Abstract
Expression of HPV E5, E6 and E7 oncogenes are likely to overcome the regulation of cell proliferation and to escape immunological control, allowing uncontrolled growth and providing the potential for malignant transformation. Thus, their three oncogenic products may represent ideal target antigens for immunotherapeutic strategies. In previous attempts, we demonstrated that genetic vaccines against recombinant HPV16 E7 antigen were able to affect the tumor growth in a pre-clinical mouse model. To improve this anti-HPV strategy we developed a novel approach in which we explored the effects of E5-based genetic immunization. We designed novel HPV16 E5 genetic vaccines based on two different gene versions: whole E5 gene and E5Multi. The last one is a long multi epitope gene designed as a harmless E5 version. Both E5 genes were codon optimized for mammalian expression. In addition, we demonstrated that HPV 16 E5 oncogene is expressed in C3 mouse cell line making it an elective model for the study of E5 based vaccine. In this mouse model the immunological and biological activity of the E5 vaccines were assessed in parallel with the activity of anti-E7 and anti-E6 vaccines already reported to be effective in an immunotherapeutic setting. These E7 and E6 vaccines were made with mutated oncogenes, the E7GGG mutant that does not bind pRb and the E6F47R mutant that is less effective in inhibiting p53, respectively. Results confirmed the immunological activity of genetic formulations based on attenuated HPV16 oncogenes and showed that E5-based genetic immunization provided notable anti-tumor effects.
Keywords: Human Papillomavirus, DNA vaccines, HPV16 E5, C3 cell line, immunologic response
Introduction
Human papillomavirus (HPV) is a well-recognized, worldwide-spread, sexually transmitted virus, counting for a severe burden for human health.1-3 In particular, HPV types classified as “high-risk” (HR) play a significant role in human carcinogenesis.4,5 In fact, persistent infection by HR-HPVs is one of the cervical cancer-associated factors best described,6 and in addition, HR-HPVs, in particular HPV16, are associated with a subset of head and neck cancers.7,8 HPVs comprise a large family of double-stranded DNA viruses, whose genome is organized into three regions: early genes (E1 to E7), late genes (L1 and L2) and long control region (LCR) containing a variety of cis-elements that regulate viral replication and gene expression.9 E5, E6 and E7 are viral oncogenes and their expression induces immortalization and transformation. In particular, the HR-E5 is able to enhance the ability of HR-E6 and HR-E7 to immortalize primary human keratinocytes and to increase the motility and invasiveness of human keratinocyte cell lines.10
Although the precise mechanism remains unclear and has been relatively less defined than that of E6 and E7, E5 may play a role in keratinocyte carcinogenesis. Indeed, while several HR-E6 and HR-E7 activities have consequences on host cell life cycle (i.e., E6 and E7 inactivate p53 and pRb, respectively)11,12 only in recent few years HR-E5 has been recognized as a possible relevant oncoprotein and as an interesting therapeutic target.13 Many, but not all papillomaviruses encode E5 proteins and the E5 ORF is often deleted in cervical carcinoma cells indicating that it may not be essential in maintaining the transformed status.14 Nevertheless HR-E5 can be involved in early phases of transformation by altering growth factor pathway, and by increasing the levels of epidermal growth factor receptor (EGFR) on the cell surface.15-17
In addition, E5 actively induces immunological escaping, favoring the persistence of infection.18,19 According to the relevance of their activities, HR-E6, HR-E7, and HR-E5 could be suitable therapeutic targets for immunotherapy against HPV-associated tumors. Although several approaches have demonstrated the potential of E6 and E7-based genetic immunization,20-22 few data are available with respect to E5.23 Hence, in premalignant lesions, where E5 is still expressed, an E5 vaccine may be a good strategy to cure and to prevent premalignant lesions from progressing into invasive cervical cancers. However, the potential of E5 protein as a tumor vaccine candidate has not been identified yet.24 In addition, the utilized challenge models are mostly based on cell lines that may not precisely simulate HPV oncogene expression patterns during natural infection, because viral gene expression is driven by non-HPV promoters.25A more accurate model of HPV associated cancer could be the C3 cell line in which the expression of all viral genes is under control of HPV promoter.26
Our previous data have demonstrated the induction of antitumor activity in a safe setting by vaccines based on two mutated, non-transforming forms of HPV16 E7 (E7GGG)27,28 and E6 (E6F47R) (De Giuli Morghen, Personal Communication). Hereby, we present the effects of novel HPV16 E5 genetic vaccines based on two different gene versions: whole E5 gene and E5Multi. The last one is a long multi epitope gene designed as a harmless E5 version.
Results
E5 protein production in bacteria
To check the activity of an E5 vaccine it is important to have at least a purified protein for ELISA tests. E5 is a very hydrophobic and small protein that it is difficult to produce in E.coli system. Nevertheless we succeeded in its production by transfection of E. coli BL21 strain with His-tag E5-recombinant pAE-plasmid. After induction and cell lysis by sonication, His-tag E5 fusion protein was purified by Ni-NTA resin. Western Blot (WB) analysis showed the presence of a protein of the expected size, indicating the correct expression of E5 by this system (Fig. 1).The yield of purified E5 protein, quantified according to the Bradford method, was 1.68 µg/µL (500 µL total) from a 500 mL bacterial culture.
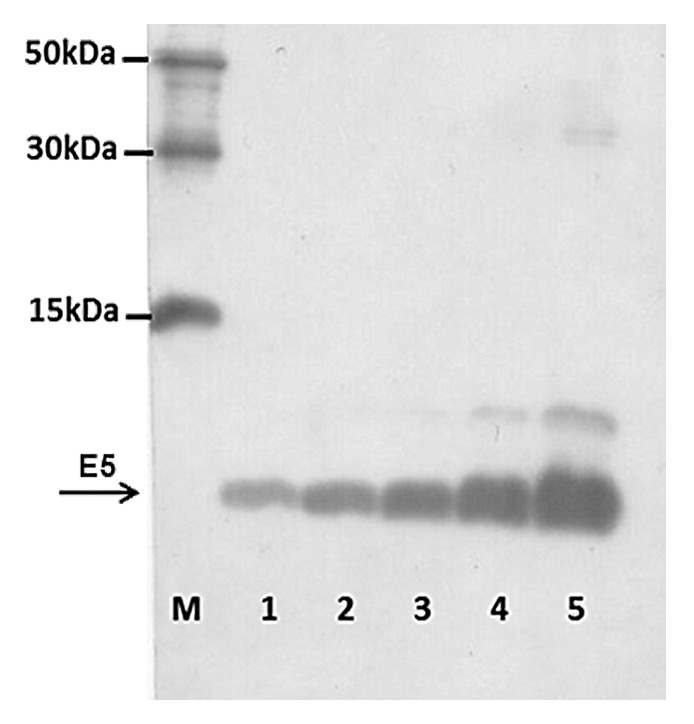
Figure 1. HPV 16 E5 peptide production. Immunoblotting of purified bacterial extracts was performed as in Material and Methods. Lanes 5 to 1–0.5 dilution of purified bacterial extracts; Lane M–6xHIS Protein Ladder (Qiagen).
