Background: Camptothecin induces replication-associated DSB formation.
Results: hMSH5-FANCJ facilitates the repair of camptothecin-induced DSBs.
Conclusion: Functional interplay between hMSH5 and FANCJ is involved in replication stress-induced DSB repair.
Significance: Understanding the mechanisms of DSB repair, induced by replication stress, is pivotal to develop new anticancer targets and therapeutic strategies.
Keywords: DNA-binding protein, DNA damage, DNA repair, DNA replication, DNA topoisomerase
Abstract
Replication stress from stalled or collapsed replication forks is a major challenge to genomic integrity. The anticancer agent camptothecin (CPT) is a DNA topoisomerase I inhibitor that causes fork collapse and double-strand breaks amid DNA replication. Here we report that hMSH5 promotes cell survival in response to CPT-induced DNA damage. Cells deficient in hMSH5 show elevated CPT-induced γ-H2AX and RPA2 foci with concomitant reduction of Rad51 foci, indicative of impaired homologous recombination. In addition, CPT-treated hMSH5-deficient cells exhibit aberrant activation of Chk1 and Chk2 kinases and therefore abnormal cell cycle progression. Furthermore, the hMSH5-FANCJ chromatin recruitment underlies the effects of hMSH5 on homologous recombination and Chk1 activation. Intriguingly, FANCJ depletion desensitizes hMSH5-deficient cells to CPT-elicited cell killing. Collectively, our data point to the existence of a functional interplay between hMSH5 and FANCJ in double-strand break repair induced by replication stress.
Introduction
Faithful DNA replication ensures the accurate distribution of genetic information from parent cells to daughter cells. Failure of accurate DNA replication creates mutations and/or chromosome rearrangements, which are the most common DNA lesions underlying cell death, cancer, and genetic disorders (1). Recent studies have not only advanced our understanding of DNA repair mechanisms that safeguard genome integrity but also identified key steps that can be therapeutically manipulated for cancer intervention. For instance, disruption of DNA replication has been commonly exploited in cancer therapy (2). This involves the use of DNA topoisomerase I inhibitors, such as camptothecin (CPT)2 and its derivatives, which specifically trap topoisomerase I at the replication fork to prevent the religation of nicked DNA strands (3). Single-strand breaks created by CPT could be converted into double-strand breaks (DSBs) during DNA replication. The repair of these so-called replication-associated DSBs or one-ended DSBs is orchestrated by the actions of DNA damage response and repair pathways (4).
Between the two predominant DSB repair pathways, i.e. homologous recombination (HR) and nonhomologous end joining, HR plays a predominant role in the repair of CPT-induced DSBs. Because CPT treatment fails to elicit Ku70/80 foci formation, the nonhomologous end joining pathway is unlikely to play a role in the processing of CPT-induced DSBs (5). HR-mediated DSB repair requires a homologous DNA template and is therefore active only in the S and G2 phases of the cell cycle. The process of HR begins with DSB recognition by the MRE11-RAD50-NBS1 complex, which, together with CtIP, initiates short end resections to generate substrates that can be further processed by two other complexes containing either DNA2 or EXO1 (6). This allows the generation of a long single-stranded DNA that is capable of initiating Rad51-dependent strand invasion of homologous DNA template (4, 7). The resulting HR intermediate structures can be channeled into one of the downstream processes such as double-Holliday junction repair, synthesis-dependent strand annealing, sister chromatid exchange, or break-induced replication (8, 9). In due course, these pathways help to maintain genomic stability, of which double-Holliday junction repair is conceived as the main process for rescuing collapsed DNA replication forks.
Successful DSB recognition and repair at the collapsed replication fork also requires the action of ATR DNA damage signaling to coordinate DSB repair with DNA replication and cell cycle regulation (10). ATR is primarily activated in the S phase by RPA-coated ssDNA that often arises during DNA replication stalling and DSB end resection. RPA-ssDNA recruits the ATR-ATRIP complex, leading to full activation of ATR together with the Rad17 clamp loader, the 9-1-1 complex, and TopBP1 (10, 11). ATR phosphorylates Chk1 to transduce an inhibitory signal to the CDC25 phosphatase, which prevents the dephosphorylation of CDKs and triggers G2/M arrest (12). The delay of cell cycle progression is essential for giving cells sufficient time to fully repair DNA lesions before entry into mitosis.
In the present study, we investigated the role of hMSH5 (human MutS homologue 5) in CPT-induced DSB repair. The MutS homologue MSH5 was initially identified as a meiotic recombination factor in yeast and mice (13–15), raising the possibility that MSH5 might have a role in the process of recombination. Previous studies have shown that hMSH5 functions in the process of DNA damage response through coordinating with c-Abl and p73 (16–18). Furthermore, hMSH5 sensitizes human cells to ionizing radiation (IR) and renders cells resistant to cisplatin (CDDP) (17, 19). Consistent with a role in recombination, hMSH5 interacts with hMRE11 (20), and hMSH5 promotes HR repair and is recruited to I-SceI generated DSBs (21). In addition, depletion of Rad51 compromises CDDP-induced hMSH5 foci formation, whereas the dominant negative hMSH5 Y742F mutation increases CDDP sensitivity and impairs Rad51 loading to I-SceI generated DSBs (19, 21). Here, we demonstrate that hMSH5 directly interacts with FANCJ, and the resulting complex promotes HR and facilitates the ATR-Chk1 signaling in response to CPT.
Experimental Procedures
Plasmids and shRNA Constructs
The generation of the mammalian expression construct Flag-hMSH5 was described previously (22). All hMSH5 fragments were subcloned into pcDNA6 vector possessing a 3× Flag tag. For GST pulldown assays, hMSH5 1–225 was subcloned into the expression vector pGEX-6p-1, and all other hMSH5 fragments were subcloned into pGEX-6p-2. pcDNA3-myc-his-BACH1 was obtained from Addgene (Cambridge, MA). The generation of shRNA constructs was performed as described previously (23). The RNAi target sequences were hMSH5 sh2 (5′-TGGGCCTGAGGGATGCCTG-3′) (17), hMSH5 sh4 (5′-ATACTAGTGACTCC ACTATCC-3′), and FANCJ sh2 (5′-GTACAGTACCTCACCTTAT-3′). The sequence of sgRNA target was 5′-TAGGGATAACAGGGTAATGG-3′ (sgRNA-HRIR3).
Antibodies
Antibodies used in the study were anti-Myc (631206; Clontech), anti-γ-H2AX (05-636; Millipore, Billerica, MA), anti-RPA2 (NB600-565; Novus, Littleton CO), anti-MRE11 (NB100-142; Novus), anti-α-tubulin (T6199; Sigma), anti-Flag M2 (F1804; Sigma), anti-BACH1 (B1310; Sigma), anti-actin (A2066; Sigma), anti-Rad51 (sc-8349; Santa Cruz, Dallas, TX), anti-PCNA (sc-56; Santa Cruz), anti-Chk1 (sc-8408; Santa Cruz), anti-GFP (sc-9996; Santa Cruz), anti-BRCA1 (sc-6954 and sc-642; Santa Cruz), anti-Chk2 (2662; Cell Signaling Technology, Danvers, MA), anti-phospho-Chk2 (Thr-68) (2661; Cell Signaling Technology), anti-phospho-Chk1 (Ser-345) (2348; Cell Signaling Technology), and anti-histone H3 (39163; Active Motif, Carlsbad, CA). Purified anti-hMSH5 antibody was described previously (22).
Cell Culture, Transfection, and Generation of Stable Cell Lines
HEK293T and U2OS cells were maintained in DMEM/high glucose (GE Healthcare HyClone, Logan, UT) supplemented with 5% FBS, 5% NBS, and 1× antibiotic-antimycotic (Invitrogen/Thermo Fisher Scientific). Plasmid DNA transfections were carried out using either a standard calcium phosphate method or an Amaxa Nucleofector (Lonza Group Ltd., Allendale, NJ). HEK293T and U2OS cells stably expressing hMSH5 sh2, hMSH5 sh4, FANCJ sh2, and Flag-hMSH5 were generated through the selection of blasticidin- or neomycin-resistant colonies.
