Abstract
The antiestrogenic drug tamoxifen is widely used in the treatment of estrogen receptor-α-positive breast cancer and substantially decreases recurrence and mortality rates. However, high interindividual variability in response is observed, calling for a personalized approach to tamoxifen treatment. Tamoxifen is bioactivated by cytochrome P450 (CYP) enzymes such as CYP2B6, CYP2C9, CYP2C19, CYP2D6 and CYP3A4/5, resulting in the formation of active metabolites, including 4-hydroxy-tamoxifen and endoxifen. Therefore, polymorphisms in the genes encoding these enzymes are proposed to influence tamoxifen and active tamoxifen metabolites in the serum and consequently affect patient response rates. To tailor tamoxifen treatment, multiple studies have been performed to clarify the influence of polymorphisms on its pharmacokinetics and pharmacodynamics. Nevertheless, personalized treatment of tamoxifen based on genotyping has not yet met consensus. This article critically reviews the published data on the effect of various genetic polymorphisms on the pharmacokinetics and pharmacodynamics of tamoxifen, and reviews the clinical implications of its findings. For each CYP enzyme, the influence of polymorphisms on pharmacokinetic and pharmacodynamic outcome measures is described throughout this review. No clear effects on pharmacokinetics and pharmacodynamics were seen for various polymorphisms in the CYP encoding genes CYP2B6, CYP2C9, CYP2C19 and CYP3A4/5. For CYP2D6, there was a clear gene-exposure effect that was able to partially explain the interindividual variability in plasma concentrations of the pharmacologically most active metabolite endoxifen; however, a clear exposure-response effect remained controversial. These controversial findings and the partial contribution of genotype in explaining interindividual variability in plasma concentrations of, in particular, endoxifen, imply that tailored tamoxifen treatment may not be fully realized through pharmacogenetics of metabolizing enzymes alone.
Key Points
| High interindividual variability in response to tamoxifen treatment of breast-cancer patients calls for a personalized approach to tailor tamoxifen treatment. |
| Various cytochrome P450 (CYP) enzymes have been proposed, and investigated, to affect the pharmacokinetics and pharmacodynamics of tamoxifen, since tamoxifen is bioactivated to more active metabolites (e.g. endoxifen) by these enzymes. |
| CYP2D6 genotype showed a clear gene-exposure effect, but can only partially explain interindividual variability. An exposure-response effect remains controversial. |
| Tailored tamoxifen treatment may not be fully realized through the pharmacogenetics of metabolizing enzymes alone. |
Introduction
Tamoxifen is an antiestrogenic drug, widely used for the treatment of estrogen receptor-α (ERα)-positive breast cancer. Adjuvant tamoxifen treatment substantially reduces breast cancer relapse and mortality rates [1]. Recently, the Adjuvant Tamoxifen: Longer Against Shorter (ATLAS) and adjuvant Tamoxifen-To offer more? (aTTom) trials have suggested the extension of tamoxifen treatment duration from 5 years to 10 years for a subpopulation of premenopausal patients, to further lower recurrence rates [2, 3]. Both pre- and postmenopausal patients are treated with tamoxifen; however, in postmenopausal patients or patients who underwent ovarian ablation, treatment with aromatase inhibitors is effective, either in a sequence, before or after tamoxifen, or for 5 years [4]. Aromatase inhibition does not work in women with active ovarian function, like in premenopausal women [5]. Inhibition of aromatase reduces feedback of estrogens to the hypothalamus-pituitary-ovary axis, leading to an increased stimulation of the ovaries via gonadotropin secretion [6]. This stimulation overrules the effect of aromatase inhibitors. Therefore, tamoxifen is currently the only drug of choice in this subpopulation. Even though a differentiation between ERα-positive and ERα-negative tumors is made prior to treatment, a high interindividual variability in response to adjuvant treatment with tamoxifen is observed [7]. Tailoring tamoxifen therapy was the main focus of an extensive number of studies with emphasis on germline genotyping as a tool to guide treatment. Bioactivation of tamoxifen is mediated by polymorphic cytochrome P450 (CYP) enzymes and may therefore be an important process causally involved in response variability [8]. Bioactivation of tamoxifen results in the formation of metabolites that have different affinity and potency towards ERα [9, 10]. The ERα receptor is known to be the main target in anti-estrogen therapy, while the role of ERβ is still under investigation [11]. The formation of the two major primary metabolites of tamoxifen, N-desmethyl-tamoxifen and 4-hydroxy-tamoxifen, is predominantly catalyzed by CYP3A4/5 and CYP2D6, respectively. The formation of the secondary metabolite 4-hydroxy-N-desmethyltamoxifen (endoxifen) is generated from N-desmethyl-tamoxifen by CYP2D6, and less substantially from 4-hydroxy-tamoxifen by CYP3A4/5 [8]. Endoxifen and 4-hydroxy-tamoxifen are potent antiestrogenic metabolites, with a 100-fold higher affinity for ER and a 30- to 100-fold higher potency in suppressing cell proliferation compared with tamoxifen, pointing towards key roles for CYP2D6 and CYP3A4/5 in the bioactivation of tamoxifen [9, 10]. Since plasma concentrations of endoxifen exceed plasma concentrations of 4-hydroxy-tamoxifen, endoxifen is proposed to be the most important metabolite of tamoxifen [9]. Nevertheless, tamoxifen metabolism has shown to be more complex than solely transformation to endoxifen via CYP2D6, depending on other factors such as serum abundance and the activity of other CYP enzymes such as CYP2B6, CYP2C9, CYP2C19 and CYP3A4/5, as depicted in Fig. 1 [8].
Fig. 1.
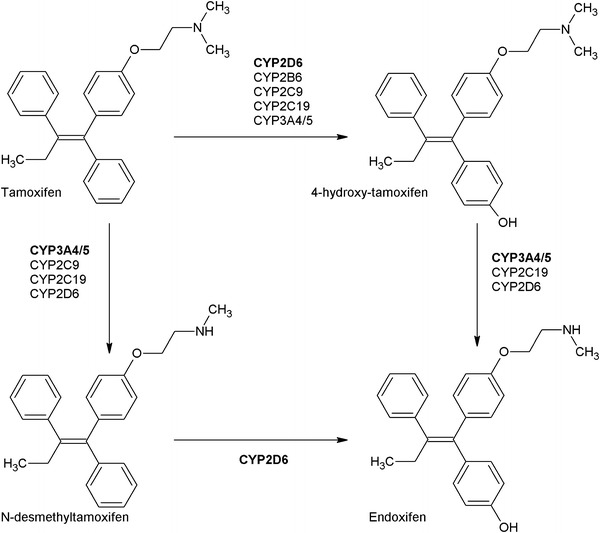
Part of the tamoxifen metabolic pathway. Bold enzymes illustrate a higher extent of contribution to the formation of the metabolite [8]. CYP cytochrome P450
Currently, only CYP2D6 genotyping is proposed to guide tamoxifen treatment, and an AmpliChip® CYP450 test for determination of the genotype has been approved by the US Food and Drug Administration (FDA). The FDA Advisory Committee recommended including pre-treatment genotyping in the drug label of tamoxifen [12]; however, such a recommendation is not included in the current label. Determination of the genotype is suggested to make treatment decisions for both postmenopausal and premenopausal women. Postmenopausal women with low metabolic activity are expected to have lower exposure to an active tamoxifen metabolite and could therefore derive more benefit from either aromatase inhibitors or a higher dose of tamoxifen, as opposed to the standard dose of 20 mg/day. Likewise, premenopausal patients can benefit from a higher dose of tamoxifen when experiencing low metabolic activity since tamoxifen is currently the only drug of choice in the premenopausal setting.
However, controversial findings of various studies, to be discussed in this review, have led to conflicting views on pharmacogenotyping as a tool to guide tamoxifen treatment. Therefore, this article critically reviews the published data regarding the effect of various genetic polymorphisms on the pharmacokinetics and pharmacodynamics of tamoxifen, and aims to review the clinical implications of these findings.
Literature Search
A literature search was performed using the PubMed/MEDLINE database. The following terms were searched in October and November 2014: [(Tamoxifen AND CYP2B6) OR (Tamoxifen AND CYP2C9) OR (Tamoxifen AND CYP2C19) OR (Tamoxifen AND CYP3A4) OR (Tamoxifen AND CYP3A5) OR (Tamoxifen AND CYP2D6)]. Studies including patients with ERα-positive breast cancer undergoing adjuvant treatment with tamoxifen for early-stage breast cancer and investigating an effect of polymorphisms in genes encoding the metabolizing enzymes CYP2B6, CYP2C9, CYP2C19, CYP3A4, CYP3A5, and/or CYP2D6 on pharmacokinetic and/or pharmacodynamic outcome measures were selected. Pharmacokinetic outcome measures included steady-state plasma concentrations of tamoxifen and its metabolites and/or associated metabolic ratios. Pharmacodynamic outcome measures included survival outcomes such as overall survival (OS), (distant or invasive) disease-free survival (DFS), (distant) recurrence-free survival (RFS), (distant) recurrence-free interval (RFI), breast cancer-free interval (BCFI), or any other measurement of breast cancer recurrence risk. Search results were limited to studies conducted in humans and full-text articles available in the English language. Various characteristics of studies and study populations were identified, such as number of patients, dose, concomitant use of CYP2D6 inhibitors and if this was accounted for, deviation from Hardy–Weinberg equilibrium, DNA derived tissue, and menopausal status.
