Abstract
Cross-reactivity of T cells is defined by recognition of two or more peptide-MHC complexes by the same T cell. Although examples of cross-reactivity have been reported, a detailed examination of cross-reactivity has not been performed. Here we take advantage of the high degree of polyclonality in the BV19 T cell repertoire responding to influenza M158–66 in HLA-A2 individuals to obtain a measure for simple cross-reactivity. We utilize an incremental epitope modulation approach to this question using the HLA-A2 restricted response to influenza M158–66 as the system. In three HLA-A2 adult subjects we identified the BV19 clonotypes in the recall response to the influenza epitope M158–66 and twelve M1 peptides substituted at TCR contact positions 63 or 65. The fraction of cross-reactive clonotypes in the M158–66 repertoire varies from 45% to 58% in the three donors. The extent of cross-reactivity, which is the additional number of peptides recognized by a single clonotype, is as high as six. We summarize the data using a graph theory approach with the cross-reactive clonotypes connecting the different HLA-A2 peptides recognized. The cross-reactive clonotypes form a well-connected network that could provide protection from escape variants. We predict that any new pathogen with an epitope whose shape corresponds to that of the peptides studied here would find a pre-existing repertoire ready to respond to it. We propose that in adult memory repertoires, previously encountered epitopes may have generated similar cross-reactive repertoires.
Keywords: Human, T cells, T cell receptor, repertoire development
Introduction
An inherent property of the interaction between T cell receptor and peptide-MHC (pMHC) complexes may be the ability of the TCR to recognize a number of related structures (1). This is referred to as cross-reactivity. Because we do not have a sufficiently detailed understanding of this interaction, it is also possible that a pMHC structure which may appear to be entirely unrelated by sequence may also be recognized. There are examples of both simple and complex cross-reactivity (2–11). Recently there have been attempts to characterize the mechanisms by which cross-reactivity can take place (reviewed in 12) to quantify to what extent cross-reactivity is observed (13), and to place the phenomenon in the context of immune responses (reviewed in 14).
Here we examine the question of cross-reactivity to structurally related epitopes by describing recall repertoires to the M158–66 epitope from influenza A virus in three HLA-A2 individuals and then measuring the recall repertoire to twelve peptides substituted at one of two TCR contact residues. The recall responses to M158–66 are observed in most HLA-A2 individuals (15, 16). The responding T cells predominantly use BV19 and a restricted CDR3 amino acid sequence (17–19). However, they are polyclonal (20, 21) and thus are able to provide significant data in terms of number of clonotypes. The approach of modifying a known epitope offers the benefit of simplicity so that the cross-reactive clonotypes can be further investigated on the basis of the structural differences of the pMHC complexes vis-à-vis M158–66. Furthermore, there has already been a description of using such peptides to map responses which proved the feasibility of this approach (22). We identified responding T cells on the basis of the focusing of the repertoire to T cells with reproducible CDR3 lengths in triplicate cultures and sequenced the CDR3 of the responders. We observe a high degree of cross-reactivity and summarize our results by generating a network graph.
Materials and methods
Peptides
Influenza A matrix M158–66 peptide and all substituted peptides were synthesized by standard solid-phase methods, purified by HPLC, and confirmed by mass spectrometry (Core Lab at Blood Research Institute, the BloodCenter of Wisconsin). Peptide sequences are listed in Table I.
Table I.
Amino acid substitutions at positions 63 and 65 in the peptides used for stimulation of human PBMC.
| Peptide | AA Sequence |
|---|---|
| M158–66 | GILGFVFTL |
| A63 | -----A--- |
| L63 | -----L--- |
| I63 | -----I--- |
| T63 | -----T--- |
| S63 | -----S--- |
| R63 | -----R--- |
| K63 | -----K--- |
| A65 | -------A- |
| S65 | -------S- |
| G65 | -------G- |
| N65 | -------N- |
| D65 | -------D- |
CTL isolation, RNA isolation, cDNA preparation
PBMC were collected from buffy coats of three healthy HLA-A2.1 donors (age 37, 38, and 47) using lymphocyte separation medium (Cellgro, Mediatech, Manassas, VA) and cryopreserved in human AB serum containing 10% DMSO at a concentration 107/ml with a control freezing system (CryoMed Freezer, Fisher Scientific, Pittsburg, PA) before use. All samples were obtained under IRB-approved protocols (IRB# BC 05–11 and BC 04–22). Frozen PBMC were stored in liquid nitrogen at −180°C. To establish CTL, PBMC were thawed and placed into complete RPMI at RT. PBMC were cultured at 2×106 cells/2 ml in the presence of 1 µM of influenza A matrix peptide M158–66 or one of the substituted peptides and 10 U/ml of recombinant IL-2 in 12-well plates for 7 days. Additional recombinant IL-2 (10U/ml) was added on day 4. All cultures were established in triplicate. Primed cells were restimulated in the presence of recombinant IL-2 (10U/ml) and an equal number of irradiated (3000 R) autologous PBMC that had been preloaded with M158–66 or a corresponding substituted peptide. After 2 weeks in culture CD8+ T cells were separated using Dynal® CD8 Positive Isolation Kit (Invitrogen, Carlsbad, CA). RNA isolation was performed using Dynabeads® mRNA DIRECT™ Kit (Invitrogen). For Donor C, RNA from cultures stimulated with peptides substituted at position 63 was isolated after three weeks of culturing with no CD8 isolation. For all donors, cDNA was synthesized immediately after RNA isolation using oligo(dT) as primer and M-MLV (Invitrogen) as reverse transcriptase.
CDR3 spectratyping
BV genes were amplified from cDNA using BV19 family-specific primer and fluorochrome-labeled TCR CB primer as described (23). 1 µl of each PCR product was used for fragment analysis (3100 Genetic Analyzer, Applied Biosystems). Data analysis was performed using proprietary software (Flynn Creek Biosciences, Hubertus, WI).
