Abstract
Vaccines dramatically reduce infection-related morbidity and mortality. Determining factors that modulate the host response is key to rational vaccine design and demands unsupervised analysis. To longitudinally resolve influenza-specific humoral immune response dynamics we constructed vaccine response profiles of influenza A- and B-specific IgM and IgG levels from 42 healthy and 31 HIV infected influenza-vaccinated individuals. Pre-vaccination antibody levels and levels at 3 predefined time points after vaccination were included in each profile. We performed hierarchical clustering on these profiles to study the extent to which HIV infection associated immune dysfunction, adaptive immune factors (pre-existing influenza-specific antibodies, T cell responses), an innate immune factor (Mannose Binding Lectin, MBL), demographic characteristics (gender, age), or the vaccine preparation (split vs. virosomal) impacted the immune response to influenza vaccination. Hierarchical clustering associated vaccine preparation and pre-existing IgG levels with the profiles of healthy individuals. In contrast to previous in vitro and animal data, MBL levels had no impact on the adaptive vaccine response. Importantly, while HIV infected subjects with low CD4 T cell counts showed a reduced magnitude of their vaccine response, their response profiles were indistinguishable from those of healthy controls, suggesting quantitative but not qualitative deficits. Unsupervised profile-based analysis ranks factors impacting the vaccine-response by relative importance, with substantial implications for comparing, designing and improving vaccine preparations and strategies. Profile similarity between HIV infected and HIV negative individuals suggests merely quantitative differences in the vaccine response in these individuals, offering a rationale for boosting strategies in the HIV infected population.
Keywords: HIV, immune dysfunction, influenza vaccination, immune response, MBL, unsupervised analysis, vaccine preparation
Each year, influenza virus infection affects 5 to 20% of the world's population.1 Annual influenza vaccination greatly contributes to reduce influenza-related morbidity and mortality.2 However, vaccine efficacy varies between 40-90%, depending on the patient group, the vaccine preparation, and the match of vaccine and circulating epidemic virus strain.1,3,4 Especially immunocompromised populations, including HIV infected individuals, often fail to mount protective vaccine responses.5,6 Therefore, understanding how immunological and non-immunological factors impact the vaccine response in humans is crucial both for the design of enhanced vaccines and improved vaccination schemes.
Pathogen-specific peak antibody levels are routinely used as surrogates of vaccine efficacy. However, a protective immune response relies on robust immune memory formation, which may not be fully captured by peak antibody titers alone, but rather by time-resolved longitudinal titer measurements, which would reflect more closely the dynamics and memory of long-term immunological protection in vivo. Moreover, conventional comparison of cross-sectional antibody peak titers is affected by confounding factors such as the sampling time-point and presence of pre-existing immunity (antigen-specific memory B cells), and thus may not accurately reflect the true immunological properties of vaccine responses.7 Recent efforts have, therefore, shifted to characterization of the vaccine response based on longitudinal data, coupled with unsupervised analyses – in order to unambiguously identify factors that impact vaccine response-dynamics.8-11
In this study, we used time-resolved measurements of the influenza-specific antibody response to generate immunological profiles – hereafter termed ‘vaccine response profiles’ – across a cohort of HIV negative and HIV infected individuals following influenza vaccination. These profiles allowed an unsupervised analysis of the association of individual vaccine responses with various factors, namely immune dysfunction associated with HIV infection, adaptive immune factors (pre-existing influenza-specific antibodies, T cell responses), an innate immune factor (Mannose Binding Lectin, MBL), demographic characteristics (gender, age), or the vaccine preparation (split vs. virosomal). The effect of MBL, a pattern recognition molecule involved in the containment of infections, was investigated since it has been previously shown to have anti-influenza effects in vitro12 and has been suggested to affect murine vaccine responses in vivo.13 The impact of MBL on the adaptive immune response to influenza vaccination in humans has, to our knowledge, not been investigated yet.
Using an unsupervised analysis of vaccine response profiles allowed us to rank the impact of the various factors on the evolution of the humoral vaccine-response, which may help guide future efforts to improve vaccines.
