Abstract
High throughput proteomics studies have identified several thousand acetylation sites on over one thousand proteins. Mitochondrial aconitase, the Krebs cycle enzyme that converts citrate to isocitrate, has been identified in many of these reports. Acetylated mitochondrial aconitase has also been identified as a target for sirtuin 3 (SIRT3) catalyzed deacetylation. However, the functional significance of mitochondrial aconitase acetylation has not been determined. Using in vitro strategies, mass spectrometric analyses, and an in vivo mouse model of obesity, we found a significant acetylation-dependent activation of aconitase. Isolated heart mitochondria subjected to in vitro chemical acetylation with either acetic anhydride or acetyl-CoA resulted in increased aconitase activity that was reversed with SIRT3 treatment. Quantitative mass spectrometry was used to measure acetylation at 21 lysine residues and found significant increases with both in vitro treatments. A high fat diet (60% kcal from fat) was used as an in vivo model and also showed significantly increased mitochondrial aconitase activity without changes in protein level. The high fat diet also produced increased aconitase acetylation at multiple sites as measured by the quantitative mass spectrometry assays. Treatment of isolated mitochondria from these mice with SIRT3 abolished the high fat diet-induced activation of aconitase and reduced acetylation. Finally, kinetic analyses found that the increase in activity was a result of increased maximal velocity and molecular modeling suggests the potential for acetylation at K144 to perturb the tertiary structure of the enzyme. The results of this study reveal a novel activation of mitochondrial aconitase by acetylation.
Keywords: protein acetylation, aconitase, SIRT3, mitochondria, mass spectrometry, heart, high fat diet, obesity
INTRODUCTION
Lysine acetylation is a reversible post-translational modification where an acetyl group is transferred to the ε-amine of lysine residues in a protein. High throughput proteomic studies have identified several thousand lysine acetylation sites distributed over greater than one thousand proteins (1-7). Bioinformatics analyses of these data reveal a potential link between acetylation, mitochondrial proteins, and metabolism (1). These studies have also shown that mitochondrial protein acetylation responds to nutritional alterations including high fat diet, fasting, and caloric restriction (1-3). There is currently some evidence of a mitochondrial protein acetyl transferase, although its exact role in the heart is not clear (8,9). However, acetyl-CoA is able to react directly with proteins to acetylate lysine residues, especially under the higher pH conditions found in the mitochondria (10). In fact, at least one group has reported that acetyl-CoA from lipid metabolism is necessary and sufficient to acetylate mitochondrial proteins (11). Conversely, deacetylation is enzymatically driven by a group of enzymes known as sirtuins. These enzymes are localized in a variety of intracellular compartments and have a range of deacylase activities, including deacetylation. Sirtuin 3 (SIRT3) is an NAD+-dependent deacetylase present in the mitochondria and is considered to be a primary regulator of mitochondrial acetylation (12). Proteomics experiments with knockout animals have shown a large number of acetylated mitochondrial proteins, but not all, are targets of SIRT3 (1-5). Overall, protein acetylation, and deacetylation, is emerging as an important regulatory post-translational modification. The term carbon stress has been used to illustrate the importance of the balance between non-enzymatic protein acetylation by acetyl-CoA and deacetylation by the sirtuins as a regulator of cellular metabolism (13-15).
Mitochondrial aconitase (ACO2) is consistently identified as an acetylation target in these high throughput proteomics experiments, with acetylation detected at multiple sites. Acetylated aconitase has also been identified as a target for SIRT3-mediated deacetylation based on both synthetic peptide microarray experiments and the analysis of acetylated proteins in Sirt3 knockout mice (1,16). Mitochondrial aconitase is a Krebs cycle enzyme that catalyzes the stereospecific dehydration-hydration reaction that converts citrate to isocitrate. This activity gives aconitase a key role in citrate metabolism with the corresponding potential to impact multiple metabolic pathways affected by citrate availability. For example, others have shown that significant amounts of citrate efflux from mitochondria into the cytosol in perfused rat hearts (17). Once exported to the cytoplasm, citrate is both a source of acetyl-CoA and has the ability to activate acetyl-CoA carboxylase. The malonyl-CoA formed by this enzyme can then act as an inhibitor of carnitine O-palmitoyltransferase 1 to control mitochondrial fatty acid uptake and β-oxidation (17,18). Therefore, an acetylation-dependent change in mitochondrial aconitase function is a potential point at which acetylation and deacetylation could alter mitochondrial fatty acid metabolism.
While these high-throughput proteomics studies show that mitochondrial aconitase is a target for acetylation and support the potential significance of the modification in mitochondrial metabolism, the exact effects on enzyme function have not been determined. In fact, functional effects of only a small number of 1000+ acetylated proteins identified in the proteomics screens have been described. In the study reported here, we found that acetylation of mitochondrial aconitase produced a unique increase in the activity of the enzyme. This activation was produced by direct treatment of isolated mitochondria with the known acetylating reagents acetic anhydride and acetyl-CoA and was reversed by treatment with recombinant SIRT3. These in vitro experiments allowed comprehensive mapping of 21 aconitase acetylation sites and the development of a quantitative proteomics method that used selected reaction monitoring (SRM) to quantify changes in acetylation at these sites. Aconitase activation and acetylation was also seen in vivo in mice fed a high fat diet. Subsequent SRM analyses showed increased acetylation in vivo at 12 of the 21 acetylation sites mapped in vitro. Treatment of the isolated mitochondria with recombinant SIRT3 reduced both the activity of the enzyme and the extent of acetylation at 6 of these sites. These data identify mitochondrial aconitase as an important target of reversible acetylation at the functional level.
MATERIAL AND METHODS
Animal model and diets
Six-week-old male C57BL/6N mice (Charles River) were housed in colony cages on a 12 h light/12 h dark cycle. In selected experiments, the mice were fed either a high fat diet (60% kcal from lard fat, 20% kcal carbohydrates, 20% kcal protein) or a control low fat diet (10% kcal from lard fat, 70% kcal carbohydrate, 20% kcal protein) (Research Diet, Inc.) for the indicated lengths of time. The mice were sacrificed by cervical dislocation and the heart harvested for analysis. All experiments were approved by the Oklahoma Medical Research Foundation Institutional Animal Care and Use Committee.
Sample processing and isolation of heart mitochondria
At sacrifice, the hearts were perfused with an isolation buffer (10 mM MOPS, 1 mM EDTA, 210 mM mannitol, 70 mM sucrose, pH 7.4) and quickly removed. Approximately 10 mg of heart tissue was flash frozen for RNA analysis. The remainder of the heart tissue was processed for mitochondrial isolation. For this isolation, the heart was homogenized in ice-cold isolation buffer using a Potter-Elvehjem homogenizer. The initial homogenate was centrifuged at 550g for 5 min at 4°C and the supernatant was filtered through cheese cloth to clarify the supernatant. Mitochondria were pelleted from the S1 supernatant by centrifugation at 10,000g for 10 min at 4°C and re-suspended in the isolation buffer. For selected treatments, the 0.05% Triton X-100 was included in the buffer to solubilize the mitochondria (19). Protein concentrations were determined using the bicinchroninic acid method with BSA as standard.
Mass Spectrometry Analysis
Mapping acetylation sites
Samples of isolated mitochondria treated with up to 200 μM acetic anhydride were used for qualitative mapping of aconitase acetylation sites. After the reaction, the mitochondrial proteins were precipitated with ice-cold acetone overnight. The precipitated proteins were dissolved in Laemmli buffer, separated by SDS-PAGE, and the gel fixed and stained. The aconitase band at approximately 85 kDa was cut from the gel, reduced, alkylated, and digested with trypsin. The samples were analyzed using data-dependent analysis on a linear ion trap mass spectrometer (ThermoScientific LTQ-XL) configured with a splitless capillary column HPLC system. The samples (10 μL aliquots) were injected onto a 10 cm × 75 μm i.d. column packed with a C18 reversed phase material (Phenomenex, Jupiter C18). The column was eluted at 150 nL/min with a 75 min linear gradient of acetonitrile in 0.1% formic acid. All collision induced dissociation (CID) spectra recorded were used to search the mouse RefSeq database with the search program Mascot. Matching CID spectra were interpreted manually to verify proper assignment as an acetylated peptide.
