Abstract
Arrestins are a small protein family with only four members in mammals. Arrestins demonstrate an amazing versatility, interacting with hundreds of different G protein-coupled receptor (GPCR) subtypes, numerous nonreceptor signaling proteins, and components of the internalization machinery, as well as cytoskeletal elements, including regular microtubules and centrosomes. Here, we focus on the structural determinants that mediate various arrestin functions. The receptor-binding elements in arrestins were mapped fairly comprehensively, which set the stage for the construction of mutants targeting particular GPCRs. The elements engaged by other binding partners are only now being elucidated and in most cases we have more questions than answers. Interestingly, even very limited and imprecise identification of structural requirements for the interaction with very few other proteins has enabled the development of signaling-biased arrestin mutants. More comprehensive understanding of the structural underpinning of different arrestin functions will pave the way for the construction of arrestins that can link the receptor we want to the signaling pathway of our choosing.
1. INTRODUCTION
As far as size is concerned, arrestins are quite average, 44–48 kDa soluble proteins. Functionally, however, arrestins are far from being average in many ways, demonstrating that evolution can pack incredible versatility into ~ 400 amino acids.
The discovery of the first member of the arrestin family (modern systematic name arrestin-1a) was rather unremarkable, except that it was discovered twice: first as S-antigen, the target of auto-antibodies in uveitis,1 then as a 48-kDa protein that binds light-activated rhodopsin,2 preferring the phosphorylated form.3 Eventually it became clear that both are the same protein, which prevents G protein activation by light-activated phosphorylated rhodopsin,4 thereby blocking (arresting) further signaling.
All this happened before the seminal discovery of striking similarity in sequence and topology between the β2-adrenergic receptor (β2AR) and rhodopsin,5 which led to the concept that there is a large family of G-protein-coupled receptors (GPCRs; also known as seven transmembrane domain receptors or 7TMRs) and fruitful ideas regarding the similarity of signaling and regulatory mechanisms in this family. The first nonvisual arrestin, termed β-arrestin because of its preference for the β2AR over rhodopsin, was cloned soon thereafter,6 followed by another nonvisual subtype (termed β-arrestin2,7 arrestin-3,8 and hTHY-ARRX,9 respectively) and cone-specific arrestin.10,11 Considering that different vertebrate species express from 800 to >3400 distinct GPCRs (SEVENS database; http://sevens.cbrc.jp/), the fact that we only have four arrestin subtypes12 is rather remarkable. Moreover, arrestin-1 and -4 are largely restricted to photoreceptors,13 whereas the two nonvisual subtypes are ubiquitously expressed and interact with hundreds of different GPCRs.12,14 Even this striking versatility is only half of the story—in addition to receptors, arrestins bind dozens,12 and possibly hundreds,15 of amazingly diverse proteins, serving as multifunctional signaling organizers in the cell (see Chapter 1).
2. WHAT THE CRYSTAL STRUCTURE REVEALS, AND WHAT IT DOES NOT
Visual arrestin-1 was the first subtype discovered,2 functionally characterized,4 cloned,16 and crystallized.17,18 The structure revealed a unique fold: an elongated molecule consisting of two cup-like domains with similar cores, each organized as a seven-strand β-sandwich (Fig. 3.1). Subsequently solved structures of arrestin-2,19,21 arrestin-3,20 and arrestin-422 and even the short splice variant of arrestin-123 showed rather disappointing similarity, offering surprisingly few clues regarding the structural underpinnings of the functional diversity of these proteins.
Figure 3.1.
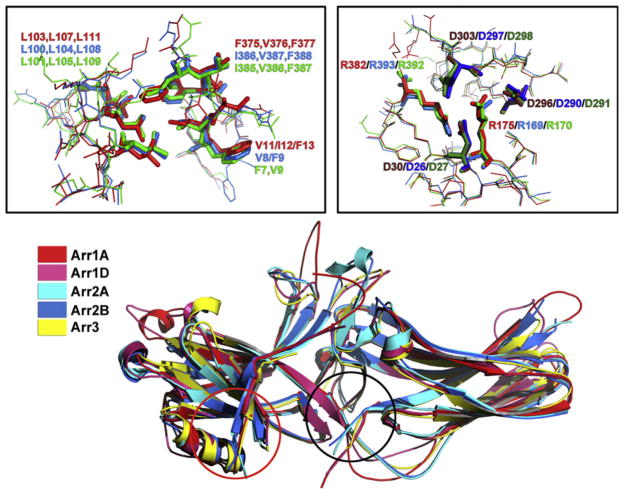
Basal conformation of different arrestin subtypes. Superimposition of the crystal structures of the two monomers in the arrestin-1 tetramer (Arr1A, red and Arr1D, pink),18 two monomers of the arrestin-2 dimer (Arr2A, light blue and Arr2B, dark blue),19 and arrestin-3 (Arr3, yellow)20 shows remarkably similar cores of both domains and variable structure of the loops and inter-domain hinge. Importantly, the variability of these elements in different monomers of the same arrestin (compare Arr1A and Arr1D, as well as Arr2A and Arr2B) is essentially as great as between arrestin subtypes, suggesting that it reflects the flexibility of these loops, rather than their subtype-specific conformations. Black and red circles show the location of the two key intramolecular interactions that hold arrestins in their basal state, the polar core in the inter-domain interface, and the three-element interaction between β-strand I, α-helix I, and β-strand XX in the C-tail, respectively. The panels above show detailed structure of these elements revealing their extremely high conservation, down to the orientation of the side chains. Right: The polar core, main phosphate sensor. Left: The three-element interaction. In both panels, residue numbers of bovine proteins are indicated as follows: arrestin-1, red; arrestin-2, blue; arrestin-3, green.
Several features revealed by X-ray crystal structure matched the predictions of previous mutagenesis studies surprisingly well. The N-terminal half of the molecule that was predicted to be a separable independently folding unit24,25 turned out to be the N-domain17,18 (Fig. 3.1). Predicted interaction between the N- and C-termini26 was also revealed.18 A molecule consisting of two domains with relatively few contacts between them appeared poised for a global conformational rearrangement predicted by the model of sequential multisite mechanism of receptor binding.25 Finally, Arg175, predicted to interact with a negatively charged partner in arrestin and function as the phosphate sensor,27,28 was found in an unusual (for a soluble protein) arrangement of solvent-excluded charged residues in the interface between the two domains, which was termed the “polar core”.18,29
However, the structures solved thus far reveal only the basal conformation of all arrestin subtypes. Several lines of evidence suggest that the structure of “active” receptor-bound arrestin is likely to be quite different (reviewed in Refs. 30,31) and indicate that arrestins can assume yet another distinct conformation favorable for the binding to microtubules.32–35 Both still remain to be elucidated.
Crystal structure of rhodopsin36 and its subsequent refinements,37–39 followed by a flurry of remarkably similar structures of nonvisual GPCRs,40–54 raised another interesting question: how does arrestin, with the long axis of ~ 75 Å, fit GPCRs with a diameter of ~ 40 Å?
3. HOW DO ARRESTINS FIT RECEPTORS?
There is an obvious caveat in fitting known arrestin and receptor structures: for the complex to form, both arrestin and receptor must be in an active conformation, and the receptor also must be phosphorylated.14,30 While the effect of receptor-attached phosphates on its conformational state is completely unknown, activation-induced changes were well characterized, first by a series of site-directed spin-labeling studies of rhodopsin55–59 then by the solution of several crystal structures of active forms of rhodopsin,60–63 β2AR,42 adenosine A2A receptor,49,64 agonist-activated β2AR stabilized by nanobody,65 and β2AR in a physiologically relevant complex with the G protein.66,67 The common theme in GPCR activation is the outward movement of helices 5 and 6, and less dramatic rearrangements of other elements, which collectively open the cavity in the middle of the cytoplasmic side of the receptor, where the C-terminus of the α-subunit of their cognate G protein binds.68–70 These movements increase the diameter of the receptor, making it larger than the short axis of arrestin, although still much smaller than its long axis (Fig. 3.2). To even begin meaningful fitting of an arrestin to a receptor, one needs to identify interacting residues in both proteins. While the arrestin-binding parts of the receptor have been identified imprecisely and only in a few GPCRs,74–76 the receptor “footprint” on different arrestins was mapped fairly comprehensively by several groups using a variety of methods (Fig. 3.2).
Figure 3.2.
Comparison of the cytoplasmic side of active receptor and arrestin. Putative complex assembled from crystal structure of active β2-adrenergic receptor (from the complex with Gs heterotrimer44) and arrestin-2.19 Phosphate-binding residues and other elements that likely come into direct contact with receptor are shown in red and blue, respectively. Darker blue shows residues in positions where the mobility of the engineered spin label is dramatically decreased upon receptor binding, whereas lighter blue denotes smaller decreases in spin label mobility (based on Ref. 71). Residue numbers correspond to bovine arrestin-2 used in.71 The comparison of these structures suggests that the receptor-binding surface of inactive arrestin-2 is greater than the cytoplasmic part of the receptor resolved in crystal. The receptor C-terminus (not resolved in any crystal structure) with attached phosphates (yellow spheres) was added manually to position the phosphates near known phosphate-binding positive charges in arrestin. The analysis of receptor-binding-induced conformational changes in arrestin-172 revealed very small shifts in relative positions of the two arrestin domains, moderate movement of the “finger loop” toward the receptor, large movement of the neighboring “139 loop” toward the N-domain and to the side (out of the way of incoming receptor), as well as the movement of two loops at the distal tips of the N- and C-domain toward the receptor. Collectively, these rearrangements would allow the finger loop to insert itself deep into the cavity between receptor helices that opens upon activation, and move the tips of the arrestin domains closer to the receptor. However, all contacts expected based on EPR studies of binding-induced changes of spin label mobility in arrestin-173 and -271 can only be readily achieved if the receptor helices move even further apart than they do in complex with Gs.44
3.1. Receptor-binding surface of arrestins
Receptor-binding residues in arrestins were identified using mutagenesis,22,25–28,71,73,77 H/D exchange,78 element swapping,79,80 peptide competition,81 epitope insertion,82 solution NMR,83–85 and site-directed spin-labeling EPR.73,86 All these experiments yielded essentially the same answer: an extensive surface on the concave sides of both arrestin domains is involved (reviewed in Ref. 14) (Fig. 3.2). The distance between the extreme positions of arrestin residues implicated in receptor binding appears to be larger than even the expanded active receptor, at least the parts visible in crystal structures. However, we should not forget that spatial localization of the phosphates necessary for high-affinity arrestin binding in vitro,25,80,87,88 in cells,89 and in living animals90,91 remains completely unknown. This is true for rhodopsin and the β2AR, where phosphorylation sites are localized in the C-terminus,92–100 which was not resolved in any of the structures. This is equally true for M2 muscarinic cholinergic receptor, where phosphorylation sites are localized on the large third cytoplasmic loop,101–103 which was deleted to obtain well-diffracting crystals.45 Thus, we do not really know the size of the arrestin-binding surface of any GPCR. We also do not know the structure of the active receptor-bound arrestin so that the notion that the receptor and arrestin do not fit will be purely theoretical until the first structure of the arrestin–receptor complex is solved. Nonetheless, the idea that arrestins and the cytoplasmic tips of GPCRs do not fit has certain merits and remains quite popular. So far two elegant models have been proposed as possible solutions for this problem.
3.2. The stoichiometry of the complex
One model is based on the high Arrhenius activation energy of the arrestin–rhodopsin interaction104 and subsequent finding that elements of arrestin-1 that do not interact either with light-activated (Rh*) or inactive phosphorylated rhodopsin (P-Rh) become engaged by the active phosphorylated form (P-Rh*).25,27 Collectively, these data suggested that a substantial conformational change in arrestin, which brings additional parts into contact with the receptor, is necessary for high-affinity binding to P-Rh*.25 The evidence that shortening of the inter-domain hinge impedes the binding of all arrestins to their cognate receptors,35,105 suggested that this conformational change could be a clam-like movement of the two arrestin domains.30 This type of movement could solve the problem of the misfit between the large receptor-binding surface of arrestin and the expected size of the arrestin-binding cytoplasmic side of GPCRs. Although this idea could still be considered plausible simply because it was not unambiguously refuted, there is no direct evidence for the large movement of the two domains relative to each other. Existing evidence suggests that while domain movement actually accompanies receptor binding, it is rather small,72,106 certainly not big enough to significantly reduce the receptor-binding “face” of arrestin. However, detected movement of the loops on both distal tips of arrestin-1 toward the center of the receptor-binding side of the molecule likely contributes to the reduction of the surface of arrestin-1 that binds rhodopsin,72 improving the fit in a different way (Fig. 3.2).