Influenza vaccine responses from previously published local vaccination cohorts were analyzed.6,14 Study participants were recruited at the University Hospital Basel, Switzerland, and vaccinated with either a trivalent virosomal vaccine (Inflexal V®, Berna Biotech) (season 2007/2008, n = 24 healthy and 31 HIV infected individuals), or an inactivated influenza virus split-vaccine (Mutagrip®, Sanofi Pasteur) (season 2008/2009, n = 18).6,14 The study was approved by the local ethics committee and all participants gave informed consent. Cohort characteristics, vaccine composition and type are summarized in Table 1. Influenza-specific IgM and IgG serum levels were available immediately prior to vaccination, and at days 7, 14 and 21 post-vaccination (Fig. S1). These have been quantified using commercially available ELISA Kits (Genzyme Virotech), as previously described.6,14 Antibody levels ≥10 Virotech Units (VE)/mL were considered protective. Influenza-specific T cells were measured using standard interferon-γ (IFN-γ) enzyme-linked immunosorbent spot (ELISpot) assays.15 Influenza antigen (Inflexal V®, Berna Biotech) was added at a final concentration of 14 μg/mL to 200.000 peripheral blood mononuclear cells (PBMC) and incubated for 16 hours. Plates were developed using an alkaline phosphatase coupled detection antibody (7-B6-1-ALP, Mabtech) and the HistoMark RED phosphatase system (KPL, Gaithersburg, Maryland, USA). Spots were counted with the ELISpot Reader System (CSR01, AID GmbH, Strassberg, Germany). Results were expressed as spot-forming cells/million PBMC (SFC/M PBMC) with a cutoff for a positive response of 50 SFC/M PBMC. All measurements were performed in duplicates and PBMC stimulated with Phytohemagglutinin (PHA) (1.8 μg/mL; REMEL, Oxoid AG, Basel, Switzerland) served as a positive control. For this study, MBL levels, which tightly correlate with MBL function, were assessed in serum of the healthy vaccinated individuals using the commercially available MBL Oligomer ELISA Kit according to the manufacturers instructions (KIT 029, BioPorto, Denmark).
Table 1.
Cohort characteristics
| Season | 2007/2008 | 2007/2008 | 2007/2008 | 2008/2009 |
|---|---|---|---|---|
| Cohort | Healthy (2007/2008) | HIV+ CD4>350/μl | HIV+ CD4≤350/μl | Healthy (2008/2009) |
| n Vaccine recipients | 24 | 22 | 9 | 18 |
| Age median (range) | 40.8 (22–76) | 46 (21–52) | 46 (30–65) | 36.8 (19–46) |
| % Female | 45.8% | 36.4% | 37.5% | 31.3% |
| Median CD4 cells/μl (range) | n.d. | 545 (364–941) | 200 (40–326) | n.d. |
| HIV Viral Load (copies/mL) | n.d. | <400 | <400 | n.d. |
| Vaccine name | Inflexal V® (Berna) | Inflexal V® (Berna) | Inflexal V® (Berna) | Mutagrip® (Sanofi Pasteur) |
| Vaccine type | Virosomal | Virosomal | Virosomal | Inactivated split |
| Strains | A/Solomon Islands/3/2007 (H1N1) A/Wisconsin/67/2005 (H3N2) B/Malaysia/2506/2004 |
A/Solomon Islands/3/2007 (H1N1) A/Wisconsin/67/2005 (H3N2) B/Malaysia/2506/2004 |
A/Solomon Islands/3/2007 (H1N1) A/Wisconsin/67/2005 (H3N2) B/Malaysia/2506/2004 |
A/Brisbane /59/2007 (H1N1) A/Brisbane/10/2007 (H3N2) B/Florida/4/2006 |
Against this background, we sought to investigate the extent to which predefined demographic, adaptive and innate immune factors influence the influenza-specific vaccine response in healthy individuals. In order to recapitulate vaccine-specific immunological characteristics on the individual level in a time-resolved fashion, we built vaccine response profiles consisting of each individual's anti-influenza A and B IgM or IgG levels measured at the 4 indicated time points (Fig. S1) in order to capture the humoral immune response at a higher dimensionality compared to traditional baseline-peak comparisons.16 These profiles were tested for similarity and clustered accordingly, without prior knowledge (i.e., unsupervised) of information on a vaccinated individual's relevant characteristics.16-18 Hierarchical clustering of vaccine response profiles was performed using the “average” clustering algorithm from the R function hclust. Employing Pearson correlation as distance metric allowed us to cluster profiles independently of baseline IgM or IgG levels, which enabled us to compare the dynamics of the humoral immune response in vivo regardless of individual variances in baseline IgM and IgG levels – as opposed to traditional baseline-peak comparisons. The significance of clusters was assessed using the pvclust R package.19 The association of vaccine response profile clustering with the following factors was assessed in healthy subjects: vaccine preparation (trivalent virosomal vs. inactivated split), demographic (age, gender), adaptive immunity (pre-existing influenza-specific IgG levels, influenza-specific T cell response), and innate immunity (circulating levels of Mannose Binding Lectin (MBL)).
