Lin M. C., Kung H.-F., Zeng Y.-X., Guan X.-Y., Xie D. (2009) Overexpression of EIF-5A2 is an independent predictor of outcome in patients of urothelial carcinoma of the bladder treated with radical cystectomy. Cancer Epidemiol. Biomarkers Prev. 18, 400–408 [DOI] [PubMed] [Google Scholar]
- 79. Tang D.-J., Dong S.-S., Ma N.-F., Xie D., Chen L., Fu L., Lau S. H., Li Y., Guan X.-Y. (2010) Overexpression of eukaryotic initiation factor 5A2 enhances cell motility and promotes tumor metastasis in hepatocellular carcinoma. Hepatology 51, 1255–1263 [DOI] [PubMed] [Google Scholar]
- 80. Reschke M., Clohessy J. G., Seitzer N., Goldstein D. P., Breitkopf S. B., Schmolze D. B., Ala U., Asara J. M., Beck A. H., Pandolfi P. P. (2013) Characterization and analysis of the composition and dynamics of the mammalian riboproteome. Cell Rep. 4, 1276–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Sievert H., Venz S., Platas-Barradas O., Dhople V. M., Schaletzky M., Nagel C. H., Braig M., Preukschas M., Pällmann N., Bokemeyer C., Brümmendorf T. H., Pörtner R., Walther R., Duncan K. E., Hauber J., et al. (2012) Protein-protein interaction network organization of the hypusine modification system. Mol. Cell. Proteomics 11, 1289–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Muller F. L., Colla S., Aquilanti E., Manzo V. E., Genovese G., Lee J., Eisenson D., Narurkar R., Deng P., Nezi L., Lee M. A., Hu B., Hu J., Sahin E., Ong D., et al. (2012) Passenger deletions generate therapeutic vulnerabilities in cancer. Nature 488, 337–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Goldberg M. S., Sharp P. A. (2012) Pyruvate kinase M2-specific siRNA induces apoptosis and tumor regression. J. Exp. Med. 209, 217–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Edgar K. A., Wallin J. J., Berry M., Lee L. B., Prior W. W., Sampath D., Friedman L. S., Belvin M. (2010) Isoform-specific phosphoinositide 3-kinase inhibitors exert distinct effects in solid tumors. Cancer Res. 70, 1164–1172 [DOI] [PubMed] [Google Scholar]
- 85. Hanazawa M., Kawasaki I., Kunitomo H., Gengyo-Ando K., Bennett K. L., Mitani S., Iino Y. (2004) The Caenorhabditis elegans eukaryotic initiation factor 5A homologue, IFF-1, is required for germ cell proliferation, gametogenesis and localization of the P-granule component PGL-1. Mech. Dev. 121, 213–224 [DOI] [PubMed] [Google Scholar]
- 86. Lee C. H., Um P. Y., Park M. H. (2001) Structure-function studies of human deoxyhypusine synthase: identification of amino acid residues critical for the binding of spermidine and NAD. Biochem. J. 355, 841–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Swiatek P. J., Lindsell C. E., del Amo F. F., Weinmaster G., Gridley T. (1994) Notch1 is essential for postimplantation development in mice. Genes Dev. 8, 707–719 [DOI] [PubMed] [Google Scholar]
- 88. Yao T. P., Oh S. P., Fuchs M., Zhou N. D., Ch'ng L. E., Newsome D., Bronson R. T., Li E., Livingston D. M., Eckner R. (1998) Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell 93, 361–372 [DOI] [PubMed] [Google Scholar]
- 89. Koleske A. J., Gifford A. M., Scott M. L., Nee M., Bronson R. T., Miczek K. A., Baltimore D. (1998) Essential roles for the Abl and Arg tyrosine kinases in neurulation. Neuron 21, 1259–1272 [DOI] [PubMed] [Google Scholar]
- 90. Lee K. F., Simon H., Chen H., Bates B., Hung M. C., Hauser C. (1995) Requirement for neuregulin receptor erbB2 in neural and cardiac development. Nature 378, 394–398 [DOI] [PubMed] [Google Scholar]
- 91. Gingras M. C., Kharitidi D., Chénard V., Uetani N., Bouchard M., Tremblay M. L., Pause A. (2009) Expression analysis and essential role of the putative tyrosine phosphatase His-domain-containing protein tyrosine phosphatase (HD-PTP). Int. J. Dev. Biol. 53, 1069–1074 [DOI] [PubMed] [Google Scholar]
- 92. Gassmann M., Casagranda F., Orioli D., Simon H., Lai C., Klein R., Lemke G. (1995) Aberrant neural and cardiac development in mice lacking the ErbB4 neuregulin receptor. Nature 378, 390–394 [DOI] [PubMed] [Google Scholar]
- 93. Thépot D., Weitzman J. B., Barra J., Segretain D., Stinnakre M. G., Babinet C., Yaniv M. (2000) Targeted disruption of the murine junD gene results in multiple defects in male reproductive function. Development 127, 143–153 [DOI] [PubMed] [Google Scholar]
- 94. Bultman S., Gebuhr T., Yee D., La Mantia C., Nicholson J., Gilliam A., Randazzo F., Metzger D., Chambon P., Crabtree G., Magnuson T. (2000) A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol. Cell 6, 1287–1295 [DOI] [PubMed] [Google Scholar]
- 95. Regan C. P., Li W., Boucher D. M., Spatz S., Su M. S., Kuida K. (2002) Erk5 null mice display multiple extraembryonic vascular and embryonic cardiovascular defects. Proc. Natl. Acad. Sci. U.S.A. 99, 9248–9253 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.