Analysis of E5 gene expression in C3 tumor cells by RT-PCR
C3 cell line was already utilized as challenge model for E6/E7 therapeutic vaccines but for E5 therapeutic vaccine no information was available regarding the E5 expression. RT-PCR was performed to detect the presence of viral transcripts encoding the E5 protein. E5-specific transcripts were revealed, as shown in Figure 2, indicating the possibility to utilize C3 cell line as pre-clinical model for E5-based vaccines.
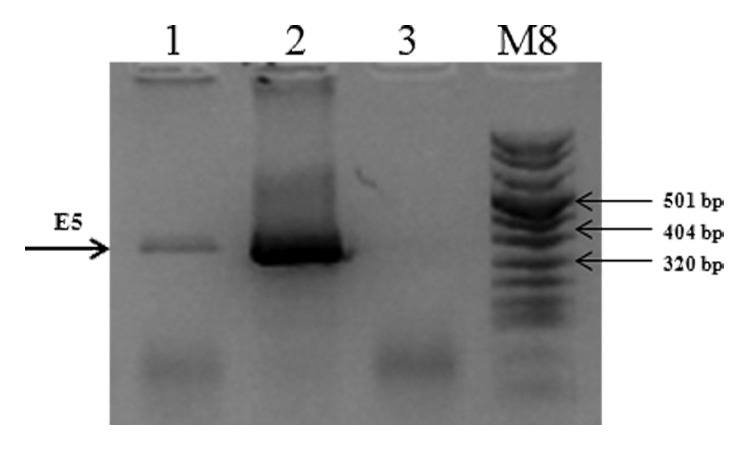
Figure 2. mRNA expression of the E5 gene in C3 cell line. Total cellular RNA was extracted from C3 cell line. cDNAs were synthesized by RT-PCR and specific amplified products were obtained with specific primers as described in “Materials and Methods”. Lane 1, E5 gene amplified from C3 RNA; Lane 2, E5 gene amplified from pCIE5 plasmid, as positive control; Lane 3, total C3 RNA without reverse transcriptase, as negative control; M, molecular weight marker VIII (Roche).
New recombinant genetic vaccines
In premalignant lesions, when E5 is still expressed, a vaccine targeted to E5-expressing cells may be a good strategy to prevent premalignant lesions from progressing to invasive cervical cancers. We designed two different constructs to be utilized as genetic vaccine: the whole E5 gene and a synthetic harmless version. Indeed, the E5 protein can be harmful in humans due its oncogenic activity, and therefore we designed an E5-based gene, the E5Multi, which contains two previous described coding sequences for immune epitopes, in duplicate.29,30 By this way, this E5Multi gene should increase the immunogenicity of the antigen and in the meantime could eliminate any possible oncogenic activity. An illustrated scheme of E5 and E5Multi genes is described in Figure 3.
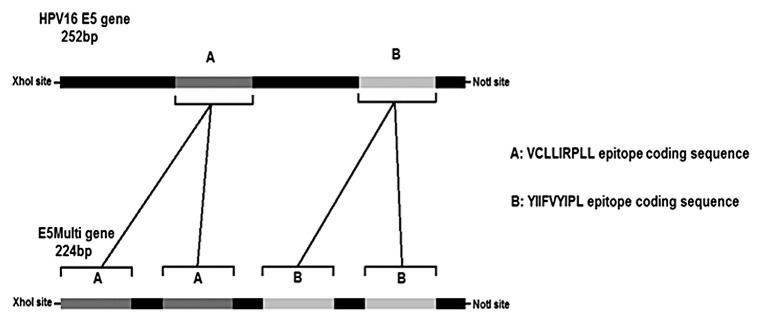
Figure 3. HPV16 E5 and E5Multi genes. The schematic representations of the E5 HPV16 gene (252 bp) with the position of CTL epitope sequences (cassettes A and B) and of the synthetic E5Multi gene (224 bp) with the position of the duplicated CTL epitope sequences (cassettes A and B) are shown together with the restriction sites for directional cloning. The aminoacid sequences of both epitopes are also indicated.
Immune response in mice immunized with E5 and E5-Multi
The HPV16 E5 gene (E5H16) and the E5Multi sequence were cloned into the pCI vector, as described in Material and Methods. The immunological effects of the E5-based vaccines were ascertained in C57BL/6 mice with the prime/boost schedule described in Figure 4A. After 14 days, serum was collected and tested for the presence of specific antibodies against E5. No circulating antibodies were detected in our ELISA assay with E.coli-produced E5 protein (Fig. 5A). Nevertheless, one week after the last boost the mice were challenged with 5x105 C3 tumor cells and the tumor volume was measured three times a week by caliper. As shown in Figure 5B, E5 gene-based vaccination, and in particular the vaccination with pCI-E5Multi plasmid, delayed significantly C3 cell-induced tumor development, while inoculation of empty pCI plasmid had no effect. Tumors were collected five weeks after challenge and their weights were measured. Tumors of E5 and E5Multi vaccinated mice were almost 2 or 4 times smaller than tumors collected from control group, respectively (Fig. 5C).
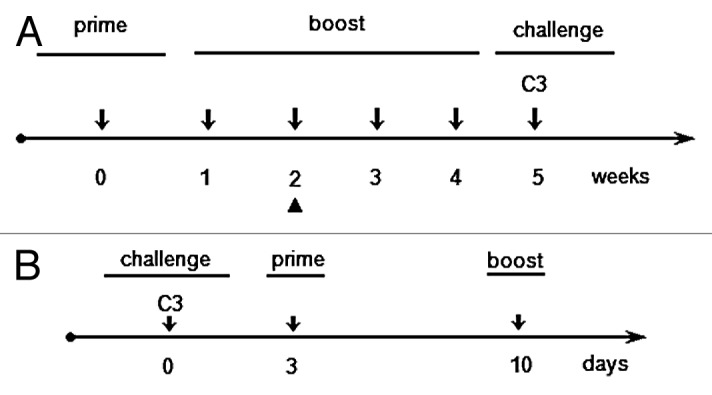
Figure 4. Prime/boost schedule for preventive or therapeutic immunizations. Five mice per group were used in both immunization schedules. (A) Preventive immunization schedule; the arrowhead (▲) indicates the day of blood sampling for ELISA test. (B) Therapeutic immunization schedule.
