Abstract
The structure of the bacterial leucine transporter from Aquifex aeolicus (LeuTAa) has been used as a model for mammalian Na+/Cl−-dependent transporters, in particular the serotonin transporter (SERT). The crystal structure of LeuTAa liganded to tricyclic antidepressants predicts simultaneous binding of inhibitor and substrate. This is incompatible with the mutually competitive inhibition of substrates and inhibitors of SERT. We explored the binding modes of tricyclic antidepressants by homology modeling and docking studies. Two approaches were used subsequently to differentiate between three clusters of potential docking poses: 1) a diagnostic SERTY95F mutation, which greatly reduced the affinity for [3H]imipramine but did not affect substrate binding; 2) competition binding experiments in the presence and absence of carbamazepine (i.e., a tricyclic imipramine analog with a short side chain that competes with [3H]imipramine binding to SERT). Binding of releasers (para-chloroamphetamine, methylene-dioxy-methamphetamine/ecstasy) and of carbamazepine were mutually exclusive, but Dixon plots generated in the presence of carbamazepine yielded intersecting lines for serotonin, MPP+, paroxetine, and ibogaine. These observations are consistent with a model, in which 1) the tricyclic ring is docked into the outer vestibule and the dimethyl-aminopropyl side chain points to the substrate binding site; 2) binding of amphetamines creates a structural change in the inner and outer vestibule that precludes docking of the tricyclic ring; 3) simultaneous binding of ibogaine (which binds to the inward-facing conformation) and of carbamazepine is indicative of a second binding site in the inner vestibule, consistent with the pseudosymmetric fold of monoamine transporters. This may be the second low-affinity binding site for antidepressants.
Introduction
The core physiological role of neurotransmitter:sodium symporters (NSS) is to terminate neurotransmission by rapid removal of neurotransmitters from the synaptic cleft and reuptake into presynaptic neurons and/or surrounding glial cells. The subfamily of NSS responsible for the reuptake of monoamines comprises the transporters for noradrenaline (NET), dopamine (DAT), and serotonin (SERT). Structures of these transporters have not yet been solved, but considerable insights have been granted by crystallographic data of their bacterial homolog LeuTAa, isolated from thermophilic bacteria Aquifex aeolicus. The first LeuTAa data revealed a high-resolution occluded structure devoid of water with a single leucine molecule in coordination with two Na+ ions (Yamashita et al., 2005). A salt bridge composed of residues Arg30 and Asp404 is suggested to act as a barrier between the bound leucine in the occluded state and the aqueous pathway connecting the primary binding site to the extracellular bulk solution.
The substrate translocation process was conceptually explained by an alternate access model (Jardetzky, 1966). This model posited a conformational cycle between inward- and outward-facing conformations and thus implicitly predicted a pseudosymmetry in the transporter molecule. This conjecture was confirmed by the structure of LeuTAa (Yamashita et al., 2005) and other unrelated transporters (Ressl et al., 2009). In LeuTAa, the pseudosymmetrical internal repeats comprise TMs 1 to 5 and TMs 6 to 10; TMs 11 and 12 represent a scaffold for this inverted repeat and provide a contact site for the dimeric interface (Yamashita et al., 2005); this is also true for SERT (Just et al., 2004). Because of its distant relation to monoamine transporters, LeuTAa has been a major source of inspiration for educated guesses on the mechanics of the translocation process in monoamine transporters (Shi et al., 2008).
In addition, LeuTAa was also used as template to understand how antidepressants bind to SERT and NET. In fact, the low-affinity interaction sufficed to allow for visualizing tricyclic antidepressants (TCAs) (i.e., clomipramine and desipramine) in the extracellular vestibule of the transporter (Singh et al., 2007; Zhou et al., 2007). The LeuTAa cocrystal structures with TCAs reveal a binding pocket harboring leucine and two Na+ ions (e.g., and ). TCA binding takes place at a site 11 Å above the central substrate binding site, thus impeding the dissociation of leucine to bulk solution via the extracellular pathway. In fact, TCAs are (low-affinity) inhibitors of substrate transport by LeuTAa, but the inhibition is uncompetitive: for instance, clomipramine, at saturating concentrations, slowed [3H]leucine dissociation from LeuTAa by up to 700-fold (Singh et al., 2007). This mode of inhibition, however, is incompatible with the competitive inhibition of antidepressant binding to SERT and NET by their substrates and vice versa, of substrate translocation by antidepressants.
It has therefore remained a matter of debate whether the observations with LeuTAa are of any relevance to understanding how antidepressants bind to the monoamine transporters SERT and NET. Here, we addressed this discrepancy by performing docking of the prototypical TCA imipramine onto a homology model of SERT based on the LeuTAa structure in the “open-to-out” conformation (Singh et al., 2008). Docking of imipramine was consistent with the hypothesis that the ring system of TCAs occupied the extracellular vestibule of the transporter, and the dimethyl-aminopropyl side chain reached deep into the substrate binding pocket. This conjecture was verified by using two approaches: 1) by introducing the Y95F mutation to remove the candidate hydrogen bond donating hydroxyl group to the basic side chain nitrogen of imipramine; and 2) by showing that carbamazepine (a tricyclic analog of imipramine devoid of the long side chain) and serotonin were accommodated simultaneously in SERT. In addition, we identified a second binding site for the tricyclic ring system in the inner vestibule of monoamine transporters, an observation that is consistent with the pseudosymmetric fold of the transporters.
Materials and Methods
Alignment and Homology Modeling
The bacterial leucine transporter (LeuTAa) in the “open-to-out” conformation (Protein Data Bank ID 3F3A; Singh et al., 2008) served as the template for building a protein homology model. The sequence alignment between LeuTAa and the primary sequence of hSERT (as given in the UniProt database, entry code P31645), based on a comprehensive study on NSS members (Beuming et al., 2006). However, initial models showed Lys399 in extracellular loop EL4b, pointing into a hydrophobic region. Thus, the alignment was slightly modified, which led to the removal of a gap in the query sequence (Supplemental Fig. 1). The final models were created using MODELLER 9.4 as implemented in Accelrys Discovery Studio 2.5 (Accelrys, San Diego, CA). Three different modeling runs generating 250 models each have been performed, which included also cocrystallized ligands of the template. These comprise the two Na+ ions, the cocrystallized tryptophan (with removed -COOH group in site 1), and the cocrystallized tetradecane, located in the extracellular vestibule. The top scored models with respect to DOPE score and PDF score were analyzed with the program PROCHECK (Laskowski et al., 1993). The most favored model showed 93.5% of the residues in allowed regions and four residues in disallowed regions. It was additionally assessed on the SWISS MODEL server (http://swissmodel.expasy.org/) Arnold et al., 2006) using the QMEAN scoring function (Benkert et al., 2008). This indicates high accuracy of the homology model in the conserved region of the binding site and parts of the extracellular vestibule (Supplemental Fig. 2). We also verified that the model was consistent with the results from previous mutagenesis studies, in particular with respect to the location and orientation of residues important for ligand interaction (Supplemental Table S1) (Barker et al., 1999; Chen and Rudnick, 2000; Henry et al., 2003, 2006; White et al., 2006; Forrest et al., 2008; Andersen et al., 2009; Field et al., 2010). After manual placement of the two Na+ ions (at similar position as in the template) (Singh et al., 2008) and the Cl− ion (Zomot et al., 2007; Forrest et al., 2007), partial charges were calculated with MOE (molecular operating environment) 2009.10. Furthermore, hydrogens were assigned with the “Protonate3d” function of MOE. Thereafter, residues within a radius of 3 Å around the ions were minimized (AMBER99 force field; RMS gradient, 0.1).