Immunoblotting and Immunoprecipitation (IP)
Cell lysates were prepared using NETN buffer (20 mm Tris-HCl, pH 8.0, 100 mm NaCl, 1 mm EDTA, and 0.5% Nonidet P-40) supplemented with 1× protease inhibitor mixture (Thermo Fisher Scientific). Bradford protein assay (Bio-Rad) was used to determine the protein concentrations of each lysate. Proteins were resolved by SDS-PAGE, transferred onto nitrocellulose membranes, and immunoblotted with indicated antibodies. For IP, cells were lysed in 400 μl of NETN buffer, and 5% of the lysates were saved as input. The rest of the lysates were incubated with primary antibodies followed by incubation with protein A/G-agarose beads (Invitrogen/Thermo Fisher Scientific), which were then washed four times with NETN buffer. Immunocomplexes were boiled in 2× Laemmli sample buffer for 10 min before they were subjected to SDS-PAGE.
Chromatin Fractionation
The assay was performed as described with modifications (24). Cell pellets were resuspended in 200 μl of solution A (10 mm HEPES, pH 7.9, 10 mm KCl, 1.5 mm MgCl2, 340 mm Sucrose, 1 mm DTT, 10 mm NaF, 1 mm Na3VO4, protease inhibitors, and 0.1% Triton X-100) and left on ice for 5 min. The nuclear (P1) and cytoplasmic fractions (S1) were separated by centrifugation at 1300 × g for 4 min. The P1 fraction was resuspended in 1 ml of solution A and left on ice for 5 min and then centrifuged at the same speed to remove remaining cytoplasmic proteins. The remaining pellet was resuspended in 200 μl of solution B (3 mm EDTA, 0.2 mm EGTA, 1 mm DTT and protease inhibitors) and left on ice for 10 min. Soluble nuclear proteins (S2) were obtained by centrifugation at 1700 × g for 4 min. The pellet was resuspended in 1 ml of solution B and centrifuged at 10,000 × g for 1 min. The insoluble chromatin fraction (P2) was resuspended in 100 μl of Laemmli sample buffer and sonicated for a total of 5 min before boiling. For micrococcal nuclease digestion, original cell preparations were split into two halves, of which one half was used for the procedure mentioned above to obtain S1, P1, S2, and P2 fractions. The other half was resuspended in 200 μl of solution A to separate P1 and S1. The P1 fraction was then resuspended in 200 μl of solution A containing 1 mm CaCl2 and 50 units of micrococcal nuclease and incubated at 37 °C for 2 min. After centrifugation at 1300 × g for 4 min, the nuclear (P1′) and cytoplasmic (S1′) fractions were obtained. The P1′ fraction was additionally washed with solution A and extracted with solution B to yield S2′ and P2′.
Immunofluorescence Microscopy
To analyze γ-H2AX and Rad51 foci, U2OS cells were grown on glass coverslips, fixed in 3% paraformaldehyde for 12 min, permeabilized in 0.5% Triton X-100 for 5 min, blocked in PBS with 3% BSA, and incubated with primary antibody for 1 h at room temperature. For RPA2 and BRCA1 foci detection, cells were pre-extracted for 5 min at 4 °C with the pre-extraction buffer (25 mm HEPES, pH 7.5, 50 mm NaCl, 1 mm EDTA, 3 mm MgCl2, 300 mm sucrose, and 0.5% Triton X-100) before being subjected to paraformaldehyde fixation. For GFP-hMSH5, FANCJ, and BRCA1 foci detection, cells were treated with pre-extraction buffer and fixed in 100% ice-cold methanol for 10 min at −20 °C. After primary antibody incubation and washes in PBS, cells were incubated with Alexa 488- or 555-conjugated secondary antibodies for 1 h at room temperature. Coverslips were then mounted onto glass slides with Prolong gold antifade reagent containing DAPI (Invitrogen). Images were captured using a Leica Leitz DMRB microscope.
GST Pulldown Assay
GST-tagged hMSH5 fragments were expressed in BL21(DE3)-RIL cells with an 8-h IPTG induction. The bacterial cells were resuspended in the sonication buffer (20 mm Tris-HCl, pH 8.0, 120 mm KCl, 10% glycerol, 1% Triton X-100, 1 mm EDTA, 1 mm DTT) supplemented with 0.2 mg/ml lysozyme and incubated at 37 °C for 5 min prior to sonication. The supernatants were incubated with glutathione-Sepharose 4B beads (GE Healthcare) at 4 °C overnight, followed by incubation with lysates from HEK293T cells expressing myc-FANCJ (precleared with glutathione-Sepharose 4B beads). Protein-bound beads were transferred to columns and washed prior to protein elution with 10 mm glutathione elution buffer. Eluted proteins were then subjected to SDS-PAGE and immunoblotting.
Clonogenic Survival Assay
Cells were seeded in triplicate in 6-cm plates at a density of 500 cells/plate. 16 h after seeding, cells were treated with CPT or CDDP for 1 or 24 h or with doxorubicin or hydroxyurea for 1 h and then released into fresh medium. After a 14-day culture, colonies with more than 50 cells were counted either directly (HEK293T cells) or after fixation and staining with 0.5% crystal violet in 20% ethanol (U2OS cells).
Homologous Recombination Assay
The generation of the 293TLα/pMMR-IR3 reporter cell line was described previously (25), in which a functional RFP gene was restored following HR. Recombination was initiated by transfecting cells with either pCBA-(I-SceI) or sgRNA-HRIR3 together with Cas9 using an Amaxa Nucleofector (Lonza Group Ltd.). Numbers of red fluorescent cells, at 7 or 4 days after transfection, were recorded and analyzed with a FACSCalibur (Becton Dickinson) using CellQuest Pro (BD Biosciences).
Results
Identification of FANCJ as an hMSH5 Binding Partner
To identify hMSH5 interacting proteins, the 293T/f5 cell line stably expressing Flag-tagged hMSH5 was utilized to perform anti-Flag IP and MS-based analysis (16). A protein at ∼140 kDa was identified as FANCJ (data not shown). FANCJ, also known as BRIP1 or BACH1, is the gene for Fanconi anemia complementation group J and participates in interstrand cross-link repair. FANCJ is also implicated to function in the processes of HR, DNA damage signaling, and DNA replication (26–30). To confirm the interaction between hMSH5 and FANCJ, reciprocal co-IP analyses were performed in the presence of DNase I (Fig. 1A), the latter was used to eliminate the potential bridging effect of DNA. We found that these two proteins interacted with each other, and this interaction was independent of DNA. This interaction was then validated by co-IP analysis of endogenous proteins (Fig. 1B). In addition, RNAi-mediated hMSH5 knockdown significantly reduced the amount of FANCJ protein that could be co-immunoprecipitated by the anti-hMSH5 antibody, attesting to the specificity of this assay (Fig. 1B).
FIGURE 1.
hMSH5 directly interacts with FANCJ. A, reciprocal co-IP analysis of the interaction between hMSH5 and FANCJ. Myc-FANCJ and Flag-hMSH5 were co-expressed in HEK293T cells and immunoprecipitated with an anti-Flag (left panel) or anti-Myc (right panel) antibody. A total of 20 units DNase I was used. B, analysis of the interaction between endogenous hMSH5 and FANCJ. Cell lysates of parental and hMSH5-depleted U2OS cells were used to perform co-IP with either IgG or the anti-hMSH5 antibody. Depletion of hMSH5 was accomplished by transfecting cells with hMSH5 shRNA. C, schematic representation of hMSH5 domain structures. D, co-IP analysis of FANCJ interaction with the full-length hMSH5 and truncated hMSH5 fragments performed with either an anti-Flag or anti-Myc antibody. E, GST pulldown analysis of the hMSH5-FANCJ interaction and the FANCJ-interacting domain on hMSH5. The full-length hMSH5 and four truncated hMSH5 fragments were expressed as GST fusion proteins in bacterial BL21-(DE3)-RIP cells and captured by glutathione-Sepharose 4B beads. Myc-FANCJ was expressed in HEK293T cells, and the resulting cell lysate was used to incubate with the protein-bound beads. IB, immunoblot.
