Abstract
Synapses mediate information flow between neurons and undergo plastic changes in response to experience, which is critical for learning and memory. Conversely, synaptic defects impair information processing and underlie many brain pathologies. Rho-family GTPases control synaptogenesis by transducing signals from extracellular stimuli to the cytoskeleton and nucleus. The Rho-GTPases Rac1 and Cdc42 promote synapse development and the growth of axons and dendrites, while RhoA antagonizes these processes. Despite its significance, many aspects of Rho-GTPase signaling remain relatively unknown. Rho-GTPases are activated by guanine nucleotide exchange factors (GEFs) and inhibited by GTPase-activating proteins (GAPs). Though the number of both GEFs and GAPs greatly exceeds that of Rho-GTPases, loss of even a single GEF or GAP often has profound effects on cognition and behavior. Here, we explore how the actions of specific GEFs and GAPs give rise to the precise spatiotemporal activation patterns of Rho-GTPases in neurons. We consider the effects of coupling GEFs and GAPs targeting the same Rho-GTPase and the modular pathways that connect specific cellular stimuli with a given Rho-GTPase via different GEFs. We discuss how the creation of sharp borders between Rho-GTPase activation zones is achieved by pairing a GEF for one Rho-GTPase with a GAP for another and the extensive crosstalk between different Rho-GTPases. Given the importance of synapses for cognition and the fundamental roles that Rho-GTPases play in regulating them, a detailed understanding of Rho-GTPase signaling is essential to the progress of neuroscience.
Introduction
The human brain contains approximately 100 billion neurons that communicate via specialized sites of contact called synapses [1]. Most excitatory synapses in the brain are situated on dendritic spines, small actin-rich protrusions on dendrites [2]. Spines undergo rapid changes in shape and number in response to stimuli [3]. This remodeling is critical for synapse formation and refinement and for the synaptic plasticity that underlies learning and memory [4]. Abnormal spine morphogenesis results in impaired information processing and is linked to numerous neurodevelopmental, neuropsychiatric, and neurodegenerative disorders [5]. Thus, uncovering the mechanisms regulating the formation and plasticity of spines and synapses will provide critical insights into brain function and the treatment of brain disorders.
Rho-family GTPases direct the actin dynamics that drive the formation and remodeling of spines and synapses [6]. Typically, the Rho-GTPases Rac1 and Cdc42 promote the formation, growth, and maintenance of spines, whereas RhoA inhibits these processes [6]. Rho-GTPases cycle between an active GTP-bound state and an inactive GDP-bound state. When active, they interact with specific effectors and initiate signaling pathways that control cytoskeletal dynamics, membrane trafficking, and gene expression [7]. To coordinate these processes properly, Rho GTPases must be regulated with great spatiotemporal precision [8]. Two classes of proteins control the on/off cycling of Rho GTPases. Guanine nucleotide exchange factors (GEFs) activate Rho GTPases by catalyzing GDP/GTP exchange, whereas GTPase-activating proteins (GAPs) inhibit Rho GTPases by stimulating GTP hydrolysis [9]. Guanine dissociation inhibitors (GDIs) also negatively regulate Rho GTPases by sequestering inactive Rho GTPases in the cytosol [10].
Considerable evidence links aberrant Rho-GTPase signaling to brain disorders associated with spine and synapse defects [5]. For instance, mutations in genes encoding Rho-GTPase regulators and effectors cause intellectual disabilities in humans [11]. Furthermore, altered Rac1 signaling is implicated in the pathogenesis of Fragile X syndrome [12, 13], Rett syndrome [14], schizophrenia [15], and substance abuse [16]. Rac1 is also downregulated in patients with major depressive disorder and in mice subjected to chronic social defeat, resulting in depression-related behaviors and abnormal spine remodeling [17]. Dysregulated RhoA signaling is likewise implicated in neurodevelopmental disorders associated with autism [18, 19]. Although precise spatiotemporal regulation of Rho-GTPase signaling is necessary for formation and maintenance of functional synapses, little is known about how this is achieved. Multiple GEFs and GAPs exist for each Rho-GTPase [9], but it is unclear how these regulatory proteins sculpt Rho-GTPase activities in space and time, specify cellular responses, and regulate crosstalk between Rho-GTPase family members. Here, we will discuss recent data that are shedding new light on how Rho-GTPase signaling is precisely regulated in cells, with emphasis on pathways essential to the formation and plasticity of excitatory synapses.
Multidimensional regulation of single Rho GTPases
GEF/GAP complexes targeting single GTPases
Fluorescent probes that report Rho-GTPase activation in live cells have revealed that Rho-GTPase signaling dynamics occur on micrometer length and subminute time scales [8]. Moreover, experiments using constitutively-active and dominant-negative Rho-GTPase mutants indicate that on/off cycling is required for Rho-GTPase-driven processes like spine morphogenesis [20–22]. These observations imply that mechanisms must exist that precisely regulate Rho-GTPase signaling at synapses to control their development, plasticity and function. One mechanism that could provide tight spatiotemporal control is GEF/GAP complexes targeting a specific Rho-GTPase.