Docking and Evaluation of the Poses
The ligands carbamazepine, dihydrocarbamazepine, and amitriptyline were built with the molecule builder tool in MOE. Imipramine, clomipramine, and desipramine were taken from the crystal structures with the Protein Data Bank IDs 2Q72, 2Q6H, and2QB4, respectively (Singh et al., 2007). Chemical structures for all compounds docked are given in Supplemental Fig. S3. Initial starting conformations were obtained by protonation of the ligands at pH 7.4 and a stochastic search with the MMFF94x force field. Conformational sampling of the ligands was achieved by short MD-simulations at 2000 K for 10 ps in MOE. Two thousand snapshots were taken every 5 fs. After energy minimization, the 100 most diverse conformations were taken as input structures for docking. The binding site was defined by using the “Site Finder” tool in MOE. Residues within a radius of 4.5 Å of the generated “dummy atoms” were selected and were shown to be are analogous to those of the template structure (Zomot et al., 2007; Singh et al., 2008). Thereafter, the conformation databases of the selected TCAs were docked into the defined binding site by keeping the protein fixed.
To avoid strong bias caused by scoring functions, we fully explored all possible poses by allowing for up to 2300 poses per docking run. Subsequent analysis combined hierarchical clustering, protein ligand interactions, and consensus scoring (Supplemental Fig. S4). First, similar poses were removed by complete linkage clustering based on the root mean square deviation values at a level of 1 Å, done with the “clv” package in R 2.11 (The Comprehensive R Archive Network; http://cran.r-project.org/). A first selection criterion was based on the “protein ligand interaction fingerprint” (PLIF), which allow the selection of poses because of their interaction pattern with the protein. The residues were chosen because of the importance for transporter function (Asp98; Barker et al., 1999), number of poses interacting (Glu493), and location of the residue (Chen and Rudnick, 2000; Henry et al., 2006; Ile179 in the extracellular vestibule; Tyr95 in the central binding site). The most frequently populated interacting residues of hSERT with imipramine were Glu493, Ile179, Trp182, Arg104, Pro403, Lys490, Tyr95, and Gly442 (decreasing order, see Supplemental Table S2). All poses showing contacts with one of the four amino acids mentioned above were extracted into separate databases and minimized with the “LigX” suite in MOE. This minimization step allowed slight adaptations of both the ligand and the protein. Thereafter, the minimized complexes were scored by the four scoring functions available in MOE and reclustered at a level of 4 Å. The centroid pose and the top scored pose of each cluster was fused into the final database. Finally, the PLIF was calculated for the minimized complexes and used as final filter: only poses with ionic interactions with Asp98 or Glu493 were considered for final assessment. The resulting 28 poses were visually analyzed. Assuming that simultaneous binding of serotonin and imipramine is not possible, placements without interactions with the central binding site were omitted. Nineteen poses were finally taken together.
Materials
Dulbecco’s modified Eagle’s medium and trypsin were purchased from PAA Laboratories GmbH (Pasching, Austria). Fetal calf serum was purchased from Invitrogen (Carlsbad, CA). -(−)-2-β-[3H]carbomethoxy-3-β-(4-fluorophenyl)tropane ([3H]WIN35,428; 85.9 and 76 Ci/mmol), [3H]imipramine (47.5 Ci/mmol), and [3H]5-hydroxytryptamine ([3H]5-HT, serotonin; 28.1 Ci/mmol) were purchased from PerkinElmer Life and Analytical Sciences (Waltham, MA). [3H]1-methyl-4-phenylpyridinium ion ([3H]MPP+) (85 Ci/mmol) was supplied by American Radiolabeled Chemicals (St. Louis, MO). Serotonin (5-HT), s-(+)-3,4-methylenedioxy-methamphetamine (MDMA), paroxetine, carbamazepine, and para-chloroamphetamine (PCA) were purchased from Sigma-Aldrich (St. Louis, MO). 1-Methyl-4-phenylpyridinium ion (MPP+) was purchased from Sigma/RBI (Natick, MA), the polyclonal rabbit antibody directed against green fluorescent protein was from Abcam PLC (Cambridge, UK). Ibogaine was kindly donated by Sacrament of Transition (Maribor, Slovenia).
Site-Directed Mutagenesis, Cell Culture, and Transfections
The SERTY95F mutation was generated in the pEYFP-C1-hSERT background (Schmid et al., 2001) using the QuikChange Lightning site-directed mutagenesis kit (Stratagene, La Jolla, CA), and confirmed with automatic sequencing (AGOWA Genomics, Berlin, Germany). The primer sequence (5′ to 3′ sense strands), with the mutated nucleotides indicated in boldface font, was CCTTCTCTCAGTGATTGGCTTTGCTGTGGACC. The generation of HEK293 T-REx cells expressing the hSERT under the control of a tetracycline inducible promoter and of the HEK293 cells stably expressing DAT has been described previously (Scholze et al., 2002). For transient expression, HEK293 cells were transfected with wild-type or mutant SERT using CaPO4 coprecipitation. For generation of a stable cell line of SERTY95F, HEK293 cells were transfected using the CaPO4 coprecipitation method, and G418 (Geneticin; Invitrogen) was added for clone selection.
Confocal Laser Scanning Microscopy
Confocal microscopy was carried out with a Zeiss Axiovert 200-LSM 510 microscope equipped with argon and helium/neon lasers (30 and 1 mW, respectively) and a 40× oil immersion objective (Zeiss Plan-Neofluar; Carl Zeiss, Thornwood, NJ). Stably transfected cells (105 cells/1.6-cm glass coverslip) were placed in a chamber containing Krebs-HEPES buffer. Yellow fluorescent protein (YFP)-tagged proteins were detected with a band pass filter (475–525 nm) using the 458-nm laser line. Images, obtained as z-stacks (slice thickness, ~1 μm), were analyzed by the LSM Image Browser to study the surface expression of the wild-type and mutant SERTs.
Radioligand Binding Assays
Membranes were prepared from HEK293 cells stably expressing wild-type or mutant SERT or DAT as in Korkhov et al., (2006); expression levels in individual cell clones varied between 4 and 40 pmol/mg; buffers used for the preparation of DAT-expressing membranes contained 10 μM ZnCl2 and were devoid of EDTA (because Zn2+ promotes the outward-facing conformation that is required for high-affinity binding of inhibitors (Scholze et al., 2002). [3H]Imipramine equilibrium binding to SERT was performed in duplicate incubations in an assay volume of 0.2 to 0.5 ml (adjusted appropriately to avoid radioligand depletion). In competition binding experiments, membrane preparations (8–25 μg/assay) were incubated with the radioligand (~2 nM [3H]imipramine), the indicated concentrations of carbamazepine, and increasing concentrations of a second competing inhibitor in buffer (20 mM Tris-HCl, 1 mM EDTA, 2 mM MgCl2, 3 mM KCl, and 120 mM NaCl, pH adjusted to 7.4). Nonspecific binding was determined in the presence of 3 μM paroxetine. Saturation experiments were done with serial dilutions of [3H]imipramine ranging from ~0.1 to 50 nM. Equilibrium binding of [3H]WIN35,428 to DAT was performed in a similar manner but in an assay volume of 0.1 ml containing ~10 nM [3H]WIN35,428 (for competition binding experiments), the indicated concentrations of carbamazepine and ibogaine and buffer (20 mM Tris-HCl, 1 mM MgCl2, 3 mM KCl, 120 mM NaCl, and 10 μM ZnCl2, pH adjusted to 7.4). In saturation experiments, the concentrations of [3H]WIN35,428 ranged from ~1 to 100 nM. Nonspecific binding was determined in parallel in the presence of 3 μM methylphenidate. Binding was allowed to proceed for 15 to 60 min at 20°C and terminated by rapid filtration onto GF/A glass microfiber filters (Whatman International Ltd, Maidstone, UK) presoaked in 0.5% polyethylenimine (Sigma-Aldrich). The radioactivity trapped on the filter was determined by liquid scintillation counting.
Uptake and Release Assays
HEK293 cells stably expressing wild-type or mutant SERT were seeded onto 48-well plates (0.5 × 105 cells/well) 24 h before the experiment. For saturation experiments, the specific activity of [3H]5-HT was varied between 30 (0.2 μM) and 200 cpm/pmol (30 μM) by the addition of unlabeled 5-HT. Assay conditions were as outlined by Korkhov et al. (2006). For release studies, HEK293 cells stably expressing wild-type SERT were grown on coverslips in 96-well plates (4 × 104 cells/well), preloaded with 0.4 μM [3H]serotonin (specific activity ~30 cpm/pmol) or 0.1 μM MPP+ (~90 cpm/pmol), for 20 min at 37°C, in a final volume of 0.1 ml/well. The coverslips were transferred into superfusion chambers, and excess radioactivity was washed out with Krebs-HEPES buffer at 25°C for 45 min with perfusion rates of 0.7 ml/min. Upon attainment of stable efflux, 2-min fractions were collected for liquid scintillation counting.
