Abstract
The development of an HIV-1 vaccine that is capable of inducing effective and broadly cross-reactive humoral and cellular immune responses remains a challenging task because of the extensive diversity of HIV-1, the difference of virus subtypes (clades) in different geographical regions, and the polymorphism of human leukocyte antigens (HLA). We performed an in silico design of 3 DNA vaccines, designated pJW4303-MEG1, pJW4303-MEG2 and pJW4303-MEG3, encoding multi-epitopes that are highly conserved within the HIV-1 subtypes most prevalent in China and can be recognized through HLA alleles dominant in China. The pJW4303-MEG1-encoded protein consisted of one Th epitope in Env, and one, 2, and 6 epitopes in Pol, Env, and Gag proteins, respectively, with a GGGS linker sequence between epitopes. The pJW4303-MEG2-encoded protein contained similar epitopes in a different order, but with the same linker as pJW4303-MEG1. The pJW4303-MEG3-encoded protein contained the same epitopes in the same order as that of pJW4303-MEG2, but with a different linker sequence (AAY). To evaluate immunogenicity, mice were immunized intramuscularly with these DNA vaccines. Both pJW4303-MEG1 and pJW4303-MEG2 vaccines induced equally potent humoral and cellular immune responses in the vaccinated mice, while pJW4303-MEG3 did not induce immune responses. These results indicate that both epitope and linker sequences are important in designing effective epitope-based vaccines against HIV-1 and other viruses.
Keywords: DNA vaccine, HLA, HIV-1, linker, multi-epitope
Abbreviations
- AIDS
acquired immunodeficiency syndrome
- BSA
bovine serum albumin
- CRF
circulating recombinant form
- CTL
cytotoxic T lymphocyte
- HIV
human immunodeficiency virus
- HLA
human leukocyte antigen
- MEG
multi-epitope genes
- MHC
major histocompatibility complex
- SFU
spot forming units
Introduction
Acquired immunodeficiency syndrome (AIDS) caused by human immunodeficiency virus type 1 (HIV-1) is a major threat to public health.1 Despite advances in antiretroviral therapy and the management of the AIDS epidemic by other measures, developing a safe and effective vaccine is still a preferred strategy to curb the global HIV pandemic. However, the development of an effectivet vaccine against HIV-1 infection is hampered by several issues, including the extensive diversity of HIV-1, the different virus subtypes (clades) in different geographical regions, and genetic polymorphism in HLA molecules in humans.2-6
Ideally, an effective vaccine with the ability to prevent HIV-1 infection would elicit both humoral and cellular immune responses.7-9 Since no candidate immunogen is capable of stimulating broadly neutralizing antibodies, a protective vaccine against HIV-1 infection based solely on the elicitation of protective antibodies is still not viable.10,11 While non-neutralizing antibodies may effectively protect against HIV-1 infection, correlates of protection have still not been defined.12 A vaccine that induces HIV-1 specific T cell responses is, therefore, an important alternative to vaccines that stimulate broadly neutralizing antibodies.13-15 To tackle the problem of viral diversity, different strategies for T-cell-based vaccines against HIV-1 infection focused on the immunogen.16 One such strategy is to design a vaccine that targets only the conserved epitopes and aims to induce a broader cross-protective immune response.17,18
Epitopes are the smallest and most important units that can elicit immune responses. As such, they are potential candidates for modern vaccine design and have unique advantages. First, epitopes are well-defined protective elements that can elicit both humoral and cellular immune responses. Second, highly conserved regions can still be found within the frequently variable viral protein.19 Because mutations in such regions would affect viral viability,20 epitopes within these regions should induce cross-protective immune responses, thus increasing the breadth of immune responses. Third, selecting immune subdominant, but protective, epitopes could avoid subdominant responses from being suppressed by dominant T-cell responses to ‘trap’ epitopes, a phenomenon which happens during natural infection.21-23
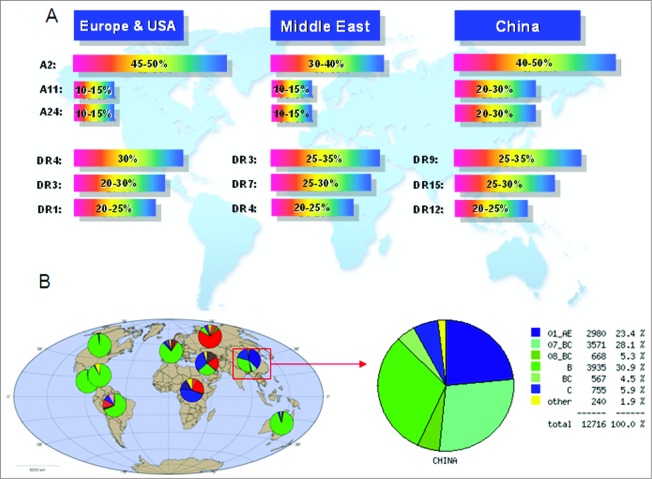
Several strategies were applied to design epitope-based vaccines, and their immunogenicity and protective efficacy were evaluated in animal models. Some of these candidates have been used in clinical trials and showed promise in the control of HIV-1 infection.24-29 However, these vaccine designs did not consider the characteristics of HIV-1 prevalent in China, HLA polymorphisms in the Chinese population, or the effect of linkers between epitopes on immunogenicity. HIV-1 prevalence and HLA alleles are different in different countries and regions. Nearly 90% of HIV-1 infections in mainland China are caused by CRF01_AE, CRF07_BC, CRF08_BC, and subtype B. In addition to these 4 subtypes, subtype C is also important, and causing about 5% of HIV-1 infections. As shown in Figure 1A, other uncommon subtypes including subtype A, subtype G, and other circulating recombinant forms are also present in China. By contrast, the epidemics in North America, Latin America, western and central Europe, and Australia are dominated by subtype B. In eastern Europe and central Asia, it is dominated by subtypes A and B, and in southern Africa and India, it is dominated by subtype C (Fig. 1B).30 The most common HLA-A alleles in the Chinese population are A*1101, *2402, *3303, and *0203, and the most common 3 haplotypes in DRB1 loci of Chinese people are DRB1*0901, *1202, and *1501, which also display unique allele frequency distributions when compared with those of other countries or regions (Fig. 1A).31,32
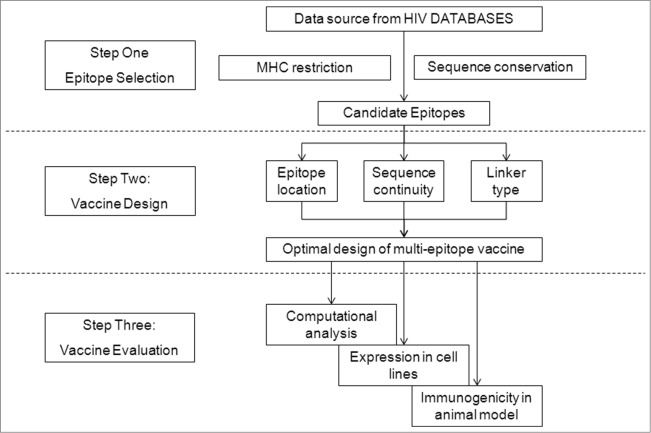
Figure 2.