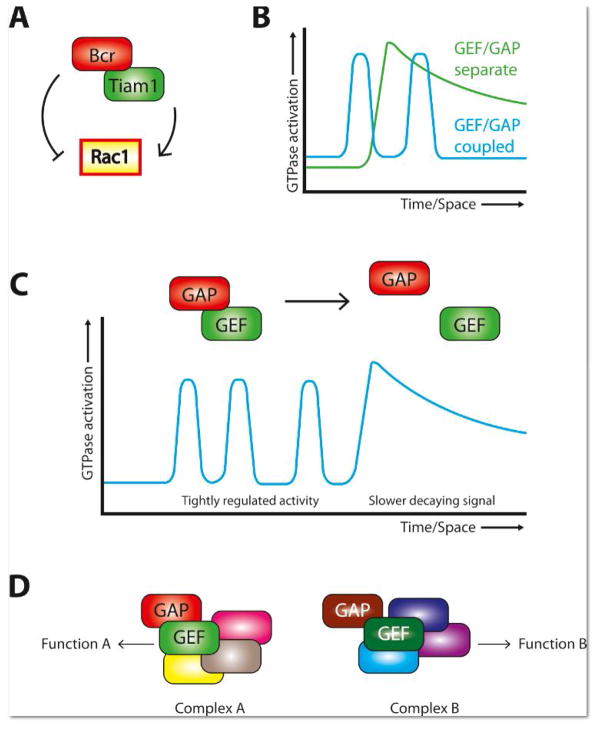
We recently reported the first functional characterization of a GEF/GAP complex targeting a single Rho-GTPase after identifying the Rac1-GAP Bcr (breakpoint cluster region) in a screen for proteins that interact with the Rac1-GEF Tiam1 (T-lymphocyte invasion and metastasis 1) (Fig. 1A) [23]. Both proteins had been implicated in excitatory synaptogenesis [24–27], but the tight association between the two in neurons was unexpected. Bcr function offsets Tiam1 function at synapses, as Bcr loss leads to increased Rac1 activation, spinogenesis, and synaptogenesis, and these defects are reversed by Tiam1 inhibition [23]. Moreover, disruption of the Tiam1/Bcr complex results in the same exuberant Rac1 activation and spine overgrowth phenotypes as either Bcr loss or competition with a GAP-dead Bcr mutant [23].
Figure 1. GEF/GAP complexes targeting single GTPases.
A. The Tiam1/BcrGEF/GAP complex regulates Rac1 activity.
B. Coupled GEF/GAP complexes tightly regulate Rho-GTPase activity by increasing the effective GTPase “off” rate and/or by reducing signal spread.
C. GEF/GAP complexes may toggle between associated and dissociated states to achieve the observed cellular patterns of GTPase activation.
D. Rho-family GEFs and GAPs are large, multidomain proteins that can scaffold functional protein complexes. Different GEF/GAP complexes could assemble different signaling modules, adding complexity and flexibility to Rho-GTPase signaling.
EphB receptor tyrosine kinases and their ephrin-B ligands utilize the Tiam1/Bcr complex to control synapse development [23]. EphBs promote spinogenesis in hippocampal pyramidal neurons via Tiam1-mediated Rac1 activation [26]. Following ephrin-B stimulation, neurons exhibit a transient increase in Tiam1’s Rac1-GEF activity coincident with a transient decrease in Bcr’s Rac1-GAP activity [23]. How this is achieved is unknown, though it could result from phosphorylation of Tiam1/Bcr and/or transient disruption of the Tiam1/Bcr complex [23]. In any case, loss of Bcr function converts the ephrin-B signal from spine-promoting to spine-eliminating due to a massive over-activation of Rac1 that triggersRac1-dependent endocytosis of EphB and AMPA receptors and leads to spine loss [23]. These data reflect the necessity of precise Rac1 regulation for neuronal development and reveal that tight coupling between Rac1 activation and inhibition is required to achieve this level of regulation.
The functions of the Tiam1/Bcr complex are not restricted to excitatory synaptogenesis in neurons. We recently showed that Bcr interacts with Tiam1 and members of the partition-defective (Par) polarity complex in cortical astrocytes [28]. The Par complex, composed of Par3, Par6, and atypical protein kinase Cζ (PKCζ), controls cell polarization in many different contexts, including directional cell migration, axon specification, and spine morphogenesis [29]. The Par complex functions in part by recruiting Tiam1 to specialized sites where it promotes local Rac1 activation [30]. We found that Bcr also associates with the Par complex and that Bcr loss in astrocytes leads to faster, more random migration and striking polarity defects in cytoskeletal organization and centrosome orientation [28]. These defects are caused by misregulation of both Rac1 and PKCζ and are rescued by wild-type Bcr, but not by a Bcr mutant that cannot associate with the Par-Tiam1 complex [28]. Thus, Bcr is a member of the Par-Tiam1 complex that controls polarized cell migration by locally restricting Rac1 and PKCζ function.
The Tiam1/Bcr complex may not be unique. Recently, Lim et al. reported a potential neuronal Rac1-specific GEF/GAP complex [31]. Searching for proteins that interact with the scaffold protein CNK2 (Connector enhancer of KSR-2), they identified a complex composed of the Rac1-GAP ARHGAP39/Vilse, the Rac1-GEFs α- and β-PIX, and other Rac1 effectors and PIX modulators as well as Rac1 itself [31]. While a complete functional dissection of this complex is not yet available, they did observe that Vilse, like Bcr, serves as a necessary counterweight against spine and synapse formation. Its interaction with CNK2 is required for proper signaling from this complex, and loss of Vilse leads to Rac1 hyperactivity and aberrant spinogenesis [31].
GEF/GAP complexes for Rho-GTPases may be more common than is appreciated, as they represent a satisfying solution to several problems encountered in Rho-GTPase signaling. First, the ability of such complexes to powerfully limit Rho-GTPase activation is an obvious mechanism for the establishment of the tightly defined regions of Rho-GTPase signaling observed in many cells [32, 33]. By coupling GEF and GAP activities, cells increase the effective GTPase ‘off’ rate, sharply reducing signal spread relative to when GTPase deactivation requires a random encounter with a GAP (Fig. 1B) [32].
Second, inherently modular GEF/GAP complexes could enable rapid alteration of Rho-GTPase signaling modes. Rho-GTPase signaling can be complex, as illustrated by RhoA and Cdc42 signaling at dendritic spines undergoing long-term potentiation (LTP) [34]. Glutamate uncaging increases RhoA and Cdc42 activation in spines [34]. The activity of both GTPases decays rapidly for ~5 minutes, then switches to a slowly decaying phase [34]. During this latter phase, activated RhoA diffuses out of the spine [34]. Though the proteins sculpting these activity patterns are unknown, a RhoA-GEF/GAP complex would be a simple solution. A tightly coupled GEF/GAP complex could account for the fast-decaying, spatially restricted phase, while complex disassembly would switch to a mode of slow decay and spatial spread (Fig. 1C). The temporal dynamics of active Cdc42 could be explained similarly, though it does not escape the spine [34], so mechanisms are required to blunt its spatial spread. Thus, assembly and disassembly of GEF/GAP complexes would allow toggling between different modes of signaling and work together with posttranslational modifications, crosstalk between different GTPases, etc. [32] to achieve the observed complex patterns of Rho-GTPase signaling.
