Abstract
Purpose
Activation of YAP1, novel oncogene in Hippo pathway, has been observed in many cancers including colorectal cancer (CRC). We investigated if activation of YAP1 is significantly associated with prognosis or treatment outcomes in CRC
Experimental Design
A gene expression signature reflecting YAP1 activation was identified in CRC cells, and CRC patients were stratified into two groups according to this signature: activated YAP1 CRC (AYCC) or inactivated YAP1 CRC (IYCC). Stratified patients in five test cohorts were evaluated to determine the effect of the signature on CRC prognosis and response to cetuximab treatment.
Results
The activated YAP1 signature was associated with poor prognosis for CRC in four independent patient cohorts with stage I–III disease (total n = 1,028). In a multivariate analysis, the impact of the YAP1 signature on the disease-free survival was independent of other clinical variables [hazard ratio (HR), 1.63; 95% confidence interval (CI), 1.25–2.13; P < 0.001]. In patients with stage IV CRC and wild-type KRAS, IYCC patients had a better disease control rate and progression-free survival (PFS) after cetuximab monotherapy than did AYCC patients; however, in patients with KRAS mutations, PFS duration after cetuximab monotherapy was not different between IYCC and AYCC patients. In multivariate analysis, the effect of YAP1 activation on PFS was independent of KRAS mutation status and other clinical variables (HR, 1.82; 95% CI, 1.05–3.16; P = 0.03).
Conclusions
Activation of YAP1 is highly associated with poor prognosis for CRC and may be useful in identifying patients with metastatic CRC resistant to cetuximab.
Introduction
Colorectal cancer (CRC) is a major contributor to cancer mortality and morbidity in developed countries and is the second leading cause of cancer deaths in the United States (1). Current prognostic models use histoclinical parameters for prognostication of individual patients but have limitation in capturing molecular heterogeneity of this disease. Recent studies identified several molecular subtypes of CRC reflecting molecular heterogeneity of CRC by using various methods of screening cancer genome (2–6). However, the biological characteristics of these subtypes are poorly understood, and the responses of these subtypes to specific treatments is unknown.
The Hippo pathway is a novel tumor suppressor pathway that is well conserved in different species (7, 8). When Hippo signaling is active, its downstream oncogene YAP1 and the related TAZ are phosphorylated and inactivated by the Hippo core complex. When Hippo signaling is absent or suppressed, however, unphosphorylated YAP1 and TAZ enter the nucleus and induce transcription of genes involved in cell proliferation and survival. Deregulation of YAP1 and TAZ has been discovered in various human cancers, including CRC (9–16).
YAP1 and TAZ play important roles in the development of CRC as evidenced by their overexpression in CRC (7, 8,10, 11, 16) which promotes proliferation and survival of CRC cells (7, 17). However, despite increasing evidence supporting the involvement of YAP1 and TAZ in CRC progression, the clinical relevance of YAP1 activation has yet to be properly examined in CRC. In the present study, we systematically characterized genomic data from multiple cohorts of CRC patients to determine the clinical significance of YAP1 activation in CRC cells. This approach led to the development of molecular signatures by which CRC patients can be stratified according to activation of YAP1. Further analysis of the data revealed that YAP1 activation is closely associated with resistance of CRC to treatment with cetuximab.
Materials and Methods
Cell culture and generation of YAP1 signatures in CRC cells
The CRC cell line NCI-H716 was purchased from the American Type Culture Collection and cultured as suggested by the supplier. A constitutively active mutant of human YAP1 (YAP1-S127A) that was described previously (18) was obtained from Addgene, non-profit organization for sharing plasmids (www.addgene.org). YAP1-S127A was expressed in NCI-H716 cells by using lentiviral vector containing YAP1-S127A coding sequence; an empty lentivirus was used as a control (mock). Overexpression of YAP1-S127A in transfected cells was confirmed via Western blotting with a mouse polyclonal antibody against human YAP1 (1:500 dilution; Santa Cruz Biotechnology) (Supplementary Fig. S1). Total RNA was extracted from NCI-H716 cells expressing exogenous YAP1-S127A and used for labeling and hybridization to human expression BeadChips (HumanHT-12 v4 Expression BeadChip Kit; Illumina) according to the manufacturer’s protocols. Untransfected and empty vector-transfected NCI-H716 cells were used as controls. All experiments were performed in triplicate. For validation of YAP1-specific signature from NCI-H716 cells, we generated additional gene expression data from MNK45 cells overexpressing same exogenous YAP1-S127A via lentiviral vector. MKN45 cells were selected because it has lowest basal level expression of YAP1 due to deletion of both alleles of YAP1 gene (19). Primary microarray data from both cell lines are available in the National Cancer for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) database (GSE41387, GSE50490). For further independent validation of YAP1 signature from NCI-H716 cells, gene expression data from MCF10A cells were downloaded and processed from GEO (GSE13861 and GSE26942).