PCR product cloning, sequencing, and colony counting
10 µl of PCR product was used for electrophoresis on a 5% polyacrylamide gel; specific bands were visualized by fluorescence detection (Typhoon TRIO+, GE Healthcare Life Sciences, Piscataway, NJ) and excised or PCR product was directly subcloned. DNA extraction was performed using QIAEX® II Gel Extraction Kit (Qiagen, Valencia, CA). 10 µl of extracted DNA was amplified for three cycles and subcloned using TOPO TA Cloning® Kit for Sequencing (Invitrogen, Carlsbad, CA). Competent cells were inoculated on LB agar plates (Difco™ Luria Agar Base, Miller, BD, Franklin Lakes, NJ) containing ampicillin (100µg/ml) and incubated overnight at 37°C. 96–144 for each sample were randomly chosen for inoculation in 400 µl LB broth (Difco Luria Broth Base, Miller, BD) containing ampicillin (100µg/ml) in deep 96-well plates and incubated overnight on a shaker (250 rpm) at 37°C. 160 µl of bacterial broth was frozen at −80°C in the presence of 40 µl of 50% glycerol. Sequences were performed by AGENCourt Bioscience Corporation (Beckman Coulter Company, Beverly, MA) or GeneWiz, Inc. (South Plainfield, NJ). Data were analyzed using FinchTV software (Geospiza, Inc., Seattle, WA). Clonotypes were named on the basis of their amino acid sequence with a numerical coding that allows reconstruction of the nucleotide sequence (24). Complete names are provided in the Supplemental Tables. When there is no chance of misunderstanding, names were shortened to omit BV CDR3 length and encoding information.
Repertoire measures and characteristics
These measures define a number of general repertoire characteristics, and are based on literature references (25, 26).
M - the number of sequences analyzed
N - the number of unique clonotypes identified
Rmax - the number of observations of the most frequent clonotype
Ns - the number of clonotypes that appeared only once (singletons)
Ncr – the number of cross-reactive clonotypes
Ns/N – the fraction of singletons
Ncr/N – the fraction of cross-reactive clonotypes
Rmax/M – the frequency of the most represented clonotype
Statistical analysis
Rank-repertoire-number summary
To analyze frequencies of cross-reactive clonotypes in different peptide repertoires, we used a rank-repertoire-number summary. Clonotype frequencies were plotted as a function of the number of peptides recognized: All clonotypes that were found only in one peptide repertoire were ranked as “1” (non-cross-reactive); all clonotypes found in M158–66 and any substituted peptides repertoire (cross-reactive) as “2”; etc. The frequency of clonotypes at each rank is plotted as the fraction of the total donor-specific repertoire (BV19 L11).
Estimation of parameters
Parameters for power law-like distribution were estimated as described in ref. (27).
pMHC structure modeling
pMHC images were generated by using the PyMol Molecular Graphics System (De Lano Scientific LLC (v0.99) San Carlos, CA).
RESULTS
Defining the recall response to M158–66 in three donors
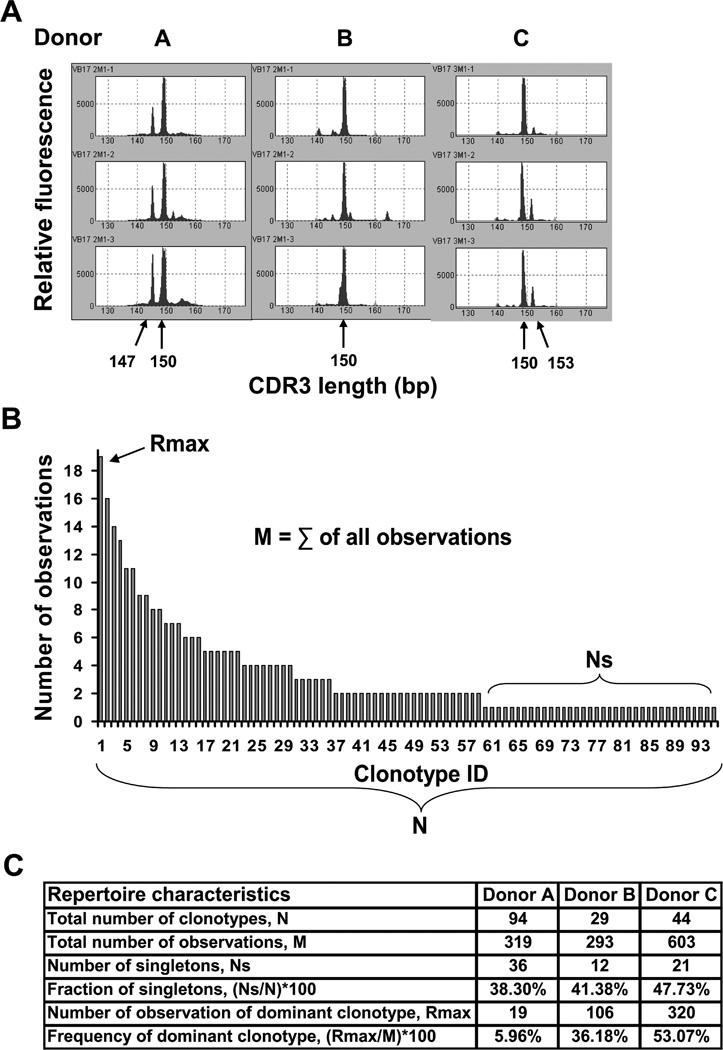
To measure the recall response we stimulated PBMC from three different HLA-A2donors with the M158–66 peptide in short-term (2 week) recall cultures in triplicate. CD8 T cells from the cultures were analyzed by spectratyping to measure repertoire skewing as a result of exposure to the M158–66. Because clonotypes that use BV19 are known to be important in this response, we focused on this BV family. The results of the BV19 spectratype analyses for all three donors are presented in Figure 1A. A band is considered specific if it was focused in two out of three cultures. The M158–66 response is characterized by focusing to a 150 bp band which corresponds to a CDR3 length of 11 aa. Additionally, shorter or longer sizes might be present in response to M158–66. Thus, a 147 bp band was identified in Donor A and 153 bp band – in Donor C.
Figure 1.