Third, GEF/GAP complexes may assist cells in creating precise Rho-GTPase signaling by coordinating the assembly of signaling complexes (Fig. 1D). GEFs/GAPs are typically large, multidomain proteins. We (and others) have argued that proper Rho-GTPase signaling requires pre-assembly of the components of particular signaling pathways and that the protein-interaction domains of GEFs/GAPs serve as scaffolds on which to do that [9, 35, 36]. A GEF/GAP complex would not only have more binding sites than either protein alone, but different complexes could hypothetically lead to diverse signals, adding another layer of complexity and flexibility to Rho-GTPase signaling.
Receptors signaling to a single GTPase through multiple pathways
Another fascinating phenomenon in Rho-GTPase signaling is modular pathways by which cell-surface receptors signal to a Rho-GTPase via different GEFs. We will consider two cases of this.
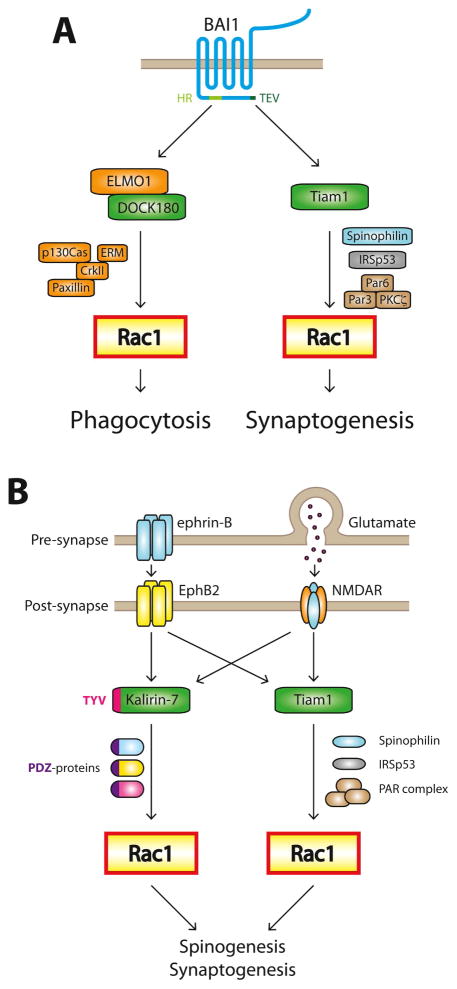
The first case involves brain-specific angiogenesis inhibitor 1 (BAI1) (Fig. 2A). BAI1 is an adhesion-G protein-coupled receptor (A-GPCR), and as such possesses a large extracellular N-terminal segment connected to its GPCR moiety via a GAIN domain [37]. BAI1 is expressed in professional phagocytes and binds to the apoptotic cell marker phosphatidylserine (PS), triggering Rac1-dependent engulfment of PS-presenting apoptotic cells [38]. BAI1 also binds lipopolysaccharide (LPS) on gram-negative bacteria, again triggering Rac1-dependent phagocytosis [39]. We demonstrated that BAI1 is present in hippocampal and cortical neurons, where it is enriched in dendritic spines and required for spine and synapse development [40]. Knockdown of BAI1 results in both spine and synapse defects and a dramatic redistribution of activated Rac1 out of spines, suggesting that BAI1 regulates spine and synapse development by inducing synaptic Rac1 activation [40].
Figure 2. Receptors signaling to single Rho-GTPases through multiple pathways.
A. Both phagocytosis and excitatory synaptogenesis require Rac1 activation. The adhesion-GPCR BAI1 regulates both processes by signaling through different Rac1-GEFs. The C-terminus of BAI1 interacts with Tiam1 through a TEV motif and DOCK180 via ELMO through a helical region (HR). Tiam1 and DOCK180 associate with divergent protein complexes capable of generating distinct Rac1 signaling that promotes synaptogenesis or phagocytosis, respectively.
B. At excitatory synapses, Rac1 is activated by EphB2 and NMDA-type glutamate receptors (NMDARs) via the Rac1-GEFs Tiam1 and Kalirin-7. These Rac1-GEFs signal through parallel pathways to promote spino- and synaptogenesis. Loss of either protein elicits a phenotype, proving that they are not redundant.
While phagocytosis and excitatory synaptogenesis both require Rac1 activation downstream of BAI1, the GEFs differ. Phagocytosis relies on the Rac1-GEF DOCK180 [38], while synaptogenesis requires Tiam1 [40]. ABAI1 mutant that cannot interact with DOCK180/ELMO1 rescues the BAI1 knockdown synapse phenotypes as well as the wild-type protein [40]. Why does a receptor require different GEFs to signal to the same Rho-GTPase? We propose that the Rac1 signals needed to regulate phagocytosis and synaptogenesis differ sufficiently such that the regulation of Rac1’s activity and downstream effectors must also differ. This could be accomplished with different GEFs, and DOCK180 and Tiam1 do differ substantially. DOCK180 interacts with CrkII [41], which binds to the focal adhesion-associated proteins p130 Cas and paxillin and to ezrin-radexin-moesin proteins that link the actin cytoskeleton to membranes [42]. These proteins likely play a role in the cytoskeletal dynamics and membrane distortion required for phagocytosis. In contrast, Tiam1 associates with the Par complex and the synaptic scaffolds spinophilin and IRSp53, which regulate actin dynamics [43–45]. Given the central role the Par complex plays in regulating cellular polarity [29], its cooperation with Tiam1 to form highly polarized excitatory synapses is not surprising. Tiam1 also has a Ras-binding domain, which links it to Ras-dependent pathways important for synapse formation and plasticity [46, 47].