As previously published,6,14 vaccination of healthy individuals induced a significant influenza-specific cellular and humoral immune response, peaking at day 14 post-vaccination in most individuals (Fig. S1). Based on the predefined cut-off for protective antibody levels, 17/42 healthy individuals (40%) had pre-existing IgG against influenza A and 25/42 (60%) against influenza B. In contrast, only 2/42 (5%) had elevated IgM levels against influenza A or B. While both vaccine regimens were found to generate robust cellular and humoral immune responses, IgM responses, in contrast to IgG, were more pronounced for the 2008/2009 cohort (Fig. S1). MBL levels ranged from 17 to 6900 ng/mL, including 6 individuals with MBL deficiency (MBL level <500 ng/mL).20 Vaccination had no impact on MBL levels as assessed 7 days post-vaccination (data not shown).
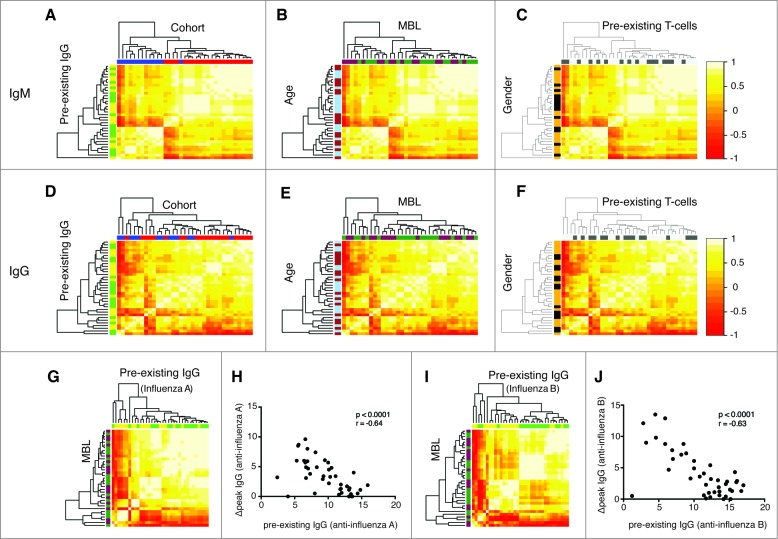
Applying our unsupervised vaccine response profile-clustering analyses, we found that IgM (Fig. 1A), but not IgG (Fig. 1B), vaccine response profiles significantly cluster by cohort –presumably relating to vaccine preparation (Fig. 1) – but not by any of the other predefined factors, including gender, age, MBL level, pre-existing IgG and pre-existing T cell responses (Fig. 1). When vaccine response profiles targeting influenza A or B were, however, analyzed separately, preexisting IgG levels to the same antigen were identified as predictors of the IgG response profile (Figs. 1G, I). This was further supported by the strong negative correlation between the pre-existing IgG and the IgG response to the same antigen (Figs. 1H, J). Importantly, we found no association between MBL levels and vaccine response profiles.
Figure 1.
Vaccine response profiles of healthy vaccinees cluster by vaccine preparation and pre-existing adaptive immunity. The hierarchical clustering of IgM (A, B, C) and IgG (D, E, F) profiles is shown. IgM and IgG profiles incorporate influenza-A and B-specific Ig levels. The heatmap depicts the pairwise Pearson correlation coefficients of all profiles determined. Factors were color-coded: Cohort (2007/2008: red, 2008/2009: blue), MBL (darkgreen: ≤ median, purple: >median), age (light blue: ≤ median, brown: >median), pre-existing IgG levels (green: ≤ median, yellow: >median), pre-existing T cells (dark gray: ≤ median, light gray: >median) and gender (black: female, yellow: male). Influenza A profiles clustered dependent on the pre-existing IgG against influenza A (G, green: ≤ median, yellow: >median), which was confirmed by a strong inverse correlation between these markers (H). The same was true for pre-existing IgG against influenza B (I, J). There was no clustering based on MBL levels (darkgreen: ≤ median, purple: >median) (G, H). Spearman Ranks correlation analysis was performed in Figures H and J. The clustering by pre-existing IgG was determined to be significant (P < 0.05).