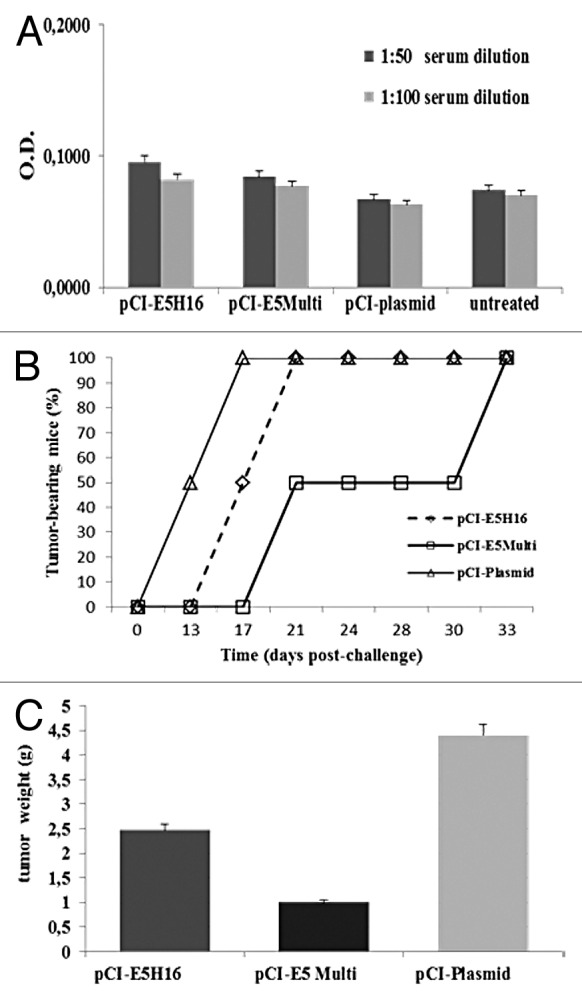
Figure 5. Preventive immunization with E5-based vaccines. (A) ELISA assay. Sera were collected from mice on day 14 after last boost and analyzed by E5 specific ELISA as described in “Materials and Methods”. Data are expressed as means for each group of mice ± SD (B) Cancer growth. Tumor development was assessed by palpation three times per week after C3 cell challenge. (C) Tumor burden. Five weeks after challenge mice were sacrificed and excised tumors weighted. Data are represented as mean of all of the mice in the groups ± SD.
E5 and E5Multi gene-based immunization elicits T cell response in vaccinated mice
Since CD8+ cytotoxic T cells have a recognized role as effectors in anti-cancer responses we hypothesized that the anti-tumor response in the animal with no specific antibodies was induced by E5 specific INF-gamma producing T cells. Five weeks after C3 challenge, spleens were collected from mice immunized with the prime/boost schedule described in Figure 4A. Collected splenocytes were tested in INF-gamma ELISPOT assay. The E.coli-produced E5 protein was used to activate anti-E5 lymphocytes. INF-gamma secreting cells were visualized and counted as spots under dissecting microscope.
The highest score was obtained in pCI-E5H16 vaccinated mice, exhibiting E5-specific T cell response approximately 30-fold higher than that recorded in control mice vaccinated with the empty vector as shown in Figure 6. Moreover, pCI-E5Multi vaccine showed 10-fold increase of E5-specific T cell response respect to the control. These results indicate that specific T cell-mediated response against the E5 protein can be generated in E5 and E5Multi-vaccinated mice.
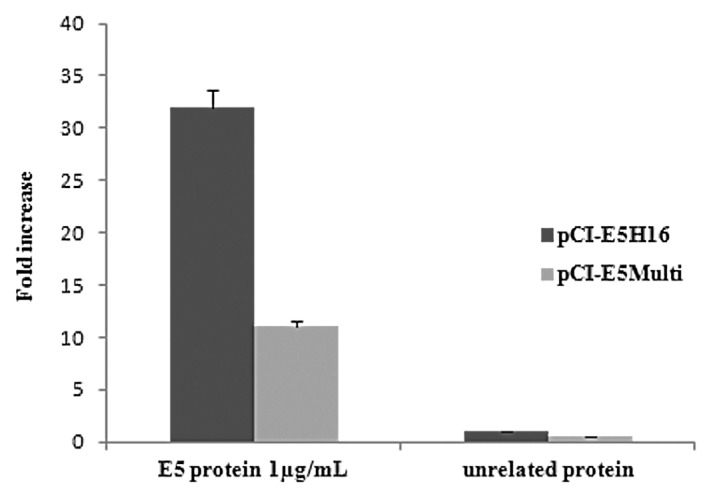
Figure 6. Cell-mediated immune responses in vaccinated mice. C57BL/6 mice were immunized by intramuscular administration of the various genetic vaccines as described in “Materials and Methods”. IFN-gamma production was measured in an ELISPOT assay after specific antigenic stimulation with E5 protein. Data are presented as fold-increase responses to the E5 protein, compared with mice vaccinated with empty vector, and they represent the means of all of the mice in the groups ± SD.