Rac1 is also activated in different ways by EphB2 and NMDA-type glutamate receptors (NMDARs) (Fig. 2B). Activation of EphB2 by ephrin-B leads to Tiam1 phosphorylation and recruitment into EphB2/NMDAR complexes [24, 26]. Tiam1 is required for EphB2-dependent spinogenesis in hippocampal neurons [26], but it is not the only Rac1-GEF in spines that associates with EphB2. The Rac1-GEF Kalirin-7 is also in a complex with EphB2 and synaptic scaffolds such as PSD-95 and SAP102, and recruitment and activation of Kalirin-7 is also necessary for spine development downstream of ephrin-B/EphB2 [48–50]. Moreover, both Tiam1 and Kalirin-7 are activated downstream of NMDARs and necessary for its positive effects on spine growth [24, 51]. However, loss of either protein elicits a phenotype, proving their non-redundancy.
What is the difference between Rac1 signals elicited by EphB2 or NMDAR via Tiam1 versus those via Kalirin-7? One probable difference is the pathways organized by these GEFs (Fig. 2B). As noted above, Tiam1 couples to the small GTPase Rasand a number of proteins that regulate cytoskeletal and membrane dynamics, including the Par polarity complex, spinophilin, and IRSp53. In contrast, Kalirin-7 interacts with a different array of proteins involved in glutamate receptor trafficking, adhesion-dependent signaling, and actin-binding, including PSD-95, SAP102, and AF-6 [52]. The seproteins could modulate Kalirin-7 activity, localization, and/or downstream pathways. Another notable difference between Tiam1 and Kalirin-7 is their expression patterns [53]. Tiam1 expression in the brain is strongly developmentally regulated: in mice, Tiam1 is widely expressed during development beginning around embryonic day E14, whereas its expression is more restricted in the adult [54]. Tiam1 expression remains particularly high in adult brain areas engaged in adult neurogenesis, suggesting an important role in neuronal development [54]. Kalirin-7, in contrast, is not detectable until approximately postnatal day P14, and its expression continues to increase throughout adolescence, suggesting a later role for Kalirin-7 in synapse maturation and maintenance [55, 56].
The presence of multiple mechanisms connecting a single receptor with a single GTPase further emphasizes the high level of specificity required for signal transduction by Rho-family GTPases. The combinatorial nature of these pathways allows cells to tailor signaling in cell-type and developmentally specified ways. It will be interesting to determine if multiple pathways linking single receptors to single Rho GTPases can mediate different effects within the same cell.
Signal coordination between different Rho GTPases
Another feature of Rho-GTPase signaling is that opposing Rho-GTPase signals are often sharply separated in space and time [32]. For example, RhoA, Cdc42 and Rac1 activity zones were spatiotemporally resolved in migrating fibroblasts using biosensors and computational multiplexing [57]. RhoA was found to be active at the leading edge of the cell, whereas Rac1 and Cdc42 activities were maintained approximately 2 μm behind with their activation times trailing that of RhoA by 40 seconds [57]. Such separation of active zones is not unique to fibroblasts. Segregated RhoA and Cdc42 zones form over the meiotic spindle during asymmetric cell division, and mutually exclusive zones occur during single-cell wound healing in Xenopus oocytes [58, 59]. These mutually exclusive zones of activity make sense in neurons, given that neuronal Rac1/Cdc42 and RhoA play opposing roles in so many cellular processes [60]. Although it is unclear how the activities of opposing Rho-GTPases are locally coordinated, crosstalk between Rho-GTPase regulators or effectors could underlie the observed signal segregation. We will discuss mechanisms by which the balance between Rac1/Cdc42 and RhoA maybe achieved.
GEF/GAP complexes that coordinate the activities of opposing Rho-GTPases
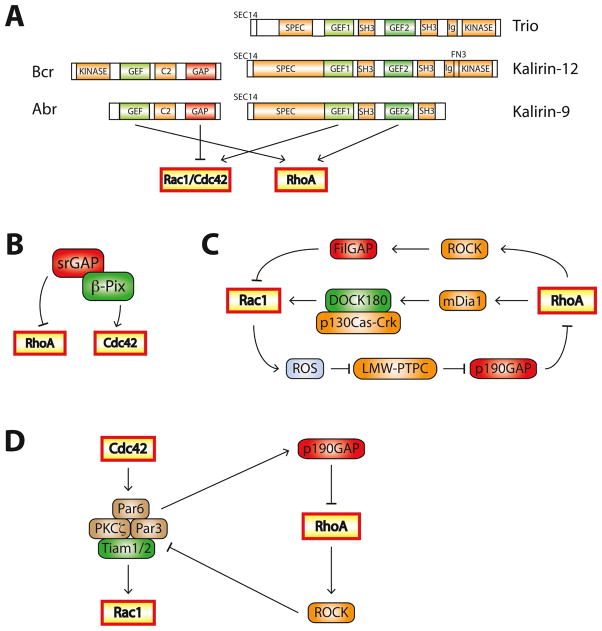
One approach to building strongly demarcated Rho-GTPase activation zones would be to tie a GEF for one Rho-GTPase to a GAP for another. This can be accomplished within a single molecule, as exemplified by Bcr and closely related Abr (active Bcr-related), which are both RhoA-GEFs andRac1/Cdc42-GAPs (Fig. 3A). Bcr and Abr are required to maintain mutually exclusive concentric zones of Cdc42 and RhoA activity during wound healing in Xenopus oocytes [61]. Active RhoA recruits Bcr/Abr to the wound edge, where they further activate RhoA and simultaneously inhibit Cdc42, segregating the signals [61].
Figure 3. Mechanisms regulating the balance between Rac1/Cdc42 and RhoA.
A. Single proteins can contain multiple Rho-family regulatory domains. Abr and Bcr are both RhoA-GEFs and Rac1/Cdc42-GAPs. Some Rho-family GEFs activate multiple Rho GTPases: Trio, Kalirin-12, and Kalirin-9 are GEFs for RhoA and Rac1/Cdc42. C2: protein kinase C conserved region 2, SEC14: domain in phosphatidylinositol transfer protein Sec14, SPEC: spectrin-like repeats, SH3: Src homology 3 domain, CC: coiled coil, Ig/FN3:Ig/fibronectin III
B. The Cdc42-GEF β-PIX exists in a complex with srGAP, a RhoA-GAP.
C. The Rac1/RhoA balance is tightly regulated through feedback networks. For example, Rac1 inhibits RhoA by inducing ROS production, leading to p190-RhoGAP activation. RhoA increases Rac1 activation through the RhoA effector mDia1, but inhibitsRac1 by activating the Rac1-GAP FilGAP through the RhoA effector ROCK.
