Background: DNA end resection is the first step in DNA double-strand break repair by homologous recombination.
Results: Ku and RPA compete for binding to ends with ssDNA. Ku blocks resection by Exo1, whereas RPA allows resection to proceed.
Conclusion: Longer ssDNA overhangs favor RPA binding and further resection.
Significance: Interplay between Ku and RPA binding to DNA ends may affect DNA repair pathway choice.
Keywords: DNA-binding protein, DNA damage, DNA damage response, homologous recombination, Saccharomyces cerevisiae, Exo1, Ku, RPA, non-homologous DNA end joining
Abstract
DNA double-strand breaks can be eliminated via non-homologous end joining or homologous recombination. Non-homologous end joining is initiated by the association of Ku with DNA ends. In contrast, homologous recombination entails nucleolytic resection of the 5′-strands, forming 3′-ssDNA tails that become coated with replication protein A (RPA). Ku restricts end access by the resection nuclease Exo1. It is unclear how partial resection might affect Ku engagement and Exo1 restriction. Here, we addressed these questions in a reconstituted system with yeast proteins. With blunt-ended DNA, Ku protected against Exo1 in a manner that required its DNA end-binding activity. Despite binding poorly to ssDNA, Ku could nonetheless engage a 5′-recessed DNA end with a 40-nucleotide (nt) ssDNA overhang, where it localized to the ssDNA-dsDNA junction and efficiently blocked resection by Exo1. Interestingly, RPA could exclude Ku from a partially resected structure with a 22-nt ssDNA tail and thus restored processing by Exo1. However, at a 40-nt tail, Ku remained stably associated at the ssDNA-dsDNA junction, and RPA simultaneously engaged the ssDNA region. We discuss a model in which the dynamic equilibrium between Ku and RPA binding to a partially resected DNA end influences the timing and efficiency of the resection process.
Introduction
DNA double-strand breaks (DSBs)4 are induced by ionizing radiation and reactive oxygen species generated by cellular metabolism and also stem from DNA replication fork collapse. Failure to correctly repair DSBs may lead to gross chromosomal rearrangements, mutagenesis, and, ultimately, cancer (1). Two pathways exist for repairing DSBs: homologous recombination (HR) and non-homologous end joining (NHEJ). NHEJ, in which the DSB ends are brought together, processed if necessary, and then religated, is initiated by the binding of Ku (a heterodimer of Ku70 and Ku80 proteins) (2). Ku is a ring-shaped molecule that threads onto and encircles the DNA (3–5). Ku is thought to provide a recruitment platform for the DNA ligase IV complex (consisting of the Dnl4 and Lif1 proteins in yeast). NHEJ relies on microhomology in the DSB overhangs to guide the repair process (6). Mutations arise during NHEJ if these overhangs contain damaged or missing bases, and chromosome rearrangements, such as translocations, ensue upon the inadvertent joining of ends from different DSBs (7, 8). Repair by HR is usually error-free but requires a homologous chromatid as a template. HR in yeast cells is initiated when nucleases, including Mre11 (a member of the Mre11-Rad50-Xrs2 or MRX complex), Exo1, and Dna2, resect the 5′-strand to create long ssDNA tails, which then become coated with replication protein A (RPA) (9, 10). RPA is subsequently replaced by the Rad51 recombinase, which catalyzes DNA joint formation with a homologous DNA sequence (11, 12). Because DNA end resection degrades the overhangs that would otherwise guide NHEJ, its onset signals a commitment to repair by HR (13, 14).
How the choice between NHEJ and HR is made has been investigated in recent years. There is direct competition between the two pathways in some circumstances, as genetic and biochemical analyses have shown that Ku binding to DSB ends blocks Exo1-dependent resection. Deletion of KU70 in yeast enhances the initiation of resection (15–17) and leads to more extensive DSB-associated ssDNA in G1 cells (18, 19). Loss of Ku also partially suppresses the ionizing radiation and methyl methanesulfonate sensitivity of resection-defective mre11 and rad50 strains by allowing Exo1 to access DNA ends (20–22). Deletion of KU70 or overexpression of EXO1 also suppresses the lethality of sae2Δ sgs1Δ cells, but deletion of DNL4 does not. Thus, suppression occurs when the Ku-imposed block of Exo1 is relieved, but not because NHEJ is inhibited (22). Finally, Ku binding to DSB ends has been shown to block Exo1 access in vitro (23, 24).
The cell cycle stage exerts a strong influence on DSB repair pathway choice. HR occurs mostly with a sister chromatid, and DNA end resection is down-regulated in G1 but becomes fully active in S/G2 (14, 25, 26). The cell cycle regulation of resection is dependent on the S/G2 cyclin-dependent kinase Cdk1, which phosphorylates Sae2, an essential cofactor for the dsDNA-specific endonuclease activity of Mre11 (27, 28). Cyclin-dependent kinases have additionally been shown to phosphorylate both EXO1 in humans and Dna2 in yeast, with these protein modifications serving a pro-resection role (29, 30).
The nature of the DSB also influences repair pathway choice. In mammalian cells, NHEJ occurs in as little as 30 min, whereas HR can take up to 7 h to complete (31). NHEJ takes longer at DSBs with damaged termini (32, 33), and because these DSBs persist in an unrepaired state longer, they may be more likely to undergo resection. The presence of ssDNA also affects how a DSB is repaired. The ssDNA content may be derived from proximal single-strand breaks that occur on opposite strands and can also be generated by limited resection. DSBs with short overhangs (<4 bases) are substrates for NHEJ, whereas longer overhangs (>10 bases) promote a single-strand annealing pathway that does not require Ku and is enhanced by Rad52 (8).
Here, we investigated how ssDNA overhang length at a DSB affects the binding of yeast Ku and RPA and how it influences Exo1 access to the DNA end. We report that, although Ku bound poorly to ssDNA, it efficiently engaged a DNA junction with a ssDNA overhang as long as 40 nucleotides (nt) and was able to restrict Exo1 access to the junction. Unlike Ku, binding of RPA to a ssDNA tail did not affect Exo1 action at an adjoining DNA terminus. Interestingly, RPA could displace Ku from a structure harboring a 22-nt overhang, leading to restoration of end processing by Exo1. We discuss these findings within the context of understanding Ku-RPA interplay at a resected DNA end germane for the timing and efficiency of further resection.
Experimental Procedures
Ku Expression and Purification
For yeast Ku expression, the coding regions of YKU70 and YKU80 were PCR-amplified using templates from the yeast ORF collection (Open Biosystems) and cloned into the BamHI/XhoI and EcoRI/SpeI sites of pESC-Ura (Agilent), placing the genes under the control of the GAL1 or GAL10 promoter, respectively. A FLAG tag was introduced to the C terminus of Yku80 to facilitate detection and purification. The yku70-R456E mutation was induced by site-directed mutagenesis. Ku and mutant Ku harboring the Yku70-R456E protein were expressed in the protease-deficient yeast strain BJ5464 (34). An overnight culture in glucose-containing medium lacking uracil was diluted 1:100 into 15 liters of raffinose medium lacking uracil. Cells were cultured at 30 °C until the A660 reached 0.8, at which time galactose was added to 2% to induce Ku expression for 12 h. Cells were harvested by centrifugation, and the pellet (∼70 g) was stored at −80 °C. Subsequent steps were carried out at 0–4 °C. After cell disruption with dry ice in a coffee grinder (Krups), 70 ml of ice-cold buffer (40 mm KH2PO4 (pH 7.4), 20% glycerol, 1 mm EDTA, 0.1% Nonidet P-40, 2 mm DTT, 200 mm KCl, and a mixture of protease inhibitors containing aprotinin, chymostatin, leupeptin, and pepstatin A (American Bioanalytical) at 5 μg/ml each and 1 mm phenylmethylsulfonyl fluoride) was added. The crude cell lysate was clarified by ultracentrifugation (100,000 × g for 45 min) and then loaded onto a 5-ml DEAE-Sepharose column. After washing the column with 50 ml of K buffer (20 mm KH2PO4 (pH 7.4), 10% glycerol, 0.5 mm EDTA, 0.01% IGEPAL, and 1 mm DTT) containing 100 mm KCl, Ku was eluted with a 100-ml gradient from 100 to 500 mm KCl. The peak fractions were pooled and then incubated with 0.5 ml of anti-FLAG M2 resin for 2 h. The matrix was washed once with 20 ml of K buffer containing 500 mm KCl and 2 mm each ATP and MgCl2 and then washed three additional times with 10 ml of K buffer containing 500 mm KCl. The Ku complex was eluted by incubating the matrix with 1 ml of K buffer containing 500 mm KCl and 200 μg/ml FLAG peptide (Sigma) for 1 h. The eluate was concentrated and loaded onto a Superdex 200 10/300 GL size exclusion column (24 ml). The peak fractions were pooled and concentrated using an Amicon microconcentrator. The purified Ku complex (∼ 500 μg) was frozen in liquid nitrogen and stored at −80 °C in small aliquots.
