Abstract
Possible persistence of bacteria in human blood as cell wall deficient forms (L-forms) represents a top research priority for microbiologists. Application of live BCG vaccine and L-form transformation of vaccine strain may display a new intriguing aspect concerning the opportunity for occurrence of unpredictable colonization inside the human body by unusual microbial life forms. L-form cultures were isolated from 141 blood samples of people previously vaccinated with BCG, none with a history of exposure to tuberculosis. Innovative methodology to access the unusual L-form elements derived from human blood was developed. The methodology outlines the path of transformation of non- cultivable L-form element to cultivable bacteria and their adaptation for growth in vitro. All isolates showed typical L-forms growth features (“fried eggs” colonies and biofilm). Electron microscopy revealed morphology evidencing peculiar characteristics of bacterial L-form population (cell wall deficient polymorphic elements of variable shape and size). Regular detection of acid fast bacteria in smears of isolated blood L-form cultures, led us to start their identification by using specific Mycobactrium spp. genetic tests. Forty five of 97 genetically tested blood cultures provided specific positive signals for mycobacteria, confirmed by at least one of the 3 specific assays (16S rRNA PCR; IS6110 Real Time PCR and spoligotyping). In conclusion, the obtained genetic evidence suggests that these L-forms are of mycobacterial origin. As the investigated people had been vaccinated with BCG, we can assume that the identified mycobacterial L-forms may be produced by persisting live BCG vaccine.
Keywords: BCG vaccinated people, human blood, L-forms, mycobacteria, persistence
Introduction
Regardless of general assumptions that the bloodstream in healthy humans is a sterile compartment under normal circumstances, series of papers over the past few decades have reported intriguing findings about observations or isolation of unidentified microorganisms in blood, called by authors “chlamidia-like,” “erythrocyte-associate,” “nanobacteria,” “human bacterial endoparasite,” primitive-phase “endobionts,” “mycoplasma-like,” L-forms, cell wall deficient bacteria and others.1-5 These unidentified pleomorphic bacteria, found in the blood of healthy people, have been described as staphylococcus-like or streptococcal-like microbes; filamentous cocco-bacillary forms; dense bodies etc.6,7 Their existence is still debated and authors' points of views are controversial.8,9 New insights into the human microbiome suggest that bacterial populations may exist not only in the areas of the body in contact with the external environment, but also likely persist in many other body tissues, inside cells of the immune system or in biofilm communities, protected by self-created polymeric matrix.10-12
In 2012, “Human Microbiome Project” has started to identify by molecular genetic sequencing the thousands of microorganisms co-existing with humans.13 Many of them defy detection by culture-based methods. Despite advances in molecular genetic techniques, the status of human blood bacteria remains unproven. Since blood is an unfavorable compartment for bacterial populations, the principal challenge is to understand whether and how bacteria may persist in it. Cell wall deficient state of bacteria, known as bacterial L-cycle, is recognized as an extreme form of life and as the ultimate tool for survival. The possible persistence of bacteria in human blood as cell wall deficient forms (L-forms) and the phenomenon of their un-cultivability represent a top research priority for microbiologists. Discovering the nature of human blood L-form persisters would give answers to many questions on their biological and medical significance.
L-forms of BCG bacilli have been isolated from the blood of people previously vaccinated against TB with BCG vaccine.14 There are also reports about the detection and isolation of BCG bacilli from patients with AIDS many years after their vaccination.15-17 The Bacille Calmette-Guérin (BCG) is a live attenuated strain of M. bovis, first introduced in 1921, and the only vaccine currently licensed for the prevention of tuberculosis. Little is known about the behavior and mechanisms, by which M. bovis BCG bacilli can persist in vaccinated persons.
In this respect, the aim of our study was to isolate L-type bacterial cultures from blood samples of people previously vaccinated with BCG in Bulgaria, determine their cultural and morphological characterization, as well as attempt their genetic identification.
Results
Blood isolates – L-form growth characteristics
Bacterial cultures were isolated from all 141 blood samples of people from Bulgaria, who had been vaccinated with BCG, but did not have a history of exposure to tuberculosis. An innovative aspect of cultivation approach principally was that we managed to adapt the “non- cultivable elements” from blood for growth in vitro. The methodology and the path to their adaptation are outlined in Materials and Methods.