Quantitative proteomics using SRM
The first type of quantitative proteomics experiment was the analysis of mitochondrial protein expression that specifically measured all Krebs cycle enzymes in isolated mitochondria and whole heart homogenates (20,21). A defined amount of bovine serum albumin (BSA, 8 pmol) was added to samples containing 60 μg total protein and the mixture precipitated with ice-cold acetone overnight. The protein pellet was dissolved at 1.0 μg/μL in Laemmli buffer and a 20 μL aliquot (20 μg protein) run just 1.5 cm into a 12.5% SDS-PAGE gel (BioRad). The gel was then fixed and stained. For each sample, the entire lane was cut, divided into smaller pieces, and washed to remove the stain. The proteins contained in the gel were reduced, alkylated, and digested with 1 μg trypsin overnight at room temperature. The peptides were extracted, dried, and reconstituted in 150 μL 1% acetic acid for analysis. The samples were analyzed on a triple quadrupole mass spectrometry system (ThermoScientific TSQ Vantage) with a splitless capillary column HPLC system (Eksigent). The samples (10μL aliquots) were injected onto a 10 cm × 75 μm i.d. column packed with a C18 reversed phase material (Phenomenex, Jupiter C18). The column was eluted at 150 nL/min with a 60 min linear gradient of acetonitrile in 0.1% formic acid. Each Krebs cycle protein was detected and quantified based on the detection of three peptides chosen and validated during a methodical development process (20). The SRM data were analyzed using the program Pinpoint (ThermoScientific) that determines the chromatographic peak areas for each peptide. The total abundance of each set of three peptides was used to calculate the abundance of the parent protein by normalizing to the total abundance of three peptides from the bovine serum albumin.
The second type of quantitative proteomics experiment measured aconitase acetylation. For this assay, 20 μg of mitochondrial protein was resolved in a full-length 12.5% SDS-PAGE gel, fixed, and stained. The aconitase band at 85kDa was cut from the gel, reduced, alkylated, and digested with 1 μg trypsin overnight at room temperature. The peptides were extracted, dried, and reconstituted in 50 μL 1% acetic acid for analysis. The samples were analyzed using SRM as described above. The detection method for each peptide was developed and optimized using samples from the in vitro treatment of permeabilized mitochondria with up to 200 μM acetic anhydride. The SRM data were analyzed using the program Pinpoint (ThermoScientific) that determine the chromatographic peak areas for each peptide. In each sample, acetylated peptide abundances were normalized to the abundance of the unmodified aconitase reference peptide VDVSPTSQR to correct for minor differences in the amount of aconitase protein among the different samples (22,23).
Western blot analysis
Aconitase was detected in homogenates of both whole heart homogenates and isolated mitochondrial proteins with antibodies for aconitase in 5% milk. Actetylated proteins were detected with and anti-acetyl lysine antibody (Cell Signaling) in 1% BSA. Primary antibody binding was visualized using Horseradish peroxidase conjugated secondary antibodies (Pierce) and SuperSignal West Pico Chemiluminescent Substrate (ThermoScientific).
Aconitase activity assay
Aconitase enzyme activity was measured as previously described (24). Briefly, aconitase activity in mitochondria samples was determined as the rate of NADP+ reduction (340 nm, ε = 6,200 M−1·cm−1) by isocitrate dehydrogenase after addition of 1 mM sodium citrate, 0.6 mM MnCl2, 0.2 mM NADP+, 1 unit/mL isocitrate dehydrogenase. Assays were performed in a buffer containing 25 mM MOPS, 0.05% Triton X-100, at pH 7.4.
Quantitative RT-PCR
Approximately 10 mg of heart tissue was immediately frozen at harvest in liquid nitrogen and stored at −80°C. RNA was extracted using Roche Tripure reagent. RNA concentration was measured by absorbance (260/280 nm ratio >1.8 for all samples) on a NanoDrop 2000 UV-Vis spectrophotometer. RNA (1.0 μg) was converted to cDNA (20 μL final volume) using the QuantiTect Reverse Transcription kit (Qiagen). Quantitative PCR was performed on a CFX96 thermocycler (BioRad) with reactions consisting of 1.0 μ L cDNA, 125 nM final concentration of each primer, and iQ SYBR Green Supermix (BioRad) in a total volume of 20 μL. Technical duplicates were performed for all samples. Relative transcript expression ratios were calculated using CFX Manager Software v2.1 and statistics were generated with REST2009 software (25). The transcript levels for target genes were normalized to three reference genes, Gapdh, Sdha, and Hsp90ab1, determined to be stable between dietary conditions using geNorm analysis (26). The respective cDNA was amplified with the following primers:
Aco2 76bp product. Forward: 5’-tgagtacatccgatatgacctgc -3’; Reverse: 5’-gagagtaagaggccggttcaa -3’ Gapdh 136bp product. Forward: 5’-atgttccagtatgactccactc-3’; Reverse: 5’-ggcctcaccccatttgatgt-3’ Sdha 106bp product. Forward: 5’-ggaacactccaaaaacagacct-3’; Reverse: 5’-ccaccactgggtattgagtagaa-3’ Hsp90ab1102bp product. Forward: 5’-tcaaacaaggagattttcctccg-3’; Reverse: 5’-actgtccaacttagaagggtc-3’
In vitro acetylation
Mitochondria isolated from heart tissue were permeabilized in 25 mM MOPS, 0.05 % Triton X-100. For the reactions with acetic anhydride, pH 7.4 was used with mitochondria at 0.25 mg/mL treated with concentrations of acetic anhydride up to 200μM for 10 min at room temperature. For reaction with acetyl-CoA, pH 7.8 was used with mitochondria at 0.5mg/mL treated with 5mM acetyl-CoA for 5min at room temperature.
SIRT3 deacetylation
The isolated and permeabilized mitochondria were incubated with recombinant human SIRT3 (10 μg) and NAD+ (1 mM) for 15 min at room temperature in a buffer of 50 mM Tris HCl, 137 mM NaCl, 2.7 mM KCl, 1 mM MgCl2, pH 8. Control reactions were performed in the absence of NAD+. The expression vector for the human SIRT3 was obtained from Dr. Eric Verdin at the Gladstone Institutes, University of California-San Francisco.
Computational Methods
The crystal structure of bovine aconitase (PDB Accession number 1AMJ) was used as a template to build a mouse model of aconitase using Modeller software. The mouse aconitase model was then used to create acetylated and unacetylated versions of aconitase using PyMOL (The PyMOL Molecular Graphics System, Version 1.7.2, Schrödinger, LLC)
Statistics
All data are presented as mean ± standard error of the mean (SEM). Statistical analyses were performed using the 2-tailed Student’s t-test. Statistically significant differences are defined as pvalues <0.05.
RESULTS
in vitro acetylation of heart mitochondrial proteins with acetic anhydride activates aconitase
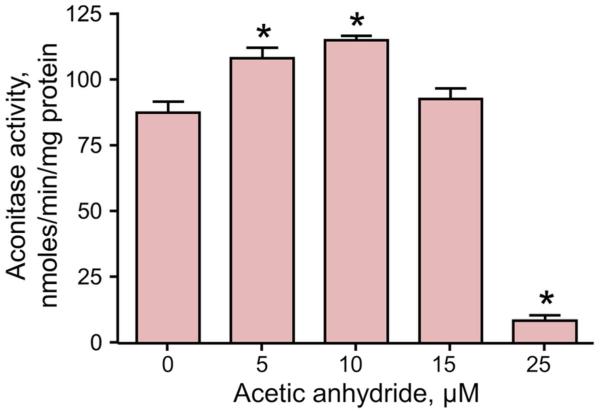
Acetylation is reported to regulate metabolic enzymes through several different mechanisms (27-33), but the role of acetylation in regulating mitochondrial aconitase remains unknown. As an initial test of the effect of acetylation on mitochondrial aconitase, isolated mitochondria were treated with acetic anhydride in vitro and aconitase enzyme activity measured. Acetic anhydride is a known acetylating reagent that we have previously shown capable of acetylating mitochondrial proteins (34). Here, a biphasic effect was seen with aconitase activity increased significantly with acetic anhydride concentrations up to 10 μM (Figure 1). The inactivation of aconitase at higher acetic anhydride concentrations shows a narrow range of effects of acetylation and the ability of protein acetylation to either activate or inhibit protein function, depending on specific reaction conditions. Nonetheless, acetic anhydride is a well-characterized lysine acetylating reagent, making the increased activity at lower concentrations a clear demonstration of the activating effect of acetylation on aconitase activity.