An alternative model attacks the problem even more radically, proposing that a single arrestin binds two GPCRs in a dimer.107 Indeed, one can simultaneously fit two cytoplasmic tips of smaller GPCRs, such as rhodopsin, into the cavities of the two arrestin domains.107 Unfortunately, to achieve this, one needs to disregard the receptor C-terminus, which is not visible in crystals, even though it is certainly involved in arrestin binding,26,28,90,108 and ignore the fact that inactive rhodopsin does not bind arrestin with high affinity.25,80,109 However, this model was proposed without mentioning these obvious caveats and has gained certain popularity, despite complete lack of supporting evidence. Unfortunately, it also did not survive experimental testing. The model predicts that the saturation point would be achieved upon binding of one arrestin molecule to two molecules of rhodopsin. This prediction was tested in mice in vivo.110 In rod photoreceptors in the dark, arrestin-1 is largely localized away from the outer segment (OS), where all rhodopsin resides. In contrast, in bright light, the bulk of arrestin-1 moves to this compartment,32,111–114 where it remains due to high-affinity binding to rhodopsin.32 Considering that the expression ratio of arrestin-1 to rhodopsin in mouse rods is ~ 0.8:1,110,115–117 if this model were true, one would never expect virtually quantitative translocation of arrestin-1 to the OS, which was reproducibly observed by many labs.32,111–114 Genetic manipulation of arrestin-1 and rhodopsin expression levels in mouse rods revealed that the amount of arrestin-1 that can move to the OS in the light is, indeed, limited by the amount of rhodopsin there, but that saturation is achieved at the ratio of translocated arrestin-1 to rhodopsin that is greater than 0.8:1, which is consistent only with 1:1 binding model.110 Obviously, in the photoreceptors of live mice, one cannot exclude the role of other proteins in arrestin-1 translocation. However, a variety of arrestin-1:rhodopsin ratios were tested in vitro using two carefully quantified pure proteins, which again yielded saturation at ~ 1:1.110 Since arrestin-1 readily self-associates, forming dimers and tetramers,118–121 this binding ratio could have been explained by an interaction of an arrestin-1 dimer with a rhodopsin dimer. However, it was shown that only monomeric arrestin-1 binds rhodopsin120 because its receptor-binding surface is shielded by sister subunits in the arrestin-1 tetramer and both possible dimers.122 Finally, monomeric rhodopsin was reconstituted into nanodiscs (HDL particles containing membrane-like lipid bilayer) and shown to bind arestin-1 not just efficiently,123 but with physiologically relevant high affinity (KD ~ 3–4 nM) and 1:1 stoichiometry.124
Thus, while the problem exists, neither model proposed so far has withstood the rigors of experimental testing. It appears very likely that in real life a single molecule of activated phosphorylated receptor fits arrestin in its active conformation well enough, but we do not know how exactly this fit is achieved. Two new ideas, which are not mutually exclusive, are suggested by the available evidence. On the arrestin side, unexpectedly large movement of the “139 loop” in the central crest, apparently out of the way of the incoming receptor,72 likely allows the “finger loop” to insert itself fairly deeply into the cavity opening in the middle of the active receptor69 (Fig. 3.2). This would result in extensive contacts between the cytoplasmic loops and extended helices of the receptor and the cavities of both arrestin domains. In addition, the loops at the tips of both arrestin-1 domains move toward the receptor,72 possibly far enough to achieve direct contact with it, as suggested by reduced mobility of several residues in these loops in the complex.71,73 As far as the receptor is concerned, the comparison of the structures of the same active β2AR in complex with an agonist,42 nanobody mimicking G protein,65 and cognate heterotrimeric G protein66 shows that, with activation, the receptor helices on the cytoplasmic side progressively move further apart. Thus, it is entirely possible that in complex with arrestin the helices move out even more, thereby increasing the size of the cytoplasmic tip of the receptor to better accommodate arrestin. Naturally, these are no more than plausible speculations and should be viewed as such. Ultimately, the issue of the arrestin–receptor fit can only be definitively resolved by the elucidation of the structure of the complex.
3.3. Phosphate-binding residues and the phosphate sensor
The fact that arrestins preferentially bind phosphorylated forms of their cognate receptors was established early on, but the first model explaining how arrestin “selects” active phosphorylated receptors from among at least four coexisting forms (active and inactive, both of which can be unphosphorylated or phosphorylated) was proposed in 1993.25 Specific binding to inactive phosphorylated (P-Rh) and light-activated unphosphorylated rhodopsin (Rh*) showed that arrestin-1 has interaction sites that recognize rhodopsin-attached phosphates and the active state of rhodopsin independently of each other.25 However, the binding to light-activated phosphorhodopsin (P-Rh*) was many times greater than either to P-Rh or Rh*,24,25 which cannot be explained by a simple cooperative two-site interaction. This led to the idea that primary binding sites engaged by Rh* and inactive P-Rh also serve as sensors. Only P-Rh* can engage both at the same time, suggesting that arrestin-1 acts as a molecular coincidence detector, where simultaneous activation of these two sensors triggers a global conformational change, which brings additional arrestin-1 elements in contact with rhodopsin, greatly increasing the energy of the interaction and therefore observed binding (reviewed in Ref. 30). This model predicts that among phosphate-binding residues in arrestin-1 and other family members, there must be at least one that not only contributes to the interaction but also serves as a sensor. The elimination of positive charges that simply bind phosphates was expected to reduce arrestin binding to P-Rh*, and even to a greater extent to inactive P-Rh, where the phosphates must be driving the interaction. In contrast, the neutralization or reversal of the charge of the putative phosphate sensor was expected to turn it on, enhancing the binding to unphosphorylated Rh*.
Since high-affinity arrestin binding was shown to require multiple phosphates on rhodopsin,25 one would expect the multiphosphorylated and therefore highly negatively charged rhodopsin C-terminus to interact with several closely spaced positively charged residues in arrestin. The first scanning mutagenesis performed targeted a 14-amino acid stretch of the primary sequence carrying six positive charges (residues 163–176).27 Considering that it was done long before crystal structure became available, this attempt proved amazingly well targeted: five out of six mutations reduced P-Rh and P-Rh* binding. The sixth, Arg175Asn, showed even more interesting phenotype: somewhat enhanced binding to P-Rh* and a dramatic ~ fivefold increase in Rh* binding.27 This is consistent with the idea that Arg175 serves as a phosphate sensor, which was artificially activated by charge neutralization, “fooling” arrestin-1 into perceiving any active form of rhodopsin as phosphorylated. However, the data with full-length protein did not prove one key point, that Arg175 actually interacts with phosphates, because the binding of Arg175Asn mutant to inactive P-Rh and P-Rh* was also increased.27 Luckily, the N-terminal half of arrestin-1, residues 1–191 containing Arg175, was previously shown to be functional and act as an unsophisticated version of arrestin; it interacts P-Rh and Rh*, and its binding to P-Rh* is essentially the sum of the two.24,25 In the context of this mini-arrestin, the Arg175Asn mutation reduced the binding to P-Rh and P-Rh* just like the other mutations, proving that Arg175 actually engages rhodopsin-attached phosphates.27 Replacement of Arg175 with all 19 alternative residues showed that positive charge is the key: Arg175Lys retained high selectivity for P-Rh*, whereas all other substitutions showed enhanced binding to Rh*, with the charge reversal mutation Arg175Glu having the most potent effect.28 The simplest interpretation of these data was that Arg175 interacts with a negatively charged partner within arrestin, and the breaking of this salt bridge by negatively charged phosphates turns the sensor on, allowing arrestin transition into high-affinity receptor-binding state.27,28 Later, the crystal structure revealed that Arg175 is part of solvent-excluded arrangement of five charged residues between the two arrestin domains (Fig. 3.1), identifying three possible negatively charged partners.17,18 Further mutagenesis proved that the salt bridge between Arg175 and Asp296 is the key phosphate sensor: charge reversal mutations of either greatly increase Rh* binding, whereas simultaneous reversal of both, restoring the salt bridge, also restores normal arrestin-1 selectivity for P-Rh*.29 Interestingly, these five charges are conserved in arrestin evolution from C. elegans to mammals,12 attesting to their important role in arrestin function. Virtually identical positions of all five side chains in different subtypes of mammalian arrestins (Fig. 3.1) support this notion. Charge reversals of homologous arginines in arrestin-2 and -3 also yield enhanced phosphorylation-independent mutants,125–130 demonstrating that this residue plays the same role in all arrestins.
Further mutagenesis identified additional phosphate-binding elements in arrestins. Arg18 in the loop between β-strands I and II is unique for arrestin-1,22 likely making is much more dependent on receptor-attached phosphates than the other subtypes.89 In contrast, two lysines in β-strand I are present in all arrestins.12 These charged residues appear to be critical for the “delivery” of phosphates to the shielded polar core.77 Importantly, they are adjacent to the bulky hydrophobic residues in β-strand I that participate in its interactions with the arrestin C-tail and α-helix I (Fig. 3.1).18 This suggested that their interaction with phosphates likely disrupts this three-element interaction, which would destabilize the basal arrestin conformation, similar to the effect of the disruption of the polar core.77 These data support the main premise of the sequential multisite binding model of the arrestin–receptor interaction25 that receptor binding is accompanied by a global conformational change in arrestin.
The action of the phosphate sensor is based on pure electrostatics; all that receptor-attached phosphates need to do to activate the sensor is to break the key salt bridge.30 This makes it essentially insensitive to the sequence context of the phosphorylated residues. This mechanism explains how just two non-visual arrestins in mammals and only one in Drosophila can interact with hundreds of different GPCRs, in which serines and threonines phosphorylated by GRKs are found within diverse sequences that can be localized in the receptor C-terminus, or any of the intracellular loops (reviewed in Ref. 14).
3.4. The conformation of the receptor-bound arrestin
Several lines of indirect evidence suggested that the conformation of receptor-bound arrestins is likely quite different from their basal state revealed by crystal structures. The first indication that this must be the case was unusually high-energy barrier of arrestin-1 binding to rhodopsin.104 Receptor-binding-induced release of the arrestin C-tail has been well documented for more than 20 years.73,86,109 In addition, the movement and/or structural rearrangement of the “finger loop” in the central crest of the receptor-binding side was also reported.83,131 Both polar core and the three-element interaction clearly support the basal conformation in all arrestin structures,18–20,22 so their destabilization by the phosphates was consistent with the idea that global rearrangement is necessary for receptor binding. Partial destabilization of the interface between the two domains enhanced arrestin binding to inactive receptor,132 again suggesting that arrestin conformation must change upon receptor binding.
Arrestin interaction with nonreceptor partners also appears to be consistent with the idea that the conformations of free and receptor-bound arrestins must be different.31 C-Raf1 and especially ERK1/2 preferentially interact with receptor-associated arrestins.133 In contrast, the ubiquitin ligases Mdm2 and parkin134,135 strongly prefer arrestins in a conformation induced by hinge deletions that impairs receptor binding.35,105,136 JNK3 also appears to prefer this conformation, although the difference in binding is less dramatic.134,136
Thus, it appeared almost certain that the conformation of receptor-bound arrestin is significantly different from the basal one observed in crystal structures, but direct evidence was missing. A recent study employing site-directed spin-labeling and long-range distance measurements using pulse EPR technique double electron–electron resonance (DEER) yielded the first experimental data on conformational rearrangements in arrestin-1 beyond the release of the C-tail.72 More than 25 distances between different residues in free and rhodopsin-associated arrestin-1 were measured. Significant changes in multiple distances combined with molecular modeling revealed binding-induced movements of several arrestin-1 elements. Some of the findings supported earlier predictions, whereas others were rather unexpected. Flexible “finger loop” (residues 67–79)73 (Fig. 3.1) in the central crest of arrestins exist in fully extended or bent conformation in different protomers in crystal oligomers.18,19 Multiple residues in this loop were shown to be immobilized upon receptor binding in both arrestin-173 and -2.71 Previous studies using fluorescent labels131 and NMR83 suggested that this loop extends and forms an α-helix upon receptor binding. Indeed, this loop was found to move in the direction of the receptor, although not as much as previously proposed,131 and the data were consistent with its helical conformation in receptor-associated arrestin-1.72 However, hypothetical movement of the two arrestin domains relative to each other, which was proposed to improve the fit between arrestins and GPCRs,30 turned out to be very small, clearly insufficient to significantly reduce the size of the receptor-binding arrestin surface.72 Two other plastic loops containing residues 157 and 344, localized at the tips of the N- and C-domain, respectively, that had different conformations in crystal strcutures,18 were found to move “inward,” in the direction of the cavities of their respective domains. These movements also slightly reduce the long axis of the receptor-binding surface, but only by a few angstroms. The most unexpected finding was a dramatic movement of the loop with residue 139 at its tip, which is adjacent to the finger loop in the basal state. This element shifts by more than 10 Å, moving in the direction of the N-domain and to the side of the molecule.72 This movement would take it out of the way of incoming receptor. Consistent with this model, spin label in position 139 was immobilized by inactive P-Rh, but in high-affinity complex with P-Rh*, its mobility increased to the level observed in free arrestin.73 Moreover, deletions in this loop, taking it out of the way without movement, increased arrestin-1 binding to rhodopsin, most dramatically to the nonpreferred forms Rh* and inactive P-Rh.72 Interestingly, the same deletions reduced the thermal stability of arrestin-1.72 Thus, it appears that the 139-loop structurally stabilizes the basal arrestin conformation and serves as a “brake”, precluding its binding to any form of rhodopsin except its preferred target, P-Rh*.
While this study clearly revealed multiple receptor-binding-induced rearrangements in arrestin-1, which are likely similar to those in nonvisual arrestins, biophysical methods cannot yield detailed atomic resolution structure of the receptor-bound arrestin. Crystal structure of the arrestin–receptor complex is necessary to obtain this information and clearly reveal the changes in arrestin molecule that underlie conformational preferences of nonreceptor binding partners.