Clonotype analysis of the 150 bp (L11) repertoire
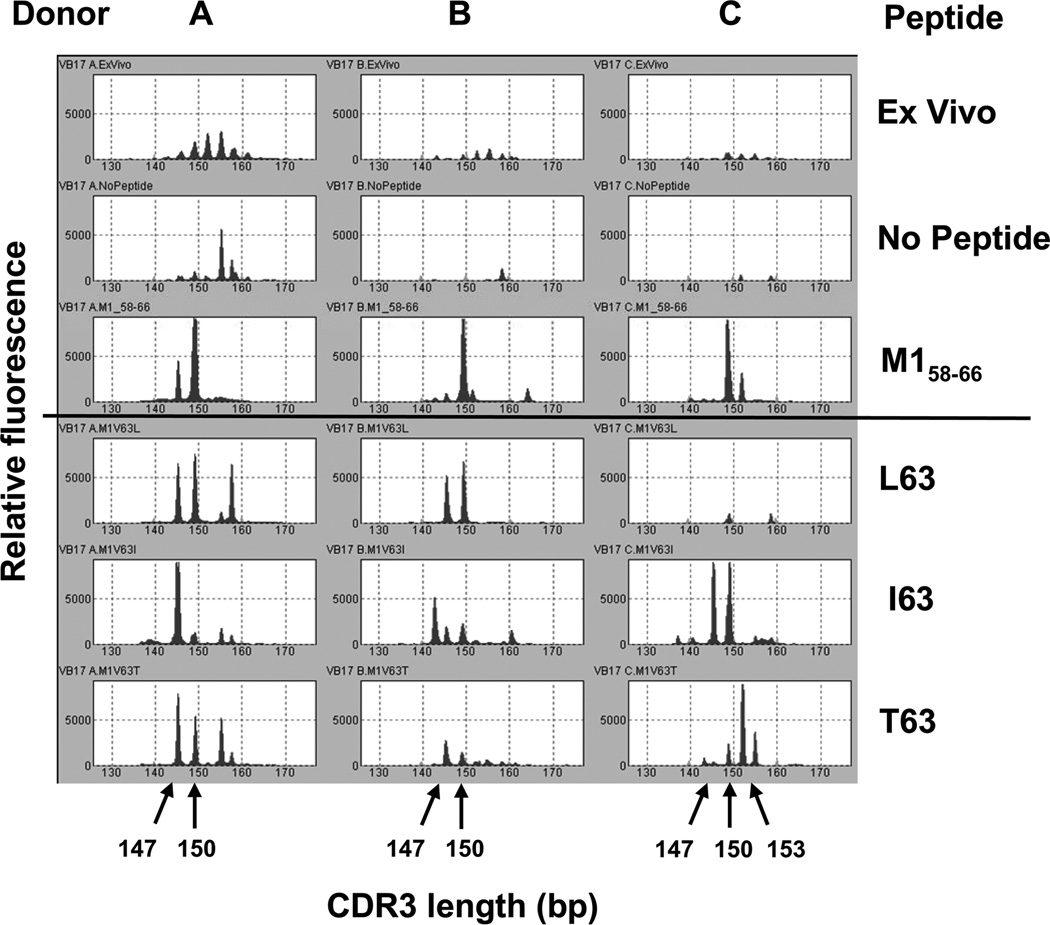
The BV19 150 bp band represents the predominant V gene and CDR3 size in the M158–66 response. To identify the clonotypes present and to measure their frequency we sequenced the CDR3 regions of the PCR product representing clonotypes of L11, from all the cultures for all three donors. An example of a clonotype distribution of the combined data from all three M158–66 cultures is shown for Donor A in Fig. 1B. The primary characteristics of the repertoire include the number of unique clonotypes identified (N), the number of sequences analyzed (M), the number of observations of the most frequent clonotype (Rmax), and the number of singletons (Ns) (as described in ref. 25, 26 and showed on Fig. 1C). These repertoire descriptors will be provided for all the repertoire analyses presented.
Repertoires from triplicate cultures for each peptide were combined into one peptide specific repertoire. For Donor C, we generated two M158–66 repertoires in different experiments and thus the number of observation (M = 603) is twice as high as for the others.
Approach to the analysis of repertoire cross-reactivity
The question we address here is to what extent clonotypes that respond to M158–66 also respond to substituted peptides that present a slight or considerable change in the epitope recognition surface. The choice of substitutions was based in part on the work of Gotch et al. (22), who evaluated different M158–66 substitutions on CTL function using established lines.
We restricted our substitutions to TCR contact residues at positions 63 and 65 (Table I). The Val at position 63 makes hydrogen bonds with Arg98 and Ser99 of CDR3 and Gln52 of CDR2 of the TCR β chain. The Thr at position 65 forms a direct hydrogen bond with Asp32 of the BV CDR1 loop and an indirect hydrogen bond interaction with Asp32 through a highly ordered water molecule that participates in a hydrogen network with the BV CDR3 loop (28). The combination of Asp32of the CDR1 and Gln52 of the CDR2 loops of the BV chain is unique to the human BV19 sequence.
We tried both conservative and non-conservative substitutions (Table I). Substitutions at these positions could result in expansion of clonotypes that react with M158–66 and one or more of the substituted peptides. Such clonotypes will define the cross-reactivity of M158–66 repertoire.
It should be noted that the three individuals studied here have generated different naïve repertoires based on their total MHC polymorphism and other polymorphisms affecting positive and negative selection. They also have had different pathogen exposure histories, which would affect their overall peripheral selected repertoire. Therefore, if we observe similar patterns we will be examining large-scale effects relatively independent of these differences.
Choosing targets for clonotype analysis
We tested each peptide in short-term (2 week) recall cultures in triplicate. CD8 T cells from the cultures were analyzed by BV19 spectratyping to measure repertoire skewing as a result of exposure to the peptides. The results of the BV19 spectratype analyses for all three donors are presented in Figure 2. A representative triplicate culture for M158–66 and the three peptides substituted at position 63 are shown. The ex vivo peak distribution is Gaussian, and the no peptide control occasionally shows some nonspecific skewing. With M158–66, all three individuals showed focusing of the 150 bp band that corresponds to an 11 amino acid CDR3 length (L11), consistent with previous data (19, 20). One individual (Donor A) showed a 147 bp band (10 aa fragment of the CDR3 region, L10) that would be expected of the J1.2 associated response to this peptide (18), and Donor C showed a weak but reproducible band of 153 bp.
Figure 2.
Substituting Val63 for Leu had a drastic effect on the response by Donor C as all BV19 responses were lost, whereas Donors A and B were unaffected, with Donor B now showing a 147 bp band. The 159 bp band in Donor A was not considered specific as there was a band of the same length in the no peptide control. Substitution of Val63 for Ile resulted in a weak but reproducible response in Donor B. Donor A showed an increase in intensity of the 147 bp band and a much reduced 150 bp band. A 147 bp band became prominent in the data from Donor C and the 150 bp band remained. A 147 bp band was predominant for the Val63 to Thr substitution in Donors A and B, and the153 bp band predominated in Donor C. The 150 bp band was reduced in all three donors after stimulation with this peptide.
Substituting Val63 for Arg, Ser, Lys, or Ala showed either a very weak BV19 response (Donor C after stimulation with R63, a 159 bp band) or no BV 19 response at all (data are not shown). Thus, three substituted peptides favor the expansion of CD8 T cells that possess TCR with similar CDR3 lengths to that observed with M158–66.There is some shifting of distribution between the CDR3 lengths and the appearance of T cells with a CDR3 length that is not observed with the M158–66 peptide.
We analyzed the peptides substituted at position 65 in a similar manner. The results for all the stimulating peptides and for the specific CDR3 lengths are summarized in Table II. We performed clonotype analysis by sequencing the PCR products. We will describe the BV19 clonotype data on the basis of the length of the CDR3.
Table II.