Figure 1. Mitochondrial aconitase is activated by acetylation using acetic anhydride.
Isolated and solubilized mitochondria (0.25 mg protein/mL) were treated with the indicated concentrations of acetic anhydride for 10 min at room temperature. Aconitase activity was measured in a coupled reaction as the rate of NADP+ reduction by isocitrate dehydrogenase with sodium citrate added as the substrate. Values are presented as the mean ± SEM (n = 3). Statistically significant differences, p<0.05, are designated with an asterisk (*)
These in vitro reactions with acetic anhydride were also a valuable resource for mapping the sites of acetylation on the enzyme and developing the quantitative SRM methods used in subsequent experiments. A total of 21 potential acetylation sites on aconitase were identified which were fairly distributed across the primary sequence of the protein (Table 1). A number of other lysine residues were targeted as potential modification sites during the development process but the respective acetylated peptides were not detected. For the acetyl-lysine peptides that were detected, the combination of the measured peptide molecular weight and fragmentation pattern seen in the CID spectra clearly supported the confident assignment of the modification site. This group of acetylation sites is also consistent with the aggregate of acetylation sites on aconitase reported in the various high-throughput proteomics experiments but represents the first comprehensive quantitative analysis of all known sites.
Table 1.
The acetylation sites in aconitase mapped by mass spectrometric analysis.
| Acetylation site | Peptide sequence | Peptide molecular ion m/z, charge |
|
|---|---|---|---|
| 1 | K31 | AK[a]VAM*SHFEPSEYIR | 608.3, +2 |
| 2 | K50 | YDLLEK[a]NINIVR | 766.4, +2 |
| 3 | K86 | GK[a]TYLR | 390.2, +2 |
| 4 | K138 | VAVPSTIHC*DHLIEAQVGGEK[a]DLR | 672.3, +4 |
| 5 | K144 | AK[a] DINQEVYNFLATAGAK | 998.0, +2 |
| 6 | K233 | VIGVK[a]LTGSLSGWTSPK | 886.5, +2 |
| 7 | K309 | YLSK[a]TGR | 433.7, +2 |
| 8 | K401 | SAAVAK[a]QALAHGLK | 469.6, +3 |
| 9 | K411 | C*K[a]SQFTITPGSEQIR | 897.4, +2 |
| 10 | K517 | FNPETDFLTGK[a]DGK | 805.9, +2 |
| 11 | K523 | FK[a] LEAPDADELPR | 771.9, +2 |
| 12 | K549 | SDFDPGQDTYQHPPK[a]DSSGQR | 802.0, +3 |
| 13 | K591 | GK[a]C*TTDHISAAGPWLK | 595.3, +3 |
| 14 | K628 | GHLDNISNNLLIGAINIENGK[a]ANSVR | 930.2, +3 |
| 15 | K689 | AIITK[a]SFAR | 524.8, +2 |
| 16 | K700 | IHETNLK[a]K | 512.8, +2 |
| 17 | K701 | K[a]QGLLPLTFADPSDYNK | 975.0, +2 |
| 18 | K723 | IHPVDK[a] LTIQGLK | 502.0, +2 |
| 19 | K730 | LTIQGLK[a]DFAPGKPLK | 590.0, +2 |
| 20 | K736 | DFAPGK[a]PLK | 507.8, +2 |
| 21 | K739 | DFAPGKPLK[a]C*VIK | 505.6, +3 |
| unmodified | VDVSPTSQR | 494.8, +2 | |
| unmodified | VAGILTVK | 400.8, +2 | |
| unmodified | NTIVTSYNR | 534.3, +2 | |
| unmodified | SQFTITPGSEQIR | 732.4, +2 |
Isolated mitochondria treated with up to 200μM acetic anhydride were reduced, alkylated, and digested with trypsin. The digests were analyzed using combinations of unbiased data dependent methods with database searches designed to find acetylated peptides and targeted analysis based on previously identified acetylation sites. For each site, the resulting collision induced dissociation (CID) spectra were interpreted manually to verify correct assignment of the peptide and acetylation position. The acetylated peptides were incorporated into the quantitative selected reaction monitoring methods use to quantify acetylation in subsequent experiments, with the additional unmodified peptide used for normalization. K[a] designates an acetylated lysine residue.
M designates an oxidized methionine residue.
C designates a carboxyamidomethylated cysteine residue
The identification experiments were the starting point for developing a method to quantify all of the acetylated peptides by SRM. This method was built and validated based on the peptide molecular weights, fragmentation patterns, and chromatographic retention times seen in the qualitative mapping experiments. A multistep process was used to develop the SRM method as described (20). Briefly, an initial version of the method was constructed using all possible b- and y- ions for the acetylated peptides, including different charge states. This initial version of the method was tested experimentally using samples of heart mitochondria treated with higher concentrations of acetic anhydride. The initial method was edited to eliminate low abundance fragment ions that did not contribute to the overall signal intensities. As a final step, the collision energy used for each fragmentation reaction was also optimized experimentally. The method included a number of unacetylated aconitase peptides that could be used for normalization as described for the native reference peptide method (22,23). The aconitase peptide (R)VDVSPSTQR was selected as the reference peptide based on the absence of lysine in the sequence that may be affected by acetylation to interfere with the tryptic digestion. The SRM method was used to quantify the complete set of acetylated peptides in all of the subsequent experiments. The abundance of each peptide was determined independently and normalized to the reference peptide. Total acetylation of aconitase was taken as the geometric mean of all acetylated peptides and normalized to the reference peptide.
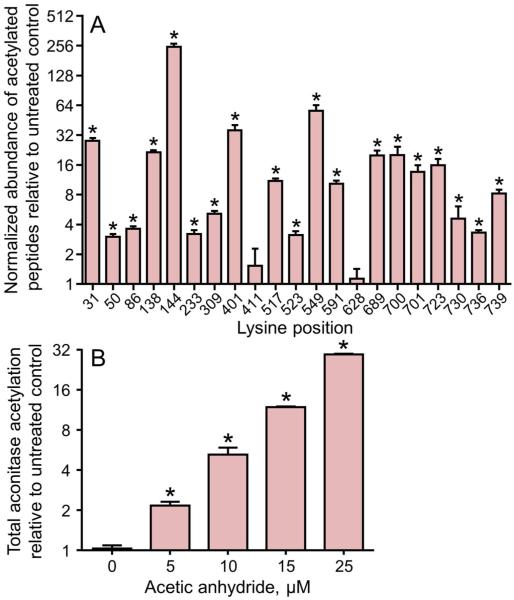
This quantitative SRM method was used to monitor the extent of acetylation on aconitase in the mitochondria treated with the increasing concentrations of acetic anhydride. Figure 2A shows the amount of acetylation for the 10μM treatment at the 21 mapped acetylation sites. For these analyses, the chromatographic peak areas for each acetylated peptide was normalized to the unmodified peptide VDVSPSTQR and shown in the figure relative to untreated controls. Individual lysine residues on aconitase exhibited differential susceptibility to this acetylation reaction. Lysine residues K31, K138, K144, K401, K549, K689, K700, K701, and K723 represent the most responsive sites with increased acetylation greater that 16-fold with 10μM acetic anhydride treatment. The quantitative mass spectrometry data was also used to measure total acetylation on aconitase and found increased total acetylation with increasing concentrations of acetic anhydride (Figure 2B). At 10μM acetic anhydride treatment, a 10-fold increase in total aconitase acetylation was observed which increased to 40-fold at 25μM concentration. The dose dependent increases in total acetylation were also confirmed through Western blot analysis using an anti-acetyl lysine antibody (data not shown). These in vitro findings show that mitochondrial aconitase can be activated through acetylation and demonstrate the effectiveness of the SRM method used to quantify acetylation on aconitase at each of the sites mapped along with the total acetylation.