Strategy of HIV-1 multi-epitope DNA vaccine design, including epitope selection, vaccine design, and vaccine evaluation.
Figure 1.
Analysis of worldwide distribution of HLA alleles and HIV-1 prevalence. (A) HIV-1 subtype distribution in China based on HIV DATABASES (www.hiv.lanl.gov). (B) HLA allele distribution.
Figure 7.
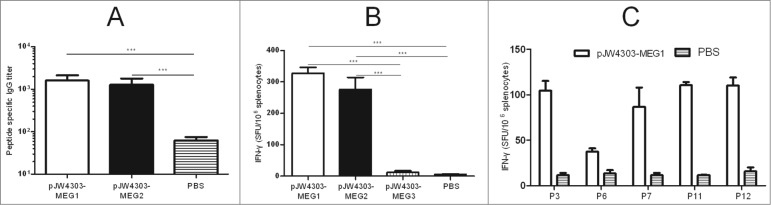
Evaluation of immune responses in BALB/c mice induced by pJW4303-MEG1, pJW4303-MEG2, and pJW4303-MEG3. BALB/c mice were intramuscularly immunized with pJW4303-MEG1, pJW4303-MEG2, and pJW4303-MEG3 at a dose of 10 μg of DNA dissolved in 200 μl of PBS according to the immunization schedule. (A) Two weeks after the final immunization, sera were collected and peptide-specific IgG was measured by ELISA. (B) On day 21 after the final immunization, mice were sacrificed, and splenocytes were stimulated with pool-peptides for 48 hours to detect cellular immune response by ELISPOT assay. (C) Splenocytes from mice injected with pJW4303-MEG1 were also stimulated with H-2d-restricted epitopes to evaluate cellular immune response to single peptides.
In this paper, we report the optimized design of a multi-epitope vaccine that takes into account all of these issues. Several elements were considered for our multiple-epitope vaccine design: epitope selection, epitope location, and linkers between epitopes. We chose 9 conserved HIV-1 CTL epitopes matching the HLA-restriction in the Chinese population, as well as a Th-epitope-rich region, and designed 3 recombinant multi-epitope genes (MEG) encoding ‘artificial’ proteins composed of these epitopes and connected by different linker sequences, followed by an evaluation of their structural characteristics in silico and their expression in vitro. Finally, their immunogenicity was evaluated in BALB/c mice in vivo (Fig. 2). Our data suggest that both the epitope and linker sequences are important in designing effective HIV-1 vaccines.
Results
Analysis and selection of HIV-1 CTL and Th epitopes
To find suitable CTL epitopes for our epitope-based vaccine construction, the online tool CTL/CD8+ Search in HIV DATABASES was used. Selection standards were set as described in materials and methods. In total, 111 epitopes with defined MHC restriction information, including human HLA molecules and the BALB/c mouse H-2d molecule, were initially obtained from the entire database, of which 49 epitopes were located in the Env protein, 45 in Gag, and 17 in Pol (data not shown). Further analysis was performed to select epitopes restricted to the predominant MHC-I alleles in Chinese population(HLA-A2/A11/A24), and 9 CTL epitopes (6 in Gag, 2 in Env, and one in Pol) were chosen as candidate epitopes for vaccine design (Table 1). When searching for Th epitopes, we noticed that almost none of the reported Th epitopes had exact MHC-II restriction information and that most of them derived from the HIV-1 infected Caucasian population whose predominant MHC-II alleles are DR01, and hence are quite different from the predominant Chinese MHC-II alleles (DR09, DR12, and DR15). Th-epitope-rich regions in the HIV protein T-Helper/CD4+ Epitope Map were selected, and MHC-II restriction was further analyzed utilizing the online software NetMHCII 2.2 server. A 36-aa-long peptide located in the V2 region of gp120 was finally chosen and predicted to contain several pan-DR epitopes. Within this peptide, 2 9-mer core sequences (KVSFEPIPI and IHYCAPAGF) were predicted to bind Chinese dominant MHC-II alleles (DRB10101, DRB10701, DRB10901, and DRB11501).
Table 1.
Characteristics of the candidate epitopes selected for epitope-based vaccine construction
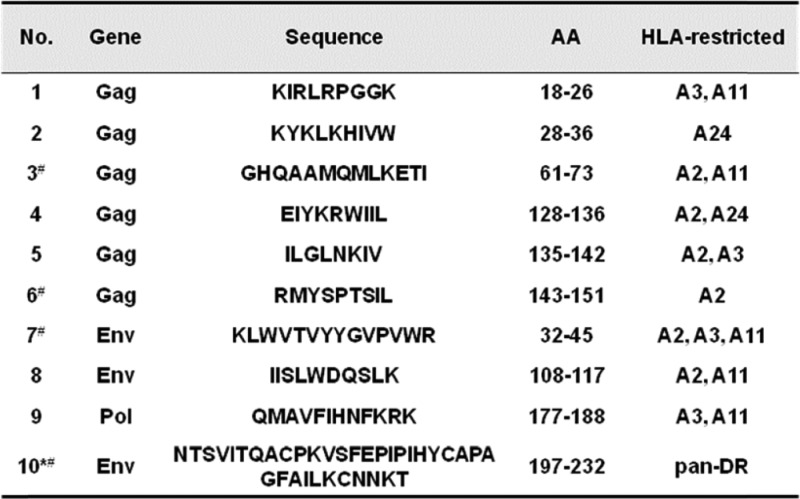
|
*T-helper epitope was predicted to contain several pan-DR epitopes after analysis by NetMHCII 2.2 server. For the stimulation experiment, this long peptide was synthesized as 3 short peptides, including P10 (NTSVITQACPKVSFEPIP), P11 (PKVSFEPIPIHYCAPAGF), and P12 (APAGFAILKCNNKT), of which P11 and P12 are also H-2d-restricted T-cell epitopes.
