Background: Rab14 regulates endosomal trafficking; however, its binding to Rab11-FIP effectors is a source of conflicting data.
Results: Rab14 interacts with the canonical Rab-binding domain of RCP, and both Rab14 and RCP function in neuritogenesis.
Conclusion: Rab11-RCP complex formation may precede recruitment of RCP by Rab14.
Significance: This study provides a conceptual framework for Rab14 and RCP in health and disease.
Keywords: cell biology, confocal microscopy, endosome, small GTPase, X-ray crystallography, effector recruitment
Abstract
Rab GTPases recruit effector proteins, via their GTP-dependent switch regions, to distinct subcellular compartments. Rab11 and Rab25 are closely related small GTPases that bind to common effectors termed the Rab11 family of interacting proteins (FIPs). The FIPs are organized into two subclasses (class I and class II) based on sequence and domain organization, and both subclasses contain a highly conserved Rab-binding domain at their C termini. Yeast two-hybrid and biochemical studies have revealed that the more distantly related Rab14 also interacts with class I FIPs. Here, we perform detailed structural, thermodynamic, and cellular analyses of the interactions between Rab14 and one of the class I FIPs, the Rab-coupling protein (RCP), to clarify the molecular aspects of the interaction. We find that Rab14 indeed binds to RCP, albeit with reduced affinity relative to conventional Rab11-FIP and Rab25-FIP complexes. However, in vivo, Rab11 recruits RCP onto biological membranes. Furthermore, biophysical analyses reveal a noncanonical 1:2 stoichiometry between Rab14-RCP in dilute solutions, in contrast to Rab11/25 complexes. The structure of Rab14-RCP reveals that Rab14 interacts with the canonical Rab-binding domain and also provides insight into the unusual properties of the complex. Finally, we show that both the Rab coupling protein and Rab14 function in neuritogenesis.
Introduction
Rab GTPases are molecular switches that belong to the Ras superfamily of small GTPases and regulate vesicle trafficking in eukaryotic cells (1). In their active GTP-bound state, membrane-localized Rabs exert their biological effects by recruiting cytosolic effector proteins to distinct subcellular compartments. Membrane attachment of active Rabs is facilitated by prenylation of one or two C-terminally situated cysteine residues by the enzyme geranylgeranyltransferase (2), thus adding a hydrophobic tail to the G-protein. This tail inserts into the lipid bilayer, and the globular domain projects into the cytosol. Upon hydrolysis of GTP, which is stimulated by GTPase-activating proteins (3), intact Rabs, together with their hydrophobic tails, are extracted from membranes by the GTP/GDP dissociation inhibitor into the cytosolic fraction (4). The switch is turned “on” by GTP/GDP exchange factors that catalyze the exchange of GDP for GTP. The functional cycle is completed by re-insertion of Rab tails into membranes.
Mammalian Rabs compose the largest family of Ras-like GTPases, with over 60 members in humans. Rabs regulate intracellular trafficking in their active GTP-bound state by recruiting one or more members of a diverse set of effector proteins. Rab structures consist of a six-stranded mixed β-sheet flanked by five α-helices. The 5′-phosphate arm of the nucleotide is bordered on one side by the P-loop (Walker A motif), which is conserved throughout the Ras superfamily, as well as in many ATPases (5, 6). The conformations of a pair of adjacent regions termed switch I and switch II are sensitive to the presence of GTP versus GDP (7), and thus convey the nucleotide specificity for effector recruitment. Additionally, a loop and strand situated between the switch motifs, termed the interswitch, also plays a role in effector recruitment. In particular, an invariant tryptophan residue that is located within an otherwise variable interswitch sequence provides a hydrophobic surface for effector binding. The overall sequence identities between mammalian Rabs range from 30 to 80% (8), with the nucleotide-proximal motifs (switch I/II, P-loop) bearing the most highly conserved (60–80%) segments.
The complexity of Rab-regulated intracellular trafficking is increased by the promiscuity of Rab/effector interactions in eukaryotic cells. Typically, a single Rab binds to multiple, often unrelated, effectors, and a single effector protein can sometimes be recognized by multiple Rab proteins. For example, Rab6 regulates Golgi traffic and interacts with effectors such as the golgin GCC185 and Rab6-interacting protein 1 (R6IP1), which are unrelated proteins (9, 10). The structures of complexes of Rab6-GCC185 and Rab6-R6IP1 have revealed that GCC185 is a dimeric coiled coil that binds two Rabs, whereas R6IP1 is a monomeric bundle of seven α-helices with a single interface for Rab6. However, the Rab6 epitope is formed along two parallel α-helices in both complexes, which have similar hydrophobic and hydrophilic features at their respective interfaces. Conversely, a single effector protein can sometimes be recognized by multiple Rabs. An example of effector promiscuity is Rabenosyn-5, which is recruited by Rab4 and Rab22 via distinct segments of coiled-coil motifs (11). In all known complexes, binding is at least partly facilitated by switch I and switch II. Comparative analyses of the known structures of Rab-effector complexes have revealed that specificity is a complex phenotype that is influenced by subtle differences in sequence and conformational diversity in switch I, switch II, and the interswitch region.
In vitro thermodynamic and kinetic studies of Rab-effector complexes reveal a variety of affinities in vitro ranging from relatively high (Rab27-Slp2a, dissociation constant Kd = 13 nm (12)) to low/weak (Rab8-OCRL, Kd = 5 μm (13)). Despite numerous structural and thermodynamic studies, it is generally difficult to discern why some complexes have higher affinity than others, or whether the differences in affinity are meaningful. A closely related issue is the question of the molecular determinants of Rab-effector specificity. Relating the structural, thermodynamic, and biological properties of Rab-effector complexes is essential for a comprehensive understanding of how Rabs cooperate to regulate vesicle trafficking.
The Rab11 subfamily comprises Rab11A, Rab11B, and Rab25 (also known as Rab11C). Rab11A and Rab11B are ubiquitously expressed, whereas Rab25 expression is restricted to epithelial tissues (14). Rab25 activity has been linked to a variety of cancers (15, 16).
Rab14 is more distantly related to the Rab11 subfamily, although it also appears to regulate overlapping endocytic pathways via interactions with class I FIPs4 (17–19). Rab14 also binds to RUFY1/Rabip4, which is a dual Rab4/Rab14 effector that regulates endosomal trafficking (20). The discrimination by Rab14 of a subset of FIPs is a unique attribute, as Rab11 and Rab25 recognize both class I and class II FIPs (21).
A high throughput yeast two-hybrid screen first reported an interaction between Rab14 and FIP2 (19). We subsequently analyzed the ability of all five FIPs to interact with Rab14 and reported that the class I FIPs (RCP, FIP2, and Rip11), but not the class II FIPs (FIP3 and FIP4), interact with Rab14. We identified the region of RCP, FIP2, and Rip11 that binds to Rab14 as their C terminus, and in the case of RCP we mapped it between residues 579 and 645 (17). This region contains the classical Rab11-binding domain (RBD), an ∼20 amino acid highly conserved motif located at the C terminus of all the Rab11-FIPs. A single amino acid mutation (I621E) in the RBD abolished the ability of RCP to interact with both Rab14 and Rab11.
In contrast to our findings, Jing et al. (22) subsequently reported that Rab14 does not bind to any of the FIPs. However, more recently, a study by Goldenring, Spearman, and co-workers (23) reported that Rab14 does indeed bind to RCP, but via a region upstream of the RBD. Furthermore, in this paper Qi et al. (23) did not observe any interaction between Rab14 and the other class I FIPs. They identified two serines (Ser-580 and Ser-582), which are not present in FIP2 and Rip11, that when mutated to asparagine and leucine, respectively, abolished the interaction with Rab14 without affecting the interaction with Rab11.
To resolve these conflicting reports, we have followed two approaches. First, we have recently reported a detailed comparison of the antibodies used by Qi et al. (23) and ourselves (24). Second, we report here the detailed structural, biophysical, and cellular analyses of Rab14 binding to the class I FIPs. We observe structurally that Rab14 does indeed bind to the conserved RBD of the Rab coupling protein (RCP), albeit as a noncanonical Rab/FIP (1:2) assembly in dilute concentrations. Our crystallographic studies provide insight into the molecular basis for Rab14-RCP binding and clarify several discrepancies involving Rab11/25 and Rab14 interactions with the FIPs. Our cell biology studies suggest that Rab11 is the major Rab that recruits RCP onto biological membranes, whereas Rab14 and RCP function in the process of neurite formation in a neuronal cell line.
Experimental Procedures
Mutagenesis and Subcloning
Rab14 was expressed by subcloning of the globular region (2–175) into pTrcHisA. PCR primers were 5′-AACTCGAGGCAACTGCACCATACAACTAC (forward) and 5′-AAGAATTCGTAGTTCTGATAGATTTTCTTGGCAG (reverse). The template pEGFP-C1/Rab14 harbored the mutation Q70L; the cDNA fragment was cloned into the XhoI/EcoRI sites of pTrcHisA. A further truncation of Rab14(7–175) was generated by inserting the corresponding cDNA into pNIC28-Bsa4 (Kan+, N-terminal His tag) using a ligation-independent cloning protocol. The template for this latter construct was pTrcHisA-Rab14, and this shorter variant was used for crystallization studies. The primers were 5′-TACTTCCAATCCATGaacta-ctcttacatc (forward) and 5′-TATCCACCTTTACTGTTAgttcagatagattttc, which generated the region corresponding to Asn-7–Asn-175 of Rab14. Following rTEV cleavage, the cloning vector introduced non-native Gly-Met residues at the N terminus. Similarly, RCPs (residues 581–649) were inserted into vector pNIC28-Bsa4 using primers 5′-TACTTCCAATCCATGccctcggaccctgca (forward) and 5′-TATCCACCTTTACTGTTAcatctttcctgcttttttg (reverse).
pTrcHisC/RCP(385–649) was generated by PCR using the following primers: 5′-CCCGGATCCAAAGGCAGCTCTCCGAATCTTCC-3′ (forward) and 5′-CCCGAATTCTTACATCTTTCCTGCTTTTTTGCC-3′ (reverse) with pEGFP-C3/RCPWT as template. The PCR product was purified and cloned into the BamHI/EcoRI sites of pTrcHisC (Invitrogen).
The NF (S580N/S582F), NL (S580N/S582L), and ΔRBD (L591/Stop) mutants were generated by QuikChange (Agilent Technologies) site-directed mutagenesis. The following sense/antisense primers were used: NF, 5′-ATGAGGTCATGATGAAGAAATACAACCCCTTCGACCCTGCATTTGCAT-3′/5′-ATGCAAATGCAGGGTCGAAGGGGTTGTATTTCTTCATCATGACCTCAT-3′; NL, 5′-TGAGGTCATGATGAAGAAATACAACCCCTTGGACCCTGCATT-3′/5′-AATGCAGGGTCCAAGGGGTTGTATTTCTTCATCATGACCTCA-3′; and ΔRBD 5′-CCCTGCATTTGCATATGCGCAGTAGTAGCACGATGAGCTGATTCAGCTGG-3′/5′-CCAGCTGAATCAGCTCATCGTGCTACTACTGCGCATATGCAAATGCAGGG-3′. pTrcHisC/RCP(385–649) and pEGFP-C3 RCPWT were used as templates.
