Abstract
Gambiense trypanosomiasis is considered an anthroponotic disease. Consequently, control programs are generally aimed at stopping transmission of Trypanosoma brucei gambiense (T. b. gambiense) by detecting and treating human cases. However, the persistence of numerous foci despite efforts to eliminate this disease questions this strategy as unique tool to pursue the eradication. The role of animals as a reservoir of T. b. gambiense is still controversial, but could partly explain maintenance of the infection at hypo-endemic levels. In the present study, we evaluated the presence of T. b. gambiense in wild animals in Equatorial Guinea. The infection rate ranged from 0.8% in the insular focus of Luba to more than 12% in Mbini, a focus with a constant trickle of human cases. The parasite was detected in a wide range of animal species including four species never described previously as putative reservoirs. Our study comes to reinforce the hypothesis that animals may play a role in the persistence of T. b. gambiense transmission, being particularly relevant in low transmission settings. Under these conditions the integration of sustained vector control and medical interventions should be considered to achieve the elimination of gambiense trypanosomiasis.
Keywords: Trypanosoma brucei gambiense, wild fauna, reservoir, human African trypanosomiasis, sleeping sickness, Equatorial Guinea
Introduction
Human African trypanosomiasis (HAT), also known as sleeping sickness, is a tropical disease caused by two subspecies of the protozoan flagellate, Trypanosoma brucei s.l.: T. b. gambiense, and T. b. rhodesiense. Both are closely related on a genetic level, but have important phenotypic differences regarding their epidemiology, transmission, clinical features, and distribution. The vector for T. brucei s.l. and other African trypanosomes is the tsetse fly (order Diptera, genus Glossina), but each subgenus of Glossina sp. has different susceptibility to diverse trypanosome species (Ashford, 2003; Kuzoe and Schofield, 2004), conditioning the distribution of both human and animal trypanosomiasis (World Health Organization, 2013).
Trypanosoma brucei gambiense is responsible for more than 97% of HAT cases and is spread over Central and West Africa (Simarro et al., 2012). In contrast with T. b. rhodesiense, which is known to circulate in domestic and wild fauna (Franco et al., 2014a), T. b. gambiense has traditionally been considered an anthroponosis, i.e., human beings are the main reservoir of the parasite (World Health Organization, 2013). Consistent with this assumption, control interventions based on detection and treatment of human cases have proven to be sufficient to drastically reduce the prevalence, even in the absence of vector control (Simarro et al., 2006; Franco et al., 2014b). Despite the effectiveness of these actions, virulent outbreaks have occurred over the past century after abandoning these control measures (Steverding, 2008). Thus, complete elimination was only rarely achieved in particular foci. The persistence of infection, even when no human cases were reported for years, has been attributed to different causes such as asymptomatic parasite carriers (Jamonneau et al., 2012), continuous reintroduction due to movements of both human and vector populations (Courtin et al., 2008), and inherent limitations of the surveillance system (Louis et al., 2008). An increasing number of studies have shown that domestic and wild fauna could also harbor T. b. gambiense even though it still remains unclear whether animals can host the parasite long enough to play an important role in the transmission of Gambiense HAT (Gibson et al., 1978; Noireau et al., 1986; Herder et al., 2002; Njiokou et al., 2006; Simo et al., 2006; Cordon-Obras et al., 2009).
Equatorial Guinea, a small country located in the Guinea Gulf, has four historical HAT foci; three in the coastal region of the continental part (Kogo, Mbini, and Campo) and one in Bioko Island (Luba) that is currently considered free of HAT (Simarro et al., 2006). Since the mid-1980s, the number of HAT cases diagnosed is very low thanks to the implementation of control campaigns based on systematic screening of endemic populations and treatment of patients. In spite of the considerable and sustained efforts, a continuous trickle of cases is reported every year in the mainland foci, Kogo, and Mbini (Franco et al., 2014a). In a previous study, we suggested that the epidemiological situation in both foci might follow different patterns. Whereas peri-domestic fauna could act as a reservoir of infection in Mbini (DNA of T. b. gambiense were detected in seven animals), we did not find T. b. gambiense in livestock from the Kogo focus (Cordon-Obras et al., 2009). In Luba, where no human cases have been reported since 1995 (Simarro et al., 2006), we were unable to detect the parasite in domestic fauna, although we found a positive tsetse fly sample, demonstrating that T. b. gambiense is still present in Bioko Island (Cordon-Obras et al., 2010). The persistence of infection in Kogo and Luba foci could not be explained by our data from domestic animal samples. Therefore, we hypothesized the existence of alternative epidemiological cycles. In such ecological context, where the forest represents more than 78.5% (22,000 km2) of the total land surface of the country, wild animals would represent potential reservoir candidates.