Figure 2. Differential acetylation by acetic anhydride is seen for the 21 acetylation sites identified in mitochondrial aconitase.
A) The abundance of the individual acetylated peptides measured by selected reaction monitoring. Data are presented for permeabilized mitochondria treated with 10 μM acetic anhydride for 10 min at room temperature. A log2 scale is used for the y-axis where a value of 1 represents no change from control. B) The dose-dependent increase in total aconitase acetylation. These values are determined as the geometric mean of the normalized abundance of the complete group of acetylated peptides at each acetic anhydride concentration. A log2 scale is used for the y-axis where a value of 1 represents no change from control. Each value is the mean ± SEM (n = 3). Statistically significant differences, p<0.05, are designated with an asterisk (*).
In vitro acetylation with acetyl-CoA activates mitochondrial aconitase
Mitochondria are the primary site of carbon utilization with a corresponding generation of acetyl-CoA through metabolism of these energetic substrates. This acetyl-CoA production supports a scenario in which acetylation of mitochondrial proteins occurs non-enzymatically using acetyl-CoA generated by lipid metabolism (10-13,27,35,36). The potential for non-enzymatic acetylation via acetyl-CoA is further enhanced by the unique pH conditions in the mitochondrial matrix the alkaline pH in the range of 7.7-8.2 is suitable for this reaction (27,37,38).
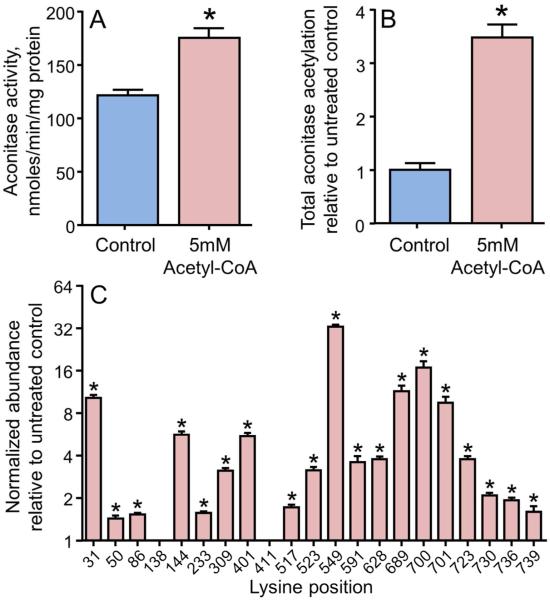
To test the ability of pH conditions in the mitochondrial matrix to activate mitochondrial aconitase via acetyl-CoA-driven acetylation, detergent-solubilized heart mitochondria were incubated at pH 7.8 with 5 mM acetyl-CoA followed by aconitase activity assays and SRM analyses. Mitochondrial aconitase activity increased by 45% with this treatment (Figure 3A). Mass spectrometric analyses found a 4-fold increase in total lysine acetylation of aconitase (Figure 3B), which was confirmed by Western blot using anti acetyl-lysine antibody (data not shown). Similar to the acetic anhydride treatment, increased acetylation was seen with acetyl-CoA for most, but not all, of the mapped lysine residues on mitochondrial aconitase. Further, the extent of modification varied considerably, with increases ranging from no change to a 40% increase in the extent of modification (Figure 3C). These data show that treatment of aconitase with physiologically relevant acetyl-CoA concentrations and an alkaline pH increased lysine acetylation at multiple sites and activated aconitase enzyme activity.
Figure 3. The metabolic intermediate acetyl-CoA acetylates mitochondrial aconitase and increases its activity.
Isolated and solubilized mitochondria were treated with 5 mM acetyl-CoA for 5min at room temperature. (A) Aconitase activity in mitochondria treated with acetyl-CoA compared to untreated controls. (B) Quantification of total acetylation status of aconitase measured by selected reaction monitoring of the acetylated peptides. The total acetylation measurement is determined as the geometric mean of the normalized abundance of all 21 acetylated peptides. Data are presented relative to the average of the untreated controls. (C) The abundance of the individual acetylated peptides measured by selected reaction monitoring. Data are presented relative to untreated controls. A log2 scale is used for the y-axis where a value of 1 represents no change from control. All values are presented as the mean ± SEM, where * indicates a significant increase in treated relative to untreated mitochondria (p < 0.05, n = 6).
Through the combined results of the in vitro acetylation experiments with acetic anhydride and acetyl- CoA, and the quantitative SRM analyses, a series of favored acetylation sites emerged. These sites are K31, K144, K401, K549, K689, K700, K701, and K723. These sites are considered favored based on the rapid dose response increase with the acetic anhydride treatment and the magnitude of the increase with the acetyl-CoA treatment as determined by the SRM analyses.
Aconitase activity and acetylation increases in hearts of mice fed a high fat diet
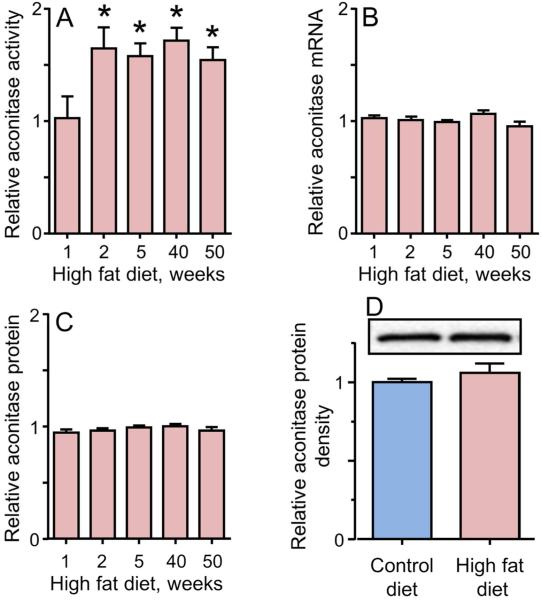
High dietary fat drives increased fatty acid oxidation and increased mitochondrial protein acetylation (9,35-40). The effect of these high fat diets on mitochondrial aconitase, however, has not been described in these reports. We have previously used a mouse model of diet-induced obesity to determine the effects of high fat diet and obesity on mitochondrial function in the heart (41-43). This mouse model was used here to examine the physiologic relevance of the changed aconitase activity and acetylation seen in the in vitro experiments. For these experiments, mice were fed either a high fat or control low fat diet for varying periods of time, heart mitochondria isolated, functional characteristics of aconitase analyzed, and the acetylation status measured. Mitochondrial aconitase activity in the hearts of mice fed a high fat diet increased at 2 weeks and remained 50% to 60% elevated for up to 50 weeks on the diet (Figure 4A). Mitochondrial aconitase message levels were measured by qRT-PCR and protein levels were measured by SRM and Western blot to determine if the increase in activity was due to increased expression. No changes were seen in either mRNA (Figure 4B) or protein (Figure 4C and 4D) at any time tested over the 50 week experiment. These data show that the mitochondrial aconitase is activated in vivo under conditions that have been linked to increased acetyl-CoA production and increase protein acetylation.
Figure 4. Mitochondrial aconitase activity increases in the cardiac tissue of mice fed a high fat diet for extended amounts of time.
Mice were fed a high fat or low fat control diet for up to 50 weeks, as indicated, and hearts were processed for functional analyses of mitochondrial aconitase. (A) Relative activity of aconitase in mitochondria from hearts of mice fed a high fat normalized to the control diet. (B) The corresponding mRNA levels by quantitative RT-PCR; and (C) the corresponding protein content determined by mass spectrometry. (D) Western blot analysis and densitometry for the expression of aconitase in high fat versus control diet for 2 weeks. All data are presented as the mean ± SEM, where * indicates a statistically significant increase in mice fed the high fat versus mice fed the control low fat diet at each time (p < 0.05, n = 5).