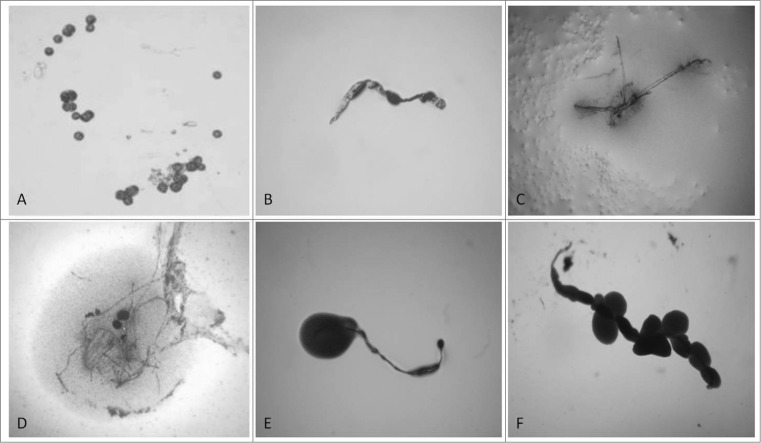
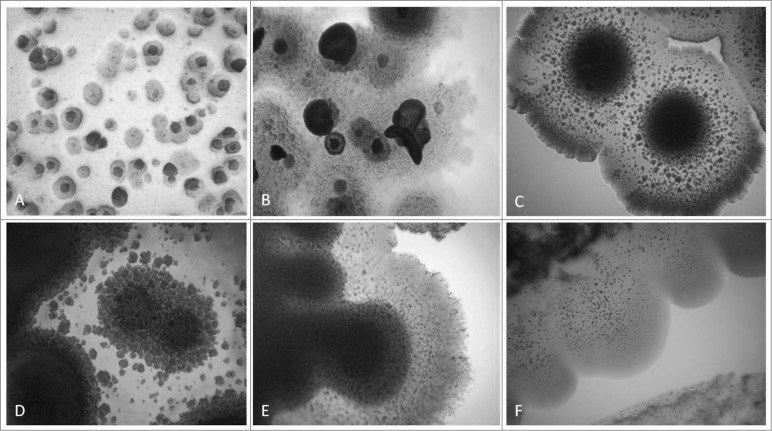
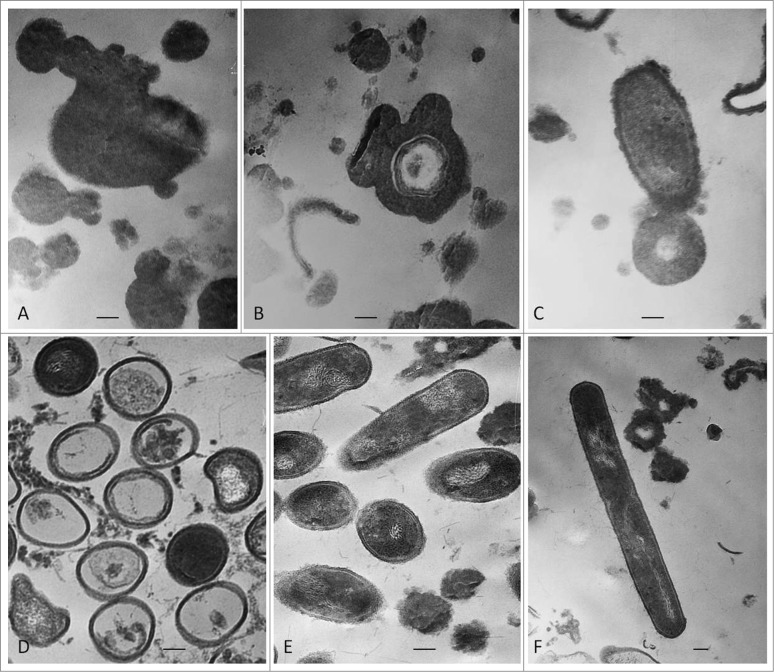
Transformation from “non- cultivable elements” into cultivable bacteria went through 2 phases of sub-cultivations in semisolid medium, each lasting one week. Presence of so called “nonculturable elements” (filamentous and spherical formations) was observed during the first phase/week of cultivation (Figs. 1A, B and C). It is important to note that these elements are distinctive for bacterial L-forms and can be seen only by light microscopy. After the so called “blind passage” and enrichment procedure, during the second sub-cultivation phase, these formations gave rise to visible growth (Figs. 1D, E and F) and formation of typical L-form colonies with “fried eggs” shape or biofilm (Fig. 2). Generally, all cultures obtained from the studied blood samples showed morphological similarities and unique growth characteristics distinctive for L-forms.
Figure 1.
“Non-cultivable elements” from blood isolates (No1; 34; 37, 48; 91; 96). Light microscopy of: (A, B, C) spherical and filamentous forms found in semisolid agar during the first phase/week of cultivation; (D, E, F) initiation of growth from filamentous elements during the second phase/week of cultivation. Magnification: 200x
Figure 2.
Development of L-form growths in semisolid agar during the second phase/week of cultivation. Light microscopy of blood isolates (No1; 34; 37, 48; 91; 96): typical “fried eggs” colonies of different size, consistence and density (A, B, C, D); formation of biofilm with gliding motility at the periphery (E, F). Magnification: 400x
Morphological features of blood L-form cultures
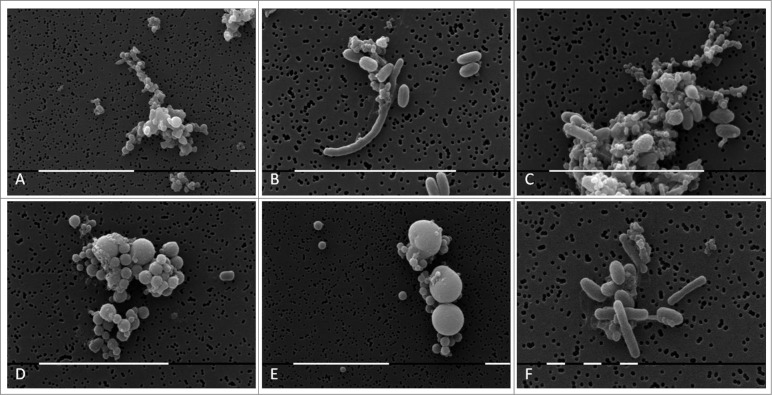
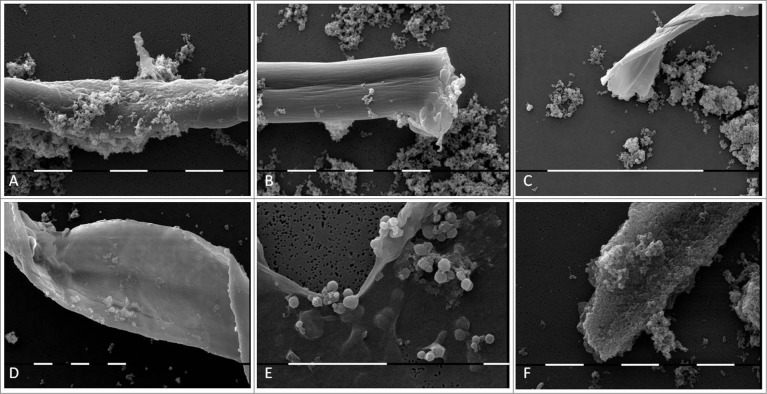
Morphological transformations of “non- cultivable elements” derived from blood and initiation of their multiplication in vitro were observed, using light and electron microscopy. All investigated 141 cultures manifested morphology evidencing peculiar characteristics of bacterial L-forms. Corresponding to the filamentous forms observed in agar during the second phase/week of cultivation and growth associated with them, scanning electron microscopy (SEM) also revealed the presence of huge filamentous and extremely thick membranous formations with many small polymorphic L-bodies fitted together (Fig. 3A–E). Groups of aggregated or tightly packed bacteria were observed, already separated from the large filamentous form (Fig. 3B, C, F). SEM examinations of L-form cultures revealed presence of polymorphic cells of variable shape and size (Fig. 4). Clusters of the smallest granular forms were most commonly found (Fig. 4A). A particularly noteworthy finding was that these granular elements continued to differentiate further into different morphological variants– oval, rod or elongated filamentous cells (Fig. 4B, C, F); coccoid or large spherical L-bodies (Fig. 4D and E).