In the present work, employing the same molecular tools previously used (Cordon-Obras et al., 2009, 2010), we performed a molecular screening of T. b. gambiense in wild fauna and considered its possible role in the maintenance of the parasite in the historical foci of Equatorial Guinea. We established potential scenarios depending on the specificity of each focus, highlighting their differences, and discussed the epidemiological implications of the occurrence of a sylvatic transmission cycle for T. b. gambiense.
Materials and Methods
Study Area
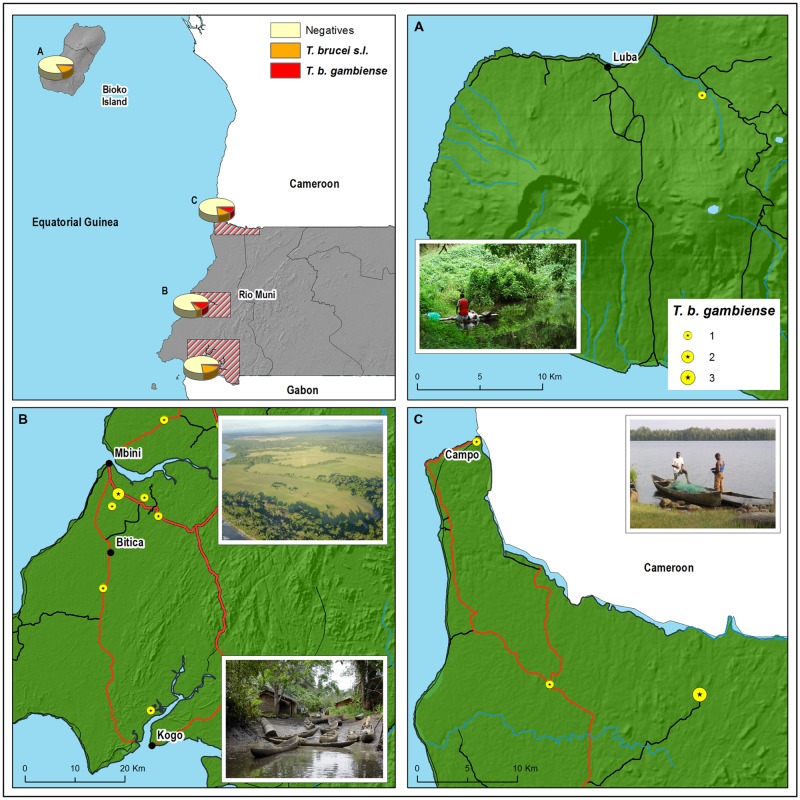
Four HAT foci from Equatorial Guinea were studied: Luba (at the southwest of Bioko Island), Rio Campo (north of the mainland region, bordering with Cameroon), Mbini (central mainland coast), and Kogo (southern mainland coast, bordering with Gabon), (Figure 1). Bioko Island is located more than 200 km from the mainland. Rainforest and abandoned cocoa plantations are widespread in Luba district (Simarro et al., 1990), covering a surface of 700 km2. Continental foci are located across the coastal region and have a typical maritime equatorial climate of four seasons. Campo extends along the Ntem River, which delineates the border between Cameroon and Equatorial Guinea, and is mostly covered by the equatorial rainforest (Simo et al., 2014). Mbini is located close to the mouth of the Wele River, whereas Kogo limits with Gabon separated by a natural boundary created by the Muni estuary. Mangrove swamp and rainforest are the predominant ecosystems in the Kogo and Mbini foci.
FIGURE 1.
Geographical distribution of the Trypanosoma brucei gambiense (T. b. gambiense)-positive animals according to the place where they were killed or trapped in (A) Luba, (B) Kogo and Mbini, and (C) Rio Campo. Main and secondary roads are represented in red and black lines, respectively.
Sampling Strategy
From June 2012 to March 2014, surveys were conducted to collect blood samples from wild fauna. Samples were obtained either in local markets (Figure 2A) or directly from hunters and trappers (Figure 2B). Our team had absolutely no control over the species selected by hunters. The blood taken by venepuncture (Figure 2C) was spread on Whatman paper, left to dry in darkness and then stored at 4°C until processing in the reference laboratory at the National Center of Tropical Medicine, Madrid (Institute of Health Carlos III). Each filter paper was labeled to link the blood sample with the additional information recorded: hunting area, date of capture, data, and place of blood sampling (Figure 2D). Each sample was stored separately to avoid cross-contamination. Ethical approval was obtained by the Ministry of Health and Social Welfare and Veterinary Service from the continental region (Ministry of Agriculture, Forestry and Environment). The study adhered to the institutions’ guidelines for animal hunter game.
FIGURE 2.
Sampling strategy. Blood samples from wild animals were collected from market (A), or directly from the hunters (B). The blood taken by venepuncture (C) was then spread on Whatman paper. (D) Filter paper with blood sample was labeled and stored separately to avoid cross-contamination.