Therapeutic immunization
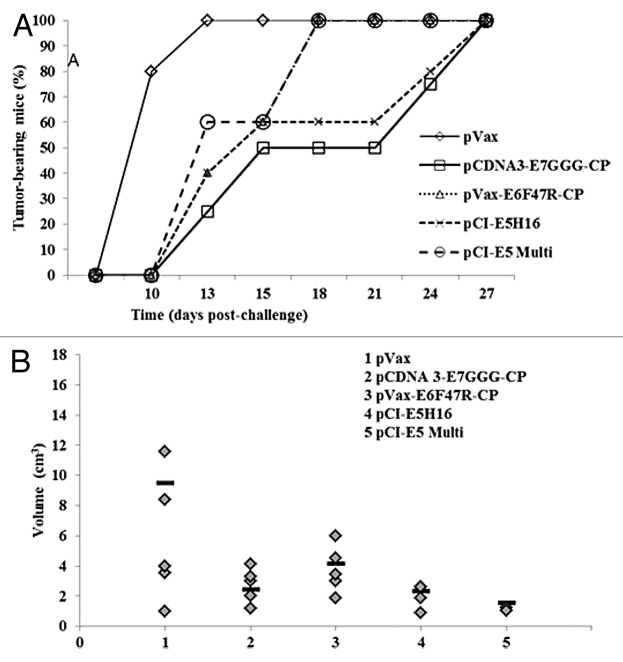
The ascertained immunological activity of these new anti-E5 vaccines prompted us to determine the therapeutic potential of these DNA vaccines in the C3 pre-clinical model. In addition, two vaccines already produced against the E7 and E6 proteins, the pcDNA3-E7GGG-CP and the pcDNA3-E6F47R-CP, respectively, were utilized for comparison. After injecting the C3 cells into naïve C57Bl6 mice, prime and boost immunizations were performed on days 3 and 10, respectively (Fig. 4B). As shown in Figure 7A, tumor appearance was delayed to day 13 post challenge (p.c.) in 75% of the pcDNA3-E7GGG-CP treated animals, in 60% of mice vaccinated with pcDNA3-E6F47R-CP and pCI-E5H16, and in 40% of the pCI-E5Multi vaccinated mice. By the same day, 100% of the mice immunized with the empty pVAX vector developed tumors. In addition, pcDNA and pCI plasmids, used as controls, produced same results with 100% of animals bearing tumors on the same day p.c. (data not shown). By day 21, tumor free mice were 50% in pcDNA3-E7GGG-CP vaccinated group and 40% in pCI-E5H16 vaccinated group (Fig. 7A). After 4 weeks all the animals were tumor affected but differences in tumor burden were detected by measuring tumor volume. The E7 vaccinated mice showed the presence of only small-volume tumors confirming data reported in a previous study27 and, interestingly, E5-vaccinated mice showed even smaller tumors (Fig. 7B).
Figure 7. Tumor growth after therapeutic E5, E6 and E7 immunization. Fifty micrograms of plasmid DNA were administered by electroporation on days 3 and 10 with either the indicated vaccine preparations or saline solution as control. (A) Cancer growth. Tumor development was assessed by palpation three times per week. (B) Tumor burden. Tumor volume was measured 4 wk p.c. and calculated as in “Material and Methods”. Bars (▬) represent the mean value per each mouse group.
Discussion
In previous reports we showed that immunization against the E7 protein of HPV16 trigger a complete immune response, which inhibited E7-expressing tumor growth in mouse models.27,31-34 Genetic vaccine with E7 recombinant DNA vaccine27,31,32 as well as plant-produced E7 recombinant protein.33,34 demonstrated the same antitumor activity. In the present study, we explored the possibility to utilize the E5 protein of HPV16 as new therapeutic target of immunization, by demonstrating that E5-based genetic immunization could induce specific CD8+ T-cell responses.
In any vaccine preparation and in particular in genetic vaccine one of the key issue is the production of possible high level of antigen in the host. To achieve a high expression of antigen after genetic vaccination, HPV 16 E5 gene was codon-optimized for the expression in mammalian cell in accordance to previous studies, showing 6 to 9-fold enhanced expression by codon optimization.35
In order to evaluate our E5-based vaccines, we needed to produce the E5 protein for the immunological testing and to identify a suitable pre-clinical mouse model. The first point was achieved by succeeding in producing the E5 protein in a bacterial system. Even without a proper codon adaptation for E. coli expression, our bacterial system was able to provide a suitable amount of E5 protein. Besides the well-known HPV E5-associated hydrophobicity,36 this protein has an intrinsic trend to aggregate as an oligo-hexamer when produced by heterologous expression systems that may represent a challenge for downstream purification procedure.37 Indeed, two chromatography steps were necessary to perform structural analysis of the HPV-16 E5 protein.38 For our goal an highly purified protein was not necessary and a single purification under denaturing conditions provided enough E5 protein for the preparation of the ELISA tests.
The second key point was achieved by identifying a suitable pre-clinical mouse model. The TC-1 cell line widely used as pre-clinical model for therapeutic vaccines25,34 is not a valid option to investigate an E5-based genetic immunization because this model does not express HPV16 E5 gene.22 Other alternative models were created by inducing an overexpression of E5 in tumor cells, which is not mimicking the low E5 expression in natural host cells.24 Thus, our analysis of C3 cell showing an expression of E5 mRNA under the control of the viral promoter indicated that C3 cell line was a suitable pre-clinical model for the study of E5 vaccines.
No anti-E5 antibody were detected in the sera of vaccinated mice, but tumor weight was smaller in E5H16 or E5Multi vaccinated mice than in controls, suggesting that cellular immune response is involved in controlling tumor growth. Moreover, tumor appearance was delayed in a considerable percentage of mice vaccinated with pCI-E5H16 and pCI-E5Multi. Furthermore in the therapeutic setting, the number of mice that remained tumor-free after three weeks was still 40% in the pCI-E5H16 group, whereas pcDNA3-E7GGG-CP vaccination group had 50% mice that were tumor-affected by the same date. Interestingly, the E5-vaccinated mice, in particular using E5Multi preparations, had even smaller-volume tumors than pcDNA3-E7GGG-CP vaccinated animals. In addition, our ELISPOT assay indicated that this anti-tumor effect is mediated by anti-E5 CD8+ T cells, which were induced by our E5-based genetic immunization. Taken together our data suggest that our E5 vaccines can produce an immunological response that in turn may affect tumor growth. However the vaccines against the other E6 and E7 oncogenes seem to be more effective in inducing an antitumor effect. These differences may account for the presence of CP sequences in the recombinant E7 and E6 vaccines. We already developed a strategy to enhance the potency of HPV DNA vaccines by fusing the attenuated HPV16 E7GGG gene to the Potato Virus X coat protein gene (pcDNA3/E7GGG-CP).27 The same HPV16 E7GGG without the CP sequence was totally ineffective against the E7-expressing tumor model, indicating that genetic vaccines require adjuvant sequences.27 The same HPV16 E7GGG gene was also fused to a DNA sequence encoding the inactive mutant of saporin protein (SAP-KQ) from Saponaria officinalis,28 increasing the CD4+/CD8+ T cell immune response against E7. Moreover, many other studies employed immune enhancing and modulating molecules, such as calreticulin,20 herpes glycoprotein,22 histone deacetylase inhibitor,39 and other molecules (for a review see ref. 40) in order to elicit a stronger immune response against HPV antigens by DNA vaccines. Thus, many different strategies can be utilized to improve the genetic vaccine activity. In this work, our E5-based genetic vaccines were produced without any fusion to adjuvant sequences, but the preparations were still able to induce CD 8+ T- mediated immune response, suggesting that these E5 antigens are particularly immunogenic. In alternative the electroporation delivering might be particularly efficacious in increasing the activity of genetic vaccines even without adjuvant molecules. Indeed, the reported data on HPV16 E7GGG were collected in mice vaccinated i.m.27 Nevertheless it is likely that a strong antitumor activity could be induced by co-expression of adjuvant molecules such as CP or saporin.27,28 The demonstrated activity of our genetic vaccines against the three HPV 16 oncogenes opens new therapeutic perspective of combinatorial vaccination such as prime-boost regimens against different HPV oncogenes. This new strategy could act in a synergistic way with other immunomodulators and could be utilized in the near future to cure patients with HPV infected cells.41 Continuing progress in these efforts may allow the cure of HPV-associated lesions.