Patient and genomic data
We assembled a multistudy microarray database of CRC expression profiles (total n = 1,108) based on the Affymetrix U133 GeneChip microarray platform. The database encompasses five different CRC cohorts for which corresponding microarray data and clinical annotations were extracted from the GEO public data repository. Cohort 1 consisted of CRC patients with stage I–III disease (n = 229) whose fresh-frozen tumor specimens had been retrieved from the tissue banks of the Royal Melbourne Hospital (Parkville, Victoria, Australia) and the H. Lee Moffitt Cancer Center and Research institute (GEO accession number GSE14333) (2). Cohort 2 was composed of 168 CRC patients with stage I–III disease whose data had been generated from fresh-frozen tumor specimens at the Institut National de la Santé et de la Recherche Médicale (Paris, France; GEO accession number GSE37892) and the Vanderbilt University Medical Center (GEO accession number GSE17538) (20). Cohort 3 was made up of 506 CRC patients with stage I–III disease from a French multicenter study (GEO accession number GSE39582) (3). The disease-free survival (DFS) duration was defined in the previous studies as the time from surgery to the first documented recurrence or death of CRC (2, 3, 20). Cohort 4 consisted of 125 CRC patients with stage I–III disease whose microarray data were generated from analysis of fresh-frozen tumor tissue at Memorial Sloan-Kettering Cancer Center (GEO accession number GSE41258) (21). In that study, the cancer-specific survival (CSS) duration was defined as the time from surgery to a documented CRC-related death. The data for cohort 5 were composed of patients with refractory metastatic CRC who received cetuximab monotherapy in a clinical trial (22). Of 110 patients who participated in the trial, 80 patients with tumor mRNA expression data (GEO accession number GSE5851) (22) were included in this cohort. In the study of this cohort, patient characteristics were presented, and the progression-free survival (PFS) duration was defined as the time from study enrollment to disease progression or death (22). KRAS mutation status in cohort 5 was determined by direct sequencing of PCR-amplified exon 2 genomic region of KRAS in previous study (22).
Gene expression data for a sixth cohort were downloaded from The Cancer Genome Atlas (TCGA; http://cancergenome.nih.gov). The full data for this cohort were described in detail previously (6). Comprehensive genetic information on 195 CRC patients in this cohort was available.
Statistical analyses of microarray data
The BRB-ArrayTools software program (http://linus.nci.nih.gov/BRB-ArrayTools.html) was used for analysis of gene expression data (23). Other statistical analyses were performed in the R language (http://www.r-project.org) or using the SPSS statistical software program (version 21; IBM Corporation). Raw data on the patient cohorts were downloaded from the GEO database and normalized using a robust multiarray averaging method (24, 25). Genes that were differentially expressed in three groups of NCI-H716 cell lines and related to YAP1 activation were identified using a t-test. Differences in gene expression among the three sample groups were considered statistically significant if the P value was less than 0.005. A heat map was generated using the Cluster and TreeView software programs (26).
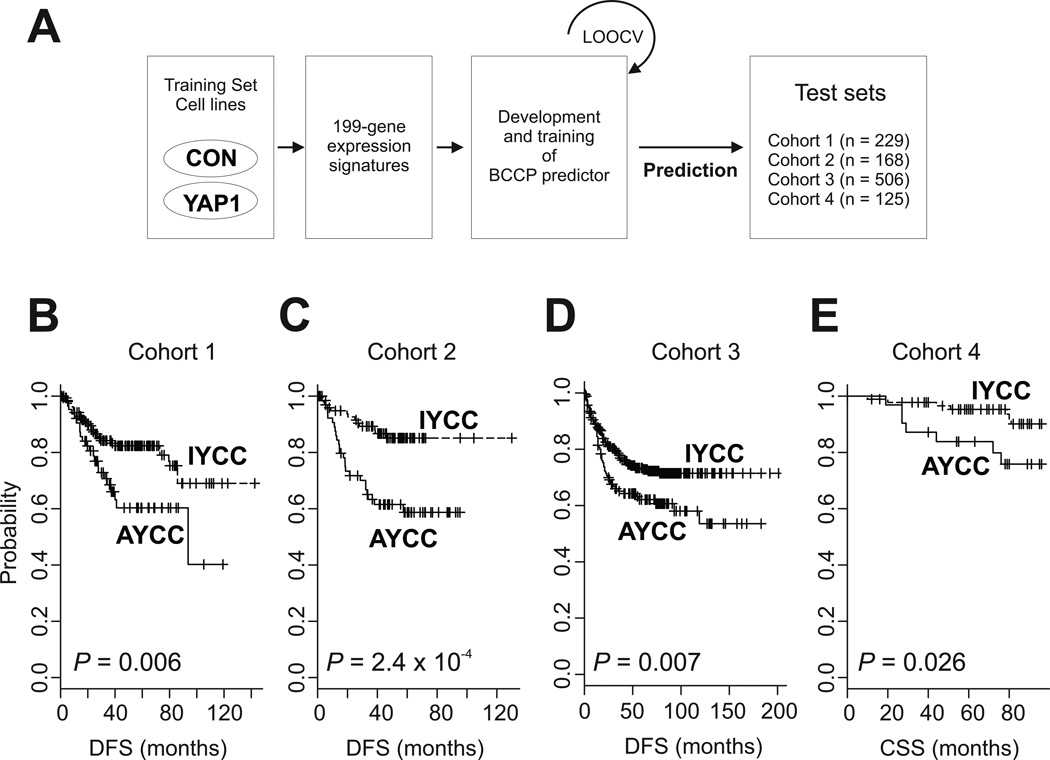
To predict a class of individual patients in the six cohorts, a previously developed approach was used (4, 27–29). Matched probes to 199 genes were selected from each data set from patients: 174 probes for cohort 1, 2, and 3 data sets (Affymetrix U133 version 2) and 142 probes for cohort 4 and 5 data sets (Affymetrix U133). Briefly, gene expression data in the training set (the YAP1 signature in NCI-H716 cells) were combined to form a classifier according to a Bayesian compound covariate predictor (BCCP) (30). The robustness of the classifier was estimated using a misclassification rate determined during leave-one-out cross-validation (LOOCV) in the training set. The BCCP classifier estimated the likelihood that an individual patient had either an activated YAP1 signature—activated YAP1 CRC (AYCC)—or an inactivated YAP1 signature—inactivated YAP1 CRC (IYCC). After the BCCP classifier was used to dichotomize the patients according to the YAP1 signature, the prognostic significance was estimated using Kaplan-Meier plots (log-rank tests). Multivariate Cox proportional hazards regression analysis was used to evaluate the effect of YAP1 signature on survival independently of other clinical parameters; the parameters included in the multivariate analyses are presented in individual tables or supplementary tables.