RPA and Exo1 Purification
The RPA complex was expressed in Escherichia coli and purified to a high degree according to a previously published procedure (35–37). Similarly, a bacmid for the expression of Exo1 was generated in DH10Bac E. coli cells (Invitrogen), after which a recombinant baculovirus was made for its expression in insect cells. It was purified to a high degree as reported previously (38).
DNA Substrates
Table 1 provides a list of the oligonucleotide sequences. To prepare the substrates, the PSOL8515 40-mer containing 3′-biotin (Integrated DNA Technologies) was 5′-labeled with [γ-32P]ATP (PerkinElmer Life Sciences) and T4 polynucleotide kinase (New England Biolabs). This was then annealed to a complementary strand by slow cooling over a period of 3 h in a model PTC-200 thermocycler (Bio-Rad). Substrates were purified by 10% native PAGE in Tris acetate/EDTA buffer. The corresponding gel bands were excised, and the DNA was electroeluted. The eluate was further concentrated using an Amicon microconcentrator and stored at −20 °C. To block the biotinylated DNA end, substrates (400 nm) were incubated in a 12-μl reaction (50 mm Tris-HCl (pH 7.1), 100 mm NaCl, 5 mm MgCl2, and 1 mm DTT) containing 2 μm streptavidin (Invitrogen) for 10 min at 20 °C. Streptavidin binding was verified by native 10% acrylamide gel electrophoresis on TBE (89 mm Tris borate (pH 8.0), and 2 mm EDTA) gels.
TABLE 1.
Oligonucleotides used to create substrates
| Name | Sequence | Description |
|---|---|---|
| PSOL8515 | ATTAAGCTCTAAGCCATGAATTCAAATGACCTCTTATCAA-3′-biotin | Labeled strand |
| PSOL8400 | TTGATAAGAGGTCATTTGAATTCATGGCTTAGAGCTTAAT | 40-bp dsDNA substrate |
| PSOL5782 | TTGATAAGAGGTCATTTGAATTCATGGCTTAGAGCTTAATTGCTGAA | 7-nt ssDNA overhang |
| PSOL8275 | TTGATAAGAGGTCATTTGAATTCATGGCTTAGAGCTTAATTGCTGAATCTGGTGCTGGGATC | 22-nt ssDNA overhang |
| PSOL8816 | TTGATAAGAGGTCATTTGAATTCATGGCTTAGAGCTTAATTGCTGAATCTGGTGCTGGGATCCAACATGTTTTAAATATG | 40-nt ssDNA overhang |
| PSOL7601 | TTGATAAGAGGTCATTTGAATTCATGGCTTAGAGCTTAATTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTT | 40-nt dT ssDNA overhang |
EMSA
Reactions (10 μl) contained 40 mm Tris-HCl (pH 7.5), 4 mm MgCl2, 100 mm KCl, 1 mm DTT, 100 μg/ml BSA, 4 nm DNA substrate, and the indicated concentrations of Ku and/or RPA. Reactions were incubated for 10 min at 30 °C. The same conditions applied for Fig. 1D (DNA competition assay), with each DNA substrate being used at 4 nm. For the competition assays (Figs. 1E, 2B, and 3G), Ku (5 nm) was incubated with radiolabeled DNA (4 nm) at 30 °C for 10 min. Unlabeled DNA (2, 4, 8, and 16 nm) was then added to the reaction and incubated at 30 °C for 10 min. Control reactions were performed by adding 0.2% SDS and 0.5 mg/ml proteinase K for 5 min at 37 °C. PAGE was performed in TBE buffer on 4% gels without any denaturant. Gels were dried on Whatman DE3 paper, and bands were quantified in a Bio-Rad PhosphorImager.
FIGURE 1.
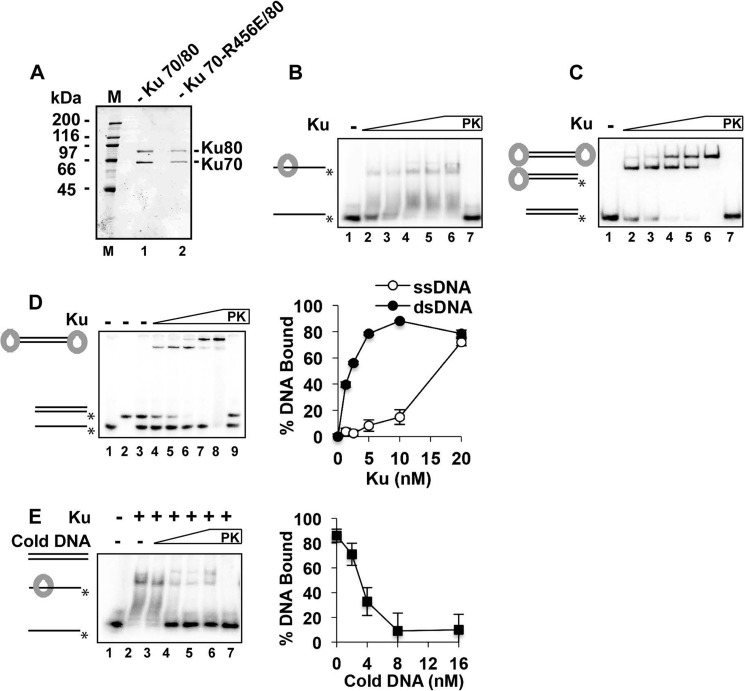
Binding of ssDNA by Ku. A, SDS-PAGE analysis of purified Ku and the Ku-RE mutant. Molecular masses are indicated in kilodaltons (M lane). B, Ku (1.25, 2.5, 5, 10, and 20 nm) was incubated with the 40-mer ssDNA (4 nm) for 10 min. C, same conditions, except that the 40-bp dsDNA was tested. D, same conditions, except that both the 40-mer ssDNA and 40-bp dsDNA substrates were included in the same reactions. Quantification of the results is shown to the right. E, Ku (5 nm) was incubated with the radiolabeled 40-mer ssDNA (4 nm) for 10 min, and unlabeled 40-bp dsDNA (0, 2, 4, 8, and 16 nm) was added, followed by a further 10 min of incubation. Quantification of the results is shown to the right. In B–E, PK lanes represent a reaction that was treated with SDS and proteinase K prior to analysis, and asterisks denote the 32P label. Error bars in D and E represent 1 S.D. based on results from three independent experiments.
FIGURE 2.
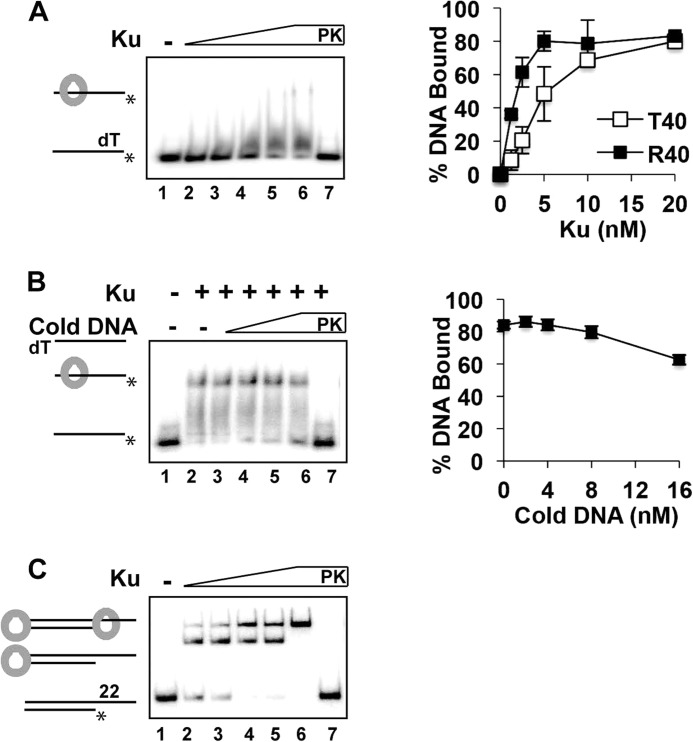
Binding of unstructured ssDNA by Ku. A, Ku (1.25, 2.5, 5, 10, and 20 nm) was incubated with oligo(dT)40 (dT; 4 nm) for 10 min. Quantification of results in Fig. 1B (R40) and those pertaining to oligo(dT)40 (T40) binding is shown to the right. B, Ku (5 nm) was incubated with the radiolabeled 40-mer ssDNA (4 nm) for 10 min, and unlabeled oligo(dT)40 (0, 2, 4, 8, and 16 nm) was added, followed by a 10-min incubation. Quantification of the results is shown to the right. C, the substrate was 40-bp dsDNA with a blunt end and a 22-nt 5′-recessed end. PK lanes represent a reaction that was treated with SDS and proteinase K prior to analysis, and asterisks denote the 32P label. Error bars in A and B represent 1 S.D. based on results from three independent experiments.