A series of constructs comprising the region 559–649 of RCP were generated by PCR amplification using the following oligonucleotides: 5′-TACTTCCAATCCATGAGCTTGGGAACTGCC (forward primer) and 5′-TATCCACCTTTACTGTTACATCTTTCCTGCTTTTTTGC (reverse primer). The underlined codons indicate the start and end of translated products. The template for these reactions were RCP constructs (residues 385–649) in pTrcHis vector that already contained the corresponding background (WT, S580N/S582F, and S580N/S582L). These WT and mutant proteins were highly soluble and also encompassed the region around Ser-580/Ser-582, which has recently been implicated in binding to Rab14. The PCR-amplified cDNA corresponding to region 559–649 of RCP was subcloned into pNIC28-Bsa4. The RCP products contained an N-terminal hexahistidine tag and an rTEV cleavage site. Table 1 is a list of the various constructs and their applications, as well as corresponding acronyms used in this study. The construct MBP-FIP2 is a fusion of maltose-binding protein (MBP) with the indicated C terminus of FIP2(410–512), and it has been described previously (25).
TABLE 1.
Constructs used for biophysical and structural analyses
The following abbreviations are used: SEC, size exclusion chromatography; MALS, multiangle light scattering; DLS, dynamic light scattering; ITC, isothermal titration calorimetry.
| Protein | Vector | Experiments |
|---|---|---|
| Rab11(1–173) | pET28b | SEC |
| Rab14(2–175) | pTrcHisA | ITC, SEC |
| Rab14(7–175)a | pNIC28-Bsa4 | DLS, MALS, crystals |
| Rab25(7–180)b | pET28b | DLS, MALS |
| RCP(581–649)a | pNIC28-Bsa4 | DLS, MALS, ITC, crystals |
| RCP(559–649) | pNIC28-Bsa4 | Fluorescence, SEC |
| RCP(590–649) | pNIC28-Bsa4 | Fluorescence |
| RCP(559–649, S580N+S582L) | pNIC28-Bsa4 | Fluorescence, SEC |
| RCP(559–649, S580N+S582F) | pNIC28-Bsa4 | Fluorescence, SEC |
| RCP(559–649, I621E) | pNIC28-Bsa4 | Fluorescence |
| RCP(559–649, S580E+S582E) | pNIC28-Bsa4 | Fluorescence |
| MBP-FIP2(410–512)b | pMal parallel | ITC, MALS |
| FIP2(410–512)b | pMal parallel | SEC, DLS |
| FIP3(612–756) | pTrcHisB | ITC |
FIP3(612–756) was generated by PCR using primers 5′-AAAGGATCCACAGAGGGACAAGGAGGCA-3′ (forward) and 5′-AAAAGAATTCCTACTTGACCTCCAGGATGG-3′ (reverse) with pEGFP-C1/FIP3WT as template. The PCR product was purified and cloned into the BamHI/EcoRI sites of pTrcHisB (Invitrogen).
Protein Expression
The following protocol was utilized for expression of all His-tagged proteins. Expression was carried out in 2× YT broth supplemented with 34 μg/ml kanamycin (FORMEDIUMTM) at 37 °C. At an A600 of 0.7, the culture was induced with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside (FORMEDIUMTM), after which cells were grown for a further 3 h at 37 °C. Cells were harvested by centrifugation, and the pellets were resuspended in His tag extraction buffer (10 mm Tris-HCl, 300 mm NaCl, 5 mm MgCl2, 20 mm imidazole, and 10 mm β-mercaptoethanol, pH 8.0) along with 0.5 mm PMSF protease inhibitor (Sigma). Cells were lysed by sonication, and the cell lysate was centrifuged at 15,000 × g for 25 min at 4 °C to remove cellular debris. The supernatants were filtered and loaded onto a nickel-agarose resin (Qiagen). The resin was washed with a 10-fold excess of extraction buffer prior to elution of the bound proteins using extraction buffer supplemented with 200 mm imidazole. Overnight incubation at 4 °C with rTEV protease was used to remove the N-terminal hexahistidine tag from each protein, and the proteins were run through a second Ni2+-agarose column. The “flow-through” fractions were collected, and the uncut proteins and rTEV (which has an uncleavable His tag) remained bound to the resin. Soluble aggregates were removed by running the eluted protein through a Superdex 200 (16/60) gel filtration column (GE Healthcare) equilibrated in column buffer (10 mm Tris-HCl, 100 mm NaCl, 5 mm MgCl2, 1 mm DTT, pH 7.5). Expression and purification of MBP-FIP2 and Rab25 have been described previously (25, 26).
For crystallization experiments, Rab14(7–175)-wtRCP(581–649) complexes were prepared by first combining the individually purified proteins. The combined samples were dialyzed against low salt buffer (10 mm Tris-Cl, 10 mm NaCl, 1 mm DTT, pH 7.5), followed by ion-exchange affinity chromatography on a Mono QTM 5/50 GL column (GE Healthcare). A salt gradient of 10–500 mm NaCl was used over 20 ml, and fractions containing the complex were further purified on a Superdex 200 (16/60) column. The final peaks were collected and concentrated between 7 and 15 mg/ml for use in crystal trials, using 10-kDa cutoff concentrators (Millipore). Prior to crystallization, Gpp(NH)p (Jena Bioscience) was added to a final concentration of 1 mm, because it appeared to improve the reproducibility of crystals. Measurements of polydispersity were performed using a DynaPro NanoStar dynamic light scattering instrument (Wyatt Corp.).
Crystallization and Structural Determination
Initial screening of Rab14(7–175)-wtRCP(581–649) with commercial sparse-matrix screens was performed using a MosquitoR robot (TTP Labtech). Subsequent optimization of crystal growth was done manually by the hanging drop method using Linbro plates. The complex was crystallized at a concentration of 8.2 mg/ml in a 1:1 ratio with reservoir containing 14% PEG 8000 and 0.1 m HEPES, pH 6.75.
Diffraction images were integrated with HKL2000 (27) and scaled using SCALEPACK. The structure was refined by first performing molecular replacement using the structure of Rab14(GDP) as a search model (PDB code 4drz). Electron density corresponding to RCP was clearly visible and was built manually using COOT (28). Alternating cycles of model building and refinement using Refmac5 (29) enabled completion of nearly the entire model of Rab14 (residues 7–175) and RCP (595–640), with R/Rf values of 0.24/0.31. At this stage, the model and data were submitted to the PDB_REDO server to optimize the refinement strategy. Further cycles of model building and refinement were performed using Refmac5 and COOT. Although the backbone density is continuous in the complex, several loops and the N/C termini are poorly ordered. The side chains of the following residues were modeled as alanine: Tyr-8–Tyr-10, Glu-33, Lys-35, Met-37, Lys-140, Leu-166, Glu-167, Lys-171, and Gln-174 in Rab14 and Leu-601, Lys-604, Gln-605, Arg-615, and Thr-640 in RCP. In addition, there is poor stereochemistry at Met-37–Ala-38, adjacent to the disulfide bridge (Cys-20–Cys-40). Phe-36 packs against an intramolecular disulfide bond, which likely affects the backbone conformations in this region. Data collection and refinement statistics are shown in Table 2. The atomic coordinates and structure factors have been deposited in the Protein Data Bank with the accession code 4d0g.
TABLE 2.
Data collection and refinement statistics
Values in parentheses correspond to the statistics for the highest resolution.
| Data collection | Rab14/RCP |
| Beamline | 24-ID-C, APS |
| Detector | ADSC |
| Space group | C2221 |
| Unit cell lengths (Å) | 95.87, 148.23, 38.81 |
| Unit cell angles (°) | 90, 90, 90 |
| Asymmetric unit | 1 × Rab14, 1 × RCP |
| Wavelength (Å) | 0.9786 |
| Low resolution limit | 80 (2.54) |
| Resolution (Å) | 2.50 (2.50) |
| Total no. of reflections | 32,904 |
| Unique reflections | 9973 |
| Multiplicity | 8.9 (5.6) |
| Completeness (%) | 96.98 (70.1) |
| Wilson B-factor (Å2) | 54.3 |
| Rmerge (%) | 9.5 (80) |
| I/σ all data | 8.5 (1.72) |
| Refinement statistics | |
| Model (chain/residues) | |
| A | 8–174; Rab14 |
| B | 595–640; RCP |
| Ramachandran map (%) | |
| Favorable + allowed | 96.6 |
| Outliers | 3.4 |
| Rwork/Rfree (%) | 22.4/29.4 |
| 2.54–2.50 Å | 30.6/36.4 |
| Non-hydrogen atoms | |
| Protein | 1,664 |
| Gpp(NH)p | 33 |
| Mg2+ | 1 |
| Waters | 16 |
| Mean isotropic B-factor (Å2) | 61.67 |
| Root mean square deviations | |
| Bond lengths (Å) | 0.008 |
| Bond angles (°) | 1.30 |
Isothermal Titration Calorimetry
Experiments were performed in duplicate on an ITC-200 calorimeter (GE Healthcare). To generate active Rabs, proteins were incubated in 10 mm EDTA for 10 min at room temperature, followed by incubation with 1 mm Gpp(NH)p, and supplemented by 15 mm MgCl2. Excess nucleotides were removed by running samples through a PD10 column (GE Healthcare). Protein concentrations were calculated based on their A280 using an ND-1000 NanoDrop spectrophotometer (Thermo Scientific). Proteins were dialyzed together in buffer (10 mm Tris-Cl, 100 mm NaCl, 5 mm MgCl2, and 1 mm DTT, pH 7.5) to minimize heats from buffer mismatch. Samples were centrifuged at 13,200 rpm for 10 min prior to concentration determination and isothermal titration calorimetry analysis. The concentrations of proteins for the injections were as follows: (i) 600 μm MBP-FIP2(410–512) fusion protein was placed in the syringe and titrated into 45 μm Rab25; (ii) 600 μm wtRCP(581–649) was placed in the syringe and 30 μm Rab14(2–175) in the cell; (iii) 600 μm Rab14(2–175) was placed in the syringe and 60 μm MBP-FIP2 in the cell; and (iv) 600 μm wtRCP(581–649) was placed in the syringe and titrated into 45 μm Rab25. All titrations were performed at 293 K.
Static and Dynamic Light Scattering
Absolute molar mass was determined by static light scattering using a miniDAWN instrument (Wyatt Technology Corp.) coupled to a Superdex 200 (10/300 GL) column. The elution buffer was 10 mm Tris-Cl, 100 mm NaCl, 5 mm MgCl2, and 1 mm DTT, pH 7.5. The concentrations of proteins injected into the system were typically 0.5 mg/ml. Data were processed using the software program Astra version 5.0 (Wyatt Corp). Rapid data acquisition (1/s) enabled several thousand independent measurements, which were averaged and used to estimate the mean molar mass and associated error (Astra). Dynamic light scattering was performed using the NanoStar instrument (Wyatt Corp.), typically using a 1 mg/ml solution of protein complex in batch mode. At least 10 independent experiments were performed on each sample to obtain a mean estimated mass from the hydrodynamic radius, along with an estimate of error.