3.5. Key determinants of receptor preference
As a general rule, arrestins preferentially bind active phosphorylated forms of their cognate receptors. Obviously, receptor-attached phosphates that activate the phosphate sensor are the common theme, so arrestin elements that bind other nonphosphorylated parts of the receptor in response to activation-induced conformational change must be responsible for receptor specificity of arrestin proteins. Two out of four arrestin subtypes in mammals are specialized and expressed primarily in photoreceptors. Interestingly, the specificity of cone arrestin-4 for cone opsins appears to be largely ensured by its selective expression in cone photoreceptors.137 In vitro, it is quite promiscuous, binding other GPCRs almost as well as nonvisual arrestins,22 both of which readily interact with dozens, if not hundreds, of different receptors.80,138,139 Thus, evolution created only one receptor-specific subtype, arrestin-1, which demonstrates clear preference for rhodopsin over other receptors.80,140
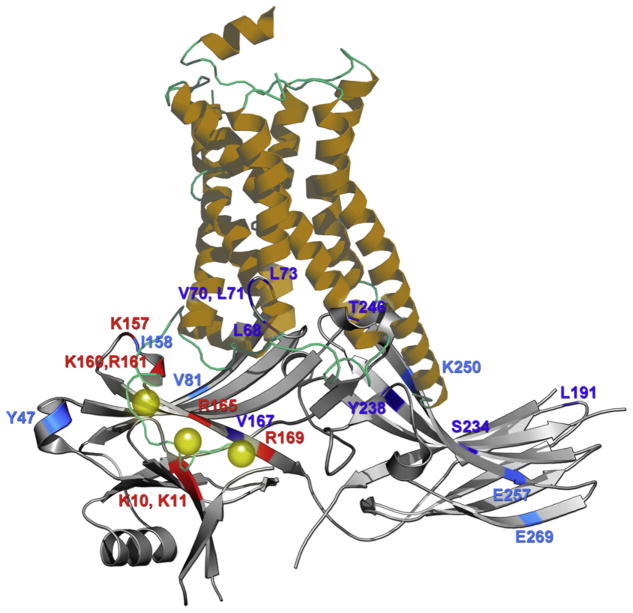
Arrestin-1 binds rhodopsin efficiently and demonstrates fairly low binding to M2 muscarinic receptor, whereas arrestin-2 has the opposite preference.80,140 Therefore, the first attempt to identify arrestin elements that determine its receptor specificity involved the construction of a series of arrestin-1/2 chimeras and testing their ability to bind these two receptors.79 The premise of these experiments was that if an element is important for receptor preference, the introduction of that part from arrestin-2 into arrestin-1 would decrease rhodopsin and increase M2 binding, whereas the introduction of the corresponding arrestin-1 part into arrestin-2 would decrease M2 and increase rhodopsin binding. N-domain residues 49–90 (β-strands V and VI with adjacent loops) and C-domain residues 237–268 (β-strands XV and XVI) of arrestin-1 and homologous parts of arrestin-2 were found to play key role in receptor preference. The exchange of these two elements between arrestin-1 and -2 completely reversed receptor specificity of both, creating one chimera that was >90% arrestin-1, yet bound M2 muscarinic receptor much better than rhodopsin, and a symmetrical chimera with >90% of arrestin-2 residues that clearly preferred rhodopsin to M2.79
Fewer than 25 residues in these two elements are nonconservative substitutions. Their individual contributions to receptor preference were tested by introducing point mutations.71 This approach led to the identification of 10 exposed side chains that collectively determine which receptor an arrestin protein prefers. Interestingly, the replacement of all 10 with alanines in arrestins-1, -2, and -3 yielded mutants that virtually lost the ability to bind any GPCRs.71 One of the nonexposed residues in the N-domain element, Val90, was found to play an unexpectedly important role; its substitution with serine (Ser86 is the homologous arrestin-2 residue) increased arrestin-1 binding to M2 muscarinic receptor more than any other point mutation.19 Arrestin-3, with similarly broad receptor specificity, has Ala87 in the same position,20 which suggests that a small side chain is important. Crystal structure shows that arrestin-1 Val90, localized between the two β-sheets, makes contacts with several other hydrophobic residues, apparently stabilizing the core of the N-domain.18 All potential partners are present in arrestin-219 and -3,20 but they do not contact the smaller side chain of Ser86 or Ala 87. This analysis indicates that the presence of the bulky Val90 increases the rigidity of the N-domain, apparently enhancing receptor specificity of arrestin-1, whereas the smaller side chains in arrestin-2 and -3, as well as in the arrestin-1-V90S mutant, make the N-domain more flexible, which appears to correlate with broad receptor specificity. These results suggest that to construct a nonvisual arrestin with narrow receptor preference, one needs to place valine in this position.
The fact that the same 10 exposed residues determine receptor specificity and largely drive the arrestin–receptor interaction71,89 suggests that the targeted manipulation of these side chains has the potential to yield nonvisual arrestin mutants with much narrower receptor specificity than their parental proteins. Since arrestins that can selectively target particular GPCRs would be very useful research tools and have clear therapeutic potential,141,142 this idea was recently tested experimentally.143 Ten different amino acid substitutions affecting 8 out of 10 identified receptor discriminator residues were introduced on the background of an arrestin-3-A87V base mutant. The binding of WT arrestin-3, A87V, and other mutant arrestins to β2AR, M2 muscarinic, as well as D1 and D2 dopamine receptors was tested in cells using a BRET-based assay.143 Seven out of 10 mutations resulted in differential changes in the binding to these receptors, yielding up to fourfold increase in GPCR selectivity over WT protein.143 This unexpectedly high success rate clearly indicates that correct targets were chosen to change the receptor preference of arrestin-3. The combination of two mutations that individually reduced β2AR binding, but did not affect the interactions with M2 or D2 receptors, yielded a variant with more than 50-fold preference for these receptors over β2AR. Similarly, two other mutations were shown to act additively; their combination yielded a version of arestin-3 with >fivefold preference for D1 over D2 receptor.143
Numerous GPCR mutations have been shown to underlie a variety of human disorders (reviewed in Ref. 144). Currently, there are no viable approaches to counteract the effects of gain-of-function mutations that result in excessive receptor signaling. Arrestins with enhanced ability to quench this signaling, such as the phosphorylation-independent mutants described above, have a potential to become a solution to this problem. However, WT nonvisual arrestins have very broad receptor specificity, so the introduction of an enhanced arrestin-2 or -3 will likely simultaneously blunt the signaling by the “bad” mutant receptor and numerous other perfectly normal GPCRs expressed in the same cell. Therefore, arrestin variants selectively targeting the overactive receptor are needed for therapeutic use in combination with other mutations changing its functional capabilities. This study was the first attempt to create something that evolution did not; a nonvisual arrestin with high receptor selectivity. Both the 70% success rate in changing receptor preference and additive effects of individual mutations show that it opened a promising new direction. However, a lot of additional experimentation is necessary to construct the arrestins that fit every GPCR whose signaling needs to be corrected to cure a particular human disorder. The same approach can be used to rein in excessive signaling by normal GPCRs that develops for various reasons in numerous human disorders.
4. INTERACTIONS WITH OTHER SIGNALING PROTEINS
Clathrin was the first nonreceptor-binding partner of arrestin proteins identified.145 Since then arrestins were shown to interact with an amazing variety of trafficking and signaling proteins (see Chapter 1). The molecular mechanisms of most of these interactions remain to be elucidated. By comparison, it appears that we know a lot about the mechanics of receptor binding, although even in this area there are more questions than answers.
4.1. Where do the other partners bind?
Only two proteins have been shown to interact with the receptor-binding surface of arrestins: microtubules35 and Ca2+-liganded calmodulin.146 In both cases, receptors bind arrestin with much higher affinity, easily winning the competition with these partners.32,146 Most other proteins interact with receptor-bound arrestins,145,147–150 which suggests that they engage nonreceptor-binding elements localized either on the convex sides of the two domains, or on the arrestin C-tail that is released upon receptor binding.
Considering the potential biological importance of these interactions, surprisingly few binding sites of nonreceptor partners have been identified, and most of those very imprecisely. The best characterized among these are clathrin and AP2 interaction sites, both of which are localized in the C-tail of arrestin-2 and -3. The main clathrin site is a short LIELD or LIEFE motif in the part of the C-tail upstream of its contact with the N-domain,151 which is not visible in any of the crystal structures of free arrestins, likely because it is inherently disordered.19–21 This interaction was resolved in the cocrystal of arrestin-2 with the clathrin β-propeller domain, where this arrestin element binds between blades 1 and 2.152 Interestingly, in the prevalent long splice variant of arrestin-2,8 another C-tail element also interacts with clathrin, binding between blades 4 and 5.152 The AP2 binding site was localized downstream of the N-domain contact in the distal C-tail, by extensive mutagenesis147,153 and cocrystallization of an arrestin peptide with the AP2 beta-appendage.154
No other partners have so far been cocrystallized with arrestins or even their elements. Only the binding sites of microtubules,35 calmodulin,146 and cAMP phosphodiesterase PDE4D5155 were mapped with any degree of precision. Particular residues in arrestin-2 were implicated in binding of MEK1156 and c-Raf1,157 and several residues in arrestin-3 were found to be critical for its ability to promote JNK3 activation.158 Thus, the structural basis of arrestin interactions with the great majority of putative partners still remains to be elucidated.
4.2. The shape of the arrestin–MAPK signaling complex
The first reports of arrestin-mediated activation of MAP kinases suggested that in each MAPKKK–MAPKK–MAPK cascade only MAPKKKs (ASK1 and c-Raf1) and MAPKs (JNK3 and ERK1/2) directly bind arrestins, whereas MAPKKs (MKK4 and MEK1) are recruited via interactions with corresponding MAPKKKs and/or MAPKs.148,149 This model further suggested that MAPKKKs bind to the N-domain of arrestins, whereas MAPKs interact with the C-domain.159 Subsequently, two MAPKKs, MEK1156 and MKK4,160 were shown to bind arrestins directly in the absence of MAPKKKs or MAPKs. Moreover, separately expressed arrestin N- and C-domains were shown to interact with all three kinases in c-Raf1–MEK1–ERK1/2 and ASK1–MKK4–JNK3 cascades.161,162 This led to an alternative model of the arrestin complex with MAP kinases, where MAPKKK, MAPKK, and MAPK have bipartite binding sites, each interacting with both arrestin domains,162 so that the three kinases are arranged on arrestin-like three hotdogs on a single bun.141 Considering that receptor binding induces a conformational change in arrestins, the dependence of arrestin scaffolding function on GPCR activation can be easily explained by this arrangement.
4.3. Conformational preferences determine outcomes
The first reports of arrestin binding to nonreceptor partners focused on GPCR-associated arrestins,145,147–149 creating an impression that these interactions are contingent on arrestin binding to receptors. This implied that only “active” receptor-bound arrestin can engage other proteins, which turned out not to be the case. In fact, accumulating evidence suggests that most arrestin-binding partners interact with arrestins in all three conformations, basal, receptor-bound, and microtubule-associated, but their conformational preferences vary widely.
ERK1/2 appears to be an extreme case; it interacts with reasonable affinity only with receptor-bound arrestins, whereas its transient interaction with free arrestins is not even detectable by coimmunoprecipitation without cross-linking.133,149 C-Raf1 also prefers receptor-bound form, although the difference in binding is not as dramatic.133 In contrast, MEK1 not only binds free arrestins156 but also does not discriminate among the three conformations.133 Unexpectedly, it was shown that both ERK1/2 and c-Raf1 bind arrestins in a conformation preferred by microtubules (which is mimicked by a 7-residue deletion in the inter-domain hinge, termed D735) better than the basal state of free WT arrestins.133 In cells, arrestins-1, -2, and -3 all bind ERK1/2 and recruit it to microtubules, reducing the overall ERK1/2 activation level, whereas arrestin-4 does not have this effect.35 Moreover, arrestin-3-D7 effectively suppresses—ERK1/2 phosphorylation induced by receptor activation, which makes it a dominant-negative form in this regard.136 The facilitation of signaling in c-Raf1–MEK1–ERK1/2 cascade appears to be strictly dependent on arrestin binding to GPCRs,133,136,157 as originally hypothesized.149 Therefore, mutant arrestins with impaired receptor binding that do interact with ERK1/2 normally recruit it away from places where it can be activated, blunting ERK1/2 activity.35,136
Arrestin-3, but not the other subtypes, promotes signaling in the ASK1–MKK4–JNK3 cascade, and this function was originally ascribed to the receptor-bound form.148 However, subsequent studies showed that free arrestin-3 effectively facilitates JNK3 phosphorylation136,158,159,162 and even localizes JNK3 to the cytoplasm independent of GPCR binding.134,136,163 An assay based on the ability of arrestins to move JNK3 from the nucleus to the cytoplasm did not detect any differences among WT arrestins, their preactivated phosphorylation-independent forms where the C-tail is forcibly detached by a triple alanine substitution (3A), and D7 mutants with impaired receptor binding.134 However, more a quantitative BRET-based assay in cells showed that arrestin-3–D7 actually binds JNK3 better.136
The ubiquitin ligase Mdm2 was first reported to interact with receptor-bound arrestins.150 However, the comparison of the ability of different forms of arrestins to remove Mdm2 from the nucleus showed that D7 mutants do it more efficiently than WT, whereas preactivated 3A mutants are the least effective,134,161 indicating that Mdm2 prefers free over receptor-bound arrestins. Yet arrestin ubiquitination by Mdm2 is clearly stimulated by receptor binding,135,150,164 suggesting that receptor-bound form is a better substrate for Mdm2. Collectively, these data suggest that Mdm2 binds free arrestins and is recruited to the receptor, where it has limited time to ubiquitinate arrestin before falling off due to reduced binding affinity. This model explains limited and fairly selective ubiquitination of arrestins by Mdm2 in response to receptor activation.164 Another E3 ubiquitin ligase, parkin, also shows strict preference for the basal and D7 forms of arrestins over 3A mutants partially mimicking receptor-bound state.135 Interestingly, in contrast to Mdm2, parkin appears to be able to shift the conformational equilibrium of arrestin to the form it prefers.135 Parkin greatly increases the binding of Mdm2 to WT arrestins, but not to D7 mutants that show enhanced Mdm2 interaction, suggesting that parkin acts by stabilizing the D7-like conformation.135 We found that parkin suppresses Mdm2-dependent arrestin ubiquitination.135 This is consistent with the idea that parkin stabilizes a basal-like arrestin conformation, whereas receptor-bound arrestins are better Mdm2 substrates.