Results of the Literature Search
The described search terms identified 451 papers, 36 of which were found to be eligible for inclusion. Of 451 papers, 102 were reviews, 10 investigated effects in animals, 60 studies were in vitro studies or investigated the metabolism of tamoxifen, 104 studies did not investigate previously described pharmacokinetic or pharmacodynamic outcome measurements, 6 studies were on bioanalytic methods, 23 studies investigated genotyping methods or tumorgenetics, 30 studies investigated drugs other than tamoxifen, and 52 hits consisted of author replies, comments, errata, or editorials. The remaining 64 studies analyzed an effect of polymorphisms on pharmacokinetics and/or pharmacokinetics. Eleven studies investigated effects in non-adjuvant-treated patients, in three studies it was unclear if receptor status was accounted for, and 13 studies did not investigate previously described pharmacokinetic or pharmacodynamic outcome measurements after reading full texts, were of poor methodological quality, or provided an insufficient amount of information; these studies were excluded from the review. Survival outcomes included mainly DFS, RFS and RFI, which were specified as time from surgery or randomization to recurrence. Event-free survival (EFS) was defined as the time from surgery or randomization to occurrence of a defined event; events were specified differently among studies. Characteristics of the 36 included studies are depicted in Table 1.
Table 1.
Characteristics of included studies
| References | Year | PK/PD | Study type | n | Menopausal status | Dose (mg/day) | CYP2D6 inhibitorsa | HWQ | DNAb |
|---|---|---|---|---|---|---|---|---|---|
| [13] | 2013 | PD | RCT | 535 | Post | 30 | − | ++ | T |
| [14] | 2006 | PK | Cohort | 158 | Both | 20 | ++ | − | G |
| [15] | 2013 | PD | Ca–Co | 57 | Both | 20 | ++ | ++ | G |
| [16] | 2013 | PK | Cohort | 135 | Both | 20 | ++ | ++ | G |
| [17] | 2005 | PD | Cohort | 223 | Post | 20 | −− | − | G+T |
| [18] | 2013 | PD | Ca–Co | 319 | Post | 20 | −− | ++ | G+T |
| [19] | 2005 | PK | Cohort | 80 | Both | 20 | ++ | + | G |
| [20] | 2008 | PD | Cohort | 67 | Both | 20 | ++ | − | G |
| [21] | 2010 | PK/PD | Cohort | 282 | Both | 20 | ++ | ++ | G |
| [22] | 2011 | PD | Ca–Co | 494 | Post | – | ++ | ++ | G |
| [23] | 2011 | PK | Cohort | 165 | Both | 20 | ++ | ++ | G |
| [24] | 2011 | PK | Cohort | 1370 | Both | – | ++ | ++ | G |
| [25] | 2011 | PD | Cohort | 190 | Post | 20 | ++ | ++ | T |
| [26] | 2011 | PK | Cohort | 236 | Post | 20 | ++ | + | G |
| [27] | 2014 | PD | Cohort | 99 | Both | – | − | ++ | G |
| [28] | 2005 | PD | Cohort | 162 | Both | – | −− | − | T |
| [29] | 2009 | PD | Cohort | 173 | Both | 20 | ++ | − | G |
| [30] | 2012 | PD | Cohort | 588 | Post | 20 | ++ | + | T |
| [31] | 2012 | PD | Cohort | 1243 | Post | 20 | −− | − | T |
| [32] | 2014 | PK/PD | Cohort | 548 | Pre | 20 | − | ++ | G |
| [33] | 2007 | PD | Cohort | 206 | Both | – | −− | + | T |
| [34] | 2009 | PD | Cohort | 1325 | Both | 20 | −− | − | T |
| [35] | 2013 | PD | Cohort | 30 | Both | – | ++ | ++ | G |
| [36] | 2013 | PK/PD | Cohort | 132 | Both | – | ++ | ++ | G |
| [37] | 2005 | PK/PD | Cohort | 98 | Post | 20 | −− | ++ | G |
| [38] | 2005 | PD | RCT | 50 | Post | 40 | − | − | T |
| [39] | 2007 | PD | Cohort | 119 | Post | 20/40 | ++ | − | T |
| [40] | 2008 | PK/PD | Ca–Co | 152 | Both | 20 | ++ | − | G |
| [41] | 2013 | PK | Cohort | 90 | Both | 20 | ++ | ++ | G |
| [42] | 2008 | PK | Cohort | 151 | Both | 20 | ++ | ++ | – |
| [43] | 2009 | PD | Cohort | 156 | Both | 20 | −− | ++ | T |
| [44] | 2010 | PD | Cohort | 493 | Both | 20 | −− | ++ | G |
| [45] | 2010 | PD | Cohort | 3155 | Both | 20 | ++ | + | G |
| [46] | 2012 | PK/PD | Cohort | 716 | Both | 20 | − | − | G |
| [47] | 2011 | PD | Cohort | 110 | Both | 20 | ++ | − | G |
| [48] | 2011 | PK | Cohort | 117 | Both | 20 | ++ | ++ | G |
PK pharmacokinetic outcomes, PD pharmacodynamic outcomes, RCT randomized controlled trial, Ca–Co case–control study, Post postmenopausal, Pre premenopausal, Both postmenopausal and premenopausal, CYP cytochrome P450, HWQ Hardy–Weinberg equilibrium, G germline DNA, T tumor tissue extracted DNA, ++ indicates yes, + indicates in part, − indicates unknown, −− indicates not
aAccounted for CYP2D6 inhibitors
bSource of DNA
Study Designs
As depicted in Table 1, a variety of study designs were used to determine the effects of polymorphisms in metabolic enzymes on pharmacokinetic and pharmacodynamic outcomes. Studies investigating the effect of polymorphisms on plasma concentrations were mostly well-designed, prospective cohort studies, while studies investigating the effect of polymorphisms on survival outcome were predominantly designed as retrospective cohort studies and, to a lesser extent, as case–control studies. Cohort studies solely included patients treated with tamoxifen and analyzed whether polymorphisms had an impact on survival in this patient group. Case–control studies compared incidences of recurrences in patients carrying variant alleles (cases) and patients carrying the wild-type genotype (controls) or compared hazard ratios (HRs) of both groups. Cases and controls were both treated with tamoxifen. Since prognosis can differ between patients, most analyses were multivariate analyses correcting for nodal status and tumor grade and stage because these factors are known to influence survival outcome. What is not known is whether CYP variant alleles can also influence prognosis. In most studies described throughout this review, only tamoxifen-treated patients have been studied. This precludes any definitive conclusion regarding either prognostic or predictive value of the CYP variant because outcome after tamoxifen is a combination of prognosis and treatment effect (prediction). In studies where the CYP variant group had a multivariate corrected, poorer outcome than the CYP wild-type group after tamoxifen treatment, any conclusion that this CYP variant was causal in lower endoxifen concentrations and therefore reduced efficacy of tamoxifen is premature. To discern the predictive effect from the prognostic effect of polymorphisms in CYP enzymes on survival outcome, a randomized controlled trial (RCT) or case–control design should be used, with four patient subgroups [49]: patients with and without the CYP polymorphism of interest, and patients with and without the treatment of interest. Studies by Beelen et al. and Wegman et al. [13, 38] investigated the prognostic value of the CYP2C19*2 and CYP2D6*4 variant alleles, respectively. Interestingly, the CYP2C19*2 variant conferred an adverse prognosis in the absence of treatment, while patients with this variant allele derived significantly more benefit from adjuvant tamoxifen than patients without this variant [13]. While reading this review, it is crucial to keep in mind that if the four subgroups are not included in the study design, conclusions regarding prognosis and/or prediction will not have any influence on patient care.
Effect of Polymorphisms on Pharmacokinetic and Pharmacodynamic Outcome Measures
For each CYP enzyme, the effect of various polymorphisms on pharmacokinetic and pharmacodynamic outcome measures will be described.
CYP2B6
CYP2B6 plays a role in the formation of the primary metabolites 4-hydroxy-tamoxifen. CYP2B6 enzymes can show different metabolic activities based on their polymorphic state [8]. Over 50 allelic variations of CYP2B6 are described, but not all associated metabolic activities are known. CYP2B6*4 shows an increased in vivo metabolic activity, and CYP2B6*6, *16 and *26 allelic variations show a decreased metabolic activity [50].
Regarding pharmacokinetic outcome measures, no association between the CYP2B6*6 genotype and endoxifen concentrations, 4-hydroxy-tamoxifen concentrations, or the metabolic ratio of tamoxifen concentration over 4-hydroxy-tamoxifen concentration (MRTAM/4OHT) was found [26, 36]. Additionally, CYP2B6*6 polymorphism was not associated with significantly different relapse-free time (RFT) [27]. The definition of RFT was in line with the definition of RFI, as described by Hudis et al. [51]. In addition, no association was found between the CYP2B6 genotype and EFS or OS [11].
CYP2C9
CYP2C9 contributes to the formation of the primary tamoxifen metabolites N-desmethyl-tamoxifen and 4-hydroxy-tamoxifen, albeit to a lesser extent than CYP2D6 and CYP3A5 isoforms. [52] The metabolic activity of CYP2C9 can be normal (*1A), decreased (*3, *5, *8, *11A, *13), or absent (*6) [50].