FIGURE 3.
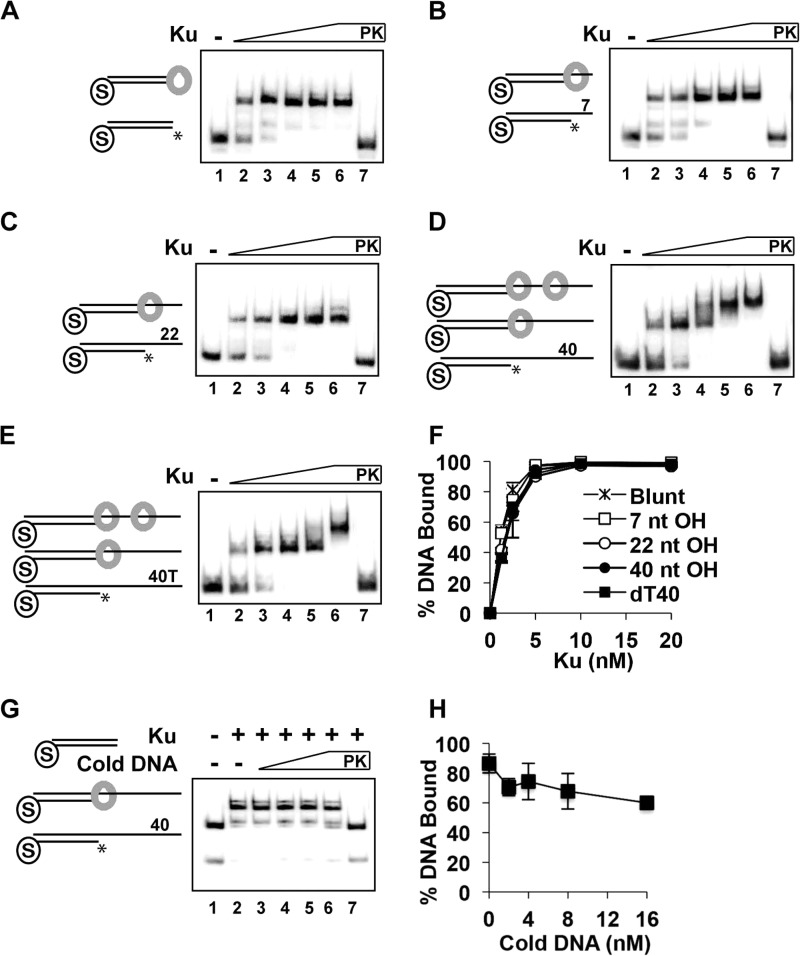
Engagement of 5′-recessed DNA ends by Ku. A–E, Ku (1.25, 2.5, 5, 10, and 20 nm) was incubated with DNA substrate (4 nm each) with one of its ends blocked by a biotin-streptavidin complex and also with a free blunt end (A) or a 7-nt (B), 22-nt (C), 40-nt (D), or oligo(dT)40 (40T or dT40; E) 5′-recessed end for 10 min. F, quantification of the results in A–E. G, Ku (5 nm) was incubated with the substrate containing a 40-nt 5′-recessed end (4 nm) for 10 min, and unlabeled 40-bp blunt-ended dsDNA (0, 2, 4, 8, and 16 nm) was added, followed by a 10-min incubation. Quantification of the results is shown to the right. In A–E and G, PK lanes represent a reaction was treated with SDS and proteinase K prior to analysis, and asterisks denote the 32P label. Error bars in F and H represent 1 S.D. based on results from three independent experiments.
Nuclease Assay
The reaction (10 μl) was assembled in 40 mm Tris-HCl (pH 7.5), 4 mm MgCl2, 100 mm KCl, 1 mm DTT, and 100 μg/ml BSA, and it contained 4 nm DNA substrate. Ku and/or RPA was preincubated with the indicated DNA substrate for 10 min at 30 °C, and the indicated amount of Exo1 was then added, followed by a 5-min incubation at 30 °C. After deproteinizing the reaction with 0.2% SDS and 0.5 mg/ml proteinase K for 5 min at 37 °C, it was analyzed on a native 10% polyacrylamide gel run in TBE buffer and then analyzed as described above.
DNase I Footprinting
The reaction (10 μl) was assembled in 10 mm Tris-HCl (pH 7.5), 2.5 mm MgCl2, and 0.5 mm CaCl2. Ku was incubated with the DNA substrate for 10 min at 30 °C before adding DNase I (50 pg), followed by an additional 3-min incubation at 37 °C. The reaction was stopped by boiling in formamide loading dye, followed by analysis on a denaturing TBE-15% polyacrylamide DNA sequencing gel, which was dried and analyzed as described above.
Results
Engagement of Recessed DNA Ends by Ku
The human Ku complex binds with high affinity to DNA ends in a sequence-independent manner (39). Within the context of DSB repair, Ku binding has been assessed with DNA containing either blunt ends or short ssDNA overhangs in the range of 4–18 nt, and the function of Ku at telomeres has been studied using DNA containing an overhang of up to 15 nt (40). The effect of appending a longer overhang on DSB engagement by Ku has not been investigated. To accomplish this goal, yeast Ku was expressed in Saccharomyces cerevisiae and purified to near homogeneity (Fig. 1A). Importantly, purified Ku could shift a 40-nt ssDNA fragment. A smear was observed, with an additional shifted band emerging at 2.5 Ku molecules/ssDNA strand (Fig. 1B). Ku was then tested with a 40-mer duplex obtained by hybridizing the 40-nt ssDNA to its complement. Consistent with previous data (40, 41), Ku bound the duplex DNA more efficiently, with the substrate fully shifted at 1.25 Ku molecules/DNA substrate (Fig. 1C). Ku binding to the DNA duplex produced two shifted bands, reflecting engagement of both DNA ends in the duplex (Fig. 1C). Titration of Ku into a mixture of dsDNA and ssDNA showed that it bound dsDNA prior to shifting the ssDNA (Fig. 1D). Thus, although Ku is able to bind ssDNA, it preferentially engages dsDNA (42). To investigate the stability of Ku binding to ssDNA, unlabeled 40-mer dsDNA was introduced into a reaction with Ku having been preloaded onto the radiolabeled ssDNA (Fig. 1E). After a 10-min incubation, most of the bound ssDNA was released upon adding an equimolar amount of the unlabeled blunt-ended duplex (Fig. 1E, lane 4), with ∼90% release seen at a 2-fold excess of the competitor (lane 5). We verified that the reaction had reached equilibrium at this time point (data not shown). These results indicate that engagement of ssDNA by Ku is dynamic.
Given that Ku has affinity for hairpin structures (43, 44), we considered the possibility that it might specifically associate with secondary structure in the ssDNA substrate. To test this idea, we monitored Ku binding to oligo(dT)40, which lacks secondary structure character (Fig. 2A). Indeed, Ku bound the oligo(dT)40 substrate less efficiently than did the ssDNA with random nucleotide content (compare Figs. 2A and 1B), suggesting that secondary structure can contribute to Ku binding to ssDNA. To investigate this further, a nucleoprotein complex of Ku and radiolabeled ssDNA was assembled and then challenged with unlabeled oligo(dT)40 (Fig. 2B). Importantly, oligo(dT)40, even at four times the concentration of the ssDNA, could not compete Ku off the latter (Fig. 2B, lane 6). Thus, it appears that Ku has only a low affinity for ssDNA, but the presence of secondary structure in the DNA leads to an enhancement of this affinity (43, 44).