Materials and Methods
Plasmids
The E5 HPV16 gene (252 bp) (NCBI Reference Sequence: NC_001526.2) was codon optimized for mammal expression using IEBD Analysis Resources. The Db-restricted CTL epitope sequence (25–33aa:VCLLIRPLL) and HLA-A 0201-restricted CTL epitope sequence (63–71aa:YIIFVYIPL) from HPV16 E5 gene were duplicated to produce an immunogenic harmless version of the E5 gene, theE5Multi (224 bp). Each sequence was inserted into the pCI-neo expression vector (Promega Corporation) between XhoI and NotI restriction sites. pCI-neo empty vector was used as negative control. Plasmid integrity was verified by DNA sequencing. The E6F47R mutant (kindly provided by G Travé; Strasburg, France) that is defective for polyubiquitination and subsequent degradation of p5342 was fused to Potato Virus X coat protein gene (CP), in order to obtain a novel recombinant genetic vaccine (De Giuli Morghen, Personal Communication). The pcDNA3-E7GGG-CP was constructed by fusing the HPV16 E7GGG sequence, mutated in the binding site for pRb,31 to the 3′ end of the CP gene as already described by Massa et al.27 In each experiment all plasmid preparations were purified by CsCl gradient to obtain endotoxin-free products.
Cell lines
The C3 tumor cells (a gift of C.J.M. Melief) are B6 embryonic mouse cells transformed with the whole HPV 16 genome and EJ ras.26 The C3 cells are syngeneic to C57BL/6 mice and were cultured in RPMI-1640 with 10% fetal bovine serum (Life Technology). Before inoculation, C3 cells were harvested by trypsinization, washed twice, and resuspended in saline solution at 5 × 105 cells/ml.
RT-PCR
RNA was extracted from C3 cells by the RNeasy Plus Mini kit (Qiagen), according to the manufacturer’s instructions. Total RNA pre-treated with deoxyribonuclease I (DNase I, Amplification grade, Invitrogen) was retro-transcribed into cDNA for 1 h at 42 °C using a random hexamer primer kit as described by the manufacturer (GeneAmp RNA PCR kit Applied Biosystem). The assay was performed both with and without reverse transcriptase to exclude the presence of contaminating DNA. The synthesized cDNA underwent PCR with Platinum TaqDNA polymerase (Invitrogen) and specific primer for the E5 gene using the following protocol: 95 °C for 3 min, then 35 cycles of denaturation at 95 °C for 50 s, annealing at 55 °C for 50 s and elongation at 72 °C for 60 s. The amplified products were resolved in ethidium bromide stained agarose gels. The E5 primers utilized were designed by the OligoAnalyzer Tool (Integrated DNA Technologies) software and synthesized according the following sequences:
Forward:5′–ATCTCGAGGCCACCATGGGATACTGCATTCACAATATAAC–3′ Reverse:5′- TAGCGGCCGCGAATTCTTATCATGTGATCAGGAATCTTG – 3′.
E5 protein
The HPV16 E5 gene was inserted into pAE bacterial expression vector43 and produced by Escherichia coli (BL21 strain) expression system in 500 mL LB medium. Induction was performed with 1mM IPTG at 23 °C and shaking at 200 rpm during 4 h. E5 protein was purified by Ni-NTA resin (Qiagen), according to the manufacturer’s instructions and analyzed by 23% SDS-PAGE. Immunoblotting was performed with primary anti-His antibody (Invitrogen) and secondary peroxidase-conjugated anti-mouse antibody (Invitrogen). The presence of reacting bands were revealed by chemoluminescence (ECL-kit,Amersham).
Enzyme-linked immune-sorbent assay (ELISA)
The anti-HPV16 E5 serum antibodies were determined by a direct ELISA assay. A 96-well plate was coated at 4 °C overnight with200 ng of E.coli-derived HPV16 His-E5 and then blocked with 150 µl of 5% (w/v) no-fat dry milk in PBS (MPBS). Sera were collected on day 14 after boost, diluted in 2% MPBS (1:100 and 1:50), and incubated for 2 h at 37 °C. Total IgGs were detected with horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG(H-L) (MP Biomedicals Life Sciences). Enzymatic activity was measured by adding 2,2-azino-bis(3-ethylbenzthiazoline sulfonate) substrate and absorbance was read at 405 nm on an ELISA microtiter plate reader.
IFN-gamma enzyme-linked immuno-spot assay (ELISPOT)
HPV16 E5-specific T-cell precursors were detected by ELISPOT for IFN-gamma-secreting cells (BDTMELISPOT BD Biosciences PharMingen) according to previous reported protocols.44 Briefly, a single-cell suspension of splenocytes harvested from each group of vaccinated mice was added to microtiter wells that were pre-coated overnight at 4 °C with anti-mouse-IFN-gamma antibody (BD Biosciences PharMingen).Lymphocyte stimulation was performed in triplicates at 37 °C for 72 h, with the E5 protein. A mixture of phorbolmyristate acetate (PMA) was used to detect cell responsiveness. The plates were incubated with a biotinylated anti-mouse IFN-gamma-antibody (BD Biosciences PharMingen) for 4 hours at room temperature. Streptavidin–HRP was then added for 1 hour at room temperature, and the cell spots stained with filtered 3-amino-9-ethylcarbazole substrate, for 1–5 min. The spots were counted under a dissecting microscope.
Animals and vaccination schedule
Charles River Laboratories supplied 6–8 weeks female C57BL/6 mice. All procedures involving handling and sacrifice of animals were performed under specific pathogen-free conditions at the Animal House of the Regina Elena National Cancer Institute. The Ethical Committee approved the protocols developed in accordance with the European guidelines no. 86/609/CEE and 116/92 for the protection of laboratory experimental animals and laboratory animal care (Ministry of Health, Department for Veterinary Public Health, Nutrition and Food Security, Protocol17/2006). Two immunization protocols were followed: the immune response protocol, in which the mice received multiple vaccinations prior tumor challenge, and the therapeutic immunization protocol, in which the mice were challenged with C3 tumor cells before administration of the vaccines. In the immunization protocol, animals were primed with the recombinant pCI-E5H16 (50 µg/mouse, i.m.) or with the recombinant pCI-E5Multi (50 µg/mouse i.m), and boosted 3 times at 1 week intervals with the same preparations. After four weeks mice were challenged in the flank by sub-cutaneous (s.c.) injection of 200 µl of saline solution containing 5 × 105 C3 tumor cells. Tumor growth was monitored by visual inspection and palpation three times a week. Animals were scored as tumor bearing when tumors reached a size of approximately 1 to 2 mm in diameter.