Immunoblotting
Membranes were prepared from HEK293 cells stably expressing wild-type or mutant SERT or DAT as in Korkhov et al. (2006). Proteins (10 μg) were resolved by denaturing gel electrophoresis on a 9% SDS-polyacrylamide gel and subsequently transferred onto a nitrocellulose membrane. The nitrocellulose membrane was blocked with 3% bovine serum albumin in Tris-buffered saline (20 mM Tris-HCl, pH 7.5, and 150 mM NaCl) containing 0.1% Tween 20, the polyclonal rabbit anti-green fluorescent protein antibody was used at 1:5000 dilution. Immunoreactive bands were visualized with horseradish peroxidase-coupled anti-rabbit antibody (diluted 1:5000) by enhanced chemiluminescence (GE Healthcare, Chalfont St. Giles, Buckinghamshire, UK).
Data Analysis
Data from binding and uptake experiments were subjected to nonlinear, least-squares curve-fitting to equations for a rectangular hyperbola using a Marquardt-Levenberg algorithm. The fit was not improved by employing a logistic equation (Hill equation). Data transformed into Dixon plots were fitted by linear regression. Statistical significance was calculated by t test or Wilcoxon test as appropriate.
Results
Ligand Docking Shows Binding of TCAs in the Changeover of Site 1 and Site 2
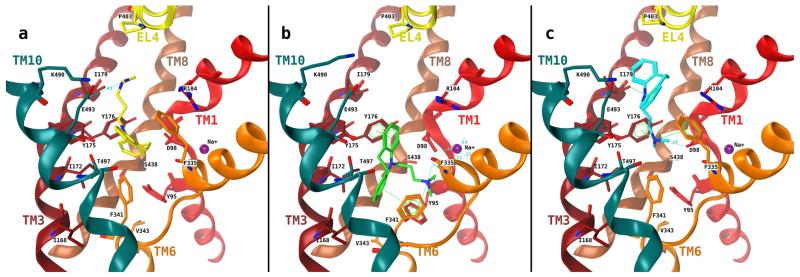
Possible binding modes for TCAs were explored by performing docking studies into a homology model of hSERT based on the crystal structure of LeuTAa. The TCA imipramine was docked into the threaded structure using a protocol that exhaustively samples the chemical space for docking poses in the defined binding site. After docking, the most popular interactions were found with residues Glu493 (Yamashita et al., 2005) forms with Arg104 part of the extracellular gate), Ile179 (one turn above Tyr176, located near the extracellular side of the central binding site; Chen and Rudnick, 2000), Trp182 (White et al., 2006; Beuming et al., 2006; and conserved in SERT, NET, DAT, and LeuT; is located in the extracellular vestibule), Pro403 (located at the kink in EL4), Arg104, Lys490 (one turn above Glu493, toward bulk solution; Andersen et al., 2009), Tyr95 (located at the intracellular side of the central binding site; Henry et al., 2006), and Gly442 (in proximity to Tyr95) of the 1916 docking poses obtained three distinct clusters of poses were retrieved by analyzing the PLIFs, clustering, and consensus scoring. Poses of the first cluster show ionic interaction between Glu493 and the charged nitrogen of the imipramine side chain as main interaction. These poses are similar to the crystal structures published in 2007 (Singh et al., 2007; Zhou et al., 2007), but the tricyclic ring is buried into the central binding site. Further interactions of the tricyclic ring with the aromatic residues Tyr175, Tyr176, and Phe335 were observed (Fig. 1a). In the second cluster, the dibenzazepine ring system is placed into the hydrophobic pocket of the central binding site surrounded by Ile168, Ile172, Tyr175, and Tyr176 of TM3, Val343 and Phe341 of TM6, and Thr497 of TM10. The nitrogen atom shows an ionic interaction with Asp98. The ring system shows π–π interactions with Tyr176 and Phe341 (Fig. 1b). The orientations in cluster 3 show the ring system in the extracellular vestibule and the amino propylene side chain rising into the central binding site with an ionic interaction between Asp98 and the charged nitrogen. Other interactions are observed between the nitrogen and Tyr176 and Phe335 (Fig. 1c). For the sake of clarity, it has to be noted that for each cluster, only one representative pose is shown. Interacting amino acids were identified with the function “ligand interactions” from MOE and varied slightly for different poses within one cluster (see Supplemental Table 3 for cluster 3 as an example). All of these docking poses are consistent with the observation of the competitive inhibition of SERT but differ in the role of Tyr95: in group 1 docking poses, the dimethyl-aminopropyl side chain points toward the bulk solution (Fig. 1a); in groups 2 and 3 poses, the hydroxyl group of Tyr95 is within a distance where hydrogen bonding to the nitrogen in the side chain side of imipramine could be possible, most probably mediated by a structural water molecule as seen in the template. To differentiate between these candidate binding modes, we mutated Tyr95 to phenylalanine. This choice also seemed justified, because there is a phenylalanine residue in the corresponding position in the NET and DAT. NET and DAT bind imipramine with an affinity that is approximately 10- and 1000-fold lower, respectively, than SERT.
Fig. 1.
Final clusters of docking poses of imipramine in the binding cavity of hSERT. TMs 1 (red), 3 (dark red), 6 (orange), 8 (brown, semitransparent), and 10 (cyan) are depicted in the absence of the remaining TMs for clarity. a, cluster 1 poses contain two distinct placements: the nitrogen atom is placed near Glu493 (a), interaction of the nitrogen with Asp98 is observed (b), and both are analogous to the crystal structure 2Q72. b, cluster 2 poses: Asp98 coordinates the charged nitrogen of imipramine, and the hydrophobic ring system is placed into the hydrophobic region of the binding site. c, cluster 3 poses: the tricyclic ring system is placed in the outer vestibule and the amino propyl side chain reaches into binding site 1. Interactions have been identified using the ligand interactions feature (distance-based) as implemented in MOE.
The TCA Dimethyl-Aminopropyl Side Chain Resides in the Primary Substrate Binding Pocket
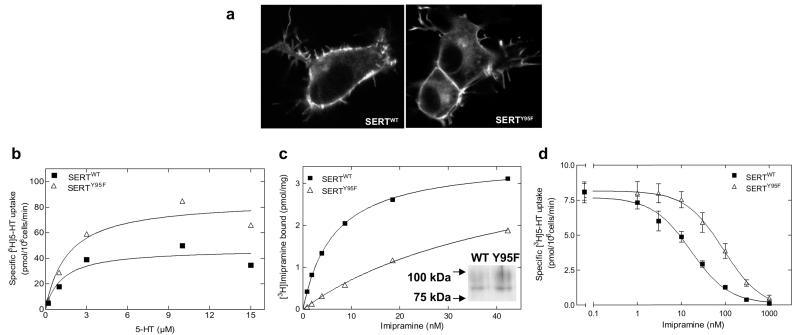
It has been shown previously that Tyr95 in the serotonin transporter coordinates high-affinity recognition of antidepressants (Henry et al., 2006); accordingly, we mutated Tyr95 to phenylalanine. We hypothesized the candidate hydrogen bond donor in the hydroxyl group in Tyr95 to mediate this high-affinity interaction with the TCA side chain. When expressed in HEK293 cells, both SERTWT and SERTY95F were found predominantly at the cell surface (Fig. 2a). This indicated the absence of gross misfolding, because incorrectly folded monoamine transporters are retained within the cell (Korkhov et al., 2008). Likewise, the affinity of SERTWT and SERTY95F for substrate uptake was comparable (Fig. 2b; KM, wt = 1.2 ± 0.8 μM, and KM, Y95F = 1.6 ± 0.8 μM). Differences in Vmax of uptake were likely to be accounted for by differences in expression levels (Fig. 2c, inset). As expected, removal of the hydroxyl group of Tyr95 resulted in a pronounced decrease in the affinity of imipramine. This was seen regardless of whether the affinity of imipramine for SERTY95F was determined directly in a binding assay using a membrane preparation (Fig. 2c, ▵) or via inhibition of substrate uptake (Fig. 2d, ▵). The affinity of SERTY95F for imipramine was lower than that of SERTWT; KD values were 6.4 ± 0.4 and 46.7 ± 4.9 nM (means ± S.D., n = 4; p < 0.01) for SERTWT and SERTY95F, respectively. This corresponds to a loss of 1.2 kcal/mol, which is consistent with the contribution of an H-bond to binding free energy (0.5–2 kcal/mol). The difference was less pronounced if imipramine-induced inhibition of serotonin uptake was examined: IC50 values were 16.3 ± 1.4 and 90.9 ± 20.9 nM (means ± S.D., n = 4; p < 0.01) for SERTWT and SERTY95F, respectively.