To test whether Ku can stably engage duplex DNA that harbors an adjoining ssDNA region, we generated a 40-bp substrate with one blunt end and a 5′-recessed end with a 22-nt ssDNA tail. Ku produced two shifted species with this substrate (Fig. 2C), just as it did with the substrate with two blunt ends (Fig. 1C). This observation suggested the possibility that Ku is able to associate with the recessed DNA end efficiently. However, to eliminate the caveat that the slower migrating nucleoprotein complex in the previous experiment (Fig. 2C) might have contained two Ku molecules at the blunt end, we constructed and tested three substrates that harbored a blunt end and also a 5′-recessed end with either a 7 or 22-nt overhang in which the blunt end had been occluded by a biotin-streptavidin complex (Fig. 3, A–C). With these substrates, Ku generated just a single nucleoprotein complex. Given that occluding both blunt ends of a DNA substrate with a biotin-streptavidin complex completely abolished Ku binding (data not shown), we concluded from the above results that Ku is able to load onto a recessed DNA end, even if the ssDNA region is 22 nt in length.
We next asked whether extending the 3′-single-stranded overhang to 40 nt would affect Ku binding. Importantly, Ku was capable of efficiently engaging substrates that harbored a blocked blunt end and a free 5′-recessed end with either a 40-nt random sequence or oligo(dT)40 overhang (Fig. 3, D and E). Moreover, a second nucleoprotein complex was generated when an excess of Ku was used (Fig. 3, D and E), suggesting that the extended ssDNA region in these substrates provides an additional site for Ku binding. To determine whether Ku associates stably with a recessed end, we challenged the nucleoprotein complex of Ku and the radiolabeled substrate containing the 40-nt recessed end with an increasing amount of unlabeled blunt-ended dsDNA (Fig. 3, G and H). Ku remained bound to the radiolabeled DNA even upon adding a 4-fold excess of competitor DNA, indicating a stable association of Ku with the recessed end (Fig. 3, G and H). Taken together, these results show that Ku is quite versatile in being able to recognize a variety of DNA end structures.
Protection of Recessed DNA Ends from Exo1 Action by Ku
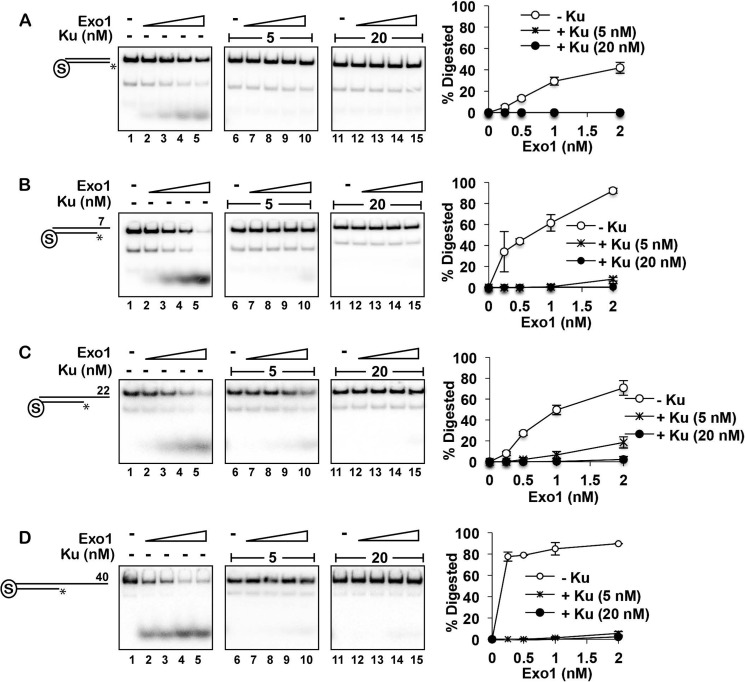
Yeast Ku is known to protect DSB ends from Exo1-mediated resection in cells (21, 22, 45–47), and human Ku can prevent human EXO1 from accessing blunt DNA ends in vitro (24). Human EXO1 is active on a variety of DNA structures in vitro, including nicked, gapped, and blunt-ended dsDNAs (48, 49). The x-ray crystallographic structure of human EXO1 has revealed that it localizes to the 5′ terminus of a 5′-recessed DNA end (50). Consistent with these observations, we found that the exonuclease activity of yeast Exo1 was significantly more robust at 5′-recessed ends (with 7, 22, or 40 nt of ssDNA) than at a blunt end (Fig. 4, A–D). We next asked whether Ku could restrict Exo1 activity on each of these substrates. Because the biotin-streptavidin complex effectively occluded DNA ends from Exo1 (data not shown), we used it to block Exo1 access from the blunt end in each substrate. In agreement with a published report that examined human Ku and human EXO1 (24), preincubation of the DNA with yeast Ku prevented degradation of the blunt-ended substrate by Exo1 (Fig. 4A). Importantly, Ku was just as capable of restricting Exo1 activity on each of the other substrates with a recessed end (Fig. 4, B–D). Notably, with the substrate that had the 40-nt recessed end, efficient restriction of Exo1 activity was observed at 5 nm Ku, which produced just a single shift in an EMSA (Fig. 3D). This implies that loading of a single Ku molecule onto a recessed end is sufficient to prevent Exo1 access. It should be noted that, because the DNA substrates harbored only a unique 5′-radiolabel, Exo1 was unable to remove even a single nucleotide when Ku was bound to either a blunt or recessed end.
FIGURE 4.
Restriction of Exo1 action by Ku at blunt and 5′-recessed DNA ends. A–D, DNA substrates (4 nm each) that had a blocked end and also a free blunt end (A) or a 7-nt (B), 22-nt (C), or 40-nt (D) 5′-recessed end were preincubated without (left panels) or with 5 (middle panels) or 20 nm (right panels) Ku for 10 min before adding Exo1 (0.25, 0.5, 1, and 2 nm), followed by a 5-min incubation. Quantification of the results is shown to the right. Error bars represent 1 S.D. based on results from three independent experiments, and asterisks denote the 32P label.