To decipher the FANCJ binding domain on hMSH5, we made a series of truncated fragments of hMSH5 (Fig. 1C). The results of the co-IP experiments indicated that all fragments with the first 225 amino acids were capable of binding to FANCJ, whereas the conserved DNA-binding and ATPase domains were not required for this interaction (Fig. 1D). The interaction between hMSH5 1–225 and FANCJ was further confirmed by a reciprocal co-IP assay (Fig. 1D, right panel). Thus, the hMSH5 1–225 fragment contains the core region for FANCJ interaction.
Next, to validate the existence of a direct physical interaction between hMSH5 and FANCJ, we carried out GST pulldown assays. GST-tagged full-length and the 1–225 fragment of hMSH5 were able to interact with FANCJ, whereas none of the other hMSH5 fragments showed an interaction with FANCJ (Fig. 1E). In addition, the hMSH5-FANCJ interaction did not require the helicase activity of FANCJ or its interaction with BRCA1 (31, 32) (data not shown). Taken together, these results indicate that FANCJ is a direct hMSH5 binding partner.
Depletion of hMSH5 Sensitized Cells to CPT Treatment
To gain insights into how hMSH5 was involved in CPT-provoked DNA damage response, especially enlightened by its interaction with FANCJ, we utilized a stable hMSH5 knockdown cell line 293T/hMSH5-sh2. The silencing effect by hMSH5 sh2 shRNA was confirmed by immunoblotting and quantification analysis (17) (Fig. 2A). Clonogenic survival assays showed that hMSH5 depleted cells were sensitive to CPT treatment, but not to doxorubicin or hydroxyurea, and this CPT sensitivity could be rescued by the expression of RNAi-resistant hMSH5 (Fig. 2B). To validate this observation and to rule out other potential nonspecific effects, hMSH5 was silenced in U2OS cells using either hMSH5 sh2 or sh4 shRNA (Fig. 2C). Depletion of hMSH5 had no effect on the levels of FANCJ and other HR proteins (Fig. 2D) (data not shown). Consistent with the role of hMSH5 in cisplatin (CDDP)-induced DNA damage repair (19), depletion of hMSH5 led to an increased sensitivity to both CDDP (1 h of treatment) and CPT (1 or 24 h of treatment) in U2OS cells (Fig. 2E; see also Fig. 7D). This sensitivity was hMSH5-dependent, because it could be reverted by the expression of RNAi-resistant hMSH5. Finally, it was evident that CPT sensitivity was related to the levels of hMSH5 (Fig. 2, C and E). Together, these results suggest that hMSH5 is involved in CPT-induced DNA damage repair.
FIGURE 2.
hMSH5 deficiency leads to CPT sensitivity. A, immunoblotting analysis of the effect of hMSH5 knockdown in 293T cells. The levels of endogenous hMSH5 were quantified and normalized by the levels of tubulin. The bar graph is used to show mean ± S.D. from three independent experiments. *, p < 0.05. B, clonogenic survival analysis of parental and hMSH5-deficient 293T cells treated with various DNA-damaging agents. All drug treatments were for 1 h. C, immunoblotting analysis of hMSH5 knockdown in U2OS cells. Quantifications were performed similarly as in A. **, p < 0.01. D, immunoblotting analysis of the levels of hMSH5 and FANCJ in hMSH5-depleted and FANCJ-depleted U2OS cells. **, p < 0.01. E, clonogenic survival analysis of parental and hMSH5-deficient U2OS cells treated with either CPT or CDDP for 1 h. Depletion of endogenous hMSH5 was achieved with either hMSH5 shRNA sh2 or sh4. Survival curves were constructed from the mean of three independent experiments. Ctrl, control. IB, immunoblot; NS, not significant.
FIGURE 7.
hMSH5 promotes CPT-induced FANCJ chromatin recruitment. A, U2OS cells expressing GFP-hMSH5 were treated with 50 nm CPT for 24 h. Cells were then analyzed by GFP and FANCJ immunostaining. Scale bar, 10 μm. The Venn diagram is used to show the percentage of hMSH5-FANCJ co-localization. The total number of foci (summation of hMSH5 only, FANCJ only, and hMSH5-FANCJ) is presented as 100%, and the percentage of co-localization is 51.8 ± 7.3%. Data is the mean ± S.D. from three independent experiments. Each experiment included 100 cells (the range of average total foci number is from 11 to 24). B, U2OS cells were transfected with a control or hMSH5 shRNA construct, and transfected cells were either left untreated or treated with 1 μm CPT for 1 h prior to their collection at 24 h. Cells were then subjected to chromatin fractionation analysis. The relative levels of chromatin-bound FANCJ (normalized by the levels of histone H3) were determined from three independent experiments. *, p < 0.05. C, U2OS cells were transfected with Flag-hMSH5, as well as a control or FANCJ shRNA construct, and transfected cells were either left untreated or treated with 1 μm CPT for 1 h prior to their collection at 24 h. Cells were then subjected to chromatin fractionation analysis. The relative levels of chromatin-bound hMSH5 (normalized by the levels of histone H3) were determined from three independent experiments. **, p < 0.01. D, clonogenic survival analysis of stable U2OS cell lines that were deficient in hMSH5, FANCJ or both. Stable U2OS knockdown cell lines were derived from U2OS cells transfected with hMSH5 sh4 and/or FANCJ sh2 constructs. Cells were treated with CPT for 24 h at indicated concentrations. Each data point represents the mean ± S.D. from three independent experiments. E, clonogenic survival analysis of FANCJ-depleted U2OS cells in response to CDDP treatment. F, immunoblotting analysis of Chk1 phosphorylation in stable hMSH5 knockdown and/or FANCJ knockdown U2OS cells treated with 1 μm CPT. Cells were collected at 24 h after 1 h CPT treatment. The graphs to the right represent the relative levels of p-Chk1 (ratio of p-Chk1/Chk1), FANCJ, and hMSH5 (normalized by the levels of actin) that were determined from four independent experiments. *, p < 0.05. **, p < 0.01. IB, immunoblot; NS, not significant; shCtrl, shRNA control; shhMSH5, shRNA hMSH5.
hMSH5 Deficiency Led to Impaired HR and the Accumulation of CPT-induced DNA Lesion
Next, we examined the process of CPT-induced DSB repair through foci analysis of well established DSB repair markers. Briefly, U2OS cells were treated with 1 μm CPT for 1 h. To determine the formation of CPT-induced γ-H2AX foci, cells were processed at 1, 4, and 8 h after CPT treatment. The pattern of γ-H2AX foci in the parental U2OS cells closely resembled those observed previously (33). Although silencing of hMSH5 had no effect on the initial γ-H2AX foci formation, the percentages of hMSH5-deficient cells positive for γ-H2AX (> 15 foci) were significantly higher than those of the controls at 4 and 8 h (Fig. 3, A and D). This suggested that the repair of CPT-induced DSBs was delayed in hMSH5-deficient cells. Likewise, in comparison to the control cells, the percentage of hMSH5-deficient cells positive for RPA2 foci (> 15 foci) increased significantly at 48 and 72 h after CPT treatment, indicating the persistence of unrepaired DNA damage, especially DSBs (Fig. 3, B and D).
FIGURE 3.
hMSH5 deficiency leads to defective HR repair of CPT-induced DSBs. A–C, representative images of γ-H2AX, RPA2, and Rad51 foci formation in response to CPT treatment. U2OS cells were transfected with either a control or hMSH5 shRNA and treated with 1 μm CPT for 1 h. Cells were fixed at 1, 4, and 8 h after treatment for γ-H2AX or Rad51 immunostaining but were fixed at 24, 48, and 72 h for RPA2 immunostaining. Scale bar, 10 μm. D, cells with at least 15 γ-H2AX, 15 RPA2, or 10 Rad51 foci were scored from 100 cells in each individual experiment. Mean and standard deviation were determined from three independent experiments. Statistically significant differences are indicated with asterisks: *, p < 0.05; **, p < 0.01. shCtrl, shRNA control; shhMSH5, shRNA hMSH5.