Fig. 2.
Cellular expression and functional characterization of wild-type and mutant SERTs. a, the cellular expression of wild-type or mutant SERTs was examined by confocal laser scanning microscopy. Representative images of HEK293 cells (105 cells/16-mm glass coverslip) stably expressing YFP-tagged transporters indicate that the expression pattern of mutated proteins are similar to those shown by the wild-type SERT. b, for saturation uptake experiments, HEK293 cells stably expressing wild-type or mutant SERTs were seeded onto 48-well plates (0.5 × 105 cells/well) 24 h before experiments. Cells were incubated with [3H]5-HT with or without 10 μM paroxetine to determine nonspecific uptake. The assay was conducted in duplicates and reaction stopped after 1 min with ice-cold Krebs-HEPES buffer. Data, plotted according to the hyperbolic model, are shown as means of a representative experiment carried out in duplicate. c, imipramine binding to SERTWT and SERTY95F: the incubation was performed in a final volume of 250 μl for 15 min at 27°C. Nonspecific binding was determined in the presence of 3 μM paroxetine and was ≤10% at the KD concentration of SERTWT. Data are from a single experiment carried out in duplicate, which was repeated three times. Average Bmax values were 3.6 ± 0.1 and 4.0 ± 0.3 pmol/mg for SERTWT and SERTY95F, respectively. The Bmax of the mutant is likely to be underestimated: because of the low affinity of [3H]imipramine, some bound ligand dissociates during the separation artifact. This conjecture was verified by immunoblotting (insert), which shows substantially higher levels of SERTY95F than of SERTWT. The immunoblot revealed two bands: the top band corresponds to the mature, fully glycosylated form, and the bottom form corresponds to the core-glycosylated (endoplasmic reticulum resident) form. d, inhibition of [3H]5-HT uptake by imipramine was determined in HEK293 stably expressing wild-type and mutant SERTs. The cells were incubated with 0.1 nM [3H]5-HT and increasing concentrations of imipramine, in the presence or absence of 10 μM paroxetine (to determine nonspecific uptake). The data (n = 3) were analyzed as described under Materials and Methods and plotted according to the sigmoidal model.
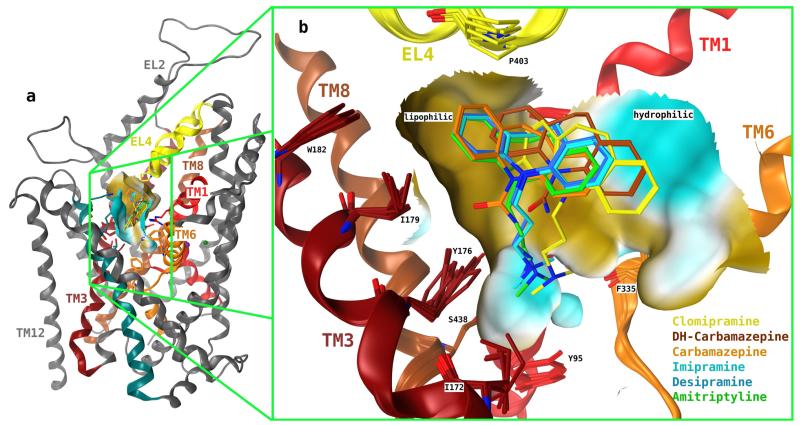
These observations are consistent with docking poses 2 and 3 but are incompatible with docking pose group 1. Specifically, the results argue for a role of hydrogen bonding between the hydroxyl group of Tyr95 to the nitrogen in the dimethylaminopropyl side chain of imipramine. In the case of cluster 2 poses, the dimethylammonium group more likely interacts via a cation/π-interaction rather than via an H-bond. This would also rule out docking poses 2. However, one has to bear in mind that both the dimethylaminopropyl side chain and the tyrosine are highly flexible moieties and thus could accommodate orientations, which also allow for H-bonding. Thus, to further validate our experimental results we sought for a complementary approach: the tricyclic compounds carbamazepine and dihydrocarbamazepine were docked into the structural model of SERT: dihydrocarbamazepine has the same ring system as imipramine but the dimethyl-aminopropyl side chain is replaced by a carboxamido group. In carbamazepine, the flexibility of the central epine ring is restricted by a double bond. In the main groups of docking poses carbamazepine and dihydrocarbamazepine were found to occupy positions similar to that of the tricyclic ring system of imipramine, desipramine, and amitriptyline (Fig. 3).
Fig. 3.
Most favored docking poses of TCAs onto hSERT. Overlay of TCAs imipramine, desipramine, amitriptyline, clomipramine, carbamazepine, and dihydrocarbamazepine as obtained by superposition of group 3 of docking poses. The ring system of the TCAs, carbamazepine, and dihydrocarbamazepine occupies the external vestibule, and the basic side chain of the TCAs is pointing toward binding site 1. Furthermore, the dibenzazepine ring system is placed near a hydrophobic region (brown) of the protein.
Carbamazepine and Imipramine Share a Common Binding Pocket in SERT
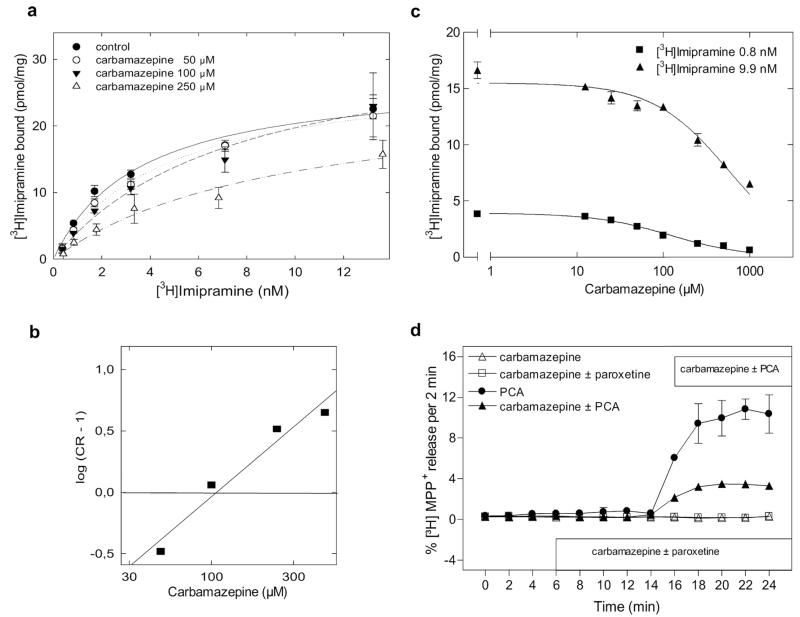
The docking studies predicted that carbamazepine should compete with imipramine for binding to SERT. This prediction was verified by two approaches. First, saturation experiments with [3H]imipramine were carried out in the presence of increasing concentrations of imipramine (Fig. 4a). Carbamazepine caused a rightward shift in the saturation curve of [3H]imipramine in a manner that was consistent with competitive inhibition. This is most readily evident from the Schild plot depicted in Fig. 4b: the slope of the Schild regression (1.13) was close to unity, and the intercept with the x-axis gave a pKB of 2.03 (i.e., an affinity estimate of 93 μM for carbamazepine). Second, carbamazepine was allowed to compete for [3H]imipramine binding at two different concentrations of the radioligand (i.e., 0.8 and 9.9 nM). The shift in the apparent IC50 values (119 ± 15 versus 563 ± 61 μM) at 0.8 and 9.9 nM [3H]imipramine, respectively; n = 3) was consistent with that predicted from the Cheng-Prusoff equation (Ki = IC50/[1 + L/KD]). The resulting affinity estimates Ki were in the range of 85 to 95 μM and thus consistent with the affinity estimate obtained from the Schild regression. We also investigated dihydrocarbamazepine: it also inhibited [3H]imipramine binding, albeit with a lower affinity than carbamazepine (data not shown).