List of BV19 CDR3 lengths chosen for clonotype analysis for the seven peptides for which a BV19 response was observed.
| Donors | A | B | C | ||||
|---|---|---|---|---|---|---|---|
| Peptides/length bp | 147 | 150 | 147 | 150 | 147 | 150 | 153 |
| M158–66 | + | + | + | + | + | + | |
| L63 | + | + | + | + | |||
| I63 | + | + | + | + | + | + | |
| T63 | + | + | + | + | + | + | |
| A65 | + | + | + | ||||
| S65 | + | + | + | + | + | ||
| G65 | + | + | + | ||||
Cross-reactivity of the BV19 repertoire in response to stimulation with substituted peptides
Cross-reactivity of the 150 bp (L11) M158–66 repertoire
The clonotypes that were present in the M158–66 repertoire and at least one of the substituted peptide repertoires are called cross-reactive. The repertoire summaries and the relative frequencies of cross-reactive clonotype for each donor are shown in Supplemental Table I. The summary at the bottom of each column shows a number of pertinent repertoire measures and characteristics. We added the repertoire measure Ncr, the number of cross-reactive clonotypes, to our repertoire summaries.
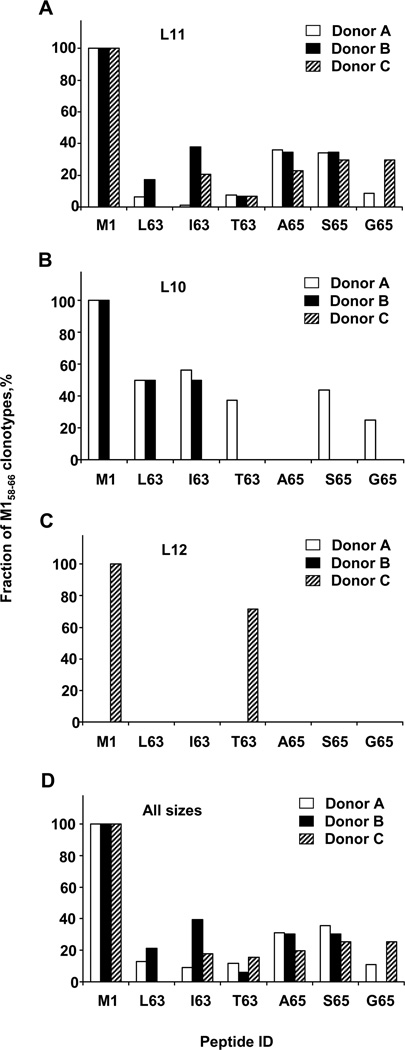
For Donor A there are 48 cross-reactive clonotypes observed in the M158–66 repertoire. Six of these are observed in the L63 repertoire (12.5%), seven in the T63 repertoire (14.6%), one clonotype in the I63 repertoire (2.1%), thirty-four (70.8%) in A65, thirty-two (66.7%) in the S65 and eight (16.7%) in G65 repertoires. These data are summarized in Figure 3A (open bars) by plotting all the clonotypes that respond to a particular substituted peptide as a percentage of those that responded to the M158–66 peptide. This shows that the differences in cross-reactivity vary by peptide and position of substitution.
Figure 3.
Data for all three donors also summarized on Figure 3A. Overall, the fraction of M158–66 cross-reactive clonotypes for position 63 substitutions was lower than for position 65. It can also be seen that cross-reactivity varies on the basis of the peptide and the donor.
CD8 T cells of all three donors similarly respond to stimulation with A65 (22.7%–36.2%), S65 (29.5%–34.5%) and T63 (6.8%–7.4%) peptides (Fig. 3A). This similarity probably reflects the ability of many M158–66-responsive clonotypes from every donor to recognize the relatively similar structural substitutions involved.
Responses to other substituted peptides are donor specific. For example, Donor A has very low (1.1%) cross-reactivity to I63, whereas Donors B and C have high cross-reactivity (37.9% and 20.5%, respectively). Donor C did not respond to the L63 peptide, but both Donors A (6.4%) and B (17.2%) did. Donor B did not respond to G65, while Donor A (8.5%) and Donor C (29.5%) demonstrated different levels of cross-reactivity. Donor A responded to all six substituted peptides. We assume that these differences in cross-reactivity in the three donors are probably due to differences in history of antigen exposure.
Cross-reactivity of the 147bp (L10) repertoire
Clonotypes with CDR3 L10 normally use BJ1.2 and have a CDR3 motif of xG(S/V/A)Y, with the Y being encoded by BJ1.2. Only Donors A and B showed L10 responses to M158–66 (Fig. 3B and Suppl. Table II). For Donor A, the L10 clonotypes showed a large extent of cross-reactivity, as five substituted peptides were recognized. The fraction of M158–66 cross-reactive clonotypes in substituted peptides repertoires varies from 25.00% to 56.25%. For Donor B the fraction of cross-reactive clonotypes was also high (~50%), however only two substituted peptide were recognized. Comparison of the L10 data with the L11 data shows that the fraction of L10 clonotypes that cross-react on the substituted peptides is more limited.
Cross-reactivity of the 153 bp (L12) repertoires
The L12 clonotypes in Donor C represent an unusual response to the M158–66 because it has never been reported before. The response at this CDR3 length is relatively oligoclonal and the clonotypes do not show any specific CDR3 amino acid motif (Suppl. Table II). However, they are also cross-reactive with the T63 peptide (71.4%) (Fig. 3C).
Summary of cross-reactivity in total BV19 repertoire
In Figure 3D we summarized data for all BV19 cross-reactive clonotypes. Overall, the data are similar to the L11 data (Fig. 3A) as a large portion of total BV19 repertoire is composed of L11 clonotypes. Overall cross-reactivity for Donor A irrespective of peptide or CDR3 length is 23.25%, for Donor B – 26.41% and for Donor C – 21.29%. Level of cross-reactivity for all three donors is very high, indicating that this is a general phenomenon.
Extent of cross-reactivity in the BV19 repertoires
The extent of cross-reactivity is a property of the clonotype and is based on the number of peptides repertoires in which the clonotype can be observed. If it is only observed once, then the clonotype is not-cross-reactive and the extent of cross-reactivity is zero.