Stopped-flow Fluorescence
Data were collected using a PiStar instrument (Applied Photophysics) and processed using associated software. Laser excitation was set to 280 nm for intrinsic tryptophan, and 1000 data points were collected over 500 ms following mixing of solutions. A filter was used for detection of fluorescence emissions at λ >320 nm. Prior to injections, proteins were dialyzed together in 10 mm Tris-Cl, 100 mm NaCl, 5 mm MgCl2, and 1 mm DTT, pH 7.5. The concentration of Rab14 was either 1 or 2 μm, and the concentrations of RCP ranged from 10 to 40 μm. All injections were performed at 273 K. These conditions enabled fitting of data to a pseudo-first order reaction. At these concentrations, there was a linear relationship between the observed rate constant (kobs) as a function of RCP concentration. The association rate (kon) was derived from the slope of a least squares fit (minimum of four concentrations), and the y-intercept provided the dissociation rate (koff). The dissociation constant (Kd) was calculated from these parameters. This experimental strategy was previously established for the study of Rab6 with its effectors (30).
Antibodies
The antibodies used were rabbit anti-Rab14 (R0656), mouse anti-α-tubulin (Thr-5168), and chicken anti-RCP (GW21574A) from Sigma. The Rab11 antibody is an “in-house” antibody raised in a rabbit that had been injected with His6-fused Rab11. A mouse monoclonal anti-Rab11 antibody from BD Biosciences (610656) was used for Western blots.
Cell Culture and Transfection
HeLa, N2a, and A431 cells were cultured in DMEM supplemented with 10% fetal bovine serum, 100 units/ml penicillin/streptomycin, and 2 mm glutamine. These cells were transfected with plasmid using TurboFect (Thermo Scientific) according to the manufacturer's instructions. For silencing experiments, Lipofectamine RNAiMax (Invitrogen) was used.
Immunofluorescence Microscopy
Cells were seeded on 10-mm glass coverslips, fixed with 4% paraformaldehyde, and blocked/permeabilized with 0.05% saponin, 0.2% bovine serum albumin. Cells were incubated with the indicated primary antibodies in the blocking solution. The secondary antibodies used were Cy3-conjugated donkey anti-rabbit from Jackson ImmunoResearch (West Grove, PA) and Alexa488-conjugated goat anti-chicken from Molecular Probes. The cells were washed extensively with phosphate-buffered saline (PBS) between antibody incubations, and the coverslips were mounted in Mowiol. Images were acquired on a Zeiss LSM510 confocal microscope (Carl Zeiss, Jena, Germany). Pearson's colocalization coefficients were calculated using the colocalization module of the Zeiss Zen 2009 software.
Neurite Outgrowth
N2a cells were transfected with the indicated constructs for 24 h using TurboFect and were then induced to differentiate by incubation in serum-free medium for a further 24 h. The cells were fixed and labeled with DAPI and anti-α-tubulin. Confocal images were recorded at ×40 magnification, and cells bearing neurites with lengths greater than 2-fold the cell body diameter were scored as neurite-bearing cells (31). Lengths were measured using ImageJ version 1.49p.
Results
Gel Filtration Analyses of Rab14 Complexes
Rab14 and FIP proteins were subjected to gel filtration analyses (Fig. 1). Purified proteins were eluted on small (10/300) and large (16/60) Superdex 200 columns to examine the formation of complexes. Rab14(7–175) (∼20 kDa) was shown to interact with the C-terminal region of FIP2 (MBP-FIP2, Fig. 1A), as well as the C terminus of RCP (wtRCP(581–649), Fig. 1B). These data show that Rab14 interacts with sufficient affinity to enable co-migration of class I FIPs by gel filtration chromatography.
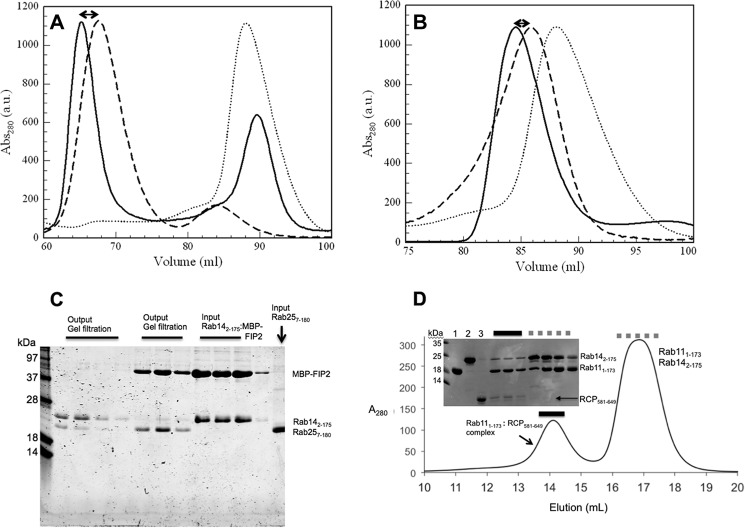
FIGURE 1.
Gel filtration chromatography analyses of Rab complexes with FIPs. A, superimposed elution profiles of MBP-FIP2(410–512) (dashed line), Rab14(2–175) (dotted line), and the complex (solid line) are shown from a Superdex 200 16/60 column. B, elution profiles of wtRCP(581–649) (dashed line), Rab14(2–175) (dotted line), and the complex (solid line) are shown. C, preformed Rab14(2–175): wtRCP(581–649) complexes (Input) were incubated with Rab25(7–180) (Input) and re-loaded onto a gel filtration column (Superdex 200 16/60, GE Healthcare). The resulting two peaks (Output lanes) reveal displacement of Rab14 and the stable Rab25(7–180), wtRCP(581–649) complex. D, excess amounts of Rab11(1–173) and Rab14(2–175( (160 μm each) were incubated with a limiting amount of wtRCP(581–649) (60 μm) in a total volume of 500 μl. The sample was subjected to gel filtration chromatography (Superdex 200 10/300GL) and analyzed by SDS-PAGE (18% gel, visualized by Coomassie Blue). Solid lines are three fractions covering the complex, and dotted lines mark the fractions covering the excess (uncomplexed) Rabs. Loading controls are shown as follows: lane 1 is purified Rab11(1–173); lane 2 is Rab14(2–175); and lane 3 is wtRCP(581–649) (7 μl of a 60 μm solution of protein were loaded in each lane).
A competition assay was performed in which Rab25(Gpp(NH)p) was incubated with Rab14(7–175)-MBP-FIP2, and the complete solution was analyzed by gel filtration chromatography (Fig. 1). Given equivalent amounts of Rab25(7–180) and Rab14(2–175), we observed that Rab25 can out-compete Rab14 for binding to FIP2. The gel filtration profile reveals that Rab25(7–180) is bound to the effector and that Rab14(2–175) is migrating alone. This competition assay reveals that Rab25 can out-compete Rab14 for binding to FIP2 in vitro.
A similar competition assay was performed for the binding of RCP with either Rab11 or Rab14. Excess amounts of Rab11(1–173) and Rab14(2–175) (160 μm each) were incubated with RCP (60 μm) in a final volume of 0.5 ml and subjected to gel filtration chromatography. Under these conditions, the majority of the complex was observed to be Rab11-RCP, with small amounts of Rab14-RCP in the “complex” peak (Fig. 1D). Overall, these results show that Rab14 binds to class I FIPs, albeit with lower affinity relative to Rab11.
Light Scattering Analyses of Rab-FIP Complexes
Multiangle light scattering analyses of Rab25(7–180)-MBP-FIP2 revealed that the molecular mass is consistent with a heterotetramer (Table 3). In contrast, the mass of Rab14-RCP was consistent with a 1:2 complex, as evidenced by multiangle light scattering. Dynamic light scattering can also provide insight into the stoichiometry of protein complexes via molecular weight estimates. In this case, the mass is inferred from the diffusion coefficient, which can be influenced by the overall shape of complexes. The stoichiometry of Rab14(7–175)-wtRCP(581–649) from dynamic light scattering revealed an intermediate ratio, which is possibly a mixture of 1:2 and 2:2 complexes. In contrast, the size of Rab25(7–180)-FIP2(410–512) was consistent with a 2:2 complex by dynamic light scattering (Table 3), in agreement with the stoichiometry from multiangle light scattering. Thus, the overall results from light scattering, in which assay conditions are 0.5–1 mg/ml protein, suggest a 1:2 stoichiometry for Rab14-RCP under dilute (noncrystallization) conditions.
TABLE 3.
Light scattering analyses of Rab effectors
The following abbreviations are used: MALS, multiangle light scattering; DLS, dynamic light scattering.
| Rab-effector complex | MALS | DLS | Ratio |
|---|---|---|---|
| kDa | kDa | ||
| Rab25(7–180), MBP-FIP2 | 135 ± 4 (140a) | 2:2 | |
| Rab25(7–180), FIP2 | 65 ± 2 (60a ) | 2:2 | |
| Rab14(7–175), wtRCP (581–649) | 49 ± 4 (40b) | Intermediate | |
| Rab14(7–175), MBP-FIP2 | 115 ± 10 (120c) | 1:2 |
a Values in parentheses indicate the expected mass of canonical 2:2 complexes of Rab25-MBP-FIP2 (multiangle light scattering) and Rab25-FIP2 (dynamic light scattering).
b For the dynamic light scattering value for Rab14(7–175), wtRCP(581–649) is best described as “intermediate” between 1:2 and 2:2 complexes. Errors in dynamic light scattering measurements were estimated from 10 data acquisitions. The mean molar mass and associate error for multiangle light scattering were calculated as described under “Experimental Procedures.”
c The multiangle light scattering value in parentheses for Rab14-MBP-FIP2 shows the expected mass of noncanonical 1:2 complexes. Ratio represents the stoichiometry, derived from experimental data that most closely fits the expected value (2:2 versus 1:2).
Isothermal Titration Calorimetry
Rab GTPases were purified, loaded with Gpp(NH)p, and titrated against various FIPs to determine their thermodynamic parameters by isothermal titration calorimetry. As described under “Experimental Procedures,” titrations were performed with the C termini of the effectors, either as isolated (hexahistidine-tagged) proteins or as fusions with MBP. Injections were performed in both directions for some binary partners, and no significant change in the measured thermodynamic parameters was observed (Fig. 2 and Table 4). The results show that Rab14 binds weakly to FIPs, relative to Rab11/25. Furthermore, the stoichiometry of the interactions are distinct, Rab11 and Rab25 complexes are 1:1, and Rab14-FIP complexes are closer to 1:2. These observations are consistent with data from light scattering.