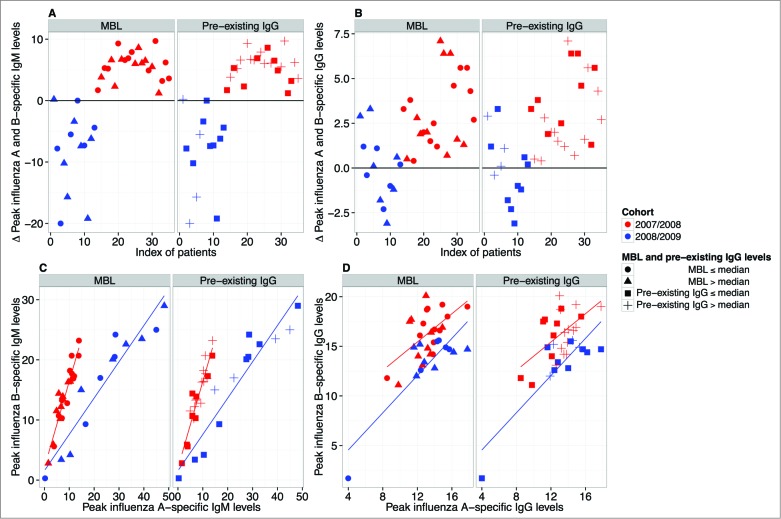
Given the difference in both the influenza vaccine response dynamics (profiles) (Fig. 1) and the magnitude of the influenza A- and B-specific immune response between the 2 vaccine cohorts (Fig. S1), we next analyzed whether the cohort effect might derive from preferential targeting of either influenza subtype. For each individual the difference in peak values of anti-influenza A and B levels for both IgM (A) and IgG (B) was calculated. Interestingly, we found that peak IgM levels showed extraordinary cohort-specific consistency: for IgM, almost all differences of the 2008/2009 (inactivated split-vaccine, Mutagrip®) cohort were negative, indicating preferential targeting of influenza A, whereas all differences of the 2007/2008 (virosomal vaccine, Inflexal®) cohort were positive, indicating preferential targeting of influenza B (Fig. 2A). Such a trend was also apparent for IgG (Fig. 2B). Moreover, we found that influenza A- and influenza B-specific IgM (Fig. 2C)—and to a lesser extent IgG (Fig. 2D)—peak levels were directly correlated within any given individual in either cohort. Neither MBL levels nor preexisting IgG had an effect on the preferential targeting of influenza A or B (Figs. 2A-D). The latter was also the case, when anti-Influenza A or B preexisting IgG levels were used rather than mean preexisting IgG levels (Fig. S2). Taken together, these findings indicate that the type of the administered vaccine dictates not only the magnitude and response dynamics of the vaccine response, but also reveals vaccine-specific preferential targeting of influenza A or B.
Figure 2.
Preferential targeting of influenza A or B depending on vaccination. For each individual the difference in peak values of anti-influenza A and B levels for both IgM (A) and IgG (B) was calculated. The 2008/2009 (inactivated split-vaccine, Mutagrip®) cohort is depicted in blue and the 2007/2008 (virosomal vaccine, Inflexal®) cohort in red. IgM (A) and IgG (B) profiles are shown. (C) Correlation analysis of influenza A- and influenza B-specific IgM (C) and IgG levels (D) is shown (red = 2007/2008 cohort; blue = 2008/2009 cohort). The effects of both low (closed circles: ≤median) and high MBL levels (triangles: >median) as well as low (closed squares: ≤median) and high (crosses: >median) pre-existing IgG levels are displayed. Pearson correlation coefficients for IgM in the 2007/2008 and 2008/2009 cohort were r = 0.94 and r = 0.95; for IgG r = 0.57 and r = 0.85, respectively. All correlations were found to be significant (p < 0.001).