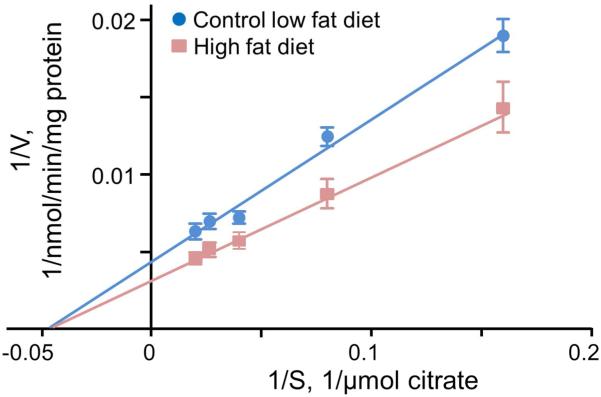
Kinetic analysis of mitochondrial aconitase isolated from mice receiving the different diets was used to examine the nature of the increased aconitase activity (Figure 5). These analyses found a 36% increase in Vmaxwith the high fat diet versus the control diet (from 242 ± 26 in control to 330 ± 35 nmol/min/mg protein with the high fat diet, p< 0.05, n=5) with no change in KM (22 ± 3 and 22 ± 3 μM). These results show that heart mitochondrial aconitase activity is increased by the high fat, without an increase in expression, due to higher catalytic efficiency of the enzyme.
Figure 5. Mitochondrial aconitase isolated from mice on a high fat diet has an increased Vmax.
Lineweaver Burk plot for the kinetic analysis of mitochondrial aconitase from hearts of mice fed a high fat versus control diet. These data were used to determine the Km and Vmax of the enzyme.
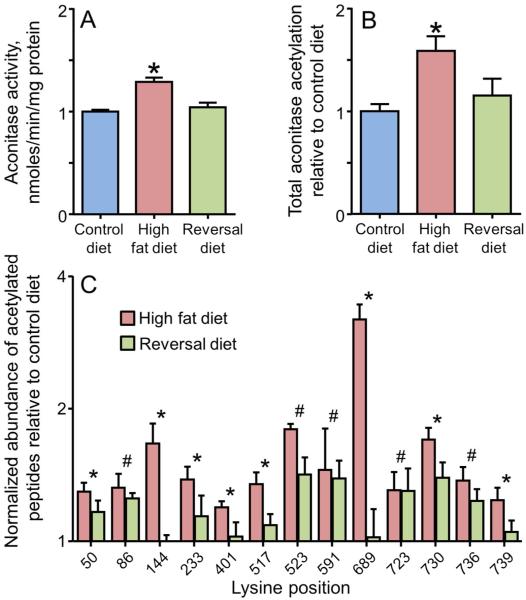
Additional experiments tested the reversibility of the aconitase activation by returning mice fed a high fat diet for 2 weeks to a control low fat diet for 1 week. With this return to the low fat diet, mitochondrial aconitase activity returned to control levels (Figure 6A) with no change in protein expression. Quantitative mass spectrometry using SRM measured a significant increase in total lysine acetylation of aconitase (Figure 6B). Individual lysine residues on aconitase exhibited differential susceptibility to acetylation, with increased acetylation at K144, K401, K523, K689 and K730 (amongst others) in response to 2 weeks on the high fat diet. Returning the animals to a low fat diet reduced the increased acetylation, with several sites returning to control levels (Figure 6C). These data provide direct evidence that heart mitochondrial aconitase is acetylated in response to high dietary fat. Further, aconitase activity returned to control when the animals were taken off the high fat diet with corresponding decreases in the acetylation at many but not all of the sites.
Figure 6. Increased mitochondrial aconitase activity and protein acetylation seen with 2 weeks on a high fat diet are reversed by return to a low fat diet.
Heart mitochondria were isolated from mice fed either a control, low fat diet for 2 weeks, a high fat diet for 2 weeks, or a reversal diet of 2 weeks high fat diet followed by 1 week low fat diet . (A) Aconitase activity in isolated mitochondria. (B) Total aconitase acetylation measured by selected reaction monitoring and determined as the geometric mean of the abundance of the 21 acetylated peptides. All values are presented as the mean ± SEM, where the asterisk (*) indicates a significant increase relative to the animals on the control low fat diet (p < 0.05, n = 5). (C) The abundance of selected acetylated peptides measured by selected reaction monitoring. For each acetylation site, the data are presented relative to the control low fat diet for the high fat diet and reversal diet. All values are presented as the mean ± SEM. A log2 scale is used for the y-axis where a value of 1 represents no change from control. Lysine acetylation sites indicated with an asterisk (*) had statistically significant increases (p<0.05, n = 5) on the high fat diet that were reduced to control values with the reversal diet. Sites indicated with a hash (#) had statistically significant increases (p<0.05) on the high fat diet that remained significantly increased with the reversal diet.
SIRT3 reverses high fat diet-induced acetylation and aconitase activation
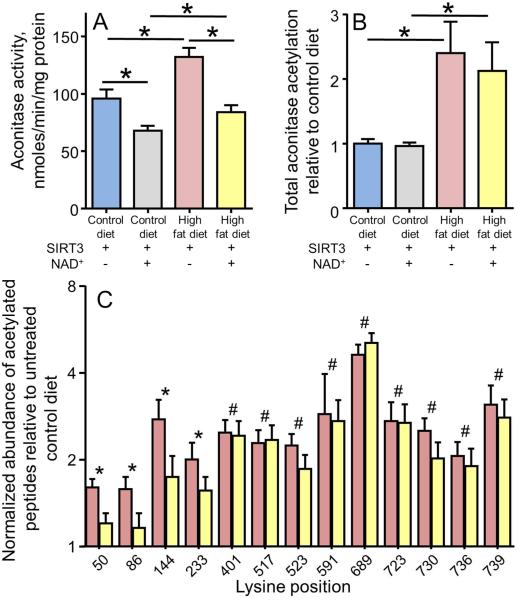
In the mitochondria, SIRT3 is the major deacetylase. SIRT3 knockout has been found to alter acetylation on a large number of mitochondrial proteins (1-6). Mitochondrial aconitase has been identified as a target of SIRT3 through peptide microarray studies (16) and SIRT3 knockout studies (1-6). However, the effect of SIRT3 deacetylation on aconitase activity in response to high dietary fat is not known. Hence, we examined the ability of SIRT3 to deacetylate mitochondrial aconitase from mice fed a high fat diet using recombinant SIRT3. Solubilized heart mitochondria isolated from mice fed high fat or control diet for 2 weeks were incubated with SIRT3 in the presence or absence of the cofactor NAD+. As seen in Figure 7A, the increased mitochondrial aconitase activity observed at 2 weeks of a high fat diet was abolished by treatment with SIRT3 and NAD+. The SIRT3 deacetylation also reduced aconitase activity in control animals consistent with a contribution of acetylation to the enzyme activity under normal diet conditions. While no measurable effect on total acetylation could be seen with the SIRT3 treatment (Figure 7B), treatment with the complete SIRT3 + NAD+ activity did reduce acetylation at a subset of the lysine acetylation sites, with K50, K86, K144, and K233 all showing significant reductions (Figure 7C). These findings confirm that the activation of aconitase seen in vivo with the high dietary fat is due to acetylation of the enzyme and that SIRT3 has the ability to reverse this activation at specific sites.
Figure 7. SIRT3-mediated deacetylation reduces the activity of mitochondrial aconitase.