Regarding pharmacokinetics, in the studies by Teft et al. (no p-values reported) and Jin et al. (p-values >0.05) no significant difference was found in mean plasma concentrations of tamoxifen or its metabolites between patients carrying two wild-type alleles or carriers of either heterozygous or homozygous variant alleles of CYP2C9*2 and CYP2C9*3 [19, 36]. Lim et al. [23] found similar results regarding CYP2C9*3 and the influence on tamoxifen and metabolite concentrations. In contrast, a significant difference in the formation of 4-hydroxy-tamoxifen from tamoxifen (p = 0.007) between homozygous wild-type carriers and carriers of CYP2C9*2 and/or *3 alleles and significant lower plasma concentrations of 4-hydroxy-tamoxifen (p = 0.0006) and endoxifen (p = 0.0024) were found [26, 32].
Regardless of the significant difference in formation of 4-hydroxy-tamoxifen and endoxifen and 4-hydroxy-tamoxifen concentration, no association between genotypes and treatment outcome, survival, or RFT has been reported [27, 33]. The definition of RFT was in line with the definition of RFI, as described by Hudis et al. [51].
CYP2C19
CYP2C19 activity could alter tamoxifen metabolism and exposure to its metabolites via catalyzation of the conversion of tamoxifen into 4-hydroxy-tamoxifen [8]. CYP2C19*2 and *3 variant alleles showed no metabolic activity, whereas CYP2C19*17 showed increased metabolic activity due to increased transcriptional activity [50].
No significant correlation between CYP2C19 genotypes and concentrations of tamoxifen or its metabolites (p > 0.05) were found by Lim et al. [23]. Mürdter et al. [26] underlined these results, finding no correlation between CYP2C19*3 or CYP2C19*17 and plasma concentrations of endoxifen and 4-hydroxy-tamoxifen or associated metabolic ratios.
Regarding survival outcome measures, Okishiro et al. [29] found no significant difference between genotypes of CYP2C19 and RFS in Japanese patients with breast cancer treated with adjuvant tamoxifen [HR 0.37, 95 % confidence interval (CI) 0.08–176; p = 0.19]. In addition, no significant impact on RFT was found for CYP2C19 variant allele carriers [27], and heterozygous carriers of a CYP2C19 variant allele did not significantly impact DFS (HR 0.93 95 % CI 0.47–1.84; p = 0.829) [14]. In addition, Moyer et al. [25] did not find a significant difference between the CYP2C19*17 genotype and DFS.
The study by Schroth et al. [33] investigated the impact of single nucleotide polymorphisms (SNPs) on RFT, EFS, and OS, but found no significant correlations between CYP2C19*2 and/or *3 carriers and these survival outcomes. However, in carriers of CYP2C19*17, improvement in RFT was found (HR 0.45, 95 % CI 0.21–0.92; p = 0.03) but this was not significant for EFS (HR 0.58, 95 % CI 0.32–1.01; p = 0.05) and OS (HR 0.61, 95 % CI 0.29–1.26; p = 0.18).
Beelen et al. [13] investigated the prognostic value of the CYP2C19*2 variant allele, comparing patients using tamoxifen with patients not using tamoxifen for both CYP2C19*2 carriers and patients with wild-type genotype. Patients carrying at least one CYP2C19*2 variant allele showed an improved RFI (HR 0.26; p = 0.001), while patients without this allele derived less benefit (HR 0.68; p = 0.18). Interestingly, breast-cancer patients carrying the CYP2C19*2 variant allele had a poor prognosis in the absence of adjuvant tamoxifen (HR 2.5) compared with patients without a variant allele. As explained by the authors, CYP2C19 exposure affects the metabolism of tamoxifen as well as estrogen catabolism. The non-functional CYP2C19*2 causes higher exposure to estrogens, leading to a possible higher susceptibility to tumors that are dependent on estrogen signaling. Therefore, these patients could be more sensitive to estrogen-inhibiting therapy, explaining the more beneficial HR in the CYP2C19*2 subgroup.
CYP3A4/5
CYP3A4/5 enzymes catalyze the formation of tamoxifen into different active metabolites, of which transformation into N-desmethyl-tamoxifen from tamoxifen and endoxifen from 4-hydroxy-tamoxifen are the most important [8]. The CYP3A4*22 polymorphism shows decreased metabolic activity, and CYP3A5*3 and CYP3A5*6 polymorphisms show no metabolic activity; therefore, lower endoxifen and N-desmethyl-tamoxifen concentrations leading to decreased response are expected to be associated with these polymorphisms [50].
Regarding the influence of CYP3A4/5 polymorphisms on the pharmacokinetics of tamoxifen, various studies have been conducted. Teft et al. unexpectedly found higher endoxifen (p < 0.05) concentrations for CYP3A4*22 carriers, as well as higher concentrations of tamoxifen (p < 0.0001), N-desmethyl-tamoxifen, 4-hydroxy-tamoxifen and other, less relevant, metabolites. Since CYP3A4*22 polymorphism shows a decreased metabolic activity, higher metabolite concentrations are not expected; however, tamoxifen concentrations were also elevated. Therefore, it is suggested that intestinal CYP3A4 activity was decreased, leading to reduced first-pass metabolism, increasing the concentration of tamoxifen and subsequently its metabolites. The study also investigated the combination of CYP2D6 and CYP3A4 polymorphisms. In patients with low CYP2D6 metabolic activity, the CYP3A4*22 allele carriers had endoxifen concentrations above a set threshold of 6.72 ng/ml compared with subtherapeutic concentrations in patients with low CYP2D6 metabolic activity and CYP3A4 wild-type. These findings indicate that CYP3A4*22 polymorphism is more important in CYP2D6 poor metabolizers [36]. This threshold was based on the 20th percentile of endoxifen concentrations in the enrolled patients because, in the study by Madlensky et al., patients with endoxifen concentrations in the lowest quintile were at the highest risk of recurrence [24, 36].
In the study by Tucker et al. [37] no significant differences were seen for tamoxifen, N-desmethyl-tamoxifen, or 4-hydroxy-tamoxifen concentrations in patients carrying at least one variant CYP3A5*3 or CYP3A5*6 allele. The influence of CYP3A5 polymorphisms on endoxifen concentrations was not investigated and possible other polymorphisms were not taken into account. Although the study by Jin et al. [19] found higher steady-state mean plasma concentrations of endoxifen in patients with at least one functional allele (82.0 nM; range 56.2–107.8) compared with patients with no functional alleles (58.1 nM; range 49.3–66.9), no significant associations were found between CYP3A5*3 homozygous carriers and any of the metabolite concentrations (tamoxifen, p = 0.98; 4-hydroxy-tamoxifen, p = 0.57; N-desmethyl-tamoxifen, p = 0.99). Additional studies did not find a correlation between carriers of CYP3A5*3 alleles and tamoxifen or tamoxifen metabolite steady-state concentrations or their metabolic ratios [8, 22, 29].
Considering pharmacodynamic survival outcomes, the study by Goetz et al. [17] found that the CYP3A5*3 variant was not associated with RFS, DFS, or OS. Furthermore, no associations between the CYP3A5*3 variant allele and treatment outcome or survival were found in the study by Schroth et al. [33].
Both multivariate and univariate analyses by Wegman et al. [39] showed unexpected improved RFS (multivariate: HR 0.13, 95 % CI 0.02–0.86; p = 0.03) in homozygous carriers of CYP3A5*3 treated with tamoxifen for 5 years.
The gene-exposure effect for CYP3A4/5 polymorphisms and tamoxifen is less clear than that for CYP2D6. The study by Teft et al. [36] investigated the relevance of the CYP3A4*22 polymorphism in different CYP2D6 genotype groups, indicating that the CYP3A pathway becomes more relevant if CYP2D6 metabolic activity is decreased.
CYP2D6
Two of the most potent metabolites of tamoxifen, 4-hydroxy-tamoxifen and endoxifen, are predominantly generated by CYP2D6 [8]. More than 100 allelic variants of CYP2D6 with different metabolic activities are currently known. Metabolic activity can either be normal (*1, *2, *33, *35), decreased (*9, *10, *17, *29, *41, *69), absent (*3, *4, *6, *7, *8, *11–*15, *18–*21, *31, *38, *40, *42, *44) or increased (*2XN, *35X2) [50]. To facilitate comparison, the predicted phenotype is derived from the genotype, enabling classification of metabolizers into four different groups: poor metabolizer (PM), intermediate metabolizer (IM), extensive metabolizer (EM), or ultrarapid metabolizer (UM).
Study results regarding the effect of CYP2D6 polymorphisms on pharmacokinetic and pharmacodynamic parameters are depicted in Tables 2 and 3, respectively.
Table 2.