D. Crosstalk between Cdc42, Rac1 and RhoA during the establishment of cell polarity. Activated Cdc42 recruits members of the Par complex (Par3, Par6 and PKCζ) to specialized sites to induce cell polarity. Par3 recruits the Rac1-GEFs Tiam1/2, resulting in local Rac1 activation. Par6 interacts with p190-RhoGAP, inhibiting RhoA activity. Conversely, RhoA can disrupt the Par complex through ROCK-dependent phosphorylation of Par3, leading to the Rac1 inhibition.
Bcr and Abr likely mediate the reciprocal relationship between Rac1 and RhoA signaling in mammalian development. We recently reported that the brains of Bcr/Abr knockout (KO) mice exhibit excessive Rac1 activity [23] and display an abnormal anterior cerebellar phenotype similar to that of RhoA KO mice [62]. Bcr/Abr KO and RhoA KO mice both possess irregular folia, ectopic granule cells, disorganizedglial scaffolding and disrupted basal lamina, suggesting that an imbalance between Rho-GTPase activities disturbs cerebellar development [62]. It is possible that, in addition to restricting Rac1 activity, Bcr and Abr serve as Rho-GEFs and their absence results in Rac1-RhoA dysregulation in a region-specific manner. Interestingly, Bcr/Abr single KO mice do not exhibit cerebellar malformation [63], suggesting that Bcr and Abr compensate for each other during cerebellar development. While the phenotypes of Bcr/Abr KO and RhoA KO mice do not completely overlap, the similarities argue that control over the Rac1/RhoA balance is necessary to achieve normal cerebellar architecture.
The GEF and GAP domains in a regulatory pair need not be encoded by the same gene. Kutys and Yamada recently identified a β-PIX/srGAP complex that regulates collagen-stimulated migration in several cell lines [64]. β-PIX mediates collagen-specific migration by activating Cdc42. Knockdown of β-PIX abolishes collagen-stimulated migration and polarized Cdc42 activity and leads to enhanced, de-localized RhoA activity [64]. srGAP1 was identified as a RhoA-GAP that associates with β-PIX at the leading edge of migrating cells [64]. The β-PIX/srGAP complex thus functions similarly to Bcr/Abr in activating its GEF target while inhibiting the opposing Rho-GTPase via its GAP functionality (Fig. 3B).
GEF/GEF complexes targeting opposing Rho-GTPases
Counter intuitively, some GEFs (Trio, UNC-73, Kalirin-9/12)contain two Rho-family GEF domains (Fig. 3A) [65]. One activates Rac1 and the closely related RhoG, while the second activates RhoA, making them dual-function GEFs for opposing Rho-GTPases. Knockdown of Kalirin-9 or Kalirin-12 in developing hippocampal cultures results in decreased dendritic length and complexity [66], while overexpression of Kalirin-9 reduces dendritic length and complexity, but only in mature neurons [66, 67]. Kalirin-9 also mediates p75/nogo receptor-dependent RhoA activation and neurite growth inhibition in response to myelin-associated glycoproteins [68]. It is unclear how these dual function Rho-family GEFs coordinate their Rac1/RhoA-GEF activities, but full functionality of both domains is inconsistent with the highly demarcated activity zones discussed thus far. It is possible that these proteins can quickly switch between their GEF activities and thus rapidly convert active Rac1 zones to active RhoA zones and vice versa and/or that some signals require simultaneous activation of canonically opposing pathways.
Downstream effectors that contribute to Rho-GTPase balance
Rac1 and RhoA signaling cascades also modulate each other’s activity(Fig. 3C). Rac1 binds to and activatesp190-RhoGAP, a RhoA-GAP that regulates dendrite and spine morphogenesis [69, 70]. Rac1 also abrogates RhoA activity by inducing reactive oxygen species(ROS) production, inhibiting LMW-PTP (low-molecular weight tyrosine phosphatase), and increasing phosphorylation and activation of p190-RhoGAP [71]. RhoA can both increase and decrease Rac1 activation. The Rac1-GAP FilGAP is phosphorylated and activated by the RhoA effector ROCK (Rho kinase), leading toRac1 inhibition [72]. In 3T3 fibroblasts, the RhoA effector mDia1 mediates Rac1 activation, which is antagonized by ROCK [73]. How mDia1 activates Rac1 is not completely understood, but appears to involve the formation of a Cas-Crk-DOCK180 Rac1-GEF complex induced by RhoA-mDia1 signaling [73].
Crosstalk between Rho-GTPases also occurs during the establishment of cell polarity(Fig. 3D). Activated Cdc42 recruits the Par complex to sites of polarity development [74], leading toRac1 activation by the Par complex-associated Rac1-GEFs Tiam1/2 [30]. RhoA-activated ROCK can disrupt the Par complex by phosphorylating Par3, thereby inhibiting its interaction with Par6 and aPKC [75]. At synapses, Rac1 activation is spatially restricted via Par3 binding to Tiam1 and confining it to spines, which is essential for proper spine morphogenesis [25]. Surprisingly, Par6 negatively regulates RhoA activity at spines through p190-RhoGAP independently of Par3 [76]. Cdc42, Rac1 and RhoA thus modulate each other through interesting local feedback mechanisms.
Conclusion
The development and function of neurons and many other cells require proper Rho-GTPase signaling. Despite the ubiquity and importance of Rac1, Cdc42, and RhoA signaling, we know relatively little about how the spatiotemporal dynamics of their activity patterns are shaped and how alterations to these patterns leads to cellular and organismal malfunction. Solving these puzzles will give insight into many brain disorders, as altered Rho-GTPase signaling is widely observed in these disorders. Recent studies of Rho-family GEFs and GAPs have yielded exciting new insights into how these signals work. Complexes between regulators of the same and different GTPases, modular signaling pathways, and crosstalk between Rho-GTPase signals are highly promising avenues along which to guide future research.