In cohort 5, differences in response of CRC to treatment of cetuximab were verified using χ2 tests. P values less than 0.05 were considered statistically significant.
Results
Correlation of YAP1 signature with clinical characteristics
Systematic comparison of the gene expression data for the three groups of NCI-H716 CRC cells identified a YAP1-specific gene expression signature comprising 199 unique genes (Supplementary Fig. S1, S2, and Supplementary Table S1). Expression of CTGF, one of the best known direct downstream targets of YAP1, was highly upregulated in YAP1-overexpressing NCI-H716 cells, providing additional confirmation that modulation of gene expression in the cells is due to activation of YAP1. To ensure authenticity of YAP1 signature, we compared 199-gene signature to gene expression data from two independent cell lines overexpressing YAP1 by using gene set enrichment approach. Vast majority of 199 genes were significantly enriched in YAP1-overexpressing MKN45 and MCF10A cells (Supplementary Fig. S3), suggesting that majority of 199 genes are direct or indirect targets of YAP1.
To examine the correlation of YAP1 activation with clinical characteristics of CRC, we compared the 199-gene YAP1 signature with gene expression data for CRC patients. Specifically, we used the BCCP algorithm to calculate the probability of YAP1 activation in CRC tissue specimens (Fig. 1A). We found that 14.9–39.3% of the patients in the six cohorts had the activated YAP1 signature (AYCC group; Table 1). Also, we analyzed the correlation of clinical characteristics with the YAP1 signature in five of the cohorts (we excluded cohort 5, which consisted of patients with stage IV CRC). Whereas AYCC patients had slightly more advanced disease than did IYCC patients in cohort 2 (P = 0.023), we did not see a clear difference in stage distribution between the AYCC and IYCC groups in the other four cohorts. In addition, we found no differences in other clinical variables between the IYCC and AYCC groups (Supplementary Tables S2–S6).
Figure 1.
Construction of a prediction model using gene expression profiles for NCI-H716 cells and analyses of survival in patient cohorts 1–4. (A) Schematic of the strategy used to construct the prediction model and evaluate predicted CRC outcomes according to gene expression signature. CON, control. (B-E) Kaplan-Meier plots of the AYCC and IYCC patients in cohorts 1–4. P values were calculated using log-rank tests. +, censored data.
Table 1.
Patient characteristics in 6 patient cohorts
| Cohort 1 (N = 229) |
Cohort 2 (N = 168) |
Cohort 3 (N = 506) |
Cohort 4 (N = 125) |
Cohort 5 (N = 80) |
Cohort 6 (N = 195) |
|
|---|---|---|---|---|---|---|
| Gender | ||||||
| Male | 123 (53.7%) | 90 (53.6%) | 279 (55.1%) | 63 (50.4%) | 44 (55.0%) | 103 (52.8%) |
| Female | 106 (46.3%) | 78 (46.4%) | 227 (44.9%) | 62 (49.6%) | 36 (45.0%) | 92 (47.2%) |
| Age | ||||||
| Median (range) | 67 (26–92) | 67 (22–97) | 68 (22–97) | 68 (23–87) | 60.5 (25–89) | 69 (35–90) |
| Location | ||||||
| Right colon | 102 (44.5%) | 57 (33.9%) | 206 (40.7%) | 50 (40.0%) | NA | 67 (34.4%) |
| Left colon | 95 (41.5%) | 72 (42.9%) | 300 (59.3%) | 65 (52.0%) | NA | 65 (33.3%) |
| Rectum | 30 (13.1%) | 0 (0.0%) | 0 (0.0%) | 10 (8.0%) | NA | 62 (31.8%) |
| Unknown | 2 (0.9%) | 39 (23.2%) | 0 (0.0%) | 0 (0.0%) | NA | 1 (0.5%) |
| Stage | ||||||
| I | 44 (19.2%) | 4 (2.4%) | 37 (7.3%) | 28 (22.4%) | 0 (0.0%) | 43 (22.1%) |
| II | 94 (41.0%) | 88 (52.4%) | 264 (52.2%) | 48 (38.4%) | 0 (0.0%) | 72 (36.9%) |
| III | 91 (39.7%) | 76 (45.2%) | 205 (40.5%) | 49 (39.2%) | 0 (0.0%) | 49 (25.1%) |
| IV | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 80 (100.0%) | 29 (14.9%) |
| Unknown | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 2 (1.0%) |
| Adjuvant chemotherapy | ||||||
| Yes | 87 (38.0%) | 23 (13.7%) | 203 (40.1%) | NA | NA | NA |
| No | 142 (62.0%) | 15 (8.9%) | 301 (59.5%) | NA | NA | NA |
| Unknown | 0 (0.0%) | 130 (77.4%) | 2 (0.4%) | NA | NA | NA |
| KRAS status | ||||||
| Wild type | NA | NA | 297 (58.7%) | NA | 43 (53.8%) | 115 (59.0%) |
| Mutant | NA | NA | 188 (37.2%) | NA | 27 (33.8%) | 80 (41.0%)1 |
| Unknown | NA | NA | 21 (4.2%) | NA | 10 (12.5%) | 0 (0.0%) |
| MSI status | ||||||
| pMMR | NA | NA | 388 (76.7%) | 87 (69.6%) | NA | 172 (88.2%) |
| dMMR | NA | NA | 73 (14.4%) | 28 (22.4%) | NA | 23 (11.8%) |
| Unknown | NA | NA | 45 (8.9%) | 10 (8.0%) | NA | 0 (0.0%) |
| YAP1 signature | ||||||
| IYCC | 163 (71.2%) | 102 (60.7%) | 366 (72.3%) | 92 (73.6%) | 57 (71.3%) | 166 (85.1%) |
| AYCC | 66 (28.8%) | 66 (39.3%) | 140 (27.7%) | 33 (26.4%) | 23 (28.8%) | 29 (14.9%) |
Abbreviations: MSI, microsatellite instability; pMMR, proficient mismatch repair; dMMR, deficient mismatch repair; IYCC, inactivated YAP1 colorectal cancer; AYCC, activated YAP1 colorectal cancer; NA, not available
Prognostic impact of YAP1 activation
We next investigated the prognostic impact of YAP1 activation using data on patients with stage I–III CRC (cohorts 1–4). Tumor recurrence and DFS data were available for cohorts 1–3, but we had to analyze CSS data for cohort 4 as DFS data for that group were not available. Kaplan-Meier curves for these patients consistently demonstrated much worse survival durations in AYCC patients than in IYCC patients (Fig. 1B–1E), indicating that the activated YAP1 signature is clearly related to poor prognosis for CRC.