Results for CYP2D6 polymorphisms and their effect on pharmacokinetic parameters
| Variant alleles | References | Outcome | Comparison | Significance | ||
|---|---|---|---|---|---|---|
| 3–8,11,14A,15,19,20,40,4x | [24] | C ss T+M1–3 | EM/EM vs. Various comb | T (NS); M1–3 (p < 0.001) M3 45 % explained by genotype |
||
| 3,4,5,6 | [19] | C ss T+M1–3 | wt/wt vs. wt/* or */* | M3 (p = 0.003) | ||
| 3,4,6,7,8,9,10,41 | [26] | C ss T+M1–3 MRDMTAM/END | EM/EM vs. Various comb | M3: 39 % explained by genotype M2: 9 % explained by genotype |
||
| 3–6,9,10,41,14,15,17 | [32] | MRDMTAM/END | CYP2D6 activity score | p < 10−77 | ||
| 3,4,8,10,41 | [36] | C ss T+M1–3 | EM/EM vs. Various comb | M3 significant | ||
| 5,10,41 | [23] | C ss T+M1–3 | wt/wt vs. wt/*5, wt/*10: *10/*10,*5/*10 | M1 (p = 0.077) and (p = 0.006) M3 (p < 0.001); M1(*10) (p = 0.011) |
||
| wt/* vs. *5/*10 | M3 (p = 0.001) | |||||
| 2–6,10,41 | [41] | C ss T+M1–3 | EM vs. PM | M1,3 (p < 0.001) | ||
| 3–6,9,10,17,41 | [16] | C ss T+M1–3 | EM/EM vs. PM/PM | M3 (p < 0.001) | ||
| 33 Alleles | [14] | MREND/DMTAM | wt/wt vs. wt/* vs */* | p < 0.001 | ||
| 4,5,10,36,41,21 | [21] | C ss T+M1–3 | wt/wt vs. wt/* or */* | M2,3 (p < 0.01) both | ||
| 2–6 | [42] | C ss T+M1–3 | EM/EM vs. EM/* vs. PM vs. UM | M1 (p = 0.001); M3 (p = 0.001) | ||
| 5,10,41 | [46] | C ss T+M1–3 | wt/wt, wt/* vs. */* | M2,3 (p < 0.001) | ||
| 2,2A,2AxN,4–6,9,10,17,41 | [48] | M1–3 | CYP2D6 activity score | M3 (p = 0.0009), Z-endoxifen (p < 0.0001) | ||
CYP cytochrome P450, C ss steady-state concentration, comb combinations, T tamoxifen, M tamoxifen metabolite; M 1 N-desmethyl-tamoxifen, M 2 4-hydroxy-tamoxifen, M 3 endoxifen, MR metabolic ratio, EM extensive metabolizer, PM poor metabolizer, UM ultrarapid metabolizer, NS not significant, MR DMTAM/END metabolic ratio of N-desmethyl-tamoxifen concentration over endoxifen concentration, MR END/DMTAM metabolic ratio of endoxifen concentration over N-desmethyl-tamoxifen concentration, wt/wt two wildtype alleles, wt/* one wildtype allele and one polymorphic allele, */* two polymorphic alleles
Table 3.
Results for CYP2D6 polymorphisms and their effect on pharmacodynamic parameters
| Variant alleles | References | Outcome | Comparison | Significancea | Remarksb | ||
|---|---|---|---|---|---|---|---|
| No significant results | |||||||
| 4 | [39] | RFS | wt/wt vs. wt/* + */* | NS | Only results for the 5-year tamoxifen treatment arm are included in this table | ||
| 3–6,10,41 | [27] | RFT | wt/wt vs. wt/* and/or */* | NS | – | ||
| 10 | [29] | RFS | wt/wt, wt/* vs. *10/*10 | NS | Lack of statistical power | ||
| 4 | [44] | TTP, PFS | wt/wt vs. wt/*4 or *4/*4 | NS | – | ||
| 5,10,41 | [46] | RFS | wt/wt vs. wt/* or */* | NS | Possible misclassification of genotypes | ||
| 4 | [22] | Recurrence | wt/wt vs. *4/*4 or *4/*1 | NS | – | ||
| 4 | [28] | OS, RFS | wt/wt vs. wt/* + */* | NS | Heterogeneous study population | ||
| 4 | [17] | RFT, DFS, OS | wt/wt, wt/ vs. *4/*4 | NS | In the univariate analysis, RFT and DFS were significantly worse for the CYP2D6 *4/*4 genotype | ||
| 10 | [43] | DFS, DDFS, BCSS, OS | wt/wt vs. wt/* or */* | NS | – | ||
| 4–6,9,10,41,UM | [45] | BCSS, OS | wt/wt vs. Any genotype | NS | In the unadjusted analysis, CYP2D6*6 (PM) were at increased risk of BCSS: HR 2.14 (95 % CI 1.05–4.36) | ||
| 2–5,10,14,18,21,41,49,52,60 | [47] | RFS, OS | EM and IM vs. PM | NS | Univariate analysis showed some significance in OS and RFS; potential lack of statistical power | ||
| Significant results | |||||||
| 4,5,10,41 | [33] | RFT, EFS, OS | EM/EM vs. wt/* or */* | RFT, EFS (p = 0.02); OS (NS) | Note: OS is not significant | ||
| 3–6,9,10,41,14,15,17 | [32] | DRFS | CYP2D activity score | p = 0.013 | Potential lack of power, potential selection bias | ||
| 4,5,10,36,41 | [15] | DFS | wt/wt vs. wt/* or */* (*10) | DFS postmenopause (p = 0.046) | Potential lack of power | ||
| 4,5,10,41,21 | [20] | RFS | wt/wt vs. *10/*10 | p = 0.0057 | Potential selection bias | ||
| 4,5,10,36,41,21 | [21] | RFS | wt/wt vs. wt/* or */* | p = 0.00036 | Potential bias in time of inclusion, cross-sectional study design | ||
| 10 | [40] | RFS | wt/wt, wt/* vs. */* | p = 0.04 | Potential bias | ||
| 4 | [38] | DRFS | wt/* or */* tamoxifen vs. wt/* or */* no tamoxifen | p = 0.0089, longer DRFS | Potential selection bias; different comparison | ||
| Most recent trials | |||||||
| 3,4,6,10,41 | [18] | IDFS | EM/EM vs. PM/IM or PM/EM | p = 0.04 and p = 0.07 | ABCSG 8 trial | ||
| 3,4,5,10,41 | [34] | TTR, EFS, DFS, OS | EM vs. EM/IM and PM | TTR (p < 0.001), EFS (p = 0.003) DFS (p = 0.005) |
– | ||
| 2,3,4,6,10,41 | [30] | Recurrence | EM vs. PM | NS | ATAC trial | ||
| 2,3,4,6,7,10,17,41 | [31] | BCFI | EM vs. PM and/or IM | NS | BIG 1-98 trial | ||
CYP cytochrome P450, IDFS invasive disease-free survival, TTR time to recurrence, BCFI breast cancer-free interval, RFT relapse-free time, EFS event-free survival, OS overall survival, RFS recurrence-free survival, DRFS distant recurrence-free survival, DFS disease-free survival, DDFS distant disease-free survival, BCSS breast cancer-specific survival, PFS progression-free survival, TTP time to tumor progression, EM extensive metabolizer, IM intermediate metabolizer, PM poor metabolizer, UM ultrarapid metabolizer, NS not significant, HR hazard ratio, CI confidence interval, ABCSG Austrian Breast and Colorectal Cancer Study Group, ATAC Armidex, Tamoxifen, Alone or in Combination, BIG Breast International Group, wt/wt two wildtype alleles, wt/* one wildtype allele and one polymorphic allele, */* two polymorphic alleles
aOutcomes of multivariate analysis, if available
bIn addition to the characteristics in Table 1
All 13 reports investigating the associations between CYP2D6 polymorphisms and pharmacokinetics found a significant effect of genotype on endoxifen concentrations and/or the formation of endoxifen from N-desmethyl-tamoxifen [14, 16, 19, 21, 23, 24, 26, 32, 36, 41, 42, 46, 48]. For N-desmethyl-tamoxifen and 4-hydroxy-tamoxifen a significant effect of CYP2D6 variant alleles was indicated by four and three studies, respectively [21, 23, 24, 41, 42, 46]. None of the studies indicated a correlation between genotype and tamoxifen concentrations.
Four studies [21, 23, 24, 41, 42, 46] estimated to what extend CYP2D6 polymorphisms could explain the variability in endoxifen concentrations by testing CYP2D6 activity as a covariate using linear models. Mürdter et al. [26] found that CYP2D6 polymorphisms explained 39 % of variability in endoxifen concentrations. Teft et al. [36] found a similar contribution of 30 %, Saladores et al. [32] found a contribution of 53 %, and Madlensky et al. [24] indicated that the CYP2D6 genotype, together with age and body mass index (BMI), explained 46 % of the variability in endoxifen concentrations.
Madlensky et al. indicated a threshold of 5.97 ng/ml for endoxifen. Patients with endoxifen concentrations above 5.97 ng/ml had lower recurrence rates (HR 0.74, 95 % CI 0.55–1.00) based on patient plasma concentrations of endoxifen and associated DFS times. Even though the majority of PMs had low endoxifen concentrations, 24 % were still able to generate endoxifen concentrations above the threshold of 5.97 ng/ml [24]. The study by Teft et al. [36] used a comparable threshold of 6.72 ng/ml. This threshold was based on the 20th percentile of endoxifen concentrations in enrolled patients, since patients with endoxifen concentrations in the lowest quintile were at highest risk of recurrence in the study conducted by Madlensky et al. The majority of PMs failed to generate an endoxifen concentration above a threshold of approximately 6.72 ng/ml.
With regard to pharmacodynamic outcomes, findings are more controversial. Various studies were conducted to clarify the influence of different polymorphisms of CYP2D6 on the pharmacodynamics of tamoxifen. The results of these studies are categorized and presented in Table 2. The first 11 studies showed no significant association between CYP2D6 polymorphisms and different types of survival outcome [17, 22, 27–29, 39, 43–47]. In contrast, seven studies indicated a significant association between CYP2D6 polymorphisms and different survival outcomes [15, 20, 21, 32, 33, 38, 40].
Only six studies investigated an effect of CYP2D6 polymorphisms on OS; however, none of these studies showed significant results [17, 28, 33, 43, 45, 47].