Host Species Identification
Wild animals captured by hunters and trappers and brought to the village markets were identified using a systematic key (Dorst and Dandelot, 1997). Molecular confirmation was performed by cytochrome b sequencing according to the conditions previously described (Steuber et al., 2005) when the species identification was problematic. This validation was also performed on T. b. gambiense-positive samples in order to accurately define the possible reservoir hosts.
Parasite Molecular Diagnosis
DNA was extracted from the samples collected employing the SpeedTools DNA Extraction kit (Biotools, Madrid, Spain) according to the manufacturer’s instructions, DNA template (1 μl) was submitted to species-specific PCR for T. brucei s.l. using the oligonucleotides described by Moser et al. (1989) that target the palindromic 177 bp satellite of mini-chromosomes. Positive samples for 177 bp PCR were submitted to a T. b. gambiense group 1-specific nested-PCR using the primers described elsewhere (Cordon-Obras et al., 2009) which amplify the known specific TgsGP gene (Berberof et al., 2001) considered to be the gold standard method for the identification of this sub-species. For both PCRs, the conditions were as follows: 1x PCR Reaction Buffer [7.5 mM Tris-HCl -pH 9-, 1.5 mM MgCl2, 5 mM KCl, and 2 mM (NH4)2SO4], 200 μM of each deoxynucleotide (dNTP), primers at 1 μM, and 1 U of Certamp Complex Enzyme Mix (Biotools, Madrid, Spain) in a final volume of 25 μl. The amplification programs were set as follows: for T. brucei s.l., 3 min at 95°C for initial DNA denaturation, 35 cycles of 95°C (30 s), 60°C (30 s), and 72°C (1 min), and a final extension step at 72°C (5 min). For the first reaction of T. b. gambiense, the fixed program was an initial denaturation step at 95°C (5 min), 35 cycles at 94°C (30 s), 55°C (1 min), and 72°C (3 min), with a final extension step at 72°C (5 min). The program for the nested reaction was identical. Negative (double distilled water) and positive [genomic DNA of T. b. gambiense ELIANE strain -MHOM/CI/52/ITMAP 2188- (Turner et al., 2004)] controls were systematically added to each assay. The amplification products were separated by electrophoresis in a 2% agarose gel stained with 1x RedSafeTM Nucleic Acid Staining Solution (Intron Biotechnology, Inc.) and photographed under UV light. All positive samples for T. b. gambiense and a number of negative samples randomly selected were submitted to a second round of diagnosis performed blindly by a different researcher. All samples yielded a second positive result, confirming the robustness and reproducibility of our diagnostic method. Rigorous measures were employed to prevent contamination of the PCR reaction by establishment of a unidirectional physical workflow (pre-PCR–post-PCR).
Statistical Analysis
Prevalence of T. brucei s.l. and T. b. gambiense infections by focus were compared through Pearson’s Chi-square test. Data analysis was conducted by using SPSS software (version 16.0.1, SPSS Inc., Chicago, IL, USA).
Results
Sample Collection
A total of 288 samples were collected within the four foci (121 in Luba, 66 in Mbini, 45 in Kogo, and 56 in Rio Campo) from June 2012 to March 2014. The complete animal records are provided as supplemental information (Supplementary Table S1). They included 26 animal species, 98.3% belonging to mammals, since they are the most frequent targets of hunting activities. Among mammals, the most represented orders were Rodentia and Artiodactyla (128 and 109 samples, respectively) given that the preferred preys in these foci are the African brush-tailed porcupine (Atherurus africanus), the African giant pouched rat (Cricetomys gambianus), and the hoofed blue duiker (Cephalophus monticola). Primates (29), Carnivora (13), and Pholidota (4) completed the mammal set. Four samples from reptiles and one from a bird was also analyzed.
Trypanosoma brucei Infection Rate
Trypanosoma brucei s.l. infection in the wild fauna was ubiquitous and homogenous in the four historical HAT foci of Equatorial Guinea, with infection rates ranging from 16.5% in Luba to 24.4% in Kogo, with 19.7% in Mbini and 23.2% in Rio Campo (Table 1). Rodentia (25) and Artiodactylia (19) represented 77.2% of the infected animals (Table 2). No statistical association was found between T. brucei prevalence and focus (X2 = 1.84, p-value = 0.61).
Table 1.
Distribution of Trypanosoma brucei s.l. and Trypanosoma brucei gambiense (T. b. gambiense) among wild fauna in different foci of Equatorial Guinea.
| Focus | T. brucei s.l. no. (%) | T. b. gambiense no. (%) |
|---|---|---|
| Luba | 20/121 (16.5) | 1/121 (0.8) |
| Mbini | 13/66 (19.7) | 8/66 (12.1) |
| Kogo | 11/45 (24.4) | 1/45 (2.2) |
| Rio Campo | 13/56 (23.2) | 5/56 (8.9) |
| Total | 57/288 (19.8) | 15/288 (5.2) |
Table 2.