We conducted further analyses to determine whether the prognostic impact of the YAP1 signature is independent of other clinical variables. We pooled the patients in cohorts 1–3 with available DFS data (n = 903) for univariate and multivariate analyses of factors affecting DFS (Table 2). In the multivariate analysis, AYCC was related to worse DFS rates than was IYCC (hazard ratio [HR], 1.63; 95% confidence interval (CI), 1.25–2.13; P < 0.001) independent of other clinical variables. When we conducted the same analyses independently after splitting the group into two (cohorts 1 and 2 versus cohort 3, as cohort 3 had more detailed clinical variables than did the other two cohorts), the independent impact of the YAP1 signature on DFS remained unchanged (Supplementary Tables S7 and S8). Furthermore, the activated YAP1 signature successfully identified patients with poor prognosis regardless of their disease stage (P < 0.05) (Supplementary Fig. S4). Taken together, these findings suggested that the prognostic relevance of the YAP1 signature to CRC patients is maintained even when taking into account the classic clinicopathological prognostic features.
Table 2.
Univariate and multivariate analyses on factors affecting DFS in stage 1–3 patients (patient data from cohorts 1, 2 and 3 were pooled together; N = 903)
| Variables | Univariate analysis1 | Multivariate analysis2 | ||||
|---|---|---|---|---|---|---|
| Patient No. |
5-year DFS |
P-value | HR | 95% CI | P-value | |
| Age3 | ||||||
| < 70 years | 475 | 70.1% | - | - | - | - |
| ≥ 70 years | 427 | 75.3% | 0.221 | - | - | - |
| Gender | ||||||
| Male | 492 | 69.3% | - | 1.00 | - | - |
| Female | 411 | 76.0% | 0.086 | 0.80 | 0.61–1.05 | 0.105 |
| Location | 0.014 | 0.070 | ||||
| Right colon | 365 | 73.3% | - | 1.00 | - | - |
| Left colon | 467 | 69.4% | - | 1.18 | 0.90–1.56 | 0.228 |
| Rectum | 30 | 77.5% | - | 1.23 | 0.56–2.67 | 0.606 |
| Unknown | 41 | 97.3% | - | 0.11 | 0.02–0.76 | 0.026 |
| Adjuvant chemotherapy | <0.001 | 0.859 | ||||
| Undone | 458 | 78.0% | - | 1.00 | - | - |
| Done | 313 | 66.6% | - | 0.94 | 0.68–1.30 | 0.716 |
| Unknown | 132 | 67.9% | - | 0.90 | 0.60–1.34 | 0.593 |
| Stage | <0.001 | <0.001 | ||||
| I | 85 | 95.7% | - | 1.00 | - | - |
| II | 446 | 79.7% | - | 4.80 | 1.50–15.29 | 0.008 |
| III | 372 | 59.0% | - | 11.88 | 3.69–38.26 | <0.001 |
| YAP1 status | ||||||
| IYCC | 631 | 77.5% | - | 1.00 | - | - |
| AYCC | 272 | 61.2% | <0.001 | 1.63 | 1.25–2.13 | <0.001 |
In univariate analyses, log-rank tests were conducted.
In multivariate Cox proportional hazard model, variables with P < 0.15 in univariate analysis were only included and the ‘enter method’ was applied.
Data on age of one patient was missing.