Four trials and a meta-analysis were of great importance in settling the controversy between positive and negative findings for an effect of CYP2D6 polymorphisms on clinical outcome: the Breast International Group (BIG) 1-98 trial [31], the Armidex, Tamoxifen, Alone or in combination (ATAC) trial [30], the Austrian Breast and Colorectal cancer Study Group (ABCSG) 8 trial [18], and the International Tamoxifen Pharmacogenomics Consortium (ITPC) meta-analysis [53]. The BIG 1-98 trial [31] and the ATAC trial [30] demonstrated no evidence for an association between CYP2D6 genotype and recurrence. However, both studies have been criticized: the BIG1-98 trial showed strong deviation from Hardy–Weinberg equilibrium, and the ATAC trial had a lack of statistical power since less than 19 % of patients randomized to tamoxifen were analyzed. However, the relevance of meeting Hardy–Weinberg equilibrium in a study reflecting clinical practice is questioned in an editorial by Berry [54]. In contrast, the ABCSG 8 trial showed that CYP2D6 PMs had a significantly higher rate of recurrence and death in patients treated with tamoxifen monotherapy for 5 years. For patients carrying two PM alleles this effect was significant (odds ratio [OR] 2.45, 95 % CI 1.05–5.73; p = 0.04), and for patients carrying one PM allele (OR 1.67, 95 % CI 0.95–2.93; p = 0.07) a trend was observed [18]. Schroth et al. found similar results; patients with reduced CYP2D6 activity, carrying either one or two PM alleles, had significantly shorter time to recurrence (HR 1.40, 95 % CI 1.04–1.90, and HR 1.90, 95 % CI 1.10–3.28, respectively). In addition, the effects on EFS (HR 1.33, 95 % CI 1.06–1.68) and DFS (HR 1.29, 95 % CI 1.03–1.61) showed significance, but the effect on OS was not significant (HR 1.15, 95 % CI 0.88–1.51), comparing EMs with heterozygous and homozygous carries of PM alleles together [34]. The ITPC meta-analysis by Provence et al. defined three groups of inclusion criteria, of which criteria 1 was the most restrictive (including ER-positive breast-cancer patients receiving tamoxifen 20 mg daily for 5 years). In this subgroup, CYP2D6 PM status was associated with shorter DFS (HR 1.25, 95 % CI 1.06–1.47; p = 0.009). However, when tamoxifen duration, menopausal status, and annual follow-up were not specified, no significant association was seen (HR 1.17, 95 % CI 0.90–1.52; p = 0.25) [criteria 2] and non-significance remained when no exclusions were applied (HR 1.07, 95 % CI 0.92–1.26; p = 0.38) [criteria 3]. The meta-analysis concluded that high restrictiveness of patient groups validates CYP2D6 genotyping [53]; however, the credibility of this study has been questioned, in part due to the lack of prospectively defining the endpoint, selection bias, and omitting OS [55].
The study by Wegman et al. [38] investigated whether or not the CYP2D6*4 variant allele was of prognostic value. Patients carrying at least one CYP2D6*4 allele had significantly improved benefit from tamoxifen treatment (p = 0.0089); for the wild-type CYP2D6, this benefit was not significant.
Thus, based on these studies it can be concluded that CYP2D6 activity has a clear effect on endoxifen concentrations, advocating a gene-exposure effect. However, interindividual variability in endoxifen concentrations can only, in part, be explained by CYP2D6 genotypes or predicted phenotypes. Whether this also translates into less efficacy of tamoxifen in CYP2D6 PMs remains controversial. As depicted in Tables 1 and 3, included studies investigating the effect of polymorphisms on survival outcome had various weaknesses and differences regarding characteristics, statistical power, methodological quality, and study design. Therefore, combining results of different studies and drawing a clear conclusion is challenging. Potential biases in a subset of studies are more extensively described in a previous review [56].
Discussion
Review of the published data on the effect of various genetic polymorphisms shows that interindividual variability in response to tamoxifen treatment cannot sufficiently be explained by genotype variability. A conclusive answer to whether genotyping is of clinical value for patients to be treated with tamoxifen is currently not available, which is mainly caused by the controversial outcomes of multiple studies, partially explained by high interstudy heterogeneity and methodological flaws in different studies. Different factors contribute to interstudy heterogeneity, such as differences in quantification of tamoxifen and metabolites, registration of co-medication, administered dose, time on tamoxifen treatment, compliance, genotype comparison, tissue used for genotyping, deviation from Hardy–Weinberg equilibrium, specification of survival outcome, statistical power, methodology, and study design. Additionally, studies are selective on what polymorphisms are taken into account, leading to potential misclassification of phenotypes.
Regardless of the extensive heterogeneity between studies, none of the conducted trials reported consistent evidence for an effect of polymorphisms in CYP2B6, CYP2C9, and CYP2C19 encoding genes on the pharmacokinetics and/or pharmacodynamics of tamoxifen. For CYP3A5 polymorphisms, there was no clear gene-exposure effect, but CYP3A4*22 showed significantly higher concentrations of endoxifen, probably attributed to higher tamoxifen concentrations. In addition, CYP2D6 PMs benefited from CYP3A4*22, resulting in higher endoxifen concentrations compared with CYP2D6 PMs lacking this genomic variation. No studies linked CYP3A4 polymorphisms to outcome. No association between CYP3A5 polymorphisms and survival outcome was found, except for the unexpected association between CYP3A5*3 homozygous carriers and improved RFS [39]. Nevertheless, further investigation is needed to determine if the CYP3A4/5 pathway in tamoxifen metabolism, and therefore its polymorphic state, becomes more important with decreasing CYP2D6 activity.
For CYP2D6, all indicated studies clearly show a significant gene-exposure effect. However, interindividual variability in endoxifen concentrations can only, in part, be attributed to the CYP2D6 genotype. This partial contribution might be a reason for the controversy seen in trials aimed at finding an association between variant allele carriers of CYP2D6 and survival outcomes. In addition, CYP enzymes are also known to play a role in estrogen metabolism. CYP3A4, for example, catalyzes the conversion of estradiol to 2-hydroxyestradiol (E2). E2 inhibits cellular proliferation, therefore SNP-induced alterations in CYP3A4 activity can affect tumor development itself, apart from its effect on tamoxifen metabolism and outcome [57, 58]. CYP2C19 polymorphisms are also known to affect estrone (E1) and E2 catabolism. High concentrations of E1 were seen in patients carrying either one or two CYP2C19*2 variant alleles, and low E2 concentrations were associated with the CYP2C19*17 genotype [59]. CYP2C19*2 variant allele carriers have been shown to be at a higher risk of developing breast cancer, and the prognosis in these patients in the absence of treatment is poor. However, these tumors are more sensitive to anti-estrogen treatment, rendering their prognosis after adjuvant tamoxifen treatment similar to breast-cancer patients with wild-type CYP2C19 [13].
While the debate continues on whether or not genotyping of CYP2D6 prior to adjuvant treatment with tamoxifen should be implemented, further validation for genotyping and other approaches to personalize treatment with tamoxifen should be explored.
To truly settle controversy on whether or not to use genotyping, previously described factors contributing to interstudy heterogeneity should be addressed in future attempts. Some selected points to consider are discussed shortly. For pharmacokinetic-oriented studies, discrepancies in quantitative analysis of tamoxifen and metabolite concentrations should be addressed. Lack of bioanalytical method selectivity can result in misinterpreting plasma concentrations. A selective liquid chromatography–tandem mass spectrometry (LC–MS/MS) method for the quantification of tamoxifen and metabolites is preferred [60]. Coadministration of CYP2D6 inhibitors, such as antidepressants, can alter exposure to active metabolites of tamoxifen and subsequently alter survival outcomes [61]. Therefore, patients using medication that interferes with CYP2D6 metabolism should be excluded, or co-medication should be registered. In addition, it is not preferable to use tumor tissue as a source for germline DNA since loss of heterozygosity at the CYP2D6 locus in breast tumors has been described [62]. Using an insensitive technique to analyse tumor tissue-derived DNA can cause misclassification of genotypes [62]. In order to prevent misclassification through incomprehensive allele coverage, validated tests should be used to ensure accurate CYP2D6 genotyping [63]. A major drawback for all studies testing an effect of polymorphisms on clinical outcome is the retrospective study design. Prospective studies, with prospectively defined endpoints and sufficient sample size, are needed to validate further recommendations [55, 64]. Post hoc analyses of prospective RCTs and case–control studies including four subgroups can be a valuable alternative for prospective studies. Since polymorphisms in metabolic enzymes can also be of prognostic value, a distinction between the prognostic and predictive value of a polymorphism in a metabolic enzyme should be made. A post hoc analysis of an RCT including an untreated control group can identify such a distinction. Once a prognostic biomarker is identified, it can be corrected for in a multivariate analysis [49].
In addition to optimization of future trials, two effects should be validated to decide upon the clinical value of genotyping: (1) a clear gene-exposure effect, and (2) a clear exposure-response effect. For CYP2D6, a clear gene-exposure effect is reported for endoxifen, as described in this review. However, the variability in plasma concentrations of endoxifen can be partially attributed to the CYP2D6 genotype, and the residual variability remains unexplained. Therefore, genotyping of CYP2D6 might not sufficiently predict exposure and, consequently, might not be applicable as a biomarker for tamoxifen treatment response. Other factors, contributing to metabolite concentration variability, should be identified and quantified. Subsequently, these factors, and the genotype, could be of clinical value to tailor tamoxifen treatment. In addition, tamoxifen and other active metabolites have different pharmacological activities and could contribute, in other extents, to treatment outcomes [48].