Fig. 4.
Carbamazepine is a low-affinity SERT ligand that shares the binding site of imipramine and precludes amphetamine action on SERT. Saturation of [3H]imipramine binding to SERT in the presence of increasing concentrations of carbamazepine, mean ± S.E.M. (n = 3). a, carbamazepine displaces [3H]imipramine at a KI predicted by the Cheng-Prusoff approximation for competitive inhibition (b) Schild plot from the data presented in a reveals the following parameters: slope = 1.13, r = 0.976 with an x-intercept of pKB = 2.03 for carbamazepine (c). For release assays (d), culture medium was removed from stably transfected HEK293 cells (40 × 104 cells/well grown in 96-well plates on coverslips). The cells were preloaded with 0.4 μM [3H]serotonin or with 0.1 μM [3H]MPP+ for 20 min at 37°C in a final volume of 100 μl of Krebs-HEPES buffer/well. The coverslips were placed into superfusing chambers, and excess radioactivity was washed out with Krebs-HEPES buffer at 25°C for 45 min at a perfusion rate of 0.7 ml/min. Upon achieving stable efflux, 2-min fractions were collected, and data were analyzed as described under Materials and Methods.
Carbamazepine was reported to cause serotonin release from brain slices in a manner independent of an action on SERT (Dailey et al., 1997). We confirmed that carbamazepine per se failed to release [3H]serotonin from SERT-expressing preloaded HEK293 cells (Fig. 4d). However, carbamazepine blunted serotonin-release induced by para-chloroamphetamine (Fig. 4d). This observation is consistent with the ability of carbamazepine to occupy the imipramine binding site of SERT and to thereby antagonize the releasing action of amphetamines.
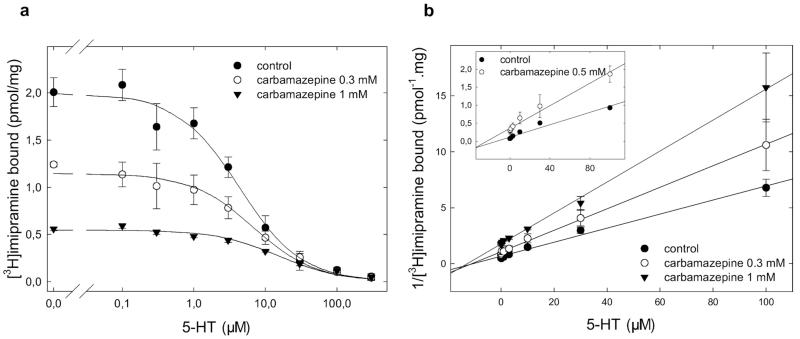
Substrates and Inhibitors Bind to SERT in a Mutually Nonexclusive Manner in the Presence of Carbamazepine
Docking pose groups 2 and 3 can thus be differentiated by the position of carbamazepine relative to the substrate binding site: in group 3, carbamazepine allows for simultaneous occupancy of the substrate binding site because it lacks the aliphatic dimethyl-aminopropyl side chain of imipramine. In contrast, in group 2 docking poses, carbamazepine would occupy the substrate binding site properly. Serotonin was allowed to compete for [3H]imipramine in the absence and presence of carbamazepine to differentiate between these two binding modes (Fig. 5a). Dixon plots (in which the reciprocal of bound radioactivity is plotted as a function of one inhibitor at a fixed concentration of the second inhibitor) allow to test whether two inhibitors can occupy the same binding site simultaneously or whether their binding is mutually exclusive (Segel, 1975). When the data summarized in Fig. 5a were replotted in a Dixon plot, they fell onto lines that pivoted around an intersection point close to the x-axis (Fig. 5b). This was also seen in membranes prepared from cells that expressed SERT at 10-fold higher density (Fig. 5b, inset). If two inhibitors compete for the same binding site, their binding is mutually exclusive, and this results in parallel lines in the Dixon plot. On the other hand, if both inhibitors can be bound simultaneously, Dixon plots are expected to yield intersecting lines (Segel, 1975). Thus, the data unequivocally show that binding of serotonin and carbamazepine is not mutually exclusive. Similar observations were also made with MPP+, another substrate of SERT (data not shown). SERT is also inhibited by a class of compounds that are collectively referred to as selective serotonin reuptake inhibitors (SSRIs). These are structurally unrelated to TCAs. Several recent modeling studies argued for binding of SSRIs to the substrate binding site (Tavoulari et al., 2009; Zhou et al., 2009; Andersen et al., 2010). If these assignments were correct, all of our experiments would be consistent with a model in which simultaneous binding of carbamazepine and paroxetine should be be possible. This conjecture was tested by measuring paroxetine-induced inhibition of [3H]imipramine binding in the absence and presence of carbamazepine. A Dixon plot of the inhibition curves again yielded a family of intersecting lines (Fig. 6a) showing that carbamazepine and paroxetine occupy mutually nonexclusive binding sites in the transporter.
Fig. 5.
Mutually nonexclusive binding of 5-HT to SERT in the presence of carbamazepine. a, competition of [3H]imipramine binding to SERT at equilibrium was performed in duplicate incubations in an assay volume of 0.2 to 0.5 ml (adjusted appropriately to avoid radioligand depletion). Membrane preparations (8–25 μg/assay) from HEK293 cells stably expressing wild-type SERT were incubated with the radioligand (~2 nM [3H]imipramine), the indicated concentrations of carbamazepine, and increasing concentrations of 5-HT in buffer (20 mM Tris-HCl, 1 mM EDTA, 2 mM MgCl2, 3 mM KCl, and 120 mM NaCl, pH adjusted to 7.4). Nonspecific binding was determined in the presence of 3 μM paroxetine. b, data in a is transformed into a Dixon plot for 5-HT by expressing the reciprocal of [3H]imipramine bound (measured in picomoles per milligram) as a function of 5-HT at a fixed concentration of carbamazepine. Data are shown as means ± S.D. of three independent experiments in duplicate.
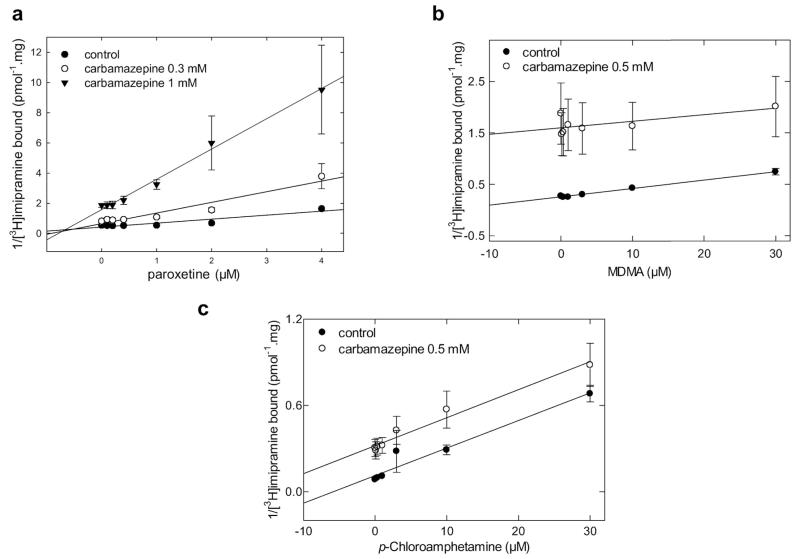
Fig. 6.