Acknowledgments
This work was supported by the National Institute of Neurological Disorders and Stroke grant R01NS062829 (K.F.T.) and the National Institute of Mental Health grant K01MH089112 (J.G.D).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Shen K, Scheiffele P. Genetics and cell biology of building specific synaptic connectivity. Annu Rev Neurosci. 2010;33:473–507. doi: 10.1146/annurev.neuro.051508.135302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bourne JN, Harris KM. Balancing structure and function at hippocampal dendritic spines. Annu Rev Neurosci. 2008;31:47–67. doi: 10.1146/annurev.neuro.31.060407.125646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhatt DH, Zhang S, Gan WB. Dendritic spine dynamics. Annu Rev Physiol. 2009;71:261–82. doi: 10.1146/annurev.physiol.010908.163140. [DOI] [PubMed] [Google Scholar]
- 4.Holtmaat A, Svoboda K. Experience-dependent structural synaptic plasticity in the mammalian brain. Nat Rev Neurosci. 2009;10(9):647–58. doi: 10.1038/nrn2699. [DOI] [PubMed] [Google Scholar]
- 5.Lai KO, Ip NY. Structural plasticity of dendritic spines: the underlying mechanisms and its dysregulation in brain disorders. Biochim Biophys Acta. 2013;1832(12):2257–63. doi: 10.1016/j.bbadis.2013.08.012. [DOI] [PubMed] [Google Scholar]
- 6.Newey SE, et al. Rho GTPases, dendritic structure, and mental retardation. J Neurobiol. 2005;64(1):58–74. doi: 10.1002/neu.20153. [DOI] [PubMed] [Google Scholar]
- 7.Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21:247–69. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- 8.Pertz O. Spatio-temporal Rho GTPase signaling - where are we now? J Cell Sci. 2010;123(Pt 11):1841–50. doi: 10.1242/jcs.064345. [DOI] [PubMed] [Google Scholar]
- 9.Tolias KF, Duman JG, Um K. Control of synapse development and plasticity by Rho GTPase regulatory proteins. Prog Neurobiol. 2011;94(2):133–48. doi: 10.1016/j.pneurobio.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DerMardirossian C, Bokoch GM. GDIs: central regulatory molecules in Rho GTPase activation. Trends Cell Biol. 2005;15(7):356–63. doi: 10.1016/j.tcb.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 11.Ba W, van der Raadt J, Nadif Kasri N. Rho GTPase signaling at the synapse: implications for intellectual disability. Exp Cell Res. 2013;319(15):2368–74. doi: 10.1016/j.yexcr.2013.05.033. [DOI] [PubMed] [Google Scholar]
- 12.De Rubeis S, et al. CYFIP1 coordinates mRNA translation and cytoskeleton remodeling to ensure proper dendritic spine formation. Neuron. 2013;79(6):1169–82. doi: 10.1016/j.neuron.2013.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen LY, et al. Physiological activation of synaptic Rac>PAK (p-21 activated kinase) signaling is defective in a mouse model of fragile X syndrome. J Neurosci. 2010;30(33):10977–84. doi: 10.1523/JNEUROSCI.1077-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Filippis B, et al. Modulation of RhoGTPases improves the behavioral phenotype and reverses astrocytic deficits in a mouse model of Rett syndrome. Neuropsychopharmacology. 2012;37(5):1152–63. doi: 10.1038/npp.2011.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayashi-Takagi A, et al. Disrupted-in-Schizophrenia 1 (DISC1) regulates spines of the glutamate synapse via Rac1. Nat Neurosci. 2010;13(3):327–32. doi: 10.1038/nn.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dietz DM, et al. Rac1 is essential in cocaine-induced structural plasticity of nucleus accumbens neurons. Nat Neurosci. 2012;15(6):891–6. doi: 10.1038/nn.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Golden SA, et al. Epigenetic regulation of RAC1 induces synaptic remodeling in stress disorders and depression. Nat Med. 2013;19(3):337–44. doi: 10.1038/nm.3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krey JF, et al. Timothy syndrome is associated with activity-dependent dendritic retraction in rodent and human neurons. Nat Neurosci. 2013;16(2):201–9. doi: 10.1038/nn.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin GN, et al. Spatiotemporal 16p11.2 Protein Network Implicates Cortical Late Mid-Fetal Brain Development and KCTD13-Cul3-RhoA Pathway in Psychiatric Diseases. Neuron. 2015;85(4):742–54. doi: 10.1016/j.neuron.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo L, et al. Differential effects of the Rac GTPase on Purkinje cell axons and dendritic trunks and spines. Nature. 1996;379(6568):837–40. doi: 10.1038/379837a0. [DOI] [PubMed] [Google Scholar]
- 21.Nakayama AY, Harms MB, Luo L. Small GTPases Rac and Rho in the maintenance of dendritic spines and branches in hippocampal pyramidal neurons. J Neurosci. 2000;20(14):5329–38. doi: 10.1523/JNEUROSCI.20-14-05329.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tashiro A, Minden A, Yuste R. Regulation of dendritic spine morphology by the rho family of small GTPases: antagonistic roles of Rac and Rho. Cereb Cortex. 2000;10(10):927–38. doi: 10.1093/cercor/10.10.927. [DOI] [PubMed] [Google Scholar]
- 23.Um K, et al. Dynamic control of excitatory synapse development by a Rac1 GEF/GAP regulatory complex. Dev Cell. 2014;29(6):701–15. doi: 10.1016/j.devcel.2014.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tolias KF, et al. The Rac1-GEF Tiam1 couples the NMDA receptor to the activity-dependent development of dendritic arbors and spines. Neuron. 2005;45(4):525–38. doi: 10.1016/j.neuron.2005.01.024. [DOI] [PubMed] [Google Scholar]
- 25.Zhang H, I, Macara G. The polarity protein PAR-3 and TIAM1 cooperate in dendritic spine morphogenesis. Nat Cell Biol. 2006;8(3):227–37. doi: 10.1038/ncb1368. [DOI] [PubMed] [Google Scholar]