An exposure-response effect can be validated by studies linking tamoxifen or metabolite concentrations to clinical outcome. This has been investigated retrospectively by Madlensky et al. [24] where endoxifen concentrations below 5.97 ng/ml correlated with more recurrences, while Saladores et al. [32] indicated that patients with endoxifen concentrations below a threshold of approximately 5.30 ng/ml were at higher risk for distant relapse or death. Additional prospective research is preferred to further validate an exposure-response relationship; however, conducting a prospective trial in the adjuvant setting is nearly impossible. Therefore, evidence from different trial settings, such as post hoc analyses of RCTs, prospectively collected cohort data in the metastatic setting, and case–control studies, should be combined in order to support an exposure-response effect.
Since there is, as yet, no conclusive predictor for exposure, measurement of plasma concentrations of tamoxifen and active metabolites could be suggested to establish exposure, ensuring the true phenotype of patients. Therapeutic drug monitoring (TDM) has advantages over the measurement of factors contributing to endoxifen exposure, such as genotype. TDM can identify EMs, or even UMs, with endoxifen concentrations below the threshold, which would have stayed unexposed using genotyping. On the other hand, not all PMs have endoxifen concentrations under the proposed threshold. This is supported by Madlensky et al. [24] who indicated that 24 % of the PMs were still able to generate therapeutic concentrations of endoxifen, and Teft et al. [36] who indicated that PMs were able to generate endoxifen, despite the lack of metabolic activity of CYP2D6. Therefore, a risk of unnecessarily high dosing might exist if treatment is only based on genotyping. In addition, TDM could identify non-compliance. However, endoxifen steady-state concentrations are only met after 1–4 months of treatment. Since steady-state endoxifen plasma concentrations are used to tailor tamoxifen treatment, a risk-period of suboptimal treatment exists between the start of treatment and the time of steady state. This short timeframe of risk will not be of clinical relevance since tamoxifen is indicated to reduce recurrence and mortality rates after years of treatment. Nevertheless, this problem could potentially be addressed by using a population pharmacokinetic model to predict steady-state plasma concentrations of endoxifen in an early stage of tamoxifen treatment [65]. Moreover, a population pharmacokinetic model could guide tamoxifen dosing from an early stage.
Both genotyping and TDM rely on the assumption that exposure is correlated with survival outcome. To anticipate either low concentrations or low metabolic activities of CYP2D6, a dose-exposure effect needs to be validated. Previous studies provide evidence for such a dose-exposure effect. An increase of tamoxifen dose from 20 mg daily to 30 or 40 mg daily, increases endoxifen concentrations [48, 66, 67]. In addition, endoxifen concentrations in CYP2D6 PMs and IMs treated with 40 mg of tamoxifen were comparable to CYP2D6 EMs treated with 20 mg, outlining the feasibility of dose adjustment based on TDM measurements [68]. Regardless of its feasibility, safety of dose adjustments should also be investigated. Several studies have investigated the toxicity of a dose increase of tamoxifen, but no data on long-term toxicity were included [69, 70].
Conclusions
No clear effects on pharmacokinetics and pharmacodynamics were seen for various polymorphisms in the CYP encoding genes CYP2B6, CYP2C9, CYP2C19, and CYP3A4/5, based on the reviewed data. For CYP2D6, there was a clear gene-exposure effect that was able to partially explain the interindividual variability in endoxifen plasma concentration; however, a clear exposure-response effect remained controversial. Even though the effects of polymorphisms on the pharmacokinetics and pharmacodynamics of tamoxifen are rationalized by its well-understood metabolism, the genotype remains a surrogate parameter for the plasma concentration of tamoxifen and its metabolites, hampering the clinical applicability of genotyping. Based on existing evidence for a link between exposure and response to tamoxifen, TDM seems to be the best approach for tailored tamoxifen treatment at the moment. However, to truly validate genotyping or any other tailored treatment of tamoxifen, additional studies linking metabolite concentrations to clinical outcome, as well as studies on toxicity, are needed, in addition to studies investigating to what extent tamoxifen and other metabolites contribute to the antiestrogenic effect of tamoxifen.
Acknowledgments
Professor S. C. Linn received research funding from A Sister’s Hope. Aurelia H. M. de Vries Schultink, Wilbert Zwart, Jos H. Beijnen and Alwin D. R. Huitema have declared no conflicts of interest.
References
- 1.Early Breast Cancer Trialists’ Collaborative Group Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005;365:1687–1717. doi: 10.1016/S0140-6736(05)66544-0. [DOI] [PubMed] [Google Scholar]
- 2.Davies C, Pan H, Godwin J, Gray R, Arriagada R, Raina V, et al. Long-term effects of continuing adjuvant tamoxifen to 10 years versus stopping at 5 years after diagnosis of oestrogen erceptor-positive breast cancer: ATLAS, a randomised trial. Lancet. 2013;381:805–816. doi: 10.1016/S0140-6736(12)61963-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gray R, Rea D, Handley K. ATTom: long-term effects of continuing adjuvant tamoxifen to 10 years versus stopping at 5 years in 6,953 women with early breast cancer. J Clin Oncol. 2013;31(suppl; abstr 5).
- 4.Regan MM, Neven P, Giobbie-Hurder A, Goldhirsch A, Ejlertsen B, Mauriac L, et al. Assessment of letrozole and tamoxifen alone and in sequence for postmenopausal women with steroid hormone receptor-positive breast cancer: the BIG 1-98 randomised clinical trial at 8·1 years median follow-up. Lancet Oncol. 2011;12:1101–1108. doi: 10.1016/S1470-2045(11)70270-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Winer EP, Hudis C, Burstein HJ, Wolff AC, Pritchard KI, Ingle JN, et al. American Society of Clinical Oncology technology assessment on the use of aromatase inhibitors as adjuvant therapy for postmenopausal women with hormone receptor-positive breast cancer: status report 2004. J Clin Oncol. 2005;23:619–629. doi: 10.1200/JCO.2005.09.121. [DOI] [PubMed] [Google Scholar]
- 6.Smith IE, Dowsett M. Aromatase inhibitors in breast cancer. N Engl J Med. 2003;348:2431–2442. doi: 10.1056/NEJMra023246. [DOI] [PubMed] [Google Scholar]
- 7.Early Breast Cancer Trialists’ Collaborative Group Relevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: patient-level meta-analysis of randomised trials. Lancet. 2011;378:771–784. doi: 10.1016/S0140-6736(11)60993-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desta Z, Ward BA, Soukhova NV, Flockhart DA. Comprehensive evaluation of tamoxifen sequential biotransformation by the human cytochrome P450 system in vitro: prominent roles for CYP3A and CYP2D6. J Pharmacol Exp Ther. 2004;310:1062–1075. doi: 10.1124/jpet.104.065607. [DOI] [PubMed] [Google Scholar]
- 9.Lim YC, Desta Z, Flockhart DA, Skaar TC. Endoxifen (4-hydroxy-N-desmethyl-tamoxifen) has anti-estrogenic effects in breast cancer cells with potency similar to 4-hydroxy-tamoxifen. Cancer Chemother Pharmacol. 2005;55:471–478. doi: 10.1007/s00280-004-0926-7. [DOI] [PubMed] [Google Scholar]
- 10.Johnson MD, Zuo H, Lee K, Trebley JP, Rae JM, Weatherman RV, et al. Pharmacological characterization of 4-hydroxy-N-desmethyl tamoxifen, a novel active metabolite of tamoxifen. Breast Cancer Res Treat. 2004;85:151–159. doi: 10.1023/B:BREA.0000025406.31193.e8. [DOI] [PubMed] [Google Scholar]
- 11.Thomas C, Gustafsson JÅ. The different roles of ER subtypes in cancer biology and therapy. Nat Rev Cancer. 2011;11:597–608. doi: 10.1038/nrc3093. [DOI] [PubMed] [Google Scholar]
- 12.Phan M, Venitz J. Summary minutes of the Advisory Committee Pharmaceutical Science Clinical Pharmacology Subcommittee. 18–19 Oct 2006. Available at: http://www.fda.gov/ohrms/dockets/ac/06/minutes/2006-4248m1.pdf. Accessed 15 Jan 2015.