Abbreviations: DFS, disease-free survival; HR, hazard ratio; CI, confidence interval; IYCC, inactivated YAP1 colorectal cancer; AYCC; activated YAP1 colorectal cancer
YAP1 activation is associated with poor response to cetuximab treatment
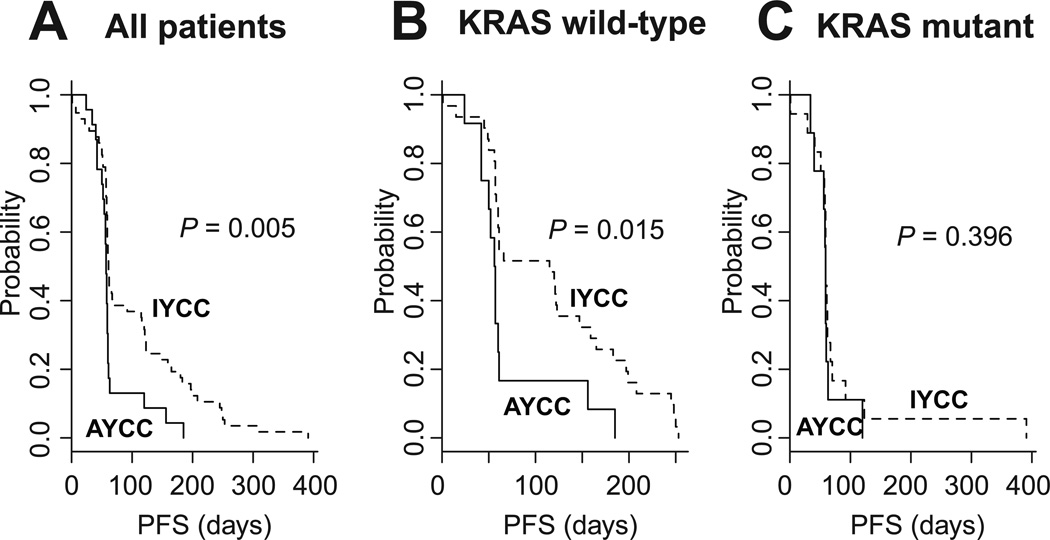
All of the patients in cohort 5 (n = 80) received cetuximab monotherapy. In the 70 patients in this cohort who had KRAS mutation status data available, we observed no difference in the KRAS mutation rates between the AYCC and IYCC groups (Supplementary Table S9). However, we did see differences in response to cetuximab between the two groups. Specifically, tumor shrinkage (complete remission [CR] or partial remission [PR]) occurred only in the IYCC group (response rate: 10.5% [IYCC] versus 0.0% [AYCC]; P = 0.175), and the disease control rate was significantly higher in the IYCC group than in the AYCC group (38.6% versus 13.0%; P = 0.026). As expected, patients with wild-type (WT) KRAS had a longer PFS duration than did patients with KRAS mutations in this cohort (22), although it didn’t reach statistical significance (P = 0.142, Supplementary Fig. S5). However, IYCC patients had a significantly longer PFS duration than did AYCC patients (P = 0.005) (Fig. 2A), more so in WT KRAS patients (P = 0.015) (Fig. 2B) than in KRAS-mutant patients (P = 0.396) (Fig. 2C). In multivariate analysis, the effect of the YAP1 signature on PFS according to other clinical variables was unchanged (Table 3).
Figure 2.
PFS according to YAP1 signature in (A) all patients (n = 80), (B) patients with WT KRAS (n = 43), and (C) patients with mutant KRAS (n = 27) in cohort 5. KRAS mutation data were not available for 10 patients.
Table 3.
Univariate and multivariate analyses on factors affecting PFS in patients who received cetuximab monotherapy (cohort 5)
| Variables | Univariate analysis1 | Multivariate analysis2 | |||||
|---|---|---|---|---|---|---|---|
| Patient No. |
PFS (median, days) |
6-month progression- free rate (%) |
P- value |
HR | 95% CI | P- value |
|
| Age3 | |||||||
| < 70 years | 54 | 59 | 18.5% | - | - | - | - |
| ≥ 70 years | 24 | 60 | 8.3% | 0.227 | - | - | - |
| Gender | |||||||
| Male | 44 | 61 | 22.7% | - | 1.00 | - | - |
| Female | 36 | 58 | 5.6% | 0.009 | 1.65 | 0.98–2.78 | 0.062 |
| KRAS status4 | |||||||
| Wild type | 43 | 61 | 20.9% | - | 1.00 | - | - |
| Mutant | 27 | 59 | 3.7% | 0.142 | 1.27 | 0.75–2.14 | 0.375 |
| YAP1 status | |||||||
| IYCC | 57 | 61 | 19.3% | - | 1.00 | - | - |
| AYCC | 23 | 57 | 4.3% | 0.005 | 1.82 | 1.05–3.16 | 0.034 |
In univariate analyses, log-rank tests were conducted.
In multivariate Cox proportional hazard model, variables with P < 0.15 in univariate analysis were only included and the ‘enter method’ was applied. In this multivariate analysis, 70 patients from cohort 6 without missing data on KRAS status were included.
Data on age of 2 patients were missing.
Data on KRAS mutational status of 10 patients were missing
Abbreviations: PFS, progression-free survival; HR, hazard ratio; CI, confidence interval; IYCC, inactivated YAP1 colorectal cancer; AYCC; activated YAP1 colorectal cancer
Relationship between the YAP1 signature and other genetic events
We further investigated the correlation between the YAP1 signature and somatic mutations in patients in cohort 6, as they had the most comprehensive genetic information, which was available from the TCGA database (Fig. 3, Supplementary Table S6). When the same genetic data are available from other cohorts, analyzed results were compared among the cohorts. For cohort 6, we compared the mutational statuses of genes involved in five pathways (WNT, mitogen-activated protein kinase, phosphoinositide 3-kinase, transforming growth factor-, and p53), which are reported to be deregulated in CRC cells (6). Of the 30 compared genes, only ACVR2A exhibited a significantly different mutation rate between the IYCC and AYCC groups (12% versus 0%; P = 0.048) (Fig. 3, Supplementary Table S6). ACVR2A is activin receptor type-2A, a member of TGF-β superfamily (31). It is interesting to point out that ACVR2A is also frequently mutated gene in stomach cancer (32), suggesting that it may play critical roles in development of gastrointestinal cancer. We observed no differences in mutation rate between the AYCC and IYCC groups for other genes, including KRAS (cohorts 3, 5, and 6), BRAF (cohorts 3 and 6), and TP53 (cohorts 3 and 6) (Supplementary Tables S4, S6, and S9). Most interestingly hypermutated tumors, which were defined by the TCGA project (6), developed more frequently in the IYCC group than the AYCC group (P = 0.05). The higher mutation rate in the IYCC group was supported by a higher frequency of nonsilent gene mutations (per 106 bases) in the IYCC group than in the AYCC group (P = 0.002, Mann-Whitney U test).
Figure 3.
Genetic changes in CRC cells according to YAP1 signature in cohort 6. Genetic data were retrieved from the TCGA database and analyzed.