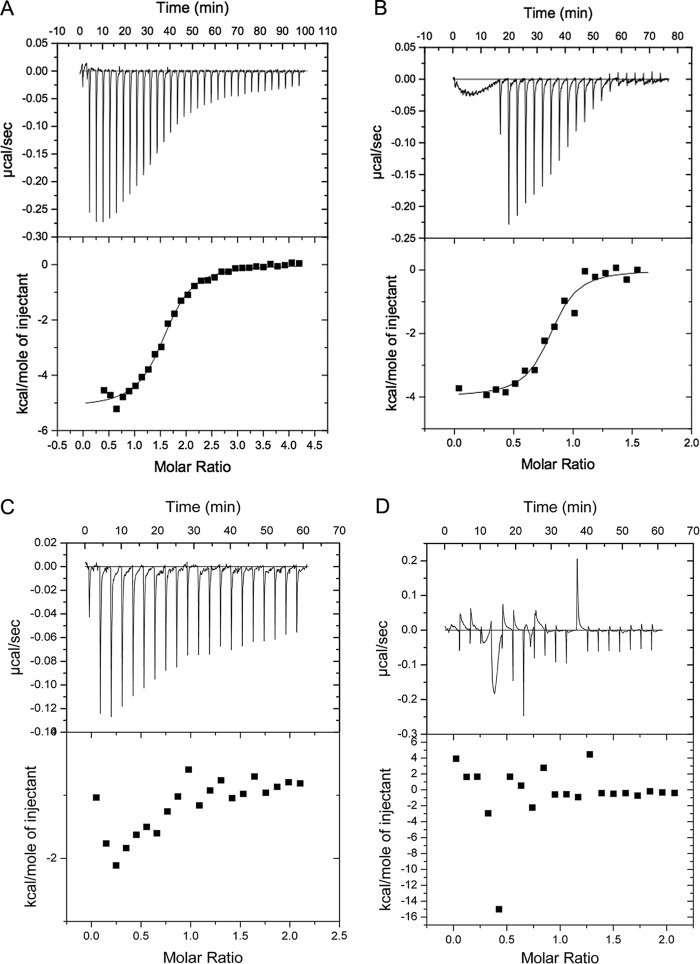
FIGURE 2.
Isothermal titration calorimetry of Rab14/effector interactions. A, raw data (top panel) and integrated heats (bottom panel) of wtRCP(581–649) (600 μm) injected into 30 μm Rab14(2–175). B, injection of 600 μm Rab14(2–175) into 60 μm MBP-FIP2(410–512). C, injection of FIP3(612–756) (600 μm) injected into 30 μm Rab14(2–175). D, control injection of FIP3(612–756) (600 μm) into buffer. Data were processed using Origin software provided by Microcal, LLC. The binding model was assumed to be 1:1, and data were fit to a quadratic binding curve using the isothermal titration calorimetry plugin for Origin (version 7.0). A blank curve (wtRCP(581–649) into buffer) that measured the heat of dilution was used as a control and subtracted from the experimental curve, prior to data processing. This small value was then estimated as a constant in the other curves and subtracted to enable fitting of the data to obtain experimental parameters (Kd, stoichiometry, ΔG, and ΔS).
TABLE 4.
Isothermal titration calorimetry of Rab-effector complexes
The direction of arrows indicates the injection of titrant. For example, wtRCP(581–649) was injected into Rab14(2–175); thus, the stoichiometry is interpreted as RCP-Rab14 = 1.6:1. The numbers are the average of at least two independent experiments.
| Kd | ΔH | ΔS | Stoichiometry | |
|---|---|---|---|---|
| μm | kcal/mol | cal/K | ||
| Rab11(GTP) | ||||
| →MBP-FIP2a | 0.25 ± 0.05 | −10.4 ± 1.5 | −4.8 ± 0.8 | 1.0 ± 0.1 |
| ←wtRCP(581–649) | 1.1 ± 0.2 | −9.1 ± 1 | −3.8 ± 0.8 | 1.0 ± 0.2 |
| Rab14(2–175)(GTP) | ||||
| →MBP-FIP2 | 2.3 ± 0.3 | −5.3 ± 0.8 | 7.9 ± 1.2 | 0.44 ± 0.1 |
| ←wtRCP(581–649) | 1.8 ± 0.3 | −5.5 ± 0.9 | 7.4 ± 1.5 | 1.6 ± 0.3 |
| Rab25(7–180)(GTP) | ||||
| →MBP-FIP2 | 0.65 ± 0.07 | −6.4 ± 0.5 | 6.6 ± 0.7 | 1.1 ± 0.2 |
| ←wtRCP(581–649) | 1.4 ± 0.3 | −6.5 ± 0.2 | 4.6 ± 1 | 1.1 ± 0.2 |
a These data were extracted from a previous study by the authors (26). The Rab25(7–180) construct is also described in this paper.
Structure of Rab14-RCP
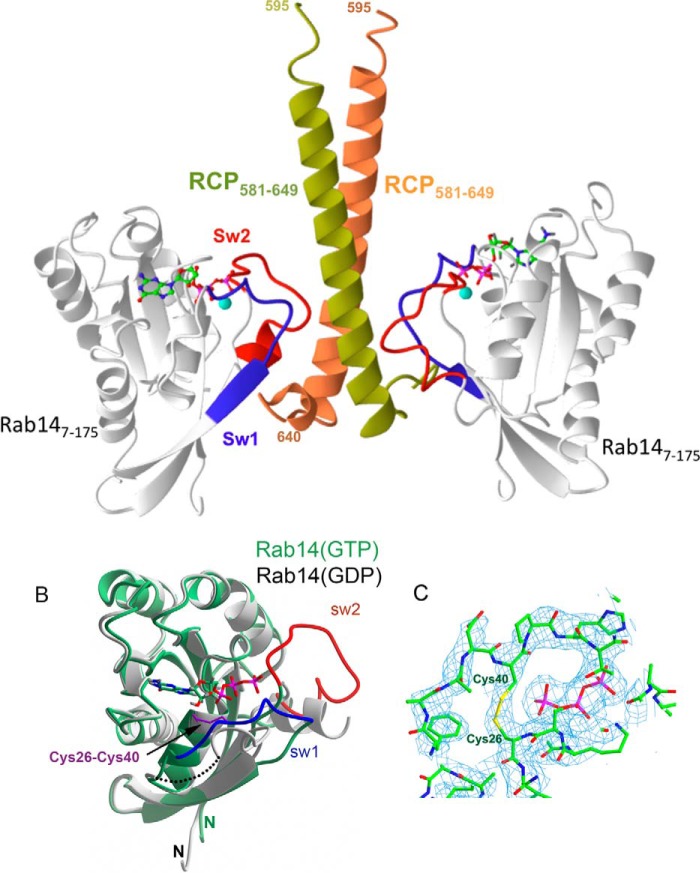
Crystals of the complex grew in C2221 space group with one molecule each of Rab14(7–175) and wtRCP(581–649) in the asymmetric unit. However, the biological assembly in solution consists of a 1:2 complex of Rab14-RCP, as evidenced by calorimetry and static and dynamic light scattering. Our interpretation is that the high concentrations of proteins in the crystallization condition enables the loading of Rab14 onto the second (symmetric) binding site, thus mediating lattice formation and crystal growth. It is not unusual to find non-native stoichiometry in crystals, and the complexes of Rab11-FIP2 and Rab11-FIP3 also deviate from the biological heterotetramer (1:1, 2:2, and 3:3 in the crystalline state (32–34)). However, in all cases of conventional Rab-FIP complexes, a 2-fold symmetric heterodimer (2:2 complex) can be identified from crystallographic or noncrystallographic symmetry.
The Rab14 interface with RCP is topologically identical to Rab11/Rab25 interactions with FIPs (Fig. 3, A and B). Ile-44 of Rab14 resides at a hydrophobic interface, packing against the C-terminal half of the α-helical coiled coil of RCP (residues 597–632). However, there are several features that are distinct. A cysteine residue in switch I, Cys-40, forms a disulfide bridge with Cys-26 in the Rab14-RCP crystal complex (Fig. 3C). This appears to be a GTP-dependent disulfide bond, because the GDP-bound Rab14 (PDB code 1Z0F) is observed in the reduced form with the switch I region distant from the nucleotide (Fig. 3B). The electron density for the disulfide bond in the crystal structure is unambiguous (Fig. 3C). In addition, there is a lack of electrostatic parity in switch I; Rab14 contains a proline (Pro-41), whereas both Rab11 and Rab25 have a positively charged residue (Lys-41 in Rab11 and Arg-42 in Rab25) that forms a salt bridge with FIP2. Thus, the negatively charged Glu-616 of RCP, conserved in class I FIPs, lacks an electrostatic partner in Rab14 (data not shown). The next residue in switch I of Rab14 is His-42, whose side chain is not positioned for a favorable interaction with FIPs. In fact, this residue is a small polar side chain in most Rabs that interacts with the γ-phosphate of GTP (Fig. 4). The switch II region of Rab14 in the RCP complex also reveals significant differences from the Rab11/25 complexes with FIPs. Met-20 in the P-loop of Rab14 protrudes into space that is generally occupied by the catalytic glutamine in active Rabs. Thus, relative to Rab11/25, the segment 70–73 in Rab14 is found in an alternative backbone conformation to avoid steric conflicts with Met-20.
FIGURE 3.
Structure of the Rab14-RCP complex. A, ribbon model of Rab14 (gray) in complex with RCP homodimer (yellow and orange). The switch regions are indicated in red and blue; GTP is a green stick model, and Mg2+ is shown as a sphere. B, superposition of Rab14 (GTP) and Rab14 (GDP; PDB code 4drz). RCP is not shown for clarity, and the disulfide bridge is represented by sticks (purple). C, refined electron density map (2Fo-Fc, 1σ) of Rab14-RCP complex around the intramolecular disulfide bond.
FIGURE 4.
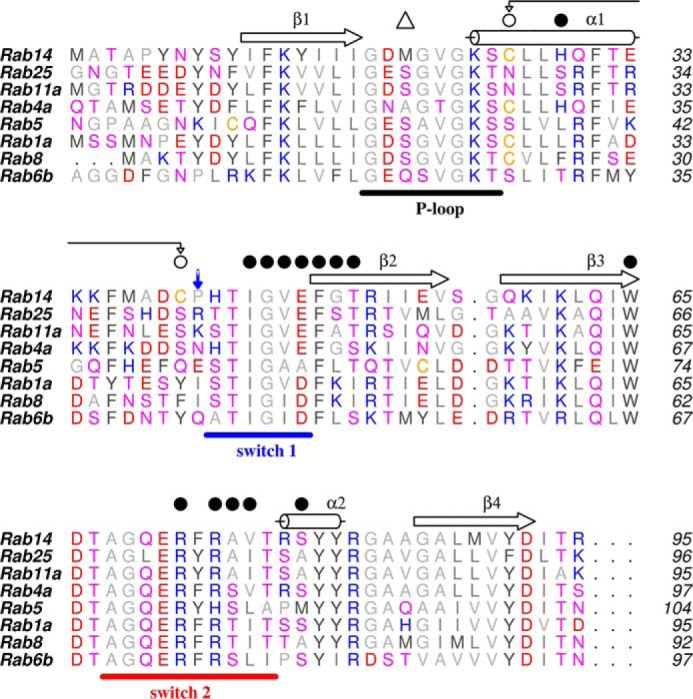
Partial sequence alignment of Rab GTPases. The sequences are truncated at β-strand 4. The secondary structure corresponds to Rab14(GTP). Closed circles are residues that interact with RCP. The disulfide bond is indicated with connecting arrows, and the open triangle indicates the position of Met20 in the P-loop. The blue downward arrow indicates a key residue that differentiates Rab14 (Pro) from Rab11/25. In structures of Rab-effector complexes, the side chains of Lys/Arg in Rab11/25 mediate an electrostatic contact with FIPs.
