Background: Surfactant phosphatidylcholine (PC) must be transported to lamellar bodies (LBs) prior to secretion from type II cells.
Results: The acyltransferase LPCAT1 interacts specifically with the phospholipid transfer protein StarD10 in vivo and in vitro.
Conclusion: The LPCAT1 and StarD10 interaction is an initial step in PC trafficking to LB.
Significance: This is the first direct evidence linking StarD10 to transport of SatPC to LB.
Keywords: lipid-binding protein, lipid trafficking, phospholipid, protein expression, protein-protein interaction
Abstract
Pulmonary surfactant, a mixture of proteins and phospholipids, plays an important role in facilitating gas exchange by maintaining alveolar stability. Saturated phosphatidylcholine (SatPC), the major component of surfactant, is synthesized both de novo and by the remodeling of unsaturated phosphatidylcholine (PC) by lyso-PC acyltransferase 1 (LPCAT1). After synthesis in the endoplasmic reticulum, SatPC is routed to lamellar bodies (LBs) for storage prior to secretion. The mechanism by which SatPC is transported to LB is not understood. The specificity of LPCAT1 for lyso-PC as an acyl acceptor suggests that formation of SatPC via LPCAT1 reacylation is a final step in SatPC synthesis prior to transport. We hypothesized that LPCAT1 forms a transient complex with SatPC and specific phospholipid transport protein(s) to initiate trafficking of SatPC from the endoplasmic reticulum to the LB. Herein we have assessed the ability of different StarD proteins to interact with LPCAT1. We found that LPCAT1 interacts with StarD10, that this interaction is direct, and that amino acids 79–271 of LPCAT1 and the steroidogenic acute regulatory protein-related lipid transfer (START) domain of START domain-containing protein 10 (StarD10) are sufficient for this interaction. The role of StarD10 in trafficking of phospholipid to LB was confirmed by the observation that knockdown of StarD10 significantly reduced transport of phospholipid to LB. LPCAT1 also interacted with one isoform of StarD7 but showed no interaction with StarD2/PC transfer protein.
Introduction
Pulmonary surfactant is a complex mixture of phospholipids and proteins that is synthesized, stored, and secreted by alveolar type II epithelial cells in the lung (1). Deficiencies and/or dysfunction of the surfactant system contribute to the pathogenesis of pulmonary diseases, including respiratory distress syndrome in infants (2) and acute lung injury in adults (3, 4). Phospholipids constitute ∼80% of the total mass of pulmonary surfactant with the remainder composed of neutral lipids (5–10%) and proteins (10%) (5). Phosphatidylcholine (PC)2 is the predominant phospholipid species in surfactant, accounting for 80% of the total phospholipids. Approximately 50–60% of surfactant PC contains two saturated fatty acid chains at the sn-1 and sn-2 positions of the glycerol backbone, a property that distinguishes it from PC found in cellular membranes that typically has an unsaturated fatty acid moiety at the sn-2 position. The high percentage of disaturated PC in surfactant is essential for its function of reducing surface tension at the air/liquid interface in the alveolus (6, 7).
Saturated phosphatidylcholine (SatPC) is synthesized de novo through the Kennedy pathway (8) or by remodeling via the Lands cycle (9). In the lung, 55–75% of SatPC is synthesized through the remodeling pathway (10, 11) where a phospholipase A2 deacylates existing unsaturated PC at the sn-2 position to generate lysophosphatidylcholine (lyso-PC), which is then reacylated with a saturated fatty acid by the lyso-PC acyltransferase 1 (LPCAT1). We and others have demonstrated previously that LPCAT1 exhibits a high affinity for palmitoyl-CoA as an acyl donor and preferentially acylates lyso-PC compared with other lysophospholipids (12–14). In the mouse lung, LPCAT1 is expressed only in surfactant-producing alveolar type II cells and is developmentally regulated, peaking at the end of gestation (12). The importance of LPCAT1 to surfactant SatPC synthesis is underscored by the previous observations that surfactant isolated from Lpcat1−/− mice or from mice bearing a hypomorphic allele of Lpcat1 exhibits a significantly decreased SatPC content and impaired surface tension-lowering capability (15, 16).
Levels of LPCAT1 can also affect surfactant homeostasis because overexpression of LPCAT1 in a lung epithelial cell line targets the enzyme CPT1 (cholinephosphotransferase), which catalyzes the terminal step in de novo PC synthesis, for degradation via the lysosomal pathway (17). LPCAT1 may also play an important role in the response of the lung epithelium to injury. LPCAT1 translocates to the nucleus in lung epithelial cells treated with LPS or LPS-containing bacteria where it regulates inducible gene expression by catalyzing the palmitoylation of histone H4 (18, 19).
Newly synthesized SatPC moves from the smooth endoplasmic reticulum (ER) to the cytoplasmic lamellar body (LB) for storage prior to secretion (for a review, see Ref. 20). Although several studies have documented the critical role of ABCA3 located on the LB surface (21, 22) in moving SatPC from the cytosol into the LB, the molecular mechanisms regulating the trafficking of SatPC from the ER to the LB have not been defined. Lipid transport can occur by various mechanisms, including diffusion between contact membranes, vesicular transport through the budding and fusion of membrane vesicles, and non-vesicular transport mediated by lipid transfer proteins (23–29). Vesicular transport is importantly involved in the trafficking of newly synthesized surfactant protein B and surfactant protein C to LB (30–32) as well as in the endocytosis-mediated recycling of surfactant protein A (33) and lipids (34–36). Initial studies suggested that transport of newly synthesized SatPC to LB also occurred via vesicles based on an electron microscopic autoradiography study of type II cells following an in vivo pulse of [3H]choline that showed the sequential appearance of radiolabeled PC in the ER followed by the Golgi and then finally LB (37). More recent studies (31, 38), however, have shown that disruption of the Golgi apparatus in type II cells by brefeldin A had no effect on both SatPC trafficking to LB and PC secretion, suggesting that a non-vesicular pathway is responsible for SatPC transport.
Because reacylation of lyso-PC is a final step in SatPC synthesis prior to its trafficking to LB, we hypothesized that LPCAT1 forms a transient complex with SatPC and a specific phospholipid transport protein(s) to initiate the movement of SatPC from the ER to the LB. The steroidogenic acute regulatory protein-related lipid transfer (START) proteins are non-vesicular lipid transporters that facilitate intracellular lipid trafficking between cellular membranes (39, 40). The START domain is a protein motif spanning ∼210 amino acids that is responsible for lipid binding. Among the 15 mammalian START domain-containing (StarD) proteins, a subfamily that comprises StarD2/PCTP, StarD7, and StarD10 has been categorized as PC-specific transporters (41–43). The fact that StarD2/PCTP, StarD7, and StarD10 are co-expressed in the lung makes them attractive candidate molecules for the transport of SatPC to LB. In this study, we demonstrate a specific and direct interaction between LPCAT1 and StarD10 both in vivo and in vitro and define regions of each molecule that are involved in this interaction. In concert with previous data (19, 44), these results provide further evidence that LPCAT1 is a multifunctional enzyme that plays important roles in diverse cellular functions.
Experimental Procedures
Animals
All protocols involving mice were reviewed and approved by the Institutional Animal Care and Use Committee at Cincinnati Children's Medical Center. The studies utilizing wild-type mice were done using strain FVB/N (Harlan Laboratories, Madison, WI). Lungs for ontogeny studies were obtained from timed fetuses on embryonic (E) days E11.5 through E18.5. Lungs were also obtained from postnatal (PN) animals on days PN0, PN1, PN7, and PN14 as well as from adults. Adult type II cells for gene expression studies were isolated by FACS from surfactant protein C/GFP mice that express green fluorescent protein under control of the human SFTPC promoter (45).
Quantitative Real Time RT-PCR (qPCR)
Total RNA was purified from lung tissue or alveolar type II cells using an RNeasy Plus Mini kit (Qiagen, Hilden, Germany) or by CsCl centrifugation (46) and reverse transcribed with the iScript kit (Bio-Rad). qPCR was performed on a 7300 Real Time PCR Machine using TaqMan probes (both from Applied Biosystems, Foster City, CA). β-Actin was used as the internal control for quantitation. Changes in gene expression are expressed as -fold changes versus controls.
LPCAT1 and StarD Expression Constructs
To generate the StarD constructs, total adult mouse liver RNA was isolated and reverse transcribed with Superscript II reverse transcriptase (Invitrogen). cDNA fragments for coding sequences with a FLAG (DYKDDDDK) or HA (YPYDVPDYA) tag added to the 3′-end were generated by PCR for StarD2/PCTP, StarD7, and StarD10 using the primers listed in Table 1; a unique restriction site was also added to the 5′-end of each primer to facilitate directional cloning. The resultant StarD2-HA, StarD7-I-HA, StarD7-II-HA, StarD10-HA, and StarD10-FLAG PCR products were digested with the appropriate restriction enzymes and ligated into the vector pcDNA3.1(+) (Invitrogen). The resulting expression constructs were sequenced in both directions to ensure that no mutations were introduced by PCR amplification. Expression of StarD10-HA, StarD10-FLAG, StarD2-HA, StarD7-I-HA, and StarD7-II-HA proteins was tested by transient transfection of HEK293 cells followed by immunoblotting using anti-HA (Santa Cruz Biotechnology, Dallas, TX) or anti-FLAG antibody (Stratagene, La Jolla, CA) to detect recombinant protein.
TABLE 1.
List of cloning primers
aa, amino acid.