Discussion
In this study, we extracted the YAP1 gene activation signature from CRC cell microarray data and classified CRC patients into two groups: AYCC and IYCC. Among four independent cohorts of patients with locoregional CRC, we found that AYCC patients had worse survival rates than did IYCC patients. Also, among those with stage IV CRC, we showed that the YAP1 signature is associated with response to cetuximab monotherapy, generating interesting hypothesis connecting YAP1 activation to potential benefit of cetuximab treatment. To our knowledge, this is the first study to suggest that the YAP1 gene signature can be used as prognostic biomarker in locoregional CRC and as potential predictive biomarker in patients with metastatic CRC receiving cetuximab treatment.
Previous studies examined the prognostic relevance of YAP1 or TAZ activation in CRC patients, but their results were inconsistent because the investigators used different methods of measuring YAP1 signal activation (11, 16, 33, 34). Wang et al. (16) measured protein expression of YAP1 in CRC to estimate YAP1 activity. However, because YAP1 is largely regulated by phosphorylation (8), this approach may not be able to fully measure YAP1 activity in CRC. In addition, Yuen et al (34), reported that the TAZ mRNA expression level was a prognostic indicator but that YAP1 mRNA expression was not related to prognosis. However, because both YAP and TAZ exist as nuclear or cytoplasmic form and activated YAP1 and TAZ function mainly in the nucleus, the total expression level cannot properly reflect the true biological activity of these proteins. Moreover, Barry et al (33), recently suggested that cytoplasmic YAP1 may have a growth-inhibitory function in CRC cells. Therefore, if YAP1 (or TAZ) is to be used as a biomarker, both the level of expression and intracellular location of this protein must be considered. Alternatively, expression patterns for downstream genes targeted by YAP1 or TAZ in CRC cells may be used as indicators of their activation because their best known molecular activity is transcription activation (7, 8). Because the cut-off point or criterion for YAP1 and TAZ activation, which is measured using mRNA or immunohistochemistry, has gone unverified until now, we developed the YAP1 signature, which reflects the transcriptional activity of YAP1 in CRC cells. Additionally, unlike previous studies that did not examine CRC patients according to disease stage (11, 16, 33, 34), we categorized patients into two classes—locoregional (stage I–III) and remote (stage IV) disease—because the therapeutic approaches for these two classes are clearly different. Using the YAP1 signature and data from four large cohorts of CRC patients (n = 1,028), we found that the activated YAP1 signature is independently predictive of poor survival in patients with stage I–III disease (Fig. 1).
Treatment with cetuximab, a monoclonal antibody against epidermal growth factor receptor, is effective against metastatic CRC, but its beneficial effect is limited to patients with WT KRAS (35, 36). However, even in patients with WT KRAS, the benefit of treatment with cetuximab is restricted to a small proportion of patients and is not sustainable (37). Therefore, selection of patients with metastatic CRC who would have the maximum benefit of this treatment is important (22, 38, 39). In the present study, using genomic data for CRC patients enrolled in a clinical trial (22), we showed that YAP1 activation is significantly associated with poor response to cetuximab therapy in CRC (Fig. 2). This observation is in good agreement with previous report showing that silencing of YAP1 sensitized ovarian cancer cells to EGFR inhibitor erlotinib (40). Furthermore, recent study also identified YAP1 as a potential biomarker for cetuximab resistance in head and neck cancer (41).
This study had a few limitations. First, our observations must be validated in prospective studies. The fact that we obtained the same results for four independent patient cohorts strongly suggests that the YAP1 signature is a reliable tool for assessing prognosis for locoregional CRC. We also demonstrated the possibility of using the YAP1 signature as a predictive marker for response of CRC to treatment with cetuximab. However, because we applied the developed YAP1 signature to retrospective patient cohorts, our observations require validation. Also necessary is investigating whether the efficacy of other chemotherapeutic agents in both the adjuvant and palliative setting is affected by the YAP1 activation status. Second, the relationship between the YAP1 signature and other clinical and genetic characteristics must be evaluated further. For example, IYCC patients in cohort 6 had a higher ACVR2A mutation rate than did AYCC patients. Whether the differences in frequency of this genetic event were real differences or resulted from random chance is unclear. The functional role of ACVR2A mutation in CRC cells is not well known. Therefore, more studies are warranted to determine the influence of YAP1 activation on the clinical characteristics of and genetic changes in CRC patients. Third, all of data in current study were generated by using microarray technology and frozen tissues that are not routinely available through general practice in clinics. Therefore, it will be necessary to identify small number of robust genes (5 to 10 genes) well reflecting YAP1 activity in CRC and use simpler but more robust methods like qRT-PCR to measure expression of these genes with RNA from formalin fixed paraffin embedded tissues in future studies. Fourth, our approach, gene expression data from tumor mass only provides very limited information on tumor heterogeneity that is very critical for understanding mechanisms of resistant to therapeutic treatment (42). Therefore, in-depth analysis of cancer cells in tumor mass by using single cell genomic approaches will be necessary to address this issues in future studies. Additional minor limitation in our prediction model includes difference in number of probes in different platforms of microarrays. However, we found no correlation between fraction of YAP1 active patients and number of probes used in prediction model.
In conclusion, our results suggest that the YAP1 signature is helpful in identifying CRC patients with poor prognoses and/or cetuximab resistance. This signature can be further developed for the tailored management of CRC.
Supplementary Material
Translational Relevance.
Colorectal cancer (CRC) is clinically heterogeneous disease. Previous studies suggested potential molecularly distinct subtypes of CRC. However, the biological characteristics of these subtypes are poorly understood, and the responses of these subtypes to specific treatments are unknown. In this study, we were able to subdivide CRC patients into two major subgroups that are characterized by activation of oncogene YAP1 and showed significant differences in disease-free survival. Furthermore, we also demonstrated that the YAP1 activation is significantly associated with lack of response to cetuximab monotherapy. Most interestingly, among patients with wild type KRAS, only patients without YAP1 activation benefited from cetuximab treatment. This study provides strong rationale for evaluating status of YAP1 activation as potential prognostic and predictive markers in future studies. This result might improve patient care by providing more practical guidance for different treatments.