Based on the insight gained from the vaccine response profile-analyses of influenza vaccine responses in healthy individuals, we proceeded to test whether HIV infection influences the relative dynamics of influenza vaccine-specific antibody levels – suggesting a qualitative difference in the response– or merely impacts the magnitude of the response, hinting at quantitative differences. HIV infected individuals with low CD4 T cell counts demonstrated influenza vaccine responses of lower magnitude than those with preserved CD4 T cell counts or uninfected controls, all vaccinated with the same vaccine (Fig. 3A).6 Using our unsupervised profile analysis approach, we therefore aimed to test whether HIV infection impacts vaccine response profiles. Notably, IgM and IgG profiles of healthy individuals and HIV infected individuals analyzed by antigen specificity (Fig. 3B), or profiles including all specificities (influenza A and B) combined (data not shown) were indistinguishable from one another. Thus, influenza-specific peak antibody levels, but not vaccine response profiles, differentiate HIV infected individuals from healthy controls, indicating that the magnitude, but not the relative influenza vaccine response dynamics, are impaired during HIV infection.
Figure 3.
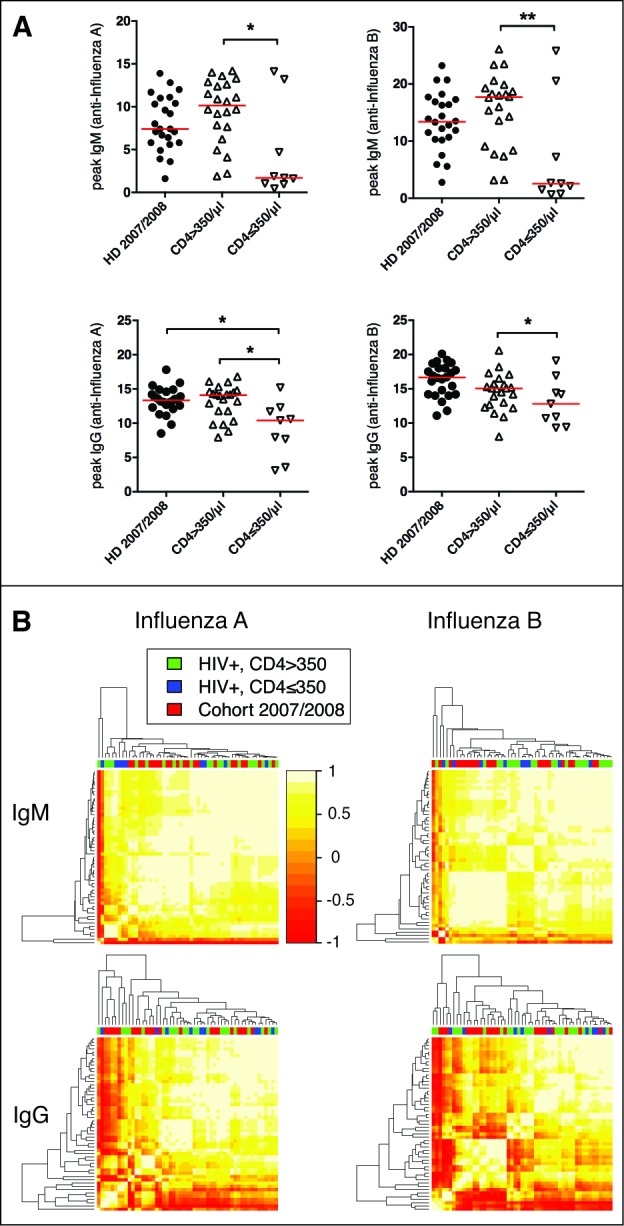
Immunoglobulin profiles do not cluster by HIV status or level of immunosuppression. (A) Peak IgM and IgG responses against influenza A and B are shown as VE/mL. HD = healthy donor, CD4 high = HIV infected individuals with CD4 counts >350/μl, CD4 low = HIV infected individuals with CD4 counts ≤350/μl. *P < 0.05, **P < 0.01. (B) The hierarchical clustering of the IgM (influenza A or B) and IgG (influenza A or B) vaccine response profiles indicated no difference between these 3 groups. The heatmap depicts the pairwise Pearson correlation coefficients of all profiles determined.