Distribution of T. brucei s.l. and T. b. gambiense according to animal species.
| Focus | Species |
Intra-species Prevalence (%) |
||
|---|---|---|---|---|
| Common name | Scientific name | T. brucei s.l. | T. b. gambiense | |
| Luba | Carnivora | |||
| Small-spotted genet | Genetta servalina | 1/2 (50) | - | |
| Rodentia | ||||
| Red-legged sun squirrel | Heliosciurus rufobrachium∗ | 3/16 (18.8) | 1/16 (6.3) | |
| Brush-tailed porcupine | Atherurus africanus | 5/34 (14.7) | - | |
| African giant pouched rat | Cricetomys gambianus | 3/25 (12) | - | |
| Artiodactyla | ||||
| Blue Duiker | Cephalophus monticola | 7/35 (20) | - | |
| Reptiles | ||||
| African Rock Python | Python sebae | 1/1 (100) | - | |
| Mbini | Primates | |||
| Red-tailed monkey | Cercopithecus ascanius∗ | 1/2 (50) | 1/2 (50) | |
| Grey-cheeked mangabey | Lophocebus albigena∗ | 1/1 (100) | 1/1 (100) | |
| Greater spot-nosed monkey | Cercopithecus nictitans | 1/5 (20) | - | |
| Carnivora | ||||
| Small-spotted genet | Genetta servalina | 1/1 (100) | 1/1 (100) | |
| African palm civet | Nandinia binotata | 2/3 (66.7) | 1/3 (33.3) | |
| Rodentia | ||||
| Brush-tailed porcupine | Atherurus africanus | 3/14 (21.4) | 3/14 (21.4) | |
| African giant pouched rat | Cricetomys gambianus | 1/6 (16.7) | 1/6 (16.7) | |
| Artiodactyla | ||||
| Blue Duiker | Cephalophus monticola | 3/19 (15.8) | - | |
| Kogo | Carnivora | |||
| Small-spotted genet | Genetta servalina | 1/1 (100) | - | |
| Rodentia | ||||
| Brush-tailed porcupine | Atherurus africanus | 4/5 (80) | - | |
| Artiodactyla | ||||
| Blue Duiker | Cephalophus monticola | 5/22 (22.7) | 1/22 (4.5) | |
| Red River Hog | Potamochoerus porcus | 1/1 (100) | - | |
| Rio Campo | Primates | |||
| Greater spot-nosed monkey | Cercopithecus nictitans | 3/4 (75) | 1 /4 (25) | |
| Carnivora | ||||
| African Lisang | Poiana richardsonii∗ | 1/2 (50) | 1/2 (50) | |
| Rodentia | ||||
| Brush-tailed porcupine | Atherurus africanus | 4/11 (36.4) | 1/11 (9.1) | |
| African giant pouched rat | Cricetomys gambianus | 2/7 (28.6) | - | |
| Artiodactyla | ||||
| Blue Duiker | Cephalophus monticola | 3/9 (33.3) | 2/9 (22.2) | |
∗Species detected as putative T. b. gambiense reservoir for the first time in this study.
A total of 15 samples were positive for the TgsGP nested-PCR. To ensure the specificity of these bands, the PCR products were sequenced. They all presented 100% homology with the TgsGP gene (DAL 972 reference strain). T. b. gambiense was detected in all foci, but with a heterogeneous pattern. Infection rate of T. b. gambiense was 12.1 and 8.8%, in Mbini, and Rio Campo, respectively, and significantly lower in Luba (0.8%) and Kogo (2.2%; X2 = 13.48, p-value <0.01). No statistical association was found between prevalence of T. brucei or T. b. gambiense and the host order (X2 = 7.91, p-value = 0.34 and X2 = 11.85, p-value = 0.1, for T. brucei and T. b. gambiense, respectively).
Overall, 10 mammals species were found to harbor T. b. gambiense, belonging to four orders (Rodentia, Artiodactyla, Carnivora, and Primates; Table 2). Similarly to T. brucei s.l. infection, 60% of the T. b. gambiense cases were distributed among rodents and artiodactyls; carnivores and primates represented the remaining 40%. Among those positives for T. b. gambiense, we identified four species so far never reported as putative reservoirs of this parasite; these were Heliosciurus rufobrachium (Red-legged sun squirrel), Cercopithecus ascanius (red-tailed monkey), Lophocebus albigena (gray-cheeked mangabey), and Poiana richardsonii (African lisang).
Discussion
The four historical foci of Equatorial Guinea depict the diversity of epidemiological settings. In the past, Luba was the most important focus of HAT in this country, reporting hundreds of cases per year (Simarro et al., 1990). The implementation of control activities based on active detection and systematic treatment of patients in 1985 drove to the elimination of the human disease one decade later (Simarro et al., 2006). Kogo and Mbini always had a marginal importance compared to Luba in terms of HAT incidence. However, despite similar control measures were implemented the complete elimination has not been yet achieved in the mainland foci. On the other hand, Campo has historically reported few cases (10 cases since 2000 according to Equatorial Guinea Ministry of Health) even in the absence of control interventions in the Guinean part of the focus (Simo et al., 2014). One might speculate that the isolation of Bioko Island would have contributed to the success of control interventions, whereas factors such as population movements, reintroduction of infected vectors, and insufficient coverage in the campaigns of active screening might be leading to a steady occurrence of cases in the mainland foci.