In the therapeutic immunization, 5 groups of 5 mice were challenged by C3 tumor cells as above described. Three days after the tumor challenge, the mice were primed with electroporation-mediated DNA vaccines. Boosting was performed one week after the priming using same plasmid preparations. Tumor size was measured with caliper, and the volume estimated by the formula (width2 × length × 0.52). Electroporation-mediated DNA vaccination was performed according to the method described by Seo et al.45 Briefly, mice were injected with 100 μL of saline solution containing 50 µg of each DNA vaccine into the tibialis muscle of the shaved left leg. DNA injection was followed immediately by square wave electroporation at the injection site using a BTX830 apparatus (BTX Harvard Apparatus). A tweezers electrode (BTX Harvard Apparatus) was used to deliver six pulses at 100 V/cm for 20 ms.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was partially supported by AIRC IG 12916 and by Project of Mutual Collaboration of the Planning Department of the National Health Service of Italian Ministry of Health and Regina Elena National Cancer Institute for Implementation of HPV-UNIT. The authors thank Silvio Flamini for maintenance of tumor cells; Giancarlo Cortese and Sabrina Germoni for animal stabling and technical assistance.
Glossary
Abbreviations:
- ELISPOT
Enzyme-linked Immuno-spot
- ELISA
Enzyme-linked immune-sorbent assay
- ORF
open reading frame
- LCR
long control region
- pRb
protein of retinoblastoma
- i.m.
intramuscularly
- p.c.
post challenge
- HPV
Human papillomavirus
- HR
high-risk
- EGFR
epidermal growth factor receptor
- WB
western blot
- RT-PCR
reverse transcription polymerase chain reaction
- INF
Interferon
- SAP
saporin
- CP
coat protein
- s.c.
sub-cutaneous
- His
histidine
- Ni-NTA
nickel-nitrilotriacetic acid
- IPTG
isopropyl-beta-D-1-thiogalactopyranoside
- MPBS
milk in Phosphate Buffered Saline
- PMA
phorbolmyristate acetate
- E5H16
HPV16 E5 gene
References
- 1.Guan P, Howell-Jones R, Li N, Bruni L, de Sanjosé S, Franceschi S, Clifford GM.. Human papillomavirus types in 115,789 HPV-positive women: a meta-analysis from cervical infection to cancer. Int J Cancer 2012; 131:2349 - 59; http://dx.doi.org/ 10.1002/ijc.27485; PMID: 22323075 [DOI] [PubMed] [Google Scholar]
- 2.Burchell AN, Winer RL, de Sanjosé S, Franco EL.. Chapter 6: Epidemiology and transmission dynamics of genital HPV infection. Vaccine 2006; 24:Suppl 3S3 - , 52-61; http://dx.doi.org/ 10.1016/j.vaccine.2006.05.031; PMID: 16950018 [DOI] [PubMed] [Google Scholar]
- 3.zur Hausen H.. Papillomaviruses and cancer: from basic studies to clinical application. [Review]Nat Rev Cancer 2002; 2:342 - 50; http://dx.doi.org/ 10.1038/nrc798; PMID: 12044010 [DOI] [PubMed] [Google Scholar]
- 4.Muñoz N, Bosch FX, de Sanjosé S, Herrero R, Castellsagué X, Shah KV, Snijders PJ, Meijer CJ, International Agency for Research on Cancer Multicenter Cervical Cancer Study Group.. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med 2003; 348:518 - 27; http://dx.doi.org/ 10.1056/NEJMoa021641; PMID: 12571259 [DOI] [PubMed] [Google Scholar]
- 5.Bzhalava D, Guan P, Franceschi S, Dillner J, Clifford G.. A systematic review of the prevalence of mucosal and cutaneous human papillomavirus types. Virology 2013; 445:224 - 31; http://dx.doi.org/ 10.1016/j.virol.2013.07.015; PMID: 23928291 [DOI] [PubMed] [Google Scholar]
- 6.zur Hausen H.. Papillomaviruses in the causation of human cancers - a brief historical account. Virology 2009; 384:260 - 5; http://dx.doi.org/ 10.1016/j.virol.2008.11.046; PMID: 19135222 [DOI] [PubMed] [Google Scholar]
- 7.Gillison ML, Koch WM, Capone RB, Spafford M, Westra WH, Wu L, Zahurak ML, Daniel RW, Viglione M, Symer DE, et al.. Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J Natl Cancer Inst 2000; 92:709 - 20; http://dx.doi.org/ 10.1093/jnci/92.9.709; PMID: 10793107 [DOI] [PubMed] [Google Scholar]
- 8.Herrero R, Castellsagué X, Pawlita M, Lissowska J, Kee F, Balaram P, Rajkumar T, Sridhar H, Rose B, Pintos J, et al. , IARC Multicenter Oral Cancer Study Group.. Human papillomavirus and oral cancer: the International Agency for Research on Cancer multicenter study. J Natl Cancer Inst 2003; 95:1772 - 83; http://dx.doi.org/ 10.1093/jnci/djg107; PMID: 14652239 [DOI] [PubMed] [Google Scholar]
- 9.Struyk L, van der Meijden E, Minnaar R, Fontaine V, Meijer I, ter Schegget J.. Transcriptional regulation of human papillomavirus type 16 LCR by different C/EBPbeta isoforms. Mol Carcinog 2000; 28:42 - 50; http://dx.doi.org/; PMID: 10820487 [DOI] [PubMed] [Google Scholar]
- 10.Barbaresi S, Cortese MS, Quinn J, Ashrafi GH, Graham SV, Campo MS.. Effects of human papillomavirus type 16 E5 deletion mutants on epithelial morphology: functional characterization of each transmembrane domain. J Gen Virol 2010; 91:521 - 30; http://dx.doi.org/ 10.1099/vir.0.016295-0; PMID: 19812262 [DOI] [PubMed] [Google Scholar]