Figure 3.
SEM of huge filamentous and membranous L-type formations with many small granular, oval or coccoid cells fit together (A-E) and tightly packed rods (F) from blood isolate No1 grown in semisolid agar during the second phase/week of cultivation. Bars: 100 μm (D); 10 μm (A, B, E, F).
Figure 4.
SEM of L-form cells from blood isolate No1 grown in semisolid agar during the second phase/week of cultivation. (A) Cluster of granular forms; (B, C, F) groups of polymorphic granular, oval, rod and elongated filamentous forms; (D, E) typical coccoid and large spherical L-form bodies of different size. Bars: 10 μm (A, B,C, D, E); 1 μm (F).
The ultrastructure of growing L-form cells and their transformations during the first and second phase of cultivation in semisolid agar was observed by transmission electron microscopy (TEM). Transition from cell wall deficient forms with unusual shapes to cells with partially or fully recovered cell wall was noted as a characteristic trend of morphological changes by adaptation in vitro. Figure 5 (A and B) shows a variety of cell wall less spheroidal and branching shapeless bodies of different size and electron density. Vesicular electron-lucent region within the central portion and formation of circular membranous rings inside the large L-bodies were observed (Fig. 5B and C). The L-form bodies in Figure 5A, B, C were taken from semisolid agar during the first week of isolation procedure. After an enrichment “blind” passage during the second week, L-forms started multiplying, going through series of intermediate forms with partly and fully recovered cell walls. A large ellipsoid body surrounded by thick cell coat is seen in Figure 5C. Reverting bacteria showed variability in internal ultrastructure and deposition of wall material. Heterogeneous population of cells with not well defined cytoplasm structure and of varying electron density is seen in Figure 5D. An interesting feature of these cells were the internal areas of “condensed material.” Notably, empty of content cells and such with fully recovered cytoplasm structure were seen as well. Almost fully differentiated cells and defined rod shape morphology are shown in Fig. 5E and F. However, presence of a few L- bodies was still noted.
Figure 5.
TEM of L-form cells from blood isolate No1 grown in semisolid agar: (A-C) cell wall deficient cells observed during the first phase/week of cultivation; (D-F) heterogeneous population of reverting bacteria with partially or fully recovered cell walls found during the second phase/week of cultivation.
Ziehl–Neelsen stained smears were prepared from primary growths and subcultures of all blood isolates and were observed by light microscopy. Although non-acid fast polymorphic bacteria were prevailing in all smears, observation of single or few acid-fast bacteria with typical for mycobacteria rod shaped morphology were noted. Acid fast bacteria were usually seen in about 1% in the observed fields. However, due to the very low presence of acid fast forms in isolated blood cultures, attempts for isolation of typical pure mycobacterial culture on Löwenstein-Jensen medium failed. Nonetheless, the regular detection of acid fast bacteria in smears of isolated blood L-form cultures, led us to begin their identification by using specific for Mycobactrium spp genetic tests.
16S rRNA gene fragment amplification
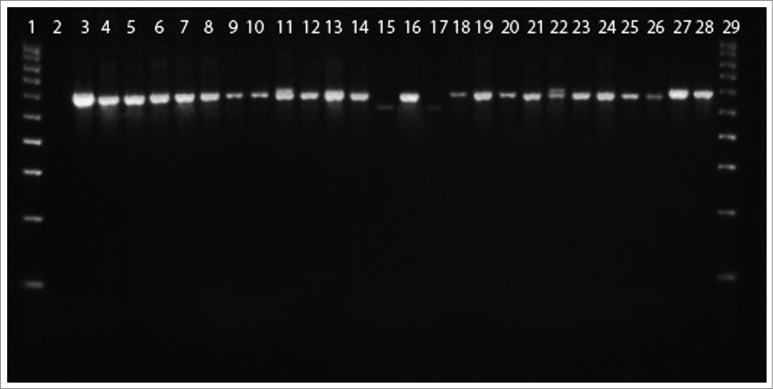
Amplification of 16S rRNA gene fragment confirmed the belonging of 24 tested blood cultures to the genus Mycobacterium. As seen in Figure 6, 22 of them showed specific bands of 650bp and 2 of cultures (no.19 and 23) - smaller bands of 600bp.
Figure 6.
16S rRNA PCR of blood L-form cultures (L) grown during the second phase/week of cultivation in semisolid agar. Legend: 1, DNA ladder 100 bp; 2,Water; 3, L1; 4, L6; 5, L14 6, L47; 7, L58; 8, L60; 9, L62; 10, L64; 11, L65; 12, L70; 13, L96; 14, L16; 15, L19; 16, L20; 17, L23; 18, L25; 19, L26; 20, L34; 21, L37; 22, L41; 23, L52; 24, L53; 25, L91; 26, L93; 27, M. tuberculosis H37Rv 10ˆ-5; 28, M. tuberculosis H37Rv 10ˆ-3; 29, DNA ladder 100 bp.