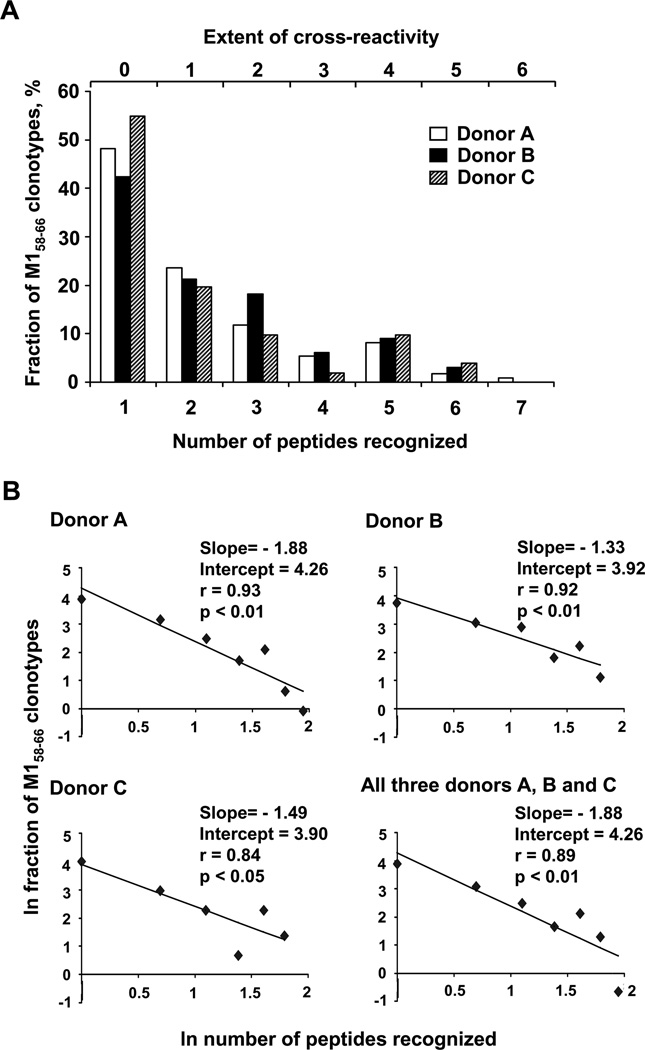
To measure the extent of cross-reactivity for each donor, we plotted the relative frequency of clonotypes as a function of the number of peptides recognized (Fig. 4A). Most of the clonotypes were only observed in the M158–66 repertoire (extent of cross-reactivity is zero). The maximum extent of cross-reactivity was six for Donor A (one clonotype) and five for Donors B and C (one and two clonotypes, respectively). When the relation between the number of clonotype in any repertoire and extent of cross-reactivity was examined we found a moderate correlation (r=0.54, 0.56, 0.51 for Donors A, B, and C, respectively) that was statistically significant (p<0.001) for each donor.
Figure 4.
When examining the data it is evident that the relative frequency drops quickly as the number of peptides recognized increases. These results could be indicative of a power law-like distribution. In a power law distribution the frequency (y) would change as a function of the number of peptide recognized (x) according to y = axb. The parameters a refers to the proportion of the clonotypes observed responding to one peptide and b to the rate at which the frequency drops. We can estimate these two parameters by plotting the natural log of the above equation, which gives ln y = ln a + b*ln x. This is the formula for a line with slope b and an intercept of ln a. We find that the ln frequency distribution of cross-reactive clonotypes in the repertoires of all three donors can be described by a power law (Fig. 4B). To summarize these data we combined the repertoires from all three donors and analyzed the distribution in a similar manner. The correlation between the data and estimated line for each data set was between 0.84 (p<0.05) to 0.93 (p<0.01). We estimated parameters for all three donors: b={−1.88±0.34 (p<0.01), −1.33±0.28 (p<0.01), −1.49±0.48 (p<0.01)}; a={4.26±0.46 (p<0.01), 3.92±0.35 (p<0.01), 3.90±0.60 (p<0.01)} and for the combined data set: b=−1.88±0.44 (p<0.01), a=4.26±0.61 (p<0.01). Data demonstrate a very good fit to a power law-like distribution. The slope (parameter b) is consistently negative and significant, indicating a rapid drop in representation of cross-reactive clonotypes as their rank increases: from 71% at the first rank to 11% at the second rank to 1.6% at the third and so forth. This behavior is observed in each donor and in a pooled repertoire.
Overall, stimulation of human PBMC with peptides substituted at TCR contact residues of M158–66 resulted in specific recall responses to at least five of the twelve peptides in all three donors studied with different degrees of clonality and cross-reactivity.
DISCUSSION
The data presented here provides a generalized measure of cross-reactivity of a well studied CD8 T cell response. Taking advantage of the polyclonality of the response to M158–66, we asked what fraction of BV19-expressing CD8 T cells that respond to influenza M158–66 will also respond to peptides that differ to varying extents at two TCR contact positions. The nature of our experiment made it likely that the cross-reactivity analyzed here represents the “structural degeneracy” model for TCR-MHC-peptide interaction (12). In keeping with this interpretation is our observation that of the twelve substitutions analyzed, the six that were recognized represented side-chains that were structurally related to the original.
The number of clonotypes in the M158–66 repertoire that can extend their recognition to this space is quite high, representing from ~45 to 58% depending on the subject analyzed. A large number clonotypes recognizing structurally related epitopes can be advantageous for recognition of novel pathogens after thymic involution and/or for protection against pathogen evasion by epitope drift.
The clonotypes that can recognize more than just the M158–66 peptide show for the most part the expected characteristics of clonotypes in this repertoire. Thus, there is not a special subset of the repertoire that shows cross-reactivity. Interestingly, two of the most cross-reactive clonotypes, sIYASd in Donor A and sTRGs in Donor B (Supplemental Table I), are not RS clonotypes. However, the most cross-reactive clonotypes in Donor C are RS clonotypes. There is also a restricted VA and JA gene association with this response (29) and it is possible that examination of cross-reactive clonotypes for α-chain usage may show specific characteristics associated with cross-reactivity.
While there are many examples of cross-reactive T cell clones (reviewed in 1, 12, 14, 30, 31) so far few studies have attempted to measure the fraction of cross-reactive clonotypes in antigen-specific repertoires. There have been well documented examples of cross-reactive clonotypes. For example, mouse T cells specific to the VV-encoded a11r198–205 epitope could be cross-reactive with three different LCMV epitopes or one other VV epitope (10). Recently, M158–66 clonotypes that recognize the structurally dissimilar EBV-BMLF1280–288 epitope were identified from a healthy donor (11). The cross-reactive and non-cross-reactive clonotypes had similar characteristics. While, tetramer staining showed that the proportion of cross-reactive T cells is low, it is not possible to calculate the fraction of BMLF1280–288 cross-reactive clonotypes in the M158–66 repertoire from the data presented. Nevertheless, these data indicate that even structurally dissimilar epitopes can generate M158–66 cross-reactive clonotypes. Ishizuka et al. (13) attempted to quantify T cell cross-reactivity to unrelated antigens by using 15 mouse and human CD8 T cell clones and exposing them to pools of peptides. They identified only a single cross-reaction for mouse TCR. Also, two cell lines, one alloreactive and the other anti-influenza, showed no cross-reactivity on the peptide pools. Their calculation of the probability of cross-reactivity for related peptides is 0.003% (1/30,000), which is much lower than our data. We can only speculate that these measures are dependent on the nature of the clones examined. One factor that may have some bearing on this issue is that there maybe a relation between the presence of cross-reactivity and the dominance of the particular epitope (10). If by dominance it is meant the high frequency of a monoclonal response our data is a counter example of this concept. In all three donors the highest frequency clonotype was cross-reactive. However, it is possible that the selection of T cell clones or hybridomas from less polyclonal repertoires favors dominant epitopes whose responding repertoires are less- or non-cross-reactive. The polyclonality itself may be a function of cross-reactivity.