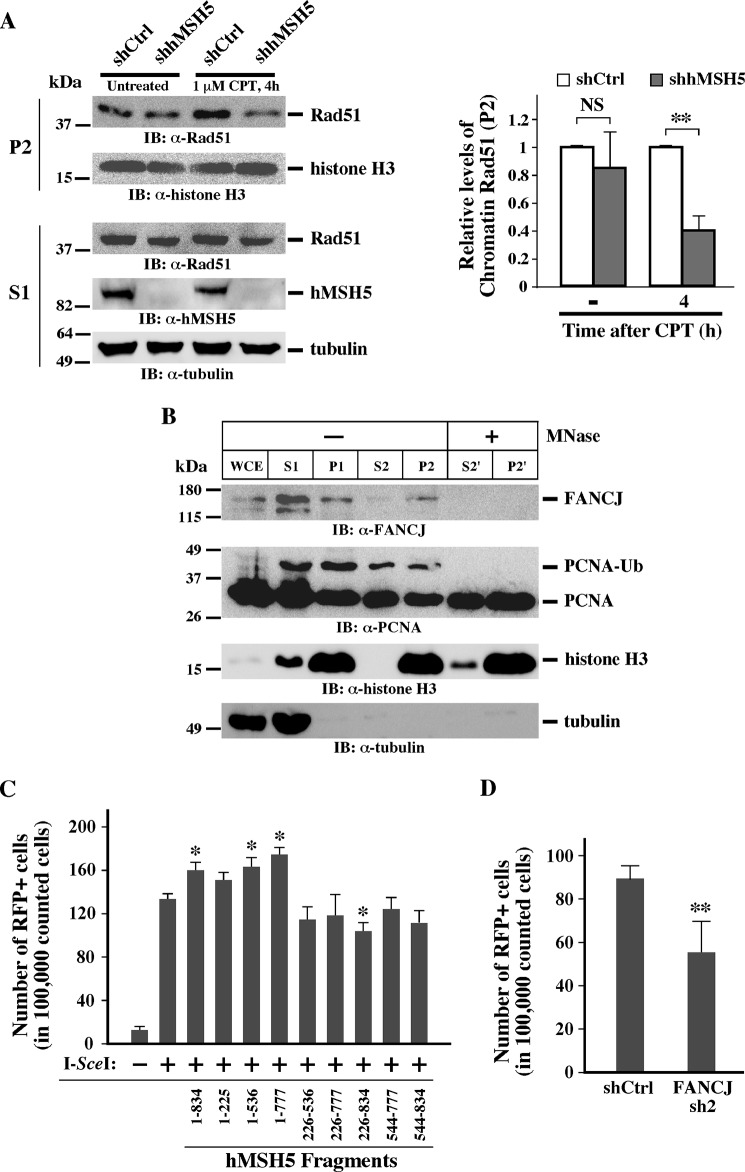
Because the persisting RPA2 foci in hMSH5-depleted cells could be a surrogate marker for defective HR, we next examined the formation of Rad51 foci at 4 and 8 h after CPT treatment. Evidently, a reduction of Rad51 foci formation was observed in hMSH5-deficient cells at both time points (Fig. 3, C and D). In these cells, the Rad51 staining was often diffused; this pattern of Rad51 staining has been previously reported and was attributed to a failure in Rad51 chromatin recruitment (34). To substantiate this observation, we analyzed CPT-provoked chromatin loading of Rad51 by carrying out a chromatin fractionation assay (24). Specifically, the levels of Rad51 in the chromatin fraction (P2) were determined by immunoblotting and analyzed by quantification (Fig. 4A). The P2 fraction designated the chromatin fraction in which FANCJ and the monoubiquitinated PCNA chromatin association could be disrupted by micrococcal nuclease treatment (Fig. 4B). In agreement with the results of Rad51 foci analysis, the level of chromatin-associated Rad51 was markedly lower at 4 h after CPT treatment in hMSH5-depleted cells (Fig. 4A), indicative of an HR defect.
FIGURE 4.
hMSH5 deficiency disrupts CPT-induced Rad51 chromatin loading and HR. A, chromatin fractionation analysis of Rad51 loading. U2OS cells were transfected with a control or hMSH5 shRNA and treated with 1 μm CPT for 1 h, and cells were collected at 4 h after treatment. Cytoplasmic (S1) and chromatin (P2) proteins were separated and subjected to immunoblotting analysis. The relative levels of chromatin Rad51 were determined by quantification and were normalized by the levels of histone H3. Four independent experiments were performed to obtain the mean and standard deviation (right panel). **, p < 0.01. B, chromatin fractionation assay performed with U2OS cells treated with CPT (see “Experimental Procedures” for details). Whole cell extract was obtained from untreated U2OS cells. Tubulin was used as a cytoplasmic protein marker, and histone H3 was used as a nuclear protein marker. P1 fraction was treated with micrococcal nuclease to obtain S2′ and P2′. Note the disappearance of PCNA-Ub and FANCJ in P2′ compared with P2 (lanes 5 and 7). C, HR reporter assay performed with the 293TLα/pMMR-IR3 reporter cell line (25). HR reporter cells were first transfected with one of the hMSH5 fragments. After 48 h, cells were transfected with the I-SceI-encoding vector pCBA-(I-SceI). FACS analysis was performed 7 days after I-SceI transfection. *, p < 0.05. D, HR reporter analysis of the effect of FANCJ deficiency. Reporter cells were first transfected with FANCJ sh2 and then, 48 h later, with Cas9 and sgRNA to generate a DSB at the I-SceI site. Cells expressing red fluorescent protein were detected and analyzed by FACS at day 4 after the transfection. **, p < 0.01. IB, immunoblot; MNase, micrococcal nuclease; NS, not significant; shCtrl, shRNA control; WCE, whole cell extract.
Consequently, we performed the HR reporter assay (25) to directly assess the role of hMSH5-FANCJ in HR. Consistent with our previous findings (21), the full-length hMSH5 promoted HR (Fig. 4C). Intriguingly, the N-terminal region of hMSH5, especially the 1–225 fragment, played an important role as the fragments lacking 1–225 failed to promote HR (Fig. 4C). Because hMSH5 1–225 interacts with FANCJ, these results raised the possibility that hMSH5 promoted HR by recruiting FANCJ. There are conflicting reports for the role of FANCJ in HR (27, 29), but we did observe a moderate reduction in DSB-induced HR when FANCJ was depleted from the HR reporter cells (Fig. 4D). Together, we found that hMSH5 facilitated the chromatin loading of Rad51 to promote HR in CPT-induced DSB repair.
hMSH5 Was Involved in the Control of Cell Cycle Checkpoints
To explore whether hMSH5 was involved in CPT-elicited DNA damage signaling, we scrutinized markers of checkpoints over a period of 48 h after CPT treatment. Depletion of hMSH5 had no effect on the initial Chk2 activation (Fig. 5A, lanes 3 and 4), indicating that the ATM signaling pathway was not affected, but hMSH5 depletion increased the levels of phospho-Chk2 at 4 h (Fig. 5A, fifth and sixth lanes, and top right bar graph). Similarly, hMSH5 deficiency had no effect initially on CPT-induced Chk1 phosphorylation, suggesting that the ATR signaling initiated normally. However, hMSH5 deficiency led to a substantial reduction of CPT-induced Chk1 phosphorylation at 16–28 h after treatment (Fig. 5, A and B). No difference in Chk1 phosphorylation was observed beyond this period. Conversely, there was no difference in RPA2 phosphorylation (Fig. 5A).