| Names | Primer sequence (5′ to 3′) |
|---|---|
| 5′HindIII/LPCAT1-aa49 | CTTAAGCTTGCAGCCATGACGTTGACGCTG |
| 5′HindIII/LPCAT1-aa79 | CTTAAGCTTGCCACCATGCCTCCTGATAAGGAG |
| 3′LPCAT1-aa211/BsrGI | GAGGCAGGTCCTATTTGTACA |
| 5′HindIII/LPCAT1-aa272 | CCCAAGCTTGCAGCCATGGAATTTCTGCCTGTGTAT |
| 3′HA/BamHI | ATGGATCCCTATGCATAGTCCGGGAC |
| 5′EcoRI/LPCAT1 | TGGAATTCGCCACCATGAGGCTGCGGGGC |
| 3′LPCAT1-aa271+HA/XhoI | CCGCTCGAGCTATGCATAGTCCGGGACGTCATAGGGATAAATTTCCACTTGGTT |
| 3′LPCAT1-aa271+FLAG/BamHI | GCGGATCCCTATTTATCATCATCATCTTTATAATCAATTTCCACTTGGTT |
| 3′LPCAT1-aa239+HA/XhoI | CCGCTCGAGCTATGCATAGTCCGGGACGTCATAGGGATATGGGTAGCGTAGCACCAC |
| 5′HindIII/LPCAT1-aa114 | CTTAAGCTTGCCACCATGCGTGTAGCTGTGAAGGG |
| 3′LPCAT1-aa239+FLAG/XhoI | CCGCTCGAGCTACCTTTTATCATCATCATCTTTATAATCTGGGTAGCGTAGCACCAC |
| 5′EcoRI/StarD2 | TGGAATTCGGAAGGATGGCGGGGGCC |
| 3′StarD2+HA/XbaI | CCTCTAGATTATGCATAGTCCGGGACGTCATAGGGATAGGTTTTCTTGTGGTA |
| 5′EcoRI/StarD10 | TGGAATTCGCCACCATGGAAAAGCCAGCT |
| 3′StarD10+FLAG/XbaI | CCTCTAGATCATTTATCATCATCATCTTTATAATCGGTGAGCGAGGTGTC |
| 3′StarD10+HA/XbaI | CCTCTAGATCATGCATAGTCCGGGACGTCATAGGGATAGGTGAGCGAGGTGTC |
| 3′StarD10-aa227+HA/XbaI | CCTCTAGATCATGCATAGTCCGGGACGTCATAGGGATACTGCTTCCACTCGGG |
| 5′BamHI/StarD10-aa19 | CGGGATCCGCCACCATGCGTGAAAGTGTCCAG |
| 3′StarD10-aa227+FLAG/XbaI | CCTCTAGATCATTTATCATCATCATCTTTATAATCCTGCTTCCACTCGGG |
| 5′EcoRI/StarD7I | TGGAATTCGCCGACATGTTCCCGA |
| 3′StarD7I+HA/XbaI | TCTAGATTATGCATAGTCCGGGACGTCATAGGGATAAGCATACTCAATCCG |
| 5′EcoRI/StarD7I-aa79 | TGGAATTCGCCGACATGGCGGCGCTCTCCGGT |
| 5′BglII/StarD7I | GGACTCAGATCTATGTTCCCGAGGAGG |
| 3′StarD7I-aa78+FLAG/BamHI | GCGGATCCTTTATCATCATCATCTTTATAATCCAAAACAGAGGCATG |
The generation and use of the LPCAT1 expression constructs LPCAT1-FLAG and LPCAT1-HA have been described previously (15). To determine the domains required for the LPCAT1 and StarD protein interaction, the following deletion constructs were generated by PCR using the appropriate full-length expression construct as template with primers listed in Table 1: LPCAT1(49–534)-HA, LPCAT1(79–534)-HA, LPCAT1(272–534)-HA, LPCAT1(1–271)-HA, LPCAT1(79–271)-HA, LPCAT1(114–239)-HA, StarD10(1–227)-HA, and StarD10(19–227)-FLAG. The truncated fragments were cloned into the vector pcDNA3.1(+), and their sequence fidelity was confirmed. The expression vector StarD7-I(1–78)-FLAG was constructed by inserting the DNA sequence coding Met1–Leu78 plus a FLAG tag into the vector pEGFP-CI (BD Biosciences).
LPCAT1 and StarD Co-immunoprecipitation (Co-IP) Assays
HEK293 cells were transfected with LPCAT1 and StarD constructs (either singly or in combination) using Lipofectamine 2000 (Invitrogen). Cells transfected with empty vector served as controls. 48 h post-transfection, cells were lysed in 1 ml of lysis/binding buffer (20 mm HEPES, pH 7.6, 150 mm NaCl, 10% glycerol, 1% Triton X-100, 1 mm EDTA, 1 mm EGTA) plus protease inhibitor mixture (Sigma-Aldrich) for 1 h at 4 °C. The lysates were cleared by centrifugation at 12,000 rpm for 15 min at 4 °C. The supernatants were incubated overnight at 4 °C with 50 μl of anti-FLAG M2 antibody bound to agarose beads (Sigma-Aldrich); resins conjugated with anti-FLAG antibody were used to circumvent detection problems caused by free immunoglobulins. The beads were washed and boiled in 25 μl of 2× Laemmli buffer. The precipitated proteins were separated by SDS-PAGE and immunoblotted with anti-HA antibody; 5% of cell lysates was used as input on a protein gel for all experiments. Expression levels of FLAG-tagged bait and HA-tagged prey proteins were demonstrated by immunoblotting using anti-FLAG and anti-HA antibody, respectively. Co-IP was also carried out by pulling down protein complexes with anti-StarD10 antibody (Santa Cruz Biotechnology) and Protein G-Sepharose (Invitrogen) followed by immunoblotting using anti-LPCAT1 antibody (15).
LPCAT1 and StarD10 Direct Protein Interaction Assays
To determine whether LPCAT1 interacts directly with StarD10, full-length LPCAT1 and StarD10 cDNAs were cloned into the expression vectors pET-24a (Novagen, Madison, WI) and pGEX-4T-1 (Ge Healthcare) to generate pET-24a/LPCAT1, pET-24a/StarD10, pGEX-4T/LPCAT1, and pGEX-4T/StarD10 constructs. The resulting constructs were used to transform bacterial strain BD21 (Novagen). Expression of GST-LPCAT1 and GST-StarD10 recombinant proteins was induced with 0.1 mm isopropyl 1-thio-β-d-galactopyranoside (Invitrogen) for 4 h, whereas LPCAT1-His and StarD10-His were induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside. The recombinant proteins were subsequently purified with GST- or His-resin (Novagen) according to the manufacturer's instructions. Briefly, bacterial pellets for GST-LPCAT1 or GST-StarD10 were lysed in GST bind/wash buffer (4.3 mm NaHPO4, 1.47 mm KH2PO4, 137 mm NaCl, 2.7 mm KCl, pH 7.2) with 0.1% Nonidet P-40 and 5 mm imidazole plus proteinase inhibitor, sonicated, and cleared. The resulting supernatants were incubated with GST-resin. Protein-bound resins were washed and eluted with GST elution buffer (50 mm Tris-HCl, pH 8.0, 10 mm reduced glutathione). Bacterial pellets for LPCAT1-His or StarD10-His were lysed in binding buffer (20 mm Tris-HCl, pH 7.9, 500 mm NaCl, 5 mm imidazole) with 0.1% Nonidet P-40 plus proteinase inhibitor, sonicated, and cleared. The resulting supernatants were incubated with nickel-charged His-resin. Protein-bound resins were washed and eluted with elution buffer (20 mm Tris-HCl, pH 7.9, 500 mm NaCl, 1 m imidazole). The concentrations of the purified proteins were determined by BCA assay (Pierce). 20 μg of GST-LPCAT1 and StarD10-His proteins were diluted in 1 ml of GST bind/wash buffer and incubated with 50 μl of GST-resin overnight at 4 °C. The precipitated proteins were separated by SDS-PAGE and immunoblotted with anti-StarD10 and anti-LPCAT1 antibodies. To examine the converse interaction, LPCAT1-His and GST-StarD10 recombinant proteins were precipitated with GST-resin and then immunoblotted with anti-LPCAT1 and anti-StarD10 antibodies.
Proximity Ligation Assays (PLAs)
Adult mouse lungs were inflation-fixed overnight in 4% paraformaldehyde, then embedded in Optimal Cutting Temperature embedding medium, and frozen. PLA was performed on 6-μm frozen sections using the Duolink In Situ kit (Olink Bioscience, Uppsala, Sweden) according to the manufacturer's instructions. Briefly, after antigen retrieval, lung sections were incubated with rabbit anti-LPCAT1 (1:150; ProteinTech, Chicago, IL) and mouse anti-StarD10 (1:50; Santa Cruz Biotechnology) antibodies overnight at 4 °C. Substitution of normal rabbit IgG (Sigma-Aldrich) and normal mouse IgG (Santa Cruz Biotechnology) for each primary antibody served as negative controls. PLA was performed with anti-rabbit PLUS and anti-mouse MINUS Duolink PLA probes. In situ ligation and subsequent amplification were carried out at 37 °C for 30 and 100 min, respectively. Images were acquired with an Olympus IX83 confocal microscope equipped with a 60× oil objective using laser excitation at 405 and 559 nm; PLA signals, which represent the sites of the protein-protein interaction, appear in red. The intensities of the PLA signals were quantified using ImageJ software.
Proteinase K Protection Assay
LPCAT1-FLAG or LPCAT1-HA proteins were expressed in HEK293 cells for 24 h. Cells were lysed, and microsomes were isolated in the absence of protease inhibitors as described previously (44). Microsomes were incubated with 50 μg/ml proteinase K (Invitrogen) in the presence or absence of 1% Triton X-100 for 60 min on ice. Treatment was stopped by the addition of 2 mm PMSF. Samples were boiled in 1× Laemmli buffer and subjected to SDS-PAGE followed by immunoblotting with anti-FLAG, anti-HA, or anti-BiP (BD Biosciences) antibody.
Mouse Type II Cell Isolation, Culture, and siRNA Treatment
Mouse alveolar type II cells were isolated from 6–7-week-old mice as described previously (47) and seeded onto Matrigel (BD Biosciences) in bronchial epithelial cell growth medium (Lonza, Basel, Switzerland) minus hydrocortisone but additionally containing 5% charcoal-stripped fetal bovine serum and 10 ng/μl FGF-7 (R&D Systems, Minneapolis, MN). Non-adherent cells were removed with a medium change after 18 h. Cells were transfected with StraD10 Stealth RNAi siRNA or control RNAi siRNA (both from Invitrogen) via HVJ-E particles (Cosmo Bio, San Diego, CA) according to the manufacturer's instructions. siRNA-transfected cells were released from Matrigel with Dispase (BD Biosciences) 72 h post-transfection and processed for protein and RNA analysis. Expression level of StarD10 was determined by qPCR and immunoblotting.
Fluorescence Assay for Phospholipid Incorporation into LB in Cultured Type II Cells
Type II cells from 16 mice were isolated, pooled, and seeded onto Matrigel. After overnight recovery, cells were transfected with StarD10 siRNA for 72 h as described above and then labeled with 250 μm NBD-palmitoyl-CoA (Avanti Polar Lipids, Alabaster, AL) for 16 h. Cells were released from Matrigel with Dispase, and then LBs were isolated as described previously (48). Briefly, type II cells were homogenized in 0.25 m sucrose/Tris-CaCl2 (STC) buffer (10 mm Tris-HCl, pH 7.4, 1 mm CaCl2, 150 mm NaCl, 0.1 mm EDTA). The homogenate was adjusted to 0.9 m sucrose by addition of 2 m STC and overlaid with 2 ml of 0.65 m and 0.45 m STC buffer. The tubes were centrifuged for 1 h at 100,000 × g at 4 °C in a Beckman L-80 ultracentrifuge with an SW55i rotor. The LB fraction, found at the 0.45 and 0.65 m interface, was collected and diluted in Tris-CaCl2 buffer and centrifuged at 35,000 × g for 20 min. The pellet was resuspended in 200 μl of Tris-CaCl2 buffer and used for fluorescence measurement and protein quantitation. The newly synthesized NBD-labeled phospholipid in LB was measured with a BioTek Synergy 2 microplate reader, and the fluorescence was quantified with Gen5 software (both from BioTek Instruments, Inc., Winooski, VT). Fluorescent palmitoyl-CoA incorporation was normalized to the LB protein content as determined by BCA. Data are presented as fluorescence intensity of NBD-labeled phospholipid formed/μg of LB protein.
Palmitic Acid (PA) Incorporation into SatPC
To determine whether siRNA knockdown of StarD10 affected SatPC synthesis, StarD10 siRNA-treated type II cells were labeled after 72 h with 1 μCi/ml [3H]PA as described previously (15). Lipids were extracted from the medium and harvested cells after 16 h of labeling (49), and SatPC was isolated according to the method of Mason et al. (50). The incorporation of PA into SatPC was determined by scintillation counting and normalized to genomic DNA content (51).