The clonotypes that constitute the M158–66 repertoire show different extents of cross-reactivity with ~50% not showing any, ~25% recognizing only one other peptide, and a deceasing number recognizing more than one. The ability to map this relation between clonotype number and extent of recognition to a power law-like distribution is in keeping with an affinity model in which the TCR structures that allow a large extent of cross-reactivity are constrained. i.e. It becomes harder to find examples in TCR structure space of instances that allow a broader recognition of peptide-MHC space. The composite of the slope and intercept of the power law-like distribution may be defining the optimum relationship between the need for pathogen epitope specificity and the need for covering possible epitope variation, without straying into self-recognition. The need for the balance between specificity and crossreactivity stems from the finite number of TCR and the number of epitopes that need to be recognized. If this relationship is a general function of TCR structure, we would expect to see similar parameters from analyses that subtly alter other previously encountered epitopes.
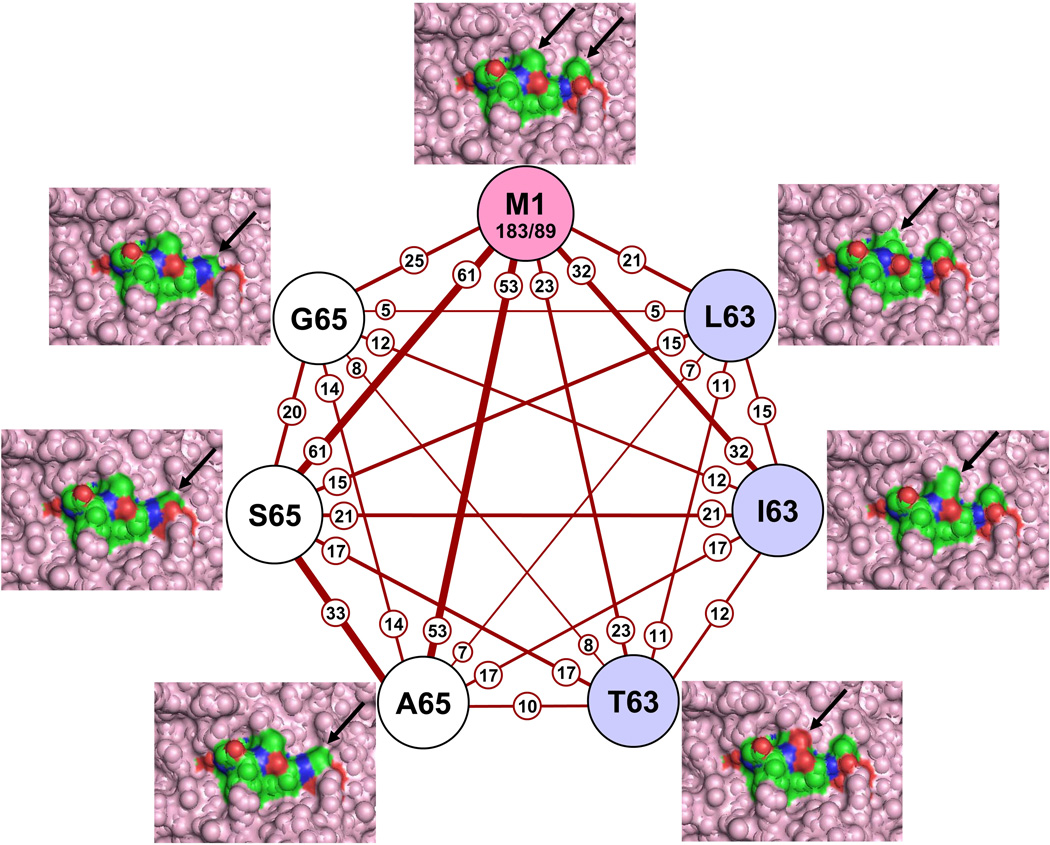
The ability of a clonotype to participate in more than one response (belong to more than one repertoire) can be envisioned as mapping out a cross-reactive network. We can gain a more detailed insight into cross-reactivity by examining the network of connections between the peptides recognized. For defining the cross-reactive network we combined the TCR data from all three donors. We borrowed the elementary node and edge concepts from graph theory to describe this network (Fig. 5). Each circle (node) defines the response to one of the peptides. Each line (edges) indicates the cross-reactive clonotype connection between any two peptides. For the M1 node we have also shown the number of clonotypes in entire repertoire (183) and in the subset of clonotypes that made these connections (89).
Figure 5.
To quantify the description of this network we add the concept of connectivity, which has two parameters—strength and breadth. The strength of a connection is the count of the number of clonotypes in a line (Fig. 5, small circles). The thickness of the lines also reflects this number. The strongest connections are between M158–66 and S65 (61), and M158–66 and A65 (53). This forms a highly connected trio in that the connectivity between A65 and S65 (33) is the third highest in the network. This implies that the M158–66 reactive clonotypes that can recognize one of these have a high chance of recognizing the other. The change of Thr65 to Ser can be considered structurally minimal, but that of Thr65 to Ala involves the loss of a solvent-exposed oxygen atom (Fig. 5, arrow pointing to changes in the structure). Nevertheless, there is sufficient similarity between these structures so that about one/sixth of the M158–66 repertoire (33/183) can recognize all three. The fourth strongest connection is M158–66 to I63 (32), although this does not define a tightly related subset as did the S65 and A65.