FIGURE 5.
hMSH5 deficiency results in aberrant cell cycle checkpoints activation. A, U2OS cells were transfected with a control or hMSH5 shRNA. Cells were then treated with 1 μm CPT for 1 h before being released into drug-free medium and collected at the indicated time points. Untreated cells were collected at 48 h after transfection. Cell lysates were used to perform immunoblotting with various antibodies as indicated. For the graphs at the right, the average ratios of p-Chk2/Chk2 and p-Chk1/Chk1 were obtained from three independent experiments. *, p < 0.05. B, the levels of p-Chk1 were determined between 16 and 28 h after CPT treatment. The average levels of p-Chk1 and standard deviations were obtained from three independent experiments and presented in the graph at the right. C, cell cycle analysis of U2OS cells that were treated the same way as described in A. Cell cycle analysis was conducted at indicated time points after CPT treatment, and percentages of cells in each phase of the cell cycle are indicated. IB, immunoblot; NS, not significant; shCtrl, shRNA control; shhMSH5, shRNA hMSH5.
These observations prompted us to examine the potential effects of hMSH5 deficiency on cell cycle regulation in response to CPT. As shown in Fig. 5C, in the absence of CPT treatment, hMSH5 deficiency posed no observable effect on cell cycle progression, whereas CPT elicited distinctive cell cycle distribution patterns in hMSH5-deficient and control cells. There were more hMSH5-deficient cells accumulated in the G1 phase and fewer cells in the S phase at 8–30 h after CPT treatment. It is known that the ATM-Chk2 signaling mainly controls the G1-S checkpoint (4); therefore, the G1 arrest could be attributed to elevated Chk2 phosphorylation (Fig. 5A). Also of note was that there were fewer hMSH5-deficient cells in the G2 phase at 24–30 h following CPT treatment (Fig. 5C). It is plausible that the lower levels of p-Chk1 resulted in a temporary partial defect at the G2-M checkpoint, but hMSH5-depleted cells eventually arrested at the G2/M phase at 60 h (Fig. 5C), presumably because of the accumulation of unrepaired DNA lesions. Taken together, hMSH5 deficiency impaired cell cycle checkpoint regulation and consequentially enhanced cell death in response to CPT treatment.
hMSH5 and FANCJ Interplay Was Important for the Maintenance of Chk1 Activation
It is known that FANCJ could also play a role in replication stress-provoked Chk1 activation (35). To scrutinize whether the role of hMSH5 in cell cycle regulation was also dependent on FANCJ, we examined the effects of various hMSH5 regions on the level of p-Chk1. We found that the reduction of p-Chk1 at 24 h after CPT treatment could be rescued by overexpression of the full-length hMSH5, as well as two hMSH5 fragments, 1–536 and 1–777, but not hMSH5 226–834 (Fig. 6A). The rescue by the full-length hMSH5 was clearly FANCJ-dependent because FANCJ-depletion significantly affected the effect of hMSH5 on CPT-induced Chk1 activation (data not shown). Together, these results indicated that the N-terminal region of hMSH5 played a key role in Chk1 activation. The partial recovery of p-Chk1 by hMSH5 226–777 could suggest that the C-terminal region of hMSH5 might have a potential inhibitory effect on Chk1 activation (Fig. 6A). Because the hMSH5 1–225 fragment lacked the DNA-binding domain (Fig. 1C), we suspected that this fragment might interfere with the function of hMSH5-FANCJ in CPT-induced DNA damage response. To test this possibility, U2OS cells expressing hMSH5 1–225 were analyzed to assess the effects of this hMSH5 fragment on CPT-induced Chk1 phosphorylation and cell cycle arrest, as well as its effect on clonogenic survival. As shown in Fig. 6B, hMSH5 1–225 caused a significant reduction of Chk1 phosphorylation at 24 h after CPT treatment. This effect of hMSH5 1–225 was also dependent on FANCJ (Fig. 6B). Furthermore, the expression of hMSH5 1–225 led to a higher percentage of cells accumulated in G1 at 24 h after CPT treatment and an increased CPT sensitivity in clonogenic survival (Fig. 6, C and D).
FIGURE 6.
The effect of hMSH5-FANCJ interaction on CPT-induced Chk1 activation and cell cycle progression. A, U2OS cells were first transfected with either a control or hMSH5 shRNA and then transfected to express various truncated hMSH5 fragments. Transfected cells were treated with 1 μm CPT for 1 h, released into drug-free medium, and collected at 24 h. The effects of hMSH5 fragments on CPT-induced Chk1 activation were assessed by immunoblotting. The relative levels of p-Chk1 (ratios of p-Chk1/Chk1) and standard deviations were determined from four independent experiments (graph below the gel images). *, p < 0.05, **, p < 0.01, ***, p < 0.005. B, parental and FANCJ-deficient U2OS cells were transfected with an empty vector or hMSH5 1–225 and then treated with 1 μm CPT for 1 h. Cells were collected at 24 h after CPT treatment. The relative levels of p-Chk1 (ratios of p-Chk1/Chk1) and standard deviations were determined from three independent experiments. **, p < 0.01, ***, p < 0.005. C, U2OS cells were transfected with either an empty vector or hMSH5 1–225. Transfected cells were treated with 1 μm CPT for 1 h and collected at 24 h for cell cycle analysis. D, clonogenic survival analysis of U2OS cells transfected with either an empty vector or hMSH5 1–225. Transfected cells were treated with CPT for 1 h at indicated concentrations. Each data point represents the mean ± S.D. from three independent experiments. Ctrl, control; IB, immunoblot; NS, not significant; shCtrl, shRNA control; shFANCJ, shRNA FANCJ.
To further test the possibility that hMSH5 directly participated in CPT-induced DNA damage repair through an interaction with FANCJ, we co-localized hMSH5 with FANCJ at CPT-induced DNA repair foci (Fig. 7A). Consistent with a previous report on FANCJ-γ-H2AX co-localization (36), we found that BRCA1 was present in most of the FANCJ foci, and the BRCA1 foci were mostly positive for γ-H2AX in CPT-treated cells (data not shown). Because hMSH5 1–225 was involved in the binding of FANCJ, it is conceivable that the attenuation of p-Chk1 at 24 h could be due to a defect in FANCJ chromatin recruitment. To this end, the results of our fractionation assay showed that FANCJ could be induced by CPT treatment, and importantly the level of chromatin-bound FANCJ was reduced in hMSH5-depleted cells at 24 h but not in untreated cells (Fig. 7B). This reduction could be rescued by the full-length hMSH5 but not hMSH5 226–834, indicating the importance of the hMSH5-FANCJ binding (data not shown). Interestingly, there was a reduction in the chromatin-bound hMSH5 in FANCJ-depleted cells (Fig. 7C). This result suggested that hMSH5 and FANCJ regulated chromatin recruitment of each other to facilitate Chk1 activation. Together, this series of experiments suggests that the hMSH5-mediated FANCJ chromatin recruitment plays an important role in the G2/M checkpoint control.
Lastly, the interplay between hMSH5 and FANCJ was further tested in cells deficient for one or both proteins. Evidently, silencing of hMSH5 showed increased sensitivity to a 24 h, low concentration CPT treatment (Fig. 7D). However, even though FANCJ knockdown cells were sensitive to cisplatin and displayed reduced levels of p-Chk1 in response to CPT treatment (Fig. 7, E and F), they were unexpectedly resistant to CPT (Fig. 7D). This observation directly contradicts previously published results obtained from FANCJ knock-out chicken cells (37). Although the exact molecular events underlying this discrepancy are not known, evolutionary differences between mammalian and the chicken FANCJ could be a likely contributing factor. In fact, available evidence tends to suggest that—at least in the process of HR—FANCJ possesses diverse functions across species (27, 38). Furthermore, silencing of FANCJ in the hMSH5-deficient background increased cell survival to the levels of the parental control cells (Fig. 7D). It is worth noting that this observation was reminiscent of FANCJ silencing in cells deficient of hMRE11; although hMRE11-deficient cells were IR-sensitive, concomitant FANCJ and hMRE11 deficiencies rendered cells resistant to IR treatment (39). Furthermore, the levels of p-Chk1 were also restored in cells deficient of both FANCJ and hMSH5 at 24 h after CPT treatment (Fig. 7F). Collectively, these results indicated that CPT sensitivity in hMSH5-deficient cells could be suppressed by depletion of FANCJ.