IS6110 Real Time PCR and spoligotyping
Both tests are specific for M. tuberculosis complex. Of the 97 blood L-form cultures tested for presence of IS6110, 25 were positive. Table 1 presents the Cycle threshold (Ct) values of the positive cultures. Of the 56 blood L-form cultures tested with spoligotyping assay, 39 showing spoligo-profiles were positive. The spoligotyping pattern presented illustratively in Table 2. Spoligo-profiles of blood L-form cultures showed great variability. Some of them were with strongly reduced number of spacers.
Table 1.
IS6110 Real Time PCR of blood L-form cultures grown during the second phase/week of cultivation in semisolid agar.
| Name | Type | Ct |
|---|---|---|
| M. tub. H37Rv | Positive Control | 11.76 |
| MG water | NTC | — |
| 1 | Unknown | 39.26 |
| 6 | Unknown | 42.94 |
| 14 | Unknown | 42.37 |
| 16 | Unknown | 44.57 |
| 19 | Unknown | 43.53 |
| 20 | Unknown | 41.09 |
| 23 | Unknown | 39.89 |
| 25 | Unknown | 41.07 |
| 26 | Unknown | 45.07 |
| 34 | Unknown | 45.30 |
| 37 | Unknown | 43.87 |
| 38 | Unknown | 48.95 |
| 41 | Unknown | 47.53 |
| 47 | Unknown | 43.99 |
| 52 | Unknown | 43.55 |
| 53 | Unknown | 39.55 |
| 58 | Unknown | 42.94 |
| 60 | Unknown | 43.19 |
| 62 | Unknown | 41.10 |
| 64 | Unknown | 42.41 |
| 65 | Unknown | 39.60 |
| 70 | Unknown | 39.32 |
| 91 | Unknown | 41.56 |
| 93 | Unknown | 48.14 |
| 96 | Unknown | 40.02 |
MG-molecular grate water; NTC – non-template control.
Table 2.
Spoligotyping profiles of blood L-form cultures grown during the second phase/week of cultivation in semisolid agar.
| Sample | Spacers (1–43) |
|---|---|
| 1 | 1111111111111101111101111110000000011111010 |
| 2 | 0001000100100000000010000000000010001011001 |
| 3 | 0000000010000000000000001001111000000000000 |
| 5 | 0100000000010000000001100110100000000001000 |
| 6 | 0111110000011000001101111010001000100000011 |
| 7 | 0000001000000000000001010000000011110100000 |
| 9 | 1100000010000000000001000000000111000000000 |
| 10 | 0011010010001001100000101000010000000000011 |
| 14 | 1111000011111000000001100000000000000000011 |
| 16 | 1111110011111001111101111111111010000101111 |
| 17 | 0000001000100000100111010000110000000000000 |
| 18 | 0100010000010000000000010111000000000001000 |
| 20 | 0111111111111101111101111111111111111101011 |
| 23 | 0010110010011100000101111011011100000000001 |
| 25 | 0010010010000000000000100010000000000000001 |
| 34 | 0011010011111000000101100001111100001000111 |
| 36 | 0000000000011101100000101111001001001000001 |
| 37 | 1111010010111101111101101001111100101101111 |
| 38 | 0000000000000000000110010001111000001000000 |
| 40 | 0000010000000000000001101001101000000100000 |
| 43 | 0111111110001001111001010011111100111001011 |
| 44 | 0000101000000000000000000110010010000000000 |
| 45 | 0000100000000000000000001110000100000100100 |
| 46 | 0000000010000000000000010110111010001100000 |
| 47 | 0011111111111100111101111111111111111101111 |
| 48 | 0001100000000000000100001110000000000000000 |
| 51 | 0000000010010100101000011111101010000101111 |
| 53 | 1101111010110000000000110111000010011100000 |
| 54 | 1101101101001000001110110011110010111100101 |
| 55 | 0111110110100001100000101011101100000111001 |
| 57 | 0001000000000000000001000110101001000100000 |
| 58 | 0000010010000000100001001000000000000000011 |
| 59 | 1101111000100000100001111000111111101000100 |
| 60 | 1111111011111101111101111111111011101101011 |
| 62 | 1111010010111101111101101111111111101101011 |
| 64 | 1111111111111101111101111111111011111101011 |
| 65 | 1111110011111101111101111111110001101101111 |
| 70 | 1111111011111100111101111111111011101101111 |
| 96 | 0011110001111101111101111111111111101100010 |
| M. bovis BCG | 1101111101111111111111111111111111111100000 |
| M. tub. H37Rv | 1111111111110011111111111111111100001111111 |
| M. tub. H37Rvˆ10–6 | 0100001111110000011001111111111100001111111 |
(1) –presence of spacer (0) absence of spacer.
The summarized data from the genetically tested 97 blood L-form cultures showed that 45 provided specific positive signals for mycobacteria confirmed at least by one of the used 3 assays. The genetically confirmed positive cultures (1*, 2*, 3*, 5*, 6*, 7, 9, 10, 14, 16, 17,18, 19, 20, 23, 25, 26, 34*, 36*, 37*, 38, 40, 41, 43, 44, 45, 46, 47, 48, 51*, 52*, 53*, 54*, 55*, 57*, 58*, 59, 60, 62, 64, 65*,70, 91, 93, 96) were obtained both from healthy persons, as marked with an asterisk, as well as from persons with non-infectious health problems.