#Epitopes marked with this symbol are also restricted to H-2d.
To confirm the conservation of the selected epitopes within viral subtypes in China, we firstly created a sequence database consisting of 52 B subtype sequences, 160 CRF01_AE subtype sequences, 33 CRF07_BC subtype sequences, and 33 CRF08_BC subtype sequences. All selected epitopes were aligned with these sequences, and most of the amino acids (AAs) were found to be highly conserved (Fig. 3). However, 5 subtype-biased variations were identified during the alignment: RMYSPT(V)SIL in CRF01_AE, GHQAAMQM(I)LKETI in CRF07_BC, KYK L(M)KHIVW in CRF01_AE, KYK(M)LKHIVW in CRF08_BC, and NTSVIT(K)QACPK in CRF01_AE.
Figure 3.
Conservation analysis of the candidate epitopes within HIV-1 viral subtypes prevalent in China. Two hundred and 7eight strains of HIV-1 (52 B subtypes, 160 CRF01_AE subtypes, 33 CRF07_BC subtypes, and 33 CRF08_BC subtypes) prevalent in mainland China were included in this study. The conservative characteristics of AAon the selected epitopes were analyzed by aligning the epitopes with the corresponding molecules on 278 strains of HIV-1, and the conservation was expressed as a percentage of the number of HIV-1 strains having the conserved AAon the epitope from the 278 strains. The core sequences of the CTL and Th epitopes are highly conserved with rare mutations.
Consequently, we obtained highly conserved epitopes within HIV-1 subtypes prevalent in China, which also allowed for ethnic-specific HLA type distributions. Three CTL epitopes and 2 potential epitopes within the Th epitope were also restricted to H-2d molecules, making it convenient to evaluate the immunogenicity of multi-epitope vaccines in BALB/c mice.
In silico design and structural analysis of the HIV-1 multi-epitope DNA vaccines
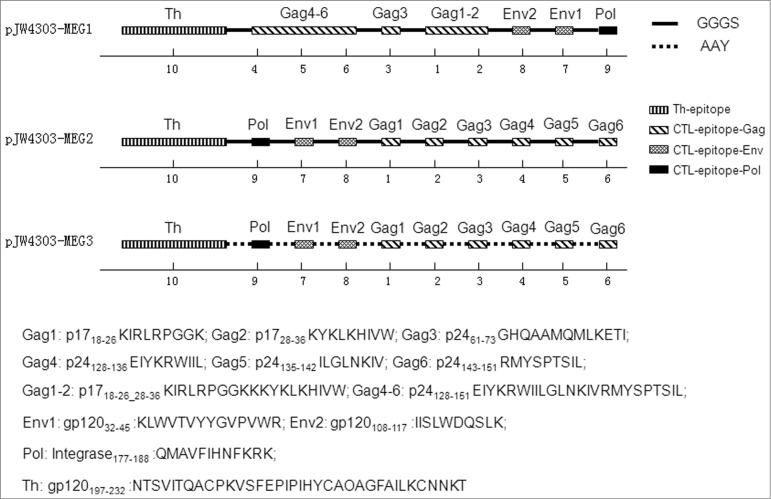
Three different multi-epitope string sequences were designed, as described in Materials and Methods, and their schematic diagrams are shown in Figure 4. We further estimated the effect of linker, epitope location, and sequence continuity on protein structural characteristics. The analysis results by PROTEAN showed that the linker between the epitopes, but not the epitope location or sequence continuity, significantly affected the characteristics. When comparing sequence MEG1 with MEG2, both of which used flexible linkers (GGGS), we found that indexes of every single epitope changed very little, notwithstanding different epitope location and/or formation (Figs. 5A and 5B). However, once linkers changed from GGGS (MEG2) to AAY (MEG3), the structural parameters changed greatly, especially indexes of α, β, turn, and coil regions, which are indications of protein secondary structure (Figs. 5B and 5C). In particular, AAY linkers reduced the number of β-, turn-, and coil-regions, but increased the number of α-regions, making sequence MEG3 less flexible and, in turn, indicating that MEG3 connected with linker ‘AAY’ is less stable in structure and possesses weaker immunogenicity.
Figure 4.
Schematic diagram of the 3 multi-epitope-based DNA vaccines. Epitopes include a 36-aa-long peptide acting as a T-helper epitope (aa 197–232), 6 CTL epitopes in Gag, 2 CTL epitopes in Env, and one epitope in Pol. Epitope locations, continuity, and linkers differ in the 3 sequences. Epitopes are connected using 2 different linkers: GGGS (solid line) was used for pJW4303-MEG1 and pJW4303-MEG2, while AAY (dashed line) was used for pJW4303-MEG3. Numbers below models stand for epitope numbers, which are shown in Table 1.
Figure 5.
Secondary structure analysis of the multi-epitope-based candidate vaccines. The secondary structure, including α region, β region, turn region and coil region, and antigenicity-related properties, such as hydrophilicity, antigenic index, surface probability, and flexible region, of the artificial proteins based on the conserved epitopes were predicted using PROTEAN pattern of LASERGENE software. Panels A, B, and C are the analytic results of the DNA vaccines pJW4303-MEG1, pJW4303-MEG2, and pJW4303-MEG3, respectively.
Construction and expression of the HIV-1 multi-epitope DNA vaccines in vitro
Three different double chain DNA sequences were inserted into vector pJW4303. DNA products were identified by restriction endonuclease cleavage analysis (Fig. 6A) and sequencing. Subsequently, DNA vaccines were transiently transfected into 293T cells to observe their expression in vitro and to assess differences between multi-epitope sequences that were predicted in the bioinformatics analysis above. Western blot showed that cells transfected with pJW4303-MEG1 or pJW4303-MEG2 expressed a protein of nearly 16kDa, whereas no signals were detected in cells transfected with pJW4303-MEG3 or negative control (Fig. 6B). Similar results were also observed when COS-7 cells were used (data not shown). These results indicate that ‘AAY’ linkers significantly influence the expression of target proteins.
Figure 6.