Requirement for the DNA End-binding Activity of Ku in Exo1 Restriction
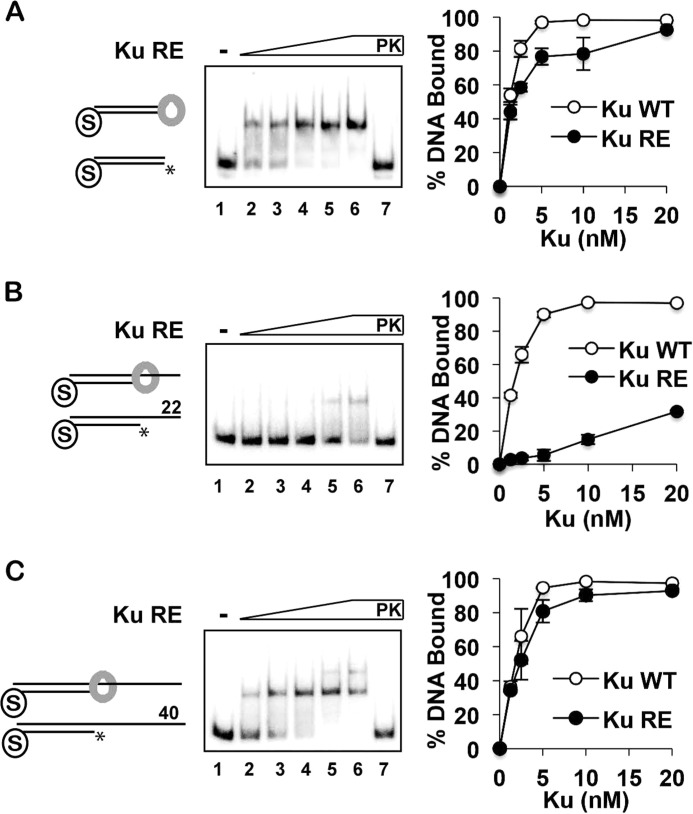
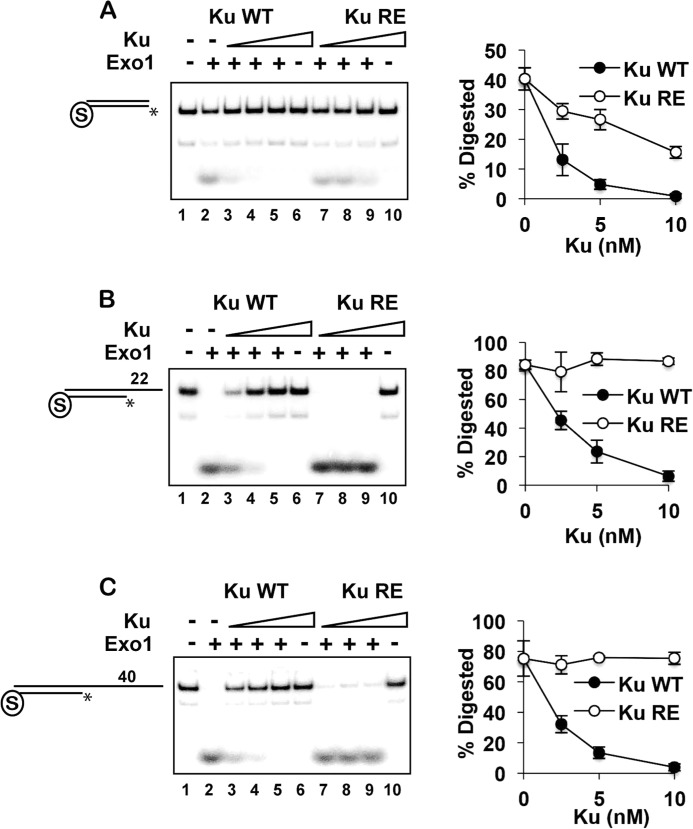
To further examine the mechanism by which Ku inhibits Exo1-dependent resection, we expressed and purified a DNA-binding mutant (referred to as Ku-RE) that harbors the yku70-R456E mutation (Fig. 1A) (41). Consistent with published results, Ku-RE was found to be partially defective in binding blunt dsDNA ends when analyzed by an EMSA (Fig. 5A). Accordingly, Ku-RE showed a reduced ability to protect blunt ends from Exo1 action (Fig. 6A). Ku-RE showed a more severe defect in binding the 22-nt 5′-recessed end (Fig. 5B) and was completely devoid of the ability to protect the end from degradation by Exo1 (Fig. 6B). Interestingly, Ku-RE bound the substrate with the 40-nt recessed end relatively well, although unlike wild-type Ku, it was unable to produce a second shift at high concentrations (Fig. 5C). A possible explanation is that Ku-RE that is free to diffuse off the 22-nt overhang is held on the DNA by secondary structure in the longer 40-nt overhang. Despite the apparent fitness of Ku-RE in engaging the 40-nt substrate (Fig. 5C), it was unable to restrict the activity of Exo1 at the recessed DNA end (Fig. 6C). Taken together, the results presented herein provide verification that the DNA end-binding activity of Ku is indispensable for the protection of different DNA end structures against Exo1 action.
FIGURE 5.
Examination of the Ku-RE mutant for DNA end binding. A–C, Ku-RE (1.25, 2.5, 5, 10, and 20 nm) was incubated with DNA substrates (4 nm each) that had a blocked end and also a free blunt end (A) or a 22-nt (B) or 40-nt (C) 5′-recessed end. The quantification compares the results from these experiments with those obtained with wild-type Ku (from Fig. 2). PK lanes represent a reaction that had been treated with SDS and proteinase K prior to analysis, and asterisks denote the 32P label. Error bars represent 1 S.D. based on results from three independent experiments.
FIGURE 6.
Requirement for Ku DNA end binding activity in the restriction of Exo1 action. A–C, Ku or Ku-RE (2.5, 5, and 10 nm) was preincubated with DNA substrates with a blocked end and also a free blunt end (A) or a 22-nt (B) or 40-nt (C) 5′ recessed end for 5 min, and Exo1 (2 nm) was added, followed by a 5-min incubation. Quantification of the results is shown to the right. Asterisks denote the 32P label. Error bars represent 1 S.D. based on results from three independent experiments.
Localization of Ku to the ssDNA-dsDNA Junction in Recessed DNA
The observation that Ku protects 5′-recessed ends from degradation by Exo1 suggests that Ku localizes to the ssDNA-dsDNA junction (44). To test this directly, we performed DNase I footprinting on the substrate with the 40-bp duplex region and a 40-nt recessed end. We first labeled the long strand at its 5′ terminus and blocked the blunt end with biotin-streptavidin to limit Ku access to only the recessed end (Fig. 7A). Protection was observed in the 40–26-nt region as the concentration of Ku was increased, corresponding to a 14-nt region of dsDNA adjacent to the ssDNA-dsDNA junction (Fig. 7A). We also conducted footprinting analysis on the same substrate that was labeled on the 3′-end of the long strand (Fig. 7B). Protection near the ssDNA-dsDNA junction was again seen, and a second area of protection in the adjoining ssDNA region was also detected at the highest concentration of Ku used (Fig. 7B). These results confirm that Ku can access a 5′-recessed DNA end and can subsequently engage ssDNA that abuts the DNA end (Fig. 3D). We note that our results are in general congruence with those in a published report showing localization of Ku to the ssDNA-dsDNA junction in DNA substrates harboring telomeric DNA sequences (40).
FIGURE 7.
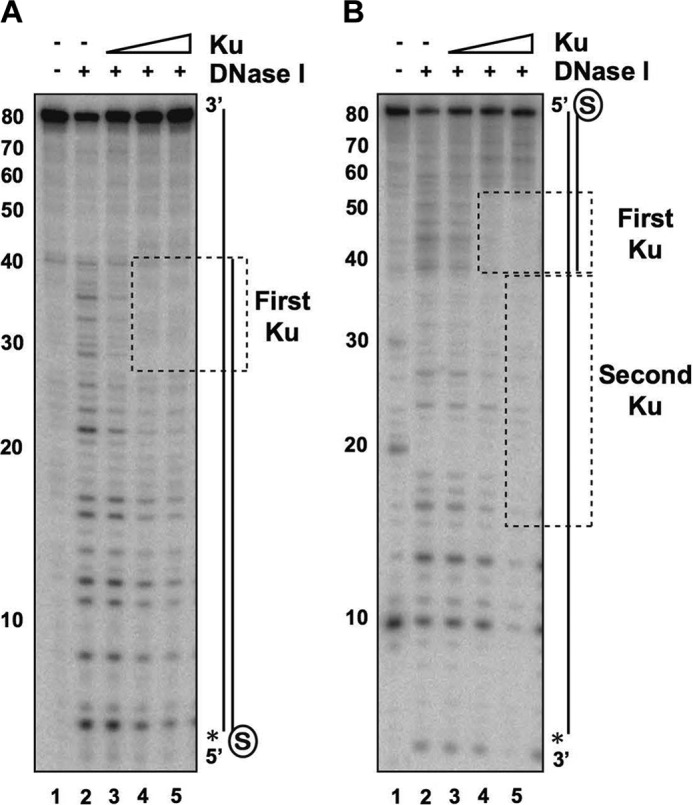
Localization of Ku to ssDNA-dsDNA junction in DNA with a recessed end. A, Ku (5, 10, or 20 nm) was incubated with the DNA substrate containing a blocked end and a free 40-nt 5′-recessed end (4 nm) for 10 min. DNase I (50 pg) was added, followed by a 3-min incubation. The asterisk denotes the 32P label, and the region protected by Ku is boxed. B, same as described for A, with the asterisk denoting the 32P label on the 3′-ssDNA overhang.
Ku-RPA Interplay at Recessed DNA Ends
The 3′-ssDNA tails generated by resection are rapidly bound by RPA, which prevents the formation of secondary structure and stimulates ongoing resection (9, 51). We next performed an EMSA to investigate whether RPA influences the loading of Ku onto a blunt or 5′-recessed end with 7, 22, or 40 nt of ssDNA. As described above, biotin-streptavidin was used to block the blunt end of each substrate to ensure that Ku and/or RPA could engage only the DNA from the free end. Each substrate was incubated with RPA alone, Ku alone, RPA followed by Ku, or Ku followed by RPA. As expected, RPA failed to shift the blunt-ended dsDNA and had no effect on Ku binding to this substrate (Fig. 8, lanes 1–5). Although RPA was able to shift the substrate with the 7-nt recessed end, it did not yield any well defined nucleoprotein species (Fig. 8, lanes 6–8). This result was fully expected considering a minimum length of >12 nt is needed for stable RPA engagement (52). Indeed, RPA had no influence on Ku binding to this substrate regardless of the order of addition of the two proteins (Fig. 8, lanes 9 and 10).