Discussion
Every individual has a unique microbiota, created over the course of their lives. Occurrence of vaccine L-forms seems an interesting fact in terms of evaluating live vaccine strain characteristics associated mostly with their behavior and features in vivo. Cell wall deficiency facilitates bacterial survival and persistence under unfavorable conditions in hosts.18-21 BCG is a live mycobacterial vaccine whose effectiveness has been the subject of various scientific debates but the persistence of the L-forms in vaccinated people has hardly ever been discussed. Application of live BCG vaccine and possible bacterial L-form transformation of the vaccine strain may display a new intriguing aspect concerning the opportunity for occurrence of unpredictable colonization inside the human body by unusual life microbial forms. Despite wide use of BCG worldwide, vaccination policies and practices vary in great range and have changed within and across countries over the years. While BCG vaccination has been never recommended in USA for routine use, BCG is given routinely to newborns in Bulgaria since 1952 to date.22
In our previous study, we found that M. bovis BCG can convert to cell wall deficient forms (L-forms) inside macrophages during infection in guinea pigs and that the L-conversion phenomenon enhanced their in vivo survival and persistence abilities significantly.23 Further, we found existence of filterable forms in commercial BCG vaccine, which are able to develop L-form populations, under the appropriate conditions.24 Therefore, we assume that L-forms of BCG vaccine may persist in the body of vaccinated people.
The current study demonstrates successful isolation of L-form cultures from blood of Bulgarian people, all vaccinated with BCG. “Non-cultivable” L-form elements derived from blood were adapted for in vitro growth by innovative enrichment methodology. Transition from unusual L-form structures (huge filaments, large and small L-bodies or very small granules) to more differentiated cells were manifested very well by electron microscopy. The growth characteristics of the isolated blood cultures from 141 persons unequivocally demonstrated typical for the bacterial L-forms transformations and developmental stages. Formation of distinctive “fried eggs” colonies corresponded to the observed by TEM polymorphic cell wall deficient cells of different shape and size. Of special interest was the formation of huge filamentous and thick membranous structures enveloping L-form population. It was observed that small cell elements, mainly granules, were released from these giant formations. It should be noted that the similar structures were found to be produced by Mycobacterium bovis L-forms under stress conditions.25 It is believed that they have a role in protection against unfavorable environment, and a role in reproduction of L-forms.18-21
It should be noted that our electron microscopy findings are very similar to the observations of Domingue and al. which first described the fine ultrastructure of pleomorphic cell wall deficient bacteria derived from blood lysates.3 Recently, Domingue hypothesized that these pleomorphic forms represent various stages in the life cycle of stressed bacteria: cell wall-deficient/defective (often called L-forms) that are difficult-to-culture or nonculturable.21 The L-form appearance of atypical, pleomorphic bacteria, derived from human blood, has attracted the interest of microbiologists in the past and continues presently. However, all researchers have had difficulties to identify and to find the origin of L-forms in the human blood. Bisset and Bartlett noted that the blood isolates may pass through different morphological stages, distinctive for the genera Mycoplasma, Mycobacterium, Streptococcus, Corynebacterium, Listeria, Micrococcus, and Bacillus.26
The observed by us acid fast bacteria, present in the isolated heterogeneous blood cultures even in minor amounts, were of special interest. The evidence suggesting that L-forms of mycobacteria are present in isolated blood cultures from BCG vaccinated persons were provided by 3 specific genetic assays (16S rRNA; IS6110 and spoligotyping). Having in mind the unusual modes of reproduction and life style of L-form cultures, their genetic identifications presented many difficulties. In total, 45 blood cultures of 97 genetically tested, were confirmed to belong to genus Mycobacterium. Sixteen of them were from healthy persons and 28 – from persons with health problems. Both IS6110 Real Time PCR and spoligotyping assays confirmed that 43 cultures were positive for M. tuberculosis complex. The spoligotyping technique proved to be the most effective tool in identifying mycobacteria L-forms. It is based on detecting of the direct-repeat (DR) region in M. tuberculosis complex members. Spoligotyping has been found to be a suitable method for analyzing minute amounts of a significantly fragmented mycobacterial DNA such as ancient DNA from mummies.27 The strongly decreased number of spacers in most of the isolated by us blood cultures indicate reduced amount of DNA or difficulties in access to it. It is known that M. bovis BCG (vaccine strain) belongs to M. tuberculosis complex. It was also found that specific genetic tests, distinguishing between M. bovis BCG and other M. tuberculosis complex strains, were inapplicable or less effective for L-form identification.28 In our previous study, we found that in result of uncoordinated L-form propagation and growth dependent processes, mutations can arise in the genome of mycobacteria.29 Therefore, we could propose that the rest of genetically unconfirmed blood L-form cultures remained unidentified, possibly due to too large changes in their genome occurring under selective pressure of strongly unfavorable blood factors, which were also causing their morphological transformations. Obviously, mutations and DNA rearrangements are associated with a cycle of morphological transformations and unusual division processes characteristic for L-form populations. L-form cells divide without peptidoglycan structures, respectively chromosome segregation does not follow normal physical and spatial rules like in normal bacterial cells, therefore chromosome segregation and cell division defects are likely to result from irregular propagation of L-forms.30 Recombination with non-coding DNA or with silent genes, as well as DNA insertion might easily occur in population of cell wall deficient bacteria.31
Occurrence of highly pleomorphic L-forms with new biologic characteristics in vivo and in vitro is assumed to be an adaptive reaction of bacteria toward unfavourable factors.18-21,29,32 Domingue states that atypical, pleomorphic L-form elements may persist in the human, including blood.21
In conclusion, we developed a methodology to access the unusual L-form elements derived from human blood and found a path to their isolation and cultivation by special techniques, as well as characterized them in detail by electron microscopy. Although the process of genetic identification of these blood isolates encountered many difficulties for technical reasons, due to the nature of L-forms, there is genetic evidence to suggest that these L-forms are of mycobacterial origin. As the investigated people had been vaccinated with BCG, we can assume that the identified mycobacterial L-forms may be produced by persisting live BCG vaccine. Because BCG has been given routinely and constantly to all newborns in Bulgaria since 1952, practically the entire population in the country is vaccinated. However, data from unvaccinated persons is absent or difficult to find, and therefore the current study is limited within a country (Bulgaria), which is with obligatory BCG vaccination policies. It should be also outlined that L-form cultures were isolated equally successfully from both healthy persons (volunteers) without any additional clinical history for treatment with antibiotics, and from persons with non-infectious health problems (oncologic, autoimmune diseases and transplantations) with history of corticosteroid, cytostatic and antibiotic treatment.