(A) Mitochondrial aconitase activity in hearts of mice fed a high fat and control diet with treatment with recombinant SIRT3 in the presence or absence of the cofactor NAD+. The four groups shown are control diet, control diet treated with both SIRT3 + NAD+, high fat diet treated with SIRT3 but without NAD+, and high fat diet treated with both SIRT3 + NAD+. (B) Quantification of the total aconitase acetylation in these same four groups. For (A) and (B) the values are presented as the mean ± SEM , where * indicates a significant difference between the indicated groups (p < 0.05, n = 5). (C) The abundance of selected acetylated peptides measured by selected reaction monitoring. For each acetylation site, the data are presented relative to the control low fat diet for the high fat diet mitochondria (red bars) and high fat diet mitochondria treated with SIRT3 + NAD+ (yellow bars). All values are presented as the mean ± SEM. A log2 scale is used for the y-axis where a value of 1 represents no change from control. Lysine acetylation sites indicated with an asterisk (*) had statistically significant increases (p<0.05, n = 5) on the high fat diet that were significantly reduced when the mitochondria were treated with the active SIRT3 system (SIRT3 + NAD+). Sites indicated with a hash (#) had statistically significant increases (p<0.05) on the high fat diet that were not significantly reduced when treated with the active SIRT3 system.
DISCUSSION
It is clear from the multiple high-throughput proteomics analyses that a large number of mitochondrial proteins are acetylated. Functional validation of these results, however, has been a vital issue with only a limited number of reports investigating the effect on function and no reports of acetylation-dependent effects on aconitase activity. Among the few proteins where function has been assessed, acetylation is most often associated with enzyme inhibition (44-49). Isocitrate dehydrogenase and succinate dehydrogenase are examples of Krebs cycle enzyme inhibited by acetylation. Increased enzyme activity with acetylation has been observed for a small number of enzymes (7,27-30). Both the cytosolic and mitochondrial forms of malate dehydrogenase, another Krebs cycle enzyme, are examples of enzymes that are activated by acetylation (7,28). This study expands the link between acetylation status and function by showing an important Krebs cycle enzyme, aconitase, is activated in response to acetylation in vivo by a high fat diet in vivo.
We used a variety of experimental conditions to build an argument for the importance of acetylation at specific sites in aconitase. In addition to those analytical data, one would speculate that if a specific lysine residue was an acetylation target needed to regulate an important enzyme activity, then that lysine would be conserved across a range of species. Sequence alignments found 7 of the 21 lysines we measured as acetylation sites are conserved across diverse animal species ranging from human to fly and including representative primate, rodent, amphibian, reptile, and fish species. These sites are K144, K411, K591, K689, K700, K701, and K736. In this group of conserved lysines, only K144 and K689 responded with acetylation changes that reflected the activity changes in all conditions tested. K144 and K689 were acetylated by treatment with both acetic anhydride and acetyl-CoA. These lysine residues also had increased acetylation in mitochondria from mice fed a high fat diet, with reduced acetylation when the mice returned to the low fat control diet. However, only K144 acetylation was reduced by SIRT3 treatment, consistent with the decreased aconitase activity that the SIRT3 produces. The sequence alignment for K144 is shown in Table 2. It has been proposed that regulatory lysines have perturbed pKa values which increase their susceptibility to acetylation (27). These perturbed pKa values could result from flanking positively charged residues. Since K144 and K689 are in close proximity to this type of flanking arginine residue, the lowered pKa theory might explain the propensity of these lysines to be acetylated. The demonstration that aconitase acetylation and activity are modulated in vivo in a mouse model of diet-induced obesity is a new contribution to the link between protein acetylation, changes in protein function, and changes in metabolism. Kinetic analysis of mitochondrial aconitase from the hearts of mice in that study showed the high fat diet produced a fundamental alteration in enzyme function as seen in the significant increase in Vmax.
Table 2.
The alignment of selected regions of the aconitase sequences to illustrate the conservation of K144 and Q541.
| Animal | K144 | Q541 |
|---|---|---|
|
| ||
| human (Homo sapiens) | GGEKDLRRAKDINQEVYNF | PKGEFDPGQDTYQHPPKDS |
| chimp (Pan troglodytes) | GGEKDLKRAKDINQEVYNF | PKAEFDPGQDTYQHPPKDS |
| mouse (Mus musculus) | GGEKDLRRAKDINQEVYNF | PRSDFDPGQDTYQHPPKDS |
| rat (Rattus norvegicus) | GGEKDLRRAKDINQEVYNF | PRSDFDPGQDTYQHPPKDS |
| dog (Canis lupus familiaris) | GGEKDLRRAKDINQEVYNF | PRAEFDPGQDTYQHPPKDS |
| pig (Sus scrofa) | GGEKDLRRAKDINQEVYNF | PRAEFDPGQDTYQHPPKDS |
| chicken (Gallus gallus) | GGEKDLRRAKDINQEVYSF | PRLDFDPGQDTYQYPPKDG |
| fish (Danio rerio) | GGAQDLQKAKEVNQEVYNF | PARDFDPGQDTYQHPPADG |
| frog (Xenopus laevis) | GGDKDLKRAKDINEEVYNF | PKSSFDPGQDTYQHPPIDG |
| snake (Python bivittatus) | GGEKDLRRAKDINQEVYSF | PKLEFDPGQDTYQYPPKDG |
| beetle (Tribolium castaneum) | GGEKDLARAKDLNQEVYDF | PKGEFDPGQDTYEAPVADG |
| fly (Drosophila melanogaster) | GGPKDLARAKDLNKEVGNL | PAKGFDPGQDTYTAPPPSG |
Regions including the K144 acetylation site and the interacting glutamine residue Q541. The numbering of the amino acids is based on the mouse sequence. Species and NCBI GI accession numbers included are: human, 4501867; chimp, 694983472; mouse, 18079339; rat, 40538860; dog, 73968966; pig, 47522738; chicken, 24637100; fish, 38707983; frog, 147904130; snake, 602637826; beetle, 91088677; and fly, 17864292.
Analysis of the three dimensional structure of aconitase predicts that K144 interacts with Q541 (Figure 8), which is in the hinge region of the protein that allows the swivel domain to change conformations as the enzyme interacts with the substrate. As shown in Table 2, Q541 is also highly conserved. Acetylation of K144 fills this space and narrows the distance from 6.1 Ǻ to 4.1 Ǻ. The resulting perturbation has the potential to alter the flexibility of this region. One would predict based on the increased Vmax seen with acetylation, that the altered flexibility enhances substrate accessibility of the active site to increase the rate of the reaction. Unfortunately, aconitase is not amenable to traditional site-directed mutagenesis experiments that might prove this link because of the difficultly in reconstituting the iron-sulfur center needed to express an active enzyme. Additionally, while these experiments monitored acetylation at a large number of lysine residues, not all lysines were amenable to mass spectrometric detection. This gap leaves the possibility that the exact picture of aconitase acetylation is incomplete.
Figure 8. Modeling of the interaction between K144 and Q541.

A portion of the hinge region of the mouse aconitase structural model based on the bovine aconitase structure (PDB:1AMI) is shown with a semi-transparent van der Waals surface except for the side chain at K144 with either lysine or acetylated-lysine, which are in a stick representations. Q541 is represented in both with semi-transparent van der Waals surface and stick representation. The filling of the gap, presumed to be in the native structure, by the acetyl group on acetylated lysing is represented by the closing of the interatomic distances as shown.
The effects of acetylation on mitochondrial aconitase raise the broader question of what is the significance of acetylation-mediated aconitase activation for metabolism in the heart. Given the central role of aconitase in citrate metabolism, it is intriguing to consider the possibility that acetylation- dependent activation of aconitase may represent a mechanism for sensing nutrient availability and modifying mitochondrial substrate selection. In the cytoplasm, citrate is a regulator of fatty acid oxidation through the allosteric activation of acetyl-CoA carboxylase (ACC) with downstream effects of mitochondrial fatty acid uptake by carnitine pamitoyltransferase (CPT1) (17,50). Thus, the increased citrate utilization driven by the 30% to 60% increase in aconitase activity in the mitochondria could reduce citrate efflux and promote β-oxidation via increased fatty acid uptake. This scenario would represent a means for mitochondria to sense changes in nutritional status and increase the use of fatty acid oxidation for energy production when lipids are available.