- 11.White EA, Sowa ME, Tan MJ, Jeudy S, Hayes SD, Santha S, Münger K, Harper JW, Howley PM.. Systematic identification of interactions between host cell proteins and E7 oncoproteins from diverse human papillomaviruses. Proc Natl Acad Sci U S A 2012; 109:E260 - 7; http://dx.doi.org/ 10.1073/pnas.1116776109; PMID: 22232672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu B, Quintero J, Baker CC.. Keratinocyte growth conditions modulate telomerase expression, senescence, and immortalization by human papillomavirus type 16 E6 and E7 oncogenes. Cancer Res 2003; 63:7815 - 24; PMID: 14633708 [PubMed] [Google Scholar]
- 13.Kim M-K, Kim HS, Kim S-H, Oh J-M, Han JY, Lim JM, Juhnn YS, Song YS.. Human papillomavirus type 16 E5 oncoprotein as a new target for cervical cancer treatment. Biochem Pharmacol 2010; 80:1930 - 5; http://dx.doi.org/ 10.1016/j.bcp.2010.07.013; PMID: 20643111 [DOI] [PubMed] [Google Scholar]
- 14.DiMaio D, Mattoon D.. Mechanisms of cell transformation by papillomavirus E5 proteins. Oncogene 2001; 20:7866 - 73; http://dx.doi.org/ 10.1038/sj.onc.1204915; PMID: 11753669 [DOI] [PubMed] [Google Scholar]
- 15.Hwang ES, Nottoli T, Dimaio D.. The HPV16 E5 protein: expression, detection, and stable complex formation with transmembrane proteins in COS cells. Virology 1995; 211:227 - 33; http://dx.doi.org/ 10.1006/viro.1995.1395; PMID: 7645215 [DOI] [PubMed] [Google Scholar]
- 16.Straight SW, Hinkle PM, Jewers RJ, McCance DJ.. The E5 oncoprotein of human papillomavirus type 16 transforms fibroblasts and effects the downregulation of the epidermal growth factor receptor in keratinocytes. J Virol 1993; 67:4521 - 32; PMID: 8392596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodríguez MI, Finbow ME, Alonso A.. Binding of human papillomavirus 16 E5 to the 16 kDa subunit c (proteolipid) of the vacuolar H+-ATPase can be dissociated from the E5-mediated epidermal growth factor receptor overactivation. Oncogene 2000; 19:3727 - 32; http://dx.doi.org/ 10.1038/sj.onc.1203718; PMID: 10949926 [DOI] [PubMed] [Google Scholar]
- 18.Ashrafi GH, Haghshenas M, Marchetti B, Campo MS.. E5 protein of human papillomavirus 16 downregulates HLA class I and interacts with the heavy chain via its first hydrophobic domain. Int J Cancer 2006; 119:2105 - 12; http://dx.doi.org/ 10.1002/ijc.22089; PMID: 16823848 [DOI] [PubMed] [Google Scholar]
- 19.Venuti A, Paolini F, Nasir L, Corteggio A, Roperto S, Campo MS, Borzacchiello G.. Papillomavirus E5: the smallest oncoprotein with many functions. Mol Cancer 2011; 10:140; http://dx.doi.org/ 10.1186/1476-4598-10-140; PMID: 22078316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng W-F, Hung C-F, Chai C-Y, Hsu K-F, He L, Ling M, Wu TC.. Tumor-specific immunity and antiangiogenesis generated by a DNA vaccine encoding calreticulin linked to a tumor antigen. J Clin Invest 2001; 108:669 - 78; http://dx.doi.org/ 10.1172/JCI200112346; PMID: 11544272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peng S, Ji H, Trimble C, He L, Tsai Y-C, Yeatermeyer J, Boyd DA, Hung CF, Wu TC.. Development of a DNA vaccine targeting human papillomavirus type 16 oncoprotein E6. J Virol 2004; 78:8468 - 76; http://dx.doi.org/ 10.1128/JVI.78.16.8468-8476.2004; PMID: 15280455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Diniz MO, Lasaro MO, Ertl HC, Ferreira LCS.. Immune responses and therapeutic antitumor effects of an experimental DNA vaccine encoding human papillomavirus type 16 oncoproteins genetically fused to herpesvirus glycoprotein D. Clin Vaccine Immunol 2010; 17:1576 - 83; http://dx.doi.org/ 10.1128/CVI.00264-10; PMID: 20739505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gill DK, Bible JM, Biswas C, Kell B, Best JM, Punchard NA, Cason J.. Proliferative T-cell responses to human papillomavirus type 16 E5 are decreased amongst women with high-grade neoplasia. J Gen Virol 1998; 79:1971 - 6; PMID: 9714245 [DOI] [PubMed] [Google Scholar]
- 24.Liu DW, Tsao YP, Hsieh CH, Hsieh JT, Kung JT, Chiang CL, Huang SJ, Chen SL.. Induction of CD8 T cells by vaccination with recombinant adenovirus expressing human papillomavirus type 16 E5 gene reduces tumor growth. J Virol 2000; 74:9083 - 9; http://dx.doi.org/ 10.1128/JVI.74.19.9083-9089.2000; PMID: 10982354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanodia S, Da Silva DM, Kast WM.. Recent advances in strategies for immunotherapy of human papillomavirus-induced lesions. Int J Cancer 2008; 122:247 - 59; http://dx.doi.org/ 10.1002/ijc.23252; PMID: 17973257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feltkamp MC, Smits HL, Vierboom MP, Minnaar RP, de Jongh BM, Drijfhout JW, ter Schegget J, Melief CJ, Kast WM.. Vaccination with cytotoxic T lymphocyte epitope-containing peptide protects against a tumor induced by human papillomavirus type 16-transformed cells. Eur J Immunol 1993; 23:2242 - 9; http://dx.doi.org/ 10.1002/eji.1830230929; PMID: 7690326 [DOI] [PubMed] [Google Scholar]
- 27.Massa S, Simeone P, Muller A, Benvenuto E, Venuti A, Franconi R.. Antitumor activity of DNA vaccines based on the human papillomavirus-16 E7 protein genetically fused to a plant virus coat protein. Hum Gene Ther 2008; 19:354 - 64; http://dx.doi.org/ 10.1089/hum.2007.122; PMID: 18439124 [DOI] [PubMed] [Google Scholar]
- 28.Massa S, Paolini F, Spanò L, Franconi R, Venuti A.. Mutants of plant genes for developing cancer vaccines. Hum Vaccin 2011; 7:Suppl147 - 55; http://dx.doi.org/ 10.4161/hv.7.0.14577; PMID: 21266841 [DOI] [PubMed] [Google Scholar]