Further investigation on people from different geographical regions with different BCG vaccination policy is needed to answer the question whether the existing cryptic L- form bacteria in human blood are only of anthropogenic vaccine (BCG) origin or if they are part of the normal blood microbiome and what is their biological and medicinal significance.
Materials and Methods
Blood samples
A total of 141 blood samples were recovered from Bulgarian people (volunteers), aged between 1 and 60 y old, as follows: 56 of them were from healthy individuals, while the remaining 85 - from patients with non- infectious health problems (autoimmune, oncologic diseases and transplantation). Since 1952, the BCG vaccine has been given routinely and obligatory to newborns in Bulgaria by intradermal application. Thus, all investigated people had been vaccinated and none of them had a history of exposure to tuberculosis. Blood sample were aseptically collected using K2E-EDTA Vacutainer tubes (BD Vacutainer, Plymouth, UK). All specimens were handled and anonymized, according to the ethical and legal guidelines. The procedures were approved by the Ethics Committee of Human Experimentation in Bulgaria.
Isolation of L-form cultures
We have developed a novel and original protocol for isolation of cell wall deficient forms (L-forms) from human blood, which includes the following steps: : (i) Blood lysis was performed with sterile distilled water at strictly fixed v/v ratio and after 30 min stay at room temperature; (ii) The lysed blood samples were inoculated in tubes with Tryptic Soy Broth ( TSB, Becton Dickinson), which were then incubated at 37°C for 72 hours; (iii) Sub-cultivation from broth on semisolid medium, prepared from TSB solidified with 0.8% (w/v) Agar (Fluca), in Petri dishes of 94×15 mm. Strictly fixed aliquots were taken from the tube bottom of the broth cultures and were plated with special technique on Petri dishes. At least 3 Petri dishes per one broth culture were plated. The Petri dishes were enveloped with parafilm and incubated at 37°C for one week. The plates were examined daily macroscopically and under light microscope for presence of “nonculturable elements” (Fig. 1A, B and C). These elements did not give rise to visible growth (colonies or biofilm formation) during the first week; (iv) Enrichment of nonculturable elements by so called “blind passage.” The “blind” passage represents an original enrichment approach including collection and merging of “nonculturable elements” from at least 3 Petri dishes and transfer of the concentrated material to one new Petri dish with semisolid medium.
The technique of “blind passage” was performed by flooding the surface of 3 Petri dishes with TSB, after that aspirating, merging and transferring the lavage fluid to the surface of a new Petri dish with semisolid agar, which was then incubated again for a period of one week at 37°C. Colonial or biofilm growth occurred by the end of the second week. The plates were examined daily macroscopically and microscopically for appearance of growth. Direct light microscopic observations of cultures were combined with Ziehl–Neelsen stained preparations.
Electron microscopy
Observations of blood L-form isolates were performed by electron microscopy. Selected samples from L-form colonies and biofilm that developed during the second phase/week of cultivation in semisolid agar, were fixed with 4% (v/v) glutaraldehyde in 0.1 M cacodylate buffer with 4.5% w/v sucrose, pH 7.2 and post-fixed in 1% (w/v) osmium tetroxide in the same buffer at room temperature for 2 h and dehydrated in serial ascending ethanol concentrations. For scanning electron microscopy (SEM), specimens were placed on membrane filters with pore size diameter of 0.22µm (Millichrom, Isopore), covered with 15–20 Å gold, visualized and photographed with scanning electron microscope Phillips SCM 515. For transmission electron microscopy (TEM), after dehydration in ethanol and propylene- oxide series, cell pellets were embedded in epoxy resin Epon-Araldite (Serva, Heidelberg, Germany). Resin blocks polymerised at 56 56°C deg;C for 48 h. Ultrathin cell sections were made with crystal glass knives on a Reichert-Jung Ultracut Microtome and were stained with 5% (w/v) uranyl acetate in 70% (v/v) methanol and 0.4% (w/v) lead citrate. Observations were made with electron microscope JEOL JEM -1011 SAP10 (Japan) at 40–100 kV.