Even though they appear almost inseparable in evolution (see Chapter 2),12 the two arrestin domains are independent folding units that can be expressed separately and retain certain functions.24–26,80 Therefore, the expression of separated domains has been repeatedly used to determine which part of arrestin contains the binding sites for different partners. Interestingly, whenever the localization of binding sites was tested by this method or using peptide arrays, it was found that the protein of interest binds both arrestin domains. This was shown for microtubules,35 the MAP kinases JNK3,161 ASK1,162 MKK4,162 c-Raf1,162 MEK1,162 ERK2,162 the cAMP phosphodiesterase PDE4D5,155 as well as the ubiquitin ligases Mdm2135,161 and parkin.135 Thus, with very few possible exceptions, the interaction with both arrestin domains appears to be a general rule. Even though receptor binding does not induce a large movement of the two arrestin domains, the domains actually shift relative to each other.72 Therefore, this rearrangement can explain conformational preference of certain partners; the relative positions of the two parts of the binding site localized on different domains actually change. The distance between these two parts and/or their relative orientation can become favorable or unfavorable for the interaction in one of the conformations.31
It is harder to reconcile the bipartite binding site with the observations that some proteins, such as MEK1, appear to bind arrestins in all conformations equally well,133 unless the interaction with one of the domains predominates, as some data suggest.156 It is even harder to explain mechanistically how ERK1/2 can prefer both receptor-bound and D7-like conformation over basal; existing data suggest that the arrestin domains shift in opposite directions in these two states.34,35 It is becoming increasingly clear that without direct structural information obtained by cocrystallization of arrestins with different partners these questions will remain unanswered.
4.4. Binding and activation do not go hand-in-hand
The original explanation for why arrestin-3 promotes JNK3 phosphorylation while the closely related arrestin-2 does not was beautifully simple; it was suggested that arrestin-2 does not bind this MAP kinase,148 and this idea appeared to be supported by the identification of putative JNK3-binding sequence RRS present in the C-domain of arrestin-3, whereas arrestin-2 has KP in homologous position.159 However, RRS is only found in rodent arrestin-3, while the prevalent sequence in mammalian species is RS.12 Subsequent studies showed that arrestin-3 from other species also binds JNK3134,163 and effectively facilitates its activation.136,158,162 In fact, all four mammalian arrestins were found to bind JNK3 comparably and remove it from the nucleus, where it spontaneously localizes, to the cytoplasm.134,161 This was the first indication that JNK3 binding and its activation are two distinct functions of arrestins. The idea that arrestin-2 might not bind the upstream kinases ASK1 and MKK4, and therefore fails to assemble the complete MAPK module necessary for JNK3 activation, also was not supported by the evidence; both nonvisual arrestins appeared to interact with all three kinases comparably in cells,162 although subsequent more precise direct binding experiments with purified proteins suggested that the affinity of arestin-3 for JNK3 and MKK4 is higher than that of arrestin-2.160 Since to assemble a productive complex arrestin must hold all three kinases simultaneously, even a subtle reduction in affinity for two of them could make a qualitative difference in the cellular environment, where the concentrations of all proteins involved are fairly low.
Arrestins-2 and -3 are 78% identical (and 88% similar),8 both bind ASK1, MKK4, and JNK3,134,162 yet only arrestin-3 facilitates JNK3 phosphorylation in cells.148,158,162 Thus, arrestin-3-specific residues must be responsible for its unique ability to promote JNK3 activation. This idea was tested by the construction of arrestin-2/3 chimeras.158 It turned out that virtually every chimera was less efficient than WT arrestin-3,158 indicating that it is much easier to destroy a function than to build it. Although these data were consistent with earlier conclusion that each kinase in the ASK1–MKK4–JNK3 module has multipartite site, interacting with both arrestin domains,162 this approach identified the C-domain as the key element in JNK3 activation.158 Interestingly, this region included residues 182–376, encompassing the Arg196–Ser197 sequence in bovine arrestin-3 that is replaced with Lys195–Pro196 in arrestin-2. To further dissect the key arrestin-3 elements necessary for JNK3 activation, the residues unique for this subtype were replaced with their arrestin-2 homologues.158 Quite a few mutations, including RS→KP, had no detectable effect, ruling out the role of those residues in JNK3 activation. Multiple other substitutions reduced arrestin-3 potency in JNK3 activation assay.158 This study identified Val343 as the most important residue, as a Val343Thr point mutation reduced the ability of arrestin-3 to activate JNK3 by two-thirds. Residues Leu278, Ser280, His350, Asp351, His352, and Ile353 were found to play supporting roles, as their substitution with arrestin-2 homologues also reduced the ability of arrestin-3 to facilitate JNK3 phosphorylation.158 Interestingly, virtually all arrestin-3 mutants with reduced or even abolished ability to activate JNK3 demonstrated essentially normal binding to all three kinases in this module, supporting the idea that the interaction with these kinases and the ability to promote JNK3 activation are different arrestin-3 functions that can be separated by targeted mutagenesis. Evolution actually separated these functions in arrestin-2, which binds the same kinases but does not facilitate JNK3 phosphorylation.162
5. DESIGNING SIGNALING-BIASED ARRESTIN MUTANTS
To a certain extent, the natural structural organization of arrestins makes targeting different aspects of their function easier. The concave sides of both domains contain allknown receptor-binding residues (Section 3.3), most other partners interact with elements on the other side of the molecule that remain accessible in the arrestin–receptor complex, whereas clathrin and AP2 engage distinct sites on the arrestin C-tail (Section 4.1) that do not appear to overlap with the binding sites of any other known partner. These sites were the first to be eliminated by targeted mutagenesis.145,147,153 Predictably, the disruption of either site precluded arrestin interactions with its respective partners without affecting GPCR binding. Moreover, arrestin mutants with disabled binding sites for clathrin, AP2, and particularly both, acted as dominant-negative, selectively suppressing arrestin-dependent GPCR internalization via coated pits.145,147,153 These findings firmly established that arrestin functions can be manipulated independently of each other. In addition, these studies showed that an arrestin can act as adominant negative in a particular regard only when a single function is destroyed in a way that preserves all other functional capabilities. Similarly, to generate signaling-biased arrestins, a single function should be enhanced or disabled by the mutations that do not significantly affect others. In reality, it turned out to be extremely difficult to change anything within the arrestin molecule without affecting more than one function. However, as long as a single aspect of arrestin functions is affected to a much greater extent than the others, the mutant can serve a particular purpose.
5.1. Enhanced phosphorylation-independent mutants
The first arrestin that binds with high affinity to any active form of its cognate receptor regardless of phosphorylation, arrestin-1–Arg175Asn, was created in the process of elucidation of the mechanism arrestins use to respond to the receptor-attached phosphates in 1995.27 This happened when arrestins were still believed to interact only with GPCRs, before arrestin binding to the first nonreceptor partner, clathrin, was discovered in 1996.145 Arg175 was later substituted by every residue in the book, and charge reversal mutation Arg175Glu was found to be the most potent.28 Later, a whole family of structurally distinct phosphorylation-independent mutants of arrestin-1 was constructed.77,165 The Arg175Glu mutant was shown to effectively suppress transducin activation by unphosphorylated light-activated rhodopsin (Rh*).166 Several mutations homologous to those that enable arrestin-1 binding to unphosphorylated Rh* were shown to yield similar phosphorylation-independent binding of both nonvisual arrestins to their cognate receptors,125–128 and to block the coupling of unphosphorylated WT and mutant GPCRs to their cognate G proteins.125,126,129
One of the phosphorylation-independent arrestin-1 mutants was recently tested for its ability to compensate for defects of rhodopsin phosphorylation in vivo.91 This enhanced arrestin-1 was shown to significantly improve survival and functional performance of rod photoreceptors lacking rhodopsin kinase, and to facilitate the rate of rod recovery ~ threefold, as compared to parental WT arrestin-1.91 This proof-of-concept study showed that one mutant protein can be used to compensate for a molecular defect in another.141 However, photoresponse recovery in “compensated” rods was much slower than in WT photoreceptors, suggesting that a significant further redesign of arrestin-1 is necessary to achieve a perfect fit and high-affinity binding to unphosphorylated Rh*.91 These results clearly showed that, even though we know about the molecular mechanism of the arrestin–receptor interaction much more than about any other arrestin function, we still need a lot of additional information to construct a perfect tool for therapeutic use.
Enhanced phosphorylation-independent versions of all arrestins have been constructed125–128 and shown to effectively desensitize several GPCRs without receptor phosphorylation.125,126,129 These mutants proved to be useful in the studies of GPCR phosphorylation129,130 and its role in particular biological processes.130,167,168 However, wider practical use of enhanced nonvisual arrestins, particularly for therapeutic purposes, is contingent on our ability to make them target the receptors of our choosing, rather than all GPCRs indiscriminately, like parental WT arrestin-2 and -3.
5.2. Constitutively monomeric arrestins
Self-association of arrestin-1 (known as S-antigen at the time) was discovered even before its role in quenching rhodopsin signaling.1 Arrestin-1 was subsequently crystallized as a tetramer (dimer of dimers) by two independent groups under different conditions.17,18 It was shown to form dimers and tetramers in solution by a variety of methods: analytical ultracentrifugation,118 small angle X-ray scattering,119 and multiangle light scattering.120 The study of arrestin-1 self-association using long-range measurements of inter-subunit distances by DEER yielded an unexpected result; the solution tetramer was found to be strikingly different from that in crystal form.120 Elucidation of its structure122 confirmed the earlier idea that arrestin-1 oligomers are storage forms.118 It also explained the earlier finding that only monomeric arrestin-1 can bind rhodopsin120; in the physiologically relevant solution tetramer and both possible dimers the rhodopsin-binding surfaces of all arestin-1 molecules are shielded by sister protomers.122
Bovine, mouse, and human arrestin-1 were shown to self-associate via the same monomer–dimer–tetramer equilibrium, although dimerization and tetramerization constants were very different in these species.121 Model-based targeted elimination of the same two phenylalanines generated self-association-deficient bovine and mouse arrestin-1.121,122 These results showed that the structure of the solution tetramer of all mammalian arrestin-1 is likely very similar, despite striking differences in dimerization and tetramerization constants. This study also generated a constitutively monomeric form of arrestin-1 that retained perfectly normal binding to rhodopsin and microtubules.121 This mutant is the molecular tool necessary to determine the biological role of robust arrestin-1 self-association at concentrations normally found in photoreceptors.110,115–117 This is not a trivial question, as only monomeric arrestin binds receptor,120 and no other signaling protein in photoreceptors has an oligomeric storage form. Interestingly, cone-specific arrestin-4 is a constitutive monomer,169 but it represents only ~ 2% of total arrestin complement in cones,170 the rest being arrestin-1 that readily self-associates. This finding, along with apparent conservation of self-association among mammalian arrestin-1 homologues,121 strongly suggests that oligomerization of this subtype has an important biological function that needs to be elucidated experimentally.
Both nonvisual arrestins were also found to self-associate171–173 and even form heterodimers.171,172 The mechanism of self-association of arrestin-1 and nonvisual subtypes appears to be different, as IP6 inhibits oligomerization of arrestin-1, while promoting self-association of nonvisual arrestins.169,172 Two IP6 binding sites in arrestin-2 were identified by crystallography and confirmed by structure-based mutagenesis.172 Self-association was shown to regulate the subcellular distribution of arrestin-2; oligomers were predominantly cytoplasmic, whereas monomers showed increased nuclear localization.172 Oligomerization of arrestin-3 was reported to be required for its interaction with the E3 ubiquitin ligase Mdm2, and elimination of IP6 binding residues inhibited Mdm2 binding and p53-dependent antiproliferative effects of arrestin-3.173 It should be noted that while self-association of arrestin-1 certainly stops at tetramers that have the shape of closed diamonds, where all potential interaction surfaces are fully engaged by sister subunits,122 it is not clear whether oligomerization of nonvisual arrestins has a natural limit. Crystallography suggests that they might form chains of any length172; biochemical data obtained with pure protein indicate that arrestin-2 can form at least tetramers but do not provide evidence that the process stops at that stage.169 Further experimentation with arrestin-2, arrestin-3, and a mixed population of the two nonvisual arrestins in the presence and absence of IP6 is necessary to resolve these issues and attempt construction of constitutively monomeric forms of these proteins with all other functions intact. These tools are necessary to determine the biological role of the oligomerization of nonvisual arrestins.