Discussion
Cellular responses to genotoxic agents are governed by the outputs of multiple processes including genotoxin-induced DNA lesion generation, DNA damage recognition, damage signaling and repair. In this study, we revealed that, in conjunction with FANCJ, hMSH5 played a dual role in CPT-induced DNA repair and damage signaling. Specifically, we found that hMSH5 was a direct FANCJ binding partner. In CPT challenged cells, hMSH5 began to promote HR repair of CPT-induced DSBs at ∼4 h and facilitated the activation of Chk1 at ∼24 h after CPT treatment. It appeared that both events were dependent on the successful chromatin recruitment of FANCJ to the sites of DNA lesions. The integrated actions of hMSH5 in CPT-induced DSB repair and Chk1/Chk2 activation were poised to have a pivotal impact on cell cycle progression. Depletion of hMSH5 affected both G1/S and G2/M checkpoints and resulted in deviant cell cycle progression, leading to the accumulation of unrepaired DNA damage in cells and therefore their increased sensitivity to CPT.
The first indication that hMSH5 was involved in HR stemmed from meiotic studies. In particular, mice lacking Msh5 gene showed defective pairing of homologous chromosomes during prophase I of meiosis (14, 15). However, the number of Rad51 foci and their intensity, although not quantified, appeared higher in Msh5 knock-out spermatocytes than that of the wild-type cells (15). By contrast, our results showed that knockdown of hMSH5 reduced the number of Rad51 foci, as well as chromatin-bound Rad51 protein, in response to CPT treatment. Our previous studies demonstrated that hMSH5 and Rad51 coexisted in the same protein complex (16), and the loading of these two proteins to proximal regions of I-SceI-induced DSBs were coordinated (21). In meiosis, Rad51 acts as an accessory factor for Dmc1-mediated meiotic recombination (40). Therefore, it is likely that Dmc1 plays a central role in meiotic HR, whereas Rad51 serves as a DSB marker in a way analogous to γ-H2AX in mitotic HR. The authors of the same report also speculated that Msh5 was required at an earlier step than Dmc1 based on phenotypes exhibited by corresponding knock-out mice (15), supporting the idea that Msh5 and Rad51 may act differently in the processes of meiotic and mitotic HR.
Defects in HR often lead to the accumulation of unrepaired DNA damage and therefore a robust G2/M arrest before entry into mitosis. This scenario has been attested in several studies on HR-directed repair of CPT-induced DNA damage by the MMS22L-TONSL complex (41–44). However, the role of hMSH5 in CPT-induced DNA damage response and repair revealed here appears to be unique. Silencing of hMSH5 led to a decreased level of p-Chk1 and attenuated G2/M arrest at 24–30 h after CPT treatment, which was accompanied by the reduction of FANCJ chromatin recruitment. Given that peculiar Chk1 (and Chk2) activation in hMSH5-depleted cells only occurred at certain time points after CPT treatment, hMSH5 is very likely to play a role in the maintenance of CPT-elicited damage signaling between the transducer kinase ATR and effector kinases Chk1 and Chk2. Our data also indicated that depletion of hMSH5 affected CPT-induced FANCJ chromatin loading. There is a possibility that FANCJ could act together with BLM to unwind DNA at the end of a DSB, creating ssDNA for loading RPA and subsequent ATR activation through TopBP1 interaction (35, 45–47). Therefore, compromised FANCJ chromatin loading could indirectly affect ATR signaling.
Inhibition of topoisomerase I can reverse replication forks to create structures resembling Holliday junction intermediates (48). It has been proposed that the binding of hMSH5 to the core Holliday junction structure may stabilize recombination intermediates (49). Thus, preventing fork collapse by stabilizing the reversed replication forks is a potential function of hMSH5 in CPT-induced DNA damage repair. This speculation is supported by the observation that, in comparison to 1-h treatment with high dose CPT, hMSH5-depleted cells were more sensitive to 24-h treatment with low dose CPT (Figs. 2E and 7D). It is conceivable that hMSH5 could be recruited to the damaged replication forks by FANCJ, and then hMSH5 becomes associated with Rad51 and FANCJ to facilitate damage-induced cell cycle arrest at G2 and fork repair (50, 51). In the absence of hMSH5, reversed replication forks are likely to be unwound by the FANCJ-BLM complex and other DNA damage response factors (1, 45, 52), leading to replication fork runoff and the formation of one-ended DSBs (48). The repair of one-ended DSBs begins with the formation of D-loops through single-end invasion (9), creating intermediate structures that can also be stabilized by hMSH5 (53). Conversely, the FANCJ-BLM complex can antagonize D-loop formation (54). Intriguingly, we found that the helicase activity of FANCJ was not required for the FANCJ-hMSH5 interaction, and it was also not important for hMSH5-mediated FANCJ chromatin loading (data not shown). Collectively, the concerted actions of the hMSH5-FANCJ and FANCJ-BLM complexes are expected to promote the repair of CPT-provoked one-ended DSBs and at the same time prevent aberrant HR events.
The cross-talk between FA and MMR proteins was first demonstrated by the direct interaction between FANCJ and MutLα (55). Recent studies suggested that the interaction between FANCJ and hMLH1 plays a role in appropriate interstrand cross-link repair damage signaling, and hMSH2 depletion suppressed the interstrand cross-link repair sensitivity exerted by cells lacking the interaction between FANCJ and hMLH1 (56, 57). In the present study, we found that the interaction between hMSH5 and FANCJ plays important roles in cellular defense to CPT challenge. Interestingly, however, depletion of FANCJ was able to not only rescue the CPT sensitivity in hMSH5-deficient cells but also render hMSH5-proficient cells resistant to CPT. This observation has raised the interesting possibility that FANCJ may exert a restrictive effect on HR repair of CPT-induced DSBs through inhibiting hMRE11 3′-to-5′ exonuclease activity (39), thus affecting the extent of the initial end resection. Because hMSH5 also interacts with hMRE11 (20), it is very plausible that the hMSH5-FANCJ and hMSH5-hMRE11 interactions will negatively regulate the interaction between FANCJ and hMRE11. The net effect of these interactions is to harness the inhibitory effect of FANCJ on the hMRE11 nuclease activity, thus promoting DNA end resection and the initiation of HR. To this end, hMSH5 deficiency is expected to strengthen the inhibition of hMRE11 nuclease by FANCJ, whereas concomitant FANCJ deficiency could antagonize this effect. However, this view may be a little simplistic because FANCJ interacts with both hMRE11 and RAD50 in the same complex (39), and the endonuclease and exonuclease activities of MRE11 are also regulated by RAD50-mediated ATP hydrolysis (58). In addition, it is possible that cells deficient in both hMSH5 and FANCJ may survive replication fork collapse by utilizing alternative pathways to activate Chk1.
G2 checkpoint inhibitors are of great clinical interest because they can sensitize p53-deficient cancer cells to chemotherapy. For instance, one such drug, CBP-93872, inhibits the maintenance but not the initiation of G2 checkpoint induced by DSBs (59). Of note, this drug is only effective in IR-treated but not UV- or hydroxyurea-treated cells. Intriguingly, hMSH5-depleted cells are sensitive to CPT, CDDP, and IR3 but not to doxorubicin or hydroxyurea, revealing that hMSH5 deficiency bears a resemblance to the effect of CBP-93872. Given the observation that hMSH5 is expressed in both primary and cancer cell lines (data not shown) and functions in both HR and the maintenance of ATR signaling, it would be interesting to explore it as a potential target for devising new strategies in cancer chemotherapy.