Paroxetine binds to SERT in a mutually nonexclusive manner in the presence of carbamazepine, but amphetamines elicit mutual exclusivity with carbamazepine. Competition of [3H]imipramine binding to SERT at equilibrium was performed in duplicate incubations in an assay volume of 0.2 to 0.5 ml (adjusted appropriately to avoid radioligand depletion). Membrane preparations (8–25 μg/assay) from HEK293 cells stably expressing wild-type SERT were incubated with the radioligand (~2 nM [3H]imipramine), the indicated concentrations of carbamazepine, and increasing concentrations of paroxetine, MDMA, and PCA in buffer (20 mM Tris-HCl, 1 mM EDTA, 2 mM MgCl2, 3 mM KCl, and 120 mM NaCl, pH adjusted to 7.4). Nonspecific binding was determined in the presence of 3 μM paroxetine. The binding data from the competition experiments is transformed into a Dixon plot for paroxetine (a), MDMA (b) and PCA (c) by expressing the reciprocal of [3H]imipramine bound (measured in picomoles per milligram) as a function of paroxetine, MDMA, and PCA at a fixed concentration of carbamazepine, respectively. Data are shown as means ± S.D. of three independent experiments in duplicate.
Mutually Exclusive Binding of para-Chloroamphetamine and MDMA to SERT but Not of Ibogaine in the Presence of Carbamazepine
Amphetamines are inwardly transported substrates of monoamine transporters that trigger serotonin efflux. Although the details remain enigmatic (Sitte and Freissmuth, 2010), it is clear that the conformation of SERT in its amphetamine bound state must differ from that of the serotonin or MPP+ liganded conformation. We therefore explored whether carbamazepine and MDMA (“ecstasy”) or para-chloroamphetamine were bound simultaneously: in the presence of carbamazepine, we observed a parallel shift in the Dixon plots for both MDMA (Fig. 6b) and para-chloroamphetamine (Fig. 6c). This is diagnostic of mutually exclusive binding of carbamazepine and amphetamines.
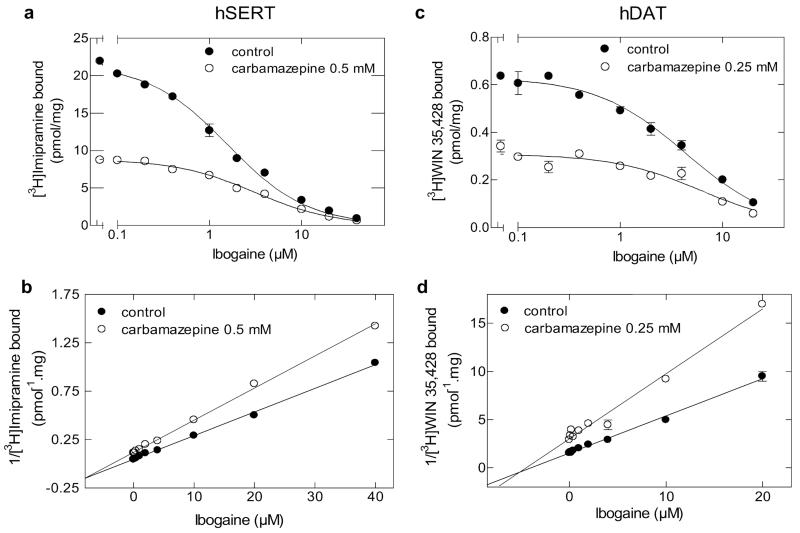
We also examined ibogaine, another ligand of SERT, which has been shown previously to stabilize SERT in the inward-facing conformation (Jacobs et al., 2007). Because binding of imipramine and carbamazepine is to the outward-facing conformation, ibogaine and carbamazepine are not predicted to be bound simultaneously. Surprisingly, this was not the case (Fig. 7). In the presence of ibogaine, we observed that the inhibition curves generated in the absence and presence of carbamazepine intersected when replotted as Dixon plots (Fig. 7, b and d). This was seen regardless of whether we examined the interaction of the two compounds in SERT (Fig. 7, a and b) or in DAT (Fig. 7, c and d). This finding can be rationalized by considering the pseudo 2-fold axis symmetry predicted to exist for NSS transport proteins (Yamashita et al., 2005; Singh et al., 2008; Ressl et al., 2009). A second binding site may exist in the inner vestibule that can accommodate the tricyclic ring of carbamazepine if ibogaine is bound to the substrate binding site in the inward-facing conformation. It is worth noting that a second (low-affinity) binding site has been described previously for imipramine in SERT (Schloss and Betz, 1995).
Fig. 7.
Mutually nonexclusive binding of ibogaine to SERT and DAT in the presence of carbamazepine. Competition of [3H]imipramine (a) or [3H]WIN35,428 (c) binding to SERT or DAT at equilibrium, respectively, was performed in duplicate incubations in an assay volume of 0.2 to 0.5 ml (SERT) or 0.1 ml (DAT). Membrane preparations (8–25 μg/assay) from HEK293 cells stably expressing wild-type SERT or DAT were incubated with the radioligand (~2 nM [3H]imipramine or ~10 nM WIN35,428), the indicated concentrations of carbamazepine, and increasing concentrations of ibogaine in buffer (20 mM Tris-HCl, 1 mM EDTA, 2 mM MgCl2, 3 mM KCl, and 120 mM NaCl, pH adjusted to 7.4). Buffers used for the preparation of DAT-expressing membranes contained 10 μM ZnCl2 and were devoid of EDTA. Nonspecific binding was determined in the presence of 3 μM paroxetine (SERT) or 3 μM methylphenidate (DAT). The binding data from the competition experiments is transformed into a Dixon plot for ibogaine (SERT) (b) and DAT (d) by expressing the reciprocal of [3H]imipramine or [3H]WIN35,428 bound (measured in picomoles per milligram) as a function of ibogaine at a fixed concentration of carbamazepine, respectively.
Discussion
Tricyclic antidepressant drugs were instrumental in the discovery of neurotransmitter reuptake and—in conjunction with other drugs and chemicals—in the dissection of the underlying mechanisms (Hertting and Axelrod, 1961). The serotonin transporter has remained a prime target for anti-depressant drugs since this seminal discovery. It has remained enigmatic how these drugs bind, because the transporter can accommodate a bewildering variety of structures. Based on the recent work with the bacterial homolog LeuTAa, two binding sites have been proposed (Supplemental Fig. 5): 1) a vestibular binding site (Singh et al., 2007; Zhou et al., 2007, 2009) that also functions as a secondary substrate binding site (Shi et al., 2008); and 2) the substrate binding site per se (Tavoulari et al., 2009; Andersen et al., 2010); the latter assignment is also supported by a combination of computer-assisted modeling and site-directed mutagenesis studies, which suggest that cocaine analogs occupy the substrate binding pocket in DAT (Beuming et al., 2008).