5.3. Manipulating MAPK signaling
MAP kinases regulate many vital cellular functions, from proliferation to apoptotic death.174 MAPK cascades consist of three kinases that sequentially activate each other by phosphorylation: upstream MAPKKKs (e.g., ASK1 and cRaf1), MAPKKs (e.g., MEK1/2, MKK4/7), and MAPKs (e.g., ERK1/2, JNK1/2/3, p38 MAPK). All MAP kinases are soluble proteins, but phosphatase activity in the cytoplasm makes it highly likely that an upstream kinase would be dephosphorylated before it finds its target by random diffusion. In addition, there are many different MAPKs, MAPKKs, and especially MAPKKKs in the cell.174 Therefore, productive complexes containing matching components are usually assembled by scaffolding proteins that bring the three kinases together, thereby facilitating signaling.175
Arrestins were shown to promote the activation of JNK3,148 ERK1/2,149 and p38 MAPK168 by scaffolding the relevant three-kinase modules. The first reports suggested that receptor activation is necessary for arrestin-dependent scaffolding of MAPK cascades.148,149,168 Subsequent studies confirmed that receptor binding is a prerequisite for arrestin-dependent activation of ERK1/2.133,136 However, arrestin-3-mediated facilitation of JNK3 phosphorylation was shown to be receptor-independent,159,162 indicating that free arrestin-3 performs this function.136,158 This was unambiguously confirmed by reconstitution of an arrestin-3–MKK4–JNK3 signaling module in vitro from purified proteins in the absence of the receptor.160
Regardless of whether free, receptor-bound, or both forms of arrestins scaffold MAPK cascades, several ways of manipulating MAPK signaling by mutant arrestins are conceivable. First, targeted mutations disrupting an interaction with one of the kinases would yield arrestins that can only form incomplete complexes, thereby sequestering the other two kinases and suppressing the activation of a particular MAPK. Second, a mutant that binds all kinases but holds them in a “wrong” configuration not conducive to signal transduction would serve this purpose even better. Third, mutations enhancing the interactions with one or more of the kinases in a particular cascade would channel signaling in that direction. Finally, mutations that create preference for ERK1/2 over JNK3 would facilitate prosurvival and suppress proapoptotic signaling, while mutations biasing arrestin toward JNK3 would have the opposite effect.
Even though the binding sites of MAP kinases remain unknown or poorly defined, several signaling-biased forms of arrestins that significantly affect MAPK signaling were created. The first in this line was arrestin-2-R307A mutant, which was shown to have impaired c-Raf1 binding, while interacting with ERK1/2 and MEK1 normally.157 In contrast to WT arrestin-2, the expression of this mutant did not rescue β2AR-dependent arrestin-mediated ERK1/2 activation in arrestin-2/3 double knockout cells.157 Whether this mutant can suppress ERK1/2 activation via other scaffolds still remains to be elucidated. Interestingly, a homologous K308A mutation in arrestin-3 did not affect c-Raf1 binding or its ability to promote ERK1/2 phosphorylation upon receptor stimulation.157 These results suggest that despite very high homology between the two nonvisual subtypes, the molecular mechanisms of scaffolding c-Raf1–MEK1–ERK1/2 cascade by arrestin-2 and -3 are subtly different.
Two signaling-biased mutants of arrestin-3 have been characterized more comprehensively.136 The ability of WT arrestin-3, its “preactivated” 3A mutant that binds GPCRs even more readily,125 and two receptor-binding-impaired forms; one with 7-residue deletion in the inter-domain hinge (D7) and the other where key receptor-binding residues were mutated to alanines (KNC),71,89 to bind JNK3 and promote its activation in the cell was compared.136 It turned out that the ability of these proteins to bind GPCRs and activate JNK3 do not correlate at all: WT arrestin-3 that binds receptors and D7 mutant that does not promoted JNK3 phosphorylation equally well, whereas both 3A and KNC did not.136 This was the most convincing evidence that arrestin-3 scaffolds ASK1–MKK4–JNK3 module independently of receptor binding. Moreover, the inactive KNC mutant was shown to interact with ASK1 and MKK4 as well as WT, and bind JNK3 even better,136 providing the strongest proof so far that recruitment of MAP kinases and the ability to promote signaling in the cascade are also separable arrestin functions. Indeed, the KNC mutant was shown to act in a dominant-negative manner, suppressing JNK3 activation in the cell. Thus, arrestin-3–KNC is a silent scaffold, inhibiting JNK3 activation by sequestering ASK1, MKK4, and JNK3 away from productive scaffolds. As could be expected, both receptor-binding-impaired forms of arrestin-3, D7 and KNC, were found to suppress ERK1/2 activation.136 Apparently, these mutants retain the ability to recruit kinases of c-Raf1–MEK1–ERK1/2 module but cannot assume the conformation conducive to ERK1/2 activation due to the defect in GPCR binding.
These studies generated tools that selectively suppress either ERK1/2 or JNK3 activity, which is believed to send prosurvival or proapoptotic signal to the cell, respectively. The ability of the arrestin-3–KNC mutant to enhance cell survival and that of arrestin-2–R307A to suppress proliferation and possibly facilitate cell death needs to be tested. Mutants where individual functions are enhanced, rather than suppressed, also must be developed. Targeted construction of distinct signaling-biased forms of arrestins would be greatly facilitated by better understanding of the molecular mechanisms involved in their binding to nonreceptor partners and the ability to scaffold MAP kinase cascades productively.
6. CONCLUSIONS: WHERE DO WE GO FROM HERE?
It might appear that as proteins go, arrestins are fairly well studied structurally and functionally. It is clear that members of this small protein family, which apparently emerged relatively late in evolution,12 likely after GPCR kinases,176 serve as multifunctional signaling organizers in the cell. However, while known phenomenology is rich and pretty well described,177 the structural basis of most arrestin functions remains obscure. The molecular mechanisms underlying arrestin interactions with most partners, as well as expected inter-dependence of these interactions, must be elucidated. This will greatly improve our understanding of cell signaling, particularly the integration of different inputs into coherent cell behavior, which is arguably the greatest challenge in modern cell biology.142 As an added bonus, elucidation of the molecular mechanisms underlying individual arrestin functions and identification of the residues involved will pave the way to intelligent design of mutant arrestins with desired signaling bias. The mutants of this type are not just valuable research tools but also hold promise of enormous therapeutic potential.
Acknowledgments
The authors are grateful to our collaborators, whose expertise and efforts made many of the studies discussed here possible. Supported by NIH Grants EY011500, GM077561, GM081756 (VVG), NS065868, and DA030103 (EVG).
Footnotes
We use systematic names of arrestin proteins: arrestin-1 (historic names S-antigen, 48-kDa protein, visual or rod arrestin); arrestin-2 (β-arrestin or β-arrestin1); arrestin-3 (β-arrestin2 or hTHY-ARRX); and arrestin-4 (cone or X-arrestin; for unclear reasons, its gene is called arrestin 3 in the HUGO database).
References
- 1.Wacker WB, Donoso LA, Kalsow CM, Yankeelov JA, Jr, Organisciak DT. Experimental allergic uveitis. Isolation, characterization, and localization of a soluble uveitopathogenic antigen from bovine retina. J Immunol. 1977;119:1949–1958. [PubMed] [Google Scholar]
- 2.Kuhn H. Light-regulated binding of rhodopsin kinase and other proteins to cattle photoreceptor membranes. Biochemistry. 1978;17:4389–4395. doi: 10.1021/bi00614a006. [DOI] [PubMed] [Google Scholar]
- 3.Kuhn H, Hall SW, Wilden U. Light-induced binding of 48-kDa protein to photoreceptor membranes is highly enhanced by phosphorylation of rhodopsin. FEBS Lett. 1984;176:473–478. doi: 10.1016/0014-5793(84)81221-1. [DOI] [PubMed] [Google Scholar]
- 4.Wilden U, Hall SW, Kühn H. Phosphodiesterase activation by photoexcited rhodopsin is quenched when rhodopsin is phosphorylated and binds the intrinsic 48-kDa protein of rod outer segments. Proc Natl Acad Sci USA. 1986;83:1174–1178. doi: 10.1073/pnas.83.5.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dixon RA, Kobilka BK, Strader DJ, et al. Cloning of the gene and cDNA for mammalian beta-adrenergic receptor and homology with rhodopsin. Nature. 1986;321:75–79. doi: 10.1038/321075a0. [DOI] [PubMed] [Google Scholar]
- 6.Lohse MJ, Benovic JL, Codina J, Caron MG, Lefkowitz RJ. Beta-arrestin: a protein that regulates beta-adrenergic receptor function. Science. 1990;248:1547–1550. doi: 10.1126/science.2163110. [DOI] [PubMed] [Google Scholar]
- 7.Attramadal H, Arriza JL, Aoki C, et al. Beta-arrestin2, a novel member of the arrestin/ beta-arrestin gene family. J Biol Chem. 1992;267:17882–17890. [PubMed] [Google Scholar]
- 8.Sterne-Marr R, Gurevich VV, Goldsmith P, et al. Polypeptide variants of beta-arrestin and arrestin3. J Biol Chem. 1993;268:15640–15648. [PubMed] [Google Scholar]
- 9.Rapoport B, Kaufman KD, Chazenbalk GD. Cloning of a member of the arrestin family from a human thyroid cDNA library. Mol Cell Endocrinol. 1992;84:R39–R43. doi: 10.1016/0303-7207(92)90038-8. [DOI] [PubMed] [Google Scholar]
- 10.Craft CM, Whitmore DH, Wiechmann AF. Cone arrestin identified by targeting expression of a functional family. J Biol Chem. 1994;269:4613–4619. [PubMed] [Google Scholar]
- 11.Sakuma H, Murakami A, Fujimaki T, Inana G. Isolation and characterization of the human X-arrestin gene. Gene. 1998;224:87–95. doi: 10.1016/s0378-1119(98)00510-1. [DOI] [PubMed] [Google Scholar]
- 12.Gurevich EV, Gurevich VV. Arrestins: ubiquitous regulators of cellular signaling pathways. Genome Biol. 2006;7:236. doi: 10.1186/gb-2006-7-9-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gurevich VV, Hanson SM, Song X, Vishnivetskiy SA, Gurevich EV. The functional cycle of visual arrestins in photoreceptor cells. Prog Retin Eye Res. 2011;30:405–430. doi: 10.1016/j.preteyeres.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gurevich VV, Gurevich EV. The structural basis of arrestin-mediated regulation of G-protein-coupled receptors. Pharm Ther. 2006;110:465–502. doi: 10.1016/j.pharmthera.2005.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiao K, McClatchy DB, Shukla AK, et al. Functional specialization of beta-arrestin interactions revealed by proteomic analysis. Proc Natl Acad Sci USA. 2007;104:12011–12016. doi: 10.1073/pnas.0704849104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shinohara T, Dietzschold B, Craft CM, et al. Primary and secondary structure of bovine retinal S antigen (48-kDa protein) Proc Natl Acad Sci USA. 1987;84:6975–6979. doi: 10.1073/pnas.84.20.6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Granzin J, Wilden U, Choe HW, Labahn J, Krafft B, Buldt G. X-ray crystal structure of arrestin from bovine rod outer segments. Nature. 1998;391:918–921. doi: 10.1038/36147. [DOI] [PubMed] [Google Scholar]
- 18.Hirsch JA, Schubert C, Gurevich VV, Sigler PB. The 2.8 A crystal structure of visual arrestin: a model for arrestin’s regulation. Cell. 1999;97:257–269. doi: 10.1016/s0092-8674(00)80735-7. [DOI] [PubMed] [Google Scholar]
- 19.Han M, Gurevich VV, Vishnivetskiy SA, Sigler PB, Schubert C. Crystal structure of beta-arrestin at 1.9 A: possible mechanism of receptor binding and membrane Translocation. Structure. 2001;9:869–880. doi: 10.1016/s0969-2126(01)00644-x. [DOI] [PubMed] [Google Scholar]
- 20.Zhan X, Gimenez LE, Gurevich VV, Spiller BW. Crystal structure of arrestin-3 reveals the basis of the difference in receptor binding between two non-visual subtypes. J Mol Biol. 2011;406:467–478. doi: 10.1016/j.jmb.2010.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Milano SK, Pace HC, Kim YM, Brenner C, Benovic JL. Scaffolding functions of arrestin-2 revealed by crystal structure and mutagenesis. Biochemistry. 2002;41:3321–3328. doi: 10.1021/bi015905j. [DOI] [PubMed] [Google Scholar]