A deeper understanding of the factors that modulate vaccine responses is key to both rational vaccine design and improving vaccination strategies. Here we applied a novel, unsupervised comparative time-resolved systems analysis to assess the impact of relevant host- and vaccine-specific factors on the influenza vaccine-induced immune response (‘vaccine response profiles’). We show that the vaccine-type (split vs. virosomal) significantly impacted both relative dynamics of the vaccine humoral immune response and antigen targeting (influenza A vs. B), suggesting that influenza vaccine preparation and design may substantially influence major traits of the vaccine response. Owing to the study design, our data does not exclude the possibility that the observed cohort-specific profile differences may in part be due to the different vaccine viral antigen composition between cohorts. However, the study design allowed probing factors impacting the vaccine response profile in 2 consecutive, independent cohorts exposed to distinct vaccine preparations. Overall, consistency of the effects within the 2 cohorts supports that the vaccine preparation, rather than individual factors such as genetic predispositions or pre-vaccination anti-influenza immune status, are the main drivers of our observations. Nonetheless, future studies should be designed to control for participant history regarding previous vaccination or natural infection, and include readouts that assess functional (e.g. hemagglutinin inhibition assays) and strain specific immune responses.
Consistent with previous reports,10 we found that across the 2 healthy cohorts pre-existing IgG levels substantially impacted the IgG profiles and correlated inversely with the peak responses. While this negative correlation is intuitive and may indicate that the vaccine is superfluous in subjects with high preexisting immunity,21,22 preexisting IgG against one influenza vaccine antigen could potentially also negatively impact immune responses against the other antigens.23 Since for our study we have no data on the strain specific immune response available, we cannot exclude that preexisting strain specific IgG might have impacted the vaccine response differentially.
When comparing HIV-infected and uninfected controls, IgG vaccine response profiles were indistinguishable from one another. Notably, this also applied for HIV infected individuals with low CD4 T cell counts that mounted reduced peak IgG levels. Thus, our data suggest a quantitative, but not a qualitative, immune dysfunction in HIV infected individuals, which may be overcome by optimizing the vaccination strategy. In support of this hypothesis, 2 recent clinical studies on influenza and hepatitis B vaccination indeed showed substantially improved vaccine responses in HIV infected individuals vaccinated more frequently and with higher antigen doses.24,25
Our approach of hierarchical clustering of vaccine response profiles, while being relatively novel, has recently been used in a similar setting.11 Whereas Bonduelle et al. merely used the fold-change for clustering, we increased the sensitivity of vaccine profiling by integrating several time points into one vaccine response profile. The main advantage of this approach is that time-resolved measurements capture the evolution of the humoral immune response better than binary measures (i.e., baseline-peak comparisons). Moreover, the approach is statistically robust, and by evocative heat map visualization superior to simple fold-change based analysis.16-18
Indeed, while fold-change analyses represent the mean of the humoral immune response, vaccine response profiles provide improved resolution of the immune response, helping distinguish responses that show a similar mean behavior. While in this study the sample size was large enough to detect substantial differences between factors impacting the vaccine response profile, we cannot exclude that we were underpowered to detect minor effects of some of the factors tested. Moreover, since our analysis focused on strain specific antibody binding titers, we cannot exclude that other parameters impact virus neutralization or hemagglutinin inhibition.
In summary, we show that vaccine response profiles—reflecting the individual dynamics of immune responses—provide clinically and immunologically valuable insight that may direct future efforts toward improving vaccine-design and -strategy.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