Identification and expression of the DNA vaccines in vitro. (A) Restriction enzyme analysis of the 3 multi-epitope DNA vaccines, with digestion by BamH I and EcoR I,follwed by 1% agarose electrophoresis. Two bands (about 5,000 bp and 500 bp) were observed for all 3 recombinant plasmids. Lane 1, pJW4303-MEG1; lane 2, pJW4303-MEG2; lane 3, pJW4303-MEG3; M, Trans 2K Plus marker. (B) Western blot analysis of DNA vaccine protein expression in vitro. 293T cells were transiently transfected with pJW4303-MEG1, pJW4303-MEG2, or pJW4303-MEG3. Expression of the proteins was identified by Western blot using rabbit anti-MEG1.E polyclonal antibody as primary antibody. A 16 kDa band was observed for both pJW4303-MEG1 and pJW4303-MEG2, but no signal was detected for pJW4303-MEG3 or the control. Lane 1, pJW4303-MEG1; lane 2, pJW4303-MEG2; lane 3, pJW4303-MEG3; lane 4, negative control. Protein marker beside the film used to illustrate protein sizes was cut from the PVDF membrane.
Validation of immunogenicity of the HIV-1 multi-epitope DNA vaccine in vivo
An animal assay was carried out to test the ability of epitope-based vaccines to induce specific immune responses. BALB/c mice were intramuscularly inoculated with the naked DNA, and antigen-specific immune responses were detected at planned time points. Mice immunized with pJW4303-MEG1 and -MEG2 developed peptide-specific antibodies of which the level reached 103 (Fig. 7A). However, peptide-specific IgG titer in mice immunized with pJW4303-MEG3 was absolutely low, and no significant difference was observed between mice immunized with pJW4303-MEG3 and PBS control when serum dilution was at 1:50.
ELISPOT assays showed that both pJW4303-MEG1 (328 SFU/106cells) and -MEG2 (276 SFU/106cells) elicited significantly higher numbers of epitope-specific IFNγ-producing cells than pJW4303-MEG3 (14 SFU/106 cells) and PBS control (5 SFU/106 cells) (Fig. 7B, p<0.001), demonstrating that both pJW4303-MEG1 and -MEG2 elicited equally strong cellular immune responses. Having proven the immunogenicity of such multi-epitope vaccines, we next investigated whether the multi-epitope vaccines connected with linker "GGGS" could elicit a T cell immune response to each epitope. To this end, we stimulated splenocytes from mice immunized with pJW4303-MEG1 using H-2d-restricted epitopes (P3, P6, P7, P11, and P12). pJW4303-MEG1-immunized mice generated cellular immune responses to all 5 mouse T-cell epitopes, suggesting that epitopes connected by linker "GGGS" could efficiently elicit immune responses to each peptide (Fig. 7C). Taken together, multi-epitope vaccines pJW4303-MEG1 and -MEG2 using linker “GGGS” induced better immune responses in vivo than pJW4303-MEG3 containing ‘AAY’, implying that ‘GGGS’ is superior to ‘AAY’ as a ‘linker’ of epitope-based vaccines.
Discussion
During the last 30 years, various strategies evolved to develop a vaccine against HIV-1, but problems still remain to be solved.33 Differences in HIV-1 prevalence and HLA subtype distribution between China and other countries and areas make it important to develop vaccines suitable for the Chinese population. In the current study, we systemically analyzed the conservation of T-cell epitopes from the most prevalent HIV-1 strains in China and analyzed the effect of rare mutations of epitopes on epitope recognition. We designed a vaccine based on epitopes highly conserved within different HIV-1 subtypes and matched the specified HLA restriction to the Chinese population through rational design considering several factors that might affect the efficacy of epitope-based vaccines, including epitope selection, epitope location, and linker sequence between epitopes. To our knowledge, this is the first report on HIV-1 vaccine design simultaneously considering Chinese HLA alleles and HIV-1 prevalence in a systematic analysis of the MHC class I-restricted epitopes of HIV-1.As HLA alleles and HIV-1 prevalence are different in different countries and regions, we considered the epidemic characteristics of HIV-1 infection in China and Chinese predominant HLA alleles for epitope selection. After analyzing epitope conservation and MHC restriction, we selected several epitopes that are conserved in most HIV-1 strains prevalent in China and restricted through MHC molecules of the Chinese population. These include 9 CTL epitopes from Gag, Pol, and Env proteins and one pan-DR Th-epitope-rich region located in Env. The nine CTL epitopes were highly conserved within the 4 dominant viral subtypes on the mainland and also conserved within the C subtype, except for rare subtype-biased mutations. However, these mutations did not affect the binding between epitope and MHC molecule. To explain, firstly, these mutations are not on the MHC anchoring sites, which are the second and ninth positions of the epitope for A02/A03/A24 molecules and the last AA of the nonamer is important for epitope binding to the A11 molecule (http://www.hiv.lanl.gov/content/immunology/motif_scan). Secondly, AAs with the same polarity do not significantly influence peptide structure, and many mutations exhibited such homopolarity exchange, for example, glutamic acid (E) changed to aspartic acid (D) in EIYKRWIIL (gag, 128–136), and lycine (K) changed to histidine (H) or arginine (R) in KYKLKHIVW (gag, 28–36). In addition, further analysis of the MHC binding affinity using online software SYFPEITHI (http://www.syfpeithi.de/index.html) and TAP binding affinity by MHCPred (version 2.0) (http://www.ddg-pharmfac.net/mhcpred/MHCPred/) demonstrated that these rare mutations did not cripple HLA recognition. The nine CTL epitopes restricted to Chinese predominant MHC-I alleles could cover the majority (at least 90%) of the Chinese population, as well as 50% of the population worldwide. This means that vaccines consisting of such epitopes might provide protection to most of the Chinese population, as well as confer protection to half of the population outside ethnic group.
Several reports indicated that Th epitopes are capable of increasing the immunogenicity of CTL epitope-based vaccines,34 and, as such, some universal Th epitopes were previously incorporated into such vaccines.34,35 In this work, Th epitopes were included as part of the multi-epitope vaccine to induce better immune responses. In contrast other studies, HIV-1-specific Th epitopes, rather than universal Th epitopes, were chosen for 2 reasons. First, T-helper lymphocytes are crucial for eliciting both humoral and cellular immune responses and are necessary to maintain memory CTL responses for HIV-1 clearance. Second, Th responses are a part of protective immunity against HIV-1 infection.36,37 However, we experienced difficulty in finding such Th epitopes, as most published work on epitope identification focused on epitopes restricted to European and American populations,38 with little information being available about epitopes that match Chinese predominant MHC-II alleles. We finally discovered a 36-aa-long peptide located in the V2 region, a T-cell epitope-rich region, and predicted that it would contain several pan-DR epitopes. Thus, we obtained 9 conserved CTL epitopes and a pan-DR Th epitope-rich region as our peptide candidates to construct a multi-epitope vaccine.