Author Contributions
Y. X. and C. H. conceived and designed the study, Y. X. and X. W. performed the experiments; Y. X. and C. H. drafted the article, and all authors contributed to the critical revision of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant GM084353 from the Public Health Service (to C. H.). The authors declare that they have no conflicts of interest with the contents of this article.
A. Al-Soodani, X. Wu, Y. Xu, and C. Her, manuscript in preparation.
- CPT
- camptothecin
- IR
- ionizing radiation
- CDDP
- cisplatin
- DSB
- double-strand break
- HR
- homologous recombination
- IP
- immunoprecipitation.
References
- 1. Branzei D., Foiani M. (2010) Maintaining genome stability at the replication fork. Nat. Rev. Mol. Cell Biol. 11, 208–219 [DOI] [PubMed] [Google Scholar]
- 2. Helleday T., Petermann E., Lundin C., Hodgson B., Sharma R. A. (2008) DNA repair pathways as targets for cancer therapy. Nat. Rev. Cancer 8, 193–204 [DOI] [PubMed] [Google Scholar]
- 3. Pommier Y. (2006) Topoisomerase I inhibitors: camptothecins and beyond. Nat. Rev. Cancer 6, 789–802 [DOI] [PubMed] [Google Scholar]
- 4. Ciccia A., Elledge S. J. (2010) The DNA damage response: making it safe to play with knives. Mol. Cell 40, 179–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Britton S., Coates J., Jackson S. P. (2013) A new method for high-resolution imaging of Ku foci to decipher mechanisms of DNA double-strand break repair. J. Cell Biol. 202, 579–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nimonkar A. V., Genschel J., Kinoshita E., Polaczek P., Campbell J. L., Wyman C., Modrich P., Kowalczykowski S. C. (2011) BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 25, 350–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Moynahan M. E., Jasin M. (2010) Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat. Rev. Mol. Cell Biol. 11, 196–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Costantino L., Sotiriou S. K., Rantala J. K., Magin S., Mladenov E., Helleday T., Haber J. E., Iliakis G., Kallioniemi O. P., Halazonetis T. D. (2014) Break-induced replication repair of damaged forks induces genomic duplications in human cells. Science 343, 88–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kowalczykowski S. C. (2000) Initiation of genetic recombination and recombination-dependent replication. Trends Biochem. Sci. 25, 156–165 [DOI] [PubMed] [Google Scholar]
- 10. Cimprich K. A., Cortez D. (2008) ATR: an essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 9, 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu S., Shiotani B., Lahiri M., Maréchal A., Tse A., Leung C. C., Glover J. N., Yang X. H., Zou L. (2011) ATR autophosphorylation as a molecular switch for checkpoint activation. Mol. Cell 43, 192–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Curtin N. J. (2012) DNA repair dysregulation from cancer driver to therapeutic target. Nat. Rev. Cancer 12, 801–817 [DOI] [PubMed] [Google Scholar]
- 13. Hollingsworth N. M., Ponte L., Halsey C. (1995) MSH5, a novel MutS homolog, facilitates meiotic reciprocal recombination between homologs in Saccharomyces cerevisiae but not mismatch repair. Genes Dev. 9, 1728–1739 [DOI] [PubMed] [Google Scholar]
- 14. de Vries S. S., Baart E. B., Dekker M., Siezen A., de Rooij D. G., de Boer P., te Riele H. (1999) Mouse MutS-like protein Msh5 is required for proper chromosome synapsis in male and female meiosis. Genes Dev. 13, 523–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Edelmann W., Cohen P. E., Kneitz B., Winand N., Lia M., Heyer J., Kolodner R., Pollard J. W., Kucherlapati R. (1999) Mammalian MutS homologue 5 is required for chromosome pairing in meiosis. Nat. Genet. 21, 123–127 [DOI] [PubMed] [Google Scholar]
- 16. Yi W., Lee T. H., Tompkins J. D., Zhu F., Wu X., Her C. (2006) Physical and functional interaction between hMSH5 and c-Abl. Cancer Res. 66, 151–158 [DOI] [PubMed] [Google Scholar]
- 17. Tompkins J. D., Wu X., Chu Y. L., Her C. (2009) Evidence for a direct involvement of hMSH5 in promoting ionizing radiation induced apoptosis. Exp. Cell Res. 315, 2420–2432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Clark N., Wu X., Her C. (2013) MutS Homologues hMSH4 and hMSH5: Genetic variations, functions, and implications in human diseases. Curr. Genomics 14, 81–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tompkins J. D., Wu X., Her C. (2012) MutS homologue hMSH5: role in cisplatin-induced DNA damage response. Mol. Cancer 11, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kato T., Sato N., Hayama S., Yamabuki T., Ito T., Miyamoto M., Kondo S., Nakamura Y., Daigo Y. (2007) Activation of Holliday junction recognizing protein involved in the chromosomal stability and immortality of cancer cells. Cancer Res. 67, 8544–8553 [DOI] [PubMed] [Google Scholar]
- 21. Wu X., Xu Y., Feng K., Tompkins J. D., Her C. (2013) MutS homologue hMSH5: recombinational DSB repair and non-synonymous polymorphic variants. PLoS One 8, e73284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yi W., Wu X., Lee T. H., Doggett N. A., Her C. (2005) Two variants of MutS homolog hMSH5: prevalence in humans and effects on protein interaction. Biochem. Biophys. Res. Commun. 332, 524–532 [DOI] [PubMed] [Google Scholar]
- 23. Vo A. T., Zhu F., Wu X., Yuan F., Gao Y., Gu L., Li G. M., Lee T. H., Her C. (2005) hMRE11 deficiency leads to microsatellite instability and defective DNA mismatch repair. EMBO Rep. 6, 438–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zou L., Cortez D., Elledge S. J. (2002) Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes Dev. 16, 198–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xu K., Wu X., Tompkins J. D., Her C. (2012) Assessment of anti-recombination and double-strand break-induced gene conversion in human cells by a chromosomal reporter. J. Biol. Chem. 287, 29543–29553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cantor S. B., Bell D. W., Ganesan S., Kass E. M., Drapkin R., Grossman S., Wahrer D. C., Sgroi D. C., Lane W. S., Haber D. A., Livingston D. M. (2001) BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell 105, 149–160 [DOI] [PubMed] [Google Scholar]
- 27. Bridge W. L., Vandenberg C. J., Franklin R. J., Hiom K. (2005) The BRIP1 helicase functions independently of BRCA1 in the Fanconi anemia pathway for DNA crosslink repair. Nat. Genet. 37, 953–957 [DOI] [PubMed] [Google Scholar]
- 28. Levitus M., Waisfisz Q., Godthelp B. C., de Vries Y., Hussain S., Wiegant W. W., Elghalbzouri-Maghrani E., Steltenpool J., Rooimans M. A., Pals G., Arwert F., Mathew C. G., Zdzienicka M. Z., Hiom K., De Winter J. P., Joenje H. (2005) The DNA helicase BRIP1 is defective in Fanconi anemia complementation group J. Nat. Genet. 37, 934–935 [DOI] [PubMed] [Google Scholar]
- 29. Litman R., Peng M., Jin Z., Zhang F., Zhang J., Powell S., Andreassen P. R., Cantor S. B. (2005) BACH1 is critical for homologous recombination and appears to be the Fanconi anemia gene product FANCJ. Cancer Cell 8, 255–265 [DOI] [PubMed] [Google Scholar]
- 30. Hiom K. (2010) FANCJ: solving problems in DNA replication. DNA Repair 9, 250–256 [DOI] [PubMed] [Google Scholar]
- 31. Yu X., Chini C. C., He M., Mer G., Chen J. (2003) The BRCT domain is a phospho-protein binding domain. Science 302, 639–642 [DOI] [PubMed] [Google Scholar]