FIGURE 8.
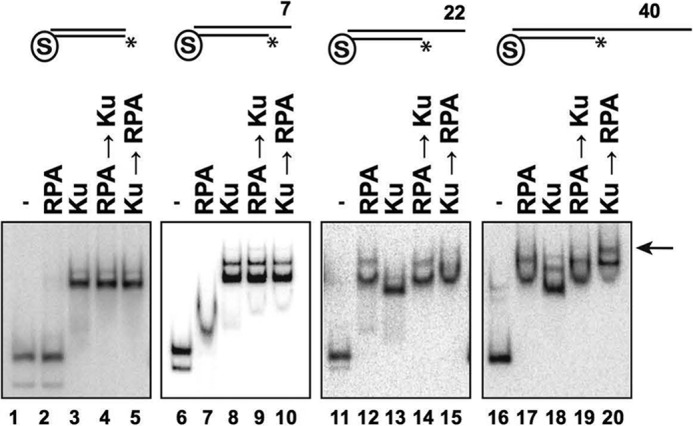
Interplay between Ku and RPA at DNA ends. EMSAs were performed using Ku (20 nm) and/or RPA (20 nm) and the indicated blocked DNA substrate (4 nm each) containing a free blunt end or a 7-, 22-, or 40-nt 5′-recessed end. In reactions in which the addition of one protein preceded the other, the DNA substrate was incubated with the first protein for 10 min, followed by the second protein and a 10-min incubation. The DNA-Ku-RPA ternary complex is indicated by the arrow. Asterisks denote the 32P label.
At the 22-nt recessed end, either Ku or RPA alone efficiently shifted the DNA, with each producing a distinctive band (Fig. 8, lanes 11–13). When the substrate was preincubated with RPA, Ku was unable to displace RPA or produce a supershift (Fig. 8, lane 14), suggesting that RPA engagement of the 22-nt overhang precludes Ku binding. Interestingly, when the DNA was first incubated with Ku followed by addition of RPA, the product ran at the same position as RPA alone (Fig. 8, lane 15). This suggests that Ku binding to this substrate is dynamic, with RPA being able to replace it via its high avidity for ssDNA.
When the single-stranded overhang at the recessed end was increased to 40 nt, incubation with RPA or Ku alone yielded a DNA shift (Fig. 8, lanes 16–18), and Ku was again unable to displace prebound RPA (lane 19). Prebinding of Ku followed by incubation with RPA led to a supershifted nucleoprotein complex, indicative of the formation of a ternary nucleoprotein complex of Ku and RPA (Fig. 8, lane 20). The increased ssDNA length in this substrate apparently prevents Ku dissociation and therefore allows for ternary complex formation.
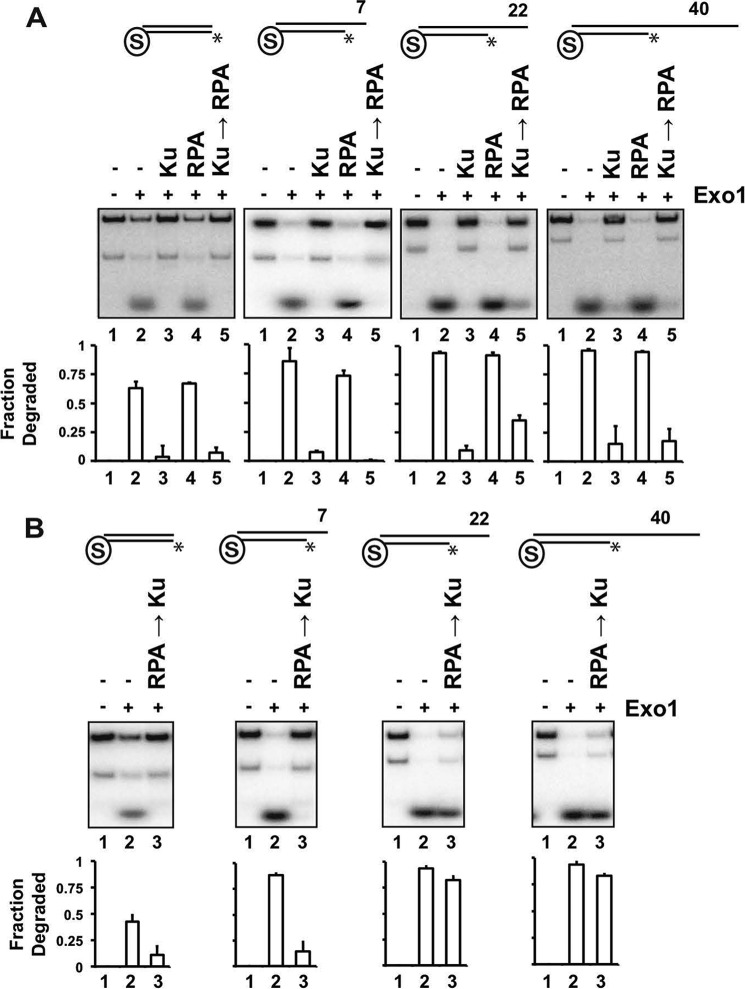
The observation that RPA can replace Ku at the 22-nt recessed end (Fig. 8, lane 15) resulted in the prediction that it would help overcome Ku-mediated inhibition of Exo1 activity. Indeed, the addition of RPA to the Ku-bound substrate partially restored access by Exo1 (Fig. 9A, third panel). RPA was not able to restore Exo1 activity at a blunt end or either a 7- or 40-nt recessed end (Fig. 9A, first, second, and fourth panels), results that are consistent with the observation that RPA did not replace Ku at these ends (Fig. 8).
FIGURE 9.
Effects of RPA and Ku on resection by Exo1. A, the various DNA substrates were incubated with RPA (20 nm), Ku (20 nm), or with Ku first and then RPA as described in the legend to Fig. 7, followed by the addition of Exo1 (2 nm for the blunt-ended substrate and 1 nm for the other substrates) and a 5-min incubation. Quantification of the results is shown below. B, same as described for A, except that the DNA substrates were incubated with RPA and then Ku before adding Exo1. Quantification of the results is shown below. Asterisks denote the 32P label. Error bars represent 1 S.D. based on results from three independent experiments.
We next preincubated DNA substrates with RPA and then added Ku followed by Exo1 (Fig. 9B). Preincubation with RPA did not affect the ability of Ku to block Exo1 action at a blunt end (Fig. 9B, first panel), which was expected because RPA had no affinity for such an end (Fig. 8, first panel). The same result was observed at the 7-nt recessed end (Fig. 9B, second panel), consistent with the ability of Ku to effectively exclude RPA from this end structure (Fig. 9A, second panel). At the 22- and 40-nt recessed ends, preincubation with RPA prevented Ku from restricting Exo1 access (Fig. 9B, third and fourth panels), also consistent with the EMSA data showing that Ku could not load onto these ends once RPA was bound (Fig. 8, third and fourth panels).
Discussion
The role of Ku as the pioneering DNA end-binding factor that marks DNA ends for rejoining via NHEJ has been studied extensively over the past 2 decades and is very well understood (53). A distinct function of Ku in the protection of DSB ends from resection by Exo1 was recently uncovered, an attribute believed to be important for providing a window of opportunity for the execution of NHEJ before any commitment to HR is permitted (15–17, 21, 47, 54, 55). However, mechanistic studies entailing reconstituted systems with purified Ku and resection proteins have been limited. Ku has been shown to engage partially resected DNA ends (i.e. 5′-recessed ends) (56), but whether it can protect them against attrition by Exo1 and whether or not RPA would influence Ku access and hence exert an impact on DNA end protection have not yet been examined.