- 26.Tolias KF, et al. The Rac1 guanine nucleotide exchange factor Tiam1 mediates EphB receptor-dependent dendritic spine development. Proc Natl Acad Sci U S A. 2007;104(17):7265–70. doi: 10.1073/pnas.0702044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oh D, et al. Regulation of synaptic Rac1 activity, long-term potentiation maintenance, and learning and memory by BCR and ABR Rac GTPase-activating proteins. J Neurosci. 2010;30(42):14134–44. doi: 10.1523/JNEUROSCI.1711-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Narayanan AS, et al. The Rac-GAP Bcr is a novel regulator of the Par complex that controls cell polarity. Mol Biol Cell. 2013;24(24):3857–68. doi: 10.1091/mbc.E13-06-0333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goldstein B, I, Macara G. The PAR proteins: fundamental players in animal cell polarization. Dev Cell. 2007;13(5):609–22. doi: 10.1016/j.devcel.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mertens AE, Pegtel DM, Collard JG. Tiam1 takes PARt in cell polarity. Trends Cell Biol. 2006;16(6):308–16. doi: 10.1016/j.tcb.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 31.Lim J, et al. The CNK2 scaffold interacts with vilse and modulates Rac cycling during spine morphogenesis in hippocampal neurons. Curr Biol. 2014;24(7):786–92. doi: 10.1016/j.cub.2014.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bement WM, Miller AL, von Dassow G. Rho GTPase activity zones and transient contractile arrays. Bioessays. 2006;28(10):983–93. doi: 10.1002/bies.20477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ngok SP, Lin WH, Anastasiadis PZ. Establishment of epithelial polarity--GEF who’s minding the GAP? J Cell Sci. 2014;127(Pt 15):3205–15. doi: 10.1242/jcs.153197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murakoshi H, Wang H, Yasuda R. Local, persistent activation of Rho GTPases during plasticity of single dendritic spines. Nature. 2011;472(7341):100–4. doi: 10.1038/nature09823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kiraly DD, et al. Synaptic plasticity, a symphony in GEF. ACS Chem Neurosci. 2010;1(5):348–365. doi: 10.1021/cn100012x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buchsbaum RJ. Rho activation at a glance. J Cell Sci. 2007;120(Pt 7):1149–52. doi: 10.1242/jcs.03428. [DOI] [PubMed] [Google Scholar]
- 37.Stephenson JR, Purcell RH, Hall RA. The BAI subfamily of adhesion GPCRs: synaptic regulation and beyond. Trends Pharmacol Sci. 2014;35(4):208–15. doi: 10.1016/j.tips.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park D, et al. BAI1 is an engulfment receptor for apoptotic cells upstream of the ELMO/Dock180/Rac module. Nature. 2007;450(7168):430–4. doi: 10.1038/nature06329. [DOI] [PubMed] [Google Scholar]
- 39.Das S, et al. Brain angiogenesis inhibitor 1 (BAI1) is a pattern recognition receptor that mediates macrophage binding and engulfment of Gram-negative bacteria. Proc Natl Acad Sci U S A. 2011;108(5):2136–41. doi: 10.1073/pnas.1014775108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duman JG, et al. The adhesion-GPCR BAI1 regulates synaptogenesis by controlling the recruitment of the Par3/Tiam1 polarity complex to synaptic sites. J Neurosci. 2013;33(16):6964–78. doi: 10.1523/JNEUROSCI.3978-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kiyokawa E, et al. Activation of Rac1 by a Crk SH3-binding protein, DOCK180. Genes Dev. 1998;12(21):3331–6. doi: 10.1101/gad.12.21.3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miyamoto Y, Yamauchi J. Cellular signaling of Dock family proteins in neural function. Cell Signal. 2010;22(2):175–82. doi: 10.1016/j.cellsig.2009.09.036. [DOI] [PubMed] [Google Scholar]
- 43.Buchsbaum RJ, Connolly BA, Feig LA. Regulation of p70 S6 kinase by complex formation between the Rac guanine nucleotide exchange factor (Rac-GEF) Tiam1 and the scaffold spinophilin. J Biol Chem. 2003;278(21):18833–41. doi: 10.1074/jbc.M207876200. [DOI] [PubMed] [Google Scholar]
- 44.Connolly BA, et al. Tiam1-IRSp53 complex formation directs specificity of rac-mediated actin cytoskeleton regulation. Mol Cell Biol. 2005;25(11):4602–14. doi: 10.1128/MCB.25.11.4602-4614.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rajagopal S, et al. Scaffold proteins IRSp53 and spinophilin regulate localized Rac activation by T-lymphocyte invasion and metastasis protein 1 (TIAM1) J Biol Chem. 2010;285(23):18060–71. doi: 10.1074/jbc.M109.051490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lambert JM, et al. Tiam1 mediates Ras activation of Rac by a PI(3)K-independent mechanism. Nat Cell Biol. 2002;4(8):621–5. doi: 10.1038/ncb833. [DOI] [PubMed] [Google Scholar]
- 47.Zhu JJ, et al. Ras and Rap control AMPA receptor trafficking during synaptic plasticity. Cell. 2002;110(4):443–55. doi: 10.1016/s0092-8674(02)00897-8. [DOI] [PubMed] [Google Scholar]
- 48.Penzes P, et al. The neuronal Rho-GEF Kalirin-7 interacts with PDZ domain-containing proteins and regulates dendritic morphogenesis. Neuron. 2001;29(1):229–42. doi: 10.1016/s0896-6273(01)00193-3. [DOI] [PubMed] [Google Scholar]
- 49.Penzes P, et al. Rapid induction of dendritic spine morphogenesis by trans-synaptic ephrinB-EphB receptor activation of the Rho-GEF kalirin. Neuron. 2003;37(2):263–74. doi: 10.1016/s0896-6273(02)01168-6. [DOI] [PubMed] [Google Scholar]