In this study, we demonstrate the presence of T. b. gambiense in different species of wild fauna in all the four endemic HAT foci of Equatorial Guinea. These findings support our previous works, where we concluded that diverse eco-epidemiological scenarios, involving different animal reservoirs, might underlie the variety of transmission patterns for T. b. gambiense in Equatorial Guinea.
We already reported that T. b. gambiense was present in domestic livestock from Mbini but not Kogo (Cordon-Obras et al., 2009). According to these results, we speculated on the occurrence of a peri-domestic transmission cycle in Mbini, involving livestock often circulating close to populations. In the present study, while the T. brucei s.l. infection rate is similar in all foci, we observed a significantly higher prevalence of T. b. gambiense in wild fauna of Mbini compared to Kogo. These data suggest an additional transmission activity in a sylvatic cycle along with the peri-domestic cycle previously hypothesized (Cordon-Obras et al., 2009). It is noteworthy that the prevalence of T. b. gambiense infection in wild fauna in Mbini focus was higher than previously reported in peri-domestic livestock (12.1% vs. 2%; Cordon-Obras et al., 2009), which could be explained by a greater preference of vector population for wild fauna or simply a greater availability of wildlife versus livestock. Further studies aiming to describe the feeding preferences of tsetse flies will be conducted to clear up this question. In Kogo, where new HAT cases are actually rare, T. b. gambiense has been detected in only one wild animal out of 45 screened, indicating that (i) a sylvatic cycle can occur in this focus, but (ii) parasite transmission level seems low.
Campo focus, shared by Equatorial Guinea and Cameroon, has particular epidemiological features. Although this focus has remained active in both sides of the border, only a handful of HAT cases have been reported over the last years in the Guinean part. Subsequently, unlike the other foci, active screening has not been undertaken in a regular basis in Campo. Some studies have reported the presence of T. b. gambiense in animals and the occurrence of human infections in the Cameroonian side of the border, even though at low level (Morlais et al., 1998; Penchenier et al., 1999; Herder et al., 2002). Our data reveal 8.9% of infection rate in wild fauna, above the prevalence reported in wild (0.6%; Njiokou et al., 2006) and domestic (4.4%; Njiokou et al., 2010) animals on the Cameroonian side. The different molecular markers used in the screening or the diverse epidemiological patterns can in part explain these contrasted results. It is noteworthy that in the Cameroonian focus of Fontem, it was described a high T. b. gambiense prevalence in pigs (14.8%; Simo et al., 2006), suggesting different epidemiological scenarios between foci from Cameroon as we noticed in Equatorial Guinea. According to our findings we might conclude that T. b. gambiense is mainly circulating in a sylvatic cycle in Campo, and the absence of human cases is likely due to the low population density in the area. However, a plan to communicate both sides of Ntem river by road (Economic Community of Central African States source, see http://www.ceeac-eccas.org) and the development of Campo municipality in the Guinean part would undoubtedly result in an increase of human presence in the area and in turn, a greater risk of exposure to T. b. gambiense infection.
The Luba focus has not reported autochthonous HAT cases since 1995 (Simarro et al., 2006). However, in a recent publication we demonstrated the persistence of T. b. gambiense in tsetse flies (Cordon-Obras et al., 2010). Given that no vertical transmission of the parasite has been reported in the tsetse fly and the apparent lack of infection in the local livestock, we suggested that a wild cycle could be the responsible for the maintenance of T. b. gambiense, even after the absence of HAT cases for decades (Simarro et al., 2006). In the present work, we found a positive sample of T. b. gambiense amongst the blood samples gathered from wild animals (0.8% prevalence). The epidemiological scenario of Luba seems to be quite similar to Kogo focus, where HAT cases are now rarely reported, T. b. gambiense is apparently absent in domestic fauna and this parasite is found at low prevalence in wild fauna. All these data together strongly suggest the existence of a sylvatic cycle with low activity in both foci which may hinder a definitive eradication of the disease.
Our data also revealed homogenous prevalence of T. brucei s.l. in all the studied foci, but this pattern is not observed in T. b. gambiense. This could be explained by the fact that control measures taken over human reservoirs and the HAT epidemiological features of each focus affect T. b. gambiense, but not T. b. brucei which is, besides, much more easily acquired and transmitted by tsetse flies (Kuzoe and Schofield, 2004).