The number of connections defines the breadth of connectivity (also referred to as the index). By definition M158–66 makes six connections to the other peptides. However, it is striking that the network is completely connected, i.e. all nodes have a line leading to all nodes. This implies a very extensive connectivity between the M158–66 structure and these six related structures, a fact that may not have been expected. For example, of the twenty-one clonotypes that recognize L63, about three quarters (15) also recognize S65. This involves the TCR accommodating a change in a different part of the interface. There were some exceptions that may have ramifications in thinking about the relation between this type of cross-reactivity and the structure and flexibility of TCR-peptide-MHC contacts. For example, a Val to Thr substitution at position 63 was tolerated, but not a Val to Ser substitution. Smaller side chains were well accepted at position 65 but not at 63. The fact that the network is completely connected also implies that there is a well-defined structural recognition boundary around this group of peptides. Not one of the 183 M158–66 clonotypes would recognize the Val63 to Ser change, even though Val63 to Thr and Thr65 to Ser were very well tolerated. This is also true for some of the other peptides analyzed. By studying these connectivity patterns, one can start to appreciate the subtlety of the patterns of cross-reactivity evidenced. To actually map the connectivity of any one clonotype, the data in the supplementary tables should be consulted.
We feel justified in pooling the data from all three donors because what is being examined here is a relation between the clonotype (TCR) and pMHC, independent of origin of the T cell. The only donor-specific changes are that the G65 and L63 responses were observed in two of three donors. This indicates that these results are generalizable to many if not all HLA-A2 individuals. Also it is important to point out that in the two donors that only recognized five substituted peptides the cross-reactive network is still completely connected.
Two of the substituted peptides that we tested were part of an alanine scan mutagenesis study by Berkhoff et al. (32). Both these substitutions decreased IFN-γ production and greatly reduced or eliminated cytotoxic responses by a M158–66 specific CTL clone. While these results are in keeping with our A65 peptide, they differ from our inability to observe cross-reactivity with the A63 peptide. Nevertheless, this study with mutant viruses indicates that cross-reactivity in this system can have functional consequences.
We have shown that for a particular highly conserved peptide epitope, the responding repertoire contains a portion of cross-reactive clonotypes recognizing a defined group structurally similar epitopes. This cross-reactivity results in a network whose connectivity can be quantified. We do not know the history of the exposures that led to the current network in these three donors, nor do we know if any of the MHC-peptide structures that we are investigating actually describe a previously encountered epitope. While the M158–66 epitope is invariant in all flu strains, one formal possibility is that some of these epitopes are generated during an influenza infection by the inherent mutagenesis of influenza which could provide such epitopes yet produce non-viable viral progeny. Nevertheless, we would predict that any newly encountered pathogen generating an epitope whose shape corresponds to that of the peptides studied here, would find a pre-existing repertoire ready to respond to it.
Our ability to define such a network in recall responses in middle-aged donors has ramifications for thinking about how novel pathogens may be recognized. If a general phenomenon, cross-reactive networks of the type described here might provide coverage for novel pathogens as long as they produced in-network epitopes. Such networks could also provide for coverage of escape variants of known epitopes. Any cross-reactivity due to apparently structurally unrelated epitopes would add to these networks.
Supplementary Material
Footnotes
This work was supported by National Institutes of Health (Grant U19 AI062627).
REFERENCES
- 1.Wucherpfennig KW. T cell receptor crossreactivity as a general property of T cell recognition. Mol. Immunol. 2004;40(14–15):1009–1017. doi: 10.1016/j.molimm.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 2.Haanen JB, Wolkers MC, Kruisbeek AM, Schumacher TN. Selective expansion of cross-reactive CD8(+) memory T cells by viral variants. J. Exp. Med. 1999;190(9):1319–1328. doi: 10.1084/jem.190.9.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hu N, D'Souza C, Cheung H, Lang H, Cheuk E, Chamberlain JW. Highly conserved pattern of recognition of influenza A wild-type and variant CD8+ CTL epitopes in HLA-A2+ humans and transgenic HLA-A2+/H2 class I-deficient mice. Vaccine. 2005;23(45):5231–5244. doi: 10.1016/j.vaccine.2005.07.032. [DOI] [PubMed] [Google Scholar]
- 4.Kasprowicz V, Ward SM, Turner A, Grammatikos A, Nolan BE, Lewis-Ximenez L, Sharp C, Woodruff J, Fleming VM, Sims S, Walker BD, Sewell AK, Lauer GM, Klenerman P. Defining the directionality and quality of influenza virus-specific CD8+ T cell cross-reactivity in individuals infected with hepatitis C virus. J. Clin. Invest. 2008;118(3):1143–1153. doi: 10.1172/JCI33082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wedemeyer H, Mizukoshi E, Davis AR, Bennink JR, Rehermann B. Cross-reactivity between hepatitis C virus and Influenza A virus determinant-specific cytotoxic T cells. J. Virol. 2001;75(23):11392–11400. doi: 10.1128/JVI.75.23.11392-11400.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friberg H, Burns L, Woda M, Kalayanarooj S, Endy TP, Stephens HA, Green S, Rothman AL, Mathew A. Memory CD8(+) T cells from naturally acquired primary dengue virus infection are highly cross-reactive. Immunol. Cell. Biol. 2010;27:1–8. doi: 10.1038/icb.2010.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen HD, Fraire AE, Joris I, Brehm MA, Welsh RM, Selin LK. Memory CD8+ T cells in heterologous antiviral immunity and immunopathology in the lung. Nat. Immunol. 2001;2(11):1067–1076. doi: 10.1038/ni727. [DOI] [PubMed] [Google Scholar]
- 8.Brehm MA, Pinto AK, Daniels KA, Schneck JP, Welsh RM, Selin LK. T cell immunodominance and maintenance of memory regulated by unexpectedly cross-reactive pathogens. Nat. Immunol. 2002;3(7):627–634. doi: 10.1038/ni806. [DOI] [PubMed] [Google Scholar]