Binding Assays
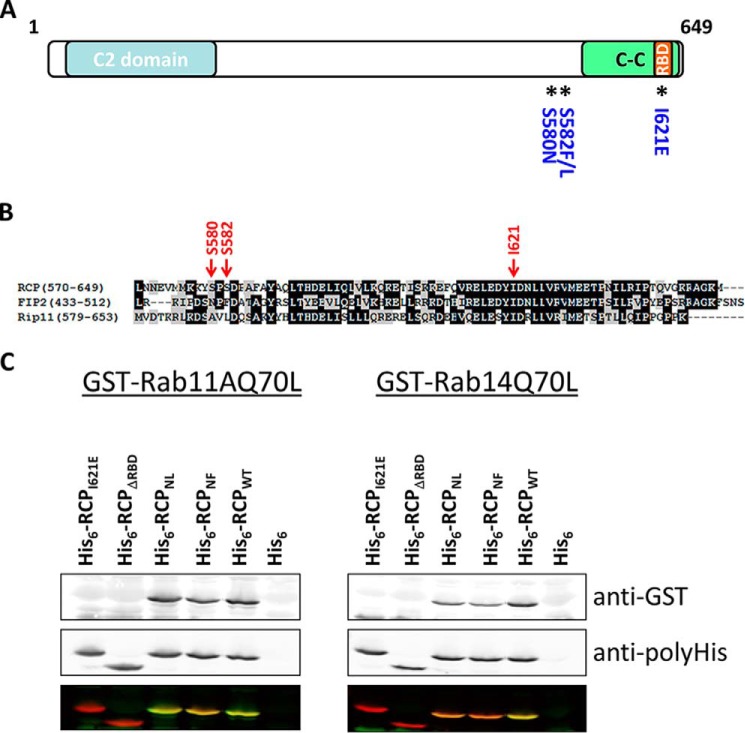
Wild-type and mutant versions of the C terminus of RCP (Fig. 5A) were subjected to an overlay assay with GST-Rab11A and GST-Rab14 to further clarify the Rab-binding region. In Qi et al. (35), Ser-580 and Ser-582 were mutated to asparagine and leucine, respectively, to match the residues of the corresponding positions in FIP2 (Fig. 5B) (35). However, inspection of this region revealed that the amino acid in FIP2, which aligns with Ser-582 of RCP, is in fact a phenylalanine and not a leucine (Fig. 5B). To address conflicting results between our previous studies (17) and that of Qi et al. (35), we analyzed two double serine mutants in RCP. These were the S580N/S582L (RCPNL) mutant used by Qi et al. (35) and a S580N/S582F (RCPNF) mutant that correctly matches FIP2. These mutations were first introduced into a truncated form of RCP (amino acids 385–649) and expressed as His fusions in Escherichia coli. We also generated a mutant with a premature stop codon at position 591, just upstream of the RBD (RCPΔRBD) (Fig. 5A). Lysates of E. coli expressing wild-type RCP and mutants were separated by SDS-PAGE, transferred to nitrocellulose, and overlaid with GST-fused Rab11A or GST-Rab14 loaded with GTPγS. Rab11 and Rab14 that bound to the His-RCP fusions were revealed with an anti-GST antibody. In contrast to the findings of Qi et al. (35), we observed that both GST-Rab11 and GST-Rab14 bound to the RCPNL and RCPNF mutants (Fig. 5C). However, consistent with our previous findings (17), neither Rab bound to the RCPΔRBD nor the RCPI621E mutant. These results confirm that in our in vitro far Western binding assays Rab14 does not require the serines at positions 580 and 582 to interact with RCP.
FIGURE 5.
Rab14 interacts with the RBD of RCP. A, schematic of RCP indicating conserved domains and the location of mutated residues. B, ClustalW alignment of the C-terminal region of the class I FIPs. C, far Western protein-protein interaction assays. Equal amounts of His6-fused RCP(385–649) and mutants were separated by SDS-PAGE, transferred to nitrocellulose, and overlaid with GTPγS-loaded GST-Rab11AQ70L (left) and GST-Rab14(Q70L) (right). Top panel shows the bound Rabs, revealed with an anti-GST antibody. Middle panel demonstrates equal loading of the His6-RCP wild-type and mutant fusion proteins, revealed with an anti-His antibody. Lower panel is an overlay of the anti-GST (green) and anti-His (red) immunoblots.
Gel Filtration Analyses of Rab14-RCP Complexes
To further assess the significance of the recently reported serine mutations in RCP on Rab14 binding, extended C-terminal constructs of RCP (residues 559–649) were generated for E. coli recombinant protein expression and subsequent in vitro biochemical studies. These fragments of wild-type and mutant RCP (wtRCP(559–649), RCPNL, and RCPNF; see Table 1) were highly soluble and encompassed amino acids 580 and 582. The RCP polypeptides were purified, incubated with Rab14(2–175) (Q70L), and analyzed by gel filtration chromatography. All of the RCP variants formed a stable complex with Rab14, and as a representative example, the results of the Rab14(2–175)-RCPNF complex are shown (Fig. 6).
FIGURE 6.
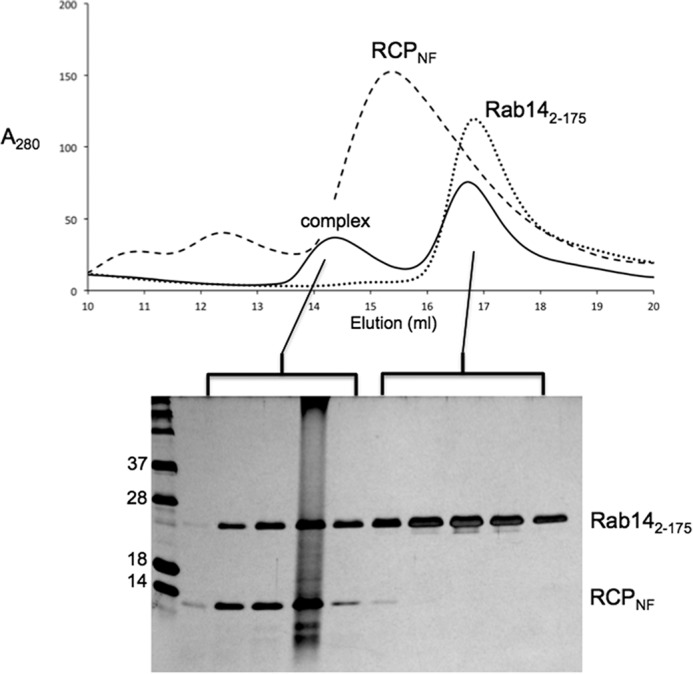
Gel filtration analysis of the complex between Rab14(2–175) and RCPNF(559–649). The proteins were purified separately, combined, and loaded onto a Superdex 200 (10/300) column (GE Healthcare). Solid lines in the elution profile represent the Rab14-RCP complex. SDS-PAGE analysis of the peaks and visualization using Coomassie stain confirm the presence of the complex, as well as an excess of Rab14. Separate gel filtration chromatography runs of RCPNF (dashes) and Rab14 (dotted lines) are overlaid, thus revealing the earlier elution volume of the Rab14-RCP complex.
Fluorescence-based Affinity Studies
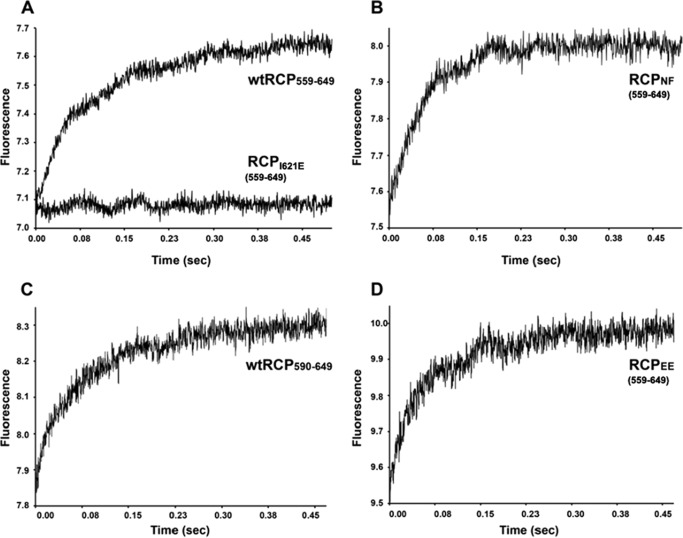
The binding affinities for Rab14 and RCP were measured using established fluorescence techniques for Rab-effector complexes (30). The proteins used in these studies are listed in Table 1. The intrinsic fluorescence changes (Trp excitation at 280 nm, emission filter at 320 nm) indicated binding of Rab14 to the wild-type and mutant RCP variants (Fig. 7A). Analyses of binding kinetics (Fig. 7B and Table 5) indicate that RCPNF binds to Rab14(2–175) with low micromolar Kd values, perhaps with slightly greater affinity than wtRCP(559–649). The phospho-mimetic RCPEE variant (S580E/S582E) and a truncated variant of RCP (wtRCP(590–649)) also bind effectively to Rab14(2–175). In summary, the various mutations at Ser-580 and Ser-582 do not disrupt binding to Rab14. As a negative control, RCPI621E, which contains a mutation in the hydrophobic binding interface (I621E), reveals no interactions with Rab14. This provides direct evidence that the fluorescence changes are specific for the Rab14-RCP interface.
FIGURE 7.
Stopped-flow fluorescence analyses of Rab14(2–175) binding to RCP (residues 559–649). The raw traces are shown as fluorescence versus time, and the RCP concentration for all traces is 20 μm, to enable direct comparisons of the magnitude of fluorescence changes upon mixing. The concentration of Rab14(2–175) is 2 μm for all traces shown. A negative control (RCPI621E, 20 μm) is superimposed with WT as evidence for specific binding. More details are available under “Experimental Procedures.”
TABLE 5.