A multi-epitope vaccine should be able to elicit immune responses to each epitope. Accordingly, 2 factors which might affect the immunogenicity of the ‘artificial’ protein were taken into account: epitope location and continuity and linker sequence between epitopes.
To assess the influence of epitope location and sequence continuity on the epitope-based vaccine, we designed 10 sets of multi-epitope molecules by randomly putting epitopes in different locations and connecting them by the same ‘GGGS’ linker. Secondary structure prediction showed that epitope location and continuity did not affect the characteristics of any epitope, indicating that epitope location is not the major factor affecting the immunogenicity of multiple-epitope vaccines (data not shown). This conclusion was further validated by experiments measuring in vitro expression and immune responses in BALB/c mice using 2 of these 10 molcules, pJW4303-MEG1 and pJW4303-MEG2.
Another major element is the ‘linker’ between epitopes. We analyzed the effect of 2 different linkers, ‘GGGS’ and ‘AAY’, on immunogenicity of the multi-epitope vaccines. DNA vaccines pJW4303-MEG1 or -MEG2 and pJW4303-MEG3, consisting of the epitopes above, were assembled by ‘GGGS’ and ‘AAY’, respectively, and their immunogenicity was analyzed in silico, in vitro, and in vivo. The results showed that the linker type connecting the epitope sequences does, indeed, have a substantial influence on protein characteristics. ‘GGGS’ was a better linker than ‘AAY’ for multi-epitope vaccines in eliciting immune responses. G-rich linkers have the capacity to improve sequence flexibility without affecting the function of proteins they attach to,39,40 while the AAY linker, as the cleavage site of proteasomes in mammalian cells, helps form natural epitopes and prevent the formation of ‘junctional epitopes’, which enhanced epitope presentation. Unexpectedly, however, linker AAY induced large changes in protein characteristics, including hydrophilicity, flexibility, α region, β region, turn region, and coil region, all of which could influence protein stability, and reduce less immunogenicity. This prediction was proved by transient transfection in cell lines and immunogenicity evaluation in BALB/c mice. DNA vaccine pJW4303-MEG3, which was connected by ‘AAY’, did not efficiently express the multiple epitopes, and failed to induce detectable humoral or cellular immune responses in mice. This finding differs from previously reported data, in which the ‘AAY’ linker increased the immunogenicity of epitope-based vaccines.41,42 Possible reasons for this phenomenon include the following: 1) the protein MEG3 was not efficiently expressed in vivo, similar to what occurred in HEK293T cells (Fig. 6B), probably because the linker AAY changed the protein structure tremendously, which lead to impaired protein expression; and 2) the peptides were not efficiently presented to immune cells. In this later scenario, the MEG3 protein might be successfully expressed in eukaryotic cells but quickly cleaved into short peptides at the AAY linker (the target of proteasomes), thus making it undetectable by Western blot in vitro and poorly immunogenic in vivo.
Collectively, we obtained several epitopes that are highly conserved in the HIV-1 subtypes prevalent in China and restricted to Chinese HLA alleles. Multi-epitope DNA vaccines pJW4303-MEG1 and pJW4303-MEG2 consisting of these epitopes assembled using ‘GGGS’ linkers were constructed and were capable of eliciting humoral and cellular immune responses in BALB/c mice. Our experimental data indicated that in addition to the epitopes themselves, the linker between the epitopes is an important factor influencing efficiency of multi-epitope-based vaccines, with G-rich linkers being particularly effective for multi-epitope vaccine design. Immunogenicity of the multi-epitope DNA vaccines was evaluated in BALB/c mice only, and future studies will evaluate the immune responses elicited in other animals such as human MHC-transgenic mice or non-human primates.
Materials and Methods
Mice
Female 6–8-week-old BALB/c mice (Beijing Experimental Animal Center, Beijing, China) were used for the mouse experiments. The experimental protocols were approved by The Laboratory Animal Center, State Key Laboratory of Pathogen and Biosecurity, Beijing Institute of Microbiology and Epidemiology IACUC's.
Cell lines
Human embryonic kidney (HEK) 293T cells were used for protein expression in vitro. They were maintained in DMEM medium (GIBCO, C12430500BT) containing 1% L-glutamine, 1% penicillin and streptomycin, and 10% or 2% fetal bovine serum (FBS) (GIBCO, 10099–141) at 37°C with 5% CO2 in a CO2 incubator.
Peptides
Twelve peptides (P1-P12) were synthesized (GenScript) and used to stimulate splenocytes in ELISPOT assays, of which P10-P12 were within peptide 10, and P1-P9 corresponded to peptides 1–9. Purity of the peptides in the form of white lyophilized powder was greater than 90%. Stock solutions of peptides (2 mg/ml) were prepared by dissolving the peptide in sterile PBS (GIBCO, C10010500BT) or in PBS containing no more than 10% DMSO (SIGMA, D2650), followed by filtration through a 0.2 μm filter (PALL, PN4612) and storage at −70°C.
Selection of candidate epitopes and establishment of HIV-1 sequence databases
All HIV-1 epitopes, including CTL and Th epitopes on HIV DATABASES (www.hiv.lanl.gov/), were summarized and analyzed to set up a basic epitope data source. Candidate CTL epitopes having the following characteristics were selected: 1) located in virus structural proteins Gag or Env or protein Pol; 2) highly conserved within virus subtypes prevalent in China; and 3) restricted to dominant Chinese MHC-I alleles. Since few epitopes have been reported to be restricted to Chinese MHC-II alleles, epitope MHC-II restriction analysis was performed, utilizing the online software NetMHCII 2.2 server to search for Th epitopes.
To evaluate epitope conservation, a virus sequence database of HIV-1 prevalent in China was constructed utilizing the Geography Search Interface pattern of HIV DATABASES. The area was focused on China, and the 4 main HIV-1 subtypes (CRF01_AE, CRF07_BC, CRF08_BC and B) were chosen. All complete sequences submitted to HIV DATABASES that fit the qualifications were selected but incomplete sequences were excluded.
Conservation of each epitope was analyzed by aligning epitopes within protein sequences utilizing MEGA 5.0 software.