Statistical Analysis
Data were analyzed and graphed using GraphPad Prism (GraphPad Software, San Diego, CA). Two group comparisons were performed by two-tailed, unpaired Student's t test. Comparisons among groups were made by one-way analysis of variance using the Turkey-Kramer multiple comparisons test. Data are presented as means ± S.E. with a p value <0.05 considered significant.
Results
Ontogeny of StarD Expression in the Developing Lung
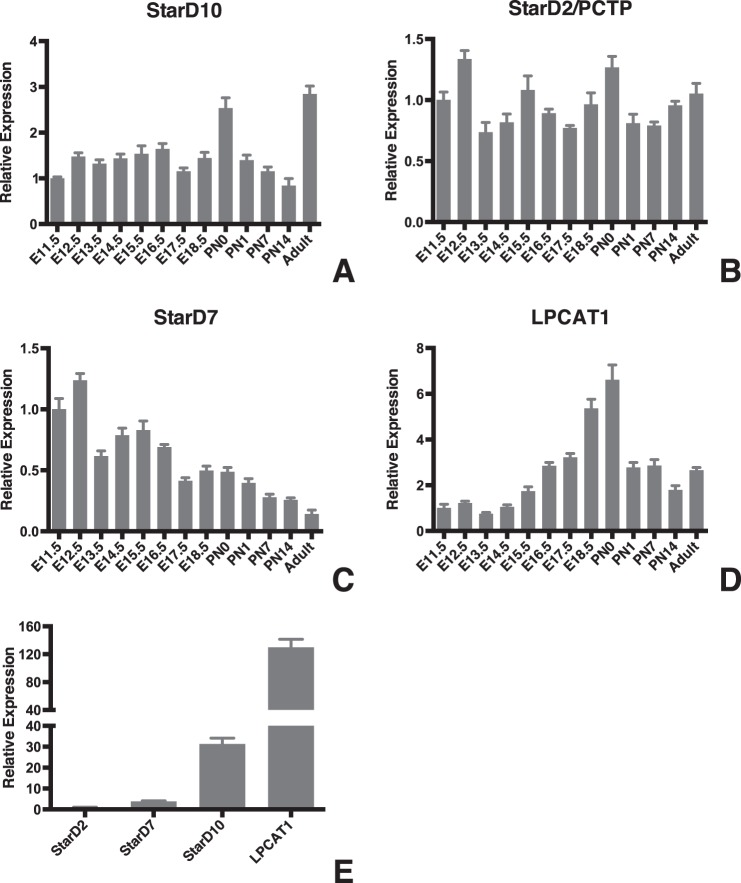
We first used qPCR to generate expression profiles for StarD2/PCTP, StarD7, and StarD10 during lung development. These patterns were compared with that of LPCAT1, which showed a progressive increase in expression during the last trimester of gestation that peaked around the time of birth (Fig. 1D). We found that StarD10 mRNA levels remained relatively constant over the course of gestation but showed a significant increase (2.5-fold versus E11.5) in expression on the day of birth (Fig. 1A). Expression decreased to prenatal levels during the first 2 weeks postnatally but then increased again in adult (2.8-fold) animals. By contrast, StarD2/PCTP expression levels showed little variation across all developmental time points examined (Fig. 1B). StarD7 expression showed an initial decrease on E13.5 to a level that was maintained through E16.5 whereupon there was a progressive decrease postnatally that continued through adulthood where levels were 15% of that seen at E11.5 (Fig. 1C). Most strikingly, when we compared expression of the different StarD proteins in adult type II cells isolated by FACS from the lungs of surfactant protein C/GFP mice, we found that StarD10 mRNA was 31 times more abundant than StarD2/PCTP and 8 times more abundant than StarD7 (Fig. 1E; p < 0.0001, n = 3).
FIGURE 1.
Ontogeny of StarD10, StarD2/PCTP, StarD7, and LPCAT1 in the developing mouse lung. A–D, qPCR analysis of StarD10 (A), StarD2/PCTP (B), StarD7 (C), and LPCAT1 (D) expression in whole mouse lungs from stages E11.5 to adult. The mRNA level of each gene was normalized to β-actin at each time point. Note the differences in scale on the graphs for relative expression of the target genes. Data were derived from three independent litters at each time point. E, comparison of StarD2/PCTP, StarD7, StarD10, and LPCAT1 expression in freshly isolated adult mouse alveolar type II cells. StarD10 abundance was 31 times that of StarD2/PCTP and 8 times that of StarD7. Note that the level of LPCAT1 mRNA was most abundant among these four genes in the type II cells. Error bars represent S.E.
LPCAT1 Interacts with StarD10
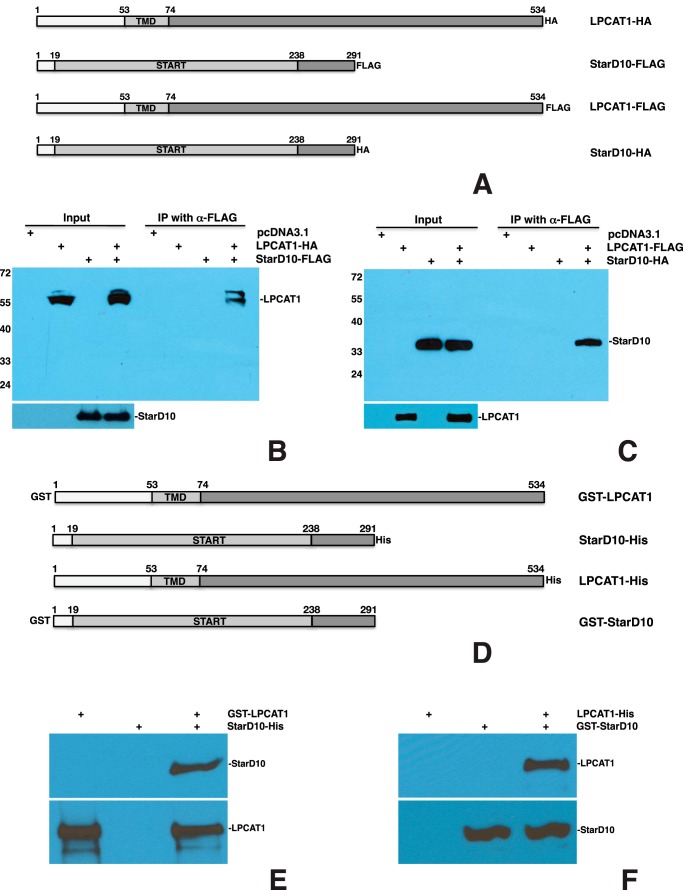
We first tested the interaction of LPCAT1 with StarD10 by co-IP assays after transfecting constructs (Fig. 2A) into HEK293 cells. We first co-transfected full-length LPCAT1-HA with full-length StarD10-FLAG. When cell lysates were immunoprecipitated using anti-FLAG antibody, electrophoresed under denaturing conditions, then blotted, and probed with anti-HA antibody, we identified a protein band at the correct molecular weight for LPCAT1 (Fig. 2B). When we performed co-IP on cells that were co-transfected with LPCAT1-FLAG and StarD10-HA, we identified a band at the correct molecular weight for StarD10 (Fig. 2C). In both cases, no bands were detected when the constructs were transfected singly. The LPCAT1-StarD10 association was also confirmed by immunoprecipitation with anti-StarD10 antibody and immunoblotting with anti-LPCAT1 antibody in transfected cells (data not shown).
FIGURE 2.
LPCAT1 interacts directly with StarD10. A, schematic of HA- or FLAG-tagged constructs for full-length mouse LPCAT1 and StarD10. Co-IP of LPCAT-HA plus StarD10-FLAG (B) and LPCAT1-FLAG plus StarD10-HA (C) is shown. Total lysates of HEK293 cells were singly transfected with empty vector, LPCAT1, or StarD10 or co-transfected with both LPCAT1 and StarD10; immunoprecipitated with resins conjugated with anti-FLAG antibody; and immunoblotted with anti-HA antibody. 5% of cell lysates was used as input; expression of HA-tagged prey and FLAG-tagged bait protein was demonstrated by immunoblotting with anti-HA and anti-FLAG antibodies, respectively. Note that HA-tagged LPCAT1 or StarD10 was pulled down by FLAG-tagged StarD10 or LPCAT1, respectively, demonstrating that co-IP with LPCAT1 and StarD10 worked in both directions. Molecular sizes are indicated in kDa. Results are representative of four independent experiments. D, schematic of GST- or His-tagged mouse LPCAT1 and StarD10 constructs used for the production of recombinant proteins. GST pulldown assays for GST-LPCAT1 plus StarD10-His (E) and LPCAT1-His plus GST-StarD10 (F) are shown. GST or His column-purified recombinant LPCAT1 and/or StarD10 was incubated with GST-resin followed by immunoblotting against LPCAT1 and StarD10. Note that StarD10-His was pulled down by GST-LPCAT1 and that LPCAT-His was pulled down by GST-StarD10, demonstrating that the interaction of LPCAT1 with StarD10 was unaffected by the position of the GST and His tags. Results are representative of four independent experiments.
Although these observations indicated a clear association between LPCAT1 and StarD10, they did not establish whether these proteins interact directly. To address this possibility, we generated constructs (Fig. 2D) for expressing GST-LPCAT1, GST-StarD10, LPCAT1-His, and StarD10-His proteins in bacteria and then purified them with either GST- or His-resin, respectively. Purified GST-LPCAT1 and StarD10-His proteins were incubated with GST-resin singly or together, and then the precipitated protein(s) was separated and immunoblotted with anti-LPCAT1 and anti-StarD10 antibodies (Fig. 2E). GST-LPCAT1 by itself bound to GST-resin and was identified with anti-LPCAT1 antibody. StarD10-His by itself did not bind to GST-resin as evidenced by the lack of detection with anti-StarD10 antibody. StarD10-His was only detected when GST-LPCAT1 was present in the same reaction, indicating a direct interaction between LPCAT1 and StarD10. This finding was confirmed by the converse experiment in which LPCAT1-His fusion protein was only detected when GST-StarD10 was also present (Fig. 2F). These studies demonstrated that recombinant LPCAT1 interacts with StarD10 in the absence of other cellular proteins or lipids.