- 22.Sutton RB, Vishnivetskiy SA, Robert J, et al. Crystal structure of cone arrestin at 2.3A: evolution of receptor specificity. J Mol Biol. 2005;354:1069–1080. doi: 10.1016/j.jmb.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 23.Granzin J, Cousin A, Weirauch M, Schlesinger R, Büldt G, Batra-Safferling R. Crystal structure of p44, a constitutively active splice variant of visual arrestin. J Mol Biol. 2012;416:611–618. doi: 10.1016/j.jmb.2012.01.028. [DOI] [PubMed] [Google Scholar]
- 24.Gurevich VV, Benovic JL. Cell-free expression of visual arrestin. Truncation mutagenesis identifies multiple domains involved in rhodopsin interaction. J Biol Chem. 1992;267:21919–21923. [PubMed] [Google Scholar]
- 25.Gurevich VV, Benovic JL. Visual arrestin interaction with rhodopsin. Sequential multisite binding ensures strict selectivity toward light-activated phosphorylated rhodopsin. J Biol Chem. 1993;268:11628–11638. [PubMed] [Google Scholar]
- 26.Gurevich VV, Chen C-Y, Kim CM, Benovic JL. Visual arrestin binding to rhodopsin. Intramolecular interaction between the basic N terminus and acidic C terminus of arrestin may regulate binding selectivity. J Biol Chem. 1994;269:8721–8727. [PubMed] [Google Scholar]
- 27.Gurevich VV, Benovic JL. Visual arrestin binding to rhodopsin. Diverse functional roles of positively charged residues within the phosphorylation-recognition region of arrestin. J Biol Chem. 1995;270:6010–6016. doi: 10.1074/jbc.270.11.6010. [DOI] [PubMed] [Google Scholar]
- 28.Gurevich VV, Benovic JL. Mechanism of phosphorylation-recognition by visual arrestin and the transition of arrestin into a high affinity binding state. Mol Pharmacol. 1997;51:161–169. doi: 10.1124/mol.51.1.161. [DOI] [PubMed] [Google Scholar]
- 29.Vishnivetskiy SA, Paz CL, Schubert C, Hirsch JA, Sigler PB, Gurevich VV. How does arrestin respond to the phosphorylated state of rhodopsin? J Biol Chem. 1999;274:11451–11454. doi: 10.1074/jbc.274.17.11451. [DOI] [PubMed] [Google Scholar]
- 30.Gurevich VV, Gurevich EV. The molecular acrobatics of arrestin activation. Trends Pharmacol Sci. 2004;25:105–111. doi: 10.1016/j.tips.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 31.Gurevich VV, Gurevich EV. The new face of active receptor bound arrestin attracts new partners. Structure. 2003;11:1037–1042. doi: 10.1016/s0969-2126(03)00184-9. [DOI] [PubMed] [Google Scholar]
- 32.Nair KS, Hanson SM, Mendez A, et al. Light-dependent redistribution of arrestin in vertebrate rods is an energy-independent process governed by protein-protein interactions. Neuron. 2005;46:555–567. doi: 10.1016/j.neuron.2005.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nair KS, Hanson SM, Kennedy MJ, Hurley JB, Gurevich VV, Slepak VZ. Direct binding of visual arrestin to microtubules determines the differential subcellular localization of its splice variants in rod photoreceptors. J Biol Chem. 2004;279:41240–41248. doi: 10.1074/jbc.M406768200. [DOI] [PubMed] [Google Scholar]
- 34.Hanson SM, Francis DJ, Vishnivetskiy SA, Klug CS, Gurevich VV. Visual arrestin binding to microtubules involves a distinct conformational change. J Biol Chem. 2006;281:9765–9772. doi: 10.1074/jbc.M510738200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hanson SM, Cleghorn WM, Francis DJ, et al. Arrestin mobilizes signaling proteins to the cytoskeleton and redirects their activity. J Mol Biol. 2007;368:375–387. doi: 10.1016/j.jmb.2007.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palczewski K, Kumasaka T, Hori T, et al. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 37.Okada T, Fujiyoshi Y, Silow M, Navarro J, Landau EM, Shichida Y. Functional role of internal water molecules in rhodopsin revealed by X-ray crystallography. Proc Natl Acad Sci USA. 2002;99:5982–5987. doi: 10.1073/pnas.082666399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li J, Edwards PC, Burghammer M, Villa C, Schertler GF. Structure of bovine rhodopsin in a trigonal crystal form. J Mol Biol. 2004;343:1409–1438. doi: 10.1016/j.jmb.2004.08.090. [DOI] [PubMed] [Google Scholar]
- 39.Standfuss J, Xie G, Edwards PC, Burghammer M, Oprian DD, Schertler GF. Crystal structure of a thermally stable rhodopsin mutant. J Mol Biol. 2007;372:1179–1188. doi: 10.1016/j.jmb.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cherezov V, Rosenbaum DM, Hanson MA, et al. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chien EYT, Liu W, Zhao Q, et al. Structure of the human dopamine D3 receptor in complex with a D2/D3 selective antagonist. Science. 2010;330:1091–1095. doi: 10.1126/science.1197410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rosenbaum DM, Zhang C, Lyons JA, et al. Structure and function of an irreversible agonist-β(2) adrenoceptor complex. Nature. 2011;469:236–240. doi: 10.1038/nature09665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu B, Chien EYT, Mol CD, et al. Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science. 2010;330:1066–1071. doi: 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rasmussen SG, Choi HJ, Rosenbaum DM, et al. Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 45.Haga K, Kruse AC, Asada H, et al. Structure of the human M2 muscarinic acetylcholine receptor bound to an antagonist. Nature. 2012;482:547–551. doi: 10.1038/nature10753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kruse AC, Hu J, Pan AC, et al. Structure and dynamics of the M3 muscarinic acetylcholine receptor. Nature. 2012;482:552–556. doi: 10.1038/nature10867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Manglik A, Kruse AC, Kobilka TS, et al. Crystal structure of the μ-opioid receptor bound to a morphinan antagonist. Nature. 2012;485:321–326. doi: 10.1038/nature10954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jaakola VP, Griffith MT, Hanson MA, et al. The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science. 2008;322:1211–1217. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu F, Wu H, Katritch V, et al. Structure of an agonist-bound human A2A adenosine receptor. Science. 2011;332:322–327. doi: 10.1126/science.1202793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shimamura T, Shiroishi M, Weyand S, et al. Structure of the human histamine H1 receptor complex with doxepin. Nature. 2011;475:65–70. doi: 10.1038/nature10236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu H, Wacker D, Mileni M, et al. Structure of the human κ-opioid receptor in complex with JDTic. Nature. 2012;485:327–332. doi: 10.1038/nature10939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moukhametzianov R, Warne T, Edwards PC, et al. Two distinct conformations of helix 6 observed in antagonist-bound structures of a beta1-adrenergic receptor. Proc Natl Acad Sci USA. 2011;108:8228–8232. doi: 10.1073/pnas.1100185108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Warne T, Serrano-Vega MJ, Baker JG, et al. Structure of a beta1-adrenergic G-protein-coupled receptor. Nature. 2008;454:486–491. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Doré AS, Robertson N, Errey JC, et al. Structure of the adenosine A(2A) receptor in complex with ZM241385 and the xanthines XAC and caffeine. Structure. 2011;19:1283–1293. doi: 10.1016/j.str.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hubbell WL, Altenbach C, Hubbell CM, Khorana HG. Rhodopsin structure, dynamics, and activation: a perspective from crystallography, site-directed spin labeling, sulf-hydryl reactivity, and disulfide cross-linking. Adv Protein Chem. 2003;63:243–290. doi: 10.1016/s0065-3233(03)63010-x. [DOI] [PubMed] [Google Scholar]
- 56.Altenbach C, Kusnetzow AK, Ernst OP, Hofmann KP, Hubbell WL. High-resolution distance mapping in rhodopsin reveals the pattern of helix movement due to activation. Proc Natl Acad Sci USA. 2008;105:7439–7444. doi: 10.1073/pnas.0802515105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Farrens DL, Altenbach C, Yang K, Hubbell WL, Khorana HG. Requirement of rigid-body motion of transmembrane helices for light activation of rhodopsin. Science. 1996;274:768–770. doi: 10.1126/science.274.5288.768. [DOI] [PubMed] [Google Scholar]
- 58.Resek JF, Farahbakhsh ZT, Hubbell WL, Khorana HG. Formation of the meta II photointermediate is accompanied by conformational changes in the cytoplasmic surface of rhodopsin. Biochemistry. 1993;32:12025–12032. doi: 10.1021/bi00096a012. [DOI] [PubMed] [Google Scholar]
- 59.Farahbakhsh ZT, Ridge KD, Khorana HG, Hubbell WL. Mapping light-dependent structural changes in the cytoplasmic loop connecting helices C and D in rhodopsin: a site-directed spin labeling study. Biochemistry. 1995;34:8812–8819. doi: 10.1021/bi00027a033. [DOI] [PubMed] [Google Scholar]
- 60.Scheerer P, Park JH, Hildebrand PW, et al. Crystal structure of opsin in its G-protein-interacting conformation. Nature. 2008;455:497–502. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- 61.Park JH, Scheerer P, Hofmann KP, Choe HW, Ernst OP. Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature. 2008;454:183–187. doi: 10.1038/nature07063. [DOI] [PubMed] [Google Scholar]
- 62.Choe HW, Kim YJ, Park JH, et al. Crystal structure of metarhodopsin II. Nature. 2011;471:651–655. doi: 10.1038/nature09789. [DOI] [PubMed] [Google Scholar]
- 63.Deupi X, Edwards P, Singhal A, et al. Stabilized G protein binding site in the structure of constitutively active metarhodopsin-II. Proc Natl Acad Sci USA. 2012;109:119–124. doi: 10.1073/pnas.1114089108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lebon G, Warne T, Edwards PC, et al. Agonist-bound adenosine A2A receptor structures reveal common features of GPCR activation. Nature. 2011;474:521–525. doi: 10.1038/nature10136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rasmussen SG, Choi HJ, Fung JJ, et al. Structure of a nanobody-stabilized active state of the β(2) adrenoceptor. Nature. 2011;469:175–180. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rasmussen SG, DeVree BT, Zou Y, et al. Crystal structure of the β2 adrenergic receptor-Gs protein complex. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chung KY, Rasmussen SG, Liu T, et al. Conformational changes in the G protein Gs induced by the β2 adrenergic receptor. Nature. 2011;477:611–615. doi: 10.1038/nature10488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kobilka BK. G protein coupled receptor structure and activation. Biochim Biophys Acta. 2007;1768:794–807. doi: 10.1016/j.bbamem.2006.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kobilka BK. Cold Spring Harbor Asia membrane protein meeting; May 16–20, 2011; p. 79. [Google Scholar]
- 70.Warne T, Moukhametzianov R, Baker JG, et al. The structural basis for agonist and partial agonist action on a β(1)-adrenergic receptor. Nature. 2011;469:241–244. doi: 10.1038/nature09746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vishnivetskiy SA, Gimenez LE, Francis DJ, et al. Few residues within an extensive binding interface drive receptor interaction and determine the specificity of arrestin proteins. J Biol Chem. 2011;286:24288–24299. doi: 10.1074/jbc.M110.213835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim M, Vishnivetskiy SA, Van Eps N, et al. Conformation of receptor-bound visual arrestin. Proc Natl Acad Sci USA. 2012;109:18407–18412. doi: 10.1073/pnas.1216304109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hanson SM, Francis DJ, Vishnivetskiy SA, et al. Differential interaction of spin-labeled arrestin with inactive and active phosphorhodopsin. Proc Natl Acad Sci USA. 2006;103:4900–4905. doi: 10.1073/pnas.0600733103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.DeGraff JL, Gurevich VV, Benovic JL. The third intracellular loop of alpha 2-adrenergic receptors determines subtype specificity of arrestin interaction. J Biol Chem. 2002;277:43247–43252. doi: 10.1074/jbc.M207495200. [DOI] [PubMed] [Google Scholar]
- 75.Gelber EI, Kroeze WK, Willins DL, et al. Structure and function of the third intracellular loop of the 5-hydroxytryptamine2A receptor: the third intracellular loop is alpha-helical and binds purified arrestins. J Neurochem. 1999;72:2206–2214. doi: 10.1046/j.1471-4159.1999.0722206.x. [DOI] [PubMed] [Google Scholar]
- 76.Raman D, Osawa S, Gurevich VV, Weiss ER. The interaction with the cytoplasmic loops of rhodopsin plays a crucial role in arrestin activation and binding. J Neurochem. 2003;84:1040–1050. doi: 10.1046/j.1471-4159.2003.01598.x. [DOI] [PubMed] [Google Scholar]
- 77.Vishnivetskiy SA, Schubert C, Climaco GC, Gurevich YV, Velez M-G, Gurevich VV. An additional phosphate-binding element in arrestin molecule. Implications for the mechanism of arrestin activation. J Biol Chem. 2000;275:41049–41057. doi: 10.1074/jbc.M007159200. [DOI] [PubMed] [Google Scholar]