Design of multi-epitope DNA candidate vaccines
Three DNA fragments consisting of MEG sequences, MEG1, MEG2 and MEG3, were designed. The main principle was to assemble the selected epitopes in a specific order with linkers. The Th epitope was placed at the N-terminus of all the combinations (Fig. 4). MEG1 contained the epitopes in the following order: Th, Gag4–6, Gag3, Gag1–2, Env2, Env1, and Pol. MEG2 had similar epitopes, but in a different order: Th, Pol, Env1, Env2, Gag1, Gag2, Gag3, Gag4, Gag5, and Gag6. The major difference between MEG1 and MEG2 was epitope location and order.In addition, MEG1 kept natural amino acid sequence continuity for some epitopes. For example, MEG1 contained Gag4–6 (p24128–151 EIYKRWIILGLNKIVRMYSPTSIL), while MEG2 had Gag4 (p24128–136), Gag5 (p24135–142) and Gag5 (p24143–151), plus 2 linkers between the epitopes. Both MEG1 and MEG2 have the flexible GGGS linker sequence. MEG3 consisted of the same epitopes in the same order as MEG2, but with an AAY linker sequence (a cleavage site for proteasomes in vivo). The three multi-epitope DNA sequences MEG1, MEG2 and MEG3 were codon-optimized for high-level expression in mice and synthesized by GenScript. They were then cloned into eukaryotic expression vector pJW4303 to form multi-epitope DNA vaccines pJW4303-MEG1, pJW4303-MEG2 and pJW4303-MEG3. Their DNA sequences were confirmed by restriction enzyme analysis and sequencing (SinoGenoMax, Beijing, China).
Analysis of antigen characteristics in the multi-epitope candidate vaccines
The PROTEAN pattern of LASERGENE (DNASTAR) was used to analyze characteristics of the 3 multi-epitope sequences. Indexes included α regions, β regions, turn regions, hydrophilicity, amphipathic regions, flexible region, and surface probability.
Transfection and Western blot
HEK293T cells cultured in T-75 flasks were transiently transfected with 40 μg of pJW4303-MEG1,pJW4303-MEG2, or pJW4303-MEG3 using Lipofectamine 2000 Transfection Reagent (Invitrogen, 11668–019). The cells were collected 48 hours post-transfection for analysis of protein expression by Western blot. SDS-PAGE gel containing protein samples was transferred onto a polyvinylidene difluoride (PVDF) membrane (MILLIPORE, IPVH00010), which was then blocked for 2 hours with 5% non-fat dry milk (APPLYGEN, P1622) in TBST buffer and incubated for 1 hour with rabbit anti-MEG1.E polyclonal antibody (home-made in our lab) diluted 1: 5000, followed by 1 hour incubation with 1:4000 diluted HRP-labeled goat anti-rabbit IgG (Santa Cruz Biotechnology, sc-2004). Finally, the membrane was soaked in chemiluminescent HRP substrate (MILLIPORE, WBKLS0100) and exposed to a suitable X-ray film (Carestream Health, 6535876).
Mouse immunization
Mice were intramuscularly injected with 10 μg of pJW4303-MEG1, pJW4303-MEG2 or pJW4303-MEG3 in 200 μl of PBS on day 0, 28, and 56. Serum samples were collected by tail-vein bleeding before the first immunization and 2 weeks after each immunization, followed by storage at −20°C for antibody detection by ELISA. Splenocytes were harvested for ELISPOT assay 21 d after final immunization.
ELISA
96-well plates coated with pooledpeptides at the concentration of 1 μg/ml of each peptide were kept at 4°C overnight and blocked with 3% bovine serum albumin (BSA) in PBS for 2 hours at 37°C. Serum samples serially diluted fold2- were added to the wells, and plates were incubated at 37°Cfor 1 hour. After washing with PBST buffer (PBS containing 0.05% Tween 20) 4 times, HRP-labeled goat anti-mouse IgG (Southern Biotech, 1030–05) was added to the plates at a 1:8000 dilution and incubated at 37°C for 1 hour. After another 4 washes with PBST, colorimetric analysis was performed using TMB substrate, and the absorbance was read at 450 nm by a spectrophotometer.
ELISPOT assay
ELISPOT assay (IFNγ and IL-4) was performed according to the manufacturer's protocol (BD Biosciences, 551083 and 551017) to evaluate cellular immune responses induced by DNA vaccines. Plates coated with capture antibody were kept at 4°C overnight. Before splenocytes were added to the wells, plates were washed with 200 μl/well blocking solution (RPMI 1640 containing 10% FBS and 1% penicillin-streptomycin-L-glutamine) once and blocked with blocking solution for 2 hours at 37°C. Cells at a final concentration of 5 × 105 cells/well in triplicate were co-stimulated with peptide-pool or single peptide, with each peptide at 10 μg/ml. Cells in culture medium only served as negative control. Plates were kept at 37°C for 48 hours, and the number of SFUs was measured by ELISPOT reader. Wells in which the number of spots was more than 3 times that of negative wells were judged to be positive.