Kinetic parameters for binding of Rab14 to RCP
Kinetic analyses of binding between Rab14 and RCP from stopped-flow fluorescence. Association (kon) and dissociation (koff) rate constants are shown, along with the derived dissociation constant (Kd). The RCP variants are as follows: RCPNF, S580N/S582F, residues 559–649; RCPEE, S580E/S582E, residues 559–649; RCPI621E, residues 559–649.
| kon × 106 | koff | Kd × 10−6 m | |
|---|---|---|---|
| m−1 s−1 | s−1 | ||
| Rab142–175: wtRCP559–649 | 3.6 (± 0.4) | 5.0 (± 0.9) | 1.6 (± 0.4) |
| Rab142–175: RCPNF | 3.7 (± 0.6) | 2.6 (± 0.4) | 0.7 (± 0.2) |
| Rab142–175: wtRCP590–649 | 4.3 (± 0.1) | 4.6 (± 0.1) | 1.1 (± 0.1) |
| Rab142–175: RCPEE | 3.6 (± 0.4) | 5.4 (± 0.5) | 1.5 (± 0.3) |
| Rab142–175: RCPI621E | Affinity below analyzable threshold |
Colocalization between RCP and Its Rab-binding Partners
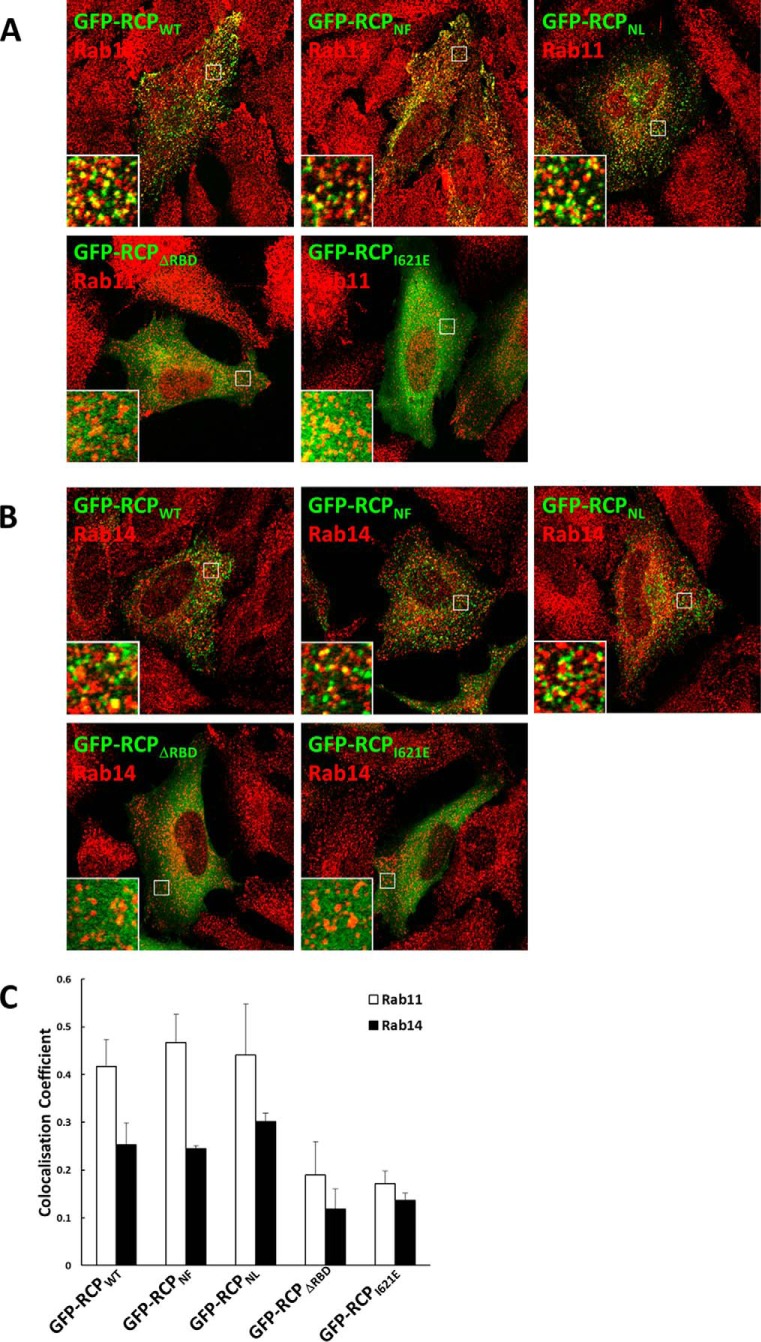
There were no obvious differences in the intracellular localization patterns of exogenously expressed full-length GFP-RCPWT or the full-length mutants GFP-RCPNF or GFP-RCPNL. All displayed a punctate vesicular pattern distributed throughout the cell (Fig. 8). In contrast, GFP-RCPΔRBD and GFP-RCPI621E were both cytosolic.
FIGURE 8.
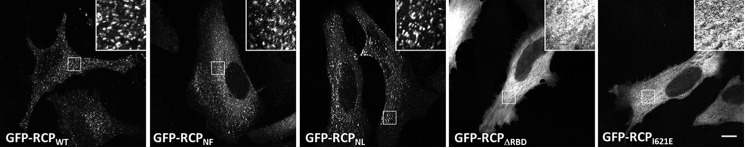
Localization of full-length RCPWT and mutants. HeLa cells expressing the indicated GFP-RCP constructs were fixed and mounted, and images were recorded on a laser scanning confocal microscope. Shown are single 0.4-μm sections. Bar, 10 μm.
Furthermore, considerable colocalization was observed between both double serine mutants and endogenous Rab11 and Rab14 (Fig. 9, A and B). Quantitative colocalization analysis revealed that there was no significant difference in the degree of Rab11 and Rab14 colocalization with either double serine mutants in comparison to GFP-RCPWT (Fig. 9C). In contrast, there was a greater than 50% reduction in the amount of colocalization between Rab11 and Rab14 with GFP-RCPΔRBD and GFP-RCPI621E when compared with GFP-RCPWT (Fig. 9C).
FIGURE 9.
Mutation of serines 580 and 582 does not affect the localization of RCP with endogenous Rab11 or Rab14. HeLa cells expressing the indicated GFP-RCP constructs were fixed and immunolabeled with antibodies that recognize endogenous Rab11 (A) and Rab14 (B). Quantitative analysis of the colocalization coefficients of each GFP-RCP fusion protein with endogenous Rab11 and Rab14 is shown (C).
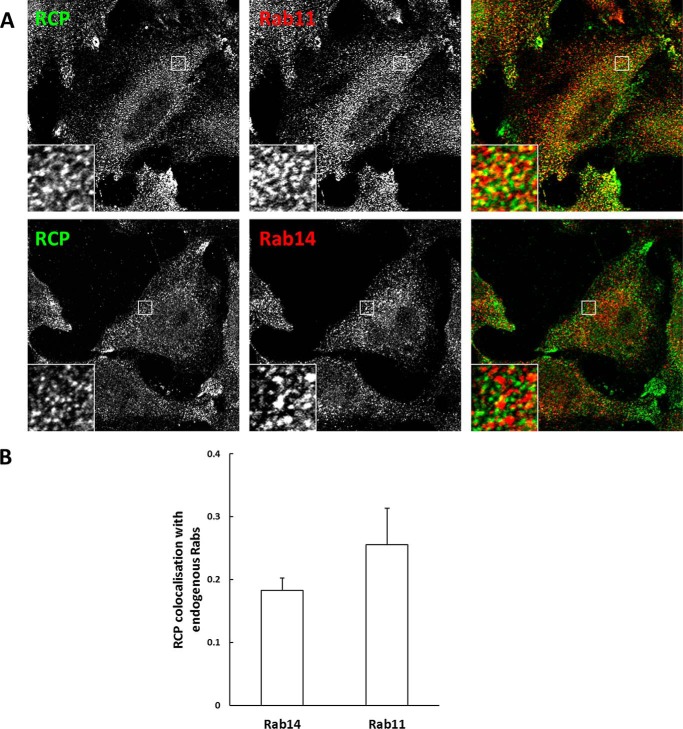
Of note, we consistently observed that GFP-RCPWT displayed greater colocalization with endogenous Rab11 than with endogenous Rab14 (Fig. 9, C and D). Similarly, endogenous RCP also showed greater colocalization with Rab11 than with Rab14 (0.25 ± 0.06 and 0.17 ± 0.01, respectively) (Fig. 10, A and B).
FIGURE 10.
RCP displays greater colocalization with Rab11. A, HeLa cells were fixed and co-labeled with antibodies that recognize endogenous RCP and Rab11 or Rab14. B, quantitative analysis of the colocalization coefficients of endogenous RCP with endogenous Rab11 and Rab14 (mean ± S.E., n = 20–30 cells from three independent experiments).
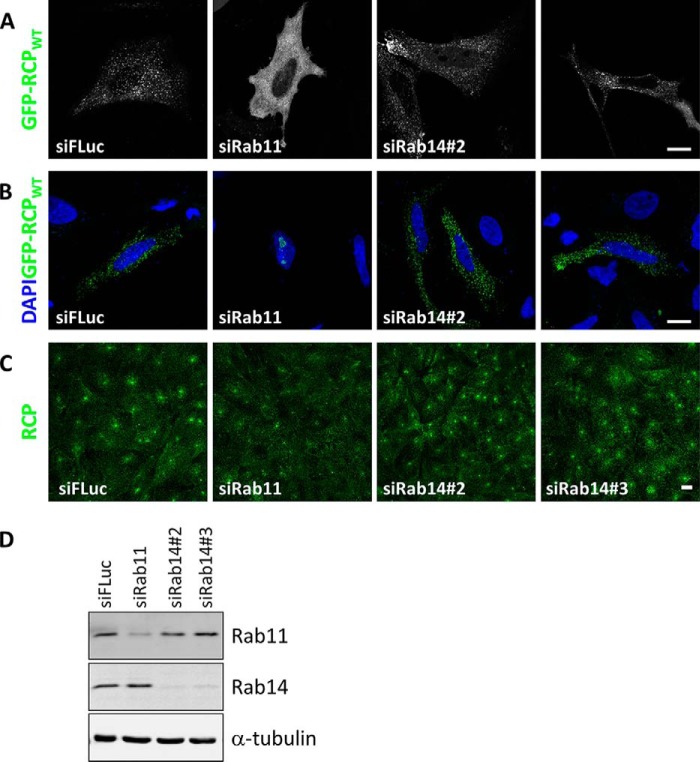
RCP Recruitment to Membranes
To examine whether Rab11 and/or Rab14 are required to recruit RCP to membranes, GFP-RCPWT was expressed in HeLa cells in which these Rabs had been individually depleted. We could efficiently deplete both Rabs by greater than 70% (Fig. 11D). The localization of GFP-RCPWT was examined in fixed cells by confocal microscopy. In cells transfected with a control siRNA (a sequence complementary to the firefly luciferase gene, siFLuc), GFP-RCPWT displayed a typical punctate pattern throughout the cell. However, in cells in which Rab11 had been depleted, GFP-RCPWT was cytosolic (Fig. 11A). In contrast, depletion of Rab14 with two independent siRNA duplexes did not affect its membrane localization. To rule out the possibility that high levels of GFP-RCPWT in the Rab11-depleted cells mask a population of RCP localized to membranes, the experiment was repeated in cells that were permeabilized prior to fixation, to wash out cytosolic proteins. Under these conditions, no GFP-RCPWT was observed localizing to cytoplasmic membranes in the Rab11-depleted cells, whereas GFP-RCPWT clearly labeled vesicles in control and Rab14-depleted cells (Fig. 11B). Surprisingly, a pool of GFP-RCPWT was observed in nucleoli in the Rab11-depleted cells that had been pre-permeabilized. The significance of this has yet to be determined.