- 78.Ohguro H, Palczewski K, Walsh KA, Johnson RS. Topographic study of arrestin using differential chemical modifications and hydrogen/deuterium exchange. Protein Sci. 1994;3:2428–2434. doi: 10.1002/pro.5560031226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vishnivetskiy SA, Hosey MM, Benovic JL, Gurevich VV. Mapping the arrestin-receptor interface. Structural elements responsible for receptor specificity of arrestin proteins. J Biol Chem. 2004;279:1262–1268. doi: 10.1074/jbc.M308834200. [DOI] [PubMed] [Google Scholar]
- 80.Gurevich VV, Dion SB, Onorato JJ, et al. Arrestin interactions with G protein-coupled receptors. Direct binding studies of wild type and mutant arrestins with rhodopsin, beta 2-adrenergic, and m2 muscarinic cholinergic receptors. J Biol Chem. 1995;270:720–731. doi: 10.1074/jbc.270.2.720. [DOI] [PubMed] [Google Scholar]
- 81.Pulvermuller A, Schroder K, Fischer T, Hofmann KP. Interactions of metarhodopsin II. Arrestin peptides compete with arrestin and transducin. J Biol Chem. 2000;275:37679–37685. doi: 10.1074/jbc.M006776200. [DOI] [PubMed] [Google Scholar]
- 82.Dinculescu A, McDowell JH, Amici SA, et al. Insertional mutagenesis and immunochemical analysis of visual arrestin interaction with rhodopsin. J Biol Chem. 2002;277:11703–11708. doi: 10.1074/jbc.M111833200. [DOI] [PubMed] [Google Scholar]
- 83.Feuerstein SE, Pulvermüller A, Hartmann R, et al. Helix formation in arrestin accompanies recognition of photoactivated rhodopsin. Biochemistry. 2009;48:10733–10742. doi: 10.1021/bi900544p. [DOI] [PubMed] [Google Scholar]
- 84.Zhuang T, Vishnivetskiy SA, Gurevich VV, Sanders CR. Elucidation of inositol hexaphosphate and heparin interaction sites and conformational changes in arrestin-1 by solution nuclear magnetic resonance. Biochemistry. 2010;49:10473–10485. doi: 10.1021/bi101596g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhuang T, Chen Q, Cho M-K, et al. Involvement of distinct arrestin-1 elements in binding to different functional forms of rhodopsin. Proc Natl Acad Sci USA. 2013;110:942–947. doi: 10.1073/pnas.1215176110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vishnivetskiy SA, Francis DJ, Van Eps N, et al. The role of arrestin alpha-helix I in receptor binding. J Mol Biol. 2010;395:42–54. doi: 10.1016/j.jmb.2009.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vishnivetskiy SA, Raman D, Wei J, Kennedy MJ, Hurley JB, Gurevich VV. Regulation of arrestin binding by rhodopsin phosphorylation level. J Biol Chem. 2007;282:32075–32083. doi: 10.1074/jbc.M706057200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Krasel C, Bunemann M, Lorenz K, Lohse MJ. Beta-arrestin binding to the beta2-adrenergic receptor requires both receptor phosphorylation and receptor activation. J Biol Chem. 2005;280:9528–9535. doi: 10.1074/jbc.M413078200. [DOI] [PubMed] [Google Scholar]
- 89.Gimenez LE, Kook S, Vishnivetskiy SA, Ahmed MR, Gurevich EV, Gurevich VV. Role of receptor-attached phosphates in binding of visual and non-visual arrestins to G protein-coupled receptors. J Biol Chem. 2012;287:9028–9040. doi: 10.1074/jbc.M111.311803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mendez A, Burns ME, Roca A, et al. Rapid and reproducible deactivation of rhodopsin requires multiple phosphorylation sites. Neuron. 2000;28:153–164. doi: 10.1016/s0896-6273(00)00093-3. [DOI] [PubMed] [Google Scholar]
- 91.Song X, Vishnivetskiy SA, Gross OP, et al. Enhanced arrestin facilitates recovery and protects rods lacking rhodopsin phosphorylation. Curr Biol. 2009;19:700–705. doi: 10.1016/j.cub.2009.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wilden U, Kuhn H. Light-dependent phosphorylation of rhodopsin: number of phosphorylation sites. Biochemistry. 1982;21:3014–3022. doi: 10.1021/bi00541a032. [DOI] [PubMed] [Google Scholar]
- 93.Bouvier M, Hausdorff WP, De Blasi A, et al. Removal of phosphorylation sites from the beta 2-adrenergic receptor delays onset of agonist-promoted desensitization. Nature. 1988;333:370–373. doi: 10.1038/333370a0. [DOI] [PubMed] [Google Scholar]
- 94.Fredericks ZL, Pitcher JA, Lefkowitz RJ. Identification of the G protein-coupled receptor kinase phosphorylation sites in the human beta2-adrenergic receptor. J Biol Chem. 1996;271:13796–13803. doi: 10.1074/jbc.271.23.13796. [DOI] [PubMed] [Google Scholar]
- 95.Hausdorff WP, Bouvier M, O’Dowd BF, Irons GP, Caron MG, Lefkowitz RJ. Phosphorylation sites on two domains of the beta 2-adrenergic receptor are involved in distinct pathways of receptor desensitization. J Biol Chem. 1989;264:12657–12665. [PubMed] [Google Scholar]
- 96.Krasel C, Zabel U, Lorenz K, Reiner S, Al-Sabah S, Lohse MJ. Dual role of the beta2-adrenergic receptor C terminus for the binding of beta-arrestin and receptor internalization. J Biol Chem. 2008;283:31840–31848. doi: 10.1074/jbc.M806086200. [DOI] [PubMed] [Google Scholar]
- 97.Seibold A, January BG, Friedman J, Hipkin RW, Clark RB. Desensitization of beta2-adrenergic receptors with mutations of the proposed G protein-coupled receptor kinase phosphorylation sites. J Biol Chem. 1998;273:7637–7642. doi: 10.1074/jbc.273.13.7637. [DOI] [PubMed] [Google Scholar]
- 98.Seibold A, Williams B, Huang ZF, et al. Localization of the sites mediating desensitization of the beta(2)-adrenergic receptor by the GRK pathway. Mol Pharmacol. 2000;58:1162–1173. doi: 10.1124/mol.58.5.1162. [DOI] [PubMed] [Google Scholar]
- 99.Trester-Zedlitz M, Burlingame A, Kobilka B, von Zastrow M. Mass spectrometric analysis of agonist effects on posttranslational modifications of the beta-2 adrenoceptor in mammalian cells. Biochemistry. 2005;44:6133–6143. doi: 10.1021/bi0475469. [DOI] [PubMed] [Google Scholar]
- 100.Vaughan DJ, Millman EE, Godines V, et al. Role of the G protein-coupled receptor kinase site serine cluster in beta2-adrenergic receptor internalization, desensitization, and beta-arrestin translocation. J Biol Chem. 2006;281:7684–7692. doi: 10.1074/jbc.M500328200. [DOI] [PubMed] [Google Scholar]
- 101.Lee KB, Ptasienski JA, Pals-Rylaarsdam R, Gurevich VV, Hosey MM. Arrestin binding to the M(2) muscarinic acetylcholine receptor is precluded by an inhibitory element in the third intracellular loop of the receptor. J Biol Chem. 2000;275:9284–9289. doi: 10.1074/jbc.275.13.9284. [DOI] [PubMed] [Google Scholar]
- 102.Pals-Rylaarsdam R, Hosey MM. Two homologous phosphorylation domains differentially contribute to desensitization and internalization of the m2 muscarinic acetylcholine receptor. J Biol Chem. 1997;272:14152–14158. doi: 10.1074/jbc.272.22.14152. [DOI] [PubMed] [Google Scholar]
- 103.Pals-Rylaarsdam R, Gurevich VV, Lee KB, Ptasienski J, Benovic JL, Hosey MM. Internalization of the m2 muscarinic acetylcholine receptor. Arrestin-independent and -dependent pathways. J Biol Chem. 1997;272:23682–23689. doi: 10.1074/jbc.272.38.23682. [DOI] [PubMed] [Google Scholar]
- 104.Schleicher A, Kuhn H, Hofmann KP. Kinetics, binding constant, and activation energy of the 48-kDa protein-rhodopsin complex by extra-metarhodopsin II. Biochemistry. 1989;28:1770–1775. doi: 10.1021/bi00430a052. [DOI] [PubMed] [Google Scholar]
- 105.Vishnivetskiy SA, Hirsch JA, Velez M-G, Gurevich YV, Gurevich VV. Transition of arrestin into the active receptor-binding state requires an extended interdomain hinge. J Biol Chem. 2002;277:43961–43968. doi: 10.1074/jbc.M206951200. [DOI] [PubMed] [Google Scholar]
- 106.Kim M, Vishnivetskiy SA, Van Eps N, et al. ASBMB meeting; 2011; pp. 750–759. [Google Scholar]
- 107.Fotiadis D, Jastrzebska B, Philippsen A, Muller DJ, Palczewski K, Engel A. Structure of the rhodopsin dimer: a working model for G-protein-coupled receptors. Curr Opin Struct Biol. 2006;16:252–259. doi: 10.1016/j.sbi.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 108.Chen J, Makino CL, Peachey NS, Baylor DA, Simon MI. Mechanisms of rhodopsin inactivation in vivo as revealed by a COOH-terminal truncation mutant. Science. 1995;267:374–377. doi: 10.1126/science.7824934. [DOI] [PubMed] [Google Scholar]
- 109.Palczewski K, Pulvermuller A, Buczylko J, Hofmann KP. Phosphorylated rhodopsin and heparin induce similar conformational changes in arrestin. J Biol Chem. 1991;266:18649–18654. [PubMed] [Google Scholar]
- 110.Hanson SM, Gurevich EV, Vishnivetskiy SA, Ahmed MR, Song X, Gurevich VV. Each rhodopsin molecule binds its own arrestin. Proc Natl Acad Sci USA. 2007;104:3125–3128. doi: 10.1073/pnas.0610886104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Broekhuyse RM, Tolhuizen EF, Janssen AP, Winkens HJ. Photoreceptor cellspecific localization of S-antigen in retina. Curr Eye Res. 1985;4:613–618. doi: 10.3109/02713688508999993. [DOI] [PubMed] [Google Scholar]
- 112.Whelan JP, McGinnis JF. Light-dependent subcellular movement of photoreceptor proteins. J Neurosci Res. 1988;20:263–270. doi: 10.1002/jnr.490200216. [DOI] [PubMed] [Google Scholar]
- 113.Elias RV, Sezate SS, Cao W, McGinnis JF. Temporal kinetics of the light/dark translocation and compartmentation of arrestin and alpha-transducin in mouse photoreceptor cells. Mol Vis. 2004;10:672–681. [PubMed] [Google Scholar]
- 114.Mendez A, Lem J, Simon M, Chen J. Light-dependent translocation of arrestin in the absence of rhodopsin phosphorylation and transducin signaling. J Neurosci. 2003;23:3124–3129. doi: 10.1523/JNEUROSCI.23-08-03124.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Strissel KJ, Sokolov M, Trieu LH, Arshavsky VY. Arrestin translocation is induced at a critical threshold of visual signaling and is superstoichiometric to bleached rhodopsin. J Neurosci. 2006;26:1146–1153. doi: 10.1523/JNEUROSCI.4289-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Song X, Vishnivetskiy SA, Seo J, Chen J, Gurevich EV, Gurevich VV. Arrestin-1 expression level in rods: balancing functional performance and photoreceptor health. Neuroscience. 2011;174:37–49. doi: 10.1016/j.neuroscience.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cleghorn WM, Tsakem EL, Song X, et al. Progressive reduction of its expression in rods reveals two pools of arrestin-1 in the outer segment with different roles in photoresponse recovery. PLoS One. 2011;6:e22797. doi: 10.1371/journal.pone.0022797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Schubert C, Hirsch JA, Gurevich VV, Engelman DM, Sigler PB, Fleming KG. How does arrestin respond to the phosphorylated state of rhodopsin? J Biol Chem. 1999;274:21186–21190. doi: 10.1074/jbc.274.17.11451. [DOI] [PubMed] [Google Scholar]
- 119.Imamoto Y, Tamura C, Kamikubo H, Kataoka M. Concentration-dependent tetramerization of bovine visual arrestin. Biophys J. 2003;85:1186–1195. doi: 10.1016/S0006-3495(03)74554-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hanson SM, Van Eps N, Francis DJ, et al. Structure and function of the visual arrestin oligomer. EMBO J. 2007;26:1726–1736. doi: 10.1038/sj.emboj.7601614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kim M, Hanson SM, Vishnivetskiy SA, et al. Robust self-association is a common feature of mammalian visual arrestin-1. Biochemistry. 2011;50:2235–2242. doi: 10.1021/bi1018607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hanson SM, Dawson ES, Francis DJ, et al. A model for the solution structure of the rod arrestin tetramer. Structure. 2008;16:924–934. doi: 10.1016/j.str.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tsukamoto H, Sinha A, Dewitt M, Farrens DL. Monomeric rhodopsin is the minimal functional unit required for arrestin binding. J Mol Biol. 2010;399:501–511. doi: 10.1016/j.jmb.2010.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bayburt TH, Vishnivetskiy SA, McLean M, et al. Monomeric rhodopsin is sufficient for normal rhodopsin kinase (GRK1) phosphorylation and arrestin-1 binding. J Biol Chem. 2011;286:1420–1428. doi: 10.1074/jbc.M110.151043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Celver J, Vishnivetskiy SA, Chavkin C, Gurevich VV. Conservation of the phosphate-sensitive elements in the arrestin family of proteins. J Biol Chem. 2002;277:9043–9048. doi: 10.1074/jbc.M107400200. [DOI] [PubMed] [Google Scholar]
- 126.Kovoor A, Celver J, Abdryashitov RI, Chavkin C, Gurevich VV. Targeted construction of phosphorylation-independent beta-arrestin mutants with constitutive activity in cells. J Biol Chem. 1999;274:6831–6834. doi: 10.1074/jbc.274.11.6831. [DOI] [PubMed] [Google Scholar]
- 127.Gurevich VV, Pals-Rylaarsdam R, Benovic JL, Hosey MM, Onorato JJ. Agonist-receptor-arrestin, an alternative ternary complex with high agonist affinity. J Biol Chem. 1997;272:28849–28852. doi: 10.1074/jbc.272.46.28849. [DOI] [PubMed] [Google Scholar]