DNA–based assays
Selected samples from L-form colonies or biofilm developed during the second phase/week of cultivation in semisolid agar, were picked up for genetic testing. Chromosomal DNA was isolated as described by Van Embden et al.33
16S rRNA
PCR was used to amplify segment of 16S rRNA gene with primers: g2R-F 5′GAGAATTCGTGCTTAACACATGCAAGTCG3′ and rM582R-R 5′ATGGATCCGTGAGATTTCACGAACAACGC3′. PCR protocol has been described by Devulder et al.34 The amplification of 16Sr RNA gene fragment was done with a 50μl reaction mixture containing 10x PCR buffer (Tris.Cl; KCl; (NH4)2SO4; 15mM MgCl2, pH 8,7, Qiagen) at final concentration of 1x; 1,25 U HotStarTaq DNA polymerase (Qiagen); 0,2 mM of each dNTPs (dNTP Mix, Qiagen); 1 mM MgCl2 (Qiagen), 0,2 μM of each primer; 5 μl template DNA and PCR-grated water up to 50 μl. The thermal profile involved initial denaturation for 10 min at 90°C, 40 cycles of denatu-ration for 25 s at 94°C, annealing for 30 s at 60°C and extension for 45 s at 72°C followed by 1 cycle at 72°C for 10 min. The PCR product (5 μL) was examined and the amount estimated visually on a 3% agarose gel. IS6110 Real Time PCR
The Real Time PCR mixtures containing a final concentration of 1X PCR buffer (Tris.Cl; KCl; (NH4)2SO4; 15mM MgCl2, pH 8,7, Qiagen), 2.5 mM MgCl2, 0,2 mM of each dNTPs (dNTP Mix, Qiagen), 1,75 U HotStarTaq DNA polymerase (Qiagen), the target specific primers and probes, were used at a final concentration of 0.5 μM and 0.5 μM respectively, finally 5 μl of template was added. The reaction mixture was performed in a final volume of 50 μl. The primers and the probe sequence were selected from a region of the IS6110: Primers IS6110 D-1 (5′- ACCTGAAAGACGTTATCCACCAT-3′) and IS6110 D-2 (5′-CGGCTAGTGCATTGTCATAGGA-3′) which amplify a 100 bp fragment; the probe: (5′- [6 FAM] TCCGACCGCGCTCCGACCGACG-[TAMRA-Q]3′), was synthesized and conjugated with the reporter dye FAM and TAMRA quencer dye, which were covalently linked to 5′ and 3′ ends oligonucleotide respectively. The reaction was optimized to obtain the best amplification kinetics and the cycle condition was performed for 1 cycle, 3 min at 95°, 30 s at 95°C and 50 s at 60°C for 50 cycles.35
Spoligotyping
The M. tuberculosis complex specific spoligotyping (spacer oligonucleotide typing) method was performed as described by Kamerbeek et al.36 Spoligotyping is used to detect mycobacteria-specific direct repeats (DRs), especially in determining the absence or presence of 43 different spacers.
Acknowledgments
We thank Albena Cherneva for excellent technical assistance.
Disclosure of Potential Conflict of Interest
No potential conflicts of interest were disclosed.
References
- 1.Pease PE, Tallack JE. A permanent endoparasite of man. The silent zoogleal/symplasm/L-form phase. Microbios 1990; 64:173-80; PMID:2084495 [PubMed] [Google Scholar]
- 2.Kajander EO, Tahvanainen E, Kuronen I, Ciftcioglu N. Comparison of Staphylococci and novel bacteria-like particles from blood Zbl Bakt 1994; 26:147-149 [Google Scholar]
- 3.Domingue GJ, Schlegel JU. Novel bacterial structures in human blood: cultural isolation. Infect Immun 1977; 15:621-7; PMID:844907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tedeschi GG, Amici D. Mycoplasma-like microorganisms probably related to L forms of bacteria in the blood of healthy persons. Cultural, morphological and histochemical data. Ann Sclavo 1972; 14:430-42; PMID:4130434 [PubMed] [Google Scholar]
- 5.Tedeschi GG, Bondi A, Paparelli M, Sprovieri G. Electron microscopical evidence of the evolution of corynebacteria-like microorganisms within human erythrocytes. Experientia 1978; 34:458-60; PMID:639937; http://dx.doi.org/ 10.1007/BF01935925 [DOI] [PubMed] [Google Scholar]
- 6.Tedeschi GG, Amici D, Sprovieri G, Vecchi A. Staphylococcus epidermidis in the circulating blood of normal and thrombocytopenic human subjects: immunological data. Experientia 1976; 32:1600-2; PMID:798697; http://dx.doi.org/ 10.1007/BF01924475 [DOI] [PubMed] [Google Scholar]
- 7.McLaughlin RW, Vali H, Lau PC, Palfree RG, De Ciccio A, Sirois M, Ahmad D, Villemur R, Desrosiers M, Chan EC. Are there naturally occurring pleomorphic bacteria in the blood of healthy humans? J Clin Microbiol 2002; 40:4771-5; PMID:12454193; http://dx.doi.org/ 10.1128/JCM.40.12.4771-4775.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aho K, Kajander EO, Raoult D. Pitfalls in detection of novel nanoorganisms. J Clin Microbiol 2003; 41: 3460-3461; PMID:12843126; http://dx.doi.org/ 10.1128/JCM.41.7.3460-3461.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kajander EO. Nanobacteria- Propagating calcifying nanoparticles. Lett Appl Microbiol 2006; 42: 549-552; PMID:16706890 [DOI] [PubMed] [Google Scholar]
- 10.Proal AD, Albert PJ, Marshall TG. Autoimmune disease in the era of the metagenome. Autoimmunity Reviews 2009; 8:677-81; PMID:19393196; http://dx.doi.org/ 10.1016/j.autrev.2009.02.016 [DOI] [PubMed] [Google Scholar]
- 11.Wirostko E, Johnson L, Wirostko B. Sarcoidosis associated uveitis. Parasitization of vitreous leucocytes by mollicute-like organisms. Acta Ophthalmol (Copenh) 1989; 67:415-24; PMID:2801045; http://dx.doi.org/ 10.1111/j.1755-3768.1989.tb01626.x [DOI] [PubMed] [Google Scholar]
- 12.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science 1999; 284:1318-22; PMID:10334980; http://dx.doi.org/ 10.1126/science.284.5418.1318 [DOI] [PubMed] [Google Scholar]