FIGURE 11.
Rab11 recruits RCP to membranes. A, HeLa cells were transfected with control siRNA (siFLuc) or siRNA-duplexes targeting Rab11 and Rab14 for 72 h. The cells were transfected with GFP-RCPWT for the final 24 h and fixed. Single 0.4-μm sections were recorded with a confocal microscope. Bar, 10 μm. B, HeLa cells were transfected with control siRNA (siFLuc) or siRNA duplexes targeting Rab11 and Rab14 for 72 h. The cells were transfected with GFP-RCPWT for the final 24 h. The cells were permeabilized, fixed, mounted, and labeled with DAPI, and single 0.4-μm sections were recorded with a confocal microscope. Bar, 10 μm. C, A431 cells were transfected for 72 h with the indicated siRNA duplexes, fixed, and labeled with an antibody that recognizes endogenous RCP. Shown are single 0.4-μm sections. Bar, 10 μm. D, lysates of HeLa cells transfected with the indicated siRNA duplexes for 72 h were separated by SDS-PAGE, transferred to nitrocellulose, and immunoblotted with the indicated antibodies.
Similar results were observed for endogenous RCP in the A431 cell line. A431 cells are a human squamous carcinoma cell line in which RCP-positive membranes primarily localize in a compact pericentriolar region. This membrane localization is considerably reduced in Rab11-depleted cells, whereas transfection with siRNAs targeting Rab14, or a control siRNA, had no effect on endogenous RCP localization (Fig. 11C).
Rab14 and RCP Are Functionally Linked in Neuritogenesis
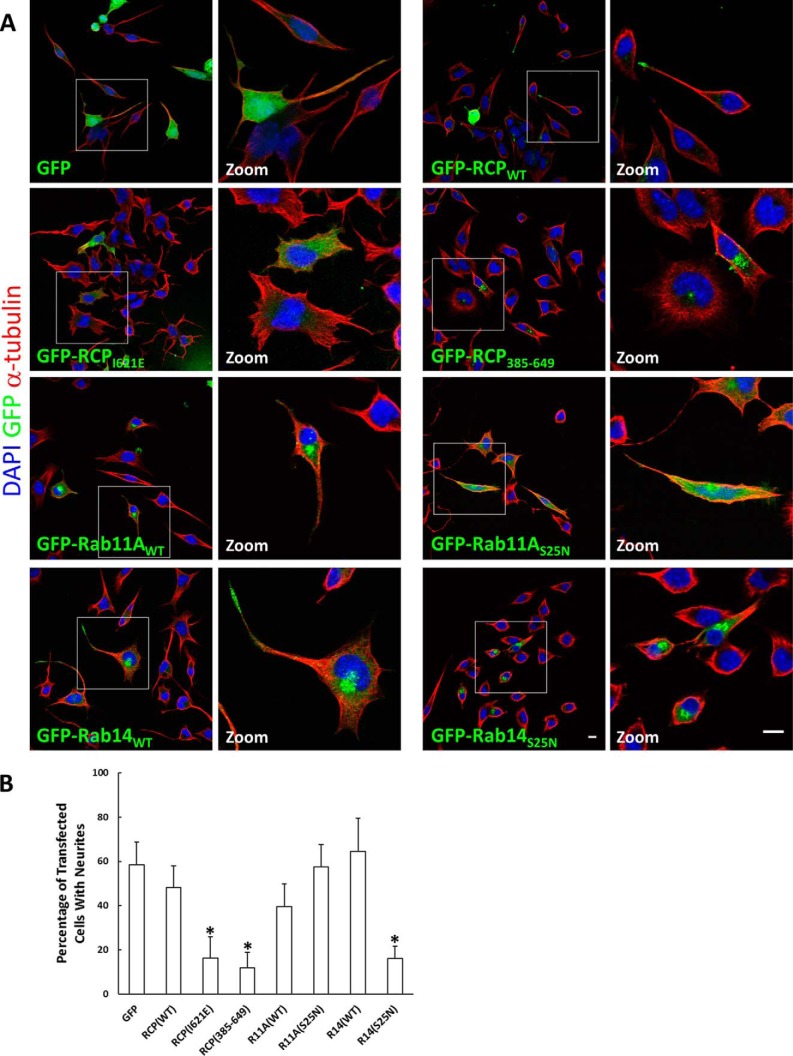
To examine the functional interplay between RCP and its Rab partners, we analyzed the effects of RCP, Rab11, and Rab14 mutants on neurite outgrowth in N2a mouse neuroblastoma cells. Overexpression of dominant-negative mutants of RCP and Rab11, or their knockdown, has been shown to inhibit neurite outgrowth in dorsal root ganglion neurons and PC12 cells (36). Neuritogenesis can be readily induced in N2a cells by withdrawing serum. We transfected these cells with wild-type and dominant-negative mutants of RCP, Rab11, and Rab14 for 24 h and then incubated them for a further 24 h in serum-free medium. Surprisingly, dominant-negative Rab11 (Rab11AS25N) had no effect on the number of N2a cells that formed neurites. In contrast, there was a greater than 70% reduction of neurite formation in transfected cells expressing dominant-negative Rab14 (Rab14S25N) or two RCP mutants (RCPI621E and RCP(385–649)) in comparison with cells expressing GFP alone or the wild-type versions of each protein (Fig. 12, A and B). Interestingly, both RCPWT and Rab14WT displayed a striking concentration at the neurite tips, whereas Rab11WT, Rab11S25N, Rab14S25N, and RCP(385–649) were predominantly localized to the cell body (Fig. 12A). RCPI621E displayed a diffuse cytosolic pattern. These results suggest that in N2a cells RCP functions in conjunction with Rab14, but independently of Rab11, to regulate neurite formation.
FIGURE 12.
RCP and Rab14 play a role in neuritogenesis in mouse N2a neuroblastoma cells. A, N2a cells were transfected with the indicated GFP fusion constructs (green) for 24 h. Cells were then incubated for a further 24 h in serum-free medium to induce cell differentiation, prior to fixation and labeling with DAPI (blue) and anti-α-tubulin (red). Bar, 10 μm. B, histogram indicates the percentage of transfected cells displaying neurites (mean ± S.E., n = 70–90 cells from three independent experiments).
Discussion
Structure of Rab14-RCP
The structure of Rab14-RCP reveals several unusual features when compared with conventional FIP complexes with Rab11 and Rab25. An intramolecular disulfide bond is observed between Cys-26 and Cys-40; Rab14 uniquely contains these two residues among the Rab family of GTPases. Upon superposition of several Rab-effector complexes (Rab8-OCRL and Rab3-Rabenosyn (11, 37)), the Cβ atom of the equivalent loci are within 6 Å distance. Thus, these two segments of Rabs, which comprise helix α1 and the subsequent loop (which precedes switch I), are close in space in the active GTP state. However, the significance of a disulfide bond and whether it forms in vivo requires further structural and functional studies. It is noteworthy that Cys-26 of Rab1 forms an intramolecular disulfide bridge with Cys-126 in the Rab1-DrrA complex (38), suggesting that it is indeed reactive in vitro. Despite the disulfide bridge in Rab14, the switch I conformation closely resembles the structures of Rab11/25 in complex with FIPs. In contrast, switch II is significantly perturbed, mainly due to the presence of Met-20, which sterically interferes with the region facing the γ-phosphate of GTP. These conformational differences along with reduced electrostatic complementarity may account for the observed weaker affinity between Rab14 and class I FIPs.
A recent study has suggested that Rab14 binding to RCP is mediated by a region upstream of the conventional Rab-binding domain. Residues Ser-580/Ser-582 were reported to be critical for RCP binding. However, our structural, biochemical, and cellular assays are consistent with binding of Rab14 to the classical RBD of the FIPs (residues 595–640 of RCP), as we have reported previously (17). To investigate whether amino acids Ser-580 and Ser-582 contribute toward Rab14 binding, we performed kinetic analyses of Rab/RCP interactions by fluorescence techniques. We found that the RCPNF mutant (S580N/S582F) was not significantly impaired in binding to Rab14. Similarly, the phospho-mimetic RCPEE mutant (S580E/S582E) bound to Rab14 at least as strongly as wild-type RCP. We cannot, however, definitively rule out a functional role for regions upstream of the conventional RBD, perhaps in modulating the strength of protein interactions. Our in vitro measurements were limited to region 559–649 of RCP, because longer fragments were unstable and either degraded or mis-folded. Nevertheless, our work demonstrates that the primary mode of the Rab14 interaction with RCP is via the C-terminal RBD.
RCP Membrane Association and Role in Neuritogenesis
Although Rab14 clearly binds to RCP and the two proteins colocalize in HeLa cells in an RBD-dependent manner, the mechanism for the subcellular association of RCP with Rab14 remains unclear. RNA interference experiments suggest that, at least in some cell types, Rab11 is the principal Rab that recruits RCP to vesicular membranes. Depletion of Rab11 results in the majority of RCP becoming cytosolic, whereas Rab14 depletion did not result in a similar effect. However, our results in N2a cells demonstrating that RCP and Rab14, but not Rab11, are important for neuritogenesis suggest that RCP also has Rab11-independent functions. Indeed, Qi et al. (23) reported that the RCP-dependent transport of the HIV-1 Env protein does not involve Rab11.
There are many examples of Rabs that share/compete for effectors. For example, Rab6 and Rab11 bind to Rab6IP1/DENND5B (10); Rab4 and Rab14 interact with RUFY1 (20), and both rabenosyn-5 (11) and myosin Va (40) bind to multiple Rabs. Further complexity arises from Rab cascades in which one Rab mediates the transport of a guanine nucleotide exchange factor that activates a downstream Rab. Such cascades regulate secretory vesicle transport (41), primary ciliogenesis (42), and intra-Golgi transport (39). Further work is required to determine whether there is an interplay between Rab11 and Rab14 in the regulation of RCP function, and we are currently investigating the possibility that RCP functions as an intermediary in a Rab11-Rab14 cascade.
Author Contributions
P. L. and A. L. designed and performed experiments and wrote the paper. S. H., T. K., E. S. T., U. M. M., and E. F. designed and performed experiments. M. W. M. and A. R. K. conceived and co-ordinated the study and wrote the paper.
Acknowledgments
This research used resources of the Advanced Photon Source, a United States Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract Number DE-AC02-06CH11357.
This work was supported by Science Foundation Ireland Principal Investigator Awards 07/IN.1/B975 (to A. R. K.) and 09/IN.1/B2629 (to M. W. M.). This work is based upon research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by National Institutes of Health Grant P41 GM103403 from NIGMS. The authors declare that they have no conflicts of interest with the contents of this article.
The atomic coordinates and structure factors (code 4d0g) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- FIP
- family of interacting proteins
- RBD
- Rab-binding domain
- RCP
- Rab coupling protein
- MBP
- maltose-binding protein
- rTEV
- recombinant tobacco etch virus
- PDB
- Protein Data Bank
- Gpp(NH)p
- guanosine 5′-(β,γ-imido)triphosphate
- GTPγS
- guanosine 5′-3-O-(thio)triphosphate.