The importance of the vestibular binding site has been a matter of debate. It can, for instance, be argued that the low affinity of TCAs for LeuTAa render these models questionable. In fact, at the concentrations range in which tricyclic antidepressants bind to LeuTAa, they also bind to many other targets including mammalian Na+ and K+ channels and numerous G protein-coupled receptors. At the current stage, it would seem doubtful that any insight can be gained from the vestibular binding of desipramine to LeuTAa that would be relevant to understand the function of the pore region of the Na+ channel. Here, we have used an approach that combines docking studies and site-directed mutagenesis with a chemical biology approach, namely, the use of the tricyclic compound carbamazepine to prioritize docking hypotheses. Carbamazepine can be viewed as a truncated version of imipramine rendered more rigid by the presence of a double bond in the central epine ring. The underlying rationale is the assumption that carbamazepine and imipramine compete for the same binding site (i.e., the site to which the tricyclic ring system docks). Our experiments provided formal proof for this assumption: in two independent assays, carbamazepine fulfilled the criteria of a (low-affinity) competitive inhibitor of [3H]imipramine binding. In addition, it blunted amphetamine-induced reverse transport. It is worth noting that there are several ways by which carbamazepine and TCAs can be docked into the SERT model: however, assuming a common binding mode for the dibenzazepine ring system, there is only one group of docking poses that allows for simultaneous binding of carbamazepine and substrate: one in which the tricyclic ring is placed in the outer vestibule and the basic side chain points into the direction of the substrate binding site (docking pose group 3; Figs. 1c and 3). This explains both the mutually exclusive binding of imipramine and serotonin and the nonexclusive binding of carbamazepine and serotonin. It is worth noting that the top-scored docking poses, which belong to group 1 (Fig. 1a), would be consistent with the orientation of desipramine and clomipramine in the vestibule of LeuTAa (Singh et al., 2007; Zhou et al., 2007). However, it is nevertheless highly likely that this is not the correct orientation: the decrease in [3H]imipramine affinity that resulted in Y95F mutation cannot be accounted for by this orientation but is readily explained by the loss of a hydrogen bond between the hydroxyl group of the tyrosine side chain at residue Tyr95 and the tertiary amine in the dimethyl-aminopropyl group of imipramine. The importance of Tyr95 of SERT for the binding of tricyclic drugs was originally appreciated based on a substitution by cysteine (Henry et al., 2006), removing the phenyl ring and introducing a side chain can alter the flexibility of the helices by hydrogen bonding to main chain amide bonds one helical turn removed. The replacement of tyrosine with phenylalanine, however, is more subtle and sufficed to greatly reduce the affinity of imipramine (but not of serotonin). We are aware that, in our model, the distance between the hydroxyl of the tyrosine residue and the tertiary amine of imipramine is a priori too large for forming a hydrogen bond. However, the LeuTAa template structure used for homology modeling shows a structural water molecule interacting with Asn21, which is analogous to Tyr95 in SERT. This structural water molecule could thus mediate the hydrogen bond between Tyr95 and imipramine. Thus, taken together, our observations are consistent with the presence of two overlapping binding sites in SERT: the tricyclic ring system is in the outer vestibule, and the dimethyl-aminopropyl side chain reaches into the substrate binding site. Other inhibitors (i.e., SSRIs) must bind in distinct modes in this region to account for the observation that binding of paroxetine and carbamazepine was not mutually exclusive. The presence of two binding communicating sites can also be inferred from the increase in affinity and blocking efficacy seen with bivalent phenethylamines (Schmitt et al., 2010).
Our approach also allowed for probing the effect of amphetamines on the conformation of the binding pocket. Amphetamines are also subject to inward transport by SERT (and the other monoamine transporters), but they alter the conformation of the transporter in a way that allows for substrate efflux (Sitte and Freissmuth, 2010). Earlier experiments suggested a counter-transport mechanism contingent on the oligomeric arrangement of transporters (Seidel et al., 2005) and an important role of serine/threonine-kinase-mediated phosphorylation of residues in the N terminus of DAT and SERT (Fog et al., 2006). Recent experiments indicate that the N terminus may possibly function as a lever (Sucic et al., 2010) in a manner analogous to that seen in BetP (Ressl et al., 2009) and to thereby afford the communication between the individual moieties within the transporter oligomers. The issue is to understand how the N terminus senses that an amphetamine—rather than serotonin—resides within the substrate binding site and the permeation pathway. Our observations show that carbamazepine cannot be accommodated by SERT in the amphetamine-bound state. The conformation of the amphetamine-bound state can also be inferred to be distinct from the inward-facing conformation: the latter is stabilized by ibogaine (Jacobs et al., 2007). The ibogaine-bound conformation, however, allowed for simultaneous binding of carbamazepine to a site that may correspond to the second, low-affinity imipramine binding site (Schloss and Betz, 1995). Thus, in the presence of amphetamine, neither the inner nor the outer vestibule is accessible to carbamazepine. This conformation is therefore fundamentally different from the serotonin-bound state. It will be interesting to explore how the change in the inner vestibule is transmitted to the N terminus of SERT (and of other monoamine transporters).
Supplementary Material
Acknowledgments
We thank Sacrament of Transition (Maribor, Slovenia) for the kind gift of ibogaine.
This work was supported by the Austrian Science Fund FWF Cell Communication in Health and Disease [Grant SFB35]; and the Medical University of Vienna, Cell Communication in Health and Disease.
ABBREVIATIONS
- NSS
neurotransmitter:sodium symporters
- WIN35,428
-(−)-2-β-carbomethoxy-3-β-(4-fluorophenyl)tropane
- 5-HT
5-hydroxytryptamine (serotonin)
- MDMA
s-(+)-3,4-methylenedioxy-methamphetamine
- MPP+
1-methyl-4-phenylpyridinium ion
- PCA
para-chloroamphetamine
- LeuTAa
leucine transporter from Aquifex aeolicus
- HEK
human embryonic kidney
- DAT
dopamine transporter
- NET
noradrenaline transporter
- SERT
serotonin transporter
- TM
transmembrane
- YFP
yellow fluorescent protein
- TCA
tricyclic antidepressant
- MOE
Molecular Operating Environment
- PLIF
protein ligand interaction fingerprint
- SSRI
selective serotonin reuptake inhibitor
Footnotes
The online version of this article (available at http://molpharm.aspetjournals.org) contains supplemental material.
References
- Andersen J, Olsen L, Hansen KB, Taboureau O, Jørgensen FS, Jørgensen AM, Bang-Andersen B, Egebjerg J, Strømgaard K, Kristensen AS. Mutational mapping and modeling of the binding site for (S)-citalopram in the human serotonin transporter. J Biol Chem. 2010;285:2051–2063. doi: 10.1074/jbc.M109.072587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen J, Taboureau O, Hansen KB, Olsen L, Egebjerg J, Strømgaard K, Kristensen AS. Location of the antidepressant binding site in the serotonin transporter: importance of Ser-438 in recognition of citalopram and tricyclic anti-depressants. J Biol Chem. 2009;284:10276–10284. doi: 10.1074/jbc.M806907200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- Barker EL, Moore KR, Rakhshan F, Blakely RD. Transmembrane domain I contributes to the permeation pathway for serotonin and ions in the serotonin transporter. J Neurosci. 1999;19:4705–4717. doi: 10.1523/JNEUROSCI.19-12-04705.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkert P, Tosatto SC, Schomburg D. QMEAN: A comprehensive scoring function for model quality assessment. Proteins. 2008;71:261–277. doi: 10.1002/prot.21715. [DOI] [PubMed] [Google Scholar]
- Beuming T, Kniazeff J, Bergmann ML, Shi L, Gracia L, Raniszewska K, Newman AH, Javitch JA, Weinstein H, Gether U, et al. The binding sites for cocaine and dopamine in the dopamine transporter overlap. Nat Neurosci. 2008;11:780–789. doi: 10.1038/nn.2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuming T, Shi L, Javitch JA, Weinstein H. A comprehensive structure-based alignment of prokaryotic and eukaryotic neurotransmitter/Na+ symporters (NSS) aids in the use of the LeuT structure to probe NSS structure and function. Mol Pharmacol. 2006;70:1630–1642. doi: 10.1124/mol.106.026120. [DOI] [PubMed] [Google Scholar]