- 9.Acierno PM, Newton DA, Brown EA, Maes LA, Baatz JE, Gattoni-Celli S. Cross-reactivity between HLA-A2-restricted FLU-M1:58–66 and HIV p17 GAG:77–85 epitopes in HIV-infected and uninfected individuals. J. Transl. Med. 2003;1(1):3–14. doi: 10.1186/1479-5876-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cornberg M, Clute SC, Watkin LB, Saccoccio FM, Kim SK, Naumov YN, Brehm MA, Aslan N, Welsh RM, Selin LK. CD8 T cell cross-reactivity networks mediate heterologous immunity in human EBV and murine vaccinia virus infections. J. Immunol. 2010;184(6):2825–2838. doi: 10.4049/jimmunol.0902168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clute SC, Naumov YN, Watkin LB, Aslan N, Sullivan JL, Thorley-Lawson DA, Luzuriaga K, Welsh RM, Puzone R, Celada F, Selin LK. Broad cross-reactive TCR repertoires recognizing dissimilar epstein-barr and influenza A virus epitopes. J. Immunol. 2010;185(11):6753–6764. doi: 10.4049/jimmunol.1000812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yin Y, Mariuzza RA. The multiple mechanisms of T cell receptor cross-reactivity. Immunity. 2009;31(6):849–851. doi: 10.1016/j.immuni.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 13.Ishizuka J, Grebe K, Shenderov E, Peters B, Chen Q, Peng Y, Wang L, Dong T, Pasquetto V, Oseroff C, Sidney J, Hickman H, Cerundolo V, Sette A, Bennink JR, McMichael A, Yewdell JW. Quantitating T cell cross-reactivity for unrelated peptide antigens. J. Immunol. 2009;183(7):4337–4345. doi: 10.4049/jimmunol.0901607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Welsh RM, Che JW, Brehm MA, Selin LK. Heterologous immunity between viruses. Immunol. Rev. 2010;235(1):244–266. doi: 10.1111/j.0105-2896.2010.00897.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bednarek MA, Sauma SY, Gammon MC, Porter G, Tamhankar S, Williamson AR, Zweerink HJ. The minimum peptide epitope from the influenza virus matrix protein. Extra and intracellular loading of HLA-A2. J. Immunol. 1991;147(12):4047–4053. [PubMed] [Google Scholar]
- 16.Morrison J, Elvin J, Latron F, Gotch F, Moots R, Strominger JL, McMichael A. Identification of the nonamer peptide from influenza A matrix protein and the role of pockets of HLA-A2 in its recognition by cytotoxic T lymphocytes. Eur. J. Immunol. 1992;22(4):903–907. doi: 10.1002/eji.1830220404. [DOI] [PubMed] [Google Scholar]
- 17.Moss PA, Moots RJ, Rosenberg WM, Rowland-Jones SJ, Bodmer HC, McMichael AJ, Bell JI. Extensive conservation of alpha and beta chains of the human T-cell antigen receptor recognizing HLA-A2 and influenza A matrix peptide. Proc. Natl. Acad. Sci. USA. 1991;88(20):8987–8990. doi: 10.1073/pnas.88.20.8987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lehner PJ, Wang EC, Moss PA, Williams S, Platt K, Friedman SM, Bell JI, Borysiewicz LK. Human HLA-A0201-restricted cytotoxic T lymphocyte recognition of influenza A is dominated by T cells bearing the V beta 17 gene segment. J. Exp. Med. 1995;181(1):79–91. doi: 10.1084/jem.181.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lawson TM, Man S, Williams S, Boon AC, Zambon M, Borysiewicz LK. Influenza A antigen exposure selects dominant Vbeta17+ TCR in human CD8+ cytotoxic T cell responses. Int. Immunol. 2001;13(11):1373–1381. doi: 10.1093/intimm/13.11.1373. [DOI] [PubMed] [Google Scholar]
- 20.Naumov YN, Hogan KT, Naumova EN, Pagel JT, Gorski J. A class I MHC-restricted recall response to a viral peptide is highly polyclonal despite stringent CDR3 selection: implications for establishing memory T cell repertoires in "real-world" conditions. J. Immunol. 1998;160(6):2842–2852. [PubMed] [Google Scholar]
- 21.Naumov YN, Naumova EN, Clute SC, Watkin LB, Kota K, Gorski J, Selin LK. Complex T cell memory repertoires participate in recall responses at extremes of antigenic load. J. Immunol. 2006;177(3):2006–2014. doi: 10.4049/jimmunol.177.3.2006. [DOI] [PubMed] [Google Scholar]
- 22.Gotch F, McMichael A, Rothbard J. Recognition of influenza A matrix protein by HLA-A2-restricted cytotoxic T lymphocytes. J.Exp.Med. 1988;168:2045–2057. doi: 10.1084/jem.168.6.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yassai M, Naumova E, Gorski J. In: Generation of TCR spectratypes by multiplex PCR for T cell repertoire analysis In: The human antigen T cell receptor: selected protocols & applications. Oksenberg J, editor. Austin, TX: Landes Company and Chapman & Hall; 1997. pp. 327–372. [Google Scholar]
- 24.Yassai MB, Naumov YN, Naumova EN, Gorski J. A clonotype nomenclature for T cell receptors. Immunogenetics. 2009;61(7):493–502. doi: 10.1007/s00251-009-0383-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Naumova EN, Gorski J, Naumov YN. Two compensatory pathways maintain long-term stability and diversity in CD8 T cell memory repertoires. J Immunol. 2009;183(4):2851–2858. doi: 10.4049/jimmunol.0900162. [DOI] [PubMed] [Google Scholar]
- 26.Naumova EN, Gorski J, Naumov YN. Simulation studies for a multistage dynamic process of immune memory response to influenza: experiment in silico. Ann. Zool. Fennici. 2008;45(5):369–384. doi: 10.5735/086.045.0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naumov YN, Naumova EN, Hogan KT, Selin LK, Gorski J. A fractal clonotype distribution in the CD8+ memory T cell repertoire could optimize potential for immune responses. J. Immunol. 2003;170(8):3994–4001. doi: 10.4049/jimmunol.170.8.3994. [DOI] [PubMed] [Google Scholar]
- 28.Stewart-Jones GB, McMichael AJ, Bell JI, Stuart DI, Jones EY. A structural basis for immunodominant human T cell receptor recognition. Nat. Immunol. 2003;4(7):657–663. doi: 10.1038/ni942. [DOI] [PubMed] [Google Scholar]
- 29.Naumov YN, Naumova EN, Yassai MB, Kota K, Welsh RM, Selin LK. Multiple glycines in TCR alpha-chains determine clonally diverse nature of human T cell memory to influenza A virus. J. Immunol. 2008;181(10):7407–7419. doi: 10.4049/jimmunol.181.10.7407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holler PD, Kranz DM. T cell receptors: affinities, cross-reactivities, and a conformer model. Mol. Immunol. 2004;40(14–15):1027–1031. doi: 10.1016/j.molimm.2003.11.013. [DOI] [PubMed] [Google Scholar]
- 31.Wilson DB, Wilson DH, Schroder K, Pinilla C, Blondelle S, Houghten RA, Garcia KC. Specificity and degeneracy of T cells. Mol. Immunol. 2004;40(14–15):1047–1055. doi: 10.1016/j.molimm.2003.11.022. [DOI] [PubMed] [Google Scholar]
- 32.Berkhoff EG, de Wit E, Geelhoed-Mieras MM, Boon AC, Symons J, Fouchier RA, Osterhaus AD, Rimmelzwaan GF. Functional constraints of influenza A virus epitopes limit escape from cytotoxic T lymphocytes. J. Virol. 2005;79(17):11239–11246. doi: 10.1128/JVI.79.17.11239-11246.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.