- 13.Beelen K, Opdam M, Severson TM, Koornstra RHT, Vincent AD, Hauptmann M, et al. CYP2C19 2 predicts substantial tamoxifen benefit in postmenopausal breast cancer patients randomized between adjuvant tamoxifen and no systemic treatment. Breast Cancer Res Treat. 2013;139:649–655. doi: 10.1007/s10549-013-2568-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borges S, Desta Z, Li L, Skaar TC, Ward BA, Nguyen A, et al. Quantitative effect of CYP2D6 genotype and inhibitors on tamoxifen metabolism: implication for optimization of breast cancer treatment. Clin Pharmacol Ther. 2006;80:61–74. doi: 10.1016/j.clpt.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 15.Chamnanphon M, Pechatanan K, Sirachainan E, Trachu N, Chantratita W, Pasomsub E, et al. Association of CYP2D6 and CYP2C19 polymorphisms and disease-free survival of Thai post-menopausal breast cancer patients who received adjuvant tamoxifen. Pharmgenomics Pers Med. 2013;6:37–48. doi: 10.2147/PGPM.S42330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fernández-Santander A, Gaibar M, Novillo A, Romero-Lorca A, Rubio M, Chicharro LM, et al. Relationship between genotypes Sult1a2 and Cyp2d6 and tamoxifen metabolism in breast cancer patients. PLoS One. 2013;8:e70183. doi: 10.1371/journal.pone.0070183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goetz MP, Rae JM, Suman VJ, Safgren SL, Ames MM, Visscher DW, et al. Pharmacogenetics of tamoxifen biotransformation is associated with clinical outcomes of efficacy and hot flashes. J Clin Oncol. 2005;23:9312–9318. doi: 10.1200/JCO.2005.03.3266. [DOI] [PubMed] [Google Scholar]
- 18.Goetz MP, Suman VJ, Hoskin TL, Gnant M, Filipits M, Safgren SL, et al. CYP2D6 metabolism and patient outcome in the Austrian Breast and Colorectal Cancer Study Group Trial (ABCSG) 8. Clin Cancer Res. 2013;19:500–507. doi: 10.1158/1078-0432.CCR-12-2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin Y, Desta Z, Stearns V, Ward B, Ho H, Lee K-H, et al. CYP2D6 genotype, antidepressant use, and tamoxifen metabolism during adjuvant breast cancer treatment. J Natl Cancer Inst. 2005;97:30–39. doi: 10.1093/jnci/dji005. [DOI] [PubMed] [Google Scholar]
- 20.Kiyotani K, Mushiroda T, Sasa M, Bando Y, Sumitomo I, Hosono N, et al. Impact of CYP2D6*10 on recurrence-free survival in breast cancer patients receiving adjuvant tamoxifen therapy. Cancer Sci. 2008;99:995–999. doi: 10.1111/j.1349-7006.2008.00780.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kiyotani K, Mushiroda T, Imamura CK, Hosono N, Tsunoda T, Kubo M, et al. Significant effect of polymorphisms in CYP2D6 and ABCC2 on clinical outcomes of adjuvant tamoxifen therapy for breast cancer patients. J Clin Oncol. 2010;28:1287–1293. doi: 10.1200/JCO.2009.25.7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lash TL, Cronin-Fenton D, Ahern TP, Rosenberg CL, Lunetta KL, Silliman RA, et al. CYP2D6 inhibition and breast cancer recurrence in a population-based study in Denmark. J Natl Cancer Inst. 2011;103:489–500. doi: 10.1093/jnci/djr010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim JSL, Chen XA, Singh O, Yap YS, Ng RCH, Wong NS, et al. Impact of CYP2D6, CYP3A5, CYP2C9 and CYP2C19 polymorphisms on tamoxifen pharmacokinetics in Asian breast cancer patients. Br J Clin Pharmacol. 2011;71:737–750. doi: 10.1111/j.1365-2125.2011.03905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Madlensky L, Natarajan L, Tchu S, Pu M, Mortimer J, Flatt SW, et al. Tamoxifen metabolite concentrations, CYP2D6 genotype, and breast cancer outcomes. Clin Pharmacol Ther. 2011;89:718–725. doi: 10.1038/clpt.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moyer AM, Suman VJ, Weinshilboum RM, Black JL, Safgren SL, Kuffel MJ, et al. SULT1A1, CYP2C19 and disease-free survival in early breast cancer patients receiving tamoxifen. Pharmacogenomics. 2011;12:1535–1543. doi: 10.2217/pgs.11.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mürdter TE, Schroth W, Bacchus-Gerybadze L, Winter S, Heinkele G, Simon W, et al. Activity levels of tamoxifen metabolites at the estrogen receptor and the impact of genetic polymorphisms of phase I and II enzymes on their concentration levels in plasma. Clin Pharmacol Ther. 2011;89:708–717. doi: 10.1038/clpt.2011.27. [DOI] [PubMed] [Google Scholar]
- 27.Mwinyi J, Vokinger K, Jetter A, Breitenstein U, Hiller C, Kullak-Ublick GA, et al. Impact of variable CYP genotypes on breast cancer relapse in patients undergoing adjuvant tamoxifen therapy. Cancer Chemother Pharmacol. 2014;73:1181–1188. doi: 10.1007/s00280-014-2453-5. [DOI] [PubMed] [Google Scholar]
- 28.Nowell SA, Ahn J, Rae JM, Scheys JO, Trovato A, Sweeney C, et al. Association of genetic variation in tamoxifen-metabolizing enzymes with overall survival and recurrence of disease in breast cancer patients. Breast Cancer Res Treat. 2005;91:249–258. doi: 10.1007/s10549-004-7751-x. [DOI] [PubMed] [Google Scholar]
- 29.Okishiro M, Taguchi T, Jin Kim S, Shimazu K, Tamaki Y, Noguchi S. Genetic polymorphisms of CYP2D6 10 and CYP2C19 2, 3 are not associated with prognosis, endometrial thickness, or bone mineral density in Japanese breast cancer patients treated with adjuvant tamoxifen. Cancer. 2009;115:952–961. doi: 10.1002/cncr.24111. [DOI] [PubMed] [Google Scholar]
- 30.Rae JM, Drury S, Hayes DF, Stearns V, Thibert JN, Haynes BP, et al. CYP2D6 and UGT2B7 genotype and risk of recurrence in tamoxifen-treated breast cancer patients. J Natl Cancer Inst. 2012;104:452–460. doi: 10.1093/jnci/djs126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Regan MM, Leyland-Jones B, Bouzyk M, Pagani O, Tang W, Kammler R, et al. CYP2D6 genotype and tamoxifen response in postmenopausal women with endocrine-responsive breast cancer: the Breast International Group 1-98 trial. J Natl Cancer Inst. 2012;104:441–451. doi: 10.1093/jnci/djs125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saladores P, Mürdter T, Eccles D, Chowbay B, Zgheib NK, Winter S, et al. Tamoxifen metabolism predicts drug concentrations and outcome in premenopausal patients with early breast cancer. Pharmacogenomics J. 2014;1:84–94. doi: 10.1038/tpj.2014.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schroth W, Antoniadou L, Fritz P, Schwab M, Muerdter T, Zanger UM, et al. Breast cancer treatment outcome with adjuvant tamoxifen relative to patient CYP2D6 and CYP2C19 genotypes. J Clin Oncol. 2007;25:5187–5193. doi: 10.1200/JCO.2007.12.2705. [DOI] [PubMed] [Google Scholar]
- 34.Schroth W, Goetz MP, Hamann U, Fasching PA, Schmidt M, Winter S, et al. Association between CYP2D6 polymorphisms and outcomes among women with early stage breast cancer treated with tamoxifen. JAMA. 2009;302:1429–1436. doi: 10.1001/jama.2009.1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sensorn I, Sirachainan E, Chamnanphon M, Pasomsub E, Trachu N, Supavilai P, et al. Association of CYP3A4/5, ABCB1 and ABCC2 polymorphisms and clinical outcomes of Thai breast cancer patients treated with tamoxifen. Pharmgenomics Pers Med. 2013;6:93–98. doi: 10.2147/PGPM.S44006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Teft WA, Gong IY, Dingle B, Potvin K, Younus J, Vandenberg TA, et al. CYP3A4 and seasonal variation in vitamin D status in addition to CYP2D6 contribute to therapeutic endoxifen level during tamoxifen therapy. Breast Cancer Res Treat. 2013;139:95–105. doi: 10.1007/s10549-013-2511-4. [DOI] [PubMed] [Google Scholar]
- 37.Tucker AN, Tkaczuk KA, Lewis LM, Tomic D, Lim CK, Flaws JA. Polymorphisms in cytochrome P4503A5 (CYP3A5) may be associated with race and tumor characteristics, but not metabolism and side effects of tamoxifen in breast cancer patients. Cancer Lett. 2005;217:61–72. doi: 10.1016/j.canlet.2004.08.027. [DOI] [PubMed] [Google Scholar]
- 38.Wegman P, Vainikka L, Stål O, Nordenskjöld B, Skoog L, Rutqvist L-E, et al. Genotype of metabolic enzymes and the benefit of tamoxifen in postmenopausal breast cancer patients. Breast Cancer Res. 2005;7:R284–R290. doi: 10.1186/bcr993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wegman P, Elingarami S, Carstensen J, Stål O, Nordenskjöld B, Wingren S. Genetic variants of CYP3A5, CYP2D6, SULT1A1, UGT2B15 and tamoxifen response in postmenopausal patients with breast cancer. Breast Cancer Res. 2007;9:R7. doi: 10.1186/bcr1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu Y, Sun Y, Yao L, Shi L, Wu Y, Ouyang T, et al. Association between CYP2D6*10 genotype and survival of breast cancer patients receiving tamoxifen treatment. Ann Oncol. 2008;19:1423–1429. doi: 10.1093/annonc/mdn155. [DOI] [PubMed] [Google Scholar]