- 13.Methe BA. Human Microbiome Project Consortium A framework for human microbiome research. Nature 2012; 486: 215-221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xalabarder C. Electron microscopy of tubercle bacilli. Excerpta Med Sect XV Chest Dis 1958; 11:467-73 [Google Scholar]
- 15.Armbruster C, Junker W, Vetter N, Jaksch G. Disseminated bacille Calmette- Guérin infection in an AIDS patient 30 years after BCG vaccination. J Infect Dis 1990; 162:1216; PMID:2230251; http://dx.doi.org/ 10.1093/infdis/162.5.1216 [DOI] [PubMed] [Google Scholar]
- 16.Smith E, Thybo S, Bennedsen J. Infection with Mycobacterium bovis in a patient with AIDS: a late complication of BCG vaccination. Scand J Infect Dis 1992; 24:109-10; PMID:1589715; http://dx.doi.org/ 10.3109/00365549209048409 [DOI] [PubMed] [Google Scholar]
- 17.Reynes J, Perez C, Lamaury I, Janbon F, Bertrand A. Bacille Calmette- Guérin is 30 years after immunization in a patient with AIDS. J Infect Dis 1989; 160:727; PMID:2794567; http://dx.doi.org/ 10.1093/infdis/160.4.727 [DOI] [PubMed] [Google Scholar]
- 18.Prozorovski SV, Kaz LN, Kagan GJ. Bacterial L-forms: Mechanisms of Formation, Structure, Role inPathology. Moscow: Taylor & Francis; 1981. [Google Scholar]
- 19.Domingue GJ. Cell-wall Deficient Bacteria: Basic Principles and Clinical Significance. Reading MA: Taylor & Francis; 1982 [Google Scholar]
- 20.Mattman LH. Cell wall Deficient Forms. Stealth Pathogens. Boca Raton, FL USA: Taylor & Francis; 2001 [Google Scholar]
- 21.Domingue GJ. Demystifying pleomorphic forms in persistence and expression of disease: Are they bacteria, and is peptidoglycan the solution? Discov Med 2010; 10:234-46; PMID:20875345 [PubMed] [Google Scholar]
- 22.Zwerling A, Behr MA, Verma A, Brewer TF, Menzies D, Pai M. The BCG World Atlas: A Database of Global BCG Vaccination Policies and Practices. PLoS Med 2011; 8(3): e1001012; PMID:21445325; http://dx.doi.org/ 10.1371/journal.pmed.1001012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Markova N, Michailova L, Kussovski V, Jourdanova M. Formation of persisting cell wall deficient forms of Mycobacterium bovis BCG during interaction with peritoneal macrophages in guinea pigs. Electronic J Biol 2008; 4:1-10 [Google Scholar]
- 24.Markova N, Slavchev G, Michailova L. Filterable forms and L-forms of Mycobacterium bovis BCG. Hum Vaccin Immunother 2012; 8:759-764; PMID:22495116; http://dx.doi.org/ 10.4161/hv.19698 [DOI] [PubMed] [Google Scholar]
- 25.Slavchev G, Michailova L, Markova N. Stress-induced L-forms of Mycobacterium bovis: a challenge to survivability. New Microbiol 2013; 36-:157-66; PMID:23686122 [PubMed] [Google Scholar]
- 26.Bisset KA, Bartlett R. The isolation and characters of L-forms and reversions of Bacillus licheniformis var. endoparasiticus (Benedck) associated with the erythrocytes of clinically normal humans. J Med Microbiol 1978; 11:335-349; PMID:682179; http://dx.doi.org/ 10.1099/00222615-11-3-335 [DOI] [PubMed] [Google Scholar]
- 27.Zink AR, Sola C, Reischl U, Grabner W, Rastogi N, Wolf H, Andreas G, Nerlich AG. Characterization of Mycobacterium tuberculosis Complex DNAs from Egyptian Mummies by Spoligotyping. J Clin Microbiol 2003; 41: 359-367; PMID:12517873; http://dx.doi.org/ 10.1128/JCM.41.1.359-367.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Slavchev G. Molecular biological and morphological aspects of L-form formation in Mycobacterium tuberculosis complex species (Dissertation; ISBN 978-954-322-782-2). Taylor & Francis, Sofia, 2014 [Google Scholar]
- 29.Slavchev G, Markova N. Genetic and morphologic variations during L-form conversion in Mycobacterium tuberculosis. Afr J Microbiol Res 2014; 8: 850-855; http://dx.doi.org/ 10.5897/AJMR2013.6321. [DOI] [Google Scholar]
- 30.Allan EJ, Hoishen C, Gumpert J. Bacterial L-forms. Adv Appl Microbiol 2009; 68: 1-39; PMID:19426852; http://dx.doi.org/ 10.1016/S0065-2164(09)01201-5 [DOI] [PubMed] [Google Scholar]
- 31.Norris V. Speculations on the initiation of chromosome replication in Escherichia coli: the dualism hypothesis. Med. Hypoth 2011; 76: 706-716; http://dx.doi.org/ 10.1016/j.mehy.2011.02.002 [DOI] [PubMed] [Google Scholar]
- 32.Markova N. Cell wall deficiency in mycobacteria: latency and persistence, in Pere Joan Cardona (ed): Understanding tuberculosis - deciphering the secret life of the bacilli. InTech, 2012; Chapter 11; pp. 193-216 [Google Scholar]
- 33.van Embden JDA, Cave MD, Crawford JT, Dale JW, Eisenach KD, Gicquel B, Hermas P, Martin C, McAdam R, Shinnick TM. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol 1993; 31: 406-409; PMID:8381814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Devulder G, Perouse DM, Flandrois JP. A multigene approach to phylogenetic analysis using the genus Mycobacterium as a model. Int J Sys Evol Microbiol 2005; 55: 293-302; http://dx.doi.org/ 10.1099/ijs.0.63222-0 [DOI] [PubMed] [Google Scholar]
- 35.Ortu S, Molicotti P, Sechi LA, Pirina P, Saba F, Vertuccio C, Deriu A, Maida I, Mura MS, Zanetti S. Rapid detection and identification of Mycobacterium tuberculosis by Real Time PCR and Bactec 960 MIG. New Microbiol 2006; 29: 75-80; PMID:16608129 [PubMed] [Google Scholar]
- 36.Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, Bunschoten A, Molhuizen H, Shaw R, Goyal M, et al.. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol 1997; 35: 907-901; PMID:9157152 [DOI] [PMC free article] [PubMed] [Google Scholar]