Acknowledgments
Grant Support
This study was supported by grants from National Cancer Institute at the National Institutes of Health (CA150229 to JSL) and the G.S. Hogan Gastrointestinal Cancer Research Fund at The University of Texas MD Anderson Cancer Center (JSL). STR DNA fingerprinting was done by the Cancer Center Support Grant-funded Characterized Cell Line core, NCI # CA016672.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Authors’ Contributions
Concept and design: K-W Lee, SS Lee, J-S Lee
Acquisition of data: K-W Lee, SS Lee, S-B Kim, Park, Sohn, H-S Lee
Analysis and interpretation of data: K-W Lee, SS Lee, S-B Kim, SS Kim, Kopetz, J-S Lee
Drafting of the manuscript: K-W Lee, SS Lee, J-S Lee
Critical revision of the manuscript for important intellectual content: All authors
Statistical analysis: K-W Lee, S-B Kim, J-S Lee
Study supervision: J-S Lee
Reference List
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Jorissen RN, Gibbs P, Christie M, Prakash S, Lipton L, Desai J, et al. Metastasis-Associated Gene Expression Changes Predict Poor Outcomes in Patients with Dukes Stage B and C Colorectal Cancer. Clin Cancer Res. 2009;15:7642–7651. doi: 10.1158/1078-0432.CCR-09-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marisa L, de RA, Duval A, Selves J, Gaub MP, Vescovo L, et al. Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and prognostic value. PLoS Med. 2013;10:e1001453. doi: 10.1371/journal.pmed.1001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oh SC, Park YY, Park ES, Lim JY, Kim SM, Kim SB, et al. Prognostic gene expression signature associated with two molecularly distinct subtypes of colorectal cancer. Gut. 2012;61:1291–1298. doi: 10.1136/gutjnl-2011-300812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park YY, Lee SS, Lim JY, Kim SC, Kim SB, Sohn BH, et al. Comparison of prognostic genomic predictors in colorectal cancer. PLoS One. 2013;8:e60778. doi: 10.1371/journal.pone.0060778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.The Cancer Genome Atlas Project. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Avruch J, Zhou D, Bardeesy N. YAP oncogene overexpression supercharges colon cancer proliferation. Cell Cycle. 2012;11:1090–1096. doi: 10.4161/cc.11.6.19453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nat Rev Cancer. 2013;13:246–257. doi: 10.1038/nrc3458. [DOI] [PubMed] [Google Scholar]
- 9.Hall CA, Wang R, Miao J, Oliva E, Shen X, Wheeler T, et al. Hippo pathway effector Yap is an ovarian cancer oncogene. Cancer Res. 2010;70:8517–8525. doi: 10.1158/0008-5472.CAN-10-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Steinhardt AA, Gayyed MF, Klein AP, Dong J, Maitra A, Pan D, et al. Expression of Yes-associated protein in common solid tumors. Hum Pathol. 2008;39:1582–1589. doi: 10.1016/j.humpath.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang L, Shi S, Guo Z, Zhang X, Han S, Yang A, et al. Overexpression of YAP and TAZ is an independent predictor of prognosis in colorectal cancer and related to the proliferation and metastasis of colon cancer cells. PLoS One. 2013;8:e65539. doi: 10.1371/journal.pone.0065539. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Wang Y, Dong Q, Zhang Q, Li Z, Wang E, Qiu X. Overexpression of yes-associated protein contributes to progression and poor prognosis of non-small-cell lung cancer. Cancer Sci. 2010;101:1279–1285. doi: 10.1111/j.1349-7006.2010.01511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu MZ, Yao TJ, Lee NP, Ng IO, Chan YT, Zender L, et al. Yes-associated protein is an independent prognostic marker in hepatocellular carcinoma. Cancer. 2009;115:4576–4585. doi: 10.1002/cncr.24495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kang W, Tong JH, Chan AW, Lee TL, Lung RW, Leung PP, et al. Yes-associated protein 1 exhibits oncogenic property in gastric cancer and its nuclear accumulation associates with poor prognosis. Clin Cancer Res. 2011;17:2130–2139. doi: 10.1158/1078-0432.CCR-10-2467. [DOI] [PubMed] [Google Scholar]
- 15.Zhang X, George J, Deb S, Degoutin JL, Takano EA, Fox SB, et al. The Hippo pathway transcriptional co-activator, YAP, is an ovarian cancer oncogene. Oncogene. 2011;30:2810–2822. doi: 10.1038/onc.2011.8. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y, Xie C, Li Q, Xu K, Wang E. Clinical and prognostic significance of Yes-associated protein in colorectal cancer. Tumour Biol. 2013;34:2169–2174. doi: 10.1007/s13277-013-0751-x. [DOI] [PubMed] [Google Scholar]
- 17.Zhou D, Zhang Y, Wu H, Barry E, Yin Y, Lawrence E, et al. Mst1 and Mst2 protein kinases restrain intestinal stem cell proliferation and colonic tumorigenesis by inhibition of Yes-associated protein (Yap) overabundance. Proc Natl Acad Sci U S A. 2011;108:E1312–E1320. doi: 10.1073/pnas.1110428108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Komuro A, Nagai M, Navin NE, Sudol M. WW domain-containing protein YAP associates with ErbB-4 and acts as a co-transcriptional activator for the carboxyl-terminal fragment of ErbB-4 that translocates to the nucleus. J Biol Chem. 2003;278:33334–33341. doi: 10.1074/jbc.M305597200. [DOI] [PubMed] [Google Scholar]
- 19.Kang W, Tong JH, Chan AW, Lee TL, Lung RW, Leung PP, et al. Yes-associated protein 1 exhibits oncogenic property in gastric cancer and its nuclear accumulation associates with poor prognosis. Clin Cancer Res. 2011;17:2130–2139. doi: 10.1158/1078-0432.CCR-10-2467. [DOI] [PubMed] [Google Scholar]