Statistics
Statistical analyses were performed using GraphPad Prism version 5.0. One-way analysis of variance (AVOVA) was used for comparison of mean or geometric mean from multiple groups. A p-value of less than 0.05 was considered statistically significant.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1. Barre-Sinoussi F, Chermann JC, Rey F, Nugeyre MT, Chamaret S, Gruest J, Dauguet C, Axler-Blin C, Vezinet-Brun F, Rouzioux C, et al. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS). Science (New York, NY) 1983; 220:868–71; http://dx.doi.org/ 10.1126/science.6189183 [DOI] [PubMed] [Google Scholar]
- 2. Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, et al. Antibody neutralization and escape by HIV-1. Nature 2003; 422:307-12; PMID:12646921; http://dx.doi.org/ 10.1038/nature01470 [DOI] [PubMed] [Google Scholar]
- 3. Klein JS, Bjorkman PJ. Few and far between: how HIV may be evading antibody avidity. PLoS Pathogens 2010; 6:e1000908; PMID:20523901; http://dx.doi.org/ 10.1371/journal.ppat.1000908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Goulder PJ, Walker BD. HIV and HLA class I: an evolving relationship. Immunity 2012; 37:426-40; PMID:22999948; http://dx.doi.org/ 10.1016/j.immuni.2012.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stern LJ, Calvo-Calle JM. HLA-DR: molecular insights and vaccine design. Curr Pharm Des 2009; 15:3249-61; PMID:19860674; http://dx.doi.org/ 10.2174/138161209789105171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Moore CB, John M, James IR, Christiansen FT, Witt CS, Mallal SA. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science (New York, NY) 2002; 296:1439-43; http://dx.doi.org/ 10.1126/science.1069660 [DOI] [PubMed] [Google Scholar]
- 7. Johnston MI, Fauci AS. An HIV vaccine–evolving concepts. N Engl J Med 2007; 356:2073-81; PMID:17507706; http://dx.doi.org/ 10.1056/NEJMra066267 [DOI] [PubMed] [Google Scholar]
- 8. Spearman P. Current progress in the development of HIV vaccines. Curr Pharm Des 2006; 12:1147-67; PMID:16515492; http://dx.doi.org/ 10.2174/138161206776055859 [DOI] [PubMed] [Google Scholar]
- 9. Li C, Shen Z, Li X, Bai J, Zeng L, Tian M, Song YJ, Ye M, Du S, Ren D, et al. Protection against SHIV-KB9 infection by combining rDNA and rFPV vaccines based on HIV multiepitope and p24 protein in Chinese rhesus macaques. Clin Dev Immunol 2012; 2012:958404; PMID:22474488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gilbert P, Wang M, Wrin T, Petropoulos C, Gurwith M, Sinangil F, D'Souza P, Rodriguez-Chavez IR, DeCamp A, Giganti M, et al. Magnitude and breadth of a nonprotective neutralizing antibody response in an efficacy trial of a candidate HIV-1 gp120 vaccine. J Infect Dis 2010; 202:595-605; PMID:20608874; http://dx.doi.org/ 10.1086/654816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McCoy LE, Weiss RA. Neutralizing antibodies to HIV-1 induced by immunization. J Exp Med 2013; 210:209-23; PMID:23401570; http://dx.doi.org/ 10.1084/jem.20121827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Karnasuta C, Paris RM, Cox JH, Nitayaphan S, Pitisuttithum P, Thongcharoen P, Brown AE, Gurunathan S, Tartaglia J, Heyward WL, et al. Antibody-dependent cell-mediated cytotoxic responses in participants enrolled in a phase I/II ALVAC-HIV/AIDSVAX B/E prime-boost HIV-1 vaccine trial in Thailand. Vaccine 2005; 23:2522-9; PMID:15752839; http://dx.doi.org/ 10.1016/j.vaccine.2004.10.028 [DOI] [PubMed] [Google Scholar]
- 13. Shacklett BL, Ferre AL. Mucosal immunity in HIV controllers: the right place at the right time. Curr Opin HIV AIDS 2011; 6:202-7; PMID:21399497; http://dx.doi.org/ 10.1097/COH.0b013e3283453e2b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mudd PA, Watkins DI. Understanding animal models of elite control: windows on effective immune responses against immunodeficiency viruses. Curr Opin HIV AIDS 2011; 6:197-201; PMID:21502922; http://dx.doi.org/ 10.1097/COH.0b013e3283453e16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Munier CM, Kelleher AD, Kent SJ, De Rose R. The role of T cell immunity in HIV-1 infection. Curr Opin Virol 2013; 3:438-46; PMID:23747036; http://dx.doi.org/ 10.1016/j.coviro.2013.05.009 [DOI] [PubMed] [Google Scholar]
- 16. Perrin H, Canderan G, Sekaly RP, Trautmann L. New approaches to design HIV-1 T-cell vaccines. Curr Opin HIV AIDS 2010; 5:368-76; PMID:20978376; http://dx.doi.org/ 10.1097/COH.0b013e32833d2cc0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kunwar P, Hawkins N, Dinges WL, Liu Y, Gabriel EE, Swan DA, Stevens CE, Maenza J, Collier AC, Mullins JI, et al. Superior control of HIV-1 replication by CD8+ T cells targeting conserved epitopes: implications for HIV vaccine design. PloS One 2013; 8:e64405; PMID:23741326; http://dx.doi.org/ 10.1371/journal.pone.0064405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Levitz L, Koita OA, Sangare K, Ardito MT, Boyle CM, Rozehnal J, Tounkara K, Dao SM, Kone Y, Koty Z, et al. Conservation of HIV-1 T cell epitopes across time and clades: validation of immunogenic HLA-A2 epitopes selected for the GAIA HIV vaccine. Vaccine 2012; 30:7547-60; PMID:23102976; http://dx.doi.org/ 10.1016/j.vaccine.2012.10.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. De Groot AS, Rivera DS, McMurry JA, Buus S, Martin W. Identification of immunogenic HLA-B7 “Achilles' heel” epitopes within highly conserved regions of HIV. Vaccine 2008; 26:3059-71; PMID:18206276; http://dx.doi.org/ 10.1016/j.vaccine.2007.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Goulder PJ, Watkins DI. HIV and SIV CTL escape: implications for vaccine design. Nat Rev Immunol 2004; 4:630-40; PMID:15286729; http://dx.doi.org/ 10.1038/nri1417 [DOI] [PubMed] [Google Scholar]