As sampling is biased due to the methods, the species that host the parasite are similar to those found in other African foci (Njiokou et al., 2006), i.e., those that are targets of hunting and trapping. In spite of this bias, we identified some species that have never been previously reported as reservoirs of T. b. gambiense. In addition, we cannot rule out that more animal species may harbor T. b. gambiense. Indeed, there is no apparent association between the rate of infection and taxonomic order in either T. b. brucei or T. b. gambiense, suggesting that these parasites can adapt to a wide range of species with no evident preference. This is consistent with previous findings that point out the ability of trypanosomes to proliferate in several species in both in vivo and in vitro experiments (Bitter et al., 1998; Herder et al., 2002; Njiokou et al., 2006; Cordon-Obras et al., 2013). Besides, it is well known that most of the other human pathogenic kinetoplastids like T. b. rhodesiense, T. cruzi, or Leishmania sp. are able to circulate in a wide range of animal species (Sobrino et al., 2008; Oda et al., 2014; Zanet et al., 2014).
Despite the provided evidences, we assume that detecting T. b. gambiense in wild fauna does not definitively prove that these animals represent relevant reservoirs for the maintenance of the parasites that are then able to infect humans. Nevertheless, epidemiological arguments such as the higher significant prevalence in Mbini (the most active HAT focus) and Campo (where no control measures were implemented), the previously detected presence of the parasite in tsetse flies in Luba (but not in humans or domestic fauna; Cordon-Obras et al., 2010), and the fact that most of the animals had already been detected previously as being positive for T. b. gambiense (Njiokou et al., 2006), support the existence of sylvatic cycles. If that is correct, it would remain to be elucidated whether both animal and human cycles are linked. To answer this question, we will further analyze by microsatellite approach the samples used in this study and others from HAT patients concurrently isolated from the same foci. The accurate genetic identification of the same populations circulating in both cycles would provide a strong argument in support of our hypothesis.
The lack of an appropriate description of possible reservoirs of T. b. gambiense is a fundamental drawback in the design of control strategies. Wild fauna is a link in the epidemiological chain that is much more difficult to control, and interventions directed against vector would probably be the way to interrupt parasite transmission. Animal reservoirs often mask the infection of a parasitic or viral disease between outbreak periods. Over the 20th century, HAT progression has followed a similar pattern (Steverding, 2008), with alternating periods of low incidence and virulent outbreaks. The intensive control efforts focused on human patients have been always very effective to control the disease, but rarely sufficient to definitively eliminate it in any focus. Recently, based on mathematical models of gambiense HAT transmission involving humans, peri-domestic and wild animals in the Cameroonian focus of Bipindi, a study suggested the crucial role of animals in the maintenance of T. b. gambiense (Funk et al., 2013). In Equatorial Guinea, if alternative reservoirs are ruled out and neglected in control programs, T. b. gambiense could be maintained in the wild cycle as has probably occurred in Luba, and may lead to virulent outbreaks in the future as has happened in the past (Steverding, 2008). As previously addressed by other authors (Gouteux and Artzrouni, 1996; Solano et al., 2013), vector control will be essential to a complete elimination and we strongly recommend its introduction in eradication programs to complement the active screening focused in the human patients.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the hunter teams of Equatorial Guinea and the staff of the National Sleeping Sickness Control Programme. We wanted to extend our thanks to the authorities of the Ministry of Health and Social Welfare of Equatorial Guinea and the Spanish Agency for International Development Cooperation (AECID). We are grateful to Dr. Vincent Jamonneau (IRD) for providing valuable comments of the manuscript. J-MB is supported by a Miguel Servet Fellowship (CP09/00300) and funded by “Fondo de Investigación Sanitaria”(FIS) PI10/01128. AB and MN are granted by ISCIII -Subdirección General de Redes y Centros de InvestigaciónCooperativa (RICET) RD12/0018/0001 and RD12/0018/0015, respectively.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.00765