In summary, this study used multiple approaches to identify and characterize a novel role of lysine acetylation in regulating aconitase activity in heart mitochondria. The key result seen throughout was an increase in aconitase activity with increased acetylation. We developed a label free quantitative mass spectrometry assay using SRM that selectively quantified the peptides containing the acetylated lysine residues and used this method to determine the dynamics of aconitase acetylation in vitro and in vivo. in vitro acetylation with acetyl-CoA detected a subset of the identified acetylation sites that increased significantly with treatment and were reversed with the SIRT3 treatments. This same quantitative pattern was also seen in aconitase isolated from heart mitochondria of mice that were fed a high fat diet. Importantly, aconitase acetylation and activity returned to control when those mice were returned to a low fat diet, strengthening the link to the high fat diet. Overall, these data directly link acetylation to a change in aconitase function in vivo that increases the activity of the enzyme.
Acknowledgment
We thank Melinda West for her work with the animals used in these experiments.
Funding sources
This work was funded by NIH grants R01AG16339 (LIS), P20GM104934 (KMH), and the Oklahoma Medical Research Foundation. The content is solely the responsibility of the authors and does not represent the official views of the National Institutes of Health
Abbreviations
- BSA
bovine serum albumin
- CID
collision induced dissociation
- SRM
selected reaction monitoring
- SIRT3
silent information regulator Sir2 homologue 3
- SEM
standard error of the mean
REFERENCES
- (1).Still AJ, Floyd BJ, Hebert AS, Bingman CA, Carson JJ, Gunderson DR, Dolan BK, Grimsrud PA, Dittenhafer-Reed KE, Stapleton DS, Keller MP, Westphall MS, Denu JM, Attie AD, Coon JJ, Pagliarini DJ. Quantification of mitochondrial acetylation dynamics highlights prominent sites of metabolic regulation. J. Biol. Chem. 2013;288:26209–26219. doi: 10.1074/jbc.M113.483396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, Grishin NV, White M, Yang XJ, Zhao Y. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- (3).Hebert AS, Dittenhafer-Reed KE, Yu W, Bailey DJ, Selen ES, Boersma MD, Carson JJ, Tonelli M, Balloon AJ, Higbee AJ, Westphall MS, Pagliarini DJ, Prolla TA, Assadi-Porter F, Roy S, Denu JM, Coon JJ. Calorie restriction and SIRT3 trigger global reprogramming of the mitochondrial protein acetylome. Mol. Cell. 2013;49:186–199. doi: 10.1016/j.molcel.2012.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Rardin MJ, Newman JC, Held JM, Cusack MP, Sorensen DJ, Li B, Schilling B, Mooney SD, Kahn CR, Verdin E, Gibson BW. Label-free quantitative proteomics of the lysine acetylome in mitochondria identifies substrates of SIRT3 in metabolic pathways. Proc. Natl. Acad. Sci. USA. 2013;110:6601–6606. doi: 10.1073/pnas.1302961110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Sol EM, Wagner SA, Weinert BT, Kumar A, Kim HS, Deng CX, Choudhary C. Proteomic investigations of lysine acetylation identify diverse substrates of mitochondrial deacetylase sirt3. PLoS One. 2012;7:e50545. doi: 10.1371/journal.pone.0050545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- (7).Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, Li Y, Shi J, An W, Hancock SM, He F, Qin L, Chin J, Yang P, Chen X, Lei Q, Xiong Y, Guan KL. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Scott I, Webster BR, Li JH, Sack MN. Identification of a molecular component of the mitochondrial acetyltransferase programme: a novel role for GCN5L1. Biochem J. 2012;443:655–661. doi: 10.1042/BJ20120118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Alrob OA, Sankaralingam S, Ma C, Wagg CS, Fillmore N, Jaswal JS, Sack MN, Lehner R, Gupta MP, Michelakis ED, Padwal RS, Johnstone DE, Sharma AM, Lopaschuk GD. Obesity-induced lysine acetylation increases cardiac fatty acid oxidation and impairs insulin signalling. Cardiovasc Res. 2014;103:485–497. doi: 10.1093/cvr/cvu156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Paik WK, Pearson D, Lee HW, Kim S. Nonenzymatic acetylation of histones with acetyl-CoA. Biochim Biophys Acta. 1970;213:513–522. doi: 10.1016/0005-2787(70)90058-4. [DOI] [PubMed] [Google Scholar]
- (11).Baeza J, Smallegan MJ, Denu JM. Site-specific reactivity of nonenzymatic lysine acetylation. ACS Chem Biol. 2015;10:122–128. doi: 10.1021/cb500848p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Schwer B, North BJ, Frye RA, Ott M, Verdin E. The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase. J. Cell Biol. 2002;158:647–657. doi: 10.1083/jcb.200205057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Wagner GR, Hirschey MD. Nonenzymatic protein acylation as a carbon stress regulated by sirtuin deacylases. Mol. Cell. 2014;54:5–16. doi: 10.1016/j.molcel.2014.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Choudhary C, Weinert BT, Nishida Y, Verdin E, Mann M. The growing landscape of lysine acetylation links metabolism and cell signalling. Nat. Rev. Mol. Cell Biol. 2014;15:536–550. doi: 10.1038/nrm3841. [DOI] [PubMed] [Google Scholar]
- (15).Newman JC, He W, Verdin E. Mitochondrial protein acylation and intermediary metabolism: regulation by sirtuins and implications for metabolic disease. J. Biol. Chem. 2012;287:42436–42443. doi: 10.1074/jbc.R112.404863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Rauh D, Fischer F, Gertz M, Lakshminarasimhan M, Bergbrede T, Aladini F, Kambach C, Becker CFW, Zerweck J, Schutkowski M, Steegborn C. An acetylome peptide microarray reveals specificities and deacetylation substrates for all human sirtuin isoforms. Nat. Commun. 2013;4:2327. doi: 10.1038/ncomms3327. [DOI] [PubMed] [Google Scholar]
- (17).Poirier M, Vincent G, Reszkom AE, Bouchard B, Kelleher JK, Brunengraber H, Des Rosiers C. Probing the link between citrate and malonyl-CoA in perfused rat hearts. Am. J. Physiol. Heart Circ. Physiol. 2002;283:H1379–H1386. doi: 10.1152/ajpheart.00244.2002. [DOI] [PubMed] [Google Scholar]
- (18).McGarry JD, Leatherman GF, Foster DW. Carnitine palmitoyltransferase I. The site of inhibition of hepatic fatty acid oxidation by malonyl-CoA. J. Biol. Chem. 1978;253:4128–4136. [PubMed] [Google Scholar]
- (19).Koley D, Bard AJ. Triton X-100 concentration effects on membrane permeability of a single HeLa cell by scanning electrochemical microscopy (SECM) Proc. Natl. Acad. Sci. U.S.A. 2010;107:16783–16787. doi: 10.1073/pnas.1011614107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Kinter M, Kinter CS. Application of Selected Reaction Monitoring to Highly Multiplexed Targeted Quantitative Proteomics. Springer Briefs in Systems Biology; NY: 2013. [Google Scholar]
- (21).Kinter CS, Lundie JM, Patel H, Rindler PM, Szweda LI, Kinter M. A quantitative proteomic profile of the Nrf2-mediated antioxidant response of macrophages to oxidized LDL determined by multiplexed selected reaction monitoring. PLoS One. 2012;7:e50016. doi: 10.1371/journal.pone.0050016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Ruse CI, Willard B, Jin JP, Haas T, Kinter M, Bond M. Quantitative dynamics of site-specific protein phosphorylation determined using liquid chromatography electrospray ionization mass spectrometry. Anal. Chem. 2002;74:1658–1664. doi: 10.1021/ac0157122. [DOI] [PubMed] [Google Scholar]