- 21. Rodriguez F, Harkins S, Slifka MK, Whitton JL. Immunodominance in virus-induced CD8(+) T-cell responses is dramatically modified by DNA immunization and is regulated by gamma interferon. J Virol 2002; 76:4251-9; PMID:11932390; http://dx.doi.org/ 10.1128/JVI.76.9.4251-4259.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Im EJ, Hong JP, Roshorm Y, Bridgeman A, Letourneau S, Liljestrom P, Potash MJ, Volsky DJ, McMichael AJ, Hanke T. Protective efficacy of serially up-ranked subdominant CD8+ T cell epitopes against virus challenges. PLoS Pathogens 2011; 7:e1002041; PMID:21625575; http://dx.doi.org/ 10.1371/journal.ppat.1002041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Manuel ER, Yeh WW, Seaman MS, Furr K, Lifton MA, Hulot SL, Autissier P, Letvin NL. Dominant CD8+ T-lymphocyte responses suppress expansion of vaccine-elicited subdominant T lymphocytes in rhesus monkeys challenged with pathogenic simian-human immunodeficiency virus. J Virol 2009; 83:10028-35; PMID:19641002; http://dx.doi.org/ 10.1128/JVI.01015-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gorse GJ, Baden LR, Wecker M, Newman MJ, Ferrari G, Weinhold KJ, Livingston BD, Villafana TL, Li H, Noonan E, et al. Safety and immunogenicity of cytotoxic T-lymphocyte poly-epitope, DNA plasmid (EP HIV-1090) vaccine in healthy, human immunodeficiency virus type 1 (HIV-1)-uninfected adults. Vaccine 2008; 26:215-23; PMID:18055072; http://dx.doi.org/ 10.1016/j.vaccine.2007.10.061 [DOI] [PubMed] [Google Scholar]
- 25. Jin X, Newman MJ, De-Rosa S, Cooper C, Thomas E, Keefer M, Fuchs J, Blattner W, Livingston BD, McKinney DM, et al. A novel HIV T helper epitope-based vaccine elicits cytokine-secreting HIV-specific CD4+ T cells in a Phase I clinical trial in HIV-uninfected adults. Vaccine 2009; 27:7080-6; PMID:19786145; http://dx.doi.org/ 10.1016/j.vaccine.2009.09.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Spearman P, Kalams S, Elizaga M, Metch B, Chiu YL, Allen M, Weinhold KJ, Ferrari G, Parker SD, McElrath MJ, et al. Safety and immunogenicity of a CTL multiepitope peptide vaccine for HIV with or without GM-CSF in a phase I trial. Vaccine 2009; 27:243-9; PMID:18996425; http://dx.doi.org/ 10.1016/j.vaccine.2008.10.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Paul S, Piontkivska H. Discovery of novel targets for multi-epitope vaccines: screening of HIV-1 genomes using association rule mining. Retrovirology 2009; 6:62; PMID:19580659; http://dx.doi.org/ 10.1186/1742-4690-6-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wilson CC, Newman MJ, Livingston BD, MaWhinney S, Forster JE, Scott J, Schooley RT, Benson CA. Clinical phase 1 testing of the safety and immunogenicity of an epitope-based DNA vaccine in human immunodeficiency virus type 1-infected subjects receiving highly active antiretroviral therapy. Clin Vaccine Immunol 2008; 15:986-94; PMID:18400976; http://dx.doi.org/ 10.1128/CVI.00492-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Paul S, Piontkivska H. Frequent associations between CTL and T-Helper epitopes in HIV-1 genomes and implications for multi-epitope vaccine designs. BMC Microbiol 2010; 10:212; PMID:20696039; http://dx.doi.org/ 10.1186/1471-2180-10-212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hemelaar J, Gouws E, Ghys PD, Osmanov S. Global trends in molecular epidemiology of HIV-1 during 2000-2007. AIDS (London, England) 2011; 25:679-89; PMID:21297424; http://dx.doi.org/ 10.1097/QAD.0b013e328342ff93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Trachtenberg E, Vinson M, Hayes E, Hsu YM, Houtchens K, Erlich H, Klitz W, Hsia Y, Hollenbach J. HLA class I (A, B, C) and class II (DRB1, DQA1, DQB1, DPB1) alleles and haplotypes in the Han from southern China. Tissue Antigens 2007; 70:455-63; PMID:17900288; http://dx.doi.org/ 10.1111/j.1399-0039.2007.00932.x [DOI] [PubMed] [Google Scholar]
- 32. Li XF, Zhang X, Chen Y, Zhang KL, Liu XJ, Li JP. An analysis of HLA-A, -B, and -DRB1 allele and haplotype frequencies of 21,918 residents living in Liaoning, China. PloS One 2014; 9:e93082; PMID:24691290; http://dx.doi.org/ 10.1371/journal.pone.0093082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Girard MP, Osmanov S, Assossou OM, Kieny MP. Human immunodeficiency virus (HIV) immunopathogenesis and vaccine development: a review. Vaccine 2011; 29:6191-218; PMID:21718747; http://dx.doi.org/ 10.1016/j.vaccine.2011.06.085 [DOI] [PubMed] [Google Scholar]
- 34. Smahel M, Polakova I, Duskova M, Ludvikova V, Kastankova I. The effect of helper epitopes and cellular localization of an antigen on the outcome of gene gun DNA immunization. Gene Ther 2014; 21:225-32; PMID:24385146; http://dx.doi.org/ 10.1038/gt.2013.81 [DOI] [PubMed] [Google Scholar]
- 35. Kumar A, Arora R, Kaur P, Chauhan VS, Sharma P. “Universal" T helper cell determinants enhance immunogenicity of a Plasmodium falciparum merozoite surface antigen peptide. J Immunol 1992; 148:1499-505; PMID:1371529 [PubMed] [Google Scholar]
- 36. Soghoian DZ, Jessen H, Flanders M, Sierra-Davidson K, Cutler S, Pertel T, Ranasinghe S, Lindqvist M, Davis I, Lane K, et al. HIV-specific cytolytic CD4 T cell responses during acute HIV infection predict disease outcome. Sci Transl Med 2012; 4:123ra25; PMID:22378925; http://dx.doi.org/ 10.1126/scitrans-lmed.3003165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Soghoian DZ, Streeck H. Cytolytic CD4(+) T cells in viral immunity. Expert Rev Vaccines 2010; 9:1453-63; PMID:21105780; http://dx.doi.org/ 10.1586/erv.10.132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chang CX, Tan AT, Or MY, Toh KY, Lim PY, Chia AS, Froesig TM, Nadua KD, Oh HL, Leong HN, et al. Conditional ligands for Asian HLA variants facilitate the definition of CD8+ T-cell responses in acute and chronic viral diseases. Eur J Immunol 2013; 43:1109-20; PMID:23280567; http://dx.doi.org/ 10.1002/eji.201243088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Reddy Chichili VP, Kumar V, Sivaraman J. Linkers in the structural biology of protein-protein interactions. Protein Sci 2013; 22:153-67; PMID:23225024; http://dx.doi.org/ 10.1002/pro.2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Deane JE, Ryan DP, Sunde M, Maher MJ, Guss JM, Visvader JE, Matthews JM. Tandem LIM domains provide synergistic binding in the LMO4:Ldb1 complex. EMBO J 2004; 23:3589-98; PMID:15343268; http://dx.doi.org/ 10.1038/sj.emboj.7600376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang QM, Sun SH, Hu ZL, Zhou FJ, Yin M, Xiao CJ, Zhang JC. Epitope DNA vaccines against tuberculosis: spacers and ubiquitin modulates cellular immune responses elicited by epitope DNA vaccine. Scand J Immunol 2004; 60:219-25; PMID:15320877; http://dx.doi.org/ 10.1111/j.0300-9475.2004.01442.x [DOI] [PubMed] [Google Scholar]
- 42. Velders MP, Weijzen S, Eiben GL, Elmishad AG, Kloetzel PM, Higgins T, Ciccarelli RB, Evans M, Man S, Smith L, et al. Defined flanking spacers and enhanced proteolysis is essential for eradication of established tumors by an epitope string DNA vaccine. J Immunol 2001; 166:5366-73; PMID:11313372; http://dx.doi.org/ 10.4049/jimmunol.166.9.5366 [DOI] [PubMed] [Google Scholar]