- 20.Smith JJ, Deane NG, Wu F, Merchant NB, Zhang B, Jiang A, et al. Experimentally derived metastasis gene expression profile predicts recurrence and death in patients with colon cancer. Gastroenterology. 2010;138:958–968. doi: 10.1053/j.gastro.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sheffer M, Bacolod MD, Zuk O, Giardina SF, Pincas H, Barany F, et al. Association of survival and disease progression with chromosomal instability: a genomic exploration of colorectal cancer. Proc Natl Acad Sci U S A. 2009;106:7131–7136. doi: 10.1073/pnas.0902232106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khambata-Ford S, Garrett CR, Meropol NJ, Basik M, Harbison CT, Wu S, et al. Expression of epiregulin and amphiregulin and K-ras mutation status predict disease control in metastatic colorectal cancer patients treated with cetuximab. J Clin Oncol. 2007;25:3230–3237. doi: 10.1200/JCO.2006.10.5437. [DOI] [PubMed] [Google Scholar]
- 23.Simon R, Lam A, Li MC, Ngan M, Menenzes S, Zhao Y. Analysis of gene expression data using BRB-ArrayTools. Cancer Inform. 2007;3:11–17. 11–17. [PMC free article] [PubMed] [Google Scholar]
- 24.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 25.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 26.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee JS, Chu IS, Mikaelyan A, Calvisi DF, Heo J, Reddy JK, et al. Application of comparative functional genomics to identify best-fit mouse models to study human cancer. Nat Genet. 2004;36:1306–1311. doi: 10.1038/ng1481. [DOI] [PubMed] [Google Scholar]
- 28.Lee JS, Heo J, Libbrecht L, Chu IS, Kaposi-Novak P, Calvisi DF, et al. A novel prognostic subtype of human hepatocellular carcinoma derived from hepatic progenitor cells. Nat Med. 2006;12:410–416. doi: 10.1038/nm1377. [DOI] [PubMed] [Google Scholar]
- 29.Lee JS, Leem SH, Lee SY, Kim SC, Park ES, Kim SB, et al. Expression signature of E2F1 and its associated genes predict superficial to invasive progression of bladder tumors. J Clin Oncol. 2010;28:2660–2667. doi: 10.1200/JCO.2009.25.0977. [DOI] [PubMed] [Google Scholar]
- 30.Ramaswamy S, Tamayo P, Rifkin R, Mukherjee S, Yeang CH, Angelo M, et al. Multiclass cancer diagnosis using tumor gene expression signatures. Proc Natl Acad Sci U S A. 2001;98:15149–15154. doi: 10.1073/pnas.211566398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wrana JL. Signaling by the TGFbeta superfamily. Cold Spring Harb Perspect Biol. 2013;5:a011197. doi: 10.1101/cshperspect.a011197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagarajan N, Bertrand D, Hillmer AM, Zang ZJ, Yao F, Jacques PE, et al. Whole-genome reconstruction and mutational signatures in gastric cancer. Genome Biol. 2012;13:R115–R113. doi: 10.1186/gb-2012-13-12-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barry ER, Morikawa T, Butler BL, Shrestha K, de la Rosa R, Yan KS, et al. Restriction of intestinal stem cell expansion and the regenerative response by YAP. Nature. 2013;493:106–110. doi: 10.1038/nature11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yuen HF, McCrudden CM, Huang YH, Tham JM, Zhang X, Zeng Q, et al. TAZ expression as a prognostic indicator in colorectal cancer. PLoS One. 2013;8:e54211. doi: 10.1371/journal.pone.0054211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karapetis CS, Khambata-Ford S, Jonker DJ, O'Callaghan CJ, Tu D, Tebbutt NC, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359:1757–1765. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 36.Van CE, Kohne CH, Hitre E, Zaluski J, Chang Chien CR, Makhson A, et al. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med. 2009;360:1408–1417. doi: 10.1056/NEJMoa0805019. [DOI] [PubMed] [Google Scholar]
- 37.Karapetis CS, Khambata-Ford S, Jonker DJ, O'Callaghan CJ, Tu D, Tebbutt NC, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359:1757–1765. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 38.Custodio A, Feliu J. Prognostic and predictive biomarkers for epidermal growth factor receptor-targeted therapy in colorectal cancer: beyond KRAS mutations. Crit Rev Oncol Hematol. 2013;85:45–81. doi: 10.1016/j.critrevonc.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 39.Tian S, Simon I, Moreno V, Roepman P, Tabernero J, Snel M, et al. A combined oncogenic pathway signature of BRAF, KRAS and PI3KCA mutation improves colorectal cancer classification and cetuximab treatment prediction. Gut. 2013;62:540–549. doi: 10.1136/gutjnl-2012-302423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang JM, Nagatomo I, Suzuki E, Mizuno T, Kumagai T, Berezov A, et al. YAP modifies cancer cell sensitivity to EGFR and survivin inhibitors and is negatively regulated by the non-receptor type protein tyrosine phosphatase 14. Oncogene. 2013;32:2220–2229. doi: 10.1038/onc.2012.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jerhammar F, Johansson AC, Ceder R, Welander J, Jansson A, Grafstrom RC, et al. YAP1 is a potential biomarker for cetuximab resistance in head and neck cancer. Oral Oncol. 2014;50:832–839. doi: 10.1016/j.oraloncology.2014.06.003. [DOI] [PubMed] [Google Scholar]
- 42.Ramos P, Bentires-Alj M. Mechanism-based cancer therapy: resistance to therapy, therapy for resistance. Oncogene. 2014;10 doi: 10.1038/onc.2014.314. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.