References
- Ashford R. W. (2003). When is a reservoir not a reservoir? Emerg. Infect. Dis. 9 1495–1496. 10.3201/eid0911.030088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berberof M., Perez-Morga D., Pays E. (2001). A receptor-like flagellar pocket glycoprotein specific to Trypanosoma brucei gambiense. Mol. Biochem. Parasitol. 113 127–138. 10.1016/S0166-6851(01)00208-0 [DOI] [PubMed] [Google Scholar]
- Bitter W., Gerrits H., Kieft R., Borst P. (1998). The role of transferrin-receptor variation in the host range of Trypanosoma brucei. Nature 391 499–502. 10.1038/35166 [DOI] [PubMed] [Google Scholar]
- Cordon-Obras C., Berzosa P., Ndong-Mabale N., Bobuakasi L., Buatiche J. N., Ndongo-Asumu P., et al. (2009). Trypanosoma brucei gambiense in domestic livestock of Kogo and Mbini foci (Equatorial Guinea). Trop. Med. Int. Health 14 535–541. 10.1111/j.1365-3156.2009.02271.x [DOI] [PubMed] [Google Scholar]
- Cordon-Obras C., Cano J., Gonzalez-Pacanowska D., Benito A., Navarro M., Bart J. M. (2013). Trypanosoma brucei gambiense adaptation to different mammalian sera is associated with VSG expression site plasticity. PLoS ONE 8:e85072 10.1371/journal.pone.0085072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordon-Obras C., Garcia-Estebanez C., Ndong-Mabale N., Abaga S., Ndongo-Asumu P., Benito A., et al. (2010). Screening of Trypanosoma brucei gambiense in domestic livestock and tsetse flies from an insular endemic focus (Luba, Equatorial Guinea). PLoS Negl. Trop. Dis. 4:e704 10.1371/journal.pntd.0000704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtin F., Jamonneau V., Duvallet G., Garcia A., Coulibaly B., Doumenge J. P., et al. (2008). Sleeping sickness in West Africa (1906-2006): changes in spatial repartition and lessons from the past. Trop. Med. Int. Health 13 334–344. 10.1111/j.1365-3156.2008.02007.x [DOI] [PubMed] [Google Scholar]
- Dorst J., Dandelot P. (1997). Guide Des Mammifêres d′Afrique 2nd Edn. Neuchatel: Delachaux & Niesflé. [Google Scholar]
- Franco J. R., Simarro P. P., Diarra A., Jannin J. G. (2014a). Epidemiology of human African trypanosomiasis. Clin. Epidemiol. 6 257–275. 10.2147/CLEP.S39728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco J. R., Simarro P. P., Diarra A., Ruiz-Postigo J. A., Jannin J. G. (2014b). The journey towards elimination of gambiense human African trypanosomiasis: not far, nor easy. Parasitology 141 748–760. 10.1017/S0031182013002102 [DOI] [PubMed] [Google Scholar]
- Funk S., Nishiura H., Heesterbeek H., Edmunds W. J., Checchi F. (2013). Identifying transmission cycles at the human-animal interface: the role of animal reservoirs in maintaining gambiense human african trypanosomiasis. PLoS Comput. Biol. 9:e1002855 10.1371/journal.pcbi.1002855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson W., Mehlitz D., Lanham S. M., Godfrey D. G. (1978). The identification of Trypanosoma brucei gambiense in Liberian pigs and dogs by isoenzymes and by resistance to human plasma. Tropenmed. Parasitol. 29 335–345. [PubMed] [Google Scholar]
- Gouteux P. J., Artzrouni M. (1996). [Is vector control needed in the fight against sleeping sickness? A biomathematical approach]. Bull. Soc. Pathol. Exot. 89 299–305. [PubMed] [Google Scholar]
- Herder S., Simo G., Nkinin S., Njiokou F. (2002). Identification of trypanosomes in wild animals from southern Cameroon using the polymerase chain reaction (PCR). Parasite 9 345–349. 10.1051/parasite/2002094345 [DOI] [PubMed] [Google Scholar]
- Jamonneau V., Ilboudo H., Kabore J., Kaba D., Koffi M., Solano P., et al. (2012). Untreated human infections by Trypanosoma brucei gambiense are not 100% fatal. PLoS Negl. Trop. Dis. 6:e1691 10.1371/journal.pntd.0001691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzoe F. A. S., Schofield C. J. (2004). Strategic Review of Traps and Targets for Tsetse and African Trypanosomiasis Control. Report of Special Programme for Research and Training in Tropical Diseases TDR/IDE/TRY/05.1. Geneva: World Health Organization; 58. [Google Scholar]
- Louis F. J., Kohagne Tongue L., Ebo O. E. V., Simarro P. P. (2008). [Organizing an active screening campaign for human African trypanosomiasis due to Trypanosoma brucei gambiense]. Med. Trop. (Mars) 68 11–16. [PubMed] [Google Scholar]
- Morlais I., Grebaut P., Bodo J. M., Djoha S., Cuny G., Herder S. (1998). Detection and identification of trypanosomes by polymerase chain reaction in wild tsetse flies in Cameroon. Acta Trop. 70 109–117. 10.1016/S0001-706X(98)00014-X [DOI] [PubMed] [Google Scholar]