- 29.Liu D-W, Yang Y-C, Lin H-F, Lin M-F, Cheng Y-W, Chu C-C, Tsao YP, Chen SL.. Cytotoxic T-lymphocyte responses to human papillomavirus type 16 E5 and E7 proteins and HLA-A*0201-restricted T-cell peptides in cervical cancer patients. J Virol 2007; 81:2869 - 79; http://dx.doi.org/ 10.1128/JVI.02256-06; PMID: 17202211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen Y-F, Lin C-W, Tsao Y-P, Chen S-L.. Cytotoxic-T-lymphocyte human papillomavirus type 16 E5 peptide with CpG-oligodeoxynucleotide can eliminate tumor growth in C57BL/6 mice. J Virol 2004; 78:1333 - 43; http://dx.doi.org/ 10.1128/JVI.78.3.1333-1343.2004; PMID: 14722288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smahel M, Síma P, Ludvíková V, Vonka V.. Modified HPV16 E7 genes as DNA vaccine against E7-containing oncogenic cells. Virology 2001; 281:231 - 8; http://dx.doi.org/ 10.1006/viro.2000.0794; PMID: 11277695 [DOI] [PubMed] [Google Scholar]
- 32.Smahel M, Síma P, Ludvíková V, Marinov I, Pokorná D, Vonka V.. Immunisation with modified HPV16 E7 genes against mouse oncogenic TC-1 cell sublines with downregulated expression of MHC class I molecules. Vaccine 2003; 21:1125 - 36; http://dx.doi.org/ 10.1016/S0264-410X(02)00519-4; PMID: 12559790 [DOI] [PubMed] [Google Scholar]
- 33.Massa S, Franconi R, Brandi R, Muller A, Mett V, Yusibov V, Venuti A.. Anti-cancer activity of plant-produced HPV16 E7 vaccine. Vaccine 2007; 25:3018 - 21; http://dx.doi.org/ 10.1016/j.vaccine.2007.01.018; PMID: 17280752 [DOI] [PubMed] [Google Scholar]
- 34.Venuti A, Massa S, Mett V, Vedova LD, Paolini F, Franconi R, Yusibov V.. An E7-based therapeutic vaccine protects mice against HPV16 associated cancer. Vaccine 2009; 27:3395 - 7; http://dx.doi.org/ 10.1016/j.vaccine.2009.01.068; PMID: 19200826 [DOI] [PubMed] [Google Scholar]
- 35.Disbrow GL, Sunitha I, Baker CC, Hanover J, Schlegel R.. Codon optimization of the HPV-16 E5 gene enhances protein expression. Virology 2003; 311:105 - 14; http://dx.doi.org/ 10.1016/S0042-6822(03)00129-6; PMID: 12832208 [DOI] [PubMed] [Google Scholar]
- 36.Oetke C, Auvinen E, Pawlita M, Alonso A.. Human papillomavirus type 16 E5 protein localizes to the Golgi apparatus but does not grossly affect cellular glycosylation. Arch Virol 2000; 145:2183 - 91; http://dx.doi.org/ 10.1007/s007050070048; PMID: 11087100 [DOI] [PubMed] [Google Scholar]
- 37.Wetherill LF, Holmes KK, Verow M, Müller M, Howell G, Harris M, Fishwick C, Stonehouse N, Foster R, Blair GE, et al.. High-risk human papillomavirus E5 oncoprotein displays channel-forming activity sensitive to small-molecule inhibitors. J Virol 2012; 86:5341 - 51; http://dx.doi.org/ 10.1128/JVI.06243-11; PMID: 22357280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang DH, Wildeman AG, Sharom FJ.. Overexpression, purification, and structural analysis of the hydrophobic E5 protein from human papillomavirus type 16. Protein Expr Purif 2003; 30:1 - 10; http://dx.doi.org/ 10.1016/S1046-5928(03)00049-4; PMID: 12821315 [DOI] [PubMed] [Google Scholar]
- 39.Lee SY, Huang Z, Kang TH, Soong R-S, Knoff J, Axenfeld E, Wang C, Alvarez RD, Chen CS, Hung CF, et al.. Histone deacetylase inhibitor AR-42 enhances E7-specific CD8⁺ T cell-mediated antitumor immunity induced by therapeutic HPV DNA vaccination. J Mol Med (Berl) 2013; 91:1221 - 31; http://dx.doi.org/ 10.1007/s00109-013-1054-9; PMID: 23715898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin K, Roosinovich E, Ma B, Hung CF, Wu TC.. Therapeutic HPV DNA vaccines. Immunol Res 2010; 47:86 - 112; http://dx.doi.org/ 10.1007/s12026-009-8141-6; PMID: 20066511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vici P, Mariani L, Pizzuti L, Sergi D, Di Lauro L, Vizza E, Tomao F, Tomao S, Cavallotti C, Paolini F, et al.. Immunologic treatments for precancerous lesions and uterine cervical cancer. J Exp Clin Cancer Res 2014; 33:29; http://dx.doi.org/ 10.1186/1756-9966-33-29; PMID: 24667138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ristriani T, Fournane S, Orfanoudakis G, Travé G, Masson M.. A single-codon mutation converts HPV16 E6 oncoprotein into a potential tumor suppressor, which induces p53-dependent senescence of HPV-positive HeLa cervical cancer cells. Oncogene 2009; 28:762 - 72; http://dx.doi.org/ 10.1038/onc.2008.422; PMID: 19015633 [DOI] [PubMed] [Google Scholar]
- 43.Ramos CR, Abreu PA, Nascimento AL, Ho PL.. A high-copy T7 Escherichia coli expression vector for the production of recombinant proteins with a minimal N-terminal His-tagged fusion peptide. Braz J Med Biol Res 2004; 37:1103 - 9; http://dx.doi.org/ 10.1590/S0100-879X2004000800001; PMID: 15273812 [DOI] [PubMed] [Google Scholar]
- 44.Miyahira Y, Murata K, Rodriguez D, Rodriguez JR, Esteban M, Rodrigues MM, Zavala F.. Quantification of antigen specific CD8+ T cells using an ELISPOT assay. J Immunol Methods 1995; 181:45 - 54; http://dx.doi.org/ 10.1016/0022-1759(94)00327-S; PMID: 7537312 [DOI] [PubMed] [Google Scholar]
- 45.Seo SH, Jin HT, Park SH, Youn JI, Sung YC.. Optimal induction of HPV DNA vaccine-induced CD8+ T cell responses and therapeutic antitumor effect by antigen engineering and electroporation. Vaccine 2009; 27:5906 - 12; http://dx.doi.org/ 10.1016/j.vaccine.2009.07.033; PMID: 19651174 [DOI] [PubMed] [Google Scholar]