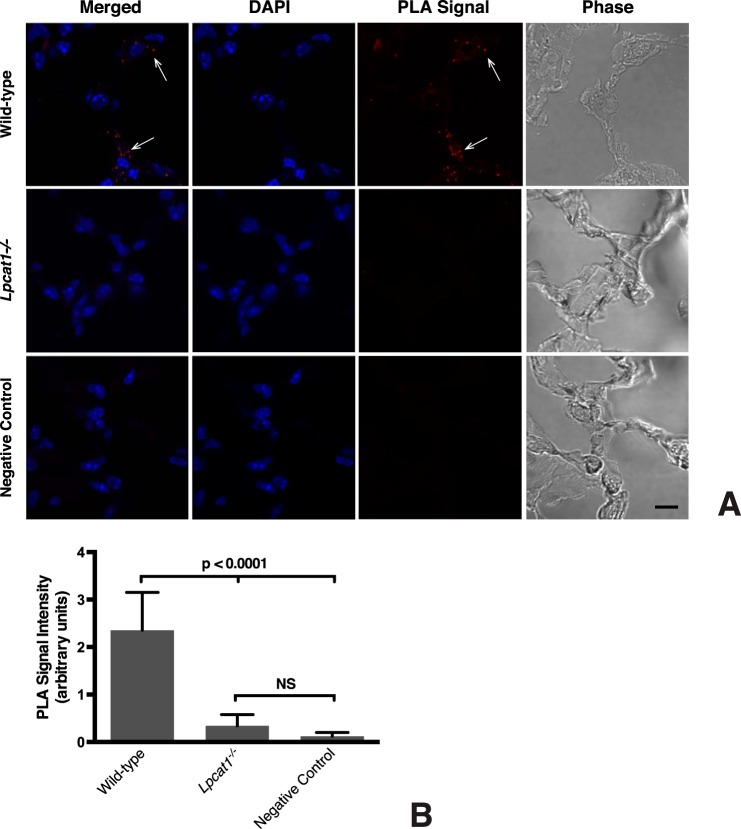
To ascertain whether the interaction between LPCAT1 and StarD10 that we observed in transfected cells also occurs in vivo, we performed in situ PLA (52, 53) on mouse lung sections. We only detected PLA signal when both anti-LPCAT1 and anti-StarD10 antibodies were present (Fig. 3A); scant signal was detected in the negative controls where anti-LPCAT1 antibody (Fig. 3A) or anti-StarD10 antibody (data not shown) was replaced with normal serum IgG. We also included a genetic negative control for these assays. As part of another study, we have generated Lpcat1−/− mice3 that exhibit the same phenotype as described previously (16, 54). Importantly, on sections of Lpcat1−/− lung, we detected very little PLA signal, which was 7-fold less than that seen in wild-type lung (Fig. 3B). These in vivo studies clearly confirmed our in vitro observation that LPCAT1 interacts with StarD10.
FIGURE 3.
LPCAT1 interacts with StarD10 in vivo. A, cryostat sections of frozen fixed adult lungs were subjected to in situ PLA using antibodies against LPCAT1 and StarD10. Confocal images of the PLA signal (red dots; arrows) shows that LPCAT1 and StarD10 interact in wild-type mouse lung (top row). No signal is apparent in sections of lungs from Lpcat1−/− mice (middle row) or in sections in which normal rabbit IgG was substituted for anti-LPCAT1 (negative control; bottom row). The images are representative of lung sections from two different mice for each group in five independent experiments. Scale bar, 10 μm. B, quantification of PLA signals. The values represent the mean ± S.E. (error bars) of five independent experiments in each group. p < 0.0001 versus Lpcat1−/− lungs and the negative control. NS, not significant (p ≥ 0.05).
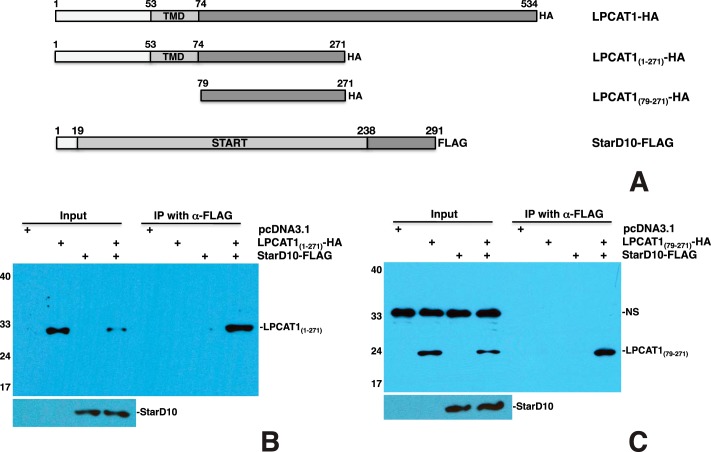
Amino Acid Residues 79–271 of LPCAT1 Are Required for Interaction with StarD10
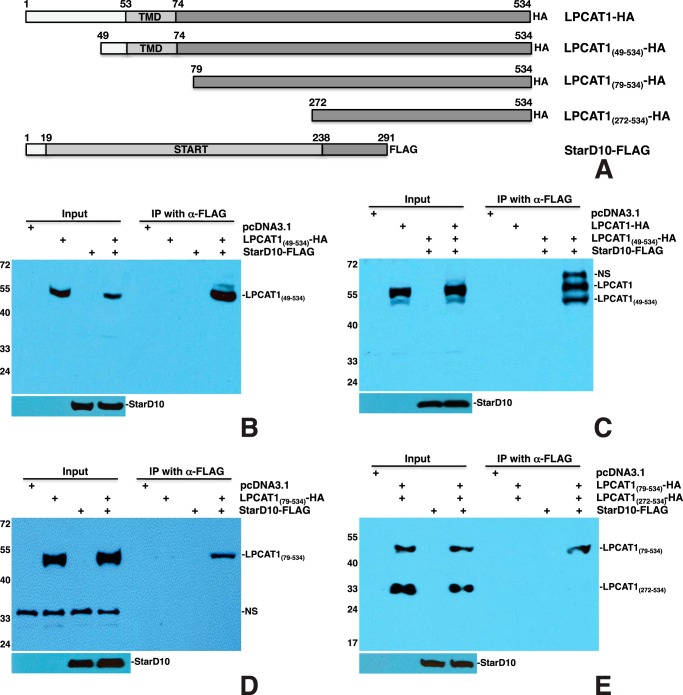
To map the LPCAT1 motif that is required for interaction with StarD10, a series of deletion constructs was generated. To determine whether the N terminus of LPCAT1 is required for interaction with StarD10, the N-terminal 48 amino acids of LPCAT1 were deleted to generate LPCAT1(49–534)-HA construct (Fig. 4A). Transfection and co-IP of LPCAT1(49–534)-HA and StarD10-FLAG showed that LPCAT1(49–534) was pulled down by StarD10 (Fig. 4B), indicating that the N terminus of LPCAT1 is not required for its interaction with StarD10. Transfection of both HA-tagged full-length LPCAT1 and LPCAT1(49–534) with StarD10-FLAG resulted in pulldown of both full-length and truncated LPCAT1 proteins by StarD10, although the expression level of full-length LPCAT1 was higher than that of the mutant (Fig. 4C), suggesting that the N terminus is required for optimal expression of LPCAT1. To determine whether the transmembrane domain (TMD) of LPCAT1 (amino acids 53–74) is required for interaction with StarD10, a construct containing the C-terminal amino acids 79–534 of LPCAT1 and an HA tag was generated (Fig. 4A). Transfection of LPCAT1(79–534)-HA with StarD10-FLAG resulted in pulldown of the truncated LPCAT1 by StarD10 (Fig. 4D), suggesting that the TMD is not required for the interaction. Several studies have also suggested that LPCAT1 has up to four TMDs, spanning amino acid residues from 53 to 271 (13, 14, 55). To eliminate all the potential TMDs, a construct containing the C-terminal amino acids 272–534 of LPCAT1 was generated (Fig. 4A). Transfection of both LPCAT1(272–534)-HA and LPCAT1(79–534)-HA with StarD10-FLAG resulted in pulldown of LPCAT1(79–534) by StarD10 but not LPCAT1(272–534) (Fig. 4E), suggesting that amino acids 79–271 of LPCAT1 are responsible for its interaction with StarD10 and that amino acid residues 272–534 of LPCAT1 are dispensable. To confirm this, a construct containing residues 79–271 of LPCAT1 was generated (Fig. 5A). Transfection of LPCAT1(79–271)-HA with StarD10-FLAG resulted in pulldown of LPCAT1(79–271) by StarD10 (Fig. 5C), suggesting that amino acids 79–271 of LPCAT1 are sufficient for interaction with StarD10. This result was further confirmed by co-IP of LPCAT1(1–271)-HA and StarD10-FLAG in which LPCAT1(1–271) was pulled down by StarD10 (Fig. 5B). Because amino acids 79–271 of LPCAT1 contains the acyltransferase domain spanning amino acids 114–239, a construct containing only the acyltransferase domain was generated to determine whether this domain is involved in interaction with StarD10. Transfection of LPCAT1(114–239)-HA with StarD10-FLAG showed no interaction between LPCAT1(114–239) and StarD10 (data not shown). Together, these studies demonstrated that amino acid residues 79–271 of LPCAT1 are required for interaction with StarD10.
FIGURE 4.
Interactions between LPCAT1 mutants and full-length StarD10. A, schematic of HA-tagged LPCAT1 truncated mutants and full-length StarD10-FLAG constructs. B, co-IP of LPCAT1(49–534)-HA plus StarD10-FLAG resulted in pulldown of LPCAT1(49–534) by StarD10. C, co-transfection of LPCAT1-HA and LPCAT1(49–534)-HA with StarD10-FLAG resulted in pulldown of both full-length and truncated LPCAT1 by StarD10. Note that the expression of full-length LPCAT1 was slightly higher than that of truncated LPCAT1(49–534). D, co-IP of LPCAT1(79–534)-HA plus StarD10-FLAG resulted in pulldown of LPCAT1(79–534) by StarD10. NS, nonspecific band (∼35 kDa). E, co-transfection of LPCAT(79–534)-HA and LPCAT1(272–534)-HA with StarD10-FLAG resulted in pulldown of only LPCAT1(79–534), suggesting that the region of LPCAT1 that interacts with StarD10 is found between amino acids 79 and 271. Note that expression of LPCAT1(272–534) is higher than that of LPCAT(79–534). Results are representative of three independent experiments.
FIGURE 5.
Amino acids 79–271 are required for interaction of LPCAT1 with StarD10. A, schematic of HA-tagged LPCAT1 mutants and full-length StarD10-FLAG constructs. B, co-IP of LPCAT1(1–271)-HA plus StarD10-FLAG showing that this fragment interacts with StarD10. C, co-IP analysis shows that mutant LPCAT1(79–271)-HA, a further truncation of LPCAT1, also interacts with StarD10-FLAG. NS, nonspecific band. Results are representative of three independent experiments.