- Chen JG, Rudnick G. Permeation and gating residues in serotonin transporter. Proc Natl Acad Sci. USA. 2000;97:1044–1049. doi: 10.1073/pnas.97.3.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dailey JW, Reith ME, Yan QS, Li MY, Jobe PC. Carbamazepine increases extracellular serotonin concentration: lack of antagonism by tetrodotoxin or zero Ca2+ Eur J Pharmacol. 1997;328:153–162. doi: 10.1016/s0014-2999(97)83041-5. [DOI] [PubMed] [Google Scholar]
- Field JR, Henry LK, Blakely RD. Transmembrane domain 6 of the human serotonin transporter contributes to an aqueously accessible binding pocket for serotonin and the psychostimulant 3,4-methylene dioxymethamphetamine. J Biol Chem. 2010;285:11270–11280. doi: 10.1074/jbc.M109.093658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fog JU, Khoshbouei H, Holy M, Owens WA, Vaegter CB, Sen N, Nikandrova Y, Bowton E, McMahon DG, Colbran RJ, et al. Calmodulin kinase II interacts with the dopamine transporter C terminus to regulate amphetamine-induced reverse transport. Neuron. 2006;51:417–429. doi: 10.1016/j.neuron.2006.06.028. [DOI] [PubMed] [Google Scholar]
- Forrest LR, Tavoulari S, Zhang YW, Rudnick G, Honig B. Identification of a chloride ion binding site in Na+/Cl -dependent transporters. Proc Natl Acad Sci USA. 2007;104:12761–12766. doi: 10.1073/pnas.0705600104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrest LR, Zhang YW, Jacobs MT, Gesmonde J, Xie L, Honig BH, Rudnick G. Mechanism for alternating access in neurotransmitter transporters. Proc Natl Acad Sci USA. 2008;105:10338–10343. doi: 10.1073/pnas.0804659105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry LK, Adkins EM, Han Q, Blakely RD. Serotonin and cocaine-sensitive inactivation of human serotonin transporters by methanethiosulfonates targeted to transmembrane domain I. J Biol Chem. 2003;278:37052–37063. doi: 10.1074/jbc.M305514200. [DOI] [PubMed] [Google Scholar]
- Henry LK, Field JR, Adkins EM, Parnas ML, Vaughan RA, Zou MF, Newman AH, Blakely RD. Tyr-95 and Ile-172 in transmembrane segments 1 and 3 of human serotonin transporters interact to establish high affinity recognition of antidepressants. J Biol Chem. 2006;281:2012–2023. doi: 10.1074/jbc.M505055200. [DOI] [PubMed] [Google Scholar]
- Hertting G, Axelrod J. Fate of tritiated noradrenaline at the sympathetic nerve-endings. Nature. 1961;192:172–173. doi: 10.1038/192172a0. [DOI] [PubMed] [Google Scholar]
- Jacobs MT, Zhang YW, Campbell SD, Rudnick G. Ibogaine, a noncompetitive inhibitor of serotonin transport, acts by stabilizing the cytoplasm-facing state of the transporter. J Biol Chem. 2007;282:29441–29447. doi: 10.1074/jbc.M704456200. [DOI] [PubMed] [Google Scholar]
- Jardetzky O. Simple allosteric model for membrane pumps. Nature. 1966;211:969–970. doi: 10.1038/211969a0. [DOI] [PubMed] [Google Scholar]
- Just H, Sitte HH, Schmid JA, Freissmuth M, Kudlacek O. Identification of an additional interaction domain in transmembrane domains 11 and 12 that supports oligomer formation in the human serotonin transporter. J Biol Chem. 2004;279:6650–6657. doi: 10.1074/jbc.M306092200. [DOI] [PubMed] [Google Scholar]
- Korkhov VM, Holy M, Freissmuth M, Sitte HH. The conserved glutamate (Glu136) in transmembrane domain 2 of the serotonin transporter is required for the conformational switch in the transport cycle. J Biol Chem. 2006;281:13439–13448. doi: 10.1074/jbc.M511382200. [DOI] [PubMed] [Google Scholar]
- Korkhov VM, Milan-Lobo L, Zuber B, Farhan H, Schmid JA, Freissmuth M, Sitte HH. Peptide-based interactions with calnexin target misassembled membrane proteins into endoplasmic reticulum-derived multilamellar bodies. J Mol Biol. 2008;378:337–352. doi: 10.1016/j.jmb.2008.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- Ressl S, Terwisscha van Scheltinga AC, Vonrhein C, Ott V, Ziegler C. Molecular basis of transport and regulation in the Na+/betaine symporter BetP. Nature. 2009;458:47–52. doi: 10.1038/nature07819. [DOI] [PubMed] [Google Scholar]
- Schloss P, Betz H. Heterogeneity of antidepressant binding sites on the recombinant rat serotonin transporter SERT1. Biochemistry. 1995;34:12590–12595. doi: 10.1021/bi00039a014. [DOI] [PubMed] [Google Scholar]
- Schmid JA, Scholze P, Kudlacek O, Freissmuth M, Singer EA, Sitte HH. Oligomerization of the human serotonin transporter and of the rat GABA transporter 1 visualized by fluorescence resonance energy transfer microscopy in living cells. J Biol Chem. 2001;276:3805–3810. doi: 10.1074/jbc.M007357200. [DOI] [PubMed] [Google Scholar]
- Schmitt KC, Mamidyala S, Biswas S, Dutta AK, Reith ME. Bivalent phenethylamines as novel dopamine transporter inhibitors: evidence for multiple substrate-binding sites in a single transporter. J Neurochem. 2010;112:1605–1618. doi: 10.1111/j.1471-4159.2010.06583.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholze P, Nørregaard L, Singer EA, Freissmuth M, Gether U, Sitte HH. The role of zinc ions in reverse transport mediated by monoamine transporters. J Biol Chem. 2002;277:21505–21513. doi: 10.1074/jbc.M112265200. [DOI] [PubMed] [Google Scholar]
- Segel IH. Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady-State Enzyme Systems. John Wiley & Sons; New York: 1975. [Google Scholar]
- Seidel S, Singer EA, Just H, Farhan H, Scholze P, Kudlacek O, Holy M, Koppatz K, Krivanek P, Freissmuth M, et al. Amphetamines take two to tango: an oligomer-based counter-transport model of neurotransmitter transport explores the amphetamine action. Mol Pharmacol. 2005;67:140–151. doi: 10.1124/mol.67.1.. [DOI] [PubMed] [Google Scholar]
- Shi L, Quick M, Zhao Y, Weinstein H, Javitch JA. The mechanism of a neurotransmitter:sodium symporter–inward release of Na+ and substrate is triggered by substrate in a second binding site. Mol Cell. 2008;30:667–677. doi: 10.1016/j.molcel.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Piscitelli CL, Yamashita A, Gouaux E. A competitive inhibitor traps LeuT in an open-to-out conformation. Science. 2008;322:1655–1661. doi: 10.1126/science.1166777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Yamashita A, Gouaux E. Antidepressant binding site in a bacterial homologue of neurotransmitter transporters. Nature. 2007;448:952–956. doi: 10.1038/nature06038. [DOI] [PubMed] [Google Scholar]
- Sitte HH, Freissmuth M. The reverse operation of Na(+)/Cl(−)-coupled neurotransmitter transporters–why amphetamines take two to tango. J Neurochem. 2010;112:340–355. doi: 10.1111/j.1471-4159.2009.06474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sucic S, Dallinger S, Zdrazil B, Weissensteiner R, Jørgensen TN, Holy M, Kudlacek O, Seidel S, Cha JH, Gether U, et al. The N terminus of monoamine transporters is a lever required for the action of amphetamines. J Biol Chem. 2010;285:10924–10938. doi: 10.1074/jbc.M109.083154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavoulari S, Forrest LR, Rudnick G. Fluoxetine (Prozac) binding to serotonin transporter is modulated by chloride and conformational changes. J Neurosci. 2009;29:9635–9643. doi: 10.1523/JNEUROSCI.0440-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White KJ, Kiser PD, Nichols DE, Barker EL. Engineered zinc-binding sites confirm proximity and orientation of transmembrane helices I and III in the human serotonin transporter. Protein Sci. 2006;15:2411–2422. doi: 10.1110/ps.062386106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A, Singh SK, Kawate T, Jin Y, Gouaux E. Crystal structure of a bacterial homologue of Na+/Cl−-dependent neurotransmitter transporters. Nature. 2005;437:215–223. doi: 10.1038/nature03978. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Zhen J, Karpowich NK, Goetz RM, Law CJ, Reith ME, Wang DN. LeuT-desipramine structure reveals how antidepressants block neurotransmitter reuptake. Science. 2007;317:1390–1393. doi: 10.1126/science.1147614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Zhen J, Karpowich NK, Law CJ, Reith ME, Wang DN. Antidepressant specificity of serotonin transporter suggested by three LeuT-SSRI structures. Nat Struct Mol Biol. 2009;16:652–657. doi: 10.1038/nsmb.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zomot E, Bendahan A, Quick M, Zhao Y, Javitch JA, Kanner BI. Mechanism of chloride interaction with neurotransmitter:sodium symporters. Nature. 2007;449:726–730. doi: 10.1038/nature06133. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.