- 50.Murata Y, Constantine-Paton M. Postsynaptic density scaffold SAP102 regulates cortical synapse development through EphB and PAK signaling pathway. J Neurosci. 2013;33(11):5040–52. doi: 10.1523/JNEUROSCI.2896-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xie Z, et al. Kalirin-7 controls activity-dependent structural and functional plasticity of dendritic spines. Neuron. 2007;56(4):640–56. doi: 10.1016/j.neuron.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mandela P, Ma XM. Kalirin, a key player in synapse formation, is implicated in human diseases. Neural Plast. 2012;2012:728161. doi: 10.1155/2012/728161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Penzes P, et al. Convergent CaMK and RacGEF signals control dendritic structure and function. Trends Cell Biol. 2008;18(9):405–13. doi: 10.1016/j.tcb.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 54.Ehler E, et al. Expression of Tiam-1 in the developing brain suggests a role for the Tiam-1-Rac signaling pathway in cell migration and neurite outgrowth. Mol Cell Neurosci. 1997;9(1):1–12. doi: 10.1006/mcne.1997.0602. [DOI] [PubMed] [Google Scholar]
- 55.Ma XM, et al. Kalirin, a multifunctional Rho guanine nucleotide exchange factor, is necessary for maintenance of hippocampal pyramidal neuron dendrites and dendritic spines. J Neurosci. 2003;23(33):10593–603. doi: 10.1523/JNEUROSCI.23-33-10593.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xie Z, et al. Coordination of synaptic adhesion with dendritic spine remodeling by AF-6 and kalirin-7. J Neurosci. 2008;28(24):6079–91. doi: 10.1523/JNEUROSCI.1170-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Machacek M, et al. Coordination of Rho GTPase activities during cell protrusion. Nature. 2009;461(7260):99–103. doi: 10.1038/nature08242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ma C, et al. Cdc42 activation couples spindle positioning to first polar body formation in oocyte maturation. Curr Biol. 2006;16(2):214–20. doi: 10.1016/j.cub.2005.11.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Benink HA, Bement WM. Concentric zones of active RhoA and Cdc42 around single cell wounds. J Cell Biol. 2005;168(3):429–39. doi: 10.1083/jcb.200411109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Govek EE, Newey SE, Van Aelst L. The role of the Rho GTPases in neuronal development. Genes Dev. 2005;19(1):1–49. doi: 10.1101/gad.1256405. [DOI] [PubMed] [Google Scholar]
- 61.Vaughan EM, et al. Control of local Rho GTPase crosstalk by Abr. Curr Biol. 2011;21(4):270–7. doi: 10.1016/j.cub.2011.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mulherkar S, et al. The small GTPases RhoA and Rac1 regulate cerebellar development by controlling cell morphogenesis, migration and foliation. Dev Biol. 2014 doi: 10.1016/j.ydbio.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kaartinen V, et al. Abnormal function of astroglia lacking Abr and Bcr RacGAPs. Development. 2001;128(21):4217–27. doi: 10.1242/dev.128.21.4217. [DOI] [PubMed] [Google Scholar]
- 64.Kutys ML, Yamada KM. An extracellular-matrix-specific GEF-GAP interaction regulates Rho GTPase crosstalk for 3D collagen migration. Nat Cell Biol. 2014;16(9):909–17. doi: 10.1038/ncb3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Miller MB, et al. Neuronal Rho GEFs in synaptic physiology and behavior. Neuroscientist. 2013;19(3):255–73. doi: 10.1177/1073858413475486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yan Y, Eipper BA, Mains RE. Kalirin-9 and Kalirin-12 Play Essential Roles in Dendritic Outgrowth and Branching. Cereb Cortex. 2014 doi: 10.1093/cercor/bhu182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Deo AJ, et al. Increased expression of Kalirin-9 in the auditory cortex of schizophrenia subjects: its role in dendritic pathology. Neurobiol Dis. 2012;45(2):796–803. doi: 10.1016/j.nbd.2011.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Harrington AW, et al. The role of Kalirin9 in p75/nogo receptor-mediated RhoA activation in cerebellar granule neurons. J Biol Chem. 2008;283(36):24690–7. doi: 10.1074/jbc.M802188200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sfakianos MK, et al. Inhibition of Rho via Arg and p190 RhoGAP in the postnatal mouse hippocampus regulates dendritic spine maturation, synapse and dendrite stability, and behavior. J Neurosci. 2007;27(41):10982–92. doi: 10.1523/JNEUROSCI.0793-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bustos RI, et al. Coordination of Rho and Rac GTPase function via p190B RhoGAP. Curr Biol. 2008;18(20):1606–11. doi: 10.1016/j.cub.2008.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nimnual AS, Taylor LJ, Bar-Sagi D. Redox-dependent downregulation of Rho by Rac. Nat Cell Biol. 2003;5(3):236–41. doi: 10.1038/ncb938. [DOI] [PubMed] [Google Scholar]
- 72.Ohta Y, Hartwig JH, Stossel TP. FilGAP, a Rho- and ROCK-regulated GAP for Rac binds filamin A to control actin remodelling. Nat Cell Biol. 2006;8(8):803–14. doi: 10.1038/ncb1437. [DOI] [PubMed] [Google Scholar]
- 73.Tsuji T, et al. ROCK and mDia1 antagonize in Rho-dependent Rac activation in Swiss 3T3 fibroblasts. J Cell Biol. 2002;157(5):819–30. doi: 10.1083/jcb.200112107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Etienne-Manneville S. Cdc42--the centre of polarity. J Cell Sci. 2004;117(Pt 8):1291–300. doi: 10.1242/jcs.01115. [DOI] [PubMed] [Google Scholar]
- 75.Nakayama M, et al. Rho-kinase phosphorylates PAR-3 and disrupts PAR complex formation. Dev Cell. 2008;14(2):205–15. doi: 10.1016/j.devcel.2007.11.021. [DOI] [PubMed] [Google Scholar]
- 76.Zhang H, I, Macara G. The PAR-6 polarity protein regulates dendritic spine morphogenesis through p190 RhoGAP and the Rho GTPase. Dev Cell. 2008;14(2):216–26. doi: 10.1016/j.devcel.2007.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]