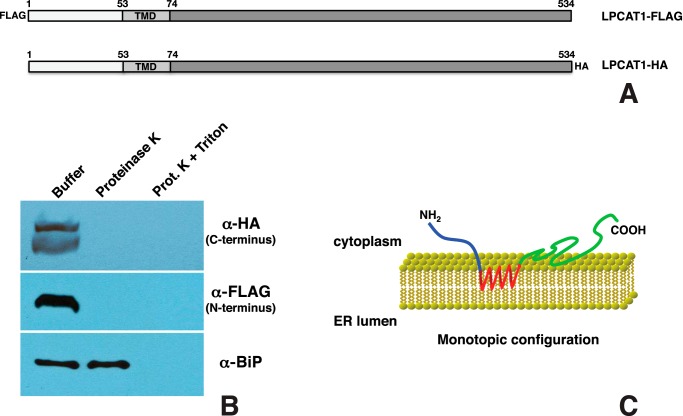
LPCAT1 Is a Monotopic Protein
A previous study from our laboratory showed that LPCAT1 is a type II transmembrane protein with the C terminus inside the ER lumen (15). This orientation would make it impossible for StarD10, a cytosolic protein, to interact with the C-terminal amino acids 79–271 of LPCAT1. A recent study by Moessinger et al. (44), however, has shown that LPCAT1 is a monotopic protein with both the N and C termini facing the cytosol. This orientation would allow an interaction between the C terminus of LPCAT1 and cytosolic StarD10. To verify the topology of LPCAT1, HEK293 cells were transfected with LPCAT1 constructs bearing either an N-terminal FLAG or a C-terminal HA tag (Fig. 6A). Microsomes were isolated and treated with proteinase K; the integrity of microsomes was confirmed by immunoblotting against BiP, an ER lumen protein. The accessibility of the N and C termini of LPCAT1 to proteinase K was assessed by immunoblotting with anti-FLAG and anti-HA antibodies, respectively. Neither the N nor the C terminus of LPCAT1 was detectable after proteinase K digestion, suggesting that both N and C termini are cytosolic (Fig. 6B). These data contradict our previous observation and confirm the observation of Moessinger et al. (44) that LPCAT1 is a monotopic protein with both the N and C termini facing the cytosol (Fig. 6C). As suggested by Moessinger et al. (44), we believe that the disparity with our previous results may be due to reduced efficacy of protein degradation by trypsin versus proteinase K.
FIGURE 6.
Topology of LPCAT1. A, schematic of N-terminal FLAG-tagged or C-terminal HA-tagged full-length LPCAT1 constructs. B, proteinase K protection assay. Microsomes isolated from HEK293 cells transiently expressing FLAG- or HA-tagged LPCAT1 were incubated with buffer only, proteinase K, or proteinase K plus Triton X-100 (Prot. K + Triton) followed by immunoblotting with anti-HA or anti-FLAG antibody. The lack of detection of the N terminus of LPCAT1 with anti-FLAG antibody and the C terminus with anti-HA antibody after proteinase K digestion indicates that both termini were exposed on the exterior of the microsome. The integrity of microsomes was demonstrated by immunoblotting against the luminal ER marker BiP. C, model depicting the monotopic orientation of LPCAT1. LPCAT1 is attached to the ER membrane through its TMD (red) with both the N and C termini facing the cytosol.
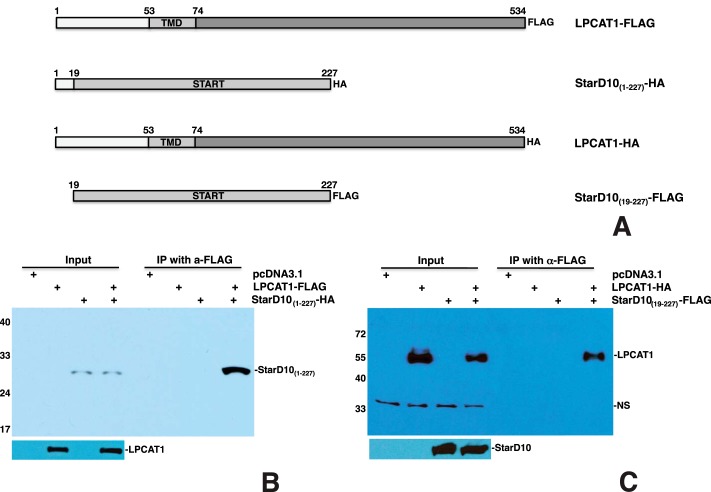
LPCAT1 Interacts with the START Domain of StarD10
We next deleted the C-terminal 64 amino acids to generate the StarD10(1–227)-HA construct (Fig. 7A) and determine whether the C terminus of StarD10 is required for interaction with LPCAT1. Transfection of LPCAT1-FLAG and StarD10(1–227)-HA resulted in pulldown of StarD10(1–227) by LPCAT1 (Fig. 7B), indicating that the C terminus of StarD10 is not required for its interaction with LPCAT1. To determine whether the N terminus is required for interaction with LPCAT1, we made a StarD10(19–227)-FLAG construct (Fig. 7A). Transfection of LPCAT1-HA and StarD10(19–227)-FLAG resulted in pulldown of LPCAT1 by truncated StarD10(19–227) (Fig. 7C), suggesting that the START domain of StarD10 is sufficient for interaction with LPCAT1.
FIGURE 7.
The START domain of StarD10 is required for interaction with LPCAT1. A, schematic of HA- or FLAG-tagged full-length LPCAT1 and mutant StarD10 constructs. B and C, co-IP of LPCAT1 and StarD10 mutants. Co-IP of LPCAT1-FLAG plus StarD10(1–227)-HA resulted in pulldown of StarD10(1–227) by LPCAT1 (B), whereas co-IP of LPCAT1-HA plus StarD10(19–227)-FLAG resulted in pulldown of LPCAT1 by StarD10(19–227) (C), demonstrating that the START domain of StarD10 is sufficient for its interaction with LPCAT1. NS, nonspecific band. Results are representative of three independent experiments.
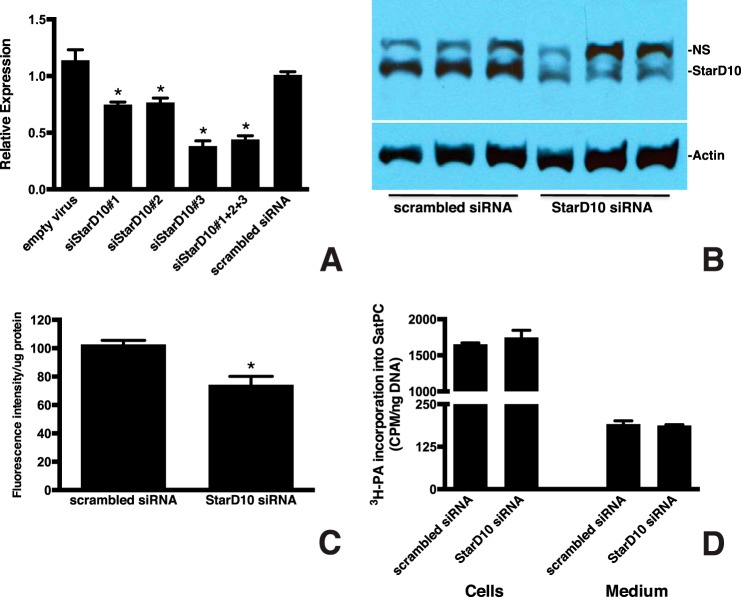
Knockdown of StarD10 Inhibits Phospholipid Trafficking to LB
To test the hypothesis that the StarD10 interaction with LPCAT1 is involved in phospholipid transport to LB, we used StarD10 siRNA to knock down StarD10 in primary cultures of mouse type II cells. We tested three different siRNAs and found a 25–62% decrease in StarD10 mRNA by qPCR 72 h after transfection (Fig. 8A), that was confirmed by immunoblotting (Fig. 8B). We chose the most effective siRNA (siStarD10 number 3) for further study. To determine the role of StarD10 in phospholipid trafficking, mouse type II cells were treated with siStarD10 number 3 for 72 h and then with NBD-palmitoyl-CoA for an additional 16 h to fluorescently label lipids. After purifying the LBs from the cultured cells by sucrose density centrifugation, the amount of fluorescence was determined and normalized to total protein. We found that suppressing StarD10 expression resulted in a significant decrease (29%; p = 0.0013, n = 6) in the amount of NBD-labeled phospholipid in LB compared with controls transfected with scrambled siRNA (Fig. 8C), suggesting that trafficking of newly synthesized phospholipid, most likely SatPC, to LB was impaired. Importantly, this decrease was not due to inhibition of overall SatPC synthesis because we observed that StarD10 siRNA treatment had no effect on the incorporation of [3H]PA into SatPC isolated from both cells and the culture medium (Fig. 8D; p > 0.05, n = 3). Collectively, these data demonstrate that StarD10 knockdown inhibits phospholipid trafficking to the LB in cultured type II cells.
FIGURE 8.
Knockdown of StarD10 results in decreased PC transport to LB. A, qPCR analysis of StarD10 mRNA in mouse type II cells treated with siRNAs targeting StarD10. Freshly isolated mouse type II cells were cultured and transfected with the viral vector alone, scrambled siRNA, and various StarD10 Stealth RNAi siRNAs. The level of endogenous StarD10 mRNA was quantitated by qPCR and normalized to β-actin. Note that StarD10 siRNA 3 was more effective in knocking down StarD10 than siRNA 1, siRNA 2, or a combination of all three. The data represent the mean ± S.E. (error bars) of three independent experiments. *, p < 0.0001 versus scrambled siRNA. B, immunoblot analysis of StarD10 protein levels in whole cell lysates treated with StarD10 siRNA. StarD10 protein was significantly reduced in cells treated with StarD10 siRNA compared with scrambled siRNA. NS, nonspecific band. Results are representative of three independent experiments. C, 72 h after siRNA treatment, type II cells were labeled for 16 h with NBD-palmitoyl-CoA, then LBs were isolated, and the fluorescent content was determined. Incorporation of fluorescent palmitoyl-CoA-labeled phospholipid into LB was normalized to LB protein content. The amount of fluorescent phospholipid that was transported to LB was significantly reduced in cells transfected with StarD10 siRNA compared with scrambled siRNA. The data represent the mean ± S.E. (error bars) of six independent experiments. *, p = 0.0013 versus scrambled siRNA. D, mouse type II cells treated with scrambled or StarD10 siRNA were labeled with [3H]PA, and incorporation into SatPC was determined. [3H]PA incorporation into SatPC was not significantly different in cells treated with scrambled or StarD10 siRNA, indicating that SatPC synthesis itself was unaffected. No change in the amount of SatPC released into the medium was detected as well. The data represent the mean ± S.E. (error bars) of three independent experiments.
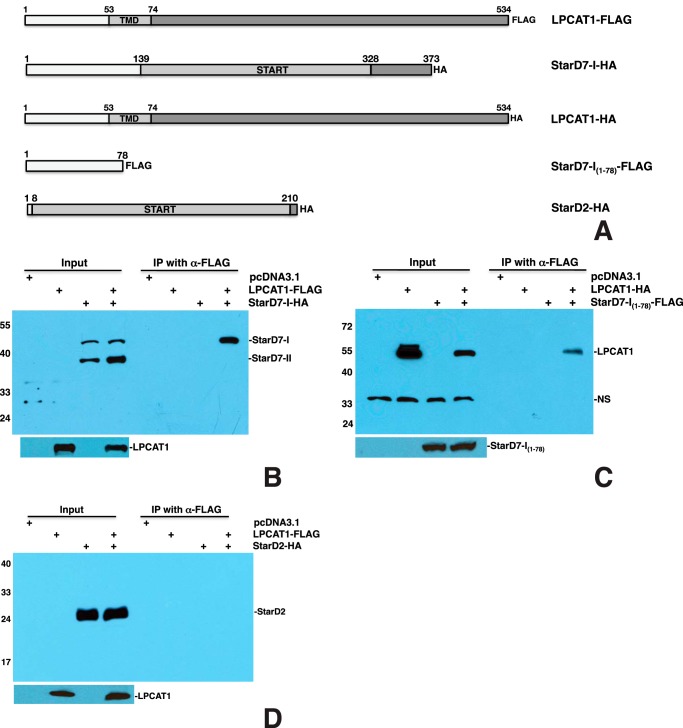
LPCAT1 Interacts with StarD7-I but Not StarD7-II or StarD2/PCTP
To determine whether StarD7 interacts with LPCAT1, we generated a full-length StarD7-I construct (Fig. 9A). Transfection and co-IP of LPCAT1-FLAG and StarD7-I-HA showed that both full-length StarD7-I and the alternative splicing variant StarD7-II, which comprises amino acids 79–373 (42), were produced by the cells but that only full-length StarD7-I was pulled down by LPCAT1 (Fig. 9B). To confirm that StarD7-II does not interact with LPCAT1, we tested a StarD7-I(79–373)-HA construct and found no detectable interaction between StarD7-I(79–373) and LPCAT1 (data not shown), suggesting that the N-terminal 78 amino acids of StarD7-I are responsible for the interaction. To confirm this, we generated a construct containing only the N-terminal 78 amino acids and a FLAG tag (Fig. 9A). Transfection of StarD7-I(1–78)-FLAG and LPCAT1-HA resulted in pulldown of LCPAT1 by StarD7-I(1–78) (Fig. 9C), demonstrating that this 78-amino acid fragment is sufficient for interaction with LPCAT1. Although StarD2/PCTP is present at much lower levels in type II cells than StarD10, we also tested its ability to interact with LPCAT1. Transfection and co-IP of StarD2-HA (Fig. 9A) and LPCAT1-FLAG showed no interaction between the two proteins (Fig. 9D).