The work was supported by the Swiss National Science Foundation (SNSF) to CTB [PZ00P3-148000] and CH [SNSF 310030_153059], and supported by the Misrock Foundation and SystemsX.ch Antibody Research Technology and Development (RTD) to STR.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1. Glezen WP. Clinical practice. Prevention and treatment of seasonal influenza. N Engl J Med 2008; 359:2579-85; PMID:19073977; http://dx.doi.org/ 10.1056/NEJMcp0807498 [DOI] [PubMed] [Google Scholar]
- 2. Kwong JC, Stukel TA, Lim J, McGeer AJ, Upshur RE, Johansen H, Sambell C, Thompson WW, Thiruchelvam D, Marra F, et al. The effect of universal influenza immunization on mortality and health care use. PLoS Med 2008; 5:e211; PMID:18959473; http://dx.doi.org/ 10.1371/journal.pmed.0050211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cox RJ, Brokstad KA, Ogra P. Influenza virus: immunity and vaccination strategies. Comparison of the immune response to inactivated and live, attenuated influenza vaccines. Scand J Immunol 2004; 59:1-15; PMID:14723616; http://dx.doi.org/ 10.1111/j.0300-9475.2004.01382.x [DOI] [PubMed] [Google Scholar]
- 4. DiazGranados CA, Denis M, Plotkin S. Seasonal influenza vaccine efficacy and its determinants in children and non-elderly adults: a systematic review with meta-analyses of controlled trials. Vaccine 2012; 31:49-57; PMID:23142300; http://dx.doi.org/ 10.1016/j.vaccine.2012.10.084 [DOI] [PubMed] [Google Scholar]
- 5. Bickel M, Wieters I, Khaykin P, Nisius G, Haberl A, Stephan C, Von Hentig N, Herrmann E, Doerr HW, Brodt HR, et al. Low rate of seroconversion after vaccination with a split virion, adjuvanted pandemic H1N1 influenza vaccine in HIV-1-infected patients. AIDS 2010; 24:F31-5; PMID:20559034; http://dx.doi.org/ 10.1097/QAD.0b013e3283398da1 [DOI] [PubMed] [Google Scholar]
- 6. Fritz S, Mossdorf E, Durovic B, Zenhaeusern G, Conen A, Steffen I, Battegay M, Nuesch R, Hess C. Virosomal influenza-vaccine induced immunity in HIV-infected individuals with high versus low CD4+ T-cell counts: clues towards a rational vaccination strategy. AIDS 2010; 24:2287-9; PMID:20625265; http://dx.doi.org/ 10.1097/QAD.0b013e32833c6f92 [DOI] [PubMed] [Google Scholar]
- 7. Beyer WE, Palache AM, Luchters G, Nauta J, Osterhaus AD. Seroprotection rate, mean fold increase, seroconversion rate: which parameter adequately expresses seroresponse to influenza vaccination? Virus Res 2004; 103:125-32; PMID:15163500; http://dx.doi.org/ 10.1016/j.virusres.2004.02.024 [DOI] [PubMed] [Google Scholar]
- 8. Laserson U, Vigneault F, Gadala-Maria D, Yaari G, Uduman M, Vander Heiden JA, Kelton W, Taek Jung S, Liu Y, Laserson J, et al. High-resolution antibody dynamics of vaccine-induced immune responses. Proc Natl Acad Sci U S A 2014; 111:4928-33; PMID:24639495; http://dx.doi.org/ 10.1073/pnas.1323862111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li S, Rouphael N, Duraisingham S, Romero-Steiner S, Presnell S, Davis C, Schmidt DS, Johnson SE, Milton A, Rajam G, et al. Molecular signatures of antibody responses derived from a systems biology study of five human vaccines. Nat Immunol 2014; 15:195-204; PMID:24336226; http://dx.doi.org/ 10.1038/ni.2789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tsang JS, Schwartzberg PL, Kotliarov Y, Biancotto A, Xie Z, Germain RN, Wang E, Olnes MJ, Narayanan M, Golding H, et al. Global analyses of human immune variation reveal baseline predictors of postvaccination responses. Cell 2014; 157:499-513; PMID:24725414; http://dx.doi.org/ 10.1016/j.cell.2014.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bonduelle O, Yahia N, Siberil S, Benhabiles N, Carrat F, Krivine A, Rozenberg F, Dimitrov J, Kaveri SV, Curjol A, et al. Longitudinal and integrative biomodeling of effector and memory immune compartments after inactivated influenza vaccination. J Immunol 2013; 191:623-31; PMID:23776176; http://dx.doi.org/ 10.4049/jimmunol.1203483 [DOI] [PubMed] [Google Scholar]