- 32. Cantor S., Drapkin R., Zhang F., Lin Y., Han J., Pamidi S., Livingston D. M. (2004) The BRCA1-associated protein BACH1 is a DNA helicase targeted by clinically relevant inactivating mutations. Proc. Natl. Acad. Sci. U.S.A. 101, 2357–2362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chowdhury D., Keogh M. C., Ishii H., Peterson C. L., Buratowski S., Lieberman J. (2005) γ-H2AX dephosphorylation by protein phosphatase 2A facilitates DNA double-strand break repair. Mol. Cell 20, 801–809 [DOI] [PubMed] [Google Scholar]
- 34. Meerang M., Ritz D., Paliwal S., Garajova Z., Bosshard M., Mailand N., Janscak P., Hübscher U., Meyer H., Ramadan K. (2011) The ubiquitin-selective segregase VCP/p97 orchestrates the response to DNA double-strand breaks. Nat. Cell Biol. 13, 1376–1382 [DOI] [PubMed] [Google Scholar]
- 35. Gong Z., Kim J. E., Leung C. C., Glover J. N., Chen J. (2010) BACH1/FANCJ acts with TopBP1 and participates early in DNA replication checkpoint control. Mol. Cell 37, 438–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Peng M., Litman R., Jin Z., Fong G., Cantor S. B. (2006) BACH1 is a DNA repair protein supporting BRCA1 damage response. Oncogene 25, 2245–2253 [DOI] [PubMed] [Google Scholar]
- 37. Schwab R. A., Nieminuszczy J., Shin-ya K., Niedzwiedz W. (2013) FANCJ couples replication past natural fork barriers with maintenance of chromatin structure. J. Cell Biol. 201, 33–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dohrn L., Salles D., Siehler S. Y., Kaufmann J., Wiesmüller L. (2012) BRCA1-mediated repression of mutagenic end-joining of DNA double-strand breaks requires complex formation with BACH1. Biochem. J. 441, 919–926 [DOI] [PubMed] [Google Scholar]
- 39. Suhasini A. N., Sommers J. A., Muniandy P. A., Coulombe Y., Cantor S. B., Masson J. Y., Seidman M. M., Brosh R. M., Jr. (2013) Fanconi anemia group J helicase and MRE11 nuclease interact to facilitate the DNA damage response. Mol. Cell Biol. 33, 2212–2227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cloud V., Chan Y. L., Grubb J., Budke B., Bishop D. K. (2012) Rad51 is an accessory factor for Dmc1-mediated joint molecule formation during meiosis. Science 337, 1222–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. O'Connell B. C., Adamson B., Lydeard J. R., Sowa M. E., Ciccia A., Bredemeyer A. L., Schlabach M., Gygi S. P., Elledge S. J., Harper J. W. (2010) A genome-wide camptothecin sensitivity screen identifies a mammalian MMS22L-NFKBIL2 complex required for genomic stability. Mol. Cell 40, 645–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Duro E., Lundin C., Ask K., Sanchez-Pulido L., MacArtney T. J., Toth R., Ponting C. P., Groth A., Helleday T., Rouse J. (2010) Identification of the MMS22L-TONSL complex that promotes homologous recombination. Mol. Cell 40, 632–644 [DOI] [PubMed] [Google Scholar]
- 43. O'Donnell L., Panier S., Wildenhain J., Tkach J. M., Al-Hakim A., Landry M. C., Escribano-Diaz C., Szilard R. K., Young J. T., Munro M., Canny M. D., Kolas N. K., Zhang W., Harding S. M., Ylanko J., Mendez M., Mullin M., Sun T., Habermann B., Datti A., Bristow R. G., Gingras A. C., Tyers M. D., Brown G. W., Durocher D. (2010) The MMS22L-TONSL complex mediates recovery from replication stress and homologous recombination. Mol. Cell 40, 619–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Piwko W., Olma M. H., Held M., Bianco J. N., Pedrioli P. G., Hofmann K., Pasero P., Gerlich D. W., Peter M. (2010) RNAi-based screening identifies the Mms22L-Nfkbil2 complex as a novel regulator of DNA replication in human cells. EMBO J. 29, 4210–4222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Suhasini A. N., Rawtani N. A., Wu Y., Sommers J. A., Sharma S., Mosedale G., North P. S., Cantor S. B., Hickson I. D., Brosh R. M., Jr. (2011) Interaction between the helicases genetically linked to Fanconi anemia group J and Bloom's syndrome. EMBO J. 30, 692–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Xie J., Peng M., Guillemette S., Quan S., Maniatis S., Wu Y., Venkatesh A., Shaffer S. A., Brosh R. M., Jr., Cantor S. B. (2012) FANCJ/BACH1 acetylation at lysine 1249 regulates the DNA damage response. PLoS Genet. 8, e1002786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Greenberg R. A., Sobhian B., Pathania S., Cantor S. B., Nakatani Y., Livingston D. M. (2006) Multifactorial contributions to an acute DNA damage response by BRCA1/BARD1-containing complexes. Genes Dev. 20, 34–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ray Chaudhuri A., Hashimoto Y., Herrador R., Neelsen K. J., Fachinetti D., Bermejo R., Cocito A., Costanzo V., Lopes M. (2012) Topoisomerase I poisoning results in PARP-mediated replication fork reversal. Nat. Struct. Mol. Biol. 19, 417–423 [DOI] [PubMed] [Google Scholar]
- 49. Snowden T., Acharya S., Butz C., Berardini M., Fishel R. (2004) hMSH4-hMSH5 recognizes Holliday junctions and forms a meiosis-specific sliding clamp that embraces homologous chromosomes. Mol. Cell 15, 437–451 [DOI] [PubMed] [Google Scholar]
- 50. Zellweger R., Dalcher D., Mutreja K., Berti M., Schmid J. A., Herrador R., Vindigni A., Lopes M. (2015) Rad51-mediated replication fork reversal is a global response to genotoxic treatments in human cells. J. Cell Biol. 208, 563–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yu X., Chen J. (2004) DNA damage-induced cell cycle checkpoint control requires CtIP, a phosphorylation-dependent binding partner of BRCA1 C-terminal domains. Mol. Cell Biol. 24, 9478–9486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Suhasini A. N., Brosh R. M., Jr. (2012) Fanconi anemia and Bloom's syndrome crosstalk through FANCJ-BLM helicase interaction. Trends Genet. 28, 7–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Shinohara M., Oh S. D., Hunter N., Shinohara A. (2008) Crossover assurance and crossover interference are distinctly regulated by the ZMM proteins during yeast meiosis. Nat. Genet. 40, 299–309 [DOI] [PubMed] [Google Scholar]
- 54. Gupta R., Sharma S., Sommers J. A., Jin Z., Cantor S. B., Brosh R. M., Jr. (2005) Analysis of the DNA substrate specificity of the human BACH1 helicase associated with breast cancer. J. Biol. Chem. 280, 25450–25460 [DOI] [PubMed] [Google Scholar]
- 55. Peng M., Litman R., Xie J., Sharma S., Brosh R. M., Jr., Cantor S. B. (2007) The FANCJ/MutLα interaction is required for correction of the cross-link response in FA-J cells. EMBO J. 26, 3238–3249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Peng M., Xie J., Ucher A., Stavnezer J., Cantor S. B. (2014) Crosstalk between BRCA-Fanconi anemia and mismatch repair pathways prevents MSH2-dependent aberrant DNA damage responses. EMBO J. 33, 1698–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Guillemette S., Branagan A., Peng M., Dhruva A., Schärer O. D., Cantor S. B. (2014) FANCJ localization by mismatch repair is vital to maintain genomic integrity after UV irradiation. Cancer Res. 74, 932–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Majka J., Alford B., Ausio J., Finn R. M., McMurray C. T. (2012) ATP hydrolysis by RAD50 protein switches MRE11 enzyme from endonuclease to exonuclease. J. Biol. Chem. 287, 2328–2341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hirokawa T., Shiotani B., Shimada M., Murata K., Johmura Y., Haruta M., Tahara H., Takeyama H., Nakanishi M. (2014) CBP-93872 inhibits NBS1-mediated ATR activation, abrogating maintenance of the DNA double-strand break-specific G2 checkpoint. Cancer Res. 74, 3880–3889 [DOI] [PubMed] [Google Scholar]