FIGURE 9.
LPCAT1 interacts with StarD7-I through its N terminus but does not interact with StarD2/PCTP. A, schematic of FLAG- or HA-tagged LPCAT1, StarD7, and StarD2/PCTP constructs. B, co-IP of LPCAT1-FLAG and StarD7-I-HA. Co-IP using LPCAT1-FLAG and StarD7-I-HA resulted in pulldown of StarD7-I by LPCAT1. The alternative splicing variant StarD7-II was detected but did not co-precipitate with LPCAT1. C, co-IP of LPCAT1-HA and StarD7-I(1–78)-FLAG. Co-IP with LPCAT1-HA and StarD7-I(1–78)-FLAG resulted in pulldown of LPCAT1 by StarD7-I(1–78), demonstrating that the N-terminal 78 amino acids of StarD7-I are sufficient for interaction with LPCAT1. NS, nonspecific band. D, StarD2/PCTP does not interact with LPCAT1. These data represent the results of five independent experiments.
Discussion
Surfactant phospholipids are synthesized in the ER and transported to mature LB where they are stored before secretion, but the mechanism by which this occurs is currently unknown. The observation that trafficking of the newly synthesized phospholipid to LB is unaffected when vesicular transport through the Golgi complex is inhibited by brefeldin A (31, 38) supports the concept that a phospholipid transfer protein(s) may be required for the surfactant lipid transport from ER to LB. In searching for a potential phospholipid transporter in alveolar type II cells, we explored the possibility that LPCAT1 partners with a lipid transfer protein to initiate SatPC transport to LB. This was based on the fact that the reacylation of lyso-PC to SatPC, which is catalyzed by LPCAT1, is a final step in SatPC synthesis by the remodeling pathway.
The START domain family of proteins comprises 15 members that are known to be involved in lipid or sterol binding (39, 56). One subfamily consists of StarD2/PCTP, StarD7, and StarD10, which are known PC transporters (41–43). Although all three proteins are expressed in alveolar type II cells, StarD10 mRNA abundance is more than 31-fold greater than StarD2/PCTP and 8-fold greater than StarD7 (Fig. 1), making it an attractive candidate as a PC transporter in type II cells. This observation let us to hypothesize that LPCAT1 forms a transient complex with SatPC and StarD10 to initiate trafficking of SatPC to LB. The data presented here support this hypothesis as we have demonstrated both that full-length LPCAT1 interacts with StarD10 in transfected HEK293 cells and in vitro (Fig. 2) and that endogenous LPCAT1 and StarD10 interact with each other in vivo (Fig. 3). Our observation that knockdown of StarD10 in primary cultures of type II cells resulted in reduced phospholipid transport to LB (Fig. 8) further supports the concept that StarD10 is involved in phospholipid trafficking in type II cells. This is the first direct evidence linking StarD10 to transport of phospholipid to LB in type II cells.
Although the mechanism by which the StarD proteins transport lipids is unclear, structural analyses have shown that the START domain forms a hydrophobic tunnel that is large enough to bind a single lipid molecule (57–59). Other recent studies have revealed that the START domain of the cholesterol transfer protein StarD3/MLN64 stimulates free cholesterol transfer from donor to acceptor mitochondrial membranes (60). Similarly, the START domain of StarD11/Ceramide Transport Protein (CERT) specifically recognizes ceramide and mediates its intermembrane transfer (61). START proteins can also contain regions other than the START domain that are involved in lipid trafficking. For example, the C terminus of StarD2/PCTP facilitates membrane binding and extraction of PC (62). Phosphorylation of serine 284 in the C terminus of StarD10 inhibits lipid transfer activity by regulating its association with cellular membranes (63). Studies have also shown that direct interactions between StarD2/PCTP and paired box gene 3 or thioesterase superfamily member 2 dictate its function as a lipid transporter through the cellular expression of paired box gene 3 or thioesterase superfamily member 2 (64). We propose that interaction between StarD10 and LPCAT1 may dictate the function of StarD10 as a lipid transporter in type II cells. Based on the fact that the START domain is required for interaction with LPCAT1 (Fig. 7) and that high homology exists among the START domains of StarD proteins, it is likely that, by interacting with LPCAT1, StarD10 is recruited to the ER to initiate SatPC transport to LB. We speculate that unsaturated PC generated from the de novo pathway is deacylated in ER by a phospholipase A2 (65) to generate lyso-PC that subsequently flips to the outer membrane of ER where the LPCAT1 reacylates it with a palmitoyl-CoA moiety to generate SatPC. After reacylating lyso-PC to SatPC, LPCAT1 recruits cytosolic StarD10 to the ER surface where it binds to newly synthesized SatPC through its START domain, extracts it from the ER membrane, and delivers it to an acceptor molecule (e.g. ABCA3 (21, 22)) in the LB.
Knockdown of StarD10 in cultured type II cells significantly (∼30%) but not totally reduced phospholipid transport to LB (Fig. 8). Furthermore, StarD10-deficient mice have normal lung function (66). These observations suggest that StarD10 is not the sole lipid transporter involved in SatPC transport. Indeed, StarD2/PCTP, StarD7, and StarD10 are all capable of PC binding (41–43) and are all expressed in type II cells. LPCAT1 interacts with StarD7-I, although the interaction between StarD7-I and LPCAT1 is mediated by the N-terminal 78 amino acids of StarD7-I and not its START domain (Fig. 9C). The fact that the N-terminal 78 amino acid residues of StarD7-I contain a mitochondrial targeting signal suggests that StarD7-I is responsible for PC transport to mitochondria (42). This observation, however, does not exclude the possibility that it can deliver PC to other cellular organelles such as LB. StarD2/PCTP is unlikely to play a compensating role for the loss of StarD10 because we observed no interaction of StarD2/PCTP with LPCAT1 (Fig. 9D). Furthermore, ablation of StarD2/PCTP has no effect on either surfactant PC composition or LB structure (67).
Most recently, Hishikawa et al. (68) have identified Sec14L3 as a phospholipid transporter in alveolar type II cells. Sec14L3, a Sec14 homolog, not only has an expression pattern similar to LPCAT1 and StarD10 but also binds to a variety of PC species. Similar to the START domain of StarD proteins, the highly conserved Sec14 domain of Sec14L3 forms a hydrophobic cavity to allow binding of a single lipid ligand (69–71). Sec14L3 interacts with 30-nm-diameter liposomes but not larger 100-nm-diameter liposomes, suggesting that the curvature of the liposome may play an important role in the ability of Sec14L3 to bind lipids. Because lipid-packing defects more likely occur in small liposomes, Sec14L3 may act as a sensor for lipid-packing defects. The fact that Sec14L3 preferentially binds to dioleoyl-PC (68), however, suggests that Sec14L3 may not be the primary phospholipid transfer protein delivering lipids to LB in type II cells where the majority of surfactant phospholipid is dipalmitoyl-PC. Although StarD10 is known to bind PC (41), its affinity for dipalmitoyl-PC has not been specifically tested. Sec14L3 is a secretory protein involved mainly in Golgi function and trafficking (72, 73). This observation also suggests that it may not be the major PC transporter in type II cells because PC trafficking to LB occurs via a route that bypasses the Golgi complex (31, 38). However, we have unpublished data3 showing that Sec14L3 interacts strongly with LPCAT1. It is therefore possible that Sec14L3 acts as a sensor for lipid-packing defects under normal conditions but can compensate to engage in PC transport if another transporter such as StarD10 is not available. Taken together, these results suggest that multiple lipid transfer proteins may exist in type II cells and play compensatory roles in PC transport.
In identifying the LPCAT1 motif/domain required for its interaction with StarD10, we generated serial deletion constructs and tested them for their ability to interact with StarD10. Our results showed that amino acid residues 79–271 of LPCAT1 are sufficient for its association with StarD10. However, according to our previous report (15), LPCAT1 is a type II transmembrane protein with C-terminal amino acids 79–534 inside of the ER lumen, which would make interaction with cytosolic StarD10 unlikely. To address this discrepancy, we repeated the proteinase protection assay using proteinase K instead of trypsin as in our previous study. The proteinase K protection assay showed that both the N and C termini were susceptible to proteinase K treatment, indicating that LPCAT1 is a monotopic protein with both the N and C termini facing the cytosol. This finding confirms the results of Moessinger et al. (44) and places the region containing amino acids 79–271 of LPCAT1 on the cytosolic surface of the ER where it is accessible to StarD10.
The concept that LPCAT1 is a protein that performs functions beyond its well documented role in lipid biosynthesis is not without precedent. As noted above, studies both in vivo and in vitro have shown that LPCAT1 can be induced to relocate into the nucleus where it participates in histone H4 palmitoylation (19). How this relocation of LPCAT1, which is normally bound to ER membranes, is accomplished is completely unknown, but such relocation is known to occur with other lipogenic enzymes (74, 75). The ability of LPCAT1 to modify histones is notable because they are known to regulate RNA polymerase II activity and hence gene transcription. This may have significant implications because LPCAT1 expression is increased in several tumors (76–78). Although this may just reflect the need for increased amounts of phospholipids in a proliferating cell population, it is also possible that increased LPCAT1 activity may give neoplastic cells a growth advantage by accelerating overall RNA synthesis.
In summary, we have demonstrated that LPCAT1 is a monotopic enzyme that interacts with StarD10 to initiate trafficking of phospholipid from the ER to LB in type II cells. The mechanism by which the interaction between LPCAT1 and StarD10 facilitates loading of the phospholipid cargo for SatPC trafficking is currently unknown. LPCAT1 also interacts with StarD7-I as well as Sec14L3,3 suggesting the possibility that multiple transporters may function in SatPC trafficking. Full elucidation of the mechanism by which SatPC is transported to LB in alveolar type II cells awaits further study.