- 12. Chang WC, White MR, Moyo P, McClear S, Thiel S, Hartshorn KL, Takahashi K. Lack of the pattern recognition molecule mannose-binding lectin increases susceptibility to influenza A virus infection. BMC Immunol 2010; 11:64; PMID:21182784; http://dx.doi.org/ 10.1186/1471-2172-11-64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ruseva M, Kolev M, Dagnaes-Hansen F, Hansen SB, Takahashi K, Ezekowitz A, Thiel S, Jensenius JC, Gadjeva M. Mannan-binding lectin deficiency modulates the humoral immune response dependent on the genetic environment. Immunology 2009; 127:279-88; PMID:19476514; http://dx.doi.org/ 10.1111/j.1365-2567.2008.03016.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mehling M, Hilbert P, Fritz S, Durovic B, Eichin D, Gasser O, Kuhle J, Klimkait T, Lindberg R, Kappos L, et al. Antigen-specific adaptive immune responses in fingolimod-treated MS patients. Annals of Neurology 2011. 69(2):408-13; PMID:21387383; http://dx.doi.org/ 10.1002/ana.22352 [DOI] [PubMed] [Google Scholar]
- 15. Berger CT, Frahm N, Price DA, Mothe B, Ghebremichael M, Hartman KL, Henry LM, Brenchley JM, Ruff LE, Venturi V, et al. High-functional-avidity cytotoxic T lymphocyte responses to HLA-B-restricted Gag-derived epitopes associated with relative HIV control. J Virol 2011; 85:9334-45; PMID:21752903; http://dx.doi.org/ 10.1128/JVI.00460-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dietz A, Gustafson MP, Lin Y, LaPlant BR, Liwski CJ, Maas ML, League SC, Bauer PR, Abraham RS, Tollefson MK, et al. Immune monitoring using the predictive power of immune profiles. Cytotherapy 2013; 15:S48-S; http://dx.doi.org/ 10.1016/j.jcyt.2013.01.183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Muraro PA, Robins H, Malhotra S, Howell M, Phippard D, Desmarais C, Sousa ADA, Griffith LM, Lim N, Nash RA, et al. T cell repertoire following autologous stem cell transplantation for multiple sclerosis. Journal of Clinical Investigation 2014; 124:1168-72; PMID:24531550; http://dx.doi.org/ 10.1172/JCI71691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nagalla S, Chou JW, Willingham MC, Ruiz J, Vaughn JP, Dubey P, Lash TL, Hamilton-Dutoit SJ, Bergh J, Sotiriou C, et al. Interactions between immunity, proliferation and molecular subtype in breast cancer prognosis. Genome Biol 2013; 14(4):R34; PMID:23618380; http://dx.doi.org/ 10.1186/gb-2013-14-4-r34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Suzuki R, Shimodaira H. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 2006; 22:1540-2; PMID:16595560; http://dx.doi.org/ 10.1093/bioinformatics/btl117 [DOI] [PubMed] [Google Scholar]
- 20. Worthley DL, Bardy PG, Gordon DL, Mullighan CG. Mannose-binding lectin and maladies of the bowel and liver. World J Gastroenterol 2006; 12:6420-8; PMID:17072973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Buricchi F, Bardelli M, Malzone C, Capecchi B, Nicolay U, Fragapane E, Castellino F, Del Giudice G, Galli G, Finco O. Impact of preexisting memory to seasonal A/H1N1 influenza virus on the immune response following vaccination against avian A/H5N1 virus. Eur J Immunol 2013; 43:641-8; PMID:23238926; http://dx.doi.org/ 10.1002/eji.201242563 [DOI] [PubMed] [Google Scholar]
- 22. Houser KV, Pearce MB, Katz JM, Tumpey TM. Impact of prior seasonal H3N2 influenza vaccination or infection on protection and transmission of emerging variants of influenza A(H3N2)v virus in ferrets. J Virol 2013; 87:13480-9; PMID:24089569; http://dx.doi.org/ 10.1128/JVI.02434-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pandey A, Singh N, Vemula SV, Couetil L, Katz JM, Donis R, Sambhara S, Mittal SK. Impact of preexisting adenovirus vector immunity on immunogenicity and protection conferred with an adenovirus-based H5N1 influenza vaccine. PLoS One 2012; 7:e33428; PMID:22432020; http://dx.doi.org/ 10.1371/journal.pone.0033428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Launay O, van der Vliet D, Rosenberg AR, Michel ML, Piroth L, Rey D, Colin de Verdiere N, Slama L, Martin K, Lortholary O, et al. Safety and immunogenicity of 4 intramuscular double doses and 4 intradermal low doses vs standard hepatitis B vaccine regimen in adults with HIV-1: a randomized controlled trial. JAMA 2011; 305:1432-40; PMID:21486976; http://dx.doi.org/ 10.1001/jama.2011.351 [DOI] [PubMed] [Google Scholar]
- 25. McKittrick N, Frank I, Jacobson JM, White CJ, Kim D, Kappes R, DiGiorgio C, Kenney T, Boyer J, Tebas P. Improved immunogenicity with high-dose seasonal influenza vaccine in HIV-infected persons: a single-center, parallel, randomized trial. Ann Intern Med 2013; 158:19-26; PMID:23277897; http://dx.doi.org/ 10.7326/0003-4819-158-1-201301010-00005 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.