- 41.Zafra-Ceres M, de Haro T, Farez-Vidal E, Blancas I, Bandres F, de Dueñas EM, et al. Influence of CYP2D6 polymorphisms on serum levels of tamoxifen metabolites in Spanish women with breast cancer. Int J Med Sci. 2013;10:932–937. doi: 10.7150/ijms.5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gjerde J, Hauglid M, Breilid H, Lundgren S, Varhaug JE, Kisanga ER, et al. Effects of CYP2D6 and SULT1A1 genotypes including SULT1A1 gene copy number on tamoxifen metabolism. Ann Oncol. 2008;19:56–61. doi: 10.1093/annonc/mdm434. [DOI] [PubMed] [Google Scholar]
- 43.Toyama T, Yamashita H, Sugiura H, Kondo N, Iwase H, Fujii Y. No association between CYP2D6*10 genotype and survival of node-negative Japanese breast cancer patients receiving adjuvant tamoxifen treatment. Jpn J Clin Oncol. 2009;39:651–656. doi: 10.1093/jjco/hyp076. [DOI] [PubMed] [Google Scholar]
- 44.Stingl JC, Parmar S, Huber-Wechselberger A, Kainz A, Renner W, Seeringer A, et al. Impact of CYP2D6*4 genotype on progression free survival in tamoxifen breast cancer treatment. Curr Med Res Opin. 2010;26:2535–2542. doi: 10.1185/03007995.2010.518304. [DOI] [PubMed] [Google Scholar]
- 45.Abraham JE, Maranian MJ, Driver KE, Platte R, Kalmyrzaev B, Baynes C, et al. CYP2D6 gene variants: association with breast cancer specific survival in a cohort of breast cancer patients from the United Kingdom treated with adjuvant tamoxifen. Breast Cancer Res. 2010;12(4):R64. doi: 10.1186/bcr2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park IH, Ro J, Park S, Lim H-S, Lee KS, Kang HS, et al. Lack of any association between functionally significant CYP2D6 polymorphisms and clinical outcomes in early breast cancer patients receiving adjuvant tamoxifen treatment. Breast Cancer Res Treat. 2012;131:455–461. doi: 10.1007/s10549-011-1425-2. [DOI] [PubMed] [Google Scholar]
- 47.Park HS, Choi J-Y, Lee M-J, Park S, Yeo C-W, Lee SS, et al. Association between genetic polymorphisms of CYP2D6 and outcomes in breast cancer patients with tamoxifen treatment. J Korean Med Sci. 2011;26:1007–1013. doi: 10.3346/jkms.2011.26.8.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barginear MF, Jaremko M, Peter I, Yu C, Kasai Y, Kemeny M, et al. Increasing tamoxifen dose in breast cancer patients based on CYP2D6 genotypes and endoxifen levels: effect on active metabolite isomers and the antiestrogenic activity score. Clin Pharmacol Ther. 2011;90:605–611. doi: 10.1038/clpt.2011.153. [DOI] [PubMed] [Google Scholar]
- 49.Beelen K, Zwart W, Linn SC. Can predictive biomarkers in breast cancer guide adjuvant endocrine therapy? Nat Rev Clin Oncol. 2012;9:529–541. doi: 10.1038/nrclinonc.2012.121. [DOI] [PubMed] [Google Scholar]
- 50.Human cytochrome P450 (CYP) allele nomenclature T. The Human Cytochrome P450 (CYP) Allele Nomenclature Database. Available at: http://www.cypalleles.ki.se/cyp2d6.htm. Accessed 28 Dec 2014.
- 51.Hudis CA, Barlow WE, Costantino JP, Gray RJ, Pritchard KI, Chapman JA, et al. Proposal for standardized definitions for efficacy end points in adjuvant breast cancer trials: the STEEP system. J Clin Oncol. 2007;25:2127–2132. doi: 10.1200/JCO.2006.10.3523. [DOI] [PubMed] [Google Scholar]
- 52.Coller JK, Krebsfaenger N, Klein K, Endrizzi K, Wolbold R, Lang T, et al. The influence of CYP2B6, CYP2C9 and CYP2D6 genotypes on the formation of the potent antioestrogen Z-4-hydroxy-tamoxifen in human liver. Br J Clin Pharmacol. 2002;54:157–167. doi: 10.1046/j.1365-2125.2002.01614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Province MA, Goetz MP, Brauch H, Flockhart DA, Hebert JM, Whaley R, et al. CYP2D6 genotype and adjuvant tamoxifen: meta-analysis of heterogeneous study populations. Clin Pharmacol Ther. 2014;95:216–227. doi: 10.1038/clpt.2013.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Berry D. CYP2D6 genotyping and the use of tamoxifen in breast cancer. J Natl Cancer Inst. 2013;105:1267–1269. doi: 10.1093/jnci/djt221. [DOI] [PubMed] [Google Scholar]
- 55.Berry DA. CYP2D6 genotype and adjuvant tamoxifen. Clin Pharmacol Ther. 2014;96:138–140. doi: 10.1038/clpt.2014.96. [DOI] [PubMed] [Google Scholar]
- 56.Lash TL, Lien EA, Sørensen HT, Hamilton-Dutoit S. Genotype-guided tamoxifen therapy: time to pause for reflection? Lancet Oncol. 2009;10:825–833. doi: 10.1016/S1470-2045(09)70030-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Blackburn HL, Ellsworth DL, Shriver CD, Ellsworth RE. Role of cytochrome P450 genes in breast cancer etiology and treatment: effects on estrogen biosynthesis, metabolism, and response to endocrine therapy. Cancer Causes Control. 2015;26(3):319–332. doi: 10.1007/s10552-014-0519-7. [DOI] [PubMed] [Google Scholar]
- 58.Tsuchiya Y, Nakajima M, Yokoi T. Cytochrome P450-mediated metabolism of estrogens and its regulation in human. Cancer Lett. 2005;227:115–124. doi: 10.1016/j.canlet.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 59.Gjerde J, Geisler J, Lundgren S, Ekse D, Varhaug JE, Mellgren G. Associations between tamoxifen, estrogens, and FSH serum levels during steady state tamoxifen treatment of postmenopausal women with breast cancer. BMC Cancer. 2010;10:313. doi: 10.1186/1471-2407-10-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jager NGL, Rosing H, Linn SC, Schellens JHM, Beijnen JH. Importance of highly selective LC–MS/MS analysis for the accurate quantification of tamoxifen and its metabolites: focus on endoxifen and 4-hydroxytamoxifen. Breast Cancer Res Treat. 2012;133:793–798. doi: 10.1007/s10549-012-2000-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Binkhorst L, Mathijssen RHJ, van Herk-Sukel MPP, Bannink M, Jager A, Wiemer EAC, et al. Unjustified prescribing of CYP2D6 inhibiting SSRIs in women treated with tamoxifen. Breast Cancer Res Treat. 2013;139:923–929. doi: 10.1007/s10549-013-2585-z. [DOI] [PubMed] [Google Scholar]
- 62.Goetz MP, Sun JX, Suman VJ, Silva GO, Perou CM, Nakamura Y, et al. Loss of heterozygosity at the CYP2D6 locus in breast cancer: implications for germline pharmacogenetic studies. J Natl Cancer Inst. 2014;107(2). doi:10.1093/jnci/dju401. [DOI] [PMC free article] [PubMed]
- 63.Schroth W, Hamann U, Fasching PA, Dauser S, Winter S, Eichelbaum M, et al. CYP2D6 polymorphisms as predictors of outcome in breast cancer patients treated with tamoxifen: expanded polymorphism coverage improves risk stratification. Clin Cancer Res. 2010;16:4468–4477. doi: 10.1158/1078-0432.CCR-10-0478. [DOI] [PubMed] [Google Scholar]
- 64.Ratain MJ, Nakamura Y, Cox NJ. CYP2D6 genotype and tamoxifen activity: understanding interstudy variability in methodological quality. Clin Pharmacol Ther. 2014;94(2):185–187. doi: 10.1038/clpt.2013.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ter Heine R, Binkhorst L, de Graan AJM, de Bruijn P, Beijnen JH, Mathijssen RHJ, et al. Population pharmacokinetic modelling to assess the impact of CYP2D6 and CYP3A metabolic phenotypes on the pharmacokinetics of tamoxifen and endoxifen. Br J Clin Pharmacol. 2014;78:572–586. doi: 10.1111/bcp.12388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jager NL, Rosing H, Schellens J, Linn S, Beijnen J. Tamoxifen dose and serum concentrations of tamoxifen and six of its metabolites in routine clinical outpatient care. Breast Cancer Res. 2014;143:447–483. doi: 10.1007/s10549-013-2824-3. [DOI] [PubMed] [Google Scholar]
- 67.Kiyotani K, Mushiroda T, Imamura CK, Tanigawara Y, Hosono N, Kubo M, et al. Dose-adjustment study of tamoxifen based on CYP2D6 genotypes in Japanese breast cancer patients. Breast Cancer Res Treat. 2012;131:137–145. doi: 10.1007/s10549-011-1777-7. [DOI] [PubMed] [Google Scholar]
- 68.Irvin WJ, Walko CM, Weck KE, Ibrahim JG, Chiu WK, Dees EC, et al. Genotype-guided tamoxifen dosing increases active metabolite exposure in women with reduced CYP2D6 metabolism: a multicenter study. J Clin Oncol. 2011;29:3232–3239. doi: 10.1200/JCO.2010.31.4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bratherton D, Brown C, Buchanan R, Hall V, Kingsley Pillers E, Wheeler T, et al. A comparison of two doses of tamoxifen (Novaldex) in postmenopausal women with advanced breast cancer: 10 mg bd versus 20 mg bd. Br J Cancer. 1984;50:199–205. doi: 10.1038/bjc.1984.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sismondi P, Biglia N, Volpi E, Giai M. Tamoxifen and endometrial cancer. Ann NY Acad Sci. 1994;734:310–321. doi: 10.1111/j.1749-6632.1994.tb21761.x. [DOI] [PubMed] [Google Scholar]