In this study, we addressed the above questions using highly purified proteins and defined DNA substrates. A number of conclusions can be made. (i) Ku efficiently engages recessed ends with as many as 40 nt of ssDNA, where it localizes to the ssDNA-dsDNA junction of the substrate. (ii) A second Ku molecule can associate with the adjoining ssDNA on the 40-nt recessed end. (iii) Ku does not stably bind ssDNA unless a ssDNA-dsDNA junction is present. (iv) Ku protects both blunt and recessed ends from digestion by Exo1. (v) The DNA end-binding activity of Ku is indispensable for DNA end protection as revealed by examining a Ku variant harboring the yku70-R456E mutation. (vi) RPA has little or no effect on the protective effect of Ku against Exo1 action at a blunt end or a 7-nt recessed end. (vii) RPA can displace Ku when the overhang length reaches 22 nt to restore end susceptibility to Exo1. (viii) Ku and RPA can simultaneously associate with a recessed end with a 40-nt overhang but only if Ku is preloaded onto the DNA substrate.
The main findings of our study are summarized in Fig. 10. Most notably, these findings help uncover a previously unrecognized functional interplay between Ku and RPA at recessed DNA ends that is likely germane for understanding DSB repair pathway choice. At DNA ends where there is insufficient ssDNA to stably engage RPA, the protective effect of Ku predominates, and the ends likely become destined to be rejoined by DNA ligase IV and associated factors (2). Importantly, the observation that RPA can displace Ku prebound to a 22-nt overhang indicates that Ku association with such a structure is dynamic, entailing its periodic departure from the end, whereupon RPA association with the ssDNA prevents its rebinding. Such an outcome is predicted to predispose the DNA end to resection by Exo1. Interestingly, when the ssDNA overhang length increases to 40 nt, RPA and Ku can engage the recessed end simultaneously, but only if Ku is present first at the ssDNA-dsDNA junction (Fig. 8). Moreover, a second Ku molecule can thread into the ssDNA region that abuts the ssDNA-dsDNA junction. In this scenario, we envision that association of Ku at the ssDNA-dsDNA junction would continue to restrict Exo1 access, so bypassing or removing the bound Ku would be necessary to allow long-range DNA end resection to occur. Recently, Sae2 has been shown to activate the endonuclease activity of the MRX complex at protein-bound DNA ends, creating a DNA nick distal to the block on the 5′-terminated strand (28). It will be of interest to investigate whether the MRX-Sae2 ensemble can likewise introduce a DNA nick distal to a Ku-bound end, so as to create an entry site for Exo1 and also for the 3′–5′ exonuclease activity of MRX. The proteasome has long been linked to DSB repair, and the proteolytic removal of Ku trapped on a recessed end would provide a means to promote resection without the need for endonucleolytic cleavage by MRX-Sae2 (28).
FIGURE 10.
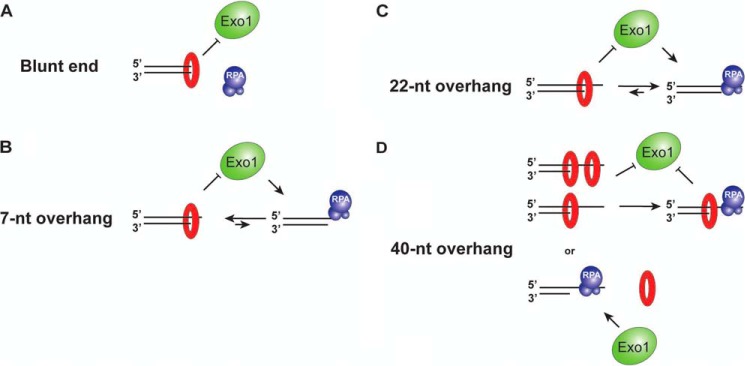
Summary of Ku-RPA interplay at DNA ends with or without a 3′-ssDNA overhang.
Author Contributions
D. S. K., J. M. D., P. S., and H. N. conceived and designed the experiments. D. S. and J. M. D. performed the experiments. D. S. and H. N. purified the proteins used in this study. D. S., J. M. D., P. S., and H. N. analyzed the data. D. S., J. M. D., and P. S. wrote the paper.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 ES007016, R01 ES015632, R01 ES015252, R01 GM057814, and K99/R00 ES021441. The authors declare that they have no conflicts of interest with the contents of this article.
- DSB
- double-strand break
- HR
- homologous recombination
- NHEJ
- non-homologous end joining
- RPA
- replication protein A
- nt
- nucleotide(s).
References
- 1. Khanna K. K., Jackson S. P. (2001) DNA double-strand breaks: signaling, repair and the cancer connection. Nat. Genet. 27, 247–254 [DOI] [PubMed] [Google Scholar]
- 2. Chiruvella K. K., Liang Z., Wilson T. E. (2013) Repair of double-strand breaks by end joining. Cold Spring Harb. Perspect. Biol. 5, a012757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Walker J. R., Corpina R. A., Goldberg J. (2001) Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature 412, 607–614 [DOI] [PubMed] [Google Scholar]
- 4. Cary R. B., Peterson S. R., Wang J., Bear D. G., Bradbury E. M., Chen D. J. (1997) DNA looping by Ku and the DNA-dependent protein kinase. Proc. Natl. Acad. Sci. U.S.A. 94, 4267–4272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. de Vries E., van Driel W., Bergsma W. G., Arnberg A. C., van der Vliet P. C. (1989) HeLa nuclear protein recognizing DNA termini and translocating on DNA forming a regular DNA-multimeric protein complex. J. Mol. Biol. 208, 65–78 [DOI] [PubMed] [Google Scholar]
- 6. Moore J. K., Haber J. E. (1996) Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol. Cell. Biol. 16, 2164–2173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wilson T. E., Lieber M. R. (1999) Efficient processing of DNA ends during yeast nonhomologous end joining. Evidence for a DNA polymerase β (Pol4)-dependent pathway. J. Biol. Chem. 274, 23599–23609 [DOI] [PubMed] [Google Scholar]
- 8. Daley J. M., Wilson T. E. (2005) Rejoining of DNA double-strand breaks as a function of overhang length. Mol. Cell. Biol. 25, 896–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mimitou E. P., Symington L. S. (2008) Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455, 770–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhu Z., Chung W. H., Shim E. Y., Lee S. E., Ira G. (2008) Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134, 981–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bizard A. H., Hickson I. D. (2014) The dissolution of double Holliday junctions. Cold Spring Harb. Perspect. Biol. 6, a016477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wyatt H. D., West S. C. (2014) Holliday junction resolvases. Cold Spring Harb. Perspect. Biol. 6, a023192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Daley J. M., Palmbos P. L., Wu D., Wilson T. E. (2005) Nonhomologous end joining in yeast. Annu. Rev. Genet. 39, 431–451 [DOI] [PubMed] [Google Scholar]
- 14. Ira G., Pellicioli A., Balijja A., Wang X., Fiorani S., Carotenuto W., Liberi G., Bressan D., Wan L., Hollingsworth N. M., Haber J. E., Foiani M. (2004) DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature 431, 1011–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee S. E., Moore J. K., Holmes A., Umezu K., Kolodner R. D., Haber J. E. (1998) Saccharomyces Ku70, Mre11/Rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell 94, 399–409 [DOI] [PubMed] [Google Scholar]
- 16. Maringele L., Lydall D. (2002) EXO1-dependent single-stranded DNA at telomeres activates subsets of DNA damage and spindle checkpoint pathways in budding yeast yku70Δ mutants. Genes Dev. 16, 1919–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Clerici M., Mantiero D., Guerini I., Lucchini G., Longhese M. P. (2008) The Yku70-Yku80 complex contributes to regulate double-strand break processing and checkpoint activation during the cell cycle. EMBO Rep. 9, 810–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang Y., Hefferin M. L., Chen L., Shim E. Y., Tseng H. M., Kwon Y., Sung P., Lee S. E., Tomkinson A. E. (2007) Role of Dnl4-Lif1 in nonhomologous end-joining repair complex assembly and suppression of homologous recombination. Nat. Struct. Mol. Biol. 14, 639–646 [DOI] [PubMed] [Google Scholar]