- Moser D. R., Cook G. A., Ochs D. E., Bailey C. P., Mckane M. R., Donelson J. E. (1989). Detection of Trypanosoma congolense and Trypanosoma brucei subspecies by DNA amplification using the polymerase chain reaction. Parasitology 99 (Pt 1) 57–66. 10.1017/S0031182000061023 [DOI] [PubMed] [Google Scholar]
- Njiokou F., Laveissiere C., Simo G., Nkinin S., Grebaut P., Cuny G., et al. (2006). Wild fauna as a probable animal reservoir for Trypanosoma brucei gambiense in Cameroon. Infect. Genet. Evol. 6 147–153. 10.1016/j.meegid.2005.04.003 [DOI] [PubMed] [Google Scholar]
- Njiokou F., Nimpaye H., Simo G., Njitchouang G. R., Asonganyi T., Cuny G., et al. (2010). Domestic animals as potential reservoir hosts of Trypanosoma brucei gambiense in sleeping sickness foci in Cameroon. Parasite 17 61–66. 10.1051/parasite/2010171061 [DOI] [PubMed] [Google Scholar]
- Noireau F., Gouteux J. P., Toudic A., Samba F., Frezil J. L. (1986). [Epidemiological importance of the animal reservoir of Trypanosoma brucei gambiense in the Congo. 1. Prevalence of animal trypanosomiasis in the foci of sleeping sickness]. Trop. Med. Parasitol. 37 393–398. [PubMed] [Google Scholar]
- Oda E., Solari A., Botto-Mahan C. (2014). Effects of mammal host diversity and density on the infection level of Trypanosoma cruzi in sylvatic kissing bugs. Med. Vet. Entomol. 28 384–390. 10.1111/mve.12064 [DOI] [PubMed] [Google Scholar]
- Penchenier L., Grebaut P., Eboo Eyenga V., Bodo J. M., Njiokou F., Binzouli J. J., et al. (1999). [The focus of human trypanosomiasis in Campo (Cameroon). History and endemic situation in 1998]. Bull. Soc. Pathol. Exot. 92 185–190. [PubMed] [Google Scholar]
- Simarro P. P., Cecchi G., Franco J. R., Paone M., Diarra A., Ruiz-Postigo J. A., et al. (2012). Estimating and mapping the population at risk of sleeping sickness. PLoS Negl. Trop. Dis. 6:e1859 10.1371/journal.pntd.0001859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simarro P. P., Franco J. R., Ndongo P., Nguema E., Louis F. J., Jannin J. (2006). The elimination of Trypanosoma brucei gambiense sleeping sickness in the focus of Luba, Bioko Island, Equatorial Guinea. Trop. Med. Int. Health 11 636–646. 10.1111/j.1365-3156.2006.01624.x [DOI] [PubMed] [Google Scholar]
- Simarro P. P., Mas J., Lancien J., Ona Sima F., Mateo M. J., Roche J. (1990). [Epidemiology of human trypanosomiasis in the Luba focus, in Equatorial Guinea]. Rev. Sanid. Hig. Publica (Madr) 64 517–534. [PubMed] [Google Scholar]
- Simo G., Asonganyi T., Nkinin S. W., Njiokou F., Herder S. (2006). High prevalence of Trypanosoma brucei gambiense group 1 in pigs from the Fontem sleeping sickness focus in Cameroon. Vet. Parasitol. 139 57–66. 10.1016/j.vetpar.2006.02.026 [DOI] [PubMed] [Google Scholar]
- Simo G., Mbida Mbida J. A., Ebo’o Eyenga V., Asonganyi T., Njiokou F., Grebaut P. (2014). Challenges towards the elimination of Human African Trypanosomiasis in the sleeping sickness focus of Campo in southern Cameroon. Parasit. Vectors 7 374 10.1186/1756-3305-7-374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobrino R., Ferroglio E., Oleaga A., Romano A., Millan J., Revilla M., et al. (2008). Characterization of widespread canine leishmaniasis among wild carnivores from Spain. Vet. Parasitol. 155 198–203. 10.1016/j.vetpar.2008.05.003 [DOI] [PubMed] [Google Scholar]
- Solano P., Torr S. J., Lehane M. J. (2013). Is vector control needed to eliminate gambiense human African trypanosomiasis? Front. Cell. Infect. Microbiol. 3:33 10.3389/fcimb.2013.00033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steuber S., Abdel-Rady A., Clausen P. H. (2005). PCR-RFLP analysis: a promising technique for host species identification of blood meals from tsetse flies (Diptera: Glossinidae). Parasitol. Res. 97 247–254. 10.1007/s00436-005-1410-y [DOI] [PubMed] [Google Scholar]
- Steverding D. (2008). The history of African trypanosomiasis. Parasit. Vectors 1 3 10.1186/1756-3305-1-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner C. M., Mclellan S., Lindergard L. A., Bisoni L., Tait A., Macleod A. (2004). Human infectivity trait in Trypanosoma brucei: stability, heritability and relationship to sra expression. Parasitology 129 445–454. 10.1017/S0031182004005906 [DOI] [PubMed] [Google Scholar]
- World Health Organization. (2013). Control and surveillance of human African trypanosomiasis. World Health Organ. Tech. Rep. Ser. 1–237. [PubMed] [Google Scholar]
- Zanet S., Sposimo P., Trisciuoglio A., Giannini F., Strumia F., Ferroglio E. (2014). Epidemiology of Leishmania infantum, Toxoplasma gondii, and Neospora caninum in Rattus rattus in absence of domestic reservoir and definitive hosts. Vet. Parasitol. 199 247–249. 10.1016/j.vetpar.2013.10.023 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.