- (23).Willard BB, Ruse CI, Keightley JA, Bond M, Kinter M. Site-specific quantitation of protein nitration using liquid chromatography/tandem mass spectrometry. Anal. Chem. 2003;75:2370–2376. doi: 10.1021/ac034033j. [DOI] [PubMed] [Google Scholar]
- (24).Nulton-Persson AC, Szweda LI. Modulation of mitochondrial function by hydrogen peroxide. J. Biol. Chem. 2001;276:23357–23361. doi: 10.1074/jbc.M100320200. [DOI] [PubMed] [Google Scholar]
- (25).Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Ghanta S, Grossmann RE, Brenner C. Mitochondrial protein acetylation as a cell-intrinsic, evolutionary driver of fat storage: Chemical and metabolic logic of acetyl-lysine modifications. Crit. Rev. Biochem. Mol. Biol. 2013;48:561–574. doi: 10.3109/10409238.2013.838204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Kim EY, Kim WK, Kang HJ, Kim JH, Chung SJ, Seo YS, Park SG, Lee SC, Bae KH. Acetylation of malate dehydrogenase 1 promotes adipogenic differentiation via activating its enzymatic activity. J. Lipid Res. 2012;53:1864–1876. doi: 10.1194/jlr.M026567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Xue L, Xu F, Meng L, Wei S, Wang J, Hao P, Bian Y, Zhang Y, Chen Y. Acetylation-dependent regulation of mitochondrial ALDH2 activation by SIRT3 mediates acute ethanol-induced eNOS activation. FEBS Lett. 2012;586:137–142. doi: 10.1016/j.febslet.2011.11.031. [DOI] [PubMed] [Google Scholar]
- (30).Li T, Liu M, Feng X, Wang Z, Das I, Xu Y, Zhou X, Sun Y, Guan KL, Xiong Y, Lei QY. Glyceraldehyde-3-phosphate dehydrogenase is activated by lysine 254 acetylation in response to glucose signal. J. Biol. Chem. 2014;289:3775–3785. doi: 10.1074/jbc.M113.531640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Bharathi SS, Zhang Y, Mohsen AW, Uppala R, Balasubramani M, Schreiber E, Uechi G, Beck ME, Rardin MJ, Vockley J, Verdin E, Gibson BW, Hirschey MD, Goetzman ES. Sirtuin 3 (SIRT3) protein regulates long-chain acyl-CoA dehydrogenase by deacetylating conserved lysines near the active site. J. Biol. Chem. 2013;288:33837–33847. doi: 10.1074/jbc.M113.510354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Guan KL, Xiong Y. Regulation of intermediary metabolism by protein acetylation. Trends Biochem. Sci. 2011;36:108–116. doi: 10.1016/j.tibs.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Tao R, Coleman MC, Pennington JD, Ozden O, Park SH, Jiang H, Kim HS, Flynn CR, Hill S, Hayes McDonald W, Olivier AK, Spitz DR, Gius D. Sirt3-mediated deacetylation of evolutionarily conserved lysine 122 regulates MnSOD activity in response to stress. Mol. Cell. 2010;40:893–904. doi: 10.1016/j.molcel.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Vadvalkar SS, Baily CN, Matsuzaki S, West M, Tesiram YA, Humphries KM. Metabolic inflexibility and protein lysine acetylation in heart mitochondria of a chronic model of type 1 diabetes. Biochem J. 2013;449:253–261. doi: 10.1042/BJ20121038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Garland PB, Shepherd D, Yates DW. Steady-state concentrations of coenzyme A, acetyl-coenzyme A and long-chain fatty acyl-coenzyme A in rat-liver mitochondria oxidizing palmitate. Biochem. J. 1965;97:587–594. doi: 10.1042/bj0970587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Pougovkina O, Te Brinke H, Ofman R, van Cruchten AG, Kulik W, Wanders RJ, Houten SM, de Boer VC. Mitochondrial protein acetylation is driven by acetyl-CoA from fatty acid oxidation. Hum. Mol. Genet. 2014;23:3513–3522. doi: 10.1093/hmg/ddu059. [DOI] [PubMed] [Google Scholar]
- (37).Wagner GR, Payne RM. Widespread and enzyme-independent Nε-acetylation and Nε-succinylation of proteins in the chemical conditions of the mitochondrial matrix. J. Biol. Chem. 2013;288:29036–29045. doi: 10.1074/jbc.M113.486753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Llopis J, McCaffery JM, Miyawaki A, Farquhar MG, Tsien RY. Measurement of cytosolic, mitochondrial, and Golgi pH in single living cells with green fluorescent proteins. Proc. Natl. Acad. Sci. USA. 1998;95:6803–6808. doi: 10.1073/pnas.95.12.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Cole MA, Murray AJ, Cochlin LE, Heather LC, McAleese S, Knight NS, Sutton E, Jamil AA, Parassol N, Clarke K. A high fat diet increases mitochondrial fatty acid oxidation and uncoupling to decrease efficiency in rat heart. Basic Res. Cardiol. 2011;106:447–457. doi: 10.1007/s00395-011-0156-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Hirschey MD, Shimazu T, Jing E, Grueter CA, Collins AM, Aouizerat B, Stancakova A, Goetzman E, Lam MM, Schwer B, Stevens RD, Muehlbauer MJ, Kakar S, Bass NM, Kuusisto J, Laakso M, Alt FW, Newgard CB, Farese RV, Jr., Kahn CR, Verdin E. SIRT3 deficiency and mitochondrial protein hyperacetylation accelerate the development of the metabolic syndrome. Mol. Cell. 2011;44:177–190. doi: 10.1016/j.molcel.2011.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Crewe C, Kinter M, Szweda LI. Rapid inhibition of pyruvate dehydrogenase: an initiating event in high dietary fat-induced loss of metabolic flexibility in the heart. PLoS One. 2013;8:e77280. doi: 10.1371/journal.pone.0077280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Rindler PM, Plafker SM, Szweda LI, Kinter M. High dietary fat selectively increases catalase expression within cardiac mitochondria. J. Biol. Chem. 2013;288:1979–1990. doi: 10.1074/jbc.M112.412890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Rindler PM, Crewe CL, Fernandes J, Kinter M, Szweda LI. Redox regulation of insulin sensitivity due to enhanced fatty acid utilization in the mitochondria. Am. J. Physiol. Heart Circ. Physiol. 2013;305:H634–H643. doi: 10.1152/ajpheart.00799.2012. [DOI] [PubMed] [Google Scholar]
- (44).Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB, Grueter CA, Harris C, Biddinger S, Ilkayeva OR, Stevens RD, Li Y, Saha AK, Ruderman NB, Bain JR, Newgard CB, Farese RV, Jr., Alt FW, Kahn CR, Verdin E. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464:121–125. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Hallows WC, Lee S, Denu JM. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc. Natl. Acad. Sci. USA. 2006;103:10230–10235. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Schwer B, Bunkenborg J, Verdin RO, Andersen JS, Verdin E. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc. Natl. Acad. Sci. USA. 2006;103:10224–10229. doi: 10.1073/pnas.0603968103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Nakagawa T, Lomb DJ, Haigis MC, Guarente L. SIRT5 Deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell. 2009;137:560–570. doi: 10.1016/j.cell.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Hallows WC, Yu W, Denu JM. Regulation of glycolytic enzyme phosphoglycerate mutase-1 by Sirt1 protein-mediated deacetylation. J. Biol. Chem. 2012;287:3850–3858. doi: 10.1074/jbc.M111.317404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Shimazu T, Hirschey MD, Hua L, Dittenhafer-Reed KE, Schwer B, Lombard DB, Li Y, Bunkenborg J, Alt FW, Denu JM, Jacobson MP, Verdin E. SIRT3 deacetylates mitochondrial 3-hydroxy-3-methylglutaryl CoA synthase 2 and regulates ketone body production. Cell. Metab. 2010;12:654–661. doi: 10.1016/j.cmet.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Vincent G, Comte B, Poirier M, Rosiers CD. Citrate release by perfused rat hearts: a window on mitochondrial cataplerosis. Am. J. Physiol. Endocrinol. Metab. 2000;278:E846–856. doi: 10.1152/ajpendo.2000.278.5.E846. [DOI] [PubMed] [Google Scholar]