- 128.Pan L, Gurevich EV, Gurevich VV. The nature of the arrestin x receptor complex determines the ultimate fate of the internalized receptor. J Biol Chem. 2003;278:11623–11632. doi: 10.1074/jbc.M209532200. [DOI] [PubMed] [Google Scholar]
- 129.Celver J, Lowe J, Kovoor A, Gurevich VV, Chavkin C. Threonine 180 is required for G-protein-coupled receptor kinase 3- and beta-arrestin 2-mediated desensitization of the mu-opioid receptor in Xenopus oocytes. J Biol Chem. 2001;276:4894–4900. doi: 10.1074/jbc.M007437200. [DOI] [PubMed] [Google Scholar]
- 130.Lowe JD, Celver JP, Gurevich VV, Chavkin C. mu-Opioid receptors desensitize less rapidly than delta-opioid receptors due to less efficient activation of arrestin. J Biol Chem. 2002;277:15729–15735. doi: 10.1074/jbc.M200612200. [DOI] [PubMed] [Google Scholar]
- 131.Sommer ME, Farrens DL, McDowell JH, Weber LA, Smith WC. Dynamics of arrestin-rhodopsin interactions: loop movement is involved in arrestin activation and receptor binding. J Biol Chem. 2007;282:25560–25568. doi: 10.1074/jbc.M702155200. [DOI] [PubMed] [Google Scholar]
- 132.Hanson SM, Gurevich VV. The differential engagement of arrestin surface charges by the various functional forms of the receptor. J Biol Chem. 2006;281:3458–3462. doi: 10.1074/jbc.M512148200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Coffa S, Breitman M, Hanson SM, et al. The effect of arrestin conformation on the recruitment of c-Raf1, MEK1, and ERK1/2 activation. PLoS One. 2011;6:e28723. doi: 10.1371/journal.pone.0028723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Song X, Raman D, Gurevich EV, Vishnivetskiy SA, Gurevich VV. Visual and both non-visual arrestins in their “inactive” conformation bind JNK3 and Mdm2 and relocalize them from the nucleus to the cytoplasm. J Biol Chem. 2006;281:21491–21499. doi: 10.1074/jbc.M603659200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ahmed MR, Zhan X, Song X, Kook S, Gurevich VV, Gurevich EV. Ubiquitin ligase parkin promotes Mdm2-arrestin interaction but inhibits arrestin ubiquitination. Biochemistry. 2011;50:3749–3763. doi: 10.1021/bi200175q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Breitman M, Kook S, Gimenez LE, et al. Silent scaffolds: inhibition of c-Jun N-terminal kinase 3 activity in cell by dominant-negative arrestin-3 mutant. J Biol Chem. 2012;287:19653–19664. doi: 10.1074/jbc.M112.358192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Smith WC, Gurevich EV, Dugger DR, et al. Cloning and functional characterization of salamander rod and cone arrestins. Invest Ophthalmol Vis Sci. 2000;41:2445–2455. [PubMed] [Google Scholar]
- 138.Barak LS, Ferguson SS, Zhang J, Caron MG. A beta-arrestin/green fluorescent protein biosensor for detecting G protein-coupled receptor activation. J Biol Chem. 1997;272:27497–27500. doi: 10.1074/jbc.272.44.27497. [DOI] [PubMed] [Google Scholar]
- 139.Walther C, Nagel S, Gimenez LE, Mörl K, Gurevich VV, Beck-Sickinger AG. Ligand-induced internalization and recycling of the human neuropeptide Y2 receptor is regulated by its carboxyl-terminal tail. J Biol Chem. 2010;285:41578–41590. doi: 10.1074/jbc.M110.162156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Gurevich VV, Richardson RM, Kim CM, Hosey MM, Benovic JL. Binding of wild type and chimeric arrestins to the m2 muscarinic cholinergic receptor. J Biol Chem. 1993;268:16879–16882. [PubMed] [Google Scholar]
- 141.Gurevich VV, Gurevich EV. Custom-designed proteins as novel therapeutic tools? The case of arrestins. Expert Rev Mol Med. 2010;12:e13. doi: 10.1017/S1462399410001444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Gurevich VV, Gurevich EV. Synthetic biology with surgical precision: targeted reengineering of signaling proteins. Cell Signal. 2012;24:899–1908. doi: 10.1016/j.cellsig.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Gimenez LE, Vishnivetskiy SA, Baameur F, Gurevich VV. Manipulation of very few receptor discriminator residues greatly enhances receptor specificity of non-visual arrestins. J Biol Chem. 2012;287:29495–29505. doi: 10.1074/jbc.M112.366674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Schöneberg T, Schulz A, Biebermann H, Hermsdorf T, Römpler H, Sangkuhl K. Mutant G-protein-coupled receptors as a cause of human diseases. Pharmacol Ther. 2004;104:173–206. doi: 10.1016/j.pharmthera.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 145.Goodman OB, Jr, Krupnick JG, Santini F, et al. Beta-arrestin acts as a clathrin adaptor in endocytosis of the beta2-adrenergic receptor. Nature. 1996;383:447–450. doi: 10.1038/383447a0. [DOI] [PubMed] [Google Scholar]
- 146.Wu N, Hanson SM, Francis DJ, et al. Arrestin binding to calmodulin: a direct interaction between two ubiquitous signaling proteins. J Mol Biol. 2006;364:955–963. doi: 10.1016/j.jmb.2006.09.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Laporte SA, Oakley RH, Zhang J, et al. The beta2-adrenergic receptor/betaarrestin complex recruits the clathrin adaptor AP-2 during endocytosis. Proc Natl Acad Sci USA. 1999;96:3712–3717. doi: 10.1073/pnas.96.7.3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.McDonald PH, Chow CW, Miller WE, et al. Beta-arrestin 2: a receptor-regulated MAPK scaffold for the activation of JNK3. Science. 2000;290:1574–1577. doi: 10.1126/science.290.5496.1574. [DOI] [PubMed] [Google Scholar]
- 149.Luttrell LM, Roudabush FL, Choy EW, et al. Activation and targeting of extracellular signal-regulated kinases by beta-arrestin scaffolds. Proc Natl Acad Sci USA. 2001;98:2449–2454. doi: 10.1073/pnas.041604898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Shenoy SK, McDonald PH, Kohout TA, Lefkowitz RJ. Regulation of receptor fate by ubiquitination of activated beta 2-adrenergic receptor and beta-arrestin. Science. 2001;294:1307–1313. doi: 10.1126/science.1063866. [DOI] [PubMed] [Google Scholar]
- 151.Krupnick JG, Goodman OB, Jr, Keen JH, Benovic JL. Arrestin/clathrin interaction. Localization of the clathrin binding domain of nonvisual arrestins to the carboxy terminus. J Biol Chem. 1997;272:15011–15016. doi: 10.1074/jbc.272.23.15011. [DOI] [PubMed] [Google Scholar]
- 152.Kang DS, Kern RC, Puthenveedu MA, von Zastrow M, Williams JC, Benovic JL. Structure of an arrestin2-clathrin complex reveals a novel clathrin binding domain that modulates receptor trafficking. J Biol Chem. 2009;284:29860–29872. doi: 10.1074/jbc.M109.023366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kim YM, Benovic JL. Differential roles of arrestin-2 interaction with clathrin and adaptor protein 2 in G protein-coupled receptor trafficking. J Biol Chem. 2002;277:30760–30768. doi: 10.1074/jbc.M204528200. [DOI] [PubMed] [Google Scholar]
- 154.Schmid EM, Ford MG, Burtey A, et al. Role of the AP2 beta-appendage hub in recruiting partners for clathrin-coated vesicle assembly. PLoS Biol. 2006;4:e262. doi: 10.1371/journal.pbio.0040262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Baillie GS, Adams DR, Bhari N, et al. Mapping binding sites for the PDE4D5 cAMP-specific phosphodiesterase to the N- and C-domains of beta-arrestin using spot-immobilized peptide arrays. Biochem J. 2007;404:71–80. doi: 10.1042/BJ20070005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Meng D, Lynch MJ, Huston E, et al. MEK1 binds directly to betaarrestin1, influencing both its phosphorylation by ERK and the timing of its isoprenaline-stimulated internalization. J Biol Chem. 2009;284:11425–11435. doi: 10.1074/jbc.M806395200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Coffa S, Breitman M, Spiller BW, Gurevich VV. A single mutation in arrestin-2 prevents ERK1/2 activation by reducing c-Raf1 binding. Biochemistry. 2011;50:6951–6958. doi: 10.1021/bi200745k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Seo J, Tsakem EL, Breitman M, Gurevich VV. Identification of arrestin-3-specific residues necessary for JNK3 kinase activation. J Biol Chem. 2011;286:27894–27901. doi: 10.1074/jbc.M111.260448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Miller WE, McDonald PH, Cai SF, Field ME, Davis RJ, Lefkowitz RJ. Identification of a motif in the carboxyl terminus of beta-arrestin2 responsible for activation of JNK3. J Biol Chem. 2001;276:27770–27777. doi: 10.1074/jbc.M102264200. [DOI] [PubMed] [Google Scholar]
- 160.Zhan X, Kaoud TS, Dalby KN, Gurevich VV. Nonvisual arrestins function as simple scaffolds assembling the MKK4-JNK3α2 signaling complex. Biochemistry. 2011;50:10520–10529. doi: 10.1021/bi201506g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Song X, Gurevich EV, Gurevich VV. Cone arrestin binding to JNK3 and Mdm2: conformational preference and localization of interaction sites. J Neurochem. 2007;103:1053–1062. doi: 10.1111/j.1471-4159.2007.04842.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Song X, Coffa S, Fu H, Gurevich VV. How does arrestin assemble MAPKs into a signaling complex? J Biol Chem. 2009;284:685–695. doi: 10.1074/jbc.M806124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Scott MG, Le Rouzic E, Perianin A, et al. Differential nucleocytoplasmic shuttling of beta-arrestins. Characterization of a leucine-rich nuclear export signal in beta-arrestin2. J Biol Chem. 2002;277:37693–37701. doi: 10.1074/jbc.M207552200. [DOI] [PubMed] [Google Scholar]
- 164.Shenoy SK, Lefkowitz RJ. Receptor-specific ubiquitination of beta-arrestin directs assembly and targeting of seven-transmembrane receptor signalosomes. J Biol Chem. 2005;280:15315–15324. doi: 10.1074/jbc.M412418200. [DOI] [PubMed] [Google Scholar]
- 165.Gurevich VV. The selectivity of visual arrestin for light-activated phosphorhodopsin is controlled by multiple nonredundant mechanisms. J Biol Chem. 1998;273:15501–15506. doi: 10.1074/jbc.273.25.15501. [DOI] [PubMed] [Google Scholar]
- 166.Gray-Keller MP, Detwiler PB, Benovic JL, Gurevich VV. Arrestin with a single amino acid substitution quenches light-activated rhodopsin in a phosphorylation-independent fashion. Biochemistry. 1997;36:7058–7063. doi: 10.1021/bi963110k. [DOI] [PubMed] [Google Scholar]
- 167.Macey TA, Lowe JD, Chavkin C. Mu opioid receptor activation of ERK1/2 is GRK3 and arrestin dependent in striatal neurons. J Biol Chem. 2006;281:34515–34524. doi: 10.1074/jbc.M604278200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Bruchas MR, Macey TA, Lowe JD, Chavkin C. Kappa opioid receptor activation of p38 MAPK is GRK3- and arrestin-dependent in neurons and astrocytes. J Biol Chem. 2006;281:18081–18089. doi: 10.1074/jbc.M513640200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Hanson SM, Vishnivetskiy SA, Hubbell WL, Gurevich VV. Opposing effects of inositol hexakisphosphate on rod arrestin and arrestin2 self-association. Biochemistry. 2008;47:1070–1075. doi: 10.1021/bi7021359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Nikonov SS, Brown BM, Davis JA, et al. Mouse cones require an arrestin for normal inactivation of phototransduction. Neuron. 2008;59:462–474. doi: 10.1016/j.neuron.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Storez H, Scott MG, Issafras H, et al. Homo- and hetero-oligomerization of beta-arrestins in living cells. J Biol Chem. 2005;280:40210–40215. doi: 10.1074/jbc.M508001200. [DOI] [PubMed] [Google Scholar]
- 172.Milano SK, Kim YM, Stefano FP, Benovic JL, Brenner C. Nonvisual arrestin oligomerization and cellular localization are regulated by inositol hexakisphosphate binding. J Biol Chem. 2006;281:9812–9823. doi: 10.1074/jbc.M512703200. [DOI] [PubMed] [Google Scholar]
- 173.Boularan C, Scott MG, Bourougaa K, et al. Beta-arrestin 2 oligomerization controls the Mdm2-dependent inhibition of p53. Proc Natl Acad Sci USA. 2007;104:18061–18066. doi: 10.1073/pnas.0705550104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Johnson GL, Dohlman HG, Graves LM. MAPK kinase kinases (MKKKs) as a target class for small-molecule inhibition to modulate signaling networks and gene expression. Curr Opin Chem Biol. 2005;9:325–331. doi: 10.1016/j.cbpa.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 175.Pearson G, Robinson F, Beers Gibson T, et al. Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr Rev. 2001;22:153–183. doi: 10.1210/edrv.22.2.0428. [DOI] [PubMed] [Google Scholar]
- 176.Mushegian A, Gurevich VV, Gurevich EV. The origin and evolution of G protein-coupled receptor kinases. PLoS One. 2012;7:e33806. doi: 10.1371/journal.pone.0033806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.DeWire SM, Ahn S, Lefkowitz RJ, Shenoy SK. Beta-arrestins and cell signaling. Annu Rev Physiol. 2007;69:483–510. doi: 10.1146/annurev.physiol.69.022405.154749. [DOI] [PubMed] [Google Scholar]