- 19. Wu D., Topper L. M., Wilson T. E. (2008) Recruitment and dissociation of nonhomologous end joining proteins at a DNA double-strand break in Saccharomyces cerevisiae. Genetics 178, 1237–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bressan D. A., Baxter B. K., Petrini J. H. (1999) The Mre11-Rad50-Xrs2 protein complex facilitates homologous recombination-based double-strand break repair in Saccharomyces cerevisiae. Mol. Cell. Biol. 19, 7681–7687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wasko B. M., Holland C. L., Resnick M. A., Lewis L. K. (2009) Inhibition of DNA double-strand break repair by the Ku heterodimer in mrx mutants of Saccharomyces cerevisiae. DNA Repair 8, 162–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mimitou E. P., Symington L. S. (2010) Ku prevents Exo1 and Sgs1-dependent resection of DNA ends in the absence of a functional MRX complex or Sae2. EMBO J. 29, 3358–3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhou Y., Paull T. T. (2013) DNA-dependent protein kinase regulates DNA end resection in concert with Mre11-Rad50-Nbs1 (MRN) and ataxia telangiectasia-mutated (ATM). J. Biol. Chem. 288, 37112–37125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang S. H., Zhou R., Campbell J., Chen J., Ha T., Paull T. T. (2013) The SOSS1 single-stranded DNA binding complex promotes DNA end resection in concert with Exo1. EMBO J. 32, 126–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Aylon Y., Liefshitz B., Kupiec M. (2004) The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. EMBO J. 23, 4868–4875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Karathanasis E., Wilson T. E. (2002) Enhancement of Saccharomyces cerevisiae end-joining efficiency by cell growth stage but not by impairment of recombination. Genetics 161, 1015–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Huertas P., Cortés-Ledesma F., Sartori A. A., Aguilera A., Jackson S. P. (2008) CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature 455, 689–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cannavo E., Cejka P. (2014) Sae2 promotes dsDNA endonuclease activity within Mre11-Rad50-Xrs2 to resect DNA breaks. Nature 514, 122–125 [DOI] [PubMed] [Google Scholar]
- 29. Ira G., Hastings P. J. (2012) DNA breakage drives nuclear search. Nat. Cell Biol. 14, 448–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tomimatsu N., Mukherjee B., Catherine Hardebeck M., Ilcheva M., Vanessa Camacho C., Louise Harris J., Porteus M., Llorente B., Khanna K. K., Burma S. (2014) Phosphorylation of EXO1 by CDKs 1 and 2 regulates DNA end resection and repair pathway choice. Nat. Commun. 5, 3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mao Z., Bozzella M., Seluanov A., Gorbunova V. (2008) Comparison of nonhomologous end joining and homologous recombination in human cells. DNA Repair 7, 1765–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shibata A., Conrad S., Birraux J., Geuting V., Barton O., Ismail A., Kakarougkas A., Meek K., Taucher-Scholz G., Löbrich M., Jeggo P. A. (2011) Factors determining DNA double-strand break repair pathway choice in G2 phase. EMBO J. 30, 1079–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Reynolds P., Anderson J. A., Harper J. V., Hill M. A., Botchway S. W., Parker A. W., O'Neill P. (2012) The dynamics of Ku70/80 and DNA-PKcs at DSBs induced by ionizing radiation is dependent on the complexity of damage. Nucleic Acids Res. 40, 10821–10831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Van Komen S., Macris M., Sehorn M. G., Sung P. (2006) Purification and assays of Saccharomyces cerevisiae homologous recombination proteins. Methods Enzymol. 408, 445–463 [DOI] [PubMed] [Google Scholar]
- 35. Krogh B. O., Symington L. S. (2004) Recombination proteins in yeast. Annu. Rev. Genet. 38, 233–271 [DOI] [PubMed] [Google Scholar]
- 36. Sung P. (1997) Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev. 11, 1111–1121 [DOI] [PubMed] [Google Scholar]
- 37. Kantake N., Sugiyama T., Kolodner R. D., Kowalczykowski S. C. (2003) The recombination-deficient mutant RPA (rfa1-t11) is displaced slowly from single-stranded DNA by Rad51 protein. J. Biol. Chem. 278, 23410–23417 [DOI] [PubMed] [Google Scholar]
- 38. Adkins N. L., Niu H., Sung P., Peterson C. L. (2013) Nucleosome dynamics regulates DNA processing. Nat. Struct. Mol. Biol. 20, 836–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dynan W. S., Yoo S. (1998) Interaction of Ku protein and DNA-dependent protein kinase catalytic subunit with nucleic acids. Nucleic Acids Res. 26, 1551–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wu T. J., Chiang Y. H., Lin Y. C., Tsai C. R., Yu T. Y., Sung M. T., Lee Y. H., Lin J. J. (2009) Sequential loading of Saccharomyces cerevisiae Ku and Cdc13p to telomeres. J. Biol. Chem. 284, 12801–12808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lopez C. R., Ribes-Zamora A., Indiviglio S. M., Williams C. L., Haricharan S., Bertuch A. A. (2011) Ku must load directly onto the chromosome end in order to mediate its telomeric functions. PLoS Genet. 7, e1002233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rathmell W. K., Chu G. (1994) A DNA end-binding factor involved in double-strand break repair and V(D)J recombination. Mol. Cell. Biol. 14, 4741–4748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Paillard S., Strauss F. (1991) Analysis of the mechanism of interaction of simian Ku protein with DNA. Nucleic Acids Res. 19, 5619–5624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Falzon M., Fewell J. W., Kuff E. L. (1993) EBP-80, a transcription factor closely resembling the human autoantigen Ku, recognizes single- to double-strand transitions in DNA. J. Biol. Chem. 268, 10546–10552 [PubMed] [Google Scholar]
- 45. Limbo O., Chahwan C., Yamada Y., de Bruin R. A., Wittenberg C., Russell P. (2007) Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol. Cell 28, 134–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Langerak P., Mejia-Ramirez E., Limbo O., Russell P. (2011) Release of Ku and MRN from DNA ends by Mre11 nuclease activity and Ctp1 is required for homologous recombination repair of double-strand breaks. PLoS Genet. 7, e1002271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tomita K., Matsuura A., Caspari T., Carr A. M., Akamatsu Y., Iwasaki H., Mizuno K., Ohta K., Uritani M., Ushimaru T., Yoshinaga K., Ueno M. (2003) Competition between the Rad50 complex and the Ku heterodimer reveals a role for Exo1 in processing double-strand breaks but not telomeres. Mol. Cell. Biol. 23, 5186–5197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Genschel J., Modrich P. (2003) Mechanism of 5′-directed excision in human mismatch repair. Mol. Cell 12, 1077–1086 [DOI] [PubMed] [Google Scholar]
- 49. Lee B. I., Wilson D. M., 3rd. (1999) The RAD2 domain of human exonuclease 1 exhibits 5′ to 3′ exonuclease and flap structure-specific endonuclease activities. J. Biol. Chem. 274, 37763–37769 [DOI] [PubMed] [Google Scholar]
- 50. Orans J., McSweeney E. A., Iyer R. R., Hast M. A., Hellinga H. W., Modrich P., Beese L. S. (2011) Structures of human exonuclease 1 DNA complexes suggest a unified mechanism for nuclease family. Cell 145, 212–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chen H., Lisby M., Symington L. S. (2013) RPA coordinates DNA end resection and prevents formation of DNA hairpins. Mol. Cell 50, 589–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Brosey C. A., Yan C., Tsutakawa S. E., Heller W. T., Rambo R. P., Tainer J. A., Ivanov I., Chazin W. J. (2013) A new structural framework for integrating replication protein A into DNA processing machinery. Nucleic Acids Res. 41, 2313–2327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Grob P., Zhang T. T., Hannah R., Yang H., Hefferin M. L., Tomkinson A. E., Nogales E. (2012) Electron microscopy visualization of DNA-protein complexes formed by Ku and DNA ligase IV. DNA Repair 11, 74–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lebel C., Rosonina E., Sealey D. C., Pryde F., Lydall D., Maringele L., Harrington L. A. (2009) Telomere maintenance and survival in Saccharomyces cerevisiae in the absence of telomerase and RAD52. Genetics 182, 671–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shim E. Y., Chung W. H., Nicolette M. L., Zhang Y., Davis M., Zhu Z., Paull T. T., Ira G., Lee S. E. (2010) Saccharomyces cerevisiae Mre11/Rad50/Xrs2 and Ku proteins regulate association of Exo1 and Dna2 with DNA breaks. EMBO J. 29, 3370–3380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Foster S. S., Balestrini A., Petrini J. H. (2011) Functional interplay of the Mre11 nuclease and Ku in the response to replication-associated DNA damage. Mol. Cell. Biol. 31, 4379–4389 [DOI] [PMC free article] [PubMed] [Google Scholar]