References
- 1. Zerial M., McBride H. (2001) Rab proteins as membrane organizers. Nat. Rev. Mol. Cell. Biol. 2, 107–117 [DOI] [PubMed] [Google Scholar]
- 2. Andres D. A., Seabra M. C., Brown M. S., Armstrong S. A., Smeland T. E., Cremers F. P., Goldstein J. L. (1993) cDNA cloning of component A of Rab geranylgeranyl transferase and demonstration of its role as a Rab escort protein. Cell 73, 1091–1099 [DOI] [PubMed] [Google Scholar]
- 3. Bernards A. (2003) GAPs galore! A survey of putative Ras superfamily GTPase activating proteins in man and Drosophila. Biochim. Biophys. Acta 1603, 47–82 [DOI] [PubMed] [Google Scholar]
- 4. Goody R. S., Rak A., Alexandrov K. (2005) The structural and mechanistic basis for recycling of Rab proteins between membrane compartments. Cell. Mol. Life Sci. 62, 1657–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Saraste M., Sibbald P. R., Wittinghofer A. (1990) The P-loop–a common motif in ATP- and GTP-binding proteins. Trends Biochem. Sci. 15, 430–434 [DOI] [PubMed] [Google Scholar]
- 6. Walker J. E., Saraste M., Runswick M. J., Gay N. J. (1982) Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1, 945–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vetter I. R., Wittinghofer A. (2001) The guanine nucleotide-binding switch in three dimensions. Science 294, 1299–1304 [DOI] [PubMed] [Google Scholar]
- 8. Pereira-Leal J. B., Seabra M. C. (2000) The mammalian Rab family of small GTPases: definition of family and subfamily sequence motifs suggests a mechanism for functional specificity in the Ras superfamily. J. Mol. Biol. 301, 1077–1087 [DOI] [PubMed] [Google Scholar]
- 9. Burguete A. S., Fenn T. D., Brunger A. T., Pfeffer S. R. (2008) Rab and Arl GTPase family members cooperate in the localization of the golgin GCC185. Cell 132, 286–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Miserey-Lenkei S., Waharte F., Boulet A., Cuif M. H., Tenza D., El Marjou A., Raposo G., Salamero J., Héliot L., Goud B., Monier S. (2007) Rab6-interacting protein 1 links Rab6 and Rab11 function. Traffic 8, 1385–1403 [DOI] [PubMed] [Google Scholar]
- 11. Eathiraj S., Pan X., Ritacco C., Lambright D. G. (2005) Structural basis of family-wide Rab GTPase recognition by rabenosyn-5. Nature 436, 415–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fukuda M. (2006) Distinct Rab27A binding affinities of Slp2-a and Slac2-a/melanophilin: hierarchy of Rab27A effectors. Biochem. Biophys. Res. Commun. 343, 666–674 [DOI] [PubMed] [Google Scholar]
- 13. Hagemann N., Hou X., Goody R. S., Itzen A., Erdmann K. S. (2012) Crystal structure of the Rab binding domain of OCRL1 in complex with Rab8 and functional implications of the OCRL1/Rab8 module for Lowe syndrome. Small GTPases 3, 107–110 [DOI] [PubMed] [Google Scholar]
- 14. Goldenring J. R., Shen K. R., Vaughan H. D., Modlin I. M. (1993) Identification of a small GTP-binding protein, Rab25, expressed in the gastrointestinal mucosa, kidney, and lung. J. Biol. Chem. 268, 18419–18422 [PubMed] [Google Scholar]
- 15. Chia W. J., Tang B. L. (2009) Emerging roles for Rab family GTPases in human cancer. Biochim. Biophys. Acta 1795, 110–116 [DOI] [PubMed] [Google Scholar]
- 16. Cheng J. M., Ding M., Aribi A., Shah P., Rao K. (2006) Loss of RAB25 expression in breast cancer. Int. J. Cancer 118, 2957–2964 [DOI] [PubMed] [Google Scholar]
- 17. Kelly E. E., Horgan C. P., Adams C., Patzer T. M., Ní Shúilleabháin D. M., Norman J. C., McCaffrey M. W. (2010) Class I Rab11-family interacting proteins are binding targets for the Rab14 GTPase. Biol. Cell 102, 51–62 [DOI] [PubMed] [Google Scholar]
- 18. Junutula J. R., De Maziére A. M., Peden A. A., Ervin K. E., Advani R. J., van Dijk S. M., Klumperman J., Scheller R. H. (2004) Rab14 is involved in membrane trafficking between the Golgi complex and endosomes. Mol. Biol. Cell 15, 2218–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fukuda M., Kanno E., Ishibashi K., Itoh T. (2008) Large scale screening for novel rab effectors reveals unexpected broad Rab binding specificity. Mol. Cell. Proteomics 7, 1031–1042 [DOI] [PubMed] [Google Scholar]
- 20. Yamamoto H., Koga H., Katoh Y., Takahashi S., Nakayama K., Shin H. W. (2010) Functional cross-talk between Rab14 and Rab4 through a dual effector, RUFY1/Rabip4. Mol. Biol. Cell 21, 2746–2755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Junutula J. R., Schonteich E., Wilson G. M., Peden A. A., Scheller R. H., Prekeris R. (2004) Molecular characterization of Rab11 interactions with members of the family of Rab11-interacting proteins. J. Biol. Chem. 279, 33430–33437 [DOI] [PubMed] [Google Scholar]
- 22. Jing J., Junutula J. R., Wu C., Burden J., Matern H., Peden A. A., Prekeris R. (2010) FIP1/RCP binding to Golgin-97 regulates retrograde transport from recycling endosomes to the trans-Golgi network. Mol. Biol. Cell 21, 3041–3053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Qi M., Williams J. A., Chu H., Chen X., Wang J. J., Ding L., Akhirome E., Wen X., Lapierre L. A., Goldenring J. R., Spearman P. (2013) Rab11-FIP1C and Rab14 direct plasma membrane sorting and particle incorporation of the HIV-1 envelope glycoprotein complex. PLoS Pathog. 9, e1003278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lindsay A. J., McCaffrey M. W. (2015) Rab antibody characterization: comparison of rab14 antibodies. Methods Mol. Biol. 1298, 161–171 [DOI] [PubMed] [Google Scholar]
- 25. Jagoe W. N., Jackson S. R., Lindsay A. J., McCaffrey M. W., Khan A. R. (2006) Purification, crystallization and preliminary x-ray diffraction studies of Rab11 in complex with Rab11-FIP2. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 62, 692–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lall P., Horgan C. P., Oda S., Franklin E., Sultana A., Hanscom S. R., McCaffrey M. W., Khan A. R. (2013) Structural and functional analysis of FIP2 binding to the endosome-localised Rab25 GTPase. Biochim. Biophys. Acta 1834, 2679–2690 [DOI] [PubMed] [Google Scholar]
- 27. Otwinoski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 28. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 29. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 30. Bergbrede T., Chuky N., Schoebel S., Blankenfeldt W., Geyer M., Fuchs E., Goody R. S., Barr F., Alexandrov K. (2009) Biophysical analysis of the interaction of Rab6a GTPase with its effector domains. J. Biol. Chem. 284, 2628–2635 [DOI] [PubMed] [Google Scholar]
- 31. Chiang M. C., Cheng Y. C., Chen H. M., Liang Y. J., Yen C. H. (2014) Rosiglitazone promotes neurite outgrowth and mitochondrial function in N2A cells via PPARγ pathway. Mitochondrion 14, 7–17 [DOI] [PubMed] [Google Scholar]
- 32. Jagoe W. N., Lindsay A. J., Read R. J., McCoy A. J., McCaffrey M. W., Khan A. R. (2006) Crystal structure of rab11 in complex with rab11 family interacting protein 2. Structure 14, 1273–1283 [DOI] [PubMed] [Google Scholar]
- 33. Eathiraj S., Mishra A., Prekeris R., Lambright D. G. (2006) Structural basis for Rab11-mediated recruitment of FIP3 to recycling endosomes. J. Mol. Biol. 364, 121–135 [DOI] [PubMed] [Google Scholar]
- 34. Shiba T., Koga H., Shin H. W., Kawasaki M., Kato R., Nakayama K., Wakatsuki S. (2006) Structural basis for Rab11-dependent membrane recruitment of a family of Rab11-interacting protein 3 (FIP3)/Arfophilin-1. Proc. Natl. Acad. Sci. U.S.A. 103, 15416–15421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Qi M., Williams J. A., Chu H., Chen X., Wang J. J., Ding L., Akhirome E., Wen X., Lapierre L. A., Goldenring J. R., Spearman P. (2013) Rab11-FIP1C and Rab14 direct plasma membrane sorting and particle incorporation of the HIV-1 envelope glycoprotein complex. PLoS Pathog. 9, e1003278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Eva R., Dassie E., Caswell P. T., Dick G., ffrench-Constant C., Norman J. C., Fawcett J. W. (2010) Rab11 and its effector Rab coupling protein contribute to the trafficking of β1 integrins during axon growth in adult dorsal root ganglion neurons and PC12 cells. J. Neurosci. 30, 11654–11669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hou X., Hagemann N., Schoebel S., Blankenfeldt W., Goody R. S., Erdmann K. S., Itzen A. (2011) A structural basis for Lowe syndrome caused by mutations in the Rab-binding domain of OCRL1. EMBO J. 30, 1659–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhu Y., Hu L., Zhou Y., Yao Q., Liu L., Shao F. (2010) Structural mechanism of host Rab1 activation by the bifunctional Legionella type IV effector SidM/DrrA. Proc. Natl. Acad. Sci. U.S.A. 107, 4699–4704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pusapati G. V., Luchetti G., Pfeffer S. R. (2012) Ric1-Rgp1 complex is a guanine nucleotide exchange factor for the late Golgi Rab6A GTPase and an effector of the medial Golgi Rab33B GTPase. J. Biol. Chem. 287, 42129–42137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lindsay A. J., Jollivet F., Horgan C. P., Khan A. R., Raposo G., McCaffrey M. W., Goud B. (2013) Identification and characterization of multiple novel Rab-myosin Va interactions. Mol. Biol. Cell 24, 3420–3434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jin Y., Sultana A., Gandhi P., Franklin E., Hamamoto S., Khan A. R., Munson M., Schekman R., Weisman L. S. (2011) Myosin V transports secretory vesicles via a Rab GTPase cascade and interaction with the exocyst complex. Dev. Cell 21, 1156–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Westlake C. J., Baye L. M., Nachury M. V., Wright K. J., Ervin K. E., Phu L., Chalouni C., Beck J. S., Kirkpatrick D. S., Slusarski D. C., Sheffield V. C., Scheller R. H., Jackson P. K. (2011) Primary cilia membrane assembly is initiated by Rab11 and transport protein particle II (TRAPPII) complex-dependent trafficking of Rabin8 to the centrosome. Proc. Natl. Acad. Sci. U.S.A. 108, 2759–2764 [DOI] [PMC free article] [PubMed] [Google Scholar]