Author Contributions
S. L. and J. M. S. designed the study and wrote the paper. S. L. performed and analyzed all experiments. M. I. partially performed and analyzed the experiments shown in Fig. 8D. C. M. and A. P. N. provided reagent, equipment, and technical assistance and contributed to the preparation of the Fig. 3. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank Mike Burhans, Gail Macke, Angelica Falcone, Kavisha Arora, and Susan Wert for excellent technical assistance and/or advice.
This work was supported, in whole or in part, by National Institutes of Health Grants HL098319 (to J. M. S.), DK080834 (to A. P. N.), and DK093045 (to A. P. N.). The authors declare no conflicts of interest with the contents of this article.
S. Lin, unpublished data.
- PC
- phosphatidylcholine
- SatPC
- saturated phosphatidylcholine
- lyso-PC
- lysophosphatidylcholine
- LPCAT1
- lyso-PC acyltransferase 1
- ER
- endoplasmic reticulum
- LB
- lamellar body
- START
- steroidogenic acute regulatory protein-related lipid transfer
- StarD
- START domain-containing protein
- PCTP
- PC transfer protein
- qPCR
- quantitative real time RT-PCR
- IP
- immunoprecipitation
- PLA
- proximity ligation assay
- NBD
- 7-nitrobenz-2-oxa-1,3-diazol-4-yl
- PA
- palmitic acid
- TMD
- transmembrane domain
- E
- embryonic
- PN
- postnatal
- STC
- sucrose/Tris-CaCl2
- BiP
- binding immunoglobulin protein.
References
- 1. Rooney S. (1985) The surfactant system and lung phospholipid biochemistry. Am. Rev. Respir. Dis. 131, 439–460 [DOI] [PubMed] [Google Scholar]
- 2. Avery M. E., Mead J. (1959) Surface properties in relation to atelectasis and hyaline membrane disease. AMA J. Dis. Child. 97, 517–523 [DOI] [PubMed] [Google Scholar]
- 3. Schmidt R., Markart P., Ruppert C., Wygrecka M., Kuchenbuch T., Walmrath D., Seeger W., Guenther A. (2007) Time-dependent changes in pulmonary surfactant function and composition in acute respiratory distress syndrome due to pneumonia or aspiration. Respir. Res. 8, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pison U., Seeger W., Buchhorn R., Joka T., Brand M., Obertacke U., Neuhof H., Schmit-Neuerburg K. P. (1989) Surfactant abnormalities in patients with respiratory failure after multiple trauma. Am. Rev. Respir. Dis. 140, 1033–1039 [DOI] [PubMed] [Google Scholar]
- 5. Possmayer F., Yu S. H., Weber J. M., Harding P. G. (1984) Pulmonary surfactant. Can. J. Biochem. Cell Biol. 62, 1121–1133 [DOI] [PubMed] [Google Scholar]
- 6. Possmayer F. (2004) in Fetal and Neonatal Physiology 2 (Polin R. A., Fox W. W., Abman S. H., eds) pp. 1014–1034, Saunders, Philadelphia [Google Scholar]
- 7. Hawco M. W., Davis P. J., Keough K. M. (1981) Lipid fluidity in lung surfactant: monolayers of saturated and unsaturated lecithins. J. Appl. Physiol. 51, 509–515 [DOI] [PubMed] [Google Scholar]
- 8. Kennedy E. P. (1961) Biosynthesis of complex lipids. Fed. Proc. 20, 934–940 [PubMed] [Google Scholar]
- 9. Lands W. E. (1958) Metabolism of glycerolipides; a comparison of lecithin and triglyceride synthesis. J. Biol. Chem. 231, 883–888 [PubMed] [Google Scholar]
- 10. den Breejen J. N., Batenburg J. J., van Golde L. M. (1989) The species of acyl-CoA in subcellular fractions of type II cells isolated from adult rat lung and their incorporation into phosphatidic acid. Biochim. Biophys. Acta 1002, 277–282 [DOI] [PubMed] [Google Scholar]
- 11. Mason R. J., Nellenbogen J. (1984) Synthesis of saturated phosphatidylcholine and phosphatidylglycerol by freshly isolated rat alveolar type II cells. Biochim. Biophys. Acta 794, 392–402 [DOI] [PubMed] [Google Scholar]
- 12. Chen X., Hyatt B. A., Mucenski M. L., Mason R. J., Shannon J. M. (2006) Identification and characterization of a lysophosphatidylcholine acyltransferase in alveolar type II cells. Proc. Natl. Acad. Sci. U.S.A. 103, 11724–11729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nakanishi H., Shindou H., Hishikawa D., Harayama T., Ogasawara R., Suwabe A., Taguchi R., Shimizu T. (2006) Cloning and characterization of mouse lung-type acyl-CoA:lysophosphatidylcholine acyltransferase 1 (LPCAT1). Expression in alveolar type II cells and possible involvement in surfactant production. J. Biol. Chem. 281, 20140–20147 [DOI] [PubMed] [Google Scholar]
- 14. Soupene E., Fyrst H., Kuypers F. A. (2008) Mammalian acyl-CoA:lysophosphatidylcholine acyltransferase enzymes. Proc. Natl. Acad. Sci. U.S.A. 105, 88–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bridges J. P., Ikegami M., Brilli L. L., Chen X., Mason R. J., Shannon J. M. (2010) LPCAT1 regulates surfactant phospholipid synthesis and is required for transitioning to air breathing in mice. J. Clin. Investig. 120, 1736–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Harayama T., Eto M., Shindou H., Kita Y., Otsubo E., Hishikawa D., Ishii S., Sakimura K., Mishina M., Shimizu T. (2014) Lysophospholipid acyltransferases mediate phosphatidylcholine diversification to achieve the physical properties required in vivo. Cell Metab. 20, 295–305 [DOI] [PubMed] [Google Scholar]
- 17. Butler P. L., Mallampalli R. K. (2010) Cross-talk between remodeling and de novo pathways maintains phospholipid balance through ubiquitination. J. Biol. Chem. 285, 6246–6258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ellis B., Kaercher L., Snavely C., Zhao Y., Zou C. (2012) Lipopolysaccharide triggers nuclear import of Lpcat1 to regulate inducible gene expression in lung epithelia. World J. Biol. Chem. 3, 159–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zou C., Ellis B. M., Smith R. M., Chen B. B., Zhao Y., Mallampalli R. K. (2011) Acyl-CoA:lysophosphatidylcholine acyltransferase I (Lpcat1) catalyzes histone protein O-palmitoylation to regulate mRNA synthesis. J. Biol. Chem. 286, 28019–28025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Goss V., Hunt A. N., Postle A. D. (2013) Regulation of lung surfactant phospholipid synthesis and metabolism. Biochim. Biophys. Acta 1831, 448–458 [DOI] [PubMed] [Google Scholar]
- 21. Mulugeta S., Gray J. M., Notarfrancesco K. L., Gonzales L. W., Koval M., Feinstein S. I., Ballard P. L., Fisher A. B., Shuman H. (2002) Identification of LBM180, a lamellar body limiting membrane protein of alveolar type II cells, as the ABC transporter protein ABCA3. J. Biol. Chem. 277, 22147–22155 [DOI] [PubMed] [Google Scholar]
- 22. Yamano G., Funahashi H., Kawanami O., Zhao L. X., Ban N., Uchida Y., Morohoshi T., Ogawa J., Shioda S., Inagaki N. (2001) ABCA3 is a lamellar body membrane protein in human lung alveolar type II cells. FEBS Lett. 508, 221–225 [DOI] [PubMed] [Google Scholar]
- 23. Voelker D. R. (2003) New perspectives on the regulation of intermembrane glycerophospholipid traffic. J. Lipid Res. 44, 441–449 [DOI] [PubMed] [Google Scholar]
- 24. Holthuis J. C., Levine T. P. (2005) Lipid traffic: floppy drives and a superhighway. Nat. Rev. Mol. Cell Biol. 6, 209–220 [DOI] [PubMed] [Google Scholar]
- 25. van Meer G., Voelker D. R., Feigenson G. W. (2008) Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9, 112–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Perez-Gil J., Weaver T. E. (2010) Pulmonary surfactant pathophysiology: current models and open questions. Physiology 25, 132–141 [DOI] [PubMed] [Google Scholar]
- 27. Lev S. (2010) Non-vesicular lipid transport by lipid-transfer proteins and beyond. Nat. Rev. Mol. Cell Biol. 11, 739–750 [DOI] [PubMed] [Google Scholar]
- 28. Prinz W. A. (2010) Lipid trafficking sans vesicles: where, why, how? Cell 143, 870–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Toulmay A., Prinz W. A. (2011) Lipid transfer and signaling at organelle contact sites: the tip of the iceberg. Curr. Opin. Cell Biol. 23, 458–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Voorhout W. F., Weaver T. E., Haagsman H. P., Geuze H. J., Van Golde L. M. (1993) Biosynthetic routing of pulmonary surfactant proteins in alveolar type II cells. Microsc. Res. Tech. 26, 366–373 [DOI] [PubMed] [Google Scholar]
- 31. Osanai K., Tsuchihara C., Hatta R., Oikawa T., Tsuchihara K., Iguchi M., Seki T., Takahashi M., Huang J., Toga H. (2006) Pulmonary surfactant transport in alveolar type II cells. Respirology 11, (suppl.) S70–S73 [DOI] [PubMed] [Google Scholar]
- 32. Conkright J. J., Apsley K. S., Martin E. P., Ridsdale R., Rice W. R., Na C. L., Yang B., Weaver T. E. (2010) Nedd4–2-mediated ubiquitination facilitates processing of surfactant protein-C. Am. J. Respir. Cell Mol. Biol. 42, 181–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Osanai K., Mason R. J., Voelker D. R. (1998) Trafficking of newly synthesized surfactant protein A in isolated rat alveolar type II cells. Am. J. Respir. Cell Mol. Biol. 19, 929–935 [DOI] [PubMed] [Google Scholar]
- 34. Kaplan M. R., Simoni R. D. (1985) Intracellular transport of phosphatidylcholine to the plasma membrane. J. Cell Biol. 101, 441–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sleight R. G., Pagano R. E. (1984) Transport of a fluorescent phosphatidylcholine analog from the plasma membrane to the Golgi apparatus. J. Cell Biol. 99, 742–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Veldhuizen R., Possmayer F. (2004) Phospholipid metabolism in lung surfactant. Subcell. Biochem. 37, 359–388 [DOI] [PubMed] [Google Scholar]
- 37. Chevalier G., Collet A. J. (1972) In vivo incorporation of choline-3H, leucine-3H and galactose-3H in alveolar type II pneumocytes in relation to surfactant synthesis. A quantitative radioautographic study in mouse by electron microscopy. Anat. Rec. 174, 289–310 [DOI] [PubMed] [Google Scholar]
- 38. Osanai K., Mason R. J., Voelker D. R. (2001) Pulmonary surfactant phosphatidylcholine transport bypasses the brefeldin A sensitive compartment of alveolar type II cells. Biochim. Biophys. Acta 1531, 222–229 [DOI] [PubMed] [Google Scholar]
- 39. Alpy F., Tomasetto C. (2005) Give lipids a START: the StAR-related lipid transfer (START) domain in mammals. J. Cell Sci. 118, 2791–2801 [DOI] [PubMed] [Google Scholar]
- 40. Alpy F., Tomasetto C. (2014) START ships lipids across interorganelle space. Biochimie 96, 85–95 [DOI] [PubMed] [Google Scholar]
- 41. Olayioye M. A., Vehring S., Müller P., Herrmann A., Schiller J., Thiele C., Lindeman G. J., Visvader J. E., Pomorski T. (2005) StarD10, a START domain protein overexpressed in breast cancer, functions as a phospholipid transfer protein. J. Biol. Chem. 280, 27436–27442 [DOI] [PubMed] [Google Scholar]
- 42. Horibata Y., Sugimoto H. (2010) StarD7 mediates the intracellular trafficking of phosphatidylcholine to mitochondria. J. Biol. Chem. 285, 7358–7365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wirtz K. W. (1991) Phospholipid transfer proteins. Annu. Rev. Biochem. 60, 73–99 [DOI] [PubMed] [Google Scholar]
- 44. Moessinger C., Kuerschner L., Spandl J., Shevchenko A., Thiele C. (2011) Human lysophosphatidylcholine acyltransferases 1 and 2 are located in lipid droplets where they catalyze the formation of phosphatidylcholine. J. Biol. Chem. 286, 21330–21339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lo B., Hansen S., Evans K., Heath J. K., Wright J. R. (2008) Alveolar epithelial type II cells induce T cell tolerance to specific antigen. J. Immunol. 180, 881–888 [DOI] [PubMed] [Google Scholar]
- 46. Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. (1979) Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry 18, 5294–5299 [DOI] [PubMed] [Google Scholar]
- 47. Rice W. R., Conkright J. J., Na C. L., Ikegami M., Shannon J. M., Weaver T. E. (2002) Maintenance of the mouse type II cell phenotype in vitro. Am. J. Physiol. Lung Cell. Mol. Physiol. 283, L256–L264 [DOI] [PubMed] [Google Scholar]
- 48. Besnard V., Matsuzaki Y., Clark J., Xu Y., Wert S. E., Ikegami M., Stahlman M. T., Weaver T. E., Hunt A. N., Postle A. D., Whitsett J. A. (2010) Conditional deletion of Abca3 in alveolar type II cells alters surfactant homeostasis in newborn and adult mice. Am. J. Physiol. Lung Cell. Mol. Physiol. 298, L646–L659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bligh E. G., Dyer W. J. (1959) A rapid method of total lipid extraction and purification. Can J. Biochem. Physiol. 37, 911–917 [DOI] [PubMed] [Google Scholar]
- 50. Mason R. J., Nellenbogen J., Clements J. A. (1976) Isolation of disaturated phosphatidylcholine with osmium tetroxide. J. Lipid Res. 17, 281–284 [PubMed] [Google Scholar]
- 51. Cesarone C. F., Bolognesi C., Santi L. (1979) Improved microfluorometric DNA determination in biological material using 33258 Hoechst. Anal. Biochem. 100, 188–197 [DOI] [PubMed] [Google Scholar]
- 52. Weibrecht I., Leuchowius K. J., Clausson C. M., Conze T., Jarvius M., Howell W. M., Kamali-Moghaddam M., Söderberg O. (2010) Proximity ligation assays: a recent addition to the proteomics toolbox. Expert Rev. Proteomics 7, 401–409 [DOI] [PubMed] [Google Scholar]
- 53. Söderberg O., Gullberg M., Jarvius M., Ridderstråle K., Leuchowius K. J., Jarvius J., Wester K., Hydbring P., Bahram F., Larsson L. G., Landegren U. (2006) Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods 3, 995–1000 [DOI] [PubMed] [Google Scholar]
- 54. Friedman J. S., Chang B., Krauth D. S., Lopez I., Waseem N. H., Hurd R. E., Feathers K. L., Branham K. E., Shaw M., Thomas G. E., Brooks M. J., Liu C., Bakeri H. A., Campos M. M., Maubaret C., Webster A. R., Rodriguez I. R., Thompson D. A., Bhattacharya S. S., Koenekoop R. K., Heckenlively J. R., Swaroop A. (2010) Loss of lysophosphatidylcholine acyltransferase 1 leads to photoreceptor degeneration in rd11 mice. Proc. Natl. Acad. Sci. U.S.A. 107, 15523–15528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Soupene E., Kuypers F. A. (2012) Phosphatidylcholine formation by LPCAT1 is regulated by Ca2+ and the redox status of the cell. BMC Biochem. 13, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Soccio R. E., Breslow J. L. (2003) StAR-related lipid transfer (START) proteins: mediators of intracellular lipid metabolism. J. Biol. Chem. 278, 22183–22186 [DOI] [PubMed] [Google Scholar]
- 57. Tsujishita Y., Hurley J. H. (2000) Structure and lipid transport mechanism of a StAR-related domain. Nat. Struct. Biol. 7, 408–414 [DOI] [PubMed] [Google Scholar]
- 58. Roderick S. L., Chan W. W., Agate D. S., Olsen L. R., Vetting M. W., Rajashankar K. R., Cohen D. E. (2002) Structure of human phosphatidylcholine transfer protein in complex with its ligand. Nat. Struct. Biol. 9, 507–511 [DOI] [PubMed] [Google Scholar]
- 59. Romanowski M. J., Soccio R. E., Breslow J. L., Burley S. K. (2002) Crystal structure of the Mus musculus cholesterol-regulated START protein 4 (StarD4) containing a StAR-related lipid transfer domain. Proc. Natl. Acad. Sci. U.S.A. 99, 6949–6954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhang M., Liu P., Dwyer N. K., Christenson L. K., Fujimoto T., Martinez F., Comly M., Hanover J. A., Blanchette-Mackie E. J., Strauss J. F., 3rd (2002) MLN64 mediates mobilization of lysosomal cholesterol to steroidogenic mitochondria. J. Biol. Chem. 277, 33300–33310 [DOI] [PubMed] [Google Scholar]
- 61. Hanada K., Kumagai K., Yasuda S., Miura Y., Kawano M., Fukasawa M., Nishijima M. (2003) Molecular machinery for non-vesicular trafficking of ceramide. Nature 426, 803–809 [DOI] [PubMed] [Google Scholar]
- 62. Feng L., Chan W. W., Roderick S. L., Cohen D. E. (2000) High-level expression and mutagenesis of recombinant human phosphatidylcholine transfer protein using a synthetic gene: evidence for a C-terminal membrane binding domain. Biochemistry 39, 15399–15409 [DOI] [PubMed] [Google Scholar]
- 63. Olayioye M. A., Buchholz M., Schmid S., Schöffler P., Hoffmann P., Pomorski T. (2007) Phosphorylation of StarD10 on serine 284 by casein kinase II modulates its lipid transfer activity. J. Biol. Chem. 282, 22492–22498 [DOI] [PubMed] [Google Scholar]
- 64. Kanno K., Wu M. K., Agate D. S., Fanelli B. J., Wagle N., Scapa E. F., Ukomadu C., Cohen D. E. (2007) Interacting proteins dictate function of the minimal START domain phosphatidylcholine transfer protein/StarD2. J. Biol. Chem. 282, 30728–30736 [DOI] [PubMed] [Google Scholar]
- 65. Fisher A. B., Dodia C., Feinstein S. I., Ho Y. S. (2005) Altered lung phospholipid metabolism in mice with targeted deletion of lysosomal-type phospholipase A2. J. Lipid Res. 46, 1248–1256 [DOI] [PubMed] [Google Scholar]
- 66. Ito M., Yamanashi Y., Toyoda Y., Izumi-Nakaseko H., Oda S., Sugiyama A., Kuroda M., Suzuki H., Takada T., Adachi-Akahane S. (2013) Disruption of Stard10 gene alters the PPARα-mediated bile acid homeostasis. Biochim. Biophys. Acta 1831, 459–468 [DOI] [PubMed] [Google Scholar]
- 67. van Helvoort A., de Brouwer A., Ottenhoff R., Brouwers J. F., Wijnholds J., Beijnen J. H., Rijneveld A., van der Poll T., van der Valk M. A., Majoor D., Voorhout W., Wirtz K. W., Elferink R. P., Borst P. (1999) Mice without phosphatidylcholine transfer protein have no defects in the secretion of phosphatidylcholine into bile or into lung airspaces. Proc. Natl. Acad. Sci. U.S.A. 96, 11501–11506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hishikawa D., Shindou H., Harayama T., Ogasawara R., Suwabe A., Shimizu T. (2013) Identification of Sec14-like 3 as a novel lipid-packing sensor in the lung. FASEB J. 27, 5131–5140 [DOI] [PubMed] [Google Scholar]
- 69. Sha B., Phillips S. E., Bankaitis V. A., Luo M. (1998) Crystal structure of the Saccharomyces cerevisiae phosphatidylinositol-transfer protein. Nature 391, 506–510 [DOI] [PubMed] [Google Scholar]
- 70. Stocker A., Tomizaki T., Schulze-Briese C., Baumann U. (2002) Crystal structure of the human supernatant protein factor. Structure 10, 1533–1540 [DOI] [PubMed] [Google Scholar]
- 71. Bankaitis V. A., Mousley C. J., Schaaf G. (2010) The Sec14 superfamily and mechanisms for crosstalk between lipid metabolism and lipid signaling. Trends Biochem. Sci. 35, 150–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Bankaitis V. A., Aitken J. R., Cleves A. E., Dowhan W. (1990) An essential role for a phospholipid transfer protein in yeast Golgi function. Nature 347, 561–562 [DOI] [PubMed] [Google Scholar]
- 73. Anantharaman V., Aravind L. (2002) The GOLD domain, a novel protein module involved in Golgi function and secretion. Genome Biol. 3, research0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Favale N. O., Fernández-Tome M. C., Pescio L. G., Sterin-Speziale N. B. (2010) The rate-limiting enzyme in phosphatidylcholine synthesis is associated with nuclear speckles under stress conditions. Biochim. Biophys. Acta 1801, 1184–1194 [DOI] [PubMed] [Google Scholar]
- 75. Agassandian M., Chen B. B., Schuster C. C., Houtman J. C., Mallampalli R. K. (2010) 14-3-3ζ escorts CCTα for calcium-activated nuclear import in lung epithelia. FASEB J. 24, 1271–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Mansilla F., da Costa K. A., Wang S., Kruhøffer M., Lewin T. M., Orntoft T. F., Coleman R. A., Birkenkamp-Demtröder K. (2009) Lysophosphatidylcholine acyltransferase 1 (LPCAT1) overexpression in human colorectal cancer. J. Mol. Med. 87, 85–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Grupp K., Sanader S., Sirma H., Simon R., Koop C., Prien K., Hube-Magg C., Salomon G., Graefen M., Heinzer H., Minner S., Izbicki J. R., Sauter G., Schlomm T., Tsourlakis M. C. (2013) High lysophosphatidylcholine acyltransferase 1 expression independently predicts high risk for biochemical recurrence in prostate cancers. Mol. Oncol. 7, 1001–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wu M., Tu T., Huang Y., Cao Y. (2013) Suppression subtractive hybridization identified differentially expressed genes in lung adenocarcinoma: ERGIC3 as a novel lung cancer-related